Abstract
Background: p53 is a well-known tumor suppressor gene involved in malignancy. Many microRNAs (miRNAs) have recently been identified as key components of p53 signaling networks, owing to the central role of p53 in many processes, these p53-regulated miRNAs may possess important role in osteosarcoma. Methods: The expression of six p53-related miRNAs (miR-34 family [including miR-34a, 34b and 34c], miR-31, miR-192, and miR-215) in 80 pairs of osteosarcoma and corresponding noncancerous bone tissues were estimated by real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR), and the associations of miRNAs expression with clinicopathological factors, p53 status, and survival of patients were analyzed. Results: We found that among all six miRNAs, miR-34 family, -192, and -215 had decreased levels, whereas the level of miR-31 was increased (p<0.05) in tumor compared with corresponding noncancerous bone tissues, and miR-192/215 in patients with p53 positive expression was lower than those with negative p53. Kaplan-Meier analysis demonstrated that osteosarcoma patients with low miR-34a (P=0.000) and miR-192 (P=0.022) expression had poorer disease-free survival (DFS). Moreover, disease-free survival (DFS) was shorter for patients with low miR-34a and miR-192 expression (P=0.007) and the combination of low miR-192 with p53 positive expression (P=0.000). Furthermore, the multivariate analysis identified that low miR-34a expression, the combination of low miR-34a and miR-192 expression levels and the combination of low miR-192 with p53 positive were independent biomarkers of shorter DFS. Conclusions: Together, these results suggest that p53-associated miR-34a and miR-192 expression could be novel prognosis biomarkers for surgically treated osteosarcoma.
Keywords: miRNA, p53, osteosarcoma, RT-qPCR, prognosis
Introduction
Osteosarcoma, as the eighth leading cancer with an incidence of 4.4 per million [1], is the most common type of primary malignancy deriving from primitive bone-forming mesenchyme, which mainly arising from the metaphysis of the long bones of adolescents and young adults [2]. Although currently patients are routinely treated with combinatorial chemotherapy, curative resection of the primary tumor, and sometimes radiotherapy, which has been shown to improve the 5-year survival rate to approximately 60-70% [3,4], a significant proportion of osteosarcoma patients still have a risk of local relapse or distant metastasis even after surgery and intensive chemotherapy. Hence, searching effective biomarkers to predict prognosis would be not only helpful for risk stratification but also offer patients more optimized therapeutic schedule.
The tumor suppressor p53 (TP53) is a central modulator of multiple biological and pathological processes, including cell cycle progression, DNA repair, epithelial-mesenchymal transition, stemness, metabolism, cell survival and angiogenesis. Upregulation of p53 is usually indicative of the presence of mutant TP53 and the p53 dysfunction, and p53 gene is reported to be mutated in sporadic osteosarcoma ranges from 30 to 40% [5]. In addition, Li-Fraumeni syndrome, which is characterized by an autosomal dominant mutation of p53, also leads to the development of osteosarcoma [6]. Thus, p53 may also have significant implications in the tumorigenesis and progression of osteosarcoma [5,7,8].
MicroRNAs (miRNAs) are short, endogenous, noncoding RNAs with 22-24 nucleotides in length, which negatively regulate RNA translation to protein by binding to the 3’ untranslated regions (3’ UTRs) of their target mRNAs. It has been demonstrated that various miRNAs can function as oncogenes or tumor suppressor genes, regulating tumor cell behaviors including proliferation, apoptosis, differentiation and metastasis in osteosarcoma [5,9]. Interestingly, p53 not only regulates the expression of protein-coding genes but also regulates the maturation of miRNAs, lead to attenuation of miRNA processing activity [10,11]. And several p53-rugulated miRNAs have been identified as important components of the p53 tumor suppressor pathway, influence various cellular biological process in human osteosarcoma [12]. For example, p53-regulated miR-34 family (miR-34a, 34b and 34c) can induce G1 arrest and apoptosis in a p53-dependent manner in osteosarcoma cells [13,14]. Similarly, p53 also induced the upregulation of miR-31, miR-192, and miR-215 in osteosarcoma cells, and these three miRNAs were reported significantly influence p53-related cell proliferation or cell cycle of osteosarcoma cell lines [15,16].
These data indicate that p53 and these six miRNAs work together to influence series of biological behavior of human osteosarcoma cells. However, the relevance of these factors in the prognosis of osteosarcoma remains need to be elucidated. In order to shed light on the potential role of p53 and these six miRNAs, in this present study, we analyzed the expression of these miRNAs and p53 in surgically resected osteosarcoma patients and their relationship with disease-free survival (DFS).
Materials and methods
Study population
A total of 80 fresh tumor samples and corresponding noncancerous bone tissue samples from osteosarcoma patients who underwent complete surgical resection, without preoperative chemoradiotherapy, in our institution were prospectively collected between June 2007 and December 2009. Approval for the study was obtained from the Ethics Committee of Shanghai Changzheng Hospital. Written informed consent was obtained from each participant.
All 80 osteosarcoma patients received follow-up. Medical records were used to ascertain patients’ medical histories, including age, gender, anatomic location, tumor size, clinical stage, distant metastasis, response to chemotherapy, and status. All patients’ slides were reviewed to confirm the diagnosis and to classify the tumor according to the sixth edition of the tumor node metastases (TNM) classification of the International Union against Cancer (UICC). Patients with evidence of other malignancies were excluded from this study. All patients completed the standard therapeutic regimen including neoadjuvant chemotherapy and surgical resection with wide or radical margin followed by adjuvant chemotherapy.
RNA extraction and miRNA quantification
Total RNA was extracted from fresh frozen tissues using Trizol (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. 10 ng of total RNA was used for each miRNA quantification. miRNA detection was performed run on the Eppendorf Mastercycler EP Gradient S (Eppendorf, Germany) using commercial assays (TaqMan microRNA assays; Applied Biosystems, Foster City, CA, USA) for miRNAs. Relative quantification was calculated using 2-ΔΔCt, where Ct is cycle threshold. Normalization was performed with universal small nuclear RNA U6 (RNU6B). Each sample was examined in triplicate, and the mean values were calculated.
Immunohistochemistry and scoring
The primary antibodies for immunohistochemistry was performed by EnVision plus System. In brief, Paraffin specimens were cut at 4-μm thickness. Sections were deparaffinized with xylene, rehydrated, and heated for 10 min to retrieve antigen. Sections were incubated with the primary rabbit antibodies for human TP53 (Santa Cruz Biotech; 1:50) at room temperature for 1 h, followed by the secondary reaction with DAKO Envision+ Reagent (DakoCytomation, Carpinteria, CA). Staining positivity was evaluated by two independent observers without any knowledge of the clinicopathological information. TP53 expression was graded as follows: -, either no immunostaining, or else only minimal immunostaining was observed; +, either weak immunoreaction was observed or strong reaction was recognized in more than 10% of the tumor cells; and ++, >50% of tumor cells showed strong immunoreaction. We recognized cases grades as + or ++, and staining of more than 10% nuclei was needed for positive cases [17].
Statistical methods
All statistical analyses were performed with SPSS 19.0 (SPSS Inc., Chicago, IL, USA). Optimal cut-off points of miRNA expression data for DFS were assessed by means of the mean expression level of miRNA in corresponding noncancerous bone tissue samples. T-test was used to compare miRNA levels with variables with two values and ANOVA for variables with more than two values. DFS was calculated from the time of surgery to the date of progression (local and/or distal tumor recurrence) or to the date of death, or last follow-up. DFS was calculated using the Kaplan-Meier method and compared using the log-rank test. All variables with a p-value <0.05 in the univariate analysis were included in a Cox multivariate analysis (proportional hazard model). Differences were considered statistically significant when P<0.05.
Results
Patient characteristics
The characteristics of patients were shown in Table 1. Their mean age was 56 years (range 12-83 years). 46 (57.5%) patients were male. Clinical follow-up was conducted up to 66 months. The median follow-up duration was 33 months (1-72 months). According to the UICC system, there were 19 tumors of stage IIA and 61 of stage IIB/III. During the follow-up period, 38 (47.5%) patients died of disease. Computed tomography scan and/or magnetic resonance imaging scan was used to confirm the diagnosis when the tumor metastasis was suspected. There was no local recurrence. 49 patients (61.2%) had distant metastases at a mean of 14.7 months (range 1-49 months) after the original diagnosis. The median disease-free survival (DFS) of patients was 35 months (95% confidence interval [CI], 23.1-42.9 months).
Table 1.
Patient characteristics
| Clinicopathological features | No. of patients | P value | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| DFS | miR-34a | miR-34b | miR-34c | miR-31 | miR-192 | miR-215 | ||
| Age (years) | 0.378 | 0.503 | 0.452 | 0.621 | 0.257 | 0.343 | 0.250 | |
| ≤55 | 43 (53.8%) | |||||||
| >55 | 37 (46.2%) | |||||||
| Gender | 0.364 | 0.593 | 0.143 | 0.221 | 0.689 | 0.506 | 0.477 | |
| Male | 46 (57.5%) | |||||||
| Female | 34 (42.5%) | |||||||
| Anatomic location | 0.435 | 0.407 | 0.217 | 0.230 | 0.241 | 0.121 | 0.730 | |
| Tibia/femur | 59 (73.8%) | |||||||
| Elsewhere | 21 (26.2%) | |||||||
| Tumor size (cm) | 0045* | 0. 025* | 0.348 | 0.022* | 0.101 | 0.237 | 0.049* | |
| ≤8 | 50 (62.5%) | |||||||
| >8 | 30 (37.5%) | |||||||
| Clinical stage | 0.003* | 0.045* | 0.204 | 0.272 | 0.067 | 0.371 | 0.769 | |
| IIA | 19 (23.8%) | |||||||
| IIB/III | 61 (76.2%) | |||||||
| Distant metastasis | 0.004* | 0. 990 | 0.125 | 0.103 | 0.034* | 0.283 | 0.106 | |
| Negative | 31 (38.8%) | |||||||
| Positive | 49 (61.2%) | |||||||
| Response to chemotherapy | 0.037* | 0.098 | 0.190 | 0.337 | 0.308 | 0.018* | 0.031* | |
| Good | 33 (41.3%) | |||||||
| Poor | 47 (58.7%) | |||||||
| P53 expression | 0.001* | 0.078 | 0.194 | 0.451 | 0.154 | 0.012* | 0.046* | |
| High | 20 (25.0%) | |||||||
| Low | 60 (75.0%) | |||||||
miRNA expression in normal and tumor tissues
The normalized real-time PCR results showed that all six miRNAs were dysregulated in tumor tissues as compared with corresponding noncancerous tissue samples. The results showed that the mean fold change (relative quantitation (RQ)=2-ΔΔCt) was 0.69 for miR-34, 0.88 for miR-34b, 0.59 for miR-34c, 1.65 for miR-31, 0.71 for miR-192, and 0.35 for miR-215 (Figure 1). Low miR-34a expression was associated with patients with larger tumor size (p=0.025) and advanced clinical stage (p=0.045); low miR-34c expression was associated with patients with larger tumor size (p=0.022); high miR-31 expression was associated with distant metastasis (p=0.034); low miR-192 expression was associated with p53 positivity (p=0.012) and response to chemotherapy (p=0.018); and low miR-215 expression was associated with larger tumor size (p=0.049), response to chemotherapy (p=0.031), and p53 positivity (p=0.046). None association between miRNAs expression and other clinical characteristics was observed (Table 1).
Figure 1.
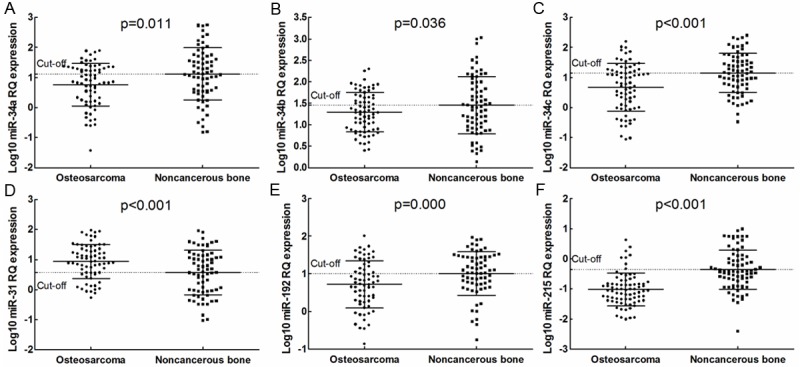
(A) miR-34a, (B) miR-34b, (C) miR-34c, (D) miR-31, (E) miR-192, and (F) miR-215 expression in tumor and normal tissue. RQ: relative quantitation.
miR-34a and miR-192 expression, individually and in combination
In order to examine the prognostic implications of the expression levels of miR-34 family, miR-31, miR-192, and miR-215, we used the cut-off point selected by means of the mean expression level of miRNAs in corresponding noncancerous bone tissue samples. All cut-off points discriminate two groups of patients: patients with high or low expression of miRNA, as observed in Figure 1. This only identified prognostic significant cut-off points for miR-34 (cut-off RQ 1.117) and miR-192 (cut-off RQ 1.012). When patients were divided according to the selected cut-off point, the median DFS for patients with low miR-34a levels was 34.3 months versus 64 months for patients with high miR-34a levels (p=0.000; Figure 2A). Median DFS for patients with low miR-192 levels was 36 months versus 61 months for patients with high miR-192 (p=0.022; Figure 2B). No other differences in DFS were observed according to the rest of four miRNAs expression levels (Data not shown).
Figure 2.
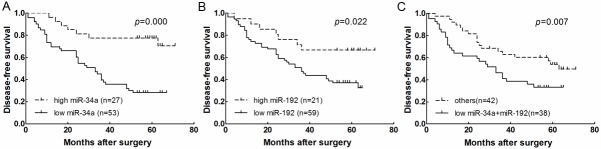
Kaplan-Meier curves for disease-free survival (DFS) according to the level of expression of (A) miR-34a, (B) miR-192 and (C) both unfavorable variables (low miR-34a and high miR-192) versus those with only one or no unfavorable variable.
In order to further explore the influence of miR-34a and miR-192, we then examined their effect in combination. When patients with both unfavorable variables (low-miR-34a and miR-192 expression), median DFS was 33 months for patients with both unfavorable variables versus 63 months for patients with only one or no unfavorable variable (p=0.007; Figure 2C).
p53 positivity and miR-192/215 expression
Immunohistochemical study revealed p53 positivity in neoplastic cells in 20 cases (13 cases showed strong immunostaining, 7 weak) (25.0%). Seventeen of 49 (34.7%) cases with distant metastasis after surgery and 13 of 45 (28.9%) cases whose final status was dead of disease showed p53 positivity. Representative immunohistochemical results are shown in Figure 3A. The normalized real-time PCR results from the 80 tumor samples showed that both patients with p53 positivity and negativity express miR-34 family and miR-31 at similar levels (data not shown). However, miR-192 and miR-215 expression in patients with p53 positivity was lower than patients with p53 negativity, with a mean fold change of 0.16 and 0.53, respectively (p=0.002 and p=0.004, respectively; Figure 3B, 3C). Median DFS was 21 months for the 20 patients with p53 positivity and 63 months for the remaining 60 with p53 negativity (p=0.013, Figure 4A). Median DFS was 11 months for the 11 patients with both low miR-192 expression and p53 positivity and 54 months for the remaining 69 patients (p=0.000, Figure 4B). In addition, median DFS of the 16 patients with both low miR-215 expression and p53 positivity was lower than that of the remaining 64 patients (31 vs. 52, p=0.047, Figure 4C).
Figure 3.
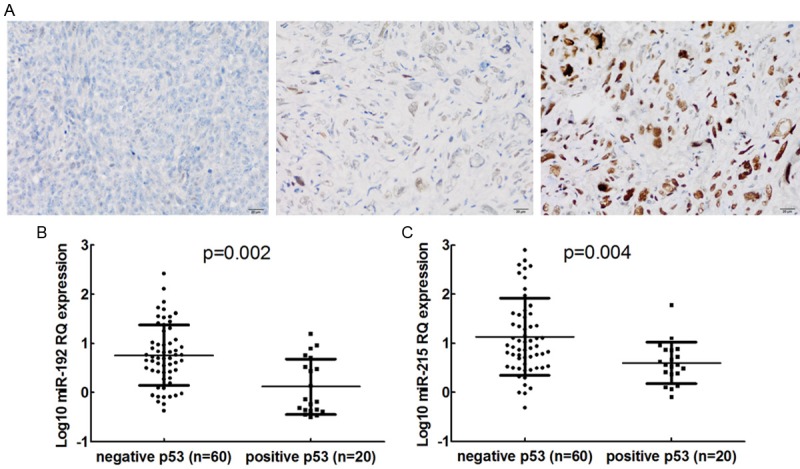
Representative immunohistochemical results of p53 expression (A) and p53 expression according to (B) miR-192 and (C) miR-215 expression levels. RQ: relative quantitation.
Figure 4.
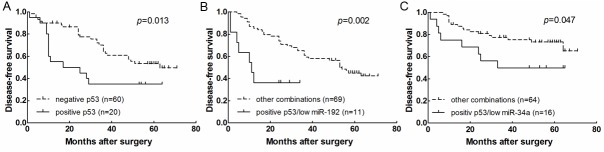
Kaplan-Meier curves for disease-free survival (DFS) in (A) patients with p53 positivity or negativity, patients with both p53 positivity and low miR-192 (B) and miR-215 (C) expression versus all other patients.
Multivariate analyses
A multivariate analysis of DFS was performed including clinical variables with univariate log-rank p<0.05: disease stage, distant metastasis, miR-34a expression, miR-192 expression and p53 expression. Low miR-34a expression (OR 3.344, 95% CI 1.434-7.779; p=0.001) and distant metastasis (OR 2.220, 95% CI 1.007-3.519; p=0.005) emerged as markers for shorter DFS, while stage IIA disease (OR 0.388, 95% CI 0.145-0.792; p=0.021) was an marker for longer DFS (Table 2).
Table 2.
Multivariate analysis of clinicopathological factors for disease-specific survival in osteosarcoma
| Variable | HR (95 % CI) | P value |
|---|---|---|
| Analysis with separately miRNA expression levels | ||
| Low miR-34a | 3.344 (1.437-7.779) | 0.001 |
| Distant metastasis | 2.200 (1.007-3.519) | 0.005 |
| Stage IIA disease | 0.388 (0.145-0.792) | 0.021 |
| Analysis including unfavorable# versus other combinations of miRNA expression and low miR-34a with or without p53 positivity | ||
| Low miR-34a and miR-192 | 1.343 (1.134-4.877) | 0.030 |
| Low miR-192 and p53 positivity | 4.183 (2.007-8.720) | 0.033 |
| Stage IIA disease | 0.492 (0.269-0.899) | 0.048 |
both unfavorable miRNA expression levels (low miR-34a and miR-192) versus those with only one or no unfavorable variable.
HR Hazard ratio, CI confidence interval, All statistical tests were 2-sided. Significance level: P<0.05.
A second multivariate analysis was then performed, including the same clinical variables, unfavorable miRNA expression levels (low miR-34a and miR-192) versus other combinations and low miR-192/215 expression with p53 positivity versus other combinations. In this second multivariate analysis, the combination of unfavorable miRNA expression levels (OR 1.343, 95% CI 1.134-4.877; p=0.030) and the combination of low miR-192 with p53 positivity (OR 4.183, 95% CI 2.007-8.720; p=0.033) were independent markers for shorter DFS, while stage IIA disease continued to be a marker for longer DFS (OR 0.492, 95% CI 0.269-0.899; p=0.048) (Table 2).
Discussion
The tumor suppressor p53 gene is one of the most frequently mutated genes in human cancers including osteosarcoma [6,18,19]. Together with the protein-coding genes, several miRNAs also act as important components of the p53 signaling cascades and thereby contribute to tumor suppression, mediate and regulate the malignant characters of multiple tumors. miR-34 family are the first miRNAs that have been found to be directly regulated by p53 [10], when ectopically expressed, miR-34 family display tumor suppressive activities in tumor biology [13]. Similarly, miR-31, miR-192, and miR-215 were also miRNAs that paly p53-associated role in inhibiting the proliferation of cancer cells [15,20-22]. These six miRNAs are all identified undergo deregulation in osteosarcoma [14-16,18]. In this present study, our results also showed significant differences in the expression levels of these six miRNAs between tumor and normal tissue from surgically resected osteosarcoma patients. While miR-31 showed a trend towards being significantly increased, miR-34 family expression was lower in tumor than in normal tissue, miR-192 and miR-215 showed heterogenous expression pattern, their expression level were all consistent with previous reports. To our surprise, although all these six miRNAs are p53-target miRNAs, only the expression of miR-192 and miR-215 is associated with p53 expression in our analysis results, this suggests that basal levels of the other four miRNAs are p53-independent, maybe some other transcriptional factors besides p53 such as SNAIL and TGF-β [23,24] are more dominant in the miRNA modulation, or other genetic and epigenetic modulations may also contribute to the altered expression of these miRNAs in osteosarcoma [14,25-27]. Further evidence is needed to prove this hypothesis, and this also provides a direction for our future study.
miR-34 family can function as tumor suppressors in human osteosarcoma cells, ectopic expression of miR-34 family leads to decrease of oncogenes, resulting in induce cell cycle arrest and apoptosis [14]; in addition, miR-34c is critical during the pathogenesis of osteosarcomas in part by regulating Notch signaling [28]. Along the same lines, miR-31 controls osteoclast formation and bone resorption by targeting RhoA [29], and overexpression of miR-31 repressed the osteogenesis of human mesenchymal stem cells (hMSCs) by targeting SATB2 [30]; and overexpression of miR-215 was shown to inhibited osteosarcoma cell proliferation and triggered cell cycle arrest at G2 phase [31]. All these findings indicated that these miRNAs maybe important regulators in tumorigenesis and progression of osteosarcoma, which also help to interpreted the results in our study that their correlation with malignant phenotype and clinical outcomes of patients with osteosarcoma.
miR-34a has been identified can be induced in response to p53 activation and mediates cell cycle arrest [22], moreover, there existence a positive feedback loop between miR-34a and p53 that target of miR-34a can increases p53 activity [32]. Like miR-34a, miR-192/215 expression also significant correlated with p53-dependent cell growth in multiple cancers [22], miR-192/215 can be transcriptionally activated by p53 and then preventing enhanced migration of plasma cells into bone marrow by modulate MDM2 expression [20], reduces cell proliferation by targeting p21 and p27 [21]. Moreover, p53 was highly overexpressed in cells that ectopic expression of miR-192 [33] or miR-215 target DTL [31]. Taken together, miR-34a and miR-192/215 may be requisite for the biological behavior modulation of cancer cells through the synergistic effect with p53, and there may exists some feed-forward mechanisms in p53-miRNAs signaling that promote the tumor progression in osteosarcoma. This is along the lines of findings in the present study, patients with both low-miR-34a and miR-192 expression, and patients with high p53 positivity and low miR-192/215 suggested a poorer prognosis, thus it is a plausible inference that the feed-forward/feedback regulatory networks based on p53 and miR-34a/192/215 play an important role in modulation of oncogenesis and deterioration.
To the best of our knowledge, while there are many recognized prognostic and predictive markers for osteosarcoma , including several protein and gene signatures, the present study is the first to explore the potential implications for tumor progression of p53-associated miRNAs related to osteosarcoma prognosis. The fact that various types of feed-forward/feedback loops exist between miRNAs, p53 and gene targets, they mediate the integrity, amplification, buffering and fine-tuning of signals that jointly contribute to the regulation of p53 signaling [11], leads us to speculate that the comprehensive analysis of p53 status with miRNAs expression may have further-reaching ramifications than that simple analysis of miRNAs expression. In order to achieve this, a better appreciation of the p53-associated miRNA-mRNA interactome is required.
In summary, our findings provide the first hints that miR-34a and miR-192 expression may be a useful prognostic marker that could be used for risk stratification and selection of osteosarcoma patients. In addition, the potential role of p53-associated miRNA in osteosarcoma warrants further investigation.
Disclosure of conflict of interest
The authors declare no conflict of interest.
References
- 1.Miao J, Wu S, Peng Z, Tania M, Zhang C. MicroRNAs in osteosarcoma: diagnostic and therapeutic aspects. Tumour Biol. 2013;34:2093–2098. doi: 10.1007/s13277-013-0940-7. [DOI] [PubMed] [Google Scholar]
- 2.Geller DS, Gorlick R. Osteosarcoma: a review of diagnosis, management, and treatment strategies. Clin Adv Hematol Oncol. 2010;8:705–718. [PubMed] [Google Scholar]
- 3.Bielack SS, Kempf-Bielack B, Delling G, Exner GU, Flege S, Helmke K, Kotz R, Salzer-Kuntschik M, Werner M, Winkelmann W, Zoubek A, Jurgens H, Winkler K. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J. Clin. Oncol. 2002;20:776–790. doi: 10.1200/JCO.2002.20.3.776. [DOI] [PubMed] [Google Scholar]
- 4.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 5.Yang J, Zhang W. New molecular insights into osteosarcoma targeted therapy. Curr Opin Oncol. 2013;25:398–406. doi: 10.1097/CCO.0b013e3283622c1b. [DOI] [PubMed] [Google Scholar]
- 6.Hauben EI, Arends J, Vandenbroucke JP, van Asperen CJ, Van Marck E, Hogendoorn PC. Multiple primary malignancies in osteosarcoma patients. Incidence and predictive value of osteosarcoma subtype for cancer syndromes related with osteosarcoma. Eur J Hum Genet. 2003;11:611–618. doi: 10.1038/sj.ejhg.5201012. [DOI] [PubMed] [Google Scholar]
- 7.Pakos EE, Kyzas PA, Ioannidis JP. Prognostic significance of TP53 tumor suppressor gene expression and mutations in human osteosarcoma: a meta-analysis. Clin Cancer Res. 2004;10:6208–6214. doi: 10.1158/1078-0432.CCR-04-0246. [DOI] [PubMed] [Google Scholar]
- 8.Fu HL, Shao L, Wang Q, Jia T, Li M, Yang DP. A systematic review of p53 as a biomarker of survival in patients with osteosarcoma. Tumour Biol. 2013;34:3817–3821. doi: 10.1007/s13277-013-0966-x. [DOI] [PubMed] [Google Scholar]
- 9.Zhou G, Shi X, Zhang J, Wu S, Zhao J. MicroRNAs in osteosarcoma: from biological players to clinical contributors, a review. J Int Med Res. 2013;41:1–12. doi: 10.1177/0300060513475959. [DOI] [PubMed] [Google Scholar]
- 10.Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer. 2012;12:613–626. doi: 10.1038/nrc3318. [DOI] [PubMed] [Google Scholar]
- 11.Krell J, Frampton AE, Colombo T, Gall TM, De Giorgio A, Harding V, Stebbing J, Castellano L. The p53 miRNA interactome and its potential role in the cancer clinic. Epigenomics. 2013;5:417–428. doi: 10.2217/epi.13.41. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi E, Hornicek FJ, Duan Z. MicroRNA Involvement in Osteosarcoma. Sarcoma. 2012;2012:359739. doi: 10.1155/2012/359739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 14.He C, Xiong J, Xu X, Lu W, Liu L, Xiao D, Wang D. Functional elucidation of MiR-34 in osteosarcoma cells and primary tumor samples. Biochem Biophys Res Commun. 2009;388:35–40. doi: 10.1016/j.bbrc.2009.07.101. [DOI] [PubMed] [Google Scholar]
- 15.Creighton CJ, Fountain MD, Yu Z, Nagaraja AK, Zhu H, Khan M, Olokpa E, Zariff A, Gunaratne PH, Matzuk MM, Anderson ML. Molecular profiling uncovers a p53-associated role for microRNA-31 in inhibiting the proliferation of serous ovarian carcinomas and other cancers. Cancer Res. 2010;70:1906–1915. doi: 10.1158/0008-5472.CAN-09-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, Orntoft TF, Andersen CL, Dobbelstein M. p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 2008;68:10094–10104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oda Y, Naka T, Takeshita M, Iwamoto Y, Tsuneyoshi M. Comparison of histological changes and changes in nm23 and c-MET expression between primary and metastatic sites in osteosarcoma: a clinicopathologic and immunohistochemical study. Hum Pathol. 2000;31:709–716. doi: 10.1053/hupa.2000.8230. [DOI] [PubMed] [Google Scholar]
- 18.Toguchida J, Yamaguchi T, Ritchie B, Beauchamp RL, Dayton SH, Herrera GE, Yamamuro T, Kotoura Y, Sasaki MS, Little JB, et al. Mutation spectrum of the p53 gene in bone and soft tissue sarcomas. Cancer Res. 1992;52:6194–6199. [PubMed] [Google Scholar]
- 19.Gokgoz N, Wunder JS, Mousses S, Eskandarian S, Bell RS, Andrulis IL. Comparison of p53 mutations in patients with localized osteosarcoma and metastatic osteosarcoma. Cancer. 2001;92:2181–2189. doi: 10.1002/1097-0142(20011015)92:8<2181::aid-cncr1561>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 20.Pichiorri F, Suh SS, Rocci A, De Luca L, Taccioli C, Santhanam R, Zhou W, Benson DM Jr, Hofmainster C, Alder H, Garofalo M, Di Leva G, Volinia S, Lin HJ, Perrotti D, Kuehl M, Aqeilan RI, Palumbo A, Croce CM. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell. 2010;18:367–381. doi: 10.1016/j.ccr.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Boni V, Bitarte N, Cristobal I, Zarate R, Rodriguez J, Maiello E, Garcia-Foncillas J, Bandres E. miR-192/miR-215 influence 5-fluorouracil resistance through cell cycle-mediated mechanisms complementary to its post-transcriptional thymidilate synthase regulation. Mol Cancer Ther. 2010;9:2265–2275. doi: 10.1158/1535-7163.MCT-10-0061. [DOI] [PubMed] [Google Scholar]
- 22.Georges SA, Biery MC, Kim SY, Schelter JM, Guo J, Chang AN, Jackson AL, Carleton MO, Linsley PS, Cleary MA, Chau BN. Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008;68:10105–10112. doi: 10.1158/0008-5472.CAN-08-1846. [DOI] [PubMed] [Google Scholar]
- 23.Siemens H, Jackstadt R, Hunten S, Kaller M, Menssen A, Gotz U, Hermeking H. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10:4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- 24.Jenkins RH, Martin J, Phillips AO, Bowen T, Fraser DJ. Transforming growth factor beta1 represses proximal tubular cell microRNA-192 expression through decreased hepatocyte nuclear factor DNA binding. Biochem J. 2012;443:407–416. doi: 10.1042/BJ20111861. [DOI] [PubMed] [Google Scholar]
- 25.Lin PC, Chiu YL, Banerjee S, Park K, Mosquera JM, Giannopoulou E, Alves P, Tewari AK, Gerstein MB, Beltran H, Melnick AM, Elemento O, Demichelis F, Rubin MA. Epigenetic repression of miR-31 disrupts androgen receptor homeostasis and contributes to prostate cancer progression. Cancer Res. 2013;73:1232–1244. doi: 10.1158/0008-5472.CAN-12-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asangani IA, Harms PW, Dodson L, Pandhi M, Kunju LP, Maher CA, Fullen DR, Johnson TM, Giordano TJ, Palanisamy N, Chinnaiyan AM. Genetic and epigenetic loss of microRNA-31 leads to feed-forward expression of EZH2 in melanoma. Oncotarget. 2012;3:1011–1025. doi: 10.18632/oncotarget.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito M, Mitsuhashi K, Igarashi H, Nosho K, Naito T, Yoshii S, Takahashi H, Fujita M, Sukawa Y, Yamamoto E, Takahashi T, Adachi Y, Nojima M, Sasaki Y, Tokino T, Baba Y, Maruyama R, Suzuki H, Imai K, Yamamoto H, Shinomura Y. MicroRNA-31 expression in relation to BRAF mutation, CpG island methylation and colorectal continuum in serrated lesions. Int J Cancer. 2014;135:2507–2515. doi: 10.1002/ijc.28920. [DOI] [PubMed] [Google Scholar]
- 28.Bae Y, Yang T, Zeng HC, Campeau PM, Chen Y, Bertin T, Dawson BC, Munivez E, Tao J, Lee BH. miRNA-34c regulates Notch signaling during bone development. Hum Mol Genet. 2012;21:2991–3000. doi: 10.1093/hmg/dds129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizoguchi F, Murakami Y, Saito T, Miyasaka N, Kohsaka H. miR-31 controls osteoclast formation and bone resorption by targeting RhoA. Arthritis Res Ther. 2013;15:R102. doi: 10.1186/ar4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie Q, Wang Z, Bi X, Zhou H, Wang Y, Gu P, Fan X. Effects of miR-31 on the osteogenesis of human mesenchymal stem cells. Biochem Biophys Res Commun. 2014;446:98–104. doi: 10.1016/j.bbrc.2014.02.058. [DOI] [PubMed] [Google Scholar]
- 31.Song B, Wang Y, Titmus MA, Botchkina G, Formentini A, Kornmann M, Ju J. Molecular mechanism of chemoresistance by miR-215 in osteosarcoma and colon cancer cells. Mol Cancer. 2010;9:96. doi: 10.1186/1476-4598-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamakuchi M, Lowenstein CJ. MiR-34, SIRT1 and p53: the feedback loop. Cell Cycle. 2009;8:712–715. doi: 10.4161/cc.8.5.7753. [DOI] [PubMed] [Google Scholar]
- 33.Song B, Wang Y, Kudo K, Gavin EJ, Xi Y, Ju J. miR-192 Regulates dihydrofolate reductase and cellular proliferation through the p53-microRNA circuit. Clin Cancer Res. 2008;14:8080–8086. doi: 10.1158/1078-0432.CCR-08-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]


