Abstract
The anillin-related protein Bud4 of Saccharomyces cerevisiae is required for axial bud site selection by linking the axial landmark to the septins, which localize at the mother bud neck. Recent studies indicate that Bud4 plays a role in septin organization during cytokinesis. Here we show that Bud4 is also involved in septin organization during bud growth prior to cytokinesis, as bud4Δ shs1Δ cells displayed an elongated bud morphology and defective septin organization at 18°C. Bud4 overexpression also affected septin organization during bud growth in shs1Δ cells at 30°C. Bud4 was previously thought to associate with the septins via its central region, while the C-terminal anillin-related region was not involved in septin association. Surprisingly, we found that the central region of Bud4 alone targets to the bud neck throughout the cell cycle, unlike full-length Bud4, which localizes to the bud neck only during G2/M phase. We identified the anillin-related region to be a second targeting domain that cooperates with the central region for proper septin association. In addition, the anillin-related region could largely mediate Bud4's function in septin organization during bud growth and bud site selection. We show that this region interacts with the C terminus of Bud3 and the two segments depend on each other for association with the septins. Moreover, like the bud4Δ mutant, the bud3Δ mutant genetically interacts with shs1Δ and cdc12-6 mutants in septin organization, suggesting that Bud4 and Bud3 may cooperate in septin organization during bud growth. These observations provide new insights into the interaction of Bud4 with the septins and Bud3.
INTRODUCTION
The correct positioning of a polarity axis in response to spatial cues is important for a variety of cellular functions, such as neuronal transmission, cell fate determination, and directional cell migration (1–3). In the budding yeast Saccharomyces cerevisiae, the spatial control of cell polarization is manifested by bud site selection, a process that takes place at late G1 stage of the cell cycle, in which the mother cell chooses a new budding site (4, 5). There are two patterns for bud site selection in budding yeast, depending on the cell type. Haploid MATa or MATα cells bud in an axial pattern, in which cells choose a new budding site next to the preceding cell division site. As a result, the bud scars are connected in a chain. Diploid MATa/MATα cells, however, bud in a bipolar pattern, in which the daughter cells choose the new budding site exclusively at the pole distal to the preceding cell division site, while the mother cells select the new budding site either adjacent to or opposite the preceding division site. As a result, the bud scars are distributed at the two poles of a cell (4–6).
Previous studies had established that a group of proteins, including Bud3, Bud4, Axl1, Axl2 (also known as Bud10), and the septins, is implicated in the control of axial bud site selection. All these proteins localize to the mother bud neck, and loss-of-function mutations in any of these proteins result in a failure to bud in an axial pattern but instead cause bipolar budding (7–13). Septins are a class of conserved GTP-binding proteins with a conserved function in cytokinesis. In budding yeast, five vegetatively expressed septins (Cdc3, Cdc10, Cdc11, Cdc12, and Shs1/Sep7) assemble into heterooligomeric complexes, which polymerize to form higher-order structure filaments at the bud neck (14, 15). Septins at the bud neck function as a scaffold involved in anchoring other proteins there. They also function as a diffusion barrier confining cortical factors in the daughter cell (16). Bud3, Bud4, Axl1, and Axl2 are thought to assemble into a protein complex (axial landmark) on the septin filaments in a sequential order at about the time of mitosis. Septin filaments first recruit Bud3 and Bud4 to the bud neck, and after that, Axl1 and Axl2 are recruited (17, 18). During cytokinesis, the septin hourglass splits into two rings and the daughter cell and the mother cell each inherit one ring. Each ring lasts until the late G1 stage of the next cell cycle (16). The four proteins on the septin ring are thought to function as a spatial memory to activate Rsr1/Bud1, a Ras-family small GTPase, which in turn activates Cdc42, a Rho-family small GTPase, leading to the assembly of a new bud at an adjacent location (19). A recent study showed that Bud3 possesses weak guanine nucleotide exchange factor (GEF) activity toward Cdc42 and can activate Cdc42 in vivo (20). This observation suggests that the axial landmark may interact with Cdc42 at multiple levels.
Apart from bud site selection, Bud4 is known to play a role in septin organization, as the septins in bud4Δ cells fail to form a discrete double ring during cytokinesis (21–23), whereas overexpression of Bud4 causes the assembly of extra septin structures (23). Bud4 is also required for normal cytokinesis in the absence of Gin4 or Cla4 (22), two protein kinases important for septin ring assembly (14).
Bud4 shares a low degree of amino acid sequence similarity (18%) with anillin, an animal protein (24). Like anillin, Bud4 contains a pleckstrin homology (PH) domain at the C terminus. In fungal anillin-related proteins, a DUF1709 domain (function unknown) is also found next to the conserved PH domain (23). The common function of anillin and fungal anillin-related proteins appears to be the regulation of cytokinesis (25), and in most cases, this function involves their interaction with the septins. For example, anillin is known to directly bind septins (26). Mid2, one of two Bud4 homologues in the fission yeast Schizosaccharomyces pombe, acts to stabilize septin rings during cytokinesis (27, 28). Int1, a Bud4 homologue in the opportunistic human pathogen Candida albicans, can interact with the septins in S. cerevisiae (29). A few anillin-related proteins regulate cytokinesis without interacting with the septins; for example, the fission yeast Mid1 plays a role in the positioning of the site for cytokinesis (30). It is currently not known whether Bud4 homologues in the filamentous fungi Neurospora crassa and Aspergillus nidulans, which are crucial for cytokinesis (31, 32), interact with the septins.
Although the functions of Bud4 in bud site selection and in septin organization during cytokinesis have been characterized by previous studies, whether Bud4 is involved in septin organization prior to cytokinesis and the exact mechanisms of how Bud4 regulates these processes are not clear. Here we report that Bud4 plays a role in septin organization during bud growth. In addition, we identified the C-terminal anillin-related region to be a second bud neck-targeting domain in Bud4. This region, which consists of DUF1709 and PH domains, interacts with the C terminus of Bud3 and plays a critical role in Bud4's function in septin organization and bud site selection.
MATERIALS AND METHODS
Strains, media, and genetic methods.
The yeast strains used in this study are listed in Table S1 in the supplemental material. Standard culture media and genetic techniques were used, except where noted (33). For GAL-driven overexpression of Bud4, cells were grown in synthetic SRG medium (0.67% yeast nitrogen base, 1% raffinose, 2% galactose). Escherichia coli strains DH12S (Life Technologies, Gaithersburg, MD) and DH5α (TaKaRa, Japan) were used as hosts for plasmid manipulation. Oligonucleotide primers for PCR were purchased from Sangon Biotech (Shanghai, China). The nucleotide sequences of the primers are available upon request.
Plasmid construction.
The pBG2 (CEN URA3 PBUD4-GFP-TCYC1) vector was generated for the expression of green fluorescent protein (GFP)-tagged Bud4 (GFP-Bud4) segments in the localization and functional complementation studies. It has the pRS316 (CEN URA3) backbone and contains the BUD4 promoter (nucleotides −1000 to +12 relative to the A residue of the start codon) amplified by PCR from plasmid pBUD4 (13) in a SacI-NotI fragment, the 728-bp NotI-BamHI fragment of GFP with an S-to-T mutation at position 65 (GFPS65T) digested from pKS-GFP, and the 263-bp SalI-KpnI fragment of the CYC1 transcription terminator from pUG36 (34). Full-length BUD4 or various BUD4 fragments were amplified by PCR from pBUD4, digested with BamHI and XhoI, and inserted into BamHI- and SalI-digested plasmid pBG2.
To generate the pBG5 (integrative, URA3 PBUD4) vector used for integrative expression of full-length Bud4 and Bud4 segments in yeast cells, the BUD4 promoter (nucleotides −1000 to +12) in a SacI-NotI fragment from pBG2 was inserted into pRS306 (integrative, URA3). Full-length BUD4 and BUD4 segments along with the TCYC1 terminator were removed from pBG2-BUD4 segments in BamHI-KpnI fragments and inserted into BamHI- and KpnI-digested pBG5, yielding pBG5-BUD4 segments. pBG5 was linearized with NcoI, whereas pBG5-BUD4 segments were linearized with ApaI for integration at the ura3-52 locus in yeast cells.
Plasmids pEGKT316-BUD4, pEGKT316-BUD4-C8, and pEGKT316-BUD4-M1 were constructed by inserting the BamHI-XhoI fragments of BUD4, the C8 segment of BUD4 (BUD4-C8), and BUD4-M1 amplified from pBUD4 by PCR into BamHI- and SalI-digested pEGKT316 (CEN URA3 UASGAL1-PCYC1-GST-TCYC1), respectively (35).
To generate the pGGFP316 vector (CEN URA3 UASGAL1-PCYC1-yEGFP3-TCYC1) used for overexpression of GFP-Bud4 segments, yEGFP3 was amplified by PCR from pUG36 (CEN URA3 PMET25-yEGFP3-TCYC1) (34) and inserted into SacI- and BamHI-digested pEGKT316 to replace the glutathione S-transferase (GST) sequence, yielding pGGFP316. Then, the BamHI-XhoI fragments of BUD4-C8 and BUD4-M1 were ligated into BamHI- and SalI-digested pGGFP316, yielding pGGFP316-BUD4-C8 and pGGFP316-BUD4-M1, respectively.
For yeast two-hybrid assay, plasmids pGBDU-BUD4-C2, pGAD-BUD4, pGAD-BUD4-C2, pGBDU-BUD3-M3/C2, pOAD-BUD3-M3/C2, and pOAD-BUD3-C1614 were constructed. Full-length BUD4 and BUD4-C2 were amplified by PCR from pBUD4, digested with BamHI and XhoI, and inserted into BamHI- and SalI-digested pGBDU-C1 (2μ URA3 GAL4-BD) and pGAD-C1 (2μ LEU2 GAL4-AD) (36), yielding plasmids pGBDU-BUD4-C2, pGAD-BUD4, and pGAD-BUD4-C2. To generate pGBDU-BUD3-M3/C2, BUD3-M3/C2 was digested from pUG36-BUD3-M3/C2 and inserted into BamHI- and SalI-digested pGBDU-C1. BUD3-M3/C2 and BUD3-C1614 were digested with SmaI and PstI from pGBDU-based plasmids and inserted into pOAD (36), yielding pOAD-BUD3-M3/C2 and pOAD-BUD3-C1614, respectively.
Yeast strain construction.
Strain JGY2737 (α bud4Δ::TRP1 shs1Δ::kanMX) was constructed by deleting BUD4 in YEF4603 (α shs1Δ::kanMX) by a PCR-based method (37). JGY2903 (α iqg1Δ::His3MX6) was constructed by deleting IQG1 in the JMY314.1-4b-derived W303 background strain JGY2482. Plasmids YIp211-CDC3-GFP, YIp128-CDC3-GFP, and YIp128-CDC3-mCherry (17) were linearized by BglII for integration at the CDC3 locus in yeast cells, generating integrated CDC3-GFP::URA3, CDC3-GFP::LEU2, and CDC3-mCherry::LEU2 alleles, respectively. Strain JGY781 (α bud3Δ bud4Δ) was generated by mating YEF3571 (α bud3Δ::HIS3) with YEF3572 (a bud4Δ::HIS3), followed by sporulation and tetrad dissection. Strain JGY2966 was generated by tagging the C terminus of Spc42 with mCherry in strain JGY2923 by a PCR-based method (37).
Microscopy.
An Olympus BX51 microscope (Tokyo, Japan) and a Retiga 2000R charge-coupled-device (CCD) camera (QImagine Corporation, Canada) were used to visualize cell morphology and GFP-tagged proteins by differential interference contrast (DIC) and fluorescence microscopy. The ImagePro Plus program (MediaCybernetics, Glen Mills, PA) was used for image processing. The budding pattern of a yeast strain was determined by staining the bud scars of cells with 0.01% calcofluor white.
RESULTS
Bud4 plays a role in septin organization during bud growth.
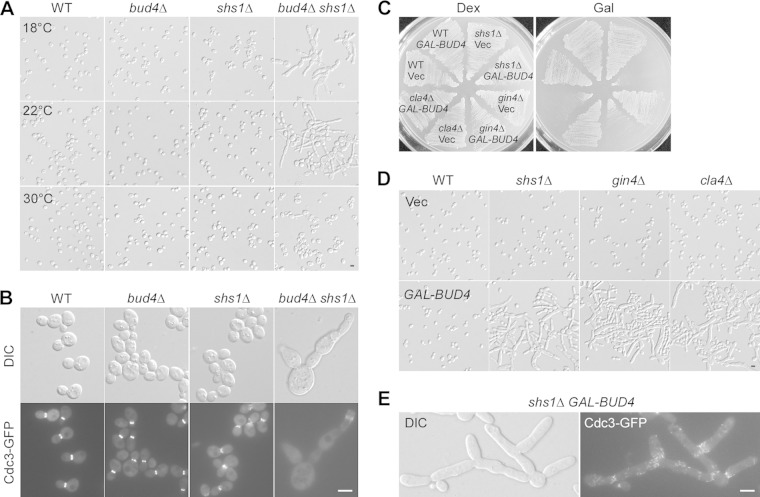
Bud4 has a role in septin organization at the end of the cell cycle, as bud4Δ cells formed a single septin ring instead of two rings at the bud neck during cytokinesis (21–23). Because bud morphogenesis occurs normally in bud4Δ cells as well as cells overexpressing Bud4 (21–23, 29), it is not clear whether Bud4 may play a role in septin organization at early stages of the cell cycle. To explore this possibility, we looked for mutants that showed a synthetic defect in bud morphology with the bud4Δ mutant. Shs1 (also known as Sep7) is one of the five vegetatively expressed septins and is not essential for cell viability (38–40). In our strain background (S288C derived), shs1Δ cells showed normal growth and cell morphology when grown on nutrient-rich yeast extract-peptone-dextrose (YPD) agar medium at 22°C to 37°C. A defective cell morphology that included slightly elongated buds could be seen only at 18°C. Interestingly, we found that bud4Δ shs1Δ cells displayed a severely defective morphology at 22°C: 70% of bud4Δ shs1Δ cells examined (n = 866) displayed elongated buds, and 13% of cells formed chains/clusters consisting of three or more cells (Fig. 1A, middle). This morphological defect was more pronounced at 18°C, where a larger population of bud4Δ shs1Δ cells (84%; n = 528) displayed an elongated bud morphology (Fig. 1A, top). The septins (indicated by Cdc3-GFP) were not well organized in these cells. Cdc3-GFP was often found to be less concentrated or absent at the bud neck in bud4Δ shs1Δ cells grown at 18°C, while it was concentrated at the bud neck in wild-type, bud4Δ, and shs1Δ cells (Fig. 1B). Some bud4Δ shs1Δ cells showed a mislocalization of Cdc3-GFP at the tips of elongated buds. These observations suggest that Bud4 is required for septin organization during bud growth, in addition to cytokinesis.
FIG 1.
Bud4 plays a role in septin organization during bud growth. (A) Cell morphology of strains YEF473A (wild type [WT]), YEF3572 (bud4Δ), YEF4603 (shs1Δ), and JGY2737 (bud4Δ shs1Δ). Cells were grown on YPD plates at 18°C and 22°C for 6 days and 30°C for 3 days. (B) Localization of Cdc3-GFP. Cells of the strains described in the legend to panel A carrying integrated CDC3-GFP::URA3 were cultivated on YPD plates at 18°C for 2 days. (C) Bud4 overexpression. Cells of strains YEF473A (wild type), YEF4603 (shs1Δ), YEF1238 (gin4Δ), and YEF1342 (cla4Δ) carrying plasmid pEGKT316 (vector [Vec]) or pEGKT316-BUD4 (GAL-BUD4) were grown on an SC-Ura medium (dextrose [Dex]) plate for 2 days and an SRG-Ura medium (Gal) plate for 3 days at 37°C. (D) Morphology of the cells described in panel C grown on an SRG-Ura medium plate at 30°C for 20 h. (E) Cdc3-GFP localization in shs1Δ cells overexpressing Bud4. Cells of strain YEF4603 (shs1Δ) carrying integrated CDC3-GFP::LEU2 and plasmid pEGKT316-BUD4 (GAL-BUD4) were grown in SRG-Ura medium at 30°C for 20 h before observation. Bars, 5 μm.
In addition to the morphological and septin organization defect, bud4Δ shs1Δ cells grew more slowly than the isogenic wild-type cells at 18°C (data not shown). Remarkably, bud4Δ shs1Δ cells did not display morphological or growth defects at either 30°C (Fig. 1A) or 37°C or when grown on synthetic complete (SC) agar medium (data not shown).
Next, we asked whether the expression of excess Bud4 could also perturb septin organization in shs1Δ cells. We observed that Bud4 overexpression in shs1Δ cells significantly impaired growth at elevated temperatures. The cells failed to grow into single colonies at 37°C (Fig. 1C). At 30°C, where the cells could grow, cell morphology was abnormal. Eighty-one percent of the cells examined (n = 833) displayed elongated buds (Fig. 1D). Cell clusters/chains, indicative of a defect in cytokinesis or cell separation, were frequently detected. This phenotype is very similar to that of cells overexpressing Int1, the Bud4 homologue in Candida albicans (29). Like the septin organization in cells overexpressing Int1, the septin organization was also abnormal in these cells overexpressing Bud4. Cdc3-GFP was not localized or organized properly at the bud neck in most cells. It was often seen as short bars that were aligned vertically at the bud neck or along the surface of elongated buds (Fig. 1E). These findings indicate that too much Bud4 in shs1Δ cells can also perturb septin organization during bud growth.
In addition to shs1Δ cells, Bud4 overexpression in gin4Δ and cla4Δ mutants, in which the septins are not well organized, also impaired growth and generated a septin mutant-like morphology similar to the effect of Bud4 overexpression in shs1Δ cells (Fig. 1C and D). Fifty-eight percent (n = 805) of gin4Δ cells and 63% (n = 763) of cla4Δ cells overexpressing Bud4 exhibited elongated buds at 30°C (Fig. 1D). Gin4 is a bud neck-associated protein kinase that can interact with Shs1 and phosphorylate it in vivo and in vitro (41). Cla4 is a p21-activated protein kinase that physically associates with and can directly phosphorylate the septins Cdc3, Cdc10, Cdc11, and, to a much lesser extent, Cdc12 (42). These findings suggest that cells defective in septin organization may be specifically sensitive to the effect of excess Bud4.
Taken together, our data suggest that Bud4 plays a role in septin organization during bud growth. Bud4 may help to stabilize the septin ring.
The C-terminal anillin-related region of Bud4 functions as a second domain that mediates Bud4's association with the septins.
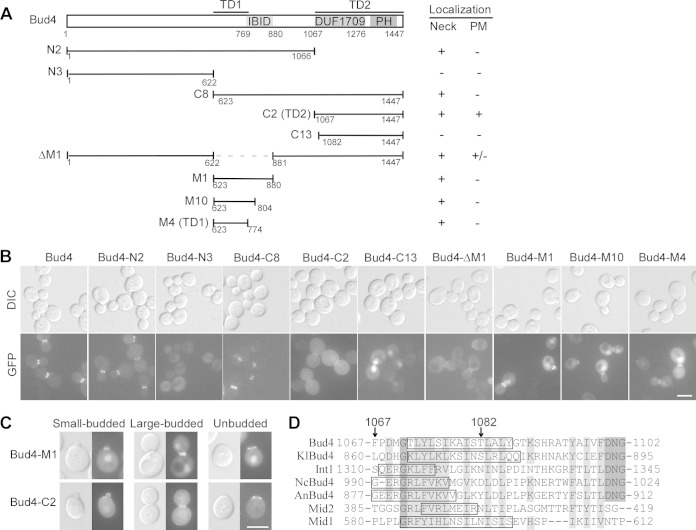
Bud4 localizes to the bud neck in cells with medium to large buds, and this localization depends on the septins at the bud neck (13). Bud4 contains an Iqg1-Bud4-interacting domain (IBID) near the middle region (43) and an anillin-related region at the C terminus. The latter consists of a DUF1709 domain (a domain of unknown function) and a pleckstrin homology (PH) domain (21, 23) (Fig. 2A). A previous study identified a Bud4 segment (amino acids [aa] 511 to 804) that could target to the bud neck, indicating that the central region of Bud4 is capable of associating with the septins (23). We confirmed this finding and found that the central bud neck-targeting region could be narrowed down to the region from aa 623 to 774, which sits next to the IBID (Fig. 2A), because three segments, Bud4-M1 (aa 623 to 880), Bud4-M10 (aa 623 to 804), and Bud4-M4 (aa 623 to 774), that carry this region all localized to the bud neck (Fig. 2B). We named the region from aa 623 to 774 targeting domain 1 (TD1). These Bud4 segments were fused to GFP and expressed in wild-type cells under the control of the BUD4 promoter. All these Bud4 segments were expressed well, as determined by immunoblotting with a monoclonal antibody against GFP (see Fig. S1 in the supplemental material). Surprisingly, we observed that TD1 localized to the bud neck in all the budded cells, irrespective of the size of the bud (Fig. 2C, Bud4-M1). The pattern of TD1 localization resembles that of the septins but apparently differs from that of full-length Bud4, which localized to the bud neck only when the buds were medium or large (13). This feature was not reported in the previous study (23). The persistent association of TD1 with the septins throughout the cell cycle suggests that Bud4 may contain a second targeting domain that could cooperate with TD1 to achieve bud neck-targeting in a temporally restricted manner.
FIG 2.
Bud4 contains two distinct targeting domains for bud neck localization. (A) Schematic representation of the Bud4 segments used for observation and summary of the localization result. PM, plasma membrane. (B) Localization of GFP-Bud4 and GFP-Bud4 segments. Cells of strain YEF473A carrying plasmid pBG2-BUD4 or pBG2-BUD4 segments were used for visualization. (C) Localization of GFP–Bud4-M1 and GFP–Bud4-C2 in cells with small buds and cells with large buds and in postdivision unbudded cells. (D) Sequence alignment of the N termini of the DUF1709 domains in fungal Bud4 homologues. The sequences included S. cerevisiae Bud4, Kluyveromyces lactis Bud4 (KlBud4), Candida albicans Int1, Neurospora crassa Bud4 (NcBud4), Aspergillus nidulans Bud4 (AnBud4), and Schizosaccharomyces pombe Mid1 and Mid2. Identical and similar residues are highlighted in dark gray and light gray shading, respectively. The starting points for Bud4-C2 (residue 1067) and Bud4-C13 (residue 1082) are indicated by arrows. The sequences marked by boxes are potential α helices determined by prediction. Bars, 5 μm.
We observed that the anillin-related region (Bud4-C2, aa 1067 to 1447) also localized to the bud neck, in addition to the plasma membrane (Fig. 2B). Interestingly, it localized to the bud neck only in cells with medium to large buds but not in cells with small buds (Fig. 2C), similar to full-length Bud4. Thus, the C-terminal anillin-related region appears to be a second bud neck-targeting domain. We named it TD2. Bud4-C8 (aa 623 to 1447), a Bud4 segment that contains both TD1 and TD2, showed a localization pattern essentially identical to that of full-length Bud4 (Fig. 2B). Because Bud4-C8 did not target to the bud neck in cells with small buds like TD1 did and not target to the entire plasma membrane like TD2 did, our results suggest that the temporally and spatially restricted association of Bud4 with the septins may require cooperation between TD1 and TD2.
In sharp contrast to our observation, the study by Kang et al. (23) reported that the anillin-related region of Bud4 is not involved in the interaction with the septins on the basis of two observations. First, Bud4 C-terminal truncation mutants still localized to the bud neck. Second, the anillin-related region did not target to the bud neck but mainly localized to the nucleus (23). The first observation can be explained by the fact that these Bud4 segments contain an intact TD1. The discrepancy in the second observation could be explained by the extra 15 amino acids (aa 1067 to 1081) in our Bud4-C2 construct, which is absent in the Bud4 segment used by Kang and colleagues (23). We generated a Bud4 segment identical to the one used by Kang et al., here designated Bud4-C13 (aa 1082 to 1447), and found that it indeed did not target to the bud neck or the plasma membrane (Fig. 2B). The extra 15-aa sequence contains part of a putative α helix and a putative nuclear export signal (NES) (1075-LSIKAISTLAL-1085) (Fig. 2D). In theory, truncation at amino acid residue 1082 would remove the majority of the α helix and destroy the NES motif, which may lead to the retention of this Bud4 segment in the nucleus. Interestingly, the regions in fungal anillin-related proteins corresponding to the 15-aa sequence in Bud4 also contain a putative α helix and show a high degree of sequence similarity (Fig. 2D).
Together, our results show that the anillin-related region of Bud4 functions as a second bud neck-targeting domain, in addition to the known targeting domain located in the central region. The spatially and temporally restricted association of Bud4 with the septins may require cooperation between these two domains.
Roles of the two bud neck-targeting domains of Bud4 in septin organization.
Because Bud4 contains two distinct domains that mediate its association with the septins, we wanted to evaluate the role of each domain in Bud4's function in septin organization. To this end, we employed two assays. First, we took advantage of the elongated bud phenotype of bud4Δ shs1Δ cells grown at a low temperature and examined which Bud4 segment could complement this morphological defect. This assay monitors the function of Bud4 in septin organization during bud growth prior to cytokinesis. Thirty-nine percent of bud4Δ shs1Δ cells carrying the empty vector displayed elongated buds (n = 1,367) at 22°C, whereas only 4% of cells expressing full-length Bud4 (n = 1,169) displayed elongated buds (Fig. 3) (for unknown reasons, cells transformed with the integrative empty vector consistently showed a decrease in the percentage of cells with elongated buds compared to that for cells carrying no vector at all). Although the central TD1 of Bud4 could associate with the septins throughout the cell cycle, we found that the expression of Bud4-N2, which carries TD1 (Fig. 2A), in bud4Δ shs1Δ cells did not complement the morphological defect, as 44% of cells examined (n = 680) displayed elongated buds. In contrast, only 7% (n = 816) and 9% (n = 869) of cells expressing Bud4-C2 and Bud4-ΔM1 displayed elongated buds, respectively (Fig. 3), indicating that the anillin-related region (TD2) could efficiently complement the elongated bud defect of bud4Δ shs1Δ cells. Since the anillin-related region is not only required but also largely sufficient for complementing the elongated bud morphology of bud4Δ shs1Δ cells, which presumably results from a defective septin organization during bud growth, this result suggests that the anillin-related region may play a pivotal role in Bud4's function in septin organization during bud growth.
FIG 3.

The anillin-related region of Bud4 is crucial for septin organization. Cells of strain JGY2737 (bud4Δ shs1Δ) carrying integrated pBG5 (vector), pBG5-BUD4, or pBG5-BUD4 segments were grown on a YPD plate at 22°C for 6 days before observation. Bar, 5 μm.
bud4Δ cells are known to form only a single septin ring instead of normally two rings at the time of cytokinesis (21, 23). Next, we examined which Bud4 segment could complement this defect. This assay monitored Bud4's function in the stabilization of two split septin rings during cytokinesis. In the course of our analysis, Kang et al. reported that Bud4 C-terminal mutants carrying deletions in the DUF1709 or PH domain failed to form a double septin ring (23). This observation indicates that the anillin-related region of Bud4 is essential for Bud4's function during cytokinesis. However, it is not clear whether this region alone is sufficient to carry out Bud4's function. Our result showed that neither TD1 nor the anillin-related region (TD2) alone is sufficient to carry out Bud4's function in the formation of two septin rings during cytokinesis (Table 1, see the results for Bud4-N2 and Bud4-M1 for the TD1 function; see the results for Bud4-C2 for the TD2 function). Interestingly, the Bud4-C8 segment, which carries both the TD1 and TD2 domains (Fig. 2A), was fully functional in the complementation of the double-septin-ring-formation defect (Table 1), suggesting that TD1 and TD2 are likely the only functional domains in Bud4 for septin organization at the end of the cell cycle. The N-terminal half of the protein, aa 1 to 622, is dispensable for this function.
TABLE 1.
Complementation of the double-septin-ring-formation defect of bud4Δ cells by full-length Bud4 and Bud4 segmentsa
| Bud4 construct | % of cells with: |
No. of cells | |
|---|---|---|---|
| Double ring | Single ring | ||
| Vector | 0 | 100 | 22 |
| Bud4 | 74 | 26 | 100 |
| Bud4-C8 | 72 | 28 | 100 |
| Bud4-N2 | 0 | 100 | 48 |
| Bud4-M1 | 0 | 100 | 45 |
| Bud4-C2 | 3 | 97 | 65 |
The pBG2 empty vector and pBG2-BUD4 segments were transformed into strain JGY2966 (bud4Δ CDC3-mCherry SPC42-mCherry). The number of cells that showed a single septin ring or double rings at telophase or anaphase (monitored by Spc42-mCherry, a component of the spindle pole body) was scored.
Together, our results show that the two bud neck-targeting domains of Bud4 are differentially required for two Bud4-regulated septin organization events. Bud4's function in septin organization during bud growth mainly requires the anillin-related region (TD2), whereas Bud4's function in the formation of two septin rings during cytokinesis requires both TD1 and TD2.
The central region of Bud4 functionally interacts with the septin ring.
Although overexpression of full-length Bud4 under the control of a galactose-inducible promoter did not affect growth, we found that overexpression of Bud4-C8 (aa 623 to 1447), which contains both the central region and the anillin-related region of Bud4, markedly reduced growth at 30°C (Fig. 4A). The central region of Bud4 appears to be responsible for causing this defect because overexpression of Bud4-M1 (aa 623 to 880), which carries only the central region of Bud4, is sufficient to severely impair growth (Fig. 4A). Cells overexpressing Bud4-C8 or Bud4-M1 also exhibited a strong defect in cell morphology. Of the cells overexpressing Bud4-C8 and Bud4-M1, 60% (n = 831) and 73% (n = 821), respectively, displayed highly elongated buds after they were grown on galactose-containing medium for 20 h (Fig. 4B). Cell clusters or cell chains could also be observed. Septin organization was also abnormal in these cells. At the bud neck and at the constrictions along the elongated buds in cells overexpressing Bud4-C8, septins were often seen as short, vertical bars instead of a ring, while in cells overexpressing Bud4-M1, septin bars longer than 5 μm, which were mostly present at a single copy per cell and which ran in the mother bud axis, could often be seen, indicating that the septins were not properly organized (Fig. 4C). Interestingly, these abnormal septin structures contained Bud4-C8 and Bud4-M1, respectively (see Fig. S2 in the supplemental material), suggesting that they are made of fully assembled septin complexes.
FIG 4.
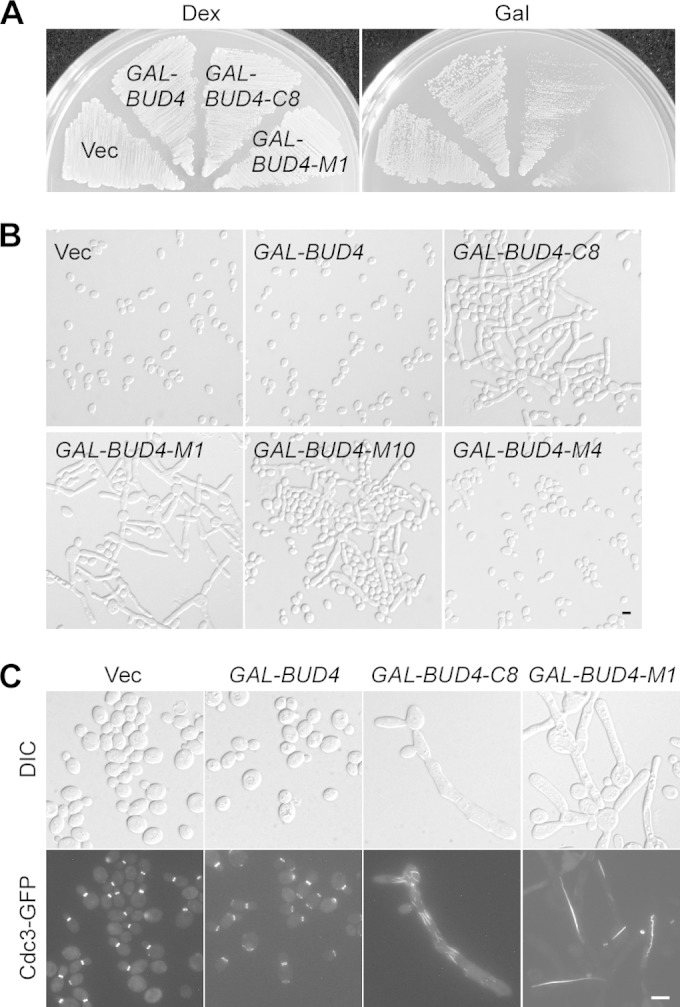
Phenotypes of Bud4 segment overexpression. (A) Cells of strain YEF3572 (bud4Δ) carrying plasmid pEGKT316 (vector), pEGKT316-BUD4, pEGKT316-BUD4-C8, or pEGKT316-BUD4-M1 were grown on an SC-Ura medium (dextrose [Dex]) plate for 2 days and an SRG-Ura medium (Gal) plate at 30°C for 3 days. (B) Morphology of the cells described in the legend to panel A and those carrying pEGKT316-BUD4-M10 or pEGKT316-BUD4-M4 grown on an SRG-Ura medium plate at 30°C for 20 h. (C) Localization of Cdc3-GFP in cells overexpressing Bud4 and Bud4 segments. Cells described in the legend to panel A carrying integrated CDC3-GFP::LEU2 were grown at 30°C for 20 h before imaging. Bars, 5 μm.
The functional domain in Bud4-M1 that is responsible for causing the septin organization defect appears to be located in the region from aa 623 to 804, which contains TD1 and the first 35 aa of IBID (Fig. 2A), since overexpression of this region caused the formation of highly elongated buds in 27% of cells (n = 862) (Fig. 4B, Bud4-M10), whereas just 2% of cells overexpressing TD1 alone (n = 982) displayed elongated buds (Fig. 4B, Bud4-M4). This finding implies that part of the IBID is required for Bud4-M1 function. The short conserved N-terminal sequence of Bud4-M1 is also critical for Bud4-M1's function since the Bud4 segment from aa 637 to 880, which lacks the N-terminal 14 aa, completely lost the ability to cause bud elongation when overexpressed (data not shown).
Because the functional region of Bud4-M1 contains part of IBID, which is known to mediate an interaction between Bud4 and Iqg1 (38), we also investigated whether Iqg1 may be required for the function of Bud4-M1. We found that overexpression of Bud4-M1 in iqg1Δ cells (W303 background) caused bud elongation to an extent similar to that in wild-type cells (data not shown), suggesting that bud elongation caused by the overexpression of Bud4-M1 may not depend on Iqg1.
Taken together, our results show that overproduction of the central region of Bud4 could disrupt septin organization. This finding suggests that the central region of Bud4 may have the ability to associate with the septins in vivo, either directly or indirectly.
The anillin-related region of Bud4 interacts with the C-terminal region of Bud3.
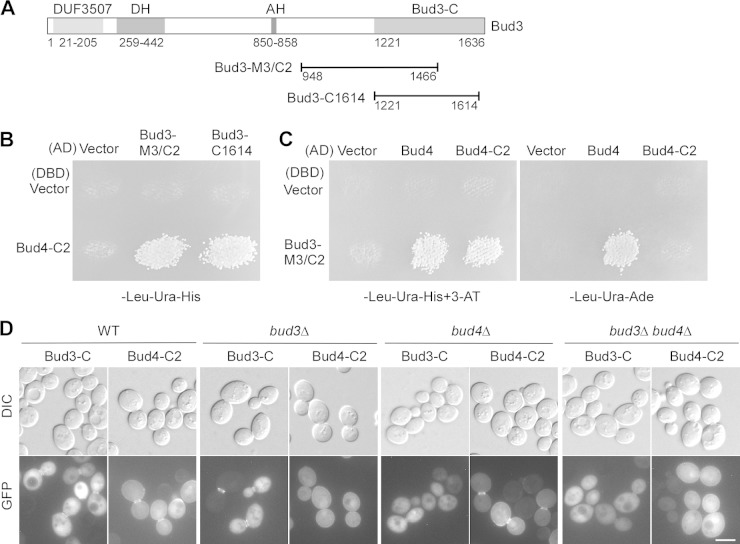
The anillin-related region of Bud4 localizes to the bud neck as well as on the plasma membrane. Because this region is critical for Bud4's function in septin organization, we wanted to know how it targets to the bud neck. Bud4 is known to associate with Bud3 in vivo (18). We therefore examined the interaction of the anillin-related region with several Bud3 segments (Fig. 5A) in a yeast two-hybrid assay. We found that the anillin-related region of Bud4 interacted with Bud3-M3/C2 (aa 948 to 1466) and Bud3-C1614 (aa 1221 to 1614) (Fig. 5B and C). Full-length Bud4 also interacted with Bud3-M3/C2 (Fig. 5C). Bud3-M3/C2 and Bud3-C1614 share the overlapping region from aa 1221 to 1466. Thus, the anillin-related region of Bud4 appears to interact with Bud3 via the C-terminal region from aa 1221 to 1466 of Bud3.
FIG 5.
Bud4-C2 interacts with the C terminus of Bud3. (A) Schematic representation of the domains of Bud3 and the locations of the Bud3-M3/C2 and Bud3-C1614 segments. The Dbl homology (DH) domain and the amphipathic helix (AH) are shown. (B) Bud4-C2 interacts with Bud3-M3/C2 and Bud3-C1614 in the yeast two-hybrid assay. Growth on the SC-Leu-Ura-His plate indicates the interaction between DBD and AD fusion proteins. Cells of strain pJ69-4α carrying pGBDU-C1 (vector, DBD) and pGBDU-BUD4-C2 were mated with cells of strain pJ69-4A carrying pOAD (vector, AD), pOAD-BUD3-M3/C2, and pOAD-BUD3-C1614. Diploids were replica plated to an SC-Leu-Ura-His plate and grown at 30°C for 3 days. (C) Bud3-M3/C2 interacts with full-length Bud4 and Bud4-C2 in the yeast two-hybrid assay. Cells of strain pJ69-4α carrying pGBDU-C1 (vector, DBD) and pGBDU-BUD3-M3/C2 were mated with cells of strain pJ69-4A carrying pGAD-C1 (vector, AD), pGAD-BUD4, and pGAD-BUD4-C2. Diploids were replica plated onto an SC-Leu-Ura-His plate containing 3-AT (3-amino-1,2,4-triazole) and an SC-Leu-Ura-Ade plate and grown at 30°C for 3 days. (D) Localization of GFP–Bud3-C and GFP–Bud4-C2 in wild-type and mutant cells. Cells of strains YEF473A (wild type), YEF3570 (bud3Δ), YEF3572 (bud4Δ), and JGY781 (bud3Δ bud4Δ) carrying pUG36-BUD3-C or pBG2-BUD4-C2 were grown on SC-Ura medium before visualization. Bar, 5 μm.
Interestingly, we found that the bud neck localization of the anillin-related region (Bud4-C2) disappeared in bud3Δ and bud3Δ bud4Δ cells, while its localization on the plasma membrane was unaffected (Fig. 5D), indicating that the bud neck localization of this region depends on Bud3. We previously reported that the C-terminal region of Bud3 (Bud3-C; aa 1221 to 1636) localized to the bud neck as a double ring in cells with medium to large buds (35). Remarkably, we found that the bud neck localization of Bud3-C was greatly diminished in bud4Δ and bud3Δ bud4Δ cells (Fig. 5D), suggesting that Bud3-C's bud neck localization also depends on Bud4.
Together, our results show that the anillin-related region of Bud4 interacts with the C-terminal region of Bud3. The C-terminal regions of these two proteins may cooperate for bud neck targeting (septin association).
Bud4 may act together with Bud3 in septin organization during bud growth.
Bud4 is thought to act together with Bud3 in the control of bud site selection at late G1 stage of the cell cycle (13, 17, 18). However, it is not clear whether Bud4 may act together with Bud3 in septin organization as well. We showed early in this study that bud4Δ shs1Δ cells displayed defects in cell morphology and septin organization (Fig. 1A and B). Interestingly, we found that bud3Δ shs1Δ cells also displayed elongated buds and cell chains when grown on YPD solid medium at a lower temperature. At 22°C, 62% of bud3Δ shs1Δ cells examined (n = 889) displayed elongated buds and 20% of cells formed cell chains (Fig. 6A). The extent of the defect was similar to that of bud4Δ shs1Δ cells. Thus, it seems that Bud4 may act together with Bud3 in septin organization during bud growth. A previous study showed that both bud4Δ cdc12-6 and bud3Δ cdc12-6 cells were defective for growth at 28°C, a temperature at which cdc12-6 cells could grow (22). We made an identical observation in our strain background (data not shown). In addition, we found that 100% of bud4Δ cdc12-6 and bud3Δ cdc12-6 cells examined (n = 400) displayed dramatically elongated buds at 28°C, whereas only 36% of cdc12-6 cells (n = 1,041) displayed elongated buds (Fig. 6B). These findings support the view that Bud4 and Bud3 may act together in septin organization. The interaction between the anillin-related region of Bud4 with Bud3's C-terminal region may provide the basis for the functional cooperation between Bud4 and Bud3 in septin organization.
FIG 6.

Bud4 may act together with Bud3 in septin organization. (A) Cell morphology of strains YEF3570 (bud3Δ), YEF4603 (shs1Δ), and JGY3035 (bud3Δ shs1Δ). Cells were grown on YPD plates at 22°C for 6 days. (B) Cell morphology of strains YEF2218 (cdc12-6), JGY3040 (bud3Δ cdc12-6), and JGY3042 (bud4Δ cdc12-6) grown on an SC medium plate at 28°C for 1 day. Bars, 5 μm.
The anillin-related region of Bud4 is also the functional domain for bud site selection.
Bud4 is essential for axial bud site selection, as haploid bud4Δ cells could not bud in the axial pattern but instead budded in a bipolar pattern (7, 13). Kang et al. (23) reported that both the central region and the C-terminal anillin-related region of Bud4 are required for Bud4's function in bud site selection. However, neither of these two regions alone is sufficient to carry out Bud4's function in bud site selection (23). In contrast to their observation, we found that the anillin-related region is largely sufficient to carry out axial budding because Bud4-C2 (aa 1067 to 1447) largely rescued the axial budding defect (Table 2), suggesting that the anillin-related region is the functional domain for Bud4 in bud site selection. We observed that 41% of cells expressing Bud4-C2 (n = 700) displayed a typical axial budding pattern, in which the bud scars were connected in a chain, while another 40% of the cells displayed an axial-like pattern, in which the bud scars were distributed at one pole but were not immediately connected. There was some space between some of the bud scars (Table 2). When the budding pattern of those cells carrying only two bud scars was scored (8), 75% of cells expressing Bud4-C2 (n = 156) exhibited the axial pattern, while 88% of cells expressing full-length Bud4 (n = 100) and 4% of cells carrying the empty vector (n = 100) exhibited the axial pattern. Because two-scar analysis monitors the fidelity of bud site selection for only one round of budding, this result supports the suggestion that Bud4-C2 is largely functional in axial budding. Similarly, Bud4-ΔM1 (aa 1 to 621 plus 881 to 1447), which contains the anillin-related region but lacks TD1, behaved like Bud4-C2. In contrast, neither Bud4-N2 (aa 1 to 1066) nor Bud4-M1 (aa 623 to 880), two segments that carry TD1 but lack the anillin-related region, rescued the axial budding defect (Table 2). We found that the Bud4 segment used by Kang et al. (23), which is here designated Bud4-C13 (aa 1082 to 1447), indeed completely failed to complement the axial budding defect of bud4Δ cells (Table 2). The lack of function for Bud4-C13, which is 15 amino acids shorter than Bud4-C2, is likely due to the truncation of the putative nuclear export signal (NES), which leads to the retention of the protein in the nucleus (Fig. 2B), causing the protein to be unable to target to the bud neck.
TABLE 2.
Complementation of axial bud site selection defect of bud4Δ cells by full-length Bud4 and Bud4 segmentsa
| Bud4 construct | % of cells expressing the following budding pattern |
No. of cells | |||
|---|---|---|---|---|---|
| Axial | Axial-like | Bipolar | Random | ||
| Vector | 8 | 72 | 20 | 200 | |
| Bud4 | 89 | 7 | 4 | 200 | |
| Bud4-N2 | 5 | 71 | 24 | 300 | |
| Bud4-C2 | 41 | 40 | 1 | 18 | 700 |
| Bud4-C13 | 5 | 63 | 32 | 300 | |
| Bud4-C8 | 84 | 10 | 6 | 400 | |
| Bud4-ΔM1 | 34 | 50 | 5 | 11 | 500 |
| Bud4-M1 | 2 | 79 | 19 | 300 | |
pBG2 empty vector and pBG2-BUD4 segments were transformed into strain YEF3572 (bud4Δ). Cells with three or more bud scars were counted for determination of the budding pattern.
Together, our results indicate that the C-terminal anillin-related region of Bud4 is the functional domain for axial bud site selection. The central TD1 in Bud4 may increase the fidelity of bud site selection by the anillin-related region via restricting its localization to the bud neck.
DISCUSSION
Targeting mechanism of Bud4's anillin-related region.
In this study, we showed that, like human anillin and the fission yeast Mid1 and Mid2 (24, 27, 28, 44), the C-terminal anillin-related region of Bud4 is capable of targeting to the cell cortex and cleavage furrow (bud neck). The only domain in the C-terminal region of anillin-related proteins with a known function is the PH domain, which could interact with the negatively charged phosphoinositides in the plasma membrane via electrostatic interaction (45, 46). For example, the PH domain of Bud4 binds specifically to phosphatidylinositol 3-phosphate in vitro (47). However, because the anillin PH domain itself is not sufficient, though it is necessary, for cleavage furrow targeting (24), as is the case for the PH domains of Bud4 and the fission yeast Mid2 (27, 28; our unpublished data), the exact mechanism by which the C-terminal regions of anillin and its fungal homologues target to the cleavage furrow is not clear. On the basis of the findings of studies on other low-affinity PH domains, the targeting of anillin and fungal homologues to the cleavage furrow may require the cooperation of the PH domain with other domains in the protein (45) or may occur via oligomerization of the PH domain, as in the case of dynamin isoforms (48). In this study, we showed that the C-terminal targeting domain in Bud4, TD2, interacts with Bud3, another member of the axial landmark, for association with the septins at the bud neck. This demonstrated, for the first time, that a fungal anillin homologue could cooperate with another protein for cleavage furrow targeting.
Role of Bud4 in septin organization.
Bud4 is essential for the formation of two septin rings during cytokinesis (21–23). In this study, we showed that Bud4 is also required for septin organization during bud growth. Thus, Bud4 emerges as a new player in the organization of septin structures. However, the detailed mechanism of how Bud4 engages in septin organization is not clear.
We showed in this study that Bud4 contains two bud neck-targeting domains, TD1 and TD2, which could potentially bind to the septins. One clue about how Bud4 regulates septin organization may come from the TD2 domain. We observed that Bud4 TD2 is required and largely sufficient for the complementation of the elongated bud defect of bud4Δ shs1Δ cells. Moreover, TD2 is absolutely required for Bud4's function in the formation of two septin rings during cytokinesis. Thus, TD2 seems to be the major functional determinant of Bud4 in septin organization. Based on our observation that Bud4-TD2 interacts with the C-terminal region of Bud3 (Bud3-C) and this interaction is crucial for targeting themselves to the bud neck, we speculate that Bud4-TD2 may initially loosely associate with the plasma membrane via the interaction of its PH domain with phosphoinositides in the plasma membrane. The interaction between Bud4-TD2 and neighboring Bud3-C may create a binding surface for the septins, which not only helps to anchor themselves to the septin filaments but also helps to stabilize the septin ring. We showed previously that Axl2, the only transmembrane protein in the axial landmark, is implicated in septin organization during bud growth and cytokinesis, as axl2Δ elm1Δ, axl2Δ gin4Δ, and axl2Δ cla4Δ cells displayed synthetic morphological defects that included bud elongation and cell chain formation as well as a severe defect in septin organization at 24°C (17). Interestingly, the function of Axl2 in septin organization is largely mediated by its cytoplasmic tail, which localized to the bud neck, and this localization completely depends on the presence of both Bud4 and Bud3 (17). Therefore, we speculate that Bud4-TD2 and Bud3-C may cooperate to recruit Axl2 to the bud neck. This may help to stabilize the septin ring by linking the septins to the plasma membrane and/or cell wall because the bulk N-terminal cadherin domain of Axl2 sticks out into the extracellular space.
Why is TD1 essential for Bud4's function in the formation of two septin rings during cytokinesis, while it is largely dispensable for septin organization during bud growth? A recent study showed that the septin hourglass-to-double ring transition is accompanied by the loss of septin subunits from the hourglass and reorganization of the remaining subunits into the two septin rings (49). This process is also accompanied by a 90° change in septin filament orientation from aligning to the mother bud axis to circling of the bud neck. During such a dramatic septin structure remodeling process, TD1 may cooperate with TD2 to provide a stronger interaction with the septin filaments. This may help to prevent the loss of septin subunits during and after the hourglass-to-double ring transition.
Role of Bud4 in axial landmark assembly.
Based on the dependency between these components for bud neck localization, we previously proposed that the axial landmark is assembled in the following order: the septins → Bud4 and Bud3 → Axl1 and Axl2 (17). This model reflects that Bud4 cooperates with Bud3 in linking the axial landmark to the septins. Bud4 is known to interact with Bud3 (18). However, how the interaction is achieved is not understood. In this study, we showed that Bud4 and Bud3 interact with each other via their C termini and thus provided a mechanism for the functional cooperation between Bud4 and Bud3 in bud site selection. The interaction between the Bud4 C terminus (Bud4-C) and Bud3-C is apparently important for Bud3's function in axial budding, because we showed previously that the N-terminal two-thirds of Bud3 is less efficient in axial budding than full-length Bud3 (35). Bud3 contains three targeting domains (aa 1 to 858, 841 to 1220, and 1221 to 1636) responsible for bud neck localization. Bud3-C (aa 1221 to 1636) happens to be the one that confers the most robust and the tightest bud neck association (35). When Bud3-C was removed, the rest of Bud3 (Bud3-NM) anchored poorly at the bud neck. The majority of protein then located on the entire plasma membrane and vacuoles (our unpublished data). We speculate that the interaction between Bud4-C and Bud3-C may create a binding surface for the septins at the bud neck, which may lead to the efficient recruitment of Bud3 to the axial landmark complex. Thus, our study highlights the importance of the C terminus of Bud4 in the assembly of the axial landmark.
Bud3-C is unlikely the only region in Bud3 that interacts with Bud4, since Bud3-C is not absolutely required for Bud3's function in axial budding (35). Bud4 may also interact with other parts of Bud3, such as the region from aa 1 to 858, which could partially function in axial budding (35). Future investigation is needed to test this possibility.
Supplementary Material
ACKNOWLEDGMENTS
We thank Erfei Bi, John R. Pringle, and Matthew Lord for kindly providing yeast strains and plasmids.
This work was supported by the National Natural Science Foundation of China (grant no. 30871347 and 31370124) and the National Infrastructure of Natural Resources for Science and Technology Program of China (no. NIMR-2014-8).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00268-14.
REFERENCES
- 1.Drubin DG, Nelson WJ. 1996. Origins of cell polarity. Cell 84:335–344. doi: 10.1016/S0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- 2.Nelson WJ. 2003. Adaptation of core mechanisms to generate cell polarity. Nature 422:766–774. doi: 10.1038/nature01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Witte H, Bradke F. 2008. The role of the cytoskeleton during neuronal polarization. Curr Opin Neurobiol 18:479–487. doi: 10.1016/j.conb.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 4.Chant J, Pringle JR. 1995. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J Cell Biol 129:751–765. doi: 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pringle JR, Bi E, Harkins HA, Zahner JE, De Virgilio C, Chant J, Corrado K, Fares H. 1995. Establishment of cell polarity in yeast. Cold Spring Harbor Symp Quant Biol 60:729–744. doi: 10.1101/SQB.1995.060.01.079. [DOI] [PubMed] [Google Scholar]
- 6.Chant J. 1999. Cell polarity in yeast. Annu Rev Cell Dev Biol 15:365–391. doi: 10.1146/annurev.cellbio.15.1.365. [DOI] [PubMed] [Google Scholar]
- 7.Chant J, Herskowitz I. 1991. Genetic control of bud site selection in yeast by a set of genes that constitute a morphogenetic pathway. Cell 65:1203–1212. doi: 10.1016/0092-8674(91)90015-Q. [DOI] [PubMed] [Google Scholar]
- 8.Flescher EG, Madden K, Snyder M. 1993. Components required for cytokinesis are important for bud site selection in yeast. J Cell Biol 122:373–386. doi: 10.1083/jcb.122.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujita A, Oka C, Arikawa Y, Katagai T, Tonouchi A, Kuhara S, Misumi Y. 1994. A yeast gene necessary for bud-site selection encodes a protein similar to insulin-degrading enzymes. Nature 372:567–570. doi: 10.1038/372567a0. [DOI] [PubMed] [Google Scholar]
- 10.Chant J, Mischke M, Mitchell E, Herskowitz I, Pringle JR. 1995. Role of Bud3p in producing the axial budding pattern of yeast. J Cell Biol 129:767–778. doi: 10.1083/jcb.129.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halme A, Michelitch M, Mitchell EL, Chant J. 1996. Bud10p directs axial cell polarization in budding yeast and resembles a transmembrane receptor. Curr Biol 6:570–579. doi: 10.1016/S0960-9822(02)00543-2. [DOI] [PubMed] [Google Scholar]
- 12.Roemer T, Madden K, Chang J, Snyder M. 1996. Selection of axial growth sites in yeast requires Axl2p, a novel plasma membrane glycoprotein. Genes Dev 10:777–793. doi: 10.1101/gad.10.7.777. [DOI] [PubMed] [Google Scholar]
- 13.Sanders SL, Herskowitz I. 1996. The Bud4 protein of yeast, required for axial budding, is localized to the mother/bud neck in a cell cycle-dependent manner. J Cell Biol 134:413–427. doi: 10.1083/jcb.134.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Versele M, Thorner J. 2005. Some assembly required: yeast septins provide the instruction manual. Trends Cell Biol 15:414–424. doi: 10.1016/j.tcb.2005.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh Y, Bi E. 2011. Septin structure and function in yeast and beyond. Trends Cell Biol 21:141–148. doi: 10.1016/j.tcb.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gladfelter AS, Pringle JR, Lew DJ. 2001. The septin cortex at the yeast mother-bud neck. Curr Opin Microbiol 4:681–689. doi: 10.1016/S1369-5274(01)00269-7. [DOI] [PubMed] [Google Scholar]
- 17.Gao X-D, Sperber LM, Kane SA, Tong Z, Tong AH, Boone C, Bi E. 2007. Sequential and distinct roles of the cadherin domain-containing protein Axl2p in cell polarization in yeast cell cycle. Mol Biol Cell 18:2542–2560. doi: 10.1091/mbc.E06-09-0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang PJ, Angerman E, Jung C-H, Park H-O. 2012. Bud4 mediates the cell-type-specific assembly of the axial landmark in budding yeast. J Cell Sci 125:3840–3849. doi: 10.1242/jcs.103697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park H-O, Bi E. 2007. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol Mol Biol Rev 71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang PJ, Lee ME, Park H-O. 2014. Bud3 activates Cdc42 to establish a proper growth site in budding yeast. J Cell Biol 206:19–28. doi: 10.1083/jcb.201402040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wloka C, Nishihama R, Onishi M, Oh Y, Hanna J, Pringle JR, Krauß M, Bi E. 2011. Evidence that a septin diffusion barrier is dispensable for cytokinesis in budding yeast. Biol Chem 392:813–829. doi: 10.1515/BC.2011.083. [DOI] [PubMed] [Google Scholar]
- 22.Eluère R, Varlet I, Bernadac A, Simon M-N. 2012. Cdk and the anillin homolog Bud4 define a new pathway regulating septin organization in yeast. Cell Cycle 11:151–158. doi: 10.4161/cc.11.1.18542. [DOI] [PubMed] [Google Scholar]
- 23.Kang PJ, Hood-DeGrenier JK, Park H-O. 2013. Coupling of septins to the axial landmark by Bud4 in budding yeast. J Cell Sci 126:1218–1226. doi: 10.1242/jcs.118521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oegema K, Savoian MS, Mitchison TJ, Field CM. 2000. Functional analysis of a human homologue of the Drosophila actin binding protein anillin suggests a role in cytokinesis. J Cell Biol 150:539–552. doi: 10.1083/jcb.150.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piekny AJ, Maddox AS. 2010. The myriad roles of anillin during cytokinesis. Semin Cell Dev Biol 21:881–891. doi: 10.1016/j.semcdb.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 26.Kinoshita M, Field CM, Coughlin ML, Straight AF, Mitchison TJ. 2002. Self- and actin-templated assembly of mammalian septins. Dev Cell 3:791–802. doi: 10.1016/S1534-5807(02)00366-0. [DOI] [PubMed] [Google Scholar]
- 27.Berlin A, Paoletti A, Chang F. 2003. Mid2p stabilizes septin rings during cytokinesis in fission yeast. J Cell Biol 160:1083–1092. doi: 10.1083/jcb.200212016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tasto JJ, Morrell JL, Gould KL. 2003. An anillin homologue, Mid2p, acts during fission yeast cytokinesis to organize the septin ring and promote cell separation. J Cell Biol 160:1093–1103. doi: 10.1083/jcb.200211126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gale C, Gerami-Nejad M, McClellan M, Vandoninck S, Longtine MS, Berman J. 2001. Candida albicans Int1p interacts with the septin ring in yeast and hyphal cells. Mol Biol Cell 12:3538–3549. doi: 10.1091/mbc.12.11.3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paoletti A, Chang F. 2000. Analysis of mid1p, a protein required for placement of the cell division site, reveals a link between the nucleus and the cell surface in fission yeast. Mol Biol Cell 11:2757–2773. doi: 10.1091/mbc.11.8.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Justa-Schuch D, Heilig Y, Richthammer C, Seiler S. 2010. Septum formation is regulated by the RHO4-specific exchange factors BUD3 and RGF3 and by the landmark protein BUD4 in Neurospora crassa. Mol Microbiol 76:220–235. doi: 10.1111/j.1365-2958.2010.07093.x. [DOI] [PubMed] [Google Scholar]
- 32.Si H, Rittenour WR, Xu K, Nicksarlian M, Calvo AM, Harris SD. 2012. Morphogenetic and developmental functions of the Aspergillus nidulans homologues of the yeast bud site selection proteins Bud4 and Axl2. Mol Microbiol 85:252–270. doi: 10.1111/j.1365-2958.2012.08108.x. [DOI] [PubMed] [Google Scholar]
- 33.Guthrie C, Fink GR. 1991. Guide to yeast genetics and molecular biology. Methods Enzymol 194:1–863. [PubMed] [Google Scholar]
- 34.Gao X-D, Caviston JP, Tcheperegine SE, Bi E. 2004. Pxl1p, a paxillin-like protein in Saccharomyces cerevisiae, may coordinate Cdc42p and Rho1p functions during polarized growth. Mol Biol Cell 15:3977–3985. doi: 10.1091/mbc.E04-01-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo J, Gong T, Gao X-D. 2011. Identification of an amphipathic helix important for the formation of ectopic septin spirals and axial budding in yeast axial landmark protein Bud3p. PLoS One 6:e16744. doi: 10.1371/journal.pone.0016744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.James P, Halladay J, Craig EA. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Longtine MS, McKenzie A III, DeMarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961. doi:. [DOI] [PubMed] [Google Scholar]
- 38.Carroll CW, Altman R, Schieltz D, Yates JR, Kellogg D. 1998. The septins are required for the mitosis-specific activation of the Gin4 kinase. J Cell Biol 143:709–717. doi: 10.1083/jcb.143.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mino A, Tanaka K, Kamei T, Umikawa M, Fujiwara T, Takai Y. 1998. Shs1p: a novel member of septin that interacts with Spa2p, involved in polarized growth in Saccharomyces cerevisiae. Biochem Biophys Res Commun 251:732–736. doi: 10.1006/bbrc.1998.9541. [DOI] [PubMed] [Google Scholar]
- 40.Iwase M, Luo J, Bi E, Toh-e A. 2007. Shs1 plays separable roles in septin organization and cytokinesis in Saccharomyces cerevisiae. Genetics 177:215–229. doi: 10.1534/genetics.107.073007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mortensen EM, McDonald H, Yates J, Kellogg DR. 2002. Cell cycle-dependent assembly of a Gin4-septin complex. Mol Biol Cell 13:2091–2105. doi: 10.1091/mbc.01-10-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Versele M, Thorner J. 2004. Septin collar formation in budding yeast requires GTP binding and direct phosphorylation by the PAK, Cla4. J Cell Biol 164:701–715. doi: 10.1083/jcb.200312070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osman MA, Konopka JB, Cerione RA. 2002. Iqg1p links spatial and secretion landmarks to polarity and cytokinesis. J Cell Biol 159:601–611. doi: 10.1083/jcb.200205084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Celton-Morizur S, Bordes N, Fraisier V, Tran PT, Paoletti A. 2004. C-terminal anchoring of mid1p to membranes stabilizes cytokinetic ring position in early mitosis in fission yeast. Mol Cell Biol 24:10621–10635. doi: 10.1128/MCB.24.24.10621-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lemmon MA, Ferguson KM, Schlessinger J. 1996. PH domains: diverse sequences with a common fold recruit signaling molecules to the cell surface. Cell 85:621–624. doi: 10.1016/S0092-8674(00)81022-3. [DOI] [PubMed] [Google Scholar]
- 46.Rameh LE, Arvidsson A-K, Carraway KL III, Couvillion AD, Rathbun G, Crompton A, VanRenterghem B, Czech MP, Ravichandran KS, Burakoff SJ, Wang D-S, Chen C-S, Cantley LC. 1997. A comparative analysis of the phosphoinositide binding specificity of pleckstrin homology domains. J Biol Chem 271:22059–22066. [DOI] [PubMed] [Google Scholar]
- 47.Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA. 2004. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell 13:677–688. doi: 10.1016/S1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- 48.Klein DE, Lee A, Frank DW, Marks MS, Lemmon MA. 1998. The pleckstrin homology domains of dynamin isoforms require oligomerization for high affinity phosphoinositide binding. J Biol Chem 273:27725–27733. doi: 10.1074/jbc.273.42.27725. [DOI] [PubMed] [Google Scholar]
- 49.Ong K, Wloka C, Okada S, Svitkina T, Bi E. 2014. Architecture and dynamic remodelling of the septin cytoskeleton during the cell cycle. Nat Commun 5:5698. doi: 10.1038/ncomms6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.