Abstract
Sandhoff disease is a rare progressive neurodegenerative genetic disorder with a high incidence among certain isolated communities and ethnic groups around the world. Previous reports have shown a high occurrence of Sandhoff disease in northern Saskatchewan. Newborn screening cards from northern Saskatchewan were retrospectively screened in order to investigate the incidence and determine the carrier frequency of Sandhoff disease in these communities. PCR-based screening was conducted for the c.115delG (p.(Val39fs)) variant in the HEXB gene that was previously found in 4 Sandhoff disease patients from this area. The carrier frequency for this allele was estimated to be ~1:27. MS/MS-based screening of hexosaminidase activity along with genetic sequencing allowed for the identification of additional variants based on low total hexosaminidase activity and high % hexosaminidase A activity relative to c.115delG carriers. In total 4 pathogenic variants were discovered in the population (c.115delG, c.619A>G, c.1601G>T, and c.1652G>A) of which two are previously unreported (c.1601G>T and c.1652G>A). The combined carrier frequency of these alleles in the study area was estimated at ~1:15. Based on the number of cases of Sandhoff disease from this area we estimate the incidence to be ~1:390 corresponding to a child being born with the disease every 1–2 years on average. The results from our study were then compared with variants in the HEXB gene from the genomes available from the 1000 Genomes project. A total of 19 HEXB variants were found in the 1092 genomes of which 5 are suspected of having a deleterious effect on hexosaminidase activity. The estimated carrier frequency of Sandhoff disease in Saskatchewan at 1:15 is more than 3 times higher than the carrier frequency in the global sample provided by the 1000 Genomes project at 1:57.
Keywords: Sandhoff, Tay–Sachs, Gangliosidosis, Hexosaminidase, HEXB, GM2
1. Introduction
Sandhoff disease (Online Mendelian Inheritance in Man no. 268800) is a rare, autosomal recessive lysosomal storage disorder caused by pathogenic variants in the hexosaminidase-B (HEXB) gene (5q13). Pathogenic variants in HEXB result in a deficiency of β-hexosaminidase A (HexA; E.C. 3.2.1.52) and β-hexosaminidase B (HexB; E.C. 3.2.1.52) due to decreased production of the β-subunit [1]. HexA is a heterodimer of α–β subunits, and HexB is a β–β homodimer [2]. A third formβ-hexosaminidase-S (HexS; E.C. 3.2.1.52) may also be present as a homodimer of α–α subunits [3]. Under normal conditions, HexA is responsible for the degradation of GM2 ganglioside. The diminished HexA activity in Sandhoff disease leads to progressive accumulation of GM2 in neuronal cells and irreversible neuronal degradation [1]. The symptoms of Sandhoff disease can manifest at different stages of life corresponding to the amount of residual enzyme activity caused by the variants present in the HEXB gene [1]. Symptoms manifest before one year and lead to death typically by four years of age for the infantile onset form of the disorder. Juvenile and adult onset forms of the disease are also possible [1].
Published estimates of Sandhoff disease carrier frequency in the general population vary from 1:310 based on the prevalence of Sandhoff disease in Australia between 1980 and 1996 [4] to 1:276 (n = 32,342) in non-Jewish Americans based on serum β-hexosaminidase levels [5]. Several isolated or highly consanguineous communities have been identified with an increased carrier frequency. The IVS-2+1 G>A splice site variant has been implicated as the predominant allele responsible for Sandhoff disease in Argentina [6]. In Saudi Arabia the high degree of consanguinity has led to a markedly high incidence for many autosomal recessive conditions including Sandhoff disease; patients are typically homozygous for a private allele [7]. Cyprus has the highest reported Sandhoff disease carrier frequency among its Christian Maronite community at 1:7 (n = 244) [8]. Sandhoff disease has also been reported among French Canadians and those of French descent [9].
In Canada, some northern Saskatchewan communities also have a high incidence of infantile onset Sandhoff disease [10]. Previously we identified the c.115delG pathogenic variant in the HEXB gene from several Sandhoff disease patients born in this area [11]. No relationship has been identified between Sandhoff disease in northern Saskatchewan where the community is largely Métis (individuals of mixed French/aboriginal descent) and the reports of Sandhoff disease among other French Canadian populations.
In this study, variants in the HEXB gene and aberrant β-hexosaminidase levels from newborn screening cards collected from individuals born in northern Saskatchewan were retrospectively investigated. Our objectives were to determine the frequency of the c.115delG variant previously found in several affected patients, investigate the possibility of other HEXB variants in the population, characterize those variants via in silico analysis, estimate the frequency of all Sandhoff disease causing variants in the population, estimate the incidence of Sandhoff disease, and compare the frequency of Sandhoff disease causing variants in our study population to the frequency in the general population. Several estimates of the frequency of Sandhoff disease causing alleles in the general population have previously been described [4,5] however, the studies have used restricted population sampling. As such a global sample of HEXB genes was considered by analyzing the data provided by the 1000 Genomes project in order to estimate the frequency of Sandhoff disease causing alleles in the global population.
2. Materials and methods
2.1. Study area and selection of newborns to screen
The high incidence of Sandhoff disease in northern Saskatchewan communities has been known since the late 1970s [10]. To establish the incidence and carrier frequency of Sandhoff disease in these northern Saskatchewan communities, we identified the communities as previously described [12]. The long-term retention of newborn screening cards in Saskatchewan began in 2000, and our retrospective analysis included all infants from the study area born between 2000 and 2012, provided their residual newborn screening card had enough blood remaining to complete both tests. Thus, our retrospective analysis was limited to newborns born between 2000 and 2012. A total of 1561 individuals were included in the study. Ethics approval for the use of residual dried blood spots was obtained from the University of Regina Research Ethics Board and the University of Saskatchewan Biomedical Research Ethics Board.
2.2. c.115delG genotyping assay
The assay for detecting the c.115delG variant has been previously described [11] and was used to determine each individual genotype for this particular variant. To summarize, this assay uses real-time PCR to detect the presence of the c.115delG allele in blood eluted from two 3 mm dried blood spot punches. The PCR assay was used to screen 1561 individuals from the study area born between 2000 and 2012. All specimens found to contain the c.115delG allele were reproducible.
2.3. β-Hexosaminidase enzyme assay
2.3.1. Substrates and internal standards
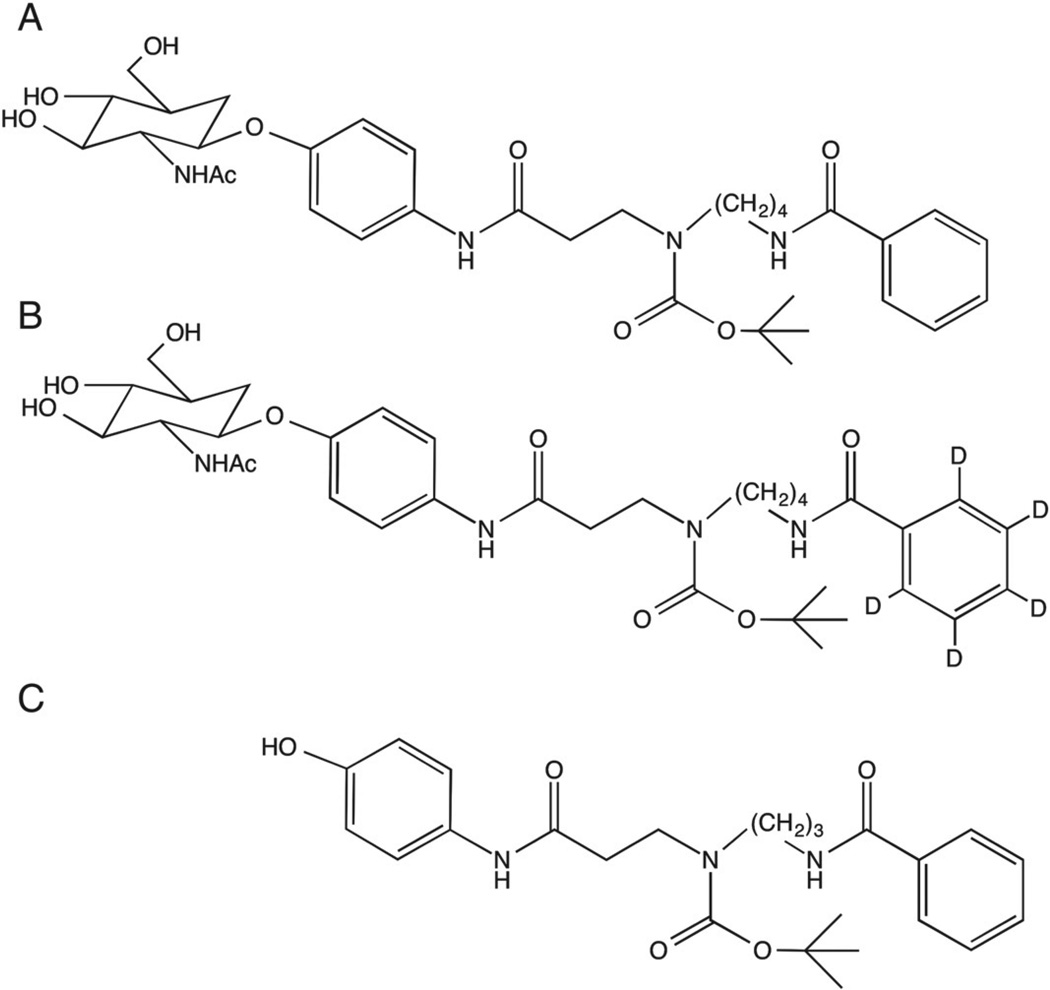
The β-hexosaminidase specific substrates and internal standards were synthesized in the laboratory of Dr. Michael Gelb at the University of Washington. The structures for the compounds can be seen in Fig. 1. A difference in molecular weight between the two substrates allowed for determination of both Total Hex and HexB in a single injection. To prepare the substrates and internal standards for use, all materials were individually dissolved in methanol. Reagent cocktails for measuring Total Hex and HexB activities were made up at 0.23 µM of their respective substrate and 0.23 µM internal standard such that each reaction would receive 3.4 × 10−3 µmol of substrate and internal standard per 15 µl of cocktail added to each 25 µl reaction. The Total Hex reagent cocktail contained the non-deuterated substrate with internal standard whereas the HexB cocktail contained deuterated substrate with internal standard. Reagent cocktails were dried under nitrogen and stored at −20 °C prior to use.
Fig. 1.
Synthetic substrates and internal standard used to assay β-hexosaminidase activity. Compounds were synthesized in the laboratory of Michael Gelb at the University of Washington. (A) Substrate added to the Total Hex reaction. (B) Substrate added to the HexB reaction. (C) Internal standard added to each reaction.
When needed the dried reagents were allowed to equilibrate to room temperature prior to being reconstituted in 8.7 mL of 0.04 M citrate-phosphate buffer (pH 4.4) and 300 µl of sodium taurocholate (120 g/l). Aliquots appropriate for the analysis of a 96-well plate were prepared and frozen at −20 °C until needed.
2.3.2. β-Hexosaminidase assay conditions
The method for assaying other lysosomal storage disorders has been described by Gelb et al. [13] and is outlined herewith relevant modifications for the β-hexosaminidase assay. From each newborn screening card a 3 mm punch was taken and deposited into an individual well of a 96-well polypropylene microplate. Blood was eluted from the punches with 70 µl of extraction buffer (20 mM sodium-phosphate monobasic, pH 7.1). The extraction plate was then incubated at 37 °C for 1 h while shaking at 850 RPM.
While incubating, a fresh plate was prepared by adding 15 µl of Total Hex reagent cocktail to each well (Total Hex plate). The Total Hex reaction was then started by adding 10 µl of each extract to the appropriate wells of the Total Hex plate. This plate was then sealed and incubated at 37 °C with shaking at 200 RPM overnight (~22 h). The dried blood spot extract plate was then re-sealed and incubated at 52 °C for 1 h with no shaking in order to heat inactivate β-Hexosaminidase A. Heat inactivation of HexA is common to other β-hexosaminidase assays [14,15] but was optimized for this assay. During the 52 °C incubation, 15 µl of the HexB reagent cocktail was added to each well of a new 96-well microplate (HexB plate). Following the 52 °C incubation, 10 µl of heat-inactivated extract was added to the corresponding wells of the HexB plate. The HexB plate was then sealed and incubated overnight at 37 °C while shaking at 200 RPM for ~22 h. After incubation the reactions in each plate were stopped by adding 100 µl of 50:50 (v/v %) methanol/ethyl acetate to each well. The 125 µl total volumes from corresponding wells of the Total Hex and HexB plates were then combined in a polypropylene deep-well plate for product purification.
2.3.3. Product purification
To the combined quenched reactions, 400 µl of ethyl acetate and 400 µl of distilled water were added and mixed to facilitate a liquid–liquid extraction. After mixing the plate was sealed and centrifuged at 14.7 G for 2 min. Following centrifugation 300 µl of the organic phase from each well was transferred to a new 96-well plate. The material in the organic phase was then dried under nitrogen then reconstituted in 200 µl of 95/5 (v/v %) ethyl acetate/methanol for solid phase extraction.
A solid phase extraction apparatus for 96-well plates was prepared by adding approximately 100 mg of silica gel to each well of the filter plate. The filter plate was washed with 1 ml of 95/5 (v/v %) ethyl acetate/ methanol under vacuum. The 200 µl volume of reconstituted product was then filtered through the plate followed by the addition of 4 × 400 µl of 95/5 (v/v %) ethyl acetate/methanol under vacuum. The filtered product was then dried down under nitrogen. Prior to analysis the material was reconstituted in 100 µl of 80/20 (v/v %) methanol/ water with 0.2% formic acid. If MS/MS analysis was not taking place the same day, the dried product was sealed and frozen at −20 °C until needed.
2.3.4. MS/MS analysis
Product detection was carried out using an API-2000 mass spectrometer (AB Sciex Concord, Ontario, CA) with Ionics Upgrade (Ionics Bolton, Ontario, CA), coupled to an Agilent HPLC. Analysis was done using flow injection with positive electrospray, and multiple reaction monitoring to identify the target compounds. The mobile phase consisted of 80/20 (v/v %) methanol/water with 0.2% formic acid and a flow rate of 150 µl per minute. Optimized instrument-specific run parameters for β-hexosaminidase product detection are summarized in Tables 1 and 2.
Table 1.
Compound specific optimized MS/MS run parameters
| Parameter | Total Hex product | HexB product | Internal standard |
|---|---|---|---|
| Transition | 456.3 > 356.3 | 461.3 > 361.3 | 442 > 342 |
| DP | 41 | 41 | 45 |
| EP | 6 | 6 | 6 |
| CE | 18 | 18 | 15 |
| CXP | 23 | 23 | 24 |
| FP | 275 | 275 | 300 |
DP: declustering potential; EP: entrance potential; CE: collision energy; CXP: collision cell exit potential; FP: focusing potential.
Table 2.
Shared MS/MS instrument settings for all experiments.
| Parameter | Setting |
|---|---|
| CAD | 6 |
| CUR | 25 |
| GS1 | 25 |
| GS2 | 50 |
| IS | 5500 |
| TEM | 300 |
CAD: collision gas; CUR curtain gas; GS1 & GS2 ion source gasses; IS ion spray voltage; TEM: temperature.
Peak areas were calculated using Analyst software (Applied Biosystems). Peak areas for the products and internal standards were used to calculate β-hexosaminidase enzyme activities. The ratio of peak area for the products to the peak area of the internal standard for each reaction was used to determine the enzyme activity relative to the amount of internal standard added to each reaction, the incubation time, and the volume of eluted blood added to each reaction. HexA enzyme activity and subsequently %HexA were calculated by subtracting the HexB activity from the Total Hex activity.
2.3.5. β-Hexosaminidase assay validation
In order to validate the β-hexosaminidase assay for detecting Sandhoff disease the following parameters were examined; 1) matrix blanks were analyzed to detect any interfering substances; 2) the possibility of spontaneous substrate degradation was examined by measuring product formation in blanks; 3) substrate depletion for each reaction (Total Hex and HexB) was evaluated by stopping the assay at time intervals from 2 to 47 h to ensure that the reaction was not going to completion; 4) precision was assessed for both inter-run and intra-run variation.
In order to distinguish individuals affected with Sandhoff disease from those unaffected it was necessary to establish a normal range for Total Hex, HexB and %HexA activities. Normal ranges were calculated using EP Evaluator® (Data Innovations, South Burlington, Vermont, USA). To assess accuracy, dried blood spots from four diagnosed Sandhoff disease patients were analyzed in order to determine the positive predictive and negative predictive values.
Results from this assay for the four Sandhoff disease patients were compared to results obtained from the Metabolic Disease Laboratory in Saskatoon, SK, CA using the standard 4-MUG fluorometric method for assaying β-hexosaminidase activity. Furthermore, the assay was used to analyze a blind panel of β-hexosaminidase deficient samples sent from an external laboratory.
Due to the long-term storage that had occurred for many of the samples included in our retrospective analysis, special consideration was taken for identifying aberrant β-hexosaminidase activity from older specimens. Total Hex activity remaining on the residual dried blood spots was found to diminish in a time dependent manner (data not shown). Hexosaminidase activity on cards collected prior to 2005 was insufficient for analysis. As such, normal ranges for Total Hex activity and %HexA for the purpose of identifying affected individuals were established for each year of our retrospective study. The MS/MS assay was used to measure β-hexosaminidase activity from 760 newborn screening cards collected between 2005 and 2009.
In order to identify potential Sandhoff disease causing variants in the population, we analyzed the MS/MS data and established a potential carrier cut-off for individuals who fell within limits of Total Hex activity and %HexA for their respective birth year. Samples that met these criteria were analyzed again by the MS/MS assay. The HEXB gene was then sequenced from samples that repeatedly fell within the cut-off range for potential carriers.
2.4. Sequencing and analysis of the HEXB gene
Sanger sequencing of the HEXB gene was carried out as previously described [11]. Exons one through fourteen of the HEXB gene and several nucleotides from the flanking intronic regions were amplified by PCR. The DNA sequence was determined using dye-terminator sequencing. Data analysis was conducted using BioNumerics v6.5 (Applied Maths). Variants were determined relative to the HEXB reference sequence NT_006713.15 from Genbank. An alignment was created for each exon and sequence differences were highlighted. Coding sequence variations and protein level variations were named according to the Human Genome Variation Society guidelines [16]. The deleterious or benign nature of non-synonymous single nucleotide polymorphisms (nsSNPs) was investigated using the online prediction tools PolyPhen-2 [17] and PROVEAN [18] in addition to further in silico analysis.
2.5. Comparison to the 1000 Genomes dataset
The publicly available genome data provided by the 1000 Genomes project was compared to our study data [19]. At the time of this analysis the 1000 Genomes data included genomes from 1092 individuals representing a global sample. Clinically relevant HEXB variants present among the 1092 individuals were analyzed and discussed. PolyPhen-2 and SIFT scores for the variants present in the 1000 Genomes dataset were considered along with in silico analysis and discussion of reports in the literature for specific variants to infer the pathogenicity of each variant. The frequency of Sandhoff disease causing alleles in the global sample was then estimated for comparison to the northern Saskatchewan population.
3. Results
3.1. c.115delG genotyping assay
The results from the c.115delG variant screening can be seen in Table 3. A total of 1561 newborns born between 2000 and 2012 were screened. We found 57 individuals who had a single copy of the c.115delG allele and 3 individuals who were homozygous for the allele and determined to be Sandhoff disease affected patients. As such, the carrier frequency of the c.115delG allele in the northern Saskatchewan communities is estimated to be approximately 1:27.
Table 3.
HEXB variants found in the northern Saskatchewan population.
| Variant | Exon | # alleles found/# alleles analyzed | Consequence | Previously described | Polyphen-2 predictiona | PROVEAN predictionb |
|---|---|---|---|---|---|---|
| c.115delG | 1 | 63/3122 | Reading frame shift | Yes [11] | N/A | N/A |
| c.362A>G | 2 | 3/48 | p.Lys121Arg | No | 0.00 benign | −0.353 neutral |
| c.619A>G | 5 | 16/48 | p.Ile207Val | Yes [20] | 0.281 benign | −0.964 neutral |
| c.1601G>T | 13 | 1/48 | p.Cys534Phe loss of C534-C551 bond | No c.1601G>A described [21] | 1.00 probably damaging | −10.029 deleterious |
| c.1652G>A | 14 | 1/48 | p.Cys551Tyr loss of C534-C551 bond | No | 1.00 probably damaging | −9.773 deleterious |
PolyPhen-2 prediction score can range from 0 (benign) to 1 (probably damaging) with a default cut-off of 0.432.
PROVEAN scores are deleterious if less than −2.5 and neutral if greater than −2.5 using default settings.
3.2. Validation of the β-hexosaminidase enzyme assay
Matrix blanks without substrate added produced no MS/MS signal. Matrix blanks with only filter paper punches and substrate added were found to produce 0.2–0.7% and 4.6–5.6% of the signal of normal patient samples for Total Hex and HexB respectively. This signal was assumed to be from spontaneous substrate degradation and a trace amount of product that may be present in the substrate stocks. As such blanks were included in every run and subtracted from test samples. The amount of product generated by Total Hex or HexB over a 47 h period was assessed. Product formation continues past the 22 h incubation time used by the assay up to 47 h.
Precision of the assay was determined by analyzing replicate dried blood spots from the same individual. Intraday precision as a result of five runs resulted in %CV values of 13.7% for Total Hex and 9.3% for HexB (n = 6). Interday precision was evaluated over two days and resulted in %CV of 10.0% for Total Hex and 18.0% for HexB (n = 6).
To establish a normal range for β-hexosaminidase levels in newborns using this assay, 400 fresh dried blood spot samples from Saskatchewan newborns were analyzed. The 400 fresh dried blood spot samples were collected and stored at room temperature for less than 1month. The normal ranges for the measured analytes are summarized in Table 4. The range for %HexA was typically 3–10 times broader than % HexB.
Table 4.
β-Hexosaminidase normal ranges and Sandhoff disease patient values.
| Parameter | Normal range | Affected patient averagea |
|---|---|---|
| Total Hex | 63.2–147.3 | 1.23 |
| HexB | 7.5–33.3 | 0.01 |
| %HexA | 72.3–91.7 | 99.75 |
β-Hexosaminidase activities are presented as µmol/h/l of blood.
Average values from 4 diagnosed Sandhoff disease patients.
β-Hexosaminidase activities from the four Sandhoff disease affected patients were about 1% of those from the unaffected population and in some cases undetectable (Table 4). For the purpose of screening, a cut-off was established such that a screen positive result for Sandhoff disease would have a Total Hex enzyme activity of less than 10% of the low end of the normal range (<6.3 µmol/h/l blood), in addition to a higher than normal %HexA (>92%). With these cut-offs the positive predictive value and negative predictive values are both 100% for detecting Sandhoff disease.
In Saskatchewan, suspected cases of Sandhoff disease are diagnosed using the standard fluorometric enzyme assay [15]. The normal ranges and average values using the fluorometric assay to determine plasma β-hexosaminidase activity are shown in Table 5. The normal ranges from the fluorometric method are established in an age-group dependent manner while normal range for the MS/MS assay described here were determined using dried blood spots from newborns. Comparison of the results for the Sandhoff patients from the MS/MS assay and the fluorometric method used in Saskatoon correlated for all four patients. When a panel of blinded samples from an outside laboratory containing dried blood spot punches from Sandhoff disease and Tay–Sachs disease patients was analyzed, the assay successfully identified deficient β-hexosaminidase activity in the Sandhoff patients but was unable to distinguish any abnormal activity in the Tay–Sachs patients.
Table 5.
β-Hexosaminidase activities using the fluorometric method on plasma.
| Parameter | Normal range (0–1 yr) | Normal range (1–3 yr) | Affected patient average |
|---|---|---|---|
| Total Hex | 390–2622 | 532–1748 | 14.5 |
| HexB | 50–1458 | 267–995 | 0 |
| %HexA | 13–56 | 24.1–52.2 | 100 |
β-Hexosaminidase activities are presented as µmol/h/l of blood.
3.3. Screening with the β-hexosaminidase enzyme assay
Of the 760 newborn screening cards screened from the study area, four of the samples fell in the cut-off range for affected individuals. Those four individuals were confirmed to be the Sandhoff disease affected patients by DNA sequencing of the HEXB gene. An additional 35 samples were below the thresholds that were established for potential carriers for each respective year.
The results from the β-hexosaminidase activity screening and the c.115delG variant screening were compared. It was found that 21 of 35 individuals in the potential carrier range possessed the c.115delG variant. A further 17 of the remaining 725 individuals had a single copy of the c.115delG variant and were missed by the original cut-offs for potential carriers. As such the cut-off for each year was adjusted so that all of the samples known to contain the c.115delG allele would be included and the data was reanalyzed. The yearly cut-off values for Total Hex and %HexA that were used are listed in Table 6. Subsequently 76 samples fell below the adjusted cut-offs of which 4 were affected individuals and 34 carried the c.115delG variant. The remaining 38 samples lacked the c.115delG allele. These 38 were repeated and the HEXB gene was sequenced from those samples that repeatedly fell below the adjusted cut-offs.
Table 6.
Cut-offs for Sandhoff disease carriers by year based on the Total Hex and %HexA of individuals found to carry the c.115delG variant.
| Year | Total Hexa,b | %HexAb |
|---|---|---|
| 2005 | 37.41 | 92.6 |
| 2006 | 32.25 | 93.1 |
| 2007 | 40.84 | 91.2 |
| 2008 | 49.18 | 89.4 |
| 2009 | 57.89 | 92.2 |
The units for Total Hex are µmol/hour/l of blood.
Cut-offs are Total Hex activity less than the values stated and %HexA greater than the values stated.
3.4. HEXB sequencing and analysis
Sequencing of the HEXB gene from individuals with low β-hexosaminidase activity as described in Section 3.3 revealed three variants in addition to the c.115delG variant previously found in the northern Saskatchewan population. These variants include c.362A>G, c.619A>G, and c.1601G>T. Furthermore, only three of the four Sandhoff disease affected individuals were homozygous for the c.115delG allele. The fourth patient was a compound heterozygote possessing the c.115delG and c.1652G>A variants. A complete list of the genetic variants and the frequency of each allele is shown in Table 3.
PolyPhen-2 and PROVEAN predictions were in agreement for all 4 nsSNPs found among the population from the study area. The c.362A>G nsSNP causes a change of Lys at position 121 of the β-hexosaminidase β subunit to Arg. Both of these amino acids share long positively charged side chains and thus have similar potential for electrostatic interactions. A PolyPhen-2 score of 0.00 (benign) and a PROVEAN score of −0.353 (neutral) correlate in predicting the c.362A>G variant to be tolerated.
The second nsSNP c.619A>G results in a conversion of Ile at position 207 to Val. The difference due to this conversion is the loss of a methyl group on the side chain of the amino acid. PolyPhen-2 and PROVEAN scores for the c.619A>G SNP were 0.281 (benign) and−0.964 (neutral), respectively.
Both c.1601G>T and c.1652G>A result in the loss of key Cys residues responsible for the formation of disulfide bonds. The c.1601G>T variant changes Cys at position 534 to Phe where as c.1652G>A results in the change of Cys at position 551 to Tyr. PolyPhen-2 and PROVEAN scores for the c.1601G>T variant were 1.00 and −10.029 respectively and for the c.1652G>A variant were 1.00 and −9.773 indicating a probably damaging or deleterious outcome for both of these nsSNPs.
Although the c.115delG allele was not analyzed by either program, its pathogenic nature is clear due to the induced reading frame shift in the coding sequence of exon 1. A summary of the variants found, the number of times each allele was detected and the PolyPhen-2 and PROVEAN predictions are listed in Table 3.
3.5. Comparison to the 1000 Genomes data set
A total of 19 variants were present in the coding region of the HEXB gene or the adjacent intronic splice regions from the 1092 individuals included in the 1000 Genomes data set. The potential pathogenicity and the frequency of those variants were analyzed. The 19 variants include 5 synonymous DNA-level variations (Table 7), one splice region variant (Table 7), and 13 missense mutations (Table 8). None of the pathogenic variants found in the Saskatchewan study population were present in the 1000 Genomes data set.
Table 7.
Synonymous and splice region HEXB variants found in the 1000 Genomes dataset.
| Variant | Global MAFa | Highest frequency population | Consequence |
|---|---|---|---|
| c.276C>T | 0.00137 | YRI | Synonymous |
| c.978G>C | 0.00046 | GBR | Synonymous |
| c.1035A>C | 0.00046 | JPT | Synonymous |
| c.1051T>C | 0.00733 | IBS | Synonymous |
| c.1251G>A | 0.00046 | YRI | Synonymous |
| c.772-4A>G | 0.04991 | IBS | Splice region variant |
Global MAF: Global minor allele frequency.
Table 8.
Missense HEXB variants found in the 1000 Genomes dataset.
| Variant | Global MAFa | Highest frequency population | Consequence | SIFTb | Polyphen-2c |
|---|---|---|---|---|---|
| c.185C>T | 0.02060 | TSI | p.Ser62Leu | 1 tolerated | 0.001 benign |
| c.251A>G | 0.00046 | CHB | p.Asn84Ser | 1 tolerated | 0 benign |
| c.362A>G | 0.20742 | LWK | p.Lys121Arg | 0.65 tolerated | 0 benign |
| c.449C>A | 0.00641 | ASW | p.Tyr150Asn | 0.18 tolerated | 0.021 benign |
| c.619A>G | 0.15064 | JPT | p.Ile207Val | 0.31 tolerated | 0.157 benign |
| c.922C>G | 0.00046 | ASW | p.Pro308Ala | 0.08 tolerated | 0.012 benign |
| c.1258A>G | 0.01236 | LWK | p.Ile420Val | 1 tolerated | 0.02 benign |
| c.1437A>C | 0.00046 | ASW | p.Gln479His | 0.15 tolerated | 0.001 benign |
| c.214C>T | 0.00595 | IBS | p.Leu72Phe | 0.001 deleterious | 0.49 possibly damaging |
| c.923C>T | 0.00046 | MXL | p.Pro308Leu | 0.001 deleterious | 0.642 possibly damaging |
| c.1066G>A | 0.00046 | CHS | p.Val356Met | 0 deleterious | 0.999 Probably Damaging |
| c.1250C>T | 0.00137 | JPT | p.Pro417Leu | 0.01 deleterious | 0.503 possibly damaging |
| c.1367A>C | 0.00046 | TSI | p.Tyr456Ser | 0.19 tolerated | 0.673 possibly damaging |
Global MAF: Global minor allele frequency.
SIFT scores are deleterious if less than or equal to 0.05 and tolerated if greater than 0.05.
PolyPhen-2 prediction score can range from 0 (benign) to 1 (probably damaging) with a default cut-off of 0.432.
The synonymous variants present in the 1000 Genomes dataset include c.276C>T, c.978G>C, c.1035A>C, c.1051T>C, and c.1251G>A. The c.722-4A>G splice region variant is located −4 nucleotides from the 5-prime end of exon 7 at the 3′ end of an intron. Given that the consensus sequence for a 3′ intron splice site is NCAG or NNAG in higher eukaryotes such as humans [22,23] it is likely that even though A is fairly conserved at the location 4 nucleotides upstream from this intron/exon boundary of the HEXB gene, the c.722-4A>G variant will be tolerated due to all four nucleotides having been observed at this location of the splice region. Therefore, all of the variants listed in Table 7 are likely benign and not included when estimating the frequency of Sandhoff disease causing alleles in the global population.
Of the 13 missense variants present in the 1000 Genomes dataset, Polyphen-2 and SIFT predictions agree that 8 are benign polymorphisms (Table 8). The 8 missense variants predicted to be benign are c.185C>T, c.251A>G, c.362A>G, c.449C>A, c.619A>G, c.922C>G, c.1258A>G, and c.1437A>C. The c.185C>T [24], c.362A>G [25], and c.619A>G [20,26,27] variants have been described in the literature. Both c.185C>T and c.362A>G have proven to be benign polymorphisms when examined in clinical samples however, c.619A>G has been implicated in adult onset GM1 gangliosidosis [20,26,27]. No reports were found for the other 5 missense variants predicted to be benign and based on the positive predictive value and accuracy for the Polyphen-2 and SIFT tools they are assumed to be as such. Therefore, of the 8missense variants, only c.619A>G was considered when calculating the frequency of pathogenic variants.
The results of the Polyphen-2 and SIFT analysis substantiate that 4 of the missense variants (c.214C>T, c.923C>T, c.1066G>A, and c.1250C>T) are probably damaging or deleterious but are discordant on 1 variant, c.1367A>C, for which the Polyphen-2 prediction is possibly damaging whereas the SIFT prediction is benign. Of the 4 missense variants for which the prediction tools outcome of potentially pathogenic was in agreement, 3 are of unknown significance and 1 has been described in clinical samples previously.
The c.214C>T variant results in the change of Leu at position 72 to Phe. Leu 72 is located at a region of the peptide chain between an alpha helix and beta sheet on the periphery of the peptide with about 42% conservation of Leu at this position. Leu 72 is not a critical residue for either the active site or protein secondary structure however, both prediction tools suggest that a change to Phe at this position will be detrimental to protein function based on the lack of Phe at this position in related sequences. The c.214C>T variant was found a total of 13 times in the American, Asian, and European super populations of the 1000 Genomes dataset but was not found in the African group.
Two variants were present at position 308 of the peptide sequence, c.922C>G (p.Pro308Ala) and c.923C>T (p.Pro308Leu). Both Polyphen-2 and SIFT agree that that the former is a benign variant due to the presence of Ala at position 308 in other closely related sequences however, both prediction tools suggest that the change of Pro 308 to Leu will be damaging to the beta subunit. At position 308 of the alignment of related HEXB sequences Pro is the most common having moderate conservation however, Ala and Ile are also present among other amino acid variants. Since the structural difference between Ile and Leu is essentially the placement of a methyl group on the side chain, the pathogenicity of the c.923C>T variant is questionable yet predicted to be damaging due to there being no Leu at position 308 in any of the aligned sequences. The c.923C>T allele was rare showing up only a single time in an individual of Mexican ancestry living in Los Angeles.
The c.1066G>A variant is also of unknown significance. The peptide locus is between an alpha helix and beta strand structures. Both prediction tools suggest that the subsequent p.Val356Met amino acid change will be detrimental to the enzymes function based on the high conservation of Val at position 356 (~80%). The c.1066G>A variant was only found in a single Southern Han Chinese individual.
The c.1250C>T variant has been described by Wakamatsu et al. and Gomez-Lira et al. in compound heterozygous patients suffering from juvenile and adult onset Sandhoff disease respectively [25,28]. This variant results in the change of Pro at position 417 to Leu. The Pro at position 417 is responsible for a turn on the periphery of the secondary structure of the beta subunit that properly orients the adjacent alpha helix and beta strands. Without Pro at position 417 it would be expected that the polypeptide would not fold properly. However, using transfection experiments Wakamatsu et al. showed that this variant actually causes the activation of a cryptic splice site which results in the loss of exon 11 from the processed transcript accounting for the loss of β-hexosaminidase activity [25]. Interestingly, this allele was observed once in each of the Finnish, Italian, and Japanese populations included in the 1000 Genomes sample which correlates well with the previous literature reports from Wakamatsu et al. (Japanese patient) and Gomez-Lira et al. (Italian patient) [25,28].
For the single variant which the two prediction tools disagreed upon the Polyphen-2 score of 0.673 indicates that amino acid variation caused by c.1367A>Cwill likely be damaging to the protein's function as a result of the Tyr to Ser change at position 456. Tyr 456 is located in a region of the peptide responsible for protein–protein interaction between the subunits that combine to form β-hexosaminidases A and B [29,30]. In fact Tyr 456, and its neighbors, Asp 452 and Tyr 450 are responsible for hydrogen bonding between the subunits that stabilize the active site of the enzyme [29,30]. Banerjee et al. have previously described and characterized the c.1367A>C variant as being non-functional by transfecting the c.1367A>C HEXB gene into COS-7 cells where they observed no functional β-hexosaminidase B formed [20,26]. The authors go on to explain how further in silico analysis of the variant predicts a dramatic change in the folding of the β-subunit. The c.1367A>C variant was observed only a single time in the 1000 Genomes dataset in an Italian individual.
Therefore, as a result of the Polyphen-2 and SIFT predictions along with the mutation analysis presented here and reports from the literature the following alleles were used to determine the frequency of potentially pathogenic alleles in the global population sample: c.619A>C, c.214C>T, c.923C>T, c.1066G>A, c.1250C>T, and c.1367A>C. Subsequently, the combined frequency for these potentially pathogenic alleles in the 1000 Genomes sample was 0.15934. If we exclude c.619A>C and only include the potentially Sandhoff disease causing alleles the frequency is 0.0087 or a Sandhoff disease carrier rate of roughly 1:57.
4. Discussion
The purpose of this study was to better understand the incidence of Sandhoff disease and the frequency of Sandhoff disease causing variants in northern Saskatchewan. When 1561 individuals were retrospectively assayed using the c.115delG allelic discrimination assay, 57 heterozygotes were discovered. Therefore, we estimate the carrier frequency for this variant to be 1:27 in the study population. As a result of finding 4 affected individuals in our retrospective study we estimate the incidence of Sandhoff disease in this population to be 1:390.
The β-hexosaminidase MS/MS assay reliably detected 4 Sandhoff disease affected individuals and proved to be semi-reliable for detecting carriers. In order to achieve a 100% positive predictive value for identifying c.115delG carriers, 32% (20/62) of the samples that repeatedly fell below the cut-offs that were established in conjunction with the c.115delG assay, did not carry the c.115delG variant. If genetic sequencing of these 20 samples was not conducted they may have been misclassified as false positives. However, upon further investigation using genetic sequencing, it was revealed that 17 of the 20 samples that did not have the c.115delG allele instead possessed one or more of the nsSNPs listed in Table 3. This left < 5% (3/62) of samples with β-hexosaminidase activity in the carrier range but no identified variant in the HEXB gene. It is possible that there are HEXA variants present in these 3 outliers however, HEXA mutation analysis was not performed. Carrier detection by enzymatic analysis in general is challenging because carriers and even non-carriers typically have a broad range for normal levels of enzyme activity. As such, carrier detection by MS/MS, fluorometric, or other enzyme activity analysis is imperfect. Identification of Tay–Sachs affected individuals was not possible using the assay conditions described here however, a modified protocol these substrates and internal standard allowed for the identification of Tay– Sachs patients.
A total of 4 nsSNPs were found in the HEXB gene among our study group in addition to the c.115delG variant. Of these 4 nsSNPs, 2 cause the loss of disulphide bonds in the resulting polypeptide. In silico analysis and online prediction tools indicate the loss of the disulphide bonds to be detrimental to β-hexosaminidase activity. No report of the c.1601G>T or c.1652G>A polymorphisms could be found in the dbSNP database or 1000 Genomes project data however, there are reports of a c.1601G>A (similar loss of disulphide bond) substitution being responsible for the infantile form of Sandhoff disease in a Japanese patient [21]. As such the available information is indicative that the c.1601G>T and c.1652G>A alleles are disease causing. Parental analysis of the compound heterozygous patient containing the c.115delG and c.1652G>A variants was not possible due to the removal of patient identifiers from samples prior to analysis. However, the presence of the 16 kb deletion common among other French-Canadian populations was ruled-out due to the presence of heterozygous SNP loci in exons 3 and 4.
The other 2 nsSNPs, c.362A>G and c.619A>G, have more ambiguous outcomes. The c.362A>G variant has been described in the online dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/) and reported to be benign based on transfection experiments [25]. In silico analysis and online prediction tools suggest that this allele is tolerated by the enzyme however, we saw reduced Total Hex activity and elevated %HexA in these individuals. It is possible that this allele causes reduced β-hexosaminidase activity within the range of tolerance so as not to cause the disease phenotype when present in a homozygous or compound heterozygous state.
Originally the c.619A>G variant was implicated as the cause of an adult form of Sandhoff disease in a compound heterozygous patient [20]. Subsequent reports of this allele have shown it to be capable of α/β-subunit dimerization, incapable of β/β-subunit dimerization [26], and tolerated in homozygous individuals producing β-hexosaminidase activity consistent with carriers [24,27] and thus, not responsible for GM2 build up due to β-hexosaminidase A deficiency. Our data correlates with the observations presented in the literature. Three c.619A>G homozygotes were detected along with 9 heterozygotes all showing β-hexosaminidase activity consistent with the range established for Sandhoff disease carriers. However, one individual possessed both c.619A>G and c.1601G>T variants. Similar to the compound heterozygote patient described by Banerjee et al. [20] the individual we found possessing the c.619A>G and c.1601G>T variants may be at risk for developing an adult form of β-hexosaminidase B deficiency with motor neuron disease.
Interestingly, since our initial report of the c.115delG variant in northern Saskatchewan it has also been detected in France [31]. The c.115delG allele was paternally inherited by a French patient whose father was of Vietnamese descent [31]. As part of future investigations it would be interesting to conduct haplotype analysis to determine the origin of the c.115delG variant.
A major advantage of using an assay to measure enzyme activity compared to single variant mutation analysis is that it can detect carriers irrespective of the variation present. The disadvantage is that the carrier range typically overlaps with the normal range leading to the possibility of missing some carriers and a high false positive rate. We observed <5% of individuals that fell in the range for Sandhoff disease carriers that had no HEXB variants and a further 5% which possessed HEXB polymorphisms predicted to be neutral. The possibility of HEXA variants present in the samples thought to be false positive carriers may bring down the false positive rate of the MS/MS assay even further. The major advantage of the PCR-based assay is that it does not produce false positives though it can only detect a single variant per primer and probe set and those variants must be known ahead of time. The use of both techniques to screen the northern Saskatchewan population has provided the opportunity to take advantage of the benefits of both methodologies.
In total the number of individuals found to carry a Sandhoff disease causing allele or c.619A>G as detected by our biochemical screening and genetic sequencing analysis was 51 (n = 760). The c.619A>G variant though not the cause of Sandhoff disease was included in this calculation since it has been implicated in adult onset β-hexosaminidase B deficiency. As such the combined carrier frequency for these variants in northern Saskatchewan is estimated at 1:15. Given the birthrate in this area and the estimated incidence of 1:390 births, a child is expected to be born with these diseases every 1–2 years on average.
Of the 19 variants found in the 1000 Genomes dataset 5 have the potential to cause Sandhoff disease. The pathogenicity of several of these variants is corroborated by reports in the literature whereas for others the exact clinical outcome has not been determined. Based on the results from prediction tools the carrier rate of Sandhoff disease causing variants may be as high as 0.0174 in the global sampling. However, due to the error associated with SIFT and Polyphen-2 predictions [17,18] and without confirming the pathogenicity of all of the potentially pathogenic alleles identified by the 1000 Genomes project the rate may be as low as 0.00549 in the global sample. The carrier rate for Sandhoff disease in the Saskatchewan sample was 0.0460. As such the carrier frequency for Sandhoff disease causing variants in Saskatchewan is about 2.5–8 times higher than that among individuals sampled by the 1000 Genomes project, and ~12–14 times higher than previous estimates made for the general population [4,5]. Given the high birthrate of Sandhoff disease affected children in northern Saskatchewan, an elevated carrier rate was expected.
A total of 19 individuals in the 1000 Genomes data set were carriers for a variant predicted to cause Sandhoff disease by either SIFT of Polyphen-2. Thus, if all of the variants predicted to be damaging do cause Sandhoff disease then the carrier rate among the 1000 Genomes sample is about 1 in 57, which is significantly higher than Sandhoff disease carrier estimates in the general population (1 in 276 to 1 in 310) [4,5]. A carrier rate of 1 in 57 in the general population along with an autosomal recessive pattern of inheritance would suggest an incidence of Sandhoff disease of roughly 1 in 13000 births. Sandhoff disease in the general population is extremely rare with an estimated incidence of 1:422,000 (n = 4.2 million) [4]. This discrepancy may be due to the error associated with SIFT and Polyphen-2 predictions, underestimates of the carrier frequency by the previously published works, or the result of randomly sampling a relatively small subset of the global population. Given the rarity of Sandhoff disease based on its incidence in the general population the carrier rate in the general population should be much lower than the Polyphen-2 and SIFT predictions suggest.
Of the 19 HEXB variants found in the 1000 Genomes data set, 5 were missense variants with deleterious predictions. Two of these have been previously described in the literature as being pathogenic whereas the other three are of questionable consequence. To determine the consequence of the three variants not found in the literature, transfection experiments could be performed to analyze the HEXB transcripts and polypeptides produced as well as measuring β-hexosaminidase activity. Determining the effect of these variants on β-hexosaminidase activity would confer a higher degree of confidence in estimating the frequency of pathogenic variants in the 1000 Genomes sample. Assuming that some of the variants predicted to be deleterious or damaging end up being tolerated then this analysis could explain why the carrier rate for suspected Sandhoff disease causing variants in the 1000 Genomes sample was so high.
Reports in the literature for the carrier frequency of Sandhoff disease range from1:310 in the general population to 1:7 in isolated communities with a high degree of consanguinity [4–9,32]. Similarly, the Saskatchewan cohort is remote and isolated, with a high degree of consanguinity [10] and has an elevated carrier frequency for Sandhoff disease causing variants relative to the general population. In other communities where there is a high carrier frequency for Sandhoff disease, programs have been established or proposed to ameliorate the impact of the disease on families and the community [7,8,32]. As such, a program aimed at preventing Sandhoff disease in northern Saskatchewan is being considered. If a carrier screening program for Sandhoff disease in Saskatchewan is to be established a multiplex PCR could be developed to detect all of the pathogenic variants discovered in this study in order to facilitate better carrier screening with a single assay. Although the c.115delG allele is the major variant responsible for the Sandhoff disease in Saskatchewan, other pathogenic variants are present in the population highlighting the need for special consideration when selecting an assay to detect this disease.
Acknowledgments
Special thanks to Jeff Eichhorst, Michele Etter, and Joyce lePage for their assistance with MS/MS assay development and troubleshooting and to Greg Horsman for supporting this work at the Saskatchewan Disease Control Laboratory. The work at the University of Washington was supported by a grant from the National Institutes of Health (DK067859).
Contributor Information
Braden Fitterer, Email: braden.fitterer@health.gov.sk.ca.
Patricia Hall, Email: patricia.l.hall@emory.edu.
Nick Antonishyn, Email: nantonishyn@health.gov.sk.ca.
Michael Gelb, Email: gelb@chem.washington.edu.
Denis Lehotay, Email: dlehotay@health.gov.sk.ca.
References
- 1.Mahuran DJ. Biochemical consequences of mutations causing the GM2 gangliosidoses. Biochim. Biophys. Acta. 1999;1455(2–3):105–138. doi: 10.1016/s0925-4439(99)00074-5. [DOI] [PubMed] [Google Scholar]
- 2.Beutler E, Kuhl W, Comings D. Hexosaminidase isozyme in type OGm2 gangliosidosis (Sandhoff-Jatzkewitz disease) Am. J. Hum. Genet. 1975;27(5):628–638. [PMC free article] [PubMed] [Google Scholar]
- 3.Ikonne JU, Rattazzi MC, Desnick RJ. Characterization of Hex S, the major residual beta hexosaminidase activity in type O Gm2 gangliosidosis (Sandhoff-Jatzkewitz disease) Am. J. Hum. Genet. 1975;27(5):639–650. [PMC free article] [PubMed] [Google Scholar]
- 4.Meikle PJ, et al. Prevalence of lysosomal storage disorders. JAMA. 1999;281(3):249–254. doi: 10.1001/jama.281.3.249. [DOI] [PubMed] [Google Scholar]
- 5.Cantor RM, et al. Sandhoff disease heterozygote detection: a component of population screening for Tay-Sachs disease carriers. II. Sandhoff disease gene frequencies in American Jewish and non-Jewish populations. Am. J. Hum. Genet. 1987;41(1):16–26. [PMC free article] [PubMed] [Google Scholar]
- 6.Kleiman FE, et al. Sandhoff disease in Argentina: high frequency of a splice site mutation in the HEXB gene and correlation between enzyme and DNA-based tests for heterozygote detection. Hum. Genet. 1994;94(3):279–282. doi: 10.1007/BF00208283. [DOI] [PubMed] [Google Scholar]
- 7.Kaya N, et al. GM2 gangliosidosis in Saudi Arabia: multiple mutations and considerations for future carrier screening. Am. J. Med. Genet. A. 2011;155(6):1281–1284. doi: 10.1002/ajmg.a.33932. [DOI] [PubMed] [Google Scholar]
- 8.Drousiotou A, et al. Sandhoff disease in Cyprus: population screening by biochemical and DNA analysis indicates a high frequency of carriers in the Maronite community. Hum. Genet. 2000;107(1):12–17. doi: 10.1007/s004390000324. [DOI] [PubMed] [Google Scholar]
- 9.Andermann E, et al. Genetic variants of Tay-Sachs disease: Tay-Sachs disease and Sandhoff's disease in French Canadians, juvenile Tay-Sachs disease in Lebanese Canadians, and a Tay-Sachs screening program in the French-Canadian population. Prog. Clin. Biol. Res. 1977;18:161–188. [PubMed] [Google Scholar]
- 10.Lowden JA, et al. Carrier detection in Sandhoff disease. Am. J. Hum. Genet. 1978;30(1):38–45. [PMC free article] [PubMed] [Google Scholar]
- 11.Fitterer BB, et al. A polymerase chain reaction-based genotyping assay for detecting a novel Sandhoff disease-causing mutation. Genet. Test. Mol. Biomarkers. 2012;16(5):401–405. doi: 10.1089/gtmb.2011.0215. [DOI] [PubMed] [Google Scholar]
- 12.Sokoro AA, et al. Diagnosis and high incidence of hyperornithinemia-hyperammonemia-homocitrullinemia (HHH) syndrome in northern Saskatchewan. J. Inherit. Metab. Dis. 2010;33(Suppl. 3):S275–S281. doi: 10.1007/s10545-010-9148-9. [DOI] [PubMed] [Google Scholar]
- 13.Gelb MH, et al. Direct multiplex assay of enzymes in dried blood spots by tandem mass spectrometry for the newborn screening of lysosomal storage disorders. J. Inherit. Metab. Dis. 2006;29(2–3):397–404. doi: 10.1007/s10545-006-0265-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Brien JS, et al. Tay-Sachs disease. N. Engl. J. Med. 1970;283(1):15–20. doi: 10.1056/NEJM197007022830104. [DOI] [PubMed] [Google Scholar]
- 15.Okada S, O'Brien JS. Tay-Sachs disease: generalized absence of a beta-D-N-acetylhexosaminidase component. Science. 1969;165(3894):698–700. doi: 10.1126/science.165.3894.698. [DOI] [PubMed] [Google Scholar]
- 16.Human Genome Variation Society. [accessed June 15, 2011]; www.hgvs.org/mutnomenrecs-DNA.html#del. [Google Scholar]
- 17.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi Y, et al. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7(10):e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerjee P, et al. Molecular basis of an adult form of beta-hexosaminidase B deficiency with motor neuron disease. Biochem. Biophys. Res. Commun. 1991;181(1):108–115. doi: 10.1016/s0006-291x(05)81388-9. [DOI] [PubMed] [Google Scholar]
- 21.Kuroki Y, et al. A novel missense mutation (C522Y) is present in the beta-hexosaminidase beta-subunit gene of a Japanese patient with infantile Sandhoff disease. Biochem. Biophys. Res. Commun. 1995;212(2):564–571. doi: 10.1006/bbrc.1995.2007. [DOI] [PubMed] [Google Scholar]
- 22.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17(4):419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 23.Iida Y. Categorical discriminant analysis of 3′-splice site signals of mRNA precursors in higher eukaryote genes. J. Theor. Biol. 1988;135(1):109–118. doi: 10.1016/s0022-5193(88)80177-2. [DOI] [PubMed] [Google Scholar]
- 24.Zhang ZX, et al. A second, large deletion in the HEXB gene in a patient with infantile Sandhoff disease. Hum. Mol. Genet. 1995;4(4):777–780. doi: 10.1093/hmg/4.4.777. [DOI] [PubMed] [Google Scholar]
- 25.Wakamatsu N, et al. A novel exon mutation in the human beta-hexosaminidase beta subunit gene affects 3′ splice site selection. J. Biol. Chem. 1992;267(4):2406–2413. [PubMed] [Google Scholar]
- 26.Banerjee P, et al. Preferential beta-hexosaminidase (Hex) A (alpha beta) formation in the absence of beta-Hex B (beta beta) due to heterozygous point mutations present in beta-Hex beta-chain alleles of a motor neuron disease patient. J. Biol. Chem. 1994;269(7):4819–4826. [PubMed] [Google Scholar]
- 27.Redonnet-Vernhet I, et al. Significance of two point mutations present in each HEXB allele of patients with adult GM2 gangliosidosis (Sandhoff disease) homozygosity for the Ile207->Val substitution is not associated with a clinical or biochemical phenotype. Biochim. Biophys. Acta. 1996;1317(2):127–133. doi: 10.1016/s0925-4439(96)00044-0. [DOI] [PubMed] [Google Scholar]
- 28.Gomez-Lira M, et al. A common beta hexosaminidase gene mutation in adult Sandhoff disease patients. Hum. Genet. 1995;96(4):417–422. doi: 10.1007/BF00191799. [DOI] [PubMed] [Google Scholar]
- 29.Maier T, et al. The X-ray crystal structure of human beta-hexosaminidase B provides new insights into Sandhoff disease. J. Mol. Biol. 2003;328(3):669–681. doi: 10.1016/s0022-2836(03)00311-5. [DOI] [PubMed] [Google Scholar]
- 30.Mark BL, et al. Crystal structure of human beta-hexosaminidase B: understanding the molecular basis of Sandhoff and Tay-Sachs disease. J. Mol. Biol. 2003;327(5):1093–1109. doi: 10.1016/s0022-2836(03)00216-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaignard P, et al. Characterization of seven novel mutations on the HEXB gene in French Sandhoff patients. Gene. 2013;512(2):521–526. doi: 10.1016/j.gene.2012.09.124. [DOI] [PubMed] [Google Scholar]
- 32.Warner TG, et al. Prenatal diagnosis of infantile GM 2 gangliosidosis type II (Sandhoff disease) by detection of N-acetylglucosaminyl-oligosaccharides in amniotic fluid with high-performance liquid chromatography. Prenat. Diagn. 1986;6(6):393–400. doi: 10.1002/pd.1970060602. [DOI] [PubMed] [Google Scholar]