Abstract
Loss of heterozygosity (LOH) plays important roles in genome dynamics, notably, during tumorigenesis. In the fungal pathogen Candida albicans, LOH contributes to the acquisition of antifungal resistance. In order to investigate the mechanisms that regulate LOH in C. albicans, we have established a novel method combining an artificial heterozygous locus harboring the blue fluorescent protein and green fluorescent protein markers and flow cytometry to detect LOH events at the single-cell level. Using this fluorescence-based method, we have confirmed that elevated temperature, treatment with methyl methanesulfonate, and inactivation of the Mec1 DNA damage checkpoint kinase triggered an increase in the frequency of LOH. Taking advantage of this system, we have searched for C. albicans genes whose overexpression triggered an increase in LOH and identified four candidates, some of which are known regulators of genome dynamics with human homologues contributing to cancer progression. Hence, the approach presented here will allow the implementation of new screens to identify genes that are important for genome stability in C. albicans and more generally in eukaryotic cells.
INTRODUCTION
Normally found as a harmless commensal organism, Candida albicans is also a major fungal pathogen of humans and is capable of causing serious and even life-threatening diseases when the immune system of the host is compromised (1). Although C. albicans is found mostly as a diploid organism, haploid and tetraploid forms have been observed in the laboratory and upon the passage of C. albicans in animal models of infection (2–4). The formation of tetraploids results from mating between diploids of opposite mating types that have undergone the so-called white-opaque phenotypic switch (5). Meiosis is thought to have been lost in C. albicans, and the formation of haploids from diploids or diploids from tetraploids results from concerted chromosome loss (2, 6). The C. albicans genome is highly plastic, undergoing a number of important genome rearrangements, such as loss-of-heterozygosity (LOH) events, aneuploidies, and the formation of isochromosomes (7). In particular, LOH has been shown to occur during commensal carriage (8) and during systemic infection in a mouse model (9) and to contribute to the acquisition of antifungal resistance (10, 11). Stressful conditions such as high temperature, oxidative stress, or azole antifungal treatment trigger an increase in the frequency of LOH, as well as changes in the mechanisms leading to LOH; while azole treatment and temperature cause an increase in LOH due to chromosome loss and reduplication, oxidative stress results in an increase in gene conversion events (12). Yet, little is known about the molecular mechanisms that control LOH in C. albicans, despite the apparent importance of LOH in the biology of this species. Overall, it has been shown that knockout mutations in genes involved in base excision repair, nucleotide excision repair, and mismatch repair had little impact on the frequency of LOH in C. albicans (13, 14). In contrast, null mutations in the MRE11 and RAD50 genes involved in homologous recombination and in the MEC1, RAD53, and DUN1 genes involved in the DNA damage checkpoint pathway result in increased frequency of LOH (13, 15, 16).
Evaluation of the frequency of LOH in C. albicans has until now relied on the use of several counterselectable marker genes, such as URA3 and GAL1, that confer distinctive phenotypes when in a heterozygous state or in a homozygous null state (17–19). URA3 encodes the orotidine-5-phosphate decarboxylase; while URA3/URA3 and URA3/ura3 strains are prototrophic for uridine and sensitive to 5-fluoroorotic acid (5-FOA), ura3/ura3 strains are auxotrophic for uridine and resistant to 5-FOA. GAL1 encodes a galactokinase; while GAL1/GAL1 and GAL1/gal1 strains are sensitive to 2-deoxygalactose (2-DG), gal1/gal1 strains are resistant to 2-DG. Hence, counting of the spontaneous 5-FOA-resistant or 2-DG-resistant clones that arise from a URA3/ura3 or a GAL1/gal1 strain, respectively, provides a measure of the frequency of LOH in these strains. Although robust, these systems are labor intensive and costly and lack the flexibility that would be required to implement a genetic screen for C. albicans genes regulating LOH. With the development of collections of knockout constructs or overexpression plasmids, such screens are becoming amenable to C. albicans (20–22) and could provide new insights into the mechanisms that control genome stability in this species and other eukaryotes.
Here, we report the development of a novel LOH reporter system that combines fluorescent markers and flow cytometry to detect LOH in C. albicans and is amenable to high-throughput screening. We show that this system can report changes in the frequency of LOH triggered by different physical and chemical stresses and knockout mutations. Moreover, using a newly developed collection of overexpression plasmids for 124 genes whose orthologs in Saccharomyces cerevisiae are involved in DNA replication, recombination, and repair, we show that the LOH reporter system can be used to identify genes whose overexpression triggers genome instability specifically through LOH events. Overall, the characterization of new players in genome maintenance will allow a better understanding of how genomic instability may contribute to the success of C. albicans as a commensal and as a pathogen.
MATERIALS AND METHODS
Strains and growth conditions.
C. albicans strains were routinely cultured at 30°C in YPD medium (1% yeast extract, 2% peptone, 2% dextrose), synthetic complete (SC) medium with the omission of the appropriate amino acids (23) or synthetic dextrose (SD) medium (0.67% yeast nitrogen base, 2% dextrose). Solid media were obtained by adding 2% agar. All of the C. albicans strains used in this work were derived from SN148 (24) and are listed in Table 1. For selection of transformants, nourseothricin was used at a final concentration of 300 μg/ml in YPD medium.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Characteristic | Reference |
|---|---|---|---|
| SN100 | his1Δ/his1Δ URA3/ura3Δ::λimm434 IRO1/iro1Δ::λimm434 | WT | 24 |
| SN148 | arg4Δ/arg4Δleu2Δ/leu2Δ his1Δ/his1Δura3::λimm434/ura3Δ::λimm434 iro1Δ::λimm434/iro1Δ::λimm434 | WT | 24 |
| CEC1027 | SN148 GAL1/gal1::CmLEU2 | GAL1 het | This study |
| CEC3998 | SN148 Ca21chr4_C_albicans_SC5314:473390-476401Δ::PTDH3-GFP-CaARG4/Ca21chr4_C_albicans_SC5314:473390-476401/Ca21chr1_C_albicans_SC5314:625304-626436 RPS1/RPS1::CIp20-Leu2 | Mono-GFP prototroph | This study |
| CEC4007 | SN148 Ca21chr4_C_albicans_SC5314:473390-476401Δ::PTDH3-BFP-CdHIS/Ca21chr4_C_albicans_SC5314:473390-476401/Ca21chr1_C_albicans_SC5314:625304-626436 RPS1/RPS1::CIp30 GAL1/gal1Δ::CmLEU2 | Mono-BFP prototroph | This study |
| CEC2683 | SN148 Ca21chr4_C_albicans_SC5314:473390-476401Δ::PTDH3-GFP-CaARG4/Ca21chr4_C_albicans_SC5314:473390-476401Δ::PTDH3-BFP-CaSAT1 | BFP-GFP | This study |
| CEC2814 | CEC1027 Ca21chr4_C_albicans_SC5314:473390-476401Δ::PTDH3-GFP-CaARG4/Ca21chr4_C_albicans_SC5314:473390-476401Δ::PTDH3-BFP-CdHIS1 | GAL1 het BFP-GFP | This study |
| CEC2824 | CEC2814 ADH1/adh1::PTDH3-cartTA::SAT1 | GAL1 het BFP-GFP/pNIMX | This study |
| CEC3172 | CEC2683 MEC1/mec1Δ::HIS1 | mec1Δ auxotroph | This study |
| CEC3183 | CEC2683 mec1Δ::LEU2/mec1Δ::HIS1 | mec1ΔΔ auxotroph | This study |
| CEC3194 | CEC2683 RPS1/RPS1::CIp30 | MEC1 prototroph | This study |
| CEC3203 | CEC3172 RPS1/RPS1::CIp30 | mec1Δ prototroph | This study |
| CEC3216 | CEC3183 RPS1/RPS1::CIp10 | mec1ΔΔ prototroph | This study |
| CEC3989 | CEC2824 RPS1/RPS1::PTET-GtwB | 3R collection control | This study |
For the LOH assay by fluorescence-activated cell sorting (FACS), aliquots of prototrophic cells were taken from the −80°C stock and grown in 96-well plates overnight in YPD medium at 30°C with rotatory shaking for recovery. The next day, these cultures were inoculated into SC-His-Arg to eliminate cells that had undergone LOH and lost the blue fluorescent protein (BFP)-HIS1 or green fluorescent protein (GFP)-ARG4 cassette as a result of the freezing-thawing step. The SC-His-Arg cultures were grown overnight at 30°C with rotatory shaking. At that point, the cells were ready for possible treatments before LOH analysis by flow cytometry.
Overexpression from PTET was induced by the addition of anhydrotetracycline (ATc, 3 μg/ml; Fisher Bioblock Scientific) in YPD or SD medium at 30°C. Overexpression experiments were carried out in the dark, as ATc is light sensitive.
For heat shock, C. albicans strain CEC3989 (Table 1) was grown in YPD medium overnight at 30, 37, or 39°C and allowed to recover overnight in YPD medium before flow cytometry analysis.
For genotoxic stress, CEC3989 was cultured for 30 min in YPD medium plus 0.01, 0.02, or 0.03% methyl methanesulfonate (MMS) at 30°C. The cells were collected, washed with fresh YPD medium, and allowed to recover in rich medium at 30°C overnight before flow cytometry analysis.
Plasmid constructions.
For the oligonucleotides used in this work, see Tables S1 and S2 in the supplemental material. For the plasmids used or generated in this work, see Table S3 in the supplemental material. For descriptions of the plasmid constructions, see Text S1 in the supplemental material. They yielded six plasmids harboring (i) the gene for either GFP or BFP placed under the control of the constitutively active promoter of the C. albicans TDH3 gene (25) and (ii) the ARG4 (26), HIS1 (27), URA3 (26), or SAT1 (27) transformation marker (see Fig. S1 in the supplemental material).
C. albicans strain construction. (i) Multiple heterozygous marker strains.
One allele of the GAL1 gene was replaced with the CmLEU2 marker gene by PCR-mediated transformation of SN148 (24). The resulting strain was named CEC1027. The PTDH3-BFP-HIS1 and PTDH3-GFP-ARG4 cassettes were PCR amplified from plasmids pECC727-BH (ECC727) and pECC596-GA (ECC596) with oligonucleotides K7 yFP-Fwd and K7 yFP-Rev carrying sequences homologous to the genomic DNA located between PGA59 and PGA62 on chromosome 4 (Ch4). The PCR products were sequentially transformed in CEC1027 as described by Gola et al. (26), yielding CEC2814. For activation of the PTET promoter, strain CEC2814 was eventually transformed with SacII- and KpnI-digested pNIMX, which encodes a tetracycline-controlled transactivator (21), yielding CEC2824. Strain CEC2683 was obtained through successive transformation of SN148 (24) with the PTDH3-BFP-SAT1 and PTDH3-GFP-ARG4 cassettes PCR amplified from plasmids pECC729-BS (ECC729) and pECC596-GA (ECC596) with oligonucleotides K7 yFP-Fwd and K7 yFP-Rev. CEC2683 was subsequently transformed with StuI-linearized CIp30 (28) to generate CEC3194, the prototrophic parental strain.
A mec1 null mutant was constructed by replacing both alleles with the LEU2 and HIS1 markers by a PCR-based method (29) in CEC2683, yielding first the heterozygous MEC1/mec1 mutant CEC3172 and then the null mec1/mec1 mutant CEC3183. CEC3172 and CEC3183 were then transformed with StuI-linearized CIp30 (28) or CIp10 (30) to generate CEC3203 and CEC3216, the prototrophic mec1 heterozygous and homozygous mutants.
(ii) C. albicans overexpression strains.
StuI-digested or I-SceI-digested CIp10-PTET-GTW derivatives were transformed into strain CEC2824 as described by Chauvel et al. (21), resulting in 124 overexpression strains (see Table S2 in the supplemental material) that, together with the CEC3989 control strain derived from CEC2824 transformed with StuI-linearized CIp10-PTET-GTW, will be referred to as the 3R overexpression collection.
All C. albicans transformants were verified by PCR with primers inside the transformed plasmid/cassette and primers in the genomic DNA regions of insertion in order to assess the proper integration of the plasmid/cassette in the C. albicans genome.
Flow cytometry analysis.
Cells from overnight cultures were diluted 1:1,000 in 1× phosphate-buffered saline (PBS) in BD Falcon tubes (product code 352054), and 106 cells were analyzed by flow cytometry in a 96-well plate format with a MACSQuant (Miltenyi) flow cytometer. We used the 405- and 488-nm lasers to excite the BFP and GFP proteins and the 425/475 and 500/550 filters to detect the BFP and GFP emission signals.
LOH screen with the BFP-GFP system. (i) Primary screen.
Strains in the 3R overexpression collection were screened for the frequency of LOH by flow cytometry following overnight growth at 30°C in YPD medium containing ATc (3 μg/ml). To evaluate the frequency of LOH at the GFP or BFP loci, we first analyzed the flow cytometry output for 14 independent cultures of the control strain (CEC3989) with FlowJo 7.6. All profiles were very similar. We created gates to define the Bfp+ Gfp− (mono-BFP) and Bfp− Gfp+ (mono-GFP) populations in one of the control samples and applied these gates to the rest of the data set for the control cultures. The percentages of Bfp+ Gfp− and Bfp− Gfp+ cells were exported in Excel format. The mean and standard deviation were calculated. The same gates were applied to all of the mutants, resulting in percentages of Bfp+ Gfp− and Bfp− Gfp+ cells for each mutant strain. We calculated a Z score for each mutant and selected mutants that showed Z scores of >5 for the Bfp− Gfp+ population (corresponding to 5 standard deviations from the mean value of the control strain).
(ii) Secondary screen.
Because the overexpression-induced morphology changes observed in some of the candidate overexpression mutants (data not shown) could distort the flow cytometry results, a secondary screen was carried out to confirm the candidate genes. All of the mutants identified in the primary screen were grown in triplicate in YPD medium and in YPD-ATc (3 μg/ml) for 8 h and reinoculated into YPD medium alone to recover overnight at 30°C. The next day, 106 cells in the yeast form, as confirmed by microscopy, were analyzed by flow cytometry and a t test was performed to validate the candidates.
Sorting of cells having undergone a LOH event.
The control strain and the candidate mutants were grown in the presence of ATc (3 μg/ml) for 8 h and allowed to recover overnight in YPD medium alone. FACS of these cultures into 1.5-ml Eppendorf tubes with a FACSAria III cell sorter (BD Biosciences) was performed at a rate of 10,000 events/s. A thousand cells from the Bfp− Gfp+ and Bfp+ Gfp− populations were sorted, recovered in 400 μl of YPD medium, and immediately plated onto YPD medium plates. The plates were incubated at 30°C for 2 days. For the Bfp− Gfp+ population, 48 colonies were inoculated into 800 μl of YPD medium in 96-well plates and grown overnight at 30°C. We observed a colony size phenotype of the Bfp+ Gfp− population, and therefore, 24 small and 24 large colonies were inoculated into 800 μl of YPD medium in 96-well plates and grown overnight at 30°C. Cultures were then analyzed by flow cytometry to validate monofluorescence and spotted onto YPD medium, SC-His-Arg, SC-His, and SC-Arg to confirm the LOH events by checking the loss of the auxotrophic marker associated with the BFP or GFP marker. We next investigated the molecular mechanisms giving rise to these LOH events by single-nucleotide polymorphism (SNP) typing of 16 isolates, when possible. Genomic DNA was extracted from the sorted clones, and PCR-restriction fragment length polymorphism (RFLP) was performed to assess the extent of the LOH. We used SNPs 95 and 156, which are part of AluI and TaqI restriction sites, respectively (31). The SNP typing analysis was carried out for both the Bfp+ Gfp− (small and large colonies) and Bfp− Gfp+ populations and was performed as described previously (31).
Whole-genome sequencing and sequence analysis.
Genomic DNAs isolated from the C. albicans isolates were processed to prepare libraries for Illumina sequencing. The TruSeq Nano DNA Sample Prep kit (Illumina) was used according to the manufacturer's recommendations. DNA was randomly fragmented by sonication to an average fragment length of 550 bp, and Illumina adapters were blunt-end ligated to the fragments. The final libraries were amplified by PCR and then sequenced on a MiSeq platform by using v3 chemistry. Three-hundred-nucleotide paired-end reads were aligned with C. albicans strain SC5314 reference genome assembly 21, which is available from the Candida Genome Database (32, 33), by Burrows-Wheeler Aligner (34). Single-nucleotide variants (SNVs) between the sequenced genomes and the reference genome were identified with the Genome Analysis Toolkit (35) at positions with a sequencing depth of ≥18×. Heterozygous SNVs were defined as positions where 15% or more of the calls showed one allele and 85% or less of the calls showed a second allele. Homozygous SNVs were defined as positions where more than 90% of the calls differed from the reference genome. Sequencing depth and heterozygous SNP density maps were constructed as described by Loll-Krippleber et al. (16). Homozygous SNP density maps were constructed by determining the number of homozygous positions per 10-kb region and plotting each value.
RESULTS
A new reporter system for LOH.
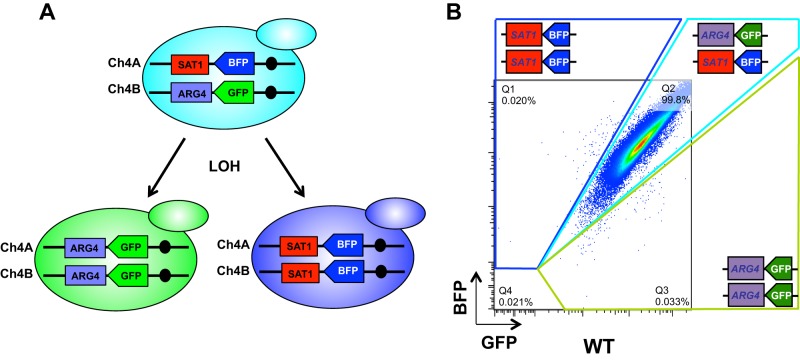
As an alternative to the GAL1 and URA3 systems routinely used for studies of LOH in C. albicans (12, 18), we developed a new system combining the use of fluorescent markers and flow cytometry that is more suitable for high-throughput studies. This system relies on the construction of an artificial locus on Ch4 in the intergenic region between the PGA59 and PGA62 open reading frames (ORFs) (36). This 9-kb region has been used previously as a platform for the integration of C. albicans two-hybrid plasmids (37), and its modification is thought to be neutral to C. albicans biology, as its deletion did not result in any phenotype (36). As shown in Fig. 1A, one of the Ch4 homologs was engineered to carry the gene for GFP (38) under the control of the promoter of the constitutively highly expressed TDH3 gene (25) and linked to an auxotrophic marker (ARG4, HIS1, URA3) or the nourseothricin resistance marker (SAT1). The other homolog was engineered to carry the gene for BFP (Evrogen) placed under the control of the TDH3 promoter and linked to another auxotrophic marker or the nourseothricin resistance marker. A series of cassettes have been developed that associate the BFP or GFP gene to different selection markers (SAT1, HIS1, URA3, and ARG4), allowing their use in various genetic backgrounds (see Fig. S1 in the supplemental material). In this setup, LOH events can be detected by flow cytometry since cells that undergo LOH at the PGA59-62 locus will produce only one of the two fluorescent proteins. As shown in Fig. 1B, FACS analysis of strain CEC2683, which carries the BFP-SAT1/GFP-ARG4 system, showed that the majority (>99%) of the cells coexpressed the two fluorescent proteins BFP and GFP. In addition, mono-GFP and mono-BFP populations were observed at frequencies of 0.020 and 0.033%, respectively. In order to verify that the cells in these populations had undergone genuine LOH events, we recovered cells from each population by cell sorting and characterized 76 and 52 cells from the mono-GFP and mono-BPF populations, respectively. Of the cells recovered from both populations, 95% (72/76) and 94% (49/52) were monofluorescent (data not shown). Moreover, spotting assays on appropriate media indicated that all of the monofluorescent cells displayed the phenotypes expected from a concomitant loss of the BFP and SAT1 genes (mono-GFP cells) or the GFP and ARG4 genes (mono-BFP cells), i.e., sensitivity to nourseothricin or arginine auxotrophy, respectively (data not shown). This initial analysis of a BFP-GFP strain revealed a spontaneous LOH frequency of 2 × 10−4 to 3 × 10−4 at this locus. Interestingly, the cells recovered from the Bfp− Gfp− population did not grow, indicating that these are nonviable cells. Fluorescence plots of mono-BFP (CEC4007), mono-GFP (CEC3998), unlabeled (SN100), and BFP-GFP-labeled strains are presented in Fig. S2A in the supplemental material.
FIG 1.
The BFP-GFP system. (A) Artificial heterozygous locus on Ch4. BFP and GFP expression cassettes were integrated into the gene-free PGA59-PGA62 intergenic region of Ch4. One homolog carries the gene coding for BFP, while the other carries the gene coding for GFP. Both the BFP and GFP genes are under the control of the promoter of the constitutively highly expressed TDH3 gene and linked to a selection marker. When LOH takes place at this locus, the cells express only one of the fluorescent proteins and LOH events can be detected by flow cytometry. (B) Flow cytometry analysis of a WT BFP-GFP strain (CEC2683). Cells were grown overnight in rich medium, diluted in 1× PBS, and analyzed on a MACSQuant cytometer (Miltenyi Biotech). One million events are displayed.
Increased LOH at the PGA59-62 locus in mec1 mutants.
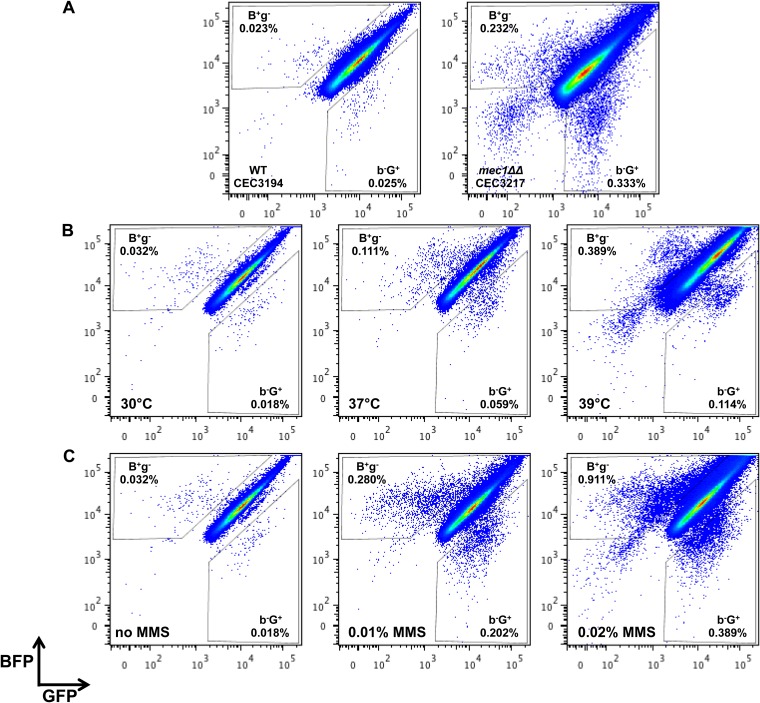
The Mec1 kinase is a central regulator of the DNA damage checkpoint in all eukaryotes (39, 40). A previous study has shown that inactivation of the C. albicans MEC1 gene results in an increase in LOH at the GAL1 locus (15). In order to test whether an increase in LOH could be detected with the BFP-GFP system, we inactivated the MEC1 gene in a strain carrying this system. Results presented in Fig. 2A and Table 2 show a statistically significantly greater proportion of mono-GFP or mono-BFP cells in the mec1 null mutant than in the wild-type (WT) strain. Interestingly, we also observed an increase in the proportion of Bfp− Gfp− cells, suggesting that inactivation of MEC1 increases cell death (Fig. 2A). Cell death in the Bfp− Gfp− population was confirmed by propidium iodide staining and microscopic analysis of these cells after cell sorting (data not shown). Hence, the BFP-GFP LOH reporter system could detect an increase in the frequency of LOH by flow cytometry upon inactivation of the MEC1 gene.
FIG 2.
Flow cytometry analysis reveals an increase in LOH in mec1 null mutants and in response to physical and chemical stresses. (A) Flow cytometry analysis of homozygous mec1 deletion strains. WT BFP-GFP strain CEC3194 and the mec1ΔΔ mutant were grown overnight in YPD medium at 30°C and analyzed by flow cytometry. (B) Flow cytometry analysis upon heat treatment. WT BFP-GFP strain CEC3989 was grown overnight in YPD medium at 30, 37, or 39°C and allowed to recover overnight in YPD medium before flow cytometry analysis. (C) Flow cytometry analysis in the presence of the DNA-damaging agent MMS. WT BFP-GFP strain CEC3989 was treated with increasing concentrations of MMS for 30 min. Cells were then allowed to recover overnight in fresh YPD medium and analyzed by flow cytometry.
TABLE 2.
LOH increase in the mec1 null mutant and in response to physical and chemical stresses
| Genotype, lab ID, and growth conditions | LOH quantification on Ch 4 (BFP loss) |
||
|---|---|---|---|
| Mean frequency of LOH ± SEM (×10−4)a | Fold change | P valued | |
| WT, CEC3194, YPD, 30°C | 2.6 ± 0.2 | ||
| mec1Δ, CEC3206, YPD, 30°C | 2.2 ± 0.1 | 1b | ≤0.005 |
| mec1ΔΔ, CEC3216, YPD, 30°C | 31.0 ± 1.0 | 12b | ≤0.005 |
| WT, CEC3989 | |||
| YPD, 30°C | 0.8 ± 0.1 | ||
| YPD, 37°C | 1.2 ± 0.1 | 1.4c | ≤0.05 |
| YPD, 39°C | 1.3 ± 0.1 | 1.5c | ≤0.01 |
| YPD + 0.01% MMS, 30°C | 19.4 ± 2.6 | 12c | ≤0.005 |
| YPD + 0.02% MMS, 30°C | 45.3 ± 1.8 | 28c | ≤0.005 |
| YPD + 0.03% MMS, 30°C | 59.0 ± 1.8 | 37c | ≤0.005 |
Six independent cultures were tested per mec1 mutant and per physical and/or chemical stress condition. Standard errors of the means were determined with GraphPad Prism software.
Ratio of the LOH frequency of a given mutant to that of the parental strain.
Ratio of the LOH frequency in the presence of the stress to that in the absence of the stress.
A Mann-Whitney test was performed to determine if the BFP loss frequency in the different mutants was statistically significantly different from the rate observed in the control strains or under the control conditions.
Increased LOH at the PGA59-62 locus in response to different stresses.
Forche et al. (12) have shown increased LOH in response to different stresses that are reminiscent of the conditions encountered by fungi during human infection, e.g., elevated temperatures, oxidative stress, and the presence of azole antifungals. We evaluated the frequency of LOH in a control strain carrying the BFP-GFP system when grown overnight at 30, 37, or 39°C. Although cells were filamenting after 7 h of growth at 37 or 39°C, microscopic analysis of cells after overnight growth at 37 or 39°C revealed that the majority of the cells reverted to the yeast form and were therefore suitable for flow cytometry analysis (data not shown). Still, we allowed the cells to recover overnight in YPD medium before flow cytometry analysis. Similar to the data obtained with the GAL1 system (12), the BFP-GFP system revealed that elevated temperatures promote LOH in C. albicans (Fig. 2B and Table 2). The fluorescence plots presented in Fig. S2B in the supplemental material confirmed that the distribution of monolabeled cells remains the same in the absence or presence of heat stress.
We also treated the control strain carrying the BFP-GFP system with the genotoxic agent MMS. After a 30-min treatment with different concentrations of MMS, cells were allowed to recover in rich medium overnight. Flow cytometry analysis revealed a frequency of LOH that increased together with MMS concentrations (Fig. 2C and Table 2).
Taken together, these data indicated that the BFP-GFP LOH reporter system was suitable for the detection of increases in the frequency of LOH resulting from physical or chemical stress.
Construction of the 3R overexpression collection.
We reasoned that the BFP-GFP LOH reporter system could provide an efficient means for the identification of genes that control LOH in C. albicans. Therefore, we implemented an overexpression screen aimed at identifying C. albicans genes whose overexpression would alter the WT frequency of LOH. To do this, we generated a collection of C. albicans overexpression strains focused on genes that, on the basis of their Gene Ontology database annotations or the function of their orthologs in S. cerevisiae, were likely to be involved in different aspects of genome maintenance such as DNA repair, DNA replication, recombination, chromosome segregation, the cell cycle, and telomere maintenance. PCR products extending from the start codon to the penultimate codon of 204 selected genes (see Table S2 in the supplemental material) were cloned into the pDONR207 donor vector. Following Sanger and Illumina/Solexa sequence validation, a total of 151 (74%) derivatives of pDONR207 were obtained. ORFs cloned into pDONR207 were subsequently transferred into uniquely barcoded CIp10-PTET-GTW plasmids (20, 21). A total of 147 (72%) CIp10-PTET-GTW derivatives were obtained and subsequently introduced at the RPS1 locus into C. albicans strain CEC2824, which harbors a GFP-HIS1/BFP-ARG4 system at the PGA59-62 locus and the pNIMX plasmid encoding a tetracycline-controlled transactivator (21). Eventually, 124 C. albicans PTET-driven overexpression strains were obtained, indicating a 60.8% overall success rate, slightly below that reported previously by Chauvel et al. (21) for a nonoverlapping set of overexpression plasmids, possibly because of the larger size of the cloned ORFs in our study (27% were >2,500 bp versus none in the study of Chauvel et al.). Together with the CEC3989 control strain harboring the empty CIp10-PTET-GTW plasmid, these 124 overexpression strains are referred to here as the 3R overexpression collection.
FACS-optimized LOH screens upon gene overexpression.
Prior to analyzing the overexpression mutants for the frequency of LOH under inducing conditions, we evaluated the frequency of LOH in control strain CEC3989. Fourteen independent YPD-ATc (3 μg/ml) cultures of the control strain were analyzed by flow cytometry. For each analysis, 1 million cells were tested and the mean frequency of LOH and the standard deviation were calculated. We found that LOH occurred at an average frequency of 2.2 × 10−4 ± 0.4 × 10−4. Subsequently, we submitted the entire 3R overexpression collection to a primary screen whereby 1 million cells of each overexpression strain were analyzed by flow cytometry after overnight induction. Thirty-three overexpression strains were selected that had a Z score (for the frequency of Bfp− Gfp+ cells) of >5 (see Fig. S3 in the supplemental material). In the secondary screen, we allowed the cells to recover in rich medium after 8 h of induction in YPD-ATc3 medium and reanalyzed the 33 overexpression strains identified in the primary screen in triplicate. While we saw no difference between Bfp+ Gfp− and Bfp− Gfp+ frequencies of the control strain and the overexpression mutants in the absence of ATc treatment, we confirmed the increase in LOH at the PGA59-62 locus after ATc treatment for four of these genes, namely, BIM1 (10-fold for Bfp+ Gfp− cells and 13-fold for Bfp− Gfp+ cells), CDC20 (32- and 38-fold, respectively), RAD51 (7- and 8-fold, respectively), and RAD53 (13-fold) (Table 3). Notably, we could not identify any overexpression strain that had a significantly reduced frequency of LOH, possibly owing to the relatively low frequency of LOH in the control strain.
TABLE 3.
LOH quantification in the four candidate overexpression mutants validated in the secondary LOH screen
| Straina | LOH quantification on Ch4 |
|||||
|---|---|---|---|---|---|---|
| Bfp+ Gfp− cells |
Bfp− Gfp+ cells |
|||||
| Mean frequency of LOH ± SEM (×10−4) | Fold changeb | P valuec | Mean frequency of LOH ± SEM (×10−4) | Fold changeb | P valuec | |
| CEC3989 (control) | 1.3 ± 0.5 | 0.7 ± 0.3 | ||||
| PTET-CDC20 | 42.0 ± 11.0 | 32 | ≤0.01 | 26.8 ± 7.9 | 38 | ≤0.01 |
| PTET-RAD53 | 17.0 ± 6.0 | 13 | ≤0.01 | 9.1 ± 1.5 | 13 | ≤0.01 |
| PTET-BIM1 | 12.5 ± 1.7 | 10 | ≤0.01 | 9.0 ± 2.9 | 13 | ≤0.01 |
| PTET-RAD51 | 8.7 ± 1.4 | 7 | ≤0.01 | 5.3 ± 1.3 | 8 | ≤0.01 |
All strains were grown on YPD + Atc.
Ratio of the LOH frequency in a given mutant to that in control strain CEC3989.
Mann-Whitney test.
Characterization of the molecular mechanisms leading to LOH.
LOH at the BFP-GFP locus could result from gene conversion, mitotic crossing over (MCO), break-induced replication (BIR), or chromosome truncation or loss. Further, to identifying genes whose overexpression increased the frequency of LOH, we tested whether the overexpression of these genes could also alter the balance between these different types of events, as observed in the control strain.
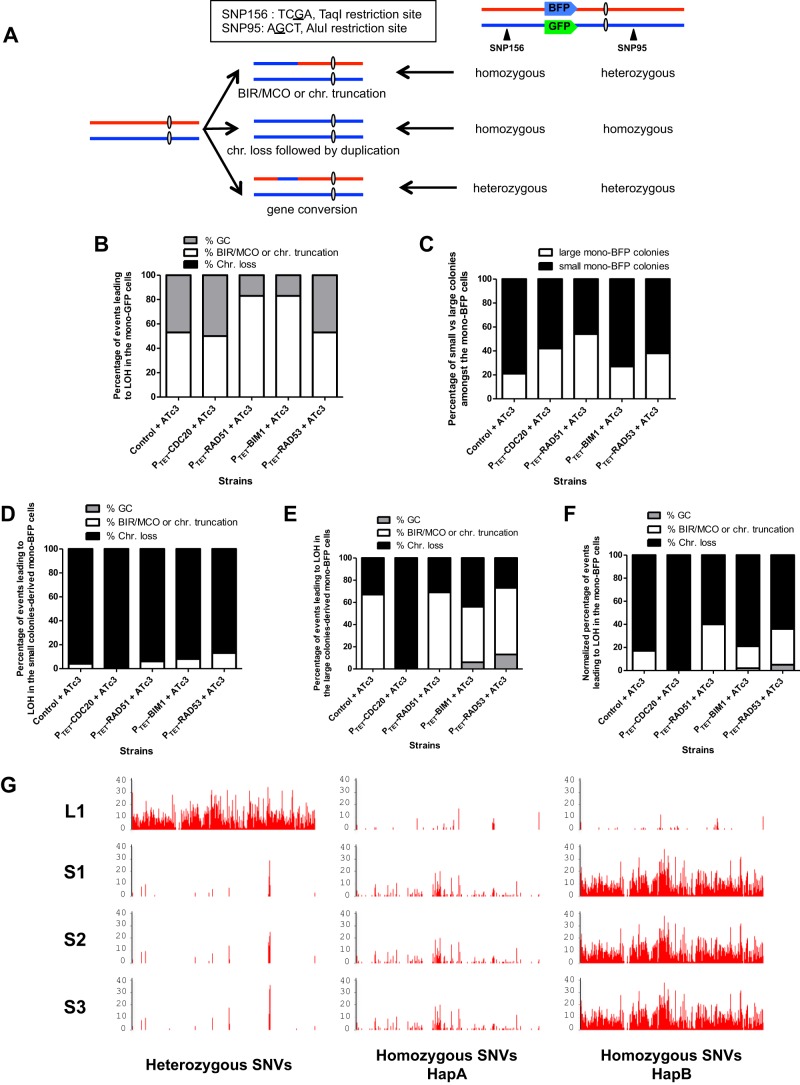
FACS was used in order to recover individual monofluorescent cells and subsequently analyze these cells by SNP-RFLP typing, which can reveal the molecular mechanism—gene conversion, MCO/BIR, or chromosome truncation or loss—underlying the LOH event responsible for loss of the BFP or GFP marker on Ch4 (Fig. 3A) (31). Cells sorted from the Bfp− Gfp+ control and mutant populations and plated on YPD medium gave rise to colonies homogeneous in size. For the control strain, SNP-RFLP typing showed that LOH in the cells that had yielded these colonies had arisen at somewhat equal frequencies through MCO/BIR or chromosome truncation, these two types of events being undistinguishable by SNP-RFLP typing, and gene conversion (Fig. 3B). Complete loss of the homolog carrying the BFP marker was never observed. A similar pattern was observed upon the overexpression of CDC20 and RAD53. In contrast, we detected an increase in the proportion of MCO/BIR or chromosome truncation events versus gene conversions resulting in loss of the BFP marker in the RAD51 and BIM1 overexpression mutants (Fig. 3B).
Fig 3.
SNP typing reveals a shift in the molecular mechanisms leading to LOH upon the overexpression of some of the candidate genes. (A) Map of Ch4 and localization of the BFP-GFP system. The SNPs used for RFLP characterization are indicated by black triangles. Telomere-proximal SNP 156 is part of a TaqI restriction site. One allele contains the TaqI site, while the other does not. Centromere-proximal SNP 95 is located in the middle of an AluI restriction site. One allele contains the AluI site, while the other does not. The heterozygosity status of these SNPs provides information on the molecular mechanisms that give rise to LOH. chr., chromosome. (B) SNP-RFLP typing of Bfp− Gfp+ cells. The histogram shows the proportion of MCO/BIR or chromosome truncation versus chromosome loss in the Bfp− Gfp+ population. MCO/BIR or chromosomal truncation events correspond to isolates that have maintained a heterozygous SNP 95 and displayed a homozygous SNP 156. Chromosome loss events correspond to isolates in which both SNPs 95 and 156 became homozygous. Gene conversion events correspond to isolates in which both SNPs 95 and 156 remained heterozygous. (C) Proportion of small versus large colonies giving rise to true mono-BFP cells. (D) SNP-RFLP typing of small-colony-derived Bfp+ Gfp− cells. (E) SNP-RFLP typing of large-colony-derived Bfp+ Gfp− cells. (F) SNP-RFLP typing of Bfp+ Gfp− cells. The normalized percentage of each molecular mechanism, for instance, gene conversion, was calculated as follows: normalized % GC = (% small colonies × % GC in small colonies) + (% large colonies × % GC in large colonies). (G) Heterozygous and homozygous SNV density maps of one large- and three small-colony-derived cells that have undergone LOH at the BFP-GFP locus of Ch4. The number of heterozygous SNVs in each 10-kb region along Ch4 was computed and is shown, revealing LOH by gene conversion in the large-colony-derived cells and by chromosome loss and reduplication on Ch4 in the small-colony-derived cells. Sequencing reads were mapped on the HapA or HapB reference sequence and yielded similar heterozygous SNP density maps. Only the result of mapping on HapB is shown (left). The number of homozygous SNVs in each 10-kb region along HapA (middle) or HapB (right) was computed and is shown. The high density of homozygous SNVs upon mapping on HapB indicates that chromosome loss in small-colony-derived cells affected Ch4B.
Notably, colonies of different sizes were observed when cells sorted from the Bfp+ Gfp− populations were plated on YPD medium (data not shown). This included colonies of a size similar to that of colonies obtained from the Bfp− Gfp+ populations and smaller colonies. This small-colony phenotype was observed for the control strain and all of the overexpression mutants tested. In the control strain, 80 and 20% of the mono-BFP cells were derived from small and large colonies, respectively (Fig. 3C). We observed an increase in the proportion of the large-colony-derived mono-BFP cells in the CDC20, RAD51, and RAD53 overexpression mutants (Fig. 3C). SNP-RFLP typing of 16 mono-BFP cells from small and large colonies revealed that, unlike in the Bfp− Gfp+ population, chromosome loss was the major mechanism responsible for loss of the GFP marker in the small-colony-derived cells of both the control strain and the overexpression mutants, other LOH events resulting from MCO/BIR, or chromosome truncation (Fig. 3D). On the other hand, SNP-RFLP typing revealed that LOH was often the result of recombination-mediated events or chromosome truncations in the larger colonies of the control strain and the RAD51, BIM1, and RAD53 overexpression mutants (Fig. 3E). In contrast, chromosome loss was the only mechanism leading to LOH in the Bfp+ Gfp− cells of the CDC20 overexpression mutants (Fig. 3D and E). SNP-RFLP typing data for both types of colonies were normalized to the proportion of small versus large colonies and combined to represent the percentage of each molecular mechanism (gene conversion, MCO/BIR, or chromosome truncation or loss) in the overall Bfp+ Gfp− population (Fig. 3F).
Although the homozygous status of both SNPs 156 and 95 is routinely recognized as a mark of LOH chromosome loss on Ch4 (31), multiple gene conversion events could also be responsible for the concomitant homozygous status of these SNPs. To determine the extent of the LOH, we performed whole-genome sequencing of three small Bfp+ Gfp− colonies obtained from an overexpression mutant that did not show any change in the frequency of LOH relative to the control and in which LOH was the result of chromosome loss, as demonstrated by SNP-RFLP typing. Heterozygous and homozygous SNP density maps against the C. albicans SC5314 HapA and HapB reference sequences (41) revealed that cells from the three small colonies had lost all of Ch4B carrying the GFP marker (Fig. 3G). The observation that Ch4 had remained disomic (data not shown) indicates that the other homolog, Ch4A, has been reduplicated. Homozygosis of Ch4A also accounts for the histidine auxotrophy displayed by Bfp+ Gfp− cells, despite the maintenance of the BFP-HIS1 cassette at the PGA59-62 locus (data not shown). Indeed, the inactive HIS4 allele reported by Gómez-Raja et al. (42) is carried by Ch4A.
Overall, our SNP-RFLP analysis revealed several interesting features. (i) The molecular mechanisms causing the LOH differed according to the Ch4 homolog undergoing the LOH event, with chromosome loss being observed only for Ch4B. (ii) This was reflected by the occurrence of small-colony clones when all or part of Ch4B was lost. (iii) When chromosome loss occurred, overexpression of CDC20 favored this mechanism. (iv) RAD51 overexpression favored MCO/BIR events. (v) BIM1 overexpression favored MCO/BIR events unless chromosome loss was possible. (vi) RAD53 overexpression favored MCO/BIR events when chromosome loss was possible.
DISCUSSION
Here, we present a FACS-optimized genetic system for the detection of LOH in C. albicans and its application to the identification of genes whose overexpression results in an increase in the frequency of LOH and changes in the frequencies of the molecular events that are at the origin of LOH.
The new BFP-GFP system displays features more appealing than those of other genetic systems currently used to study LOH in C. albicans. Indeed, we provide a powerful and robust tool that allows rare-event analysis and high-throughput LOH detection since up to 96 samples can be processed in a single run. This system is robust and reproducible because LOH is studied on a cellular scale and a large number of cells are analyzed. Unlike the systems based on the GAL1 or URA3 marker, which require large amounts of expensive drugs, the BFP-GFP system does not require any costly consumables. The use of drugs such as 2-DG or 5-FOA also raises the issue of exposing cells to selective pressure that might distort the evaluation of the frequency of LOH. In contrast, the BFP-GFP system allows measurement of the frequency of spontaneous LOH since cells are not exposed to any stress. Like the other LOH reporter systems, the BFP-GFP system is, in theory, flexible since the markers can be inserted anywhere in the genome, thus enabling determination of the frequency of LOH at different genomic locations and therefore the study of site-specific LOH events.
The BFP-GFP system was used to screen a collection of 124 C. albicans overexpression mutants to identify genes whose overexpression would result in an increase in the frequency of LOH. Our screen identified four candidates as regulators of genome stability in C. albicans. Null or conditional deletion mutants have been obtained and studied for three of these genes, CDC20 (43), RAD51 (44), and RAD53 (45), yet the role of these genes in genome integrity was never addressed, except for RAD53, whose deletion increases the frequency of LOH (16). Reports of C. albicans mutants overexpressing the four candidate genes could not be found in the literature. In contrast, S. cerevisiae mutants null for the four candidate genes, containing conditional deletions of them, or overexpressing them have been described. Although genome stability was investigated in the deletion mutants, the question of genome integrity in the overexpression mutants was not addressed.
Our screen identified CDC20 as the gene whose overexpression triggers the greatest increase in the frequency of LOH. In S. cerevisiae, the essential CDC20 gene encodes an activator of ubiquitin-protein ligase activity and is required for the metaphase-anaphase transition. At this stage of the mitotic cycle, Cdc20p enables the anaphase-promoting complex to properly degrade securin, allowing the degradation of the cohesin rings that link the two sister chromatids. Several studies have shown that degradation of Cdc20p is an essential and conserved mechanism of spindle assembly checkpoint maintenance (46, 47). Therefore, the sustained presence of the Cdc20 protein could perturb genome integrity and our observation that overexpression of C. albicans CDC20 favors LOH, especially through chromosome loss when these types of events can occur, is consistent with this hypothesis. Conditional CDC20 deletion mutants of C. albicans have been characterized by Chou et al. (43). Their work suggested that, similar to ScCDC20, CaCDC20 was important for the metaphase-to-anaphase transition and mitotic exit. Interestingly, CDC20 has a distinct role in morphogenesis in C. albicans, which is not the case in S. cerevisiae.
Rad53 and Rad51, two proteins involved in DNA damage checkpoint maintenance and DNA repair, were identified as causing an increase in LOH upon overexpression. Rad53 is a kinase involved in DNA damage checkpoint maintenance whose deletion causes a growth defect, an increased sensitivity to several DNA-damaging agents, and a defect in genotoxic-stress-induced filamentation in C. albicans (45). An increase in LOH could also be observed in CaRAD53 deletion mutants (16). The recombinase Rad51 plays a major role in homologous recombination during DNA double-strand break repair by searching for sequence homology and promoting strand pairing. Deletion of RAD51 in C. albicans results in a decreased growth rate and increased sensitivity to various DNA-damaging agents (44). On the other hand, genome integrity has not been investigated in CaRAD51 deletion mutants. In S. cerevisiae, RAD51 overexpression has been shown to promote genome instability, probably by inhibiting the accurate repair of double-strand breaks by homologous recombination (48). Similarly, RAD51 overexpression in the mouse leads to an increase in genome instability, notably, LOH events (49). Our observation that, in addition to increasing the frequency of LOH, overexpression of RAD51 and RAD53 favored MCO/BIR events is consistent with their role in DNA double-strand break repair, their overexpression being likely to unbalance the DNA damage response and favor recombination-mediated events.
In this work, a structural component of the microtubule skeleton of yeast encoded by BIM1 has been identified as increasing the frequency of LOH upon overexpression. While ScBIM1 deletion has been shown to result in increased sensitivity to various DNA-damaging agents (50) and increased chromosome instability (51), the effect of BIM1 overexpression on genome integrity has not been investigated, probably in part because of the strong growth defect associated with BIM1 overexpression in S. cerevisiae (52–54). Similarly, we also observed a growth defect associated with BIM1 overexpression in C. albicans (data not shown). Our observation that BIM1 overexpression, in addition to promoting LOH, favors MCO/BIR events was unexpected, as Bim1 is involved in chromosome segregation, and therefore its overexpression would be more likely to cause chromosome loss. We hypothesized that the overexpression-induced reduced growth rate associated with BIM1 overexpression could favor the occurrence of DNA breaks and thus explain the increase in MCO/BIR events or chromosome truncations in the BIM1 overexpression mutant.
Our SNP typing analysis of sorted cells revealed an additional important aspect. Indeed, we have observed that the molecular mechanisms giving rise to LOH can be homolog specific, with chromosome loss never affecting Ch4A. Notably, loss of all or part of Ch4B was accompanied by variations in colony size. As cells in small colonies have almost all arisen through chromosome loss (Fig. 3D) and yield large colonies (data not shown), we hypothesize that small colonies reflect a lower growth rate of cells that are monosomic for Ch4A and eventually duplicate this chromosome. Chromosome homozygosis was long thought to be hampered in C. albicans by the presence of recessive lethal mutations dispersed throughout the genome. The existence of haploids (2), completely homozygous rad52-derived isolates (55), and partly homozygous diploid parasexual progenies (56) demonstrated that homozygosis of each one of the eight chromosomes can occur. Nevertheless, a bias in the homolog being retained was reported for certain chromosomes (2, 55). This was the case for Ch4, one homolog of which could never be lost (2, 55). Our observation that Ch4A could never be lost suggests the presence of at least one recessive lethal allele on Ch4B. Because MCO/BIR or chromosome truncation-mediated LOH occurred in mono-GFP cells, we can narrow the recessive lethal allele(s)-carrying region to the right arm of Ch4B or the left arm of Ch4B between the PGA59/62 locus and the centromere. Genome sequencing of these mono-GFP cells will reveal the extent of the LOH and allow refinement of the position of this hypothetical recessive lethal allele(s). While homozygosis along the entire smaller chromosomes, Ch5, Ch6, and Ch7, was often observed, MCO/BIR events or chromosome truncations have been reported to be responsible for most LOH in both Ch1 homologs (12, 13). Altogether, these observations suggest that the mechanisms underlying LOH in C. albicans could be chromosome specific, as well as homolog specific. The presence of essential alleles that cannot be lost might indeed select for cells that have repaired double-strand breaks through BIR, especially in the case of Ch1, one of the largest chromosomes in C. albicans (3.2 Mb). On the other hand, the loss of one Ch4 homolog, which is half the size of Ch1 (1.6 Mb), might be less detrimental to the cell. Alternatively, Ch4 might be more prone to mitotic nondisjunction events or to unrepaired DNA lesions.
Some expected genes, such as TUB2, were not recovered in our screen (57). This may reflect the inherent limitations of a genetic screen. First, a number of the usual suspects were shown to play a role in genome stability on the basis of deletion mutant study, which does not imply that their overexpression triggers an increase in the frequency of LOH. Second, the level of overexpression achieved in this study might be insufficient to cause a significant phenotype. Because the proteins are produced in an untagged form, we have no means of verifying their production. Third, the lack of phenotype could result from the presence of peptides encoded by the att sequences at the amino- and carboxy-terminal ends of the proteins, which could alter their proper folding and activity.
Living cells have evolved different mechanisms in order to ensure genome integrity (58). Interestingly, these mechanisms are conserved across species, and cancer genetic studies have linked malfunctions in these processes to the genome instability observed in human cancer cells (59, 60). Therefore, the characterization of all of the players involved in genome integrity maintenance and a better understanding of the associated signaling pathways are very important from the human perspective. In this respect, functional genomic approaches to model eukaryotic species such as S. cerevisiae have uncovered many components required for genome maintenance and integrity (61). Genome-wide genetic screens of this species have examined deletion mutants for a growth defect upon exposure to different DNA-damaging agents (62–65) or for synthetic genetic interaction with other mutant genes important for the cellular response to DNA damage (66–69). Other screens identified mutants with increased genomic instability by investigating the mutation rates within a specific gene (70) or the rates of gross chromosomal rearrangements (51, 71–75). While several studies have used S. cerevisiae to study genome stability, drastic genome changes are not well tolerated by this organism, thus correlating with a high fitness cost (76). Interestingly, C. albicans is more tolerant of genome changes, which is reminiscent of the situation in cancer cells that continue to divide rapidly despite undergoing massive genome rearrangements. The tools presented here, in association with the C. albicans ORFeome we are currently constructing with the group of Carol Munro in Aberdeen (20), will allow C. albicans to become a model of choice for the study of mechanisms involved in eukaryotic genome integrity.
Supplementary Material
ACKNOWLEDGMENTS
We thank Christiane Bouchier and Laurence Ma from the Pasteur Plateforme Genomique (PF1) for Illumina sequencing of the overexpression plasmids. We are thankful to members of the BIOASTER genomics and transcriptomics core facility, namely, Lilia Boucinha and Frédéric Reynier, for help with genome sequencing. We are also grateful to other members of Fungal Biology and Pathogenicity Unit for helpful discussions.
This work was supported by the French Government's Investissement d'Avenir program, Laboratoire d'Excellence Integrative Biology of Emerging Infectious Diseases (grant ANR-10-LABX-62-IBEID). R.L.-K. was the recipient of a Ph.D. grant from INRA and the Institut Pasteur. M.L. was the recipient of a postdoctoral fellowship from the Institut Pasteur (Bourse Roux).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00286-14.
REFERENCES
- 1.Edmond MB, Wallace SE, McClish DK, Pfaller MA, Jones RN, Wenzel RP. 1999. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin Infect Dis 29:239–244. doi: 10.1086/520192. [DOI] [PubMed] [Google Scholar]
- 2.Hickman MA, Zeng G, Forche A, Hirakawa MP, Abbey D, Harrison BD, Wang YM, Su CH, Bennett RJ, Wang Y, Berman J. 2013. The ‘obligate diploid’ Candida albicans forms mating-competent haploids. Nature 494:55–59. doi: 10.1038/nature11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hull CM, Raisner RM, Johnson AD. 2000. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science 289:307–310. doi: 10.1126/science.289.5477.307. [DOI] [PubMed] [Google Scholar]
- 4.Magee BB, Magee PT. 2000. Induction of mating in Candida albicans by construction of MTLa and MTLalpha strains. Science 289:310–313. doi: 10.1126/science.289.5477.310. [DOI] [PubMed] [Google Scholar]
- 5.Miller MG, Johnson AD. 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110:293–302. doi: 10.1016/S0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- 6.Bennett RJ, Johnson AD. 2003. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J 22:2505–2515. doi: 10.1093/emboj/cdg235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selmecki A, Forche A, Berman J. 2010. Genomic plasticity of the human fungal pathogen Candida albicans. Eukaryot Cell 9:991–1008. doi: 10.1128/EC.00060-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diogo D, Bouchier C, d'Enfert C, Bougnoux ME. 2009. Loss of heterozygosity in commensal isolates of the asexual diploid yeast Candida albicans. Fungal Genet Biol 46:159–168. doi: 10.1016/j.fgb.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Forche A, Magee PT, Selmecki A, Berman J, May G. 2009. Evolution in Candida albicans populations during a single passage through a mouse host. Genetics 182:799–811. doi: 10.1534/genetics.109.103325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coste A, Turner V, Ischer F, Morschhauser J, Forche A, Selmecki A, Berman J, Bille J, Sanglard D. 2006. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 172:2139–2156. doi: 10.1534/genetics.105.054767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunkel N, Blass J, Rogers PD, Morschhauser J. 2008. Mutations in the multi-drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol Microbiol 69:827–840. doi: 10.1111/j.1365-2958.2008.06309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forche A, Abbey D, Pisithkul T, Weinzierl MA, Ringstrom T, Bruck D, Petersen K, Berman J. 2011. Stress alters rates and types of loss of heterozygosity in Candida albicans. mBio 2:e00129-11. doi: 10.1128/mBio.00129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Legrand M, Chan CL, Jauert PA, Kirkpatrick DT. 2007. Role of DNA mismatch repair and double-strand break repair in genome stability and antifungal drug resistance in Candida albicans. Eukaryot Cell 6:2194–2205. doi: 10.1128/EC.00299-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Legrand M, Chan CL, Jauert PA, Kirkpatrick DT. 2008. Analysis of base excision and nucleotide excision repair in Candida albicans. Microbiology 154:2446–2456. doi: 10.1099/mic.0.2008/017616-0. [DOI] [PubMed] [Google Scholar]
- 15.Legrand M, Chan CL, Jauert PA, Kirkpatrick DT. 2011. The contribution of the S-phase checkpoint genes MEC1 and SGS1 to genome stability maintenance in Candida albicans. Fungal Genet Biol 48:823–830. doi: 10.1016/j.fgb.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loll-Krippleber R, d'Enfert C, Feri A, Diogo D, Perin A, Marcet-Houben M, Bougnoux ME, Legrand M. 2014. A study of the DNA damage checkpoint in Candida albicans: uncoupling of the functions of Rad53 in DNA repair, cell cycle regulation and genotoxic stress-induced polarized growth. Mol Microbiol 91:452–471. doi: 10.1111/mmi.12471. [DOI] [PubMed] [Google Scholar]
- 17.Boeke JD, LaCroute F, Fink GR. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet 197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 18.Forche A, May G, Beckerman J, Kauffman S, Becker J, Magee PT. 2003. A system for studying genetic changes in Candida albicans during infection. Fungal Genet Biol 39:38–50. doi: 10.1016/S1087-1845(02)00585-6. [DOI] [PubMed] [Google Scholar]
- 19.Gorman JA, Gorman JW, Koltin Y. 1992. Direct selection of galactokinase-negative mutants of Candida albicans using 2-deoxy-galactose. Curr Genet 21:203–206. doi: 10.1007/BF00336842. [DOI] [PubMed] [Google Scholar]
- 20.Cabral V, Chauvel M, Firon A, Legrand M, Nesseir A, Bachellier-Bassi S, Chaudhari Y, Munro CA, d'Enfert C. 2012. Modular gene over-expression strategies for Candida albicans. Methods Mol Biol 845:227–244. doi: 10.1007/978-1-61779-539-8_15. [DOI] [PubMed] [Google Scholar]
- 21.Chauvel M, Nesseir A, Cabral V, Znaidi S, Goyard S, Bachellier-Bassi S, Firon A, Legrand M, Diogo D, Naulleau C, Rossignol T, d'Enfert C. 2012. A versatile overexpression strategy in the pathogenic yeast Candida albicans: identification of regulators of morphogenesis and fitness. PLoS One 7:e45912. doi: 10.1371/journal.pone.0045912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noble SM, French S, Kohn LA, Chen V, Johnson AD. 2010. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet 42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rose M, Winston F, Hieter P. 1990. Methods in yeast genetics. a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor Laboratory, NY. [Google Scholar]
- 24.Noble SM, Johnson AD. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell 4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delgado ML, Gil ML, Gozalbo D. 2003. Candida albicans TDH3 gene promotes secretion of internal invertase when expressed in Saccharomyces cerevisiae as a glyceraldehyde-3-phosphate dehydrogenase-invertase fusion protein. Yeast 20:713–722. doi: 10.1002/yea.993. [DOI] [PubMed] [Google Scholar]
- 26.Gola S, Martin R, Walther A, Dunkler A, Wendland J. 2003. New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast 20:1339–1347. doi: 10.1002/yea.1044. [DOI] [PubMed] [Google Scholar]
- 27.Schaub Y, Dunkler A, Walther A, Wendland J. 2006. New pFA-cassettes for PCR-based gene manipulation in Candida albicans. J Basic Microbiol 46:416–429. doi: 10.1002/jobm.200510133. [DOI] [PubMed] [Google Scholar]
- 28.Dennison PM, Ramsdale M, Manson CL, Brown AJ. 2005. Gene disruption in Candida albicans using a synthetic, codon-optimised Cre-loxP system. Fungal Genet Biol 42:737–748. doi: 10.1016/j.fgb.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 29.Wilson RB, Davis D, Mitchell AP. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol 181:1868–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murad AM, Lee PR, Broadbent ID, Barelle CJ, Brown AJ. 2000. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16:325–327. doi:. [DOI] [PubMed] [Google Scholar]
- 31.Forche A, Steinbach M, Berman J. 2009. Efficient and rapid identification of Candida albicans allelic status using SNP-RFLP. FEMS Yeast Res 9:1061–1069. doi: 10.1111/j.1567-1364.2009.00542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Binkley J, Arnaud MB, Inglis DO, Skrzypek MS, Shah P, Wymore F, Binkley G, Miyasato SR, Simison M, Sherlock G. 2014. The Candida Genome Database: the new homology information page highlights protein similarity and phylogeny. Nucleic Acids Res 42:D711–D716. doi: 10.1093/nar/gkt1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van het Hoog M, Rast TJ, Martchenko M, Grindle S, Dignard D, Hogues H, Cuomo C, Berriman M, Scherer S, Magee BB, Whiteway M, Chibana H, Nantel A, Magee PT. 2007. Assembly of the Candida albicans genome into sixteen supercontigs aligned on the eight chromosomes. Genome Biol 8:R52. doi: 10.1186/gb-2007-8-4-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreno-Ruiz E, Ortu G, de Groot PW, Cottier F, Loussert C, Prevost MC, de Koster C, Klis FM, Goyard S, d'Enfert C. 2009. The GPI-modified proteins Pga59 and Pga62 of Candida albicans are required for cell wall integrity. Microbiology 155:2004–2020. doi: 10.1099/mic.0.028902-0. [DOI] [PubMed] [Google Scholar]
- 37.Stynen B, Van Dijck P, Tournu H. 2010. A CUG codon adapted two-hybrid system for the pathogenic fungus Candida albicans. Nucleic Acids Res 38:e184. doi: 10.1093/nar/gkq725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cormack BP, Bertram G, Egerton M, Gow NA, Falkow S, Brown AJ. 1997. Yeast-enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. Microbiology 143(Pt 2):303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- 39.Kato R, Ogawa H. 1994. An essential gene, ESR1, is required for mitotic cell growth, DNA repair and meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res 22:3104–3112. doi: 10.1093/nar/22.15.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou BB, Elledge SJ. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 41.Muzzey D, Schwartz K, Weissman JS, Sherlock G. 2013. Assembly of a phased diploid Candida albicans genome facilitates allele-specific measurements and provides a simple model for repeat and indel structure. Genome Biol 14:R97. doi: 10.1186/gb-2013-14-9-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gómez-Raja J, Andaluz E, Magee B, Calderone R, Larriba G. 2008. A single SNP, G929T (Gly310Val), determines the presence of a functional and a non-functional allele of HIS4 in Candida albicans SC5314: detection of the non-functional allele in laboratory strains. Fungal Genet Biol 45:527–541. doi: 10.1016/j.fgb.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chou H, Glory A, Bachewich C. 2011. Orthologues of the anaphase-promoting complex/cyclosome coactivators Cdc20p and Cdh1p are important for mitotic progression and morphogenesis in Candida albicans. Eukaryot Cell 10:696–709. doi: 10.1128/EC.00263-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.García-Prieto F, Gómez-Raja J, Andaluz E, Calderone R, Larriba G. 2010. Role of the homologous recombination genes RAD51 and RAD59 in the resistance of Candida albicans to UV light, radiomimetic and anti-tumor compounds and oxidizing agents. Fungal Genet Biol 47:433–445. doi: 10.1016/j.fgb.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi QM, Wang YM, Zheng XD, Lee RT, Wang Y. 2007. Critical role of DNA checkpoints in mediating genotoxic-stress-induced filamentous growth in Candida albicans. Mol Biol Cell 18:815–826. doi: 10.1091/mbc.E06-05-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stevenson LF, Kennedy BK, Harlow E. 2001. A large-scale overexpression screen in Saccharomyces cerevisiae identifies previously uncharacterized cell cycle genes. Proc Natl Acad Sci U S A 98:3946–3951. doi: 10.1073/pnas.051013498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu L, Pena Castillo L, Mnaimneh S, Hughes TR, Brown GW. 2006. A survey of essential gene function in the yeast cell division cycle. Mol Biol Cell 17:4736–4747. doi: 10.1091/mbc.E06-04-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paffett KS, Clikeman JA, Palmer S, Nickoloff JA. 2005. Overexpression of Rad51 inhibits double-strand break-induced homologous recombination but does not affect gene conversion tract lengths. DNA Repair 4:687–698. doi: 10.1016/j.dnarep.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 49.Richardson C, Stark JM, Ommundsen M, Jasin M. 2004. Rad51 overexpression promotes alternative double-strand break repair pathways and genome instability. Oncogene 23:546–553. doi: 10.1038/sj.onc.1207098. [DOI] [PubMed] [Google Scholar]
- 50.Kapitzky L, Beltrao P, Berens TJ, Gassner N, Zhou C, Wuster A, Wu J, Babu MM, Elledge SJ, Toczyski D, Lokey RS, Krogan NJ. 2010. Cross-species chemogenomic profiling reveals evolutionarily conserved drug mode of action. Mol Syst Biol 6:451. doi: 10.1038/msb.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuen KW, Warren CD, Chen O, Kwok T, Hieter P, Spencer FA. 2007. Systematic genome instability screens in yeast and their potential relevance to cancer. Proc Natl Acad Sci U S A 104:3925–3930. doi: 10.1073/pnas.0610642104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwartz K, Richards K, Botstein D. 1997. BIM1 encodes a microtubule-binding protein in yeast. Mol Biol Cell 8:2677–2691. doi: 10.1091/mbc.8.12.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR, Boone C, Andrews B. 2006. Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell 21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 54.Yoshikawa K, Tanaka T, Ida Y, Furusawa C, Hirasawa T, Shimizu H. 2011. Comprehensive phenotypic analysis of single-gene deletion and overexpression strains of Saccharomyces cerevisiae. Yeast 28:349–361. doi: 10.1002/yea.1843. [DOI] [PubMed] [Google Scholar]
- 55.Andaluz E, Bellido A, Gómez-Raja J, Selmecki A, Bouchonville K, Calderone R, Berman J, Larriba G. 2011. Rad52 function prevents chromosome loss and truncation in Candida albicans. Mol Microbiol 79:1462–1482. doi: 10.1111/j.1365-2958.2011.07532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Forche A, Alby K, Schaefer D, Johnson AD, Berman J, Bennett RJ. 2008. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol 6:e110. doi: 10.1371/journal.pbio.0060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burke D, Gasdaska P, Hartwell L. 1989. Dominant effects of tubulin overexpression in Saccharomyces cerevisiae. Mol Cell Biol 9:1049–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harper JW, Elledge SJ. 2007. The DNA damage response: ten years after. Mol Cell 28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 59.Pfau SJ, Amon A. 2012. Chromosomal instability and aneuploidy in cancer: from yeast to man. EMBO Rep 13:515–527. doi: 10.1038/embor.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Negrini S, Gorgoulis VG, Halazonetis TD. 2010. Genomic instability—an evolving hallmark of cancer. Nat Rev Mol Cell Biol 11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 61.Kolodner RD, Putnam CD, Myung K. 2002. Maintenance of genome stability in Saccharomyces cerevisiae. Science 297:552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- 62.Bennett CB, Lewis LK, Karthikeyan G, Lobachev KS, Jin YH, Sterling JF, Snipe JR, Resnick MA. 2001. Genes required for ionizing radiation resistance in yeast. Nat Genet 29:426–434. doi: 10.1038/ng778. [DOI] [PubMed] [Google Scholar]
- 63.Birrell GW, Giaever G, Chu AM, Davis RW, Brown JM. 2001. A genome-wide screen in Saccharomyces cerevisiae for genes affecting UV radiation sensitivity. Proc Natl Acad Sci U S A 98:12608–12613. doi: 10.1073/pnas.231366398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang M, Bellaoui M, Boone C, Brown GW. 2002. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc Natl Acad Sci U S A 99:16934–16939. doi: 10.1073/pnas.262669299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hanway D, Chin JK, Xia G, Oshiro G, Winzeler EA, Romesberg FE. 2002. Previously uncharacterized genes in the UV- and MMS-induced DNA damage response in yeast. Proc Natl Acad Sci U S A 99:10605–10610. doi: 10.1073/pnas.152264899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bellaoui M, Chang M, Ou J, Xu H, Boone C, Brown GW. 2003. Elg1 forms an alternative RFC complex important for DNA replication and genome integrity. EMBO J 22:4304–4313. doi: 10.1093/emboj/cdg406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mayer ML, Pot I, Chang M, Xu H, Aneliunas V, Kwok T, Newitt R, Aebersold R, Boone C, Brown GW, Hieter P. 2004. Identification of protein complexes required for efficient sister chromatid cohesion. Mol Biol Cell 15:1736–1745. doi: 10.1091/mbc.E03-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, Boone C. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 69.Warren CD, Eckley DM, Lee MS, Hanna JS, Hughes A, Peyser B, Jie C, Irizarry R, Spencer FA. 2004. S-phase checkpoint genes safeguard high-fidelity sister chromatid cohesion. Mol Biol Cell 15:1724–1735. doi: 10.1091/mbc.E03-09-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang ME, Rio AG, Nicolas A, Kolodner RD. 2003. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc Natl Acad Sci U S A 100:11529–11534. doi: 10.1073/pnas.2035018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alabrudzinska M, Skoneczny M, Skoneczna A. 2011. Diploid-specific [corrected] genome stability genes of S. cerevisiae: genomic screen reveals haploidization as an escape from persisting DNA rearrangement stress. PLoS One 6:e21124. doi: 10.1371/journal.pone.0021124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Andersen MP, Nelson ZW, Hetrick ED, Gottschling DE. 2008. A genetic screen for increased loss of heterozygosity in Saccharomyces cerevisiae. Genetics 179:1179–1195. doi: 10.1534/genetics.108.089250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kanellis P, Gagliardi M, Banath JP, Szilard RK, Nakada S, Galicia S, Sweeney FD, Cabelof DC, Olive PL, Durocher D. 2007. A screen for suppressors of gross chromosomal rearrangements identifies a conserved role for PLP in preventing DNA lesions. PLoS Genet 3:e134. doi: 10.1371/journal.pgen.0030134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smith S, Hwang JY, Banerjee S, Majeed A, Gupta A, Myung K. 2004. Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 101:9039–9044. doi: 10.1073/pnas.0403093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Strome ED, Wu X, Kimmel M, Plon SE. 2008. Heterozygous screen in Saccharomyces cerevisiae identifies dosage-sensitive genes that affect chromosome stability. Genetics 178:1193–1207. doi: 10.1534/genetics.107.084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.