Abstract
Plant growth depends on stem cell niches in meristems. In the root apical meristem, the quiescent center (QC) cells form a niche together with the surrounding stem cells. Stem cells produce daughter cells that are displaced into a transit-amplifying (TA) domain of the root meristem. TA cells divide several times to provide cells for growth. SHORTROOT (SHR) and SCARECROW (SCR) are key regulators of the stem cell niche. Cytokinin controls TA cell activities in a dose-dependent manner. Although the regulatory programs in each compartment of the root meristem have been identified, it is still unclear how they coordinate one another. Here, we investigate how PHABULOSA (PHB), under the posttranscriptional control of SHR and SCR, regulates TA cell activities. The root meristem and growth defects in shr or scr mutants were significantly recovered in the shr phb or scr phb double mutant, respectively. This rescue in root growth occurs in the absence of a QC. Conversely, when the modified PHB, which is highly resistant to microRNA, was expressed throughout the stele of the wild-type root meristem, root growth became very similar to that observed in the shr; however, the identity of the QC was unaffected. Interestingly, a moderate increase in PHB resulted in a root meristem phenotype similar to that observed following the application of high levels of cytokinin. Our protoplast assay and transgenic approach using ARR10 suggest that the depletion of TA cells by high PHB in the stele occurs via the repression of B-ARR activities. This regulatory mechanism seems to help to maintain the cytokinin homeostasis in the meristem. Taken together, our study suggests that PHB can dynamically regulate TA cell activities in a QC-independent manner, and that the SHR-PHB pathway enables a robust root growth system by coordinating the stem cell niche and TA domain.
Author Summary
Plant roots are programmed to grow continuously into the soil, searching for nutrients and water. The iterative process of cell division, elongation, and differentiation contributes to root growth. The quiescent center (QC) is known to maintain the root meristem, and thus ensure root growth. In this study, we report a novel aspect of root growth regulation controlled independently of the QC by PHABULOSA (PHB). In shr mutant plants, PHB, which in the meristem is actively restricted to the central region of the stele by SHORTROOT (SHR) via miR165/6, suppresses root meristem activity leading to root growth arrest. A high concentration of PHB in the stele does this by modulating B-ARR activity through a QC-independent pathway. Accordingly, we observed a significant recovery of root meristem activity and growth in the shr phb double mutant, while the QC remained absent. However, the presence of QC may be required to sustain continuous root growth. On the basis of our results, we propose that SHR maintains root growth via two separate pathways: by modulating PHB levels in the root stele, and by maintaining the QC identity.
Introduction
Plants, unlike animals, grow continuously and dynamically adjust their architecture. Meristems at the apices of shoots and roots harbor stem cells and serve as the center of cell division and growth. In the root apical meristem (RAM), the quiescent center (QC) maintains a stem cell population to form a stem cell niche [1], [2]. There are two main pools of stem cells, the proximal and distal stem cells, which are named based on their position relative to the QC (reviewed by [3]). Distal stem cells produce the root cap. Post-embryonic root growth in an apical direction, hereafter referred to as root growth, occurs via iterative divisions of the transit-amplifying cells (TA cells), derived from the proximal stem cells, and their subsequent cell elongation.
Two main regulatory programs are crucial for the root stem cell niche: one directed by PLETHORAs (PLTs) and the other by SHR and SCR. The PLTs play an essential role in establishing the QC and the RAM during embryogenesis [4]. SHR and SCR, the GRAS family transcription factors, act together to maintain the stem cell niche and root growth [5], [6], [7]. When SHR or SCR is knocked out, QC cells are not maintained and root growth terminates prematurely. The essential role of SHR and SCR in QC specification was further evident in a regeneration study of QC cells. In contrast to wild-type plants, QC cells failed to recover in shr and scr mutants after laser ablation [8].
The role of the QC within the stem cell niche has been extensively studied in Arabidopsis. Laser ablation of QC cells first indicated that the QC maintains the adjacent stem cell population [2]. In this study, the role of the QC in distal stem cell maintenance is evident; however, the extent to which the QC contributes to the maintenance of the proximal stem cells and the TA cell population is unclear [3]. In wox5 mutants, which affects the identity and morphology of the QC cells, the size of the TA cell population is unaltered and root growth is relatively normal even though the distal stem cells are not maintained [9]. Furthermore, root regeneration experiments have suggested that the QC is not required for the regeneration competency of the meristem cells [10].
In addition to the stem cell niche, cell division activities of the TA cell population significantly affect root growth. Cytokinin signaling plays a key role in this process. In the absence of cytokinin, TA cell proliferation is strongly inhibited. As a result, severe defects in root growth are observed in the triple mutant of ARR1, 10, and 12, the type B Arabidopsis response regulator (B-ARR) genes, and the triple mutant of AHK2, 3 and 4, cytokinin receptor genes [11], [12]. Under high cytokinin levels, the transition of TA cell status from dividing to differentiating is promoted, reducing the size of the meristem and slowing down root growth [13]. This process is mediated by ARR1 and 12, which up-regulate the expression of SHY2, an inhibitor of auxin signaling [14]. A recent study indicated that SCR in the QC regulates this process in a non-cell autonomous manner [15].
The HOMEODOMAIN-LEUCINE ZIPPER Class III (HD-ZIP III) genes are key regulators of shoot and root meristems (reviewed by [16], [17]). In Arabidopsis, the HD-ZIP III family consists of five members: PHABULOSA (PHB), PHAVOLUTA (PHV), CORONA (CNA), REVOLUTA (REV), and ARABIDOPSIS THALIANA HOMEOBOX8 (ATHB8) [18]. These genes are post-transcriptionally regulated by microRNA (miRNA) 165/6 [19], [20]. Studies have shown that during root meristem establishment, HD-ZIP III proteins are excluded from the basal embryo pole [21], [22]. SERRATE (SE), a zinc-finger (ZnF) domain-containing protein, is a component of dicing bodies that process pre-miRNAs together with DICER-LIKE1 (DCL1) [23]. In se mutants, the production of mature miRNAs is reduced, thus PHB and PHV expand to the basal embryo pole, and formation of the root meristem fails [21]. SHR and SCR also suppress HD-ZIP III in the root by directly regulating the transcription of miR165A and 166B [24]. However, unlike SE, SHR and SCR mainly affect post-embryonic root development.
In summary, significant progress has been made to identify key transcription factors and signaling molecules in the function and maintenance of the root stem cell niche and TA cells. However, it remains unclear how these factors and molecules are integrated to control root growth. Here, we show that PHB, the concentration of which is controlled by SHR, governs TA cell activities by repressing B-ARRs. Such a repressive effect of PHB on B-ARR activities is enhanced by high cytokinin. Therefore, restricting the PHB level via the SHR-microRNA pathway is a critical step for maintaining root growth.
Results
Elevated PHB levels suppress root proximal meristem activity
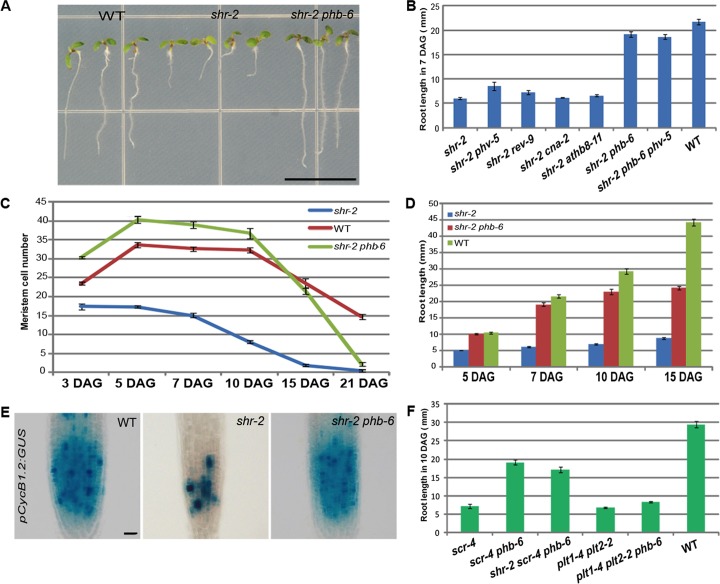
Previously, we reported that SHR-dependent expression of miR165/6 in the ground tissue layer quantitatively and spatially restricts HD-ZIP III expression within the stele, which is necessary for normal xylem tissue patterning [24]. We also observed that the short root phenotype of shr mutants could be partially recovered when miR165/6 was miss-expressed in the ground tissue or stele, suggesting that ectopic HD-ZIP III expression reduced root growth in this mutant. However, at the time, it was not known which of the HD-ZIP III genes was responsible for root growth. We tested this by analyzing double mutants of shr and each of the five HD-ZIP III genes. In the double mutant of shr and phb-6 (hereafter phb), a loss-of-function mutant allele of PHB with a Ds insertion, the roots were significantly longer than the shr mutant roots and more or less comparable to the wild type 7 days after germination (DAG) (Fig. 1A; S1A Fig.). Under the growth conditions used in this experiment, phb single mutant roots were slightly shorter than the wild type (S1A Fig.), which excluded the possibility that the rescue of root length in shr phb mutants is an additive effect. The negative influence of PHB on root length in the shr mutant was further supported in our suppressor screen of EMS-mutagenized shr plants. In this screen, we identified two new loss-of-function phb alleles, which are phb-15 and phb-16 (S1A Fig.). Mutations in CORONA (CNA) or ATHB8, two other HD-ZIP III genes, however, failed to rescue root length in the shr mutant, whereas PHAVOLUTA (PHV) and REVOLUTA (REV) elicited only a weak recovery (Fig. 1B). Introducing a knockout mutant of PHV, the HD-ZIP III family member most closely related to PHB [19], to the shr phb did not yield a further increase in shr phb root length (Fig. 1B).
Fig 1. PHB regulates root meristem activity downstream of SHR-SCR.
(A) Seedlings of shr phb (7 DAG) mutants display significant recovery in root length. (B) Growth restoration in shr phb roots compared with other shr HD-ZIP III mutants and shr phb phv-5 mutants. (C) Meristem size over time in the shr, shr phb, and wild-type roots. (D) Root length measured over a time course. The shr phb roots grow in a more determinate manner than do the wild-type roots. (E) Expression of pCycB1.2:GUS in the shr, shr phb, and wild-type roots (5 DAG). (F) phb-mediated recovery in root meristem/growth is specific to the SHR-SCR pathway. The error bars represent the standard error (n = 20–40 plants). Scale bars: A, 1 cm; E, 20 μm.
To understand how root length is rescued in shr phb mutants, we analyzed root growth and meristem activities in comparison with the wild type and shr mutant (Fig. 1C-1E). The shr mutant roots barely grew beyond 5 DAG, whereas shr phb roots displayed growth similar to, or faster than, wild-type roots until 7 DAG. After 7 DAG, however, the growth of these plants decelerated and had almost ceased by 15 DAG (Fig. 1D). Consistent with the recovery of root growth, the meristem in the shr phb roots was significantly larger than the shr meristem (S1B Fig.). A time-course analysis of root meristem [25] showed that both wild-type and shr phb roots displayed an increase in meristem size between 3 and 5 DAG, whereas shr roots showed a steady decrease (Fig. 1C). At 5 DAG, the meristem of shr phb roots was larger than that of the wild type (Student’s t-test; P < 0.001, α = 0.05). We also examined meristem cell division using a G2-M cell cycle marker, pCycB1.2:GUS [26]. GUS expression at 5 DAG indicated full recovery of cell division in the proximal meristem of the shr phb roots (Fig. 1E). Interestingly, pCycB1.2:GUS expression was comparable in the wild-type and shr phb roots at 15 DAG, despite the deceleration of root growth in the shr phb plants (S1C and 1D Fig.). These data suggest that the slowing of growth is not caused by a complete loss of cell cycle activity but rather it may be due to some other defects that are not directly related to meristematic function. For example, the shr phb mutant has defective development of the phloem sieve elements, which could affect the transport of nutrients required for root growth (S2A Fig.).
SCR regulates miR165/6 expression together with SHR [24]. Thus, we examined whether PHB also affects root growth activity controlled by SCR, and indeed, scr-4 phb roots displayed a significant recovery in the size of the proximal meristem (S1B Fig.) and root length compared with scr-4 roots (Fig. 1F). Consistent with these findings, shr scr-4 phb triple mutant plants also recovered root length (Fig. 1F). Finally, we investigated whether phb could suppress the short root phenotype of plt1 plt2 mutants. However, plt1-4 plt2-2 phb triple mutants did not exhibit any noticeable recovery in root length (Fig. 1F). Taken together, these data suggest that PHB is a key downstream factor of SHR and SCR in the regulation of the meristem and root growth.
TA cell proliferation and root growth in the absence of QC
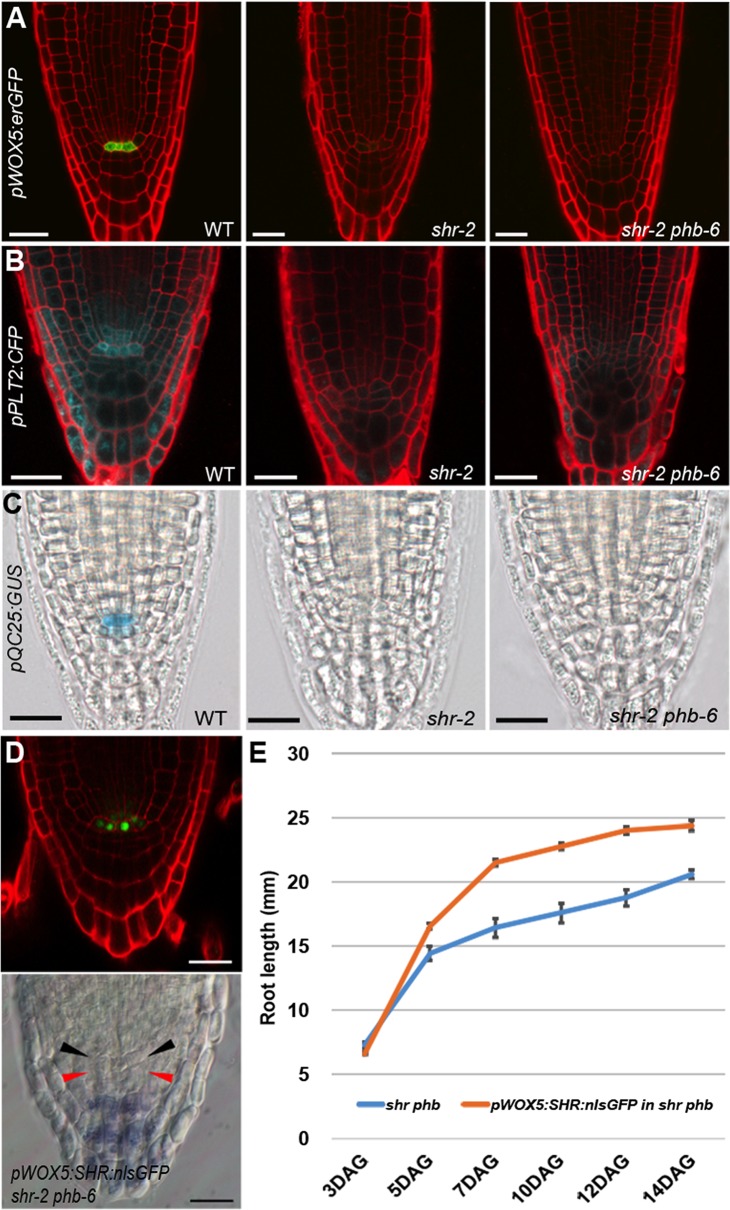
The short root phenotype of the shr and scr mutants has been ascribed to the failure of stem cell maintenance in the absence of a QC. The lack of stem cell maintenance in shr mutants is supported by a steady decrease in root meristem size upon germination (Fig. 1C). Because shr phb mutants displayed both an active increase in root meristem size and a rescue in root length, we examined whether the QC is rescued in the shr phb roots. To our surprise, none of the QC markers analyzed in actively growing shr phb roots indicated QC recovery (Fig. 2; S2B Fig.). The expression of pWOX5:erGFP (endoplasmic reticulum-localized GFP) and pQC25:GUS, which is strong and specific to the QC cells in wild-type roots, was below the level of detection in the QC regions of both shr phb and shr roots (Fig. 2A and 2C) [9], [27]. In the wild-type roots, expression of pPLT2:CFP, pPLT1:CFP, pSCR:erGFP, pAGL42:erGFP, and pQC6:erGFP [6], [28], [29], [30], are at their highest in the QC and are lower in the neighboring cell types. Analysis of these markers also indicated the lack of a QC in the shr phb roots (Fig. 2B; S2B Fig.). We also analyzed pWOX5:erGFP in the wild-type, shr and shr phb embryos to determine whether the different WOX5 expression patterns in the shr and shr phb embryos could explain the recovery of root growth. Up to the mid-torpedo stage, we detected GFP expression in both wild-type and shr embryos (S3A Fig.). Thereafter, contrary to that in the wild-type embryos, pWOX5:erGFP expression declined dramatically in the shr mutants and was no longer detectable (S3B Fig.). The dynamics of pWOX5:erGFP expression in the shr phb mutant were indistinguishable from those in the shr mutant.
Fig 2. Recovery of root meristem/growth activity in the absence of a functional QC.
(A–C) Expression analysis of QC markers in the wild-type, shr, and shr phb roots (5 DAG). (A) pWOX5:erGFP. (B) pPLT2:CFP. (C) pQC25:GUS. (D) Analysis of QC status in a shr phb root expressing pWOX5:SHR:nlsGFP, by confocal microscopy (upper panel) and Lugol staining (lower panel). Black arrowhead indicates the QC where SHR-nlsGFP is observed in the upper panel. Red arrowhead indicates the recovered columella stem cell layer where starch granules are absent. (E) The comparison of root length between shr-2 phb-6 and pWOX5:SHR:GFP;shr-2 phb-6. Root lengths were measured from seedlings 3–14 DAG. Scale bars: A–C, 25 μm; D, 20 μm. The error bars represent the standard error (n = 5–10 plants).
Without the QC, the distal stem cells cannot be maintained. Lugol staining indicated that distal stem cells differentiate prematurely in shr phb roots, similar to that in shr roots visualized by the presence of starch granules that marks differentiated columella cells (S3C Fig.). This is consistent with the lack of QC marker expression in the shr phb mutant, which supports the observation that the QC is absent in these plants.
Recently, it was suggested that the QC promotes root growth by regulating the transition between cell proliferation and differentiation in a non-cell autonomous manner [15]. Therefore, we asked whether the presence of the QC in the shr phb mutant could fully restore root growth to wild-type levels. To investigate this, SHR fused to nuclear-localized GFP was expressed in shr phb roots under the WOX5 promoter (pWOX5:SHR:nlsGFP; shr phb). Unlike the shr phb roots in which WOX5 expression diminished in the late stage of embryogenesis, three independent shr phb transgenic lines maintained SHR-nlsGFP expression in the QC region of post-embryonic roots, suggesting at least a partial recovery of QC driven by WOX5 (Fig. 2D). Furthermore, the meristem of the shr phb roots expressing WOX5 appeared to be more organized than that of the shr phb plants, and the columella stem cells were restored as indicated by Lugol staining (Fig. 2D). Expression of WOX5 in the QC position increased the growth potential of the shr phb roots; however, their lengths were significantly shorter than those of the wild-type roots (Fig. 2E). Together these data suggest that proliferation of TA cells is controlled by the level of PHB, and that the PHB level plays a significant role in root growth, in combination with the QC maintenance and other unknown factors that affect root growth.
PHB regulates TA cell proliferation and root growth from the stele
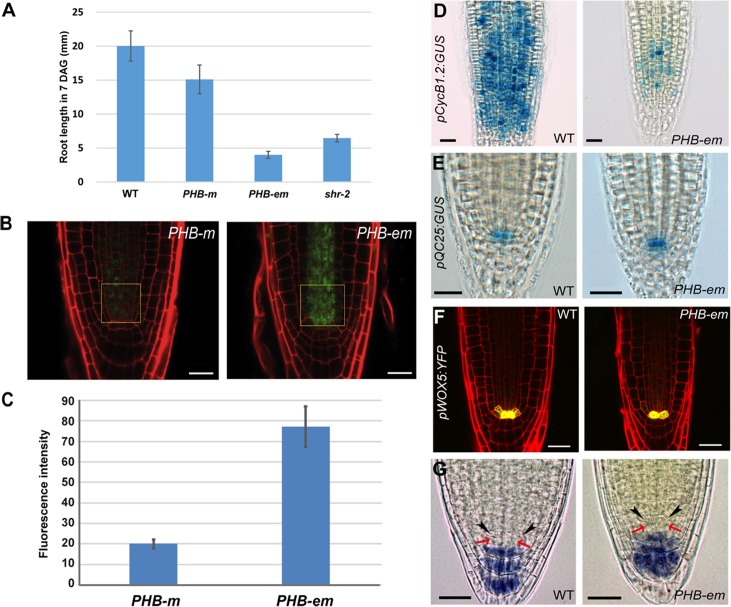
SHR posttranscriptionally restricts HD-ZIP III mRNA to the center of the stele [24]. In the root meristem of shr mutants, PHB is observed throughout the stele (S4B Fig.), which results in the short root phenotype that can be rescued by the misexpression of miR165/6 in the ground tissue. Hence, high PHB levels in the stele of the root meristem probably influence root growth. To confirm this, we increased the dosage of PHB in a specific manner throughout the stele in the wild-type root meristem. We modified PHB so that it was not efficiently targeted for degradation by miR165/6, and then expressed it under the WOODEN LEG (WOL) promoter, which drives gene expression in stele cells in the root meristem [31] (S5 Fig.). Two types of modified PHB were used: one with a single silent mutation that partially interferes with miR165/6 binding (PHB-m) [24], and the other with four silent mutations that strongly block miR165/6 binding (PHB-em) (S4A Fig.). Driving PHB-m under the PHB promoter results in the broadening of PHB domain throughout the stele as we previously reported (S4B Fig.). Expression analysis using confocal microscopy showed that PHB-GFP is present at a significantly higher level in the stele cells of roots expressing pWOL:PHB-em:GFP NLS than in those expressing pWOL:PHB-m:GFP NLS (Fig. 3B and 3C). Consistent with high PHB-GFP levels inhibiting normal root growth, roots expressing pWOL:PHB-em:GFP NLS were much shorter than those expressing pWOL:PHB-m:GFP NLS, and were either similar to or shorter than the shr mutant roots (Fig. 3A). Accordingly, the meristem size of the pWOL:PHB-em:GFP NLS roots was also dramatically reduced (S4C Fig., upper panel). Furthermore, these root meristems had significantly fewer stele cells compared to wild-type root meristems (S4C Fig., lower panel). TA cell proliferation, as detected by pCycB1.2:GUS expression, decreased dramatically in these roots compared to the wild-type ones (Fig. 3D). Collectively, these data suggest that high levels of PHB in the stele cells of the root meristem suppress TA cell proliferation and root growth, and therefore signaling from the stele is a rate-limiting factor in root growth.
Fig 3. PHB in the stele regulates root meristem and growth activity in a QC-independent manner.
(A) A comparison of root lengths in wild-type, pWOL:PHB-m:GFPNLS, pWOL:PHB-em:GFPNLS and shr-2 plants (7 DAG). The error bars represent the standard error (n = 9–14 plants). (B, C) Quantitative comparison of PHB-GFP levels in the root stele cells expressing pWOL:PHB-m:GFP NLS and pWOL:PHB-em:GFP NLS. Fluorescence intensity was measured for GFP in the boxed area of the panel (B). Error bars represent standard error (n = 11) (D) pCycB1.2:GUS expression shows drastic reduction in cell division potential in the proximal meristem. Expression of pQC25:GUS (E) and pWOX5:YFP (F) in the PHB-em roots. (G) Starch granule accumulation as visualized by Lugol’s staining in the pWOL:PHB-em:GFP NLS roots. The black arrowheads and red arrows indicate the QC and columella stem cells, respectively. Scale bars: B, 20 μm; D-G, 25 μm.
Next, we examined the QC status in the pWOL:PHB-em:GFP NLS lines that displayed root meristem and growth phenotypes similar to that of the shr mutant. The expression patterns of pQC25:GUS and pWOX5:YFP in pWOL:PHB-em:GFP NLS lines were similar to that in the wild type (Fig. 3E and 3F); their expression persisted even in older roots (15 DAG; S4D and S4E Fig.). Lugol staining of roots at 5 and 7 DAG also supported the presence of a QC (Fig. 3G). Nevertheless, in the pWOL:PHB-em:GFP NLS roots, the cells in the QC position were often enlarged and showed aberrant cell divisions (S4F Fig.). This phenotype was more frequent in older roots (7 DAG or older). Furthermore, in situ analysis indicated that there is a decline in WOX5 mRNA levels in these transgenic roots (S4G Fig.). These data further confirm that PHB regulation of the TA cell proliferation and root growth is independent of the QC status.
A genome-wide survey of downstream pathways of PHB in the root stele
To determine how high levels of PHB in the stele suppress root growth in shr mutant plants, we generated stele-specific genome-wide gene expression data from wild-type, shr-2, and shr-2 phb-6 roots. In this experiment, we used a protoplast/cell-sorting technique to isolate and collect the stele cells from roots expressing pWOL:erGFP in three genotypes, and mRNAs in the stele were profiled using the Arabidopsis Tiling 1.0R arrays from Affymetrix [32] (S5 and S6A Fig.; S1 Table). To identify genes or pathways that are regulated by PHB, we identified genes that are differentially expressed between the shr mutant and the wild type (Corrected p-value, FDR <0.05; fold difference >2). Under the given criteria, 1933 genes were revealed as differentially expressed between the two (S2 Table). We then investigated the expression patterns of these genes in the shr, shr phb, and wild-type plants using principal component analysis (PCA) and QT clustering [33] (S6B and S6C Fig.; S2 Table). These analyses indicated that most changes in gene expression in the shr mutant (92% represented by 1781 genes in clusters 1–3) were due to high levels of PHB. In total, 1170 genes were activated (cluster 1) and 611 genes were repressed (clusters 2 and 3) by PHB in shr mutant plants (S6C Fig.).
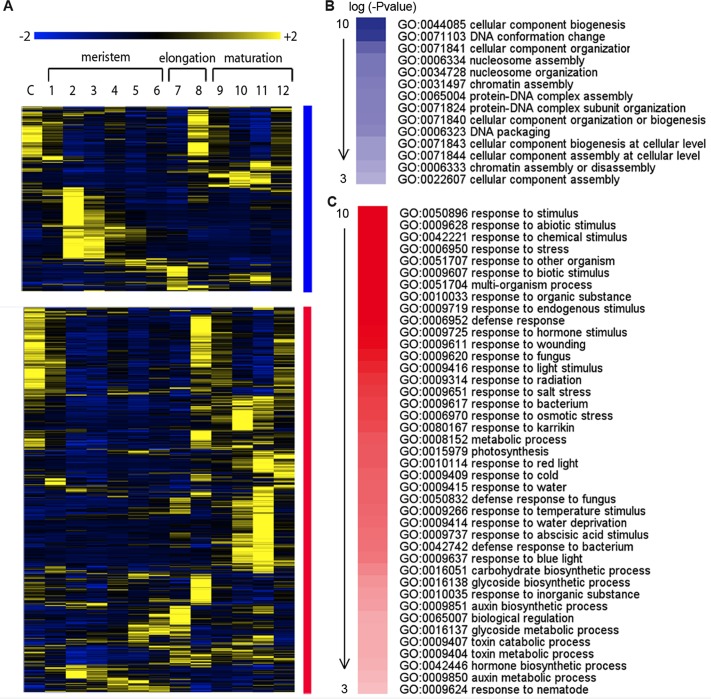
Next, we analyzed the spatial expression of genes that are activated or repressed by high levels of PHB in the shr mutant using the expression data generated following micro-dissection of a root along the longitudinal axis and the cell type-specific root expression data (Fig. 4A; S7 Fig.) [24], [29], [34], [35], [36]. Hierarchical clustering of genes present in the root micro-dissection data indicates that ∼40% of the genes repressed by PHB are highly expressed in the TA domain, whereas those activated by PHB are enriched in the maturation zone. GO term enrichment analysis suggested that the pathways repressed by high levels of PHB are related to cell proliferation processes [37] (S3 Table; Fig. 4B). This suggests that in the stele, PHB actively represses genes involved in TA cell activities. In contrast, pathways activated by PHB are biotic and abiotic stress responses, and those involved in hormone production and signaling. This includes genes in auxin, ABA, and cytokinin biosynthesis and response (S3 Table; Fig. 4B). Cell-type specific expression of nearly 40% of genes repressed by PHB is enriched in the xylem precursor cells (S7 Fig.) [38]. This is consistent with the recovery of xylem patterning in shr phb mutant plants [24]. In contrast to the genes repressed by PHB, those activated by PHB showed enriched expression in multiple cell types. The overrepresentation of genes involved in multiple signaling processes might explain the lack of enriched patterns of gene activation in response to PHB in particular cell types.
Fig 4. Downstream genes of SHR-PHB in the root stele.
(A) Hierarchical clustering of expression in the Arabidopsis root along the longitudinal axis for genes that are repressed (blue; genes in cluster 2 and 3 in S6C Fig.) or activated (red; genes in cluster 1 in S6C Fig.) by a high level of PHB in shr mutants. Expression values are normalized by row. (B) Over-represented biological functions of genes that are repressed (P-value marked in blue) or (C) activated (P-value marked in red) by a high level of PHB in shr mutants.
A balance between auxin and cytokinin signaling is important for root growth and meristem activity [39]. Our stele-specific mRNA analysis suggests that the SHR-PHB pathway might affect the balance of these two hormones. We further examined this by analyzing the expression pattern of pDR5:YFP venus and pPIN1:PIN1:GFP, in shr, shr phb, and wild-type roots (S8A–S8C Fig.). As reported recently [40], pPIN1:PIN1:GFP expression declined in both 5- and 10-day-old shr roots. Its expression was moderately restored in the roots of shr phb plants, which is consistent with our data from stele profiling. A similar expression pattern was observed for pDR5:YFP venus. These results suggest that high PHB levels in shr mutant plants reduce auxin signaling (transport/biosynthesis) in the root meristem, but its effect appears to be relatively minor.
PHB locally regulates genes involved in cytokinin biosynthesis in the root meristem
Previous studies have suggested the existence of a homeostatic feedback loop mechanism between auxin and cytokinin in the root, in which cytokinin positively regulates a subset of genes involved in auxin biosynthesis [41], [42]. Our tissue-specific stele expression profiling revealed that high PHB levels in shr mutants up-regulate genes involved in auxin biosynthesis/metabolism. In addition, the level of PIN1 protein, which is degraded by high levels of cytokinin, was reduced in the shr mutant and was then moderately restored in the shr phb mutant (S8B and S8C Fig.) [43]. Moreover, cytokinin levels are increased in shr roots [44]. These results led us to examine whether high PHB in shr mutants affects the cytokinin status in the roots.
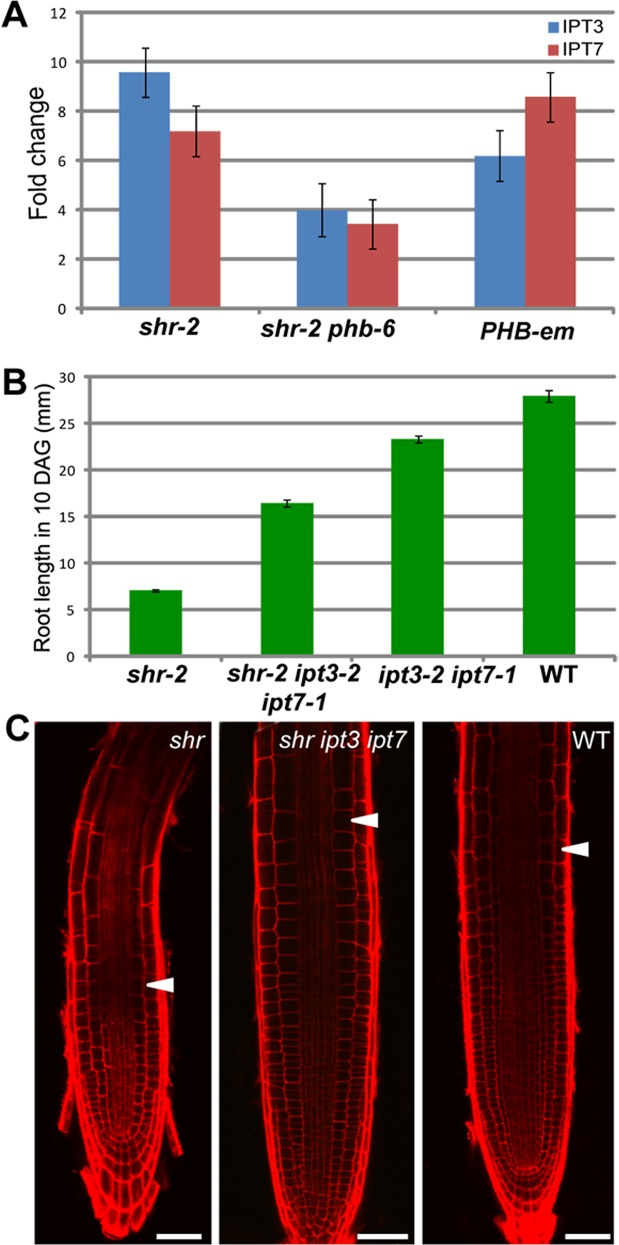
ISOPENTENYL TRANSFERASE (IPT) 1, 3, 5, and 7 promote cytokinin biosynthesis in Arabidopsis roots [45]. Our stele profiling data indicated that IPT5 and 7 are upregulated by PHB in the shr stele. Previous expression data in the shr and wild-type root tips revealed the up-regulation of IPT3 and 7 in shr mutants [35]. We corroborated these findings by quantifying the expression levels of IPT3, 5, and 7 in the wild-type, shr, and shr phb roots using qRT-PCR. Consistently, mRNA levels of IPT3 and 7 were found to increase in shr roots compared with wild-type roots, and their levels were partially restored in shr phb roots (Fig. 5A). These results suggest that high levels of PHB enhance the expression of IPT3 and 7. We further confirmed this by measuring the mRNA levels of IPT3 and 7 in the root tips of transgenic plants expressing pWOL:PHB-em:GFP NLS (Fig. 5A).
Fig 5. The SHR-PHB pathway controls root growth in a cytokinin-dependent manner (A) Relative mRNA levels of IPT3 and IPT7 in shr, shr phb, and pWOL:PHB-em:GFP NLS (PHB-em) roots.
Data are normalized to Expression levels in wild-type roots. (B) Root lengths (10 DAG) and (C) meristem images (5 DAG) in shr, shr ipt3-2 ipt7-1 mutants. The error bars represent the standard error (n = 20 plants). Scale bar: 50 μm. The white arrowhead marks the end of the meristem.
We then asked whether PHB regulation of IPT3 and 7 influences root meristem/growth activity by measuring the size of the root meristem and the length of the root in shr ipt3-2 ipt7-1 triple mutant plants. The shr ipt3-2 ipt7-1 plants showed a recovery in the root length and meristem size compared to shr plants, but to a much lesser degree than the shr phb roots (Fig. 5B and 5C; P < 0.001, α = 0.05). Under our growth conditions, the ipt3-2 ipt7-1 double mutant roots were slightly shorter than those of wild-type plants. Collectively, these data indicated that PHB influences root meristem and growth activity in the shr plants, at least partly via the regulation of cytokinin biosynthesis.
PHB in the shr mutant interferes with B-ARR activities
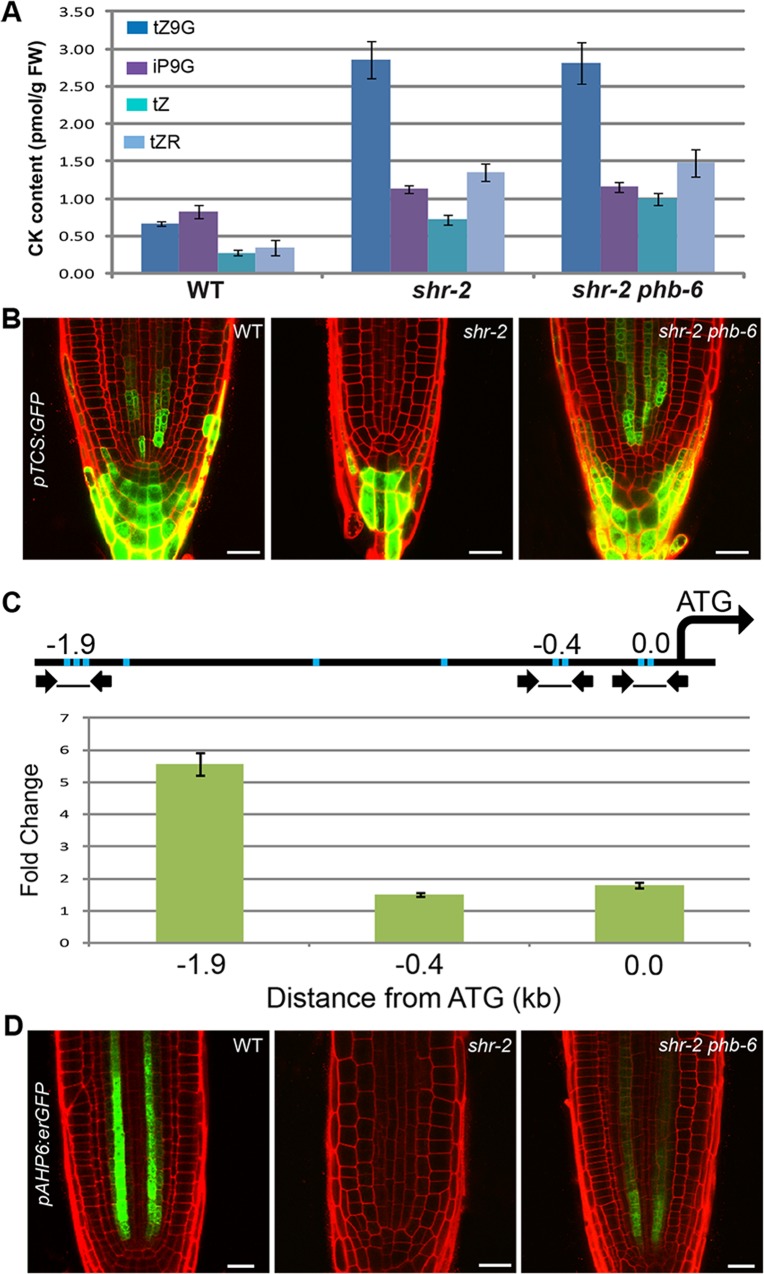
To determine the significance of cytokinin biosynthesis regulation by PHB in the root meristem, we measured cytokinin levels in wild-type, shr, and shr phb roots. Consistent with the previous study, an increase in cytokinin level was observed in the shr roots (Fig. 6A; S9A Fig.). Various forms of trans-zeatin increased by approximately four-fold in shr plants in comparison to the wild type. However, we failed to detect a decline in global cytokinin levels in shr phb roots, as compared with those in shr roots. This suggests that there is another layer of regulation mediated by PHB in the cytokinin pathway, which is probably involved in restoring the shr phb root growth.
Fig 6. The SHR-PHB pathway and cytokinin signaling.
(A) Cytokinin (CK) expression in wild-type, shr, and shr phb roots. tZ, Transzeatin; tZR, tZ Riboside; iP9G, N6-(Δ2-isopentenyl) adenine-9-glucoside; tZ9G, tZ-9-glucoside. (B) Expression of pTCS:GFP in wild-type, shr, and shr phb roots. (C) Real-time PCR and ChIP showing PHB binding to the ARR7 promoter. Line diagram represents the ARR7 promoter region. Arrows and blue bars indicate primers and B-ARR binding elements (GGATT/AATCT), respectively. (D) Recovery in pAHP6:erGFP expression in the shr phb roots in comparison with shr roots. The error bars represent the standard deviation (A) and standard error (C) (n = 3 biological replicates). Scale bars: 25 μm. CK, cytokinin.
In our stele profiling data, the expression of multiple A-ARRs, such as ARR5, 6, 7, 8, 9, and 15, was slightly induced in shr mutants but was further upregulated in shr phb mutants (S9B Fig.). We validated this finding by qRT-PCR analysis (S9C Fig.). Considering that global cytokinin levels were comparable in both shr and shr phb roots, this result implied that high PHB levels in the shr mutant might be actively suppressing the expression of A-ARRs.
All A-ARR genes have cis-regulatory elements that are directly bound by B-ARRs [46]. pTCS:erGFP is a synthetic cytokinin reporter construct with B-ARR binding elements, and its expression therefore reflects the transcriptional activity of B-ARRs. When the in vivo cytokinin level increases in the wild-type root meristem, more B-ARRs are activated and thereby enhance pTCS:erGFP expression [47]. Thus, we investigated the involvement of PHB in B-ARR function by analyzing the expression of pTCS:erGFP in the wild-type, shr, and shr phb roots [47]. Despite a high cytokinin level, shr roots displayed a dramatic reduction in pTCS:erGFP expression (Fig. 6B). In contrast, TCS expression was restored in shr phb mutants to levels higher than those in wild-type plants. Thus, high levels of PHB appear to interfere with B-ARRs that induce TCS transcription in the root stele. Consistent with this theory, TCS expression was found to be absent or very weak in the root stele of phb-7d mutants carrying a semi-dominant point mutation in the microRNA target site of PHB [24] (S9E Fig.).
A high level of PHB could interfere with B-ARR activity directly or indirectly. To determine how PHB affects B-ARR activity, we developed a transgenic line expressing PHB-em fused to the glucocorticoid receptor (GR) under the control of the PHB promoter, and crossed a line expressing this construct with pTCS:erGFP (S10A and S10B Fig.). The resulting F1 seedlings were treated with dexamethasone for 3 hours and pTCS:erGFP expression in roots was analyzed in comparison with that of the untreated control plants. GFP expression in roots harboring both pTCS:erGFP and pPHB:PHB-em:GR was much weaker than that of roots carrying pTCS:erGFP alone, suggesting that the PHB-em:GR is leaky. Nevertheless, with dexamethasone treatment, we found a significant reduction in GFP expression in roots with both pTCS:erGFP and pPHB:PHB-em:GR, not in roots with pTCS:erGFP alone. Since the response of pTCS:erGFP occurs only after 3 hours, we investigated the possibility that PHB expressed at high levels can interact with the promoters of A-ARR genes. ARR7, an A-ARR that is highly responsive to PHB in the root meristem, was selected and used to perform a chromatin immunoprecipitation (ChIP) assay (Fig. 6C; S9B and S9C Fig.). The ChIP assay, combined with quantitative real-time PCR, indicated enrichment of PHB binding to the ARR7 promoter region where B-ARR binding elements are present. We then tested whether an enhanced ARR7 expression contributes to root length recovery in shr phb mutants by analyzing root length in shr phb arr7 triple mutant plants (S9D Fig.). The shr phb arr7 roots showed a partial reduction in root length compared with that of shr phb, suggesting that A-ARRs may influence the restoration of root growth in shr phb mutant plants (Student’s t-test; P < 0.001, α = 0.05).
The suppression of negative regulators of cytokinin signaling was recently shown to enhance cytokinin sensitivity in Arabidopsis roots [48]. Consistent with this, our expression analysis of two cytokinin-sensitive markers, AHP6 [49] and S32 [30], [44], indicated the enhanced cytokinin sensitivity in the shr. The expression of pAHP6:erGFP, which is inhibited by high levels of cytokinin, was absent in shr mutants and was partially restored in shr phb roots (Fig. 6D). The expression of pS32:erGFP, which is induced by high levels of cytokinin, increased and expanded in the shr roots but was consistent with the wild-type pattern in shr phb roots (S9F Fig.). Our stele-specific microarray data also supported the dynamics of these markers. Thus, it appears that the increased cytokinin sensitivity in shr mutants is due to the suppression of A-ARR expression by PHB.
PHB in the shr suppresses root growth by blocking B-ARR activity
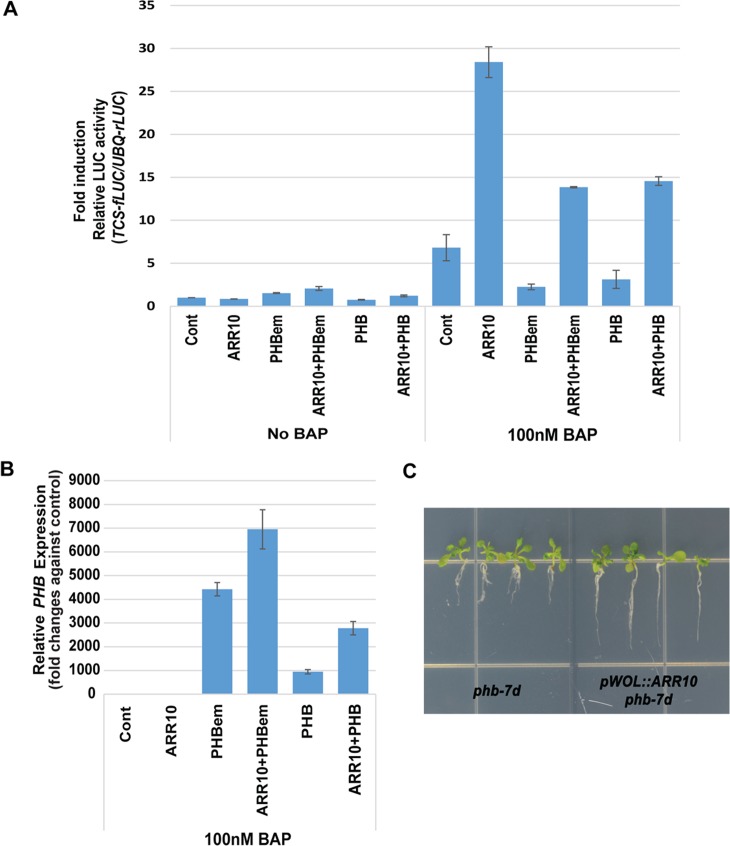
Our data so far indicate that high levels of PHB inhibit B-ARR activity. Since ChIP data suggest that PHB acts on B-ARR binding targets, we hypothesized that PHB might block B-ARRs in a quantifiable manner. To test this hypothesis, we employed a protoplast transient assay system (Fig. 7A; S11A Fig.). We measured luciferase activity driven by the TCS promoter (pTCS:LUC) in protoplasts transfected with 1) ARR10 alone, 2) PHB-em alone, 3) ARR10 and PHB-em, 4) PHB alone, and 5) ARR10 and PHB which were all driven by the 35S promoter. In two independent experiments, PHB mRNA levels expressed from p35S:PHB-em were found higher than PHB from p35S:PHB (Fig. 7B; S11B Fig.). In the absence of exogenous cytokinin, we did not detect a dramatic difference in luciferase activity between the protoplasts transfected with different constructs. However, in the presence of 6-benzylaminopurine (BAP), PHB protein expressed by p35S:PHB-em strongly suppressed ARR10 activity. This repressive effect was also observed in protoplasts transfected with PHB-em alone. However, the effect of PHB expressed by p35S:PHB on ARR10 activities was inconsistent. This seems to be due to the influence of miRNA 165/6 that targets PHB mRNA but not PHB-em (Fig. 7B; S11B Fig.). Collectively, these data confirm that high levels of PHB repress B-ARR activities, more effectively under high cytokinin.
Fig 7. PHB suppresses B-ARR activities in the presence of high cytokinin.
A protoplast assay that measures the ARR10 activities in the presence of PHB. (A) A high dosage of PHB (p35S:PHB-em and p35S:PHB) suppressed ARR10 activities under the high cytokinin. For the reporter assay, pUBQ:rLUC was co-transfected as an internal transfection control. Relative TCS activities with effectors (fold induction) were inferred by measuring the ratios between luminescence from fire fly luciferase (pTCS:fLUC) and luminescence from renilla luciferase (pUBQ:rLUC) and then dividing those values with the ratio obtained from control without an effector and BAP treatment. The error bar represents standard deviation. (B) PHB expression from 100 nM BAP-treated protoplasts is analyzed by real-time RT-PCR. Relative PHB expression was obtained by measuring fold changes against PHB expression in the control protoplasts. The error bar represents standard deviation (n = 3). (C) Increasing ARR10 levels by expressing it under WOL promoter restores the root growth in the phb-7D. BAP, 6-benzylaminopurine.
Our protoplast assay further indicated that the root growth defect in shr-2, phb-7d and pWOL:PHB-em:GFP NLS plants might be due to depleted, as opposed to enhanced, cytokinin signaling. If this mechanism were important in regulating root growth, supplying more B-ARR to the shr or phb-7d mutant would overcome the inhibitory effect of PHB on root growth. To test this hypothesis, we introduced pWOL:ARR10 into the phb-7d mutant. Analysis of four T2 plants showed a noticeable restoration in root growth, supporting our hypothesis (Fig. 7C).
Discussion
PHB in the root stele regulates TA cell proliferation
Controlling the spatial domains/levels of HD-ZIP III genes is critical for multiple developmental processes: ad/abaxial organization of lateral organs [50], specification of apical/basal axis in embryos [22], and vascular patterning [24], [51]. In this study, we demonstrate that this regulation is also essential for proper organization of the root meristem and root growth. Among the five HD-ZIP III genes, PHB seems to be the major player, because deletion of the other HD-ZIP III genes did not restore root growth in the same way as phb did in the shr mutant. Consistent with the results of a previous study showing that SCR acts in conjunction with SHR to posttranscriptionally suppress PHB and other HD-ZIP IIIs, the scr phb mutant is also able to restore root length.
Despite recovery of root growth, we did not detect any visible sign (based on cell morphology and QC marker expression) of an accompanying recovery of QC cells in the shr phb mutant. TA cell proliferation becomes very active in shr phb, as indicated by the expression of pCycB1.2:GUS, and the size of the meristem over time, displayed very similar dynamics between wild-type and shr phb roots. These findings indicate that the proliferation of TA cells is prolonged in the absence of QC cells in shr phb mutants. Conversely, pWOL:PHB-em:GFP NLS plants expressing a high level of PHB in the root stele cannot maintain TA cell proliferation and root growth while sustaining a QC. Therefore, PHB in the stele cells regulates TA cell activities required for root growth, independently of the QC in a non-cell autonomous manner. Interestingly, once the TA cells stop functioning in the pWOL:PHB-em:GFP NLS roots, the QC starts to lose its identity as a secondary effect. In animal systems, the maintenance of stem cell niches requires a communication between stem cells and their differentiated progenies, or TA cells [52], [53], [54]. When TA cells in the intestine are damaged, mitotically inactive cells in the stem cell niche become active and show stem cell characteristics [55], [56]. These features suggest that the mechanisms of stem cell niche functioning in animals and plants might be similar, even though the components involved are distinct.
Root growth is a dynamic process. Growth accelerates to a certain time point and then decelerates to terminate (reviewed by [57]). During this process, the meristem structure also changes. Toward the end of the root growth period, the QC degenerates, which is defined by its cell division. This results in the transition of the closed meristem to an open meristem in the Arabidopsis root [58], [59]. Therefore, the presence of a QC appears to be important for maintaining root growth over time. In this context, the RAM might operate in a similar way to the shoot apical meristem, where the organizing center is important for prolonging the meristem activity [60]. shr phb roots have open meristems and grow in a determinate manner. This determinate root growth does not appear to be caused by the exhaustion of TA cells because the root meristem size of shr phb plants does not change from that of the wild type until 15 DAG. Therefore, the early termination of root growth in shr phb mutants might be due to the lack of QC cells. Consistent with this theory, when the QC is recovered in shr phb roots by expressing pWOX5::SHR:nlsGFP, the growth becomes more active and less determinate than the shr phb. However, the overall root growth in the presence of QC in these plants remained below the wild-type level. Together, these data suggest that there is another aspect of root growth regulation, or that the QC is not fully restored. It has been reported that root growth in shr mutants can be partially rescued via the expression of cell-autonomous SHR expression in the stele [24], [61]. Our further investigation suggests that this restores the defect in phloem development and accompanies a partial recovery in root growth (manuscript in preparation).
PHB regulates root meristem/growth activity via alteration of B-ARR activities
Cytokinin affects root meristem and growth in a concentration-dependent manner [62]. Our study suggests that PHB regulates the activity of the root meristem and growth by two mechanisms: one involving cytokinin synthesis and the other involving changes in B-ARR activities. Our gene expression analysis indicated an up-regulation of IPT3 and IPT7 expression in accordance with elevated levels of PHB, and vice versa. Direct regulation of IPT7 expression by PHB was recently demonstrated [63]. However, activation of this pathway is not sufficient to produce a change in global cytokinin levels in the root, as we could not detect a change in cytokinin levels in the shr phb roots compared to that in shr plants. Nevertheless, when we prevented the up-regulation of IPT3 and IPT7 by PHB in shr mutants by generating shr ipt3 ipt7 triple mutants, root length and meristem size were partially restored. This is consistent with the results from our protoplast assay, which showed that the alteration of B-ARR activity by PHB is enhanced under high cytokinin levels.
Parallel to local cytokinin production, PHB regulates cytokinin signaling, and this process appears to be the main factor affecting root meristem and growth activity in the shr mutant. Despite the fact that cytokinin levels were similarly high in the shr and shr phb mutants, the expression of cytokinin-responsive markers was not consistent in shr and shr phb roots. Expression patterns of pAHP6:GFP and pS32:GFP indicated a higher cytokinin status in the shr root stele, while the shr phb mutants were similar to wild-type plants. On the other hand, pTCS:GFP, which reflects the transcriptional activities of B-ARRs is strongly suppressed in the shr and phb-7d mutants, but is recovered in the shr phb plants, demonstrating the effect of elevated PHB on cytokinin signaling pathway. Furthermore, consistent with the pTCS:GFP results, our stele-specific gene expression data obtained from the shr, shr phb, and wild-type roots indicated that a high level of PHB in the shr stele actively suppresses the expression of at least 50% of A-ARRs in the genome. On the basis of these data, together with the ChIP-PCR result suggesting that PHB can bind to the ARR7 promoter, we propose that increased PHB expression suppresses the cytokinin-mediated transcriptional activation by B-ARRs. This hindrance in B-ARR activity on one hand seems to increase cytokinin sensitivity in the shr roots, but on the other hand it affects TA cell proliferation leading to root growth arrest in a manner comparable to a cytokinin depleted mutant such as the arr1 10 12 triple mutant. Data from shr phb arr7 and pWOL:ARR10 provide further support for the role of PHB in B-ARR activities.
The behavior of PHB in the shr mutant was different to that reported previously for PHB in the cytokinin-signaling pathway. Our analysis of PHB in the protoplast assay suggests that PHB can suppress B-ARR activities. High cytokinin levels somehow augment the action of PHB on B-ARRs. The effect of cytokinin in this context might be related to changes in the phosphorylation status of B-ARRs, which somehow enhances repressive activities of PHB via physical interaction, competition for the same promoter element, or alteration of protein stability.
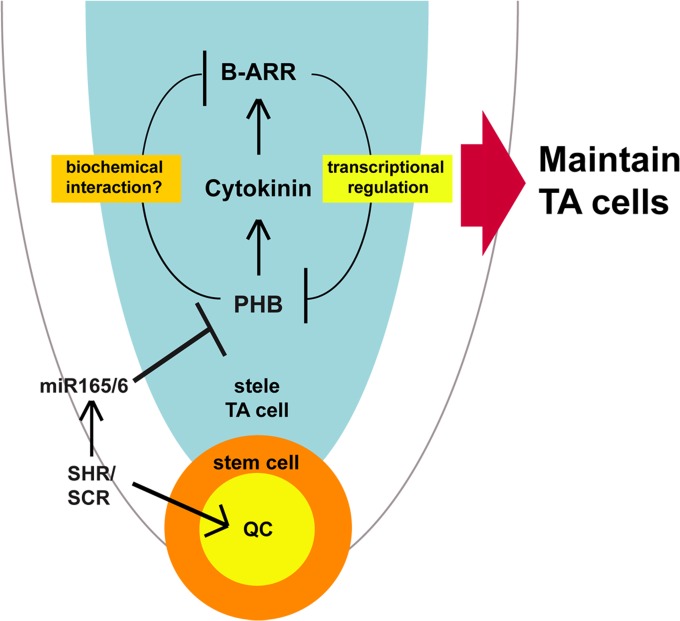
On the basis of the results presented in this study, we propose a model explaining how the stem cell niche and TA cell activities are coordinated via the SHR, SCR, PHB, and cytokinin pathways (Fig. 8). In this model, SHR and SCR regulate root meristem and growth via two pathways. One pathway, which maintains the QC, is required for stem cell maintenance and the subsequent extension of root growth, and the other pathway, mediated by posttranscriptional suppression of PHB, is critical for TA cell proliferation. In the latter, PHB regulates the root meristem and growth activities by modulating local cytokinin production and B-ARR activities in a context dependent manner. Such a dynamic regulatory system might enable the root meristem to respond appropriately to the ever-changing growth environment. Future investigations into the detailed mechanisms underlying how PHB alters B-ARRs, could help to determine how these pathways orchestrate root meristem activity and growth.
Fig 8. A model of SHR-PHB-cytokinin signaling in post-embryonic root growth.
SHR/SCR posttranscriptionally regulates PHB expression in the stele via the miRNA 165/6. PHB in the stele promotes cytokinin biosynthesis, thereby enhancing B-ARR activities in one hand. On the other hand, PHB interferes with B-ARR activities depending on its concentration. Such regulation generates a negative feedback loop, which likely contributes to the homeostasis of cytokinin signaling.
Methods
Plant materials and treatment
The Arabidopsis thaliana mutant/marker lines used are listed in S4 Table. Ecotype Columbia, or lines introgressed into Columbia, were used in this study unless stated otherwise. Seeds were surface-sterilized and germinated on MS agar plates supplemented with 1% sucrose at 22–23°C under a photoperiod of 16-h light/8-h dark. The various mutant combinations used were generated via genetic crossing, and in each case, the genotypes were verified by polymerase chain reaction (PCR). The oligonucleotide sequences used are provided in S5 Table. For the suppressor screen, shr homozygous seeds were treated with EMS as previously described [64]. The phb-15 and phb-16 alleles were identified in reciprocal crosses with shr-2 phb-6 mutants and subsequently confirmed by PCR sequencing. Both the phb-15 and 16 alleles harbor a stop codon (C>>T) at amino acid positions 149 (exon 3) and 384 (exon 8), respectively.
Histology, microscopy, RNA expression, and ChIP analysis
Primary roots of 5 to 21-day-old vertically grown seedlings were used for various analyses. Root lengths (from the base of the hypocotyl to the tip of the primary root) were measured using Image J software (www.rsbweb.nih.gov). The results presented here are the average data of 15–50 seedlings. GUS staining, Lugol staining, stereo (Leica DM5500B) and confocal microscopy (Leica TCS SP5 Laser Scanning Confocal Microscope) were conducted, with excitation (Ex) and emission (Em) wavelengths (Ex/Em) as follows: 405/461–504 nm for CFP, 488 nm/505–530 nm for GFP, 514 nm/525–555 nm for YFP, and 488–514 nm/613–679 nm for propidium iodide. Observation of callose on sieve plates in the phloem was performed as previously described [65]. In addition, qRT-PCR (using 2-mm-long segments of root tips, ABI 7900HT, Applied Biosystems), embryo analysis and whole-mount RNA in situ hybridization were performed as previously described [24], [66], [67], [68], [69], [70],. ChIP experiments (three biological replicates per experiment) were performed as previously described [7]. We used 5-day-old roots from seedlings expressing pWOL:PHB-em:GFP NLS. The oligonucleotide sequences used are listed in S5 Table.
Microarray experiments
Wild-type, shr and shr phb plants harboring pWOL:erGFP were germinated and grown for 6 days on solid MS medium under the same conditions as described above. Apical half portions of roots were cut and harvested. Protoplasts were prepared from root tissues as previously described [32], and pWOL:erGFP expressing protoplasts were collected with an Aria high-speed flow cell sorter (BD Biosciences) and were subsequently used for microarray analysis. Total RNA was isolated from collected protoplasts using an RNeasy Plant Mini Kit (Qiagen). RNA integrity was determined using a bioanalyzer (Agilent BioAnalyzer 2100). Poly-adenylated RNA was isolated from total RNA using a RiboMinus Plant Kit for RNA-Seq (Invitrogen). Probe preparation was conducted according to the manufacturer’s instructions (GeneChip Whole Transcript Double-Stranded Target Assay Manual from Affymetrix Inc.), and then biotinylated double-stranded DNA probes were hybridized to the Arabidopsis Tiling 1.0R arrays (Affymetrix Inc.) at Cornell Life Sciences Core Laboratories Center. Two (shr and shr phb mutants) to three (wild-type plants) biological replicate datasets with high correlation coefficients were generated for analysis from each genetic line.
Microarray data normalization and analysis
Microarray data were normalized using the RMA algorithm [71] in BIOCONDUCTOR, with Tiling 1.0R array CDF that contains gene-specific single-copy exonic probe sets [72]. Genes that were differentially expressed between the wild-type plant and the shr mutant were detected using the LIMMA package [73]. Principal component analysis (PCA) and clustering of differentially expressed genes were performed and visualized using MultiExperimental Viewer [74].
Plasmids and constructs
PHB-em, generated via site-directed mutagenesis following the manufacturer’s protocol (QuikChange II Site-Directed Mutagenesis Kit, Stratagene), was cloned into pDONR221 (Invitrogen). The 4.5-kb upstream sequence of WOX5 was PCR amplified from Arabidopsis genomic DNA and cloned into pDONRP4_P1R. pWOX5:SHR:nlsGFP was generated by fusing SHR cDNA to nlsGFP and the WOX5 promoter. Transgenes were placed into dpGreen vectors using the Invitrogen MultiSite Gateway system and were mobilized into plants using the floral dip method, as previously described [75], [30]. For protoplast assays, we used a pTCS:fLUC (Firefly Luciferase) construct as a reporter system and a pUBQ10:rLUC (Renilla Luciferase) construct as an internal transfection control. For the effector construct, a region containing the coding sequences of target genes (ARR10, PHB, PHB-em) was inserted downstream of the 35S promoter using a modified pHBT-sGFP(S65T)-NOS vector. As empty vector, pSPYCE, a binary vector with cauliflower mosaic virus 35S promoter and the C-terminal (156–239 aa) domain of YFP, was used.
Cytokinin measurement
Cytokinin quantification was performed as previously described [76], with some modifications. Briefly, ∼140 mg of the fresh half-bottom part of roots was collected from 6-day-old seedlings for each experimental replicate. Root tissues that were frozen in liquid nitrogen were processed in a vibration mill with tungsten carbide beads and 1-mL Bieleski buffer [77]. Deuterium-labeled cytokinin internal standards (Olchemim Ltd., Czech Republic) were added at the extraction stage, at 3 pmol per sample, to evaluate the recovery during purification and validate the quantification. The crude extract was then centrifuged (15 000 × g at 4°C), and the pellets were re-extracted in the same way. The combined supernatants were purified using a 1-g SCX column, evaporated to water phase in a vacuum followed by immunoaffinity chromatography using an immobilized wide-range anti-cytokinin monoclonal antibody as previously described [78]. The fraction containing cytokinin 9-glucosides, bases, and ribosides was evaporated to dryness, re-dissolved in the mobile phase and analyzed by UPLC-MS/MS [79]. The data were analyzed using Masslynx 4.1 software (Waters, Milford, MA, USA) and quantified by the standard isotope-dilution method.
Arabidopsis protoplast transient expression: Reporter assay and real-time RT-PCR
Arabidopsis mesophyll protoplasts were isolated and transfected as previously described [80], and DNA was prepared using a large-scale preparation of high-quality plasmid DNA [81]. Protoplasts (6 × 104 cells) were transfected with 40 μg of plasmid DNA with different combinations of reporter (pTCS:fLUC), effector (p35S:ARR10, p35S:PHB, p35S:PHBem), and internal control (pUBQ:rLUC) plasmids (S6 Table). Protoplasts were incubated in WI [0.5 M mannitol, 4 mM MES (pH 5.7), 20 mM KCl] at room temperature for 1 h in the light. After 1-h incubation, protoplasts were treated with 100 nM of 6-benzylaminopurine (BAP) and incubated for 5 h under the same conditions. Luciferase reporter activity was determined by the Dual Luciferase Assay System (Promega) and detected by Microlumat 96V Luminometer (Berthold, Bad Wildbad, Germany). To analyze PHB expression, quantitative RT-PCR analyses were carried out using total RNAs extracted from protoplasts (3 × 104 cells), which were collected from protoplast transient assay. Total RNA extraction was performed with PicoPure RNA Isolation Kit (Arcturus). 20μL of reverse transcript reaction was performed for the first cDNA strand synthesis using 500ng of total RNAs and Superscript III reverse transcriptase (Invitrogen). For quantitative PCR reactions, a master mix was prepared using an iQ SYBR Green supermix (Bio-Rad) and PCR condition was programmed according to the manufacturer’s instructions for CFX96 Real-Time PCR machine (Bio-Rad). Three technical replicate reactions were performed. Primer information for RT-PCR is available in S5 Table. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as internal control gene.
Accession numbers
Microarray data have been submitted to the GEO database (accession number GSE33015). Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: SHR (At4G37650), PHB (At2G34710), PHV (At1G30490), CNA (At1G52150), ATHB8 (At4G32880), REV (At5G60690), SCR (At3G54220), PLT1 (At3G20840), PLT2 (At1G51190), S32 (At2g18380), AGL42 (At5g62165), IPT3 (At3G63110), IPT7 (At3G23630), ARR7 (At1G19050), ARR10 (At4G31920) and GAPDH (At3g26650).
Supporting Information
(A) Restoration in root length in shr phb double mutants. New phb alleles; phb-15 & 16 also resulted in a significant recovery in the shr root growth. (B) Root meristem size in wild-type, shr-2, phb-6, shr-2 phb-6, scr-4, and scr-4 phb-6 plants (5 DAG). (C) Comparable levels of pCycB1.2:GUS expression in the proximal meristem cells in wild-type and shr phb roots (15 DAG). (D) Quantification of pCycB1.2:GUS expression is shown in the panel (C). White arrowheads marks the end of the meristem. Error bars represent the standard error (n = 15–30 plants). Scale bars: B, 50 μm; C, 20 μm.
(TIF)
(A) Aniline blue staining of sieve plates in phloem sieve elements in wild-type, shr-2, and shr-2 phb-6 roots. Two sieve plates are organized in parallel in the wild-type root, whereas only one is found in the shr-2 and shr-2 phb-6 roots. (B) Expression patterns of known QC marker genes in wild-type, shr-2 and shr-2 phb-6 roots. Red arrowheads mark position of sieve plates and white arrowheads mark QC. Scale bars represent 25 μm.
(TIF)
(A) Similar levels of pWOX5:erGFP expression in wild-type, shr-2, and shr-2 phb-6 embryos at the late heart stage. (B) A significant decrease in pWOX5:erGFP expression in shr-2 and shr-2 phb-6 embryos in contrast to the wild type at the bent-cotyledon stage. (C) Lugol’s staining shows precocious differentiation of columella stem cells in the shr-2 phb-6 roots (5 DAG). Black and red arrowheads indicate QC and columella stem cells respectively. Scale bars represent A and B, 25 μm; C, 20 μm.
(TIF)
(A) Four synonymous mutations introduced into the miRNA 165/6 binding site of PHB via site-directed mutagenesis. Mutated bases are shown in purple. (B) A comparison of PHB-GFP expression domains in pPHB:PHB:GFP, pPHB:PHB-m:GFP and pPHB:PHB:GFP shr-2 roots (5 DAG). Scale bars represent 20 μm. (C) A comparison of root meristem size in wild-type, pWOL:PHB-m:GFP NLS and pWOL:PHB-em:GFP NLS plants (upper panel: 7 DAG, lower panel: 5 DAG). Scale bars represent 20 μm (upper panel) and 10 μm (lower panel). White arrowheads mark where meristem ends. Asterisks indicate cortex cells. (D, E) Expression of pWOX5:YFP (D) and pQC25:GUS (E) in 15 day-old pWOL:PHB-em:GFP NLS roots. (F) Aberrant cell divisions in the QC position of pWOL:PHB-em:GFP NLS roots (10 DAG). (G) WOX5 in situ suggests decline in its expression levels in the QC region of pWOL:PHB-em:GFP NLS roots. White arrows indicate QC cells undergoing abnormal cell divisions; Scale bars represent 25 μm for D-G.
(TIF)
Expression pattern of pWOL:erGFP in the wild-type, shr-2, and shr-2 phb-6 roots. Scale bars represent 25 μm.
(TIF)
(A) Correlation coefficients (R2) between cell sorting/tiling array data. Biological replicates are highlighted in yellow. (B, C) Expression dynamics of differentially expressed genes between the shr and wild-type plants, summarized with PCA (B) and QT clustering (C). shr-2, SHR1 and 2; wild type, WT1, 2 and 3; shr-2 phb-6, DM1 and 2.
(TIF)
Expression patterns are classified by Hierarchical Clustering.
(TIF)
(A) pDR5:YFPvenus shows a reduced signal in shr-2 mutants in comparison to wild-type and shr-2 phb-6 plants (7 DAG). (B, C) Reduced expression of pPIN1:PIN1:GFP in 5- (B) and 10- (C) day-old shr-2 roots in comparison to the wild type. Expression is partially restored in shr-2 phb-6. Scale bars represent 25 μm.
(TIF)
(A) Cytokinin content in the wild-type, shr-2 and shr-2 phb-6 roots. tZ, Transzeatin; tZR, tZ riboside; cZ, cis-zeatin; cZR, cZ riboside; iP, N6-(Δ2-isopentenyl) adenine; iPR, iP riboside; DHZ, Dihydrozeatin; DHZR, Dihydrozeatin riboside; DHZ9G, DHZ-9-glucoside; iP9G, iP-9-glucoside; tZ9G, tZ-9-glucoside; cZ9G, cZ-9-glucoside. (B) Heat map showing expression levels of A-ARRs in the wild-type (WT1, 2 and 3), shr (SHR1 and 2), and shr phb (DM1 and 2) root stele. (C) Relative mRNA levels of ARR5, ARR7 and ARR15 in the shr-2 and shr-2 phb-6 roots. Data are normalized to the wild-type plants. (D) Root growth in shr-2 phb-6 arr7 mutant. (E) Expression pattern of pTCS:erGFP in phb-7d roots. (F) pS32:erGFP expression in the wild-type, shr-2 and shr-2 phb-6 roots. Scale bars represent 25 μm. SD: standard deviation (n = 3 biological replicates).
(TIF)
(A) Quantitative comparison of TCS-GFP levels in the stele and root cap expressing the wild type (WT) and transgenic line expressing PHB-em fused to the glucocorticoid receptor (GR) under the PHB promoter. (B) Expression patterns of pTCS:erGFP in WT and transgenic line crossed with pPHB:PHBem:GR. All seedlings grew under the same conditions. After 5 DAG, seedlings were transferred to new media including no DEX or 10 uM DEX and were incubated for 3 h. GFP expression is reduced following treatment with 10 uM DEX in transgenic lines.
(TIF)
(A) A high dosage of PHB (p35S:PHB-em) suppressed ARR10 activities under the high cytokinin. In contrast, PHB expressed by p35S:PHB was not effective on ARR10 activities. (B) PHB expression is analyzed by qRT-PCR using 100 nM BAP-treated protoplasts. The error bar represents standard deviation. BAP, 6-benzylaminopurine.
(TIF)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLSX)
Acknowledgments
We thank Gustavo Acevedo, Ah-reum Jeong, Susan Li, and Changxi Yin for assisting with experiments at various stages; Philip Benfey, Ben Scheres, Renze Heidstra, Ikram Blilou, Tatsuo Kakimoto, Frederic Berger, Bruno Muller, Joe Kieber, and Ottoline Leyser for providing seeds and DNA; and John Bowman, Maria Harrison, Jose Dinneny, and anonymous reviewers for their valuable comments on this manuscript.
Funding Statement
This work was supported by National Science Foundation (IOS-0818071, www.nsf.gov), Triad Foundation, National Research Foundation of Korea (2013R1A1A3009272, www.nrf.re.kr/nrf_eng_cms), and Seoul National University Research grants to JYL and Ministry of Education, Youth and Sports of the Czech Republic (MSM 6198959216) and ED0007/01/01 Centre of the Region Haná for Biotechnological and Agricultural Research grants to PT. KHR and ESK are supported by Brain Korea 21 Plus Program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, et al. (1993) Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84. [DOI] [PubMed] [Google Scholar]
- 2. van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B (1997) Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390: 287–289. [DOI] [PubMed] [Google Scholar]
- 3. Bennett T, Scheres B (2010) Root development-two meristems for the price of one? Curr Top in Dev Biol 91: 67–102. 10.1016/S0070-2153(10)91003-X [DOI] [PubMed] [Google Scholar]
- 4. Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, et al. (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120. [DOI] [PubMed] [Google Scholar]
- 5. Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, et al. (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433. [DOI] [PubMed] [Google Scholar]
- 6. Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, et al. (2000) The SHORT-ROOT Gene Controls Radial Patterning of the Arabidopsis Root through Radial Signaling. Cell 101: 555–567. [DOI] [PubMed] [Google Scholar]
- 7. Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, et al. (2007) An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316: 421–425. [DOI] [PubMed] [Google Scholar]
- 8. Xu J, Hofhuis H, Heidstra R, Sauer M, Friml J, et al. (2006) A molecular framework for plant regeneration. Science 311: 385–388. [DOI] [PubMed] [Google Scholar]
- 9. Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, et al. (2007) Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814. [DOI] [PubMed] [Google Scholar]
- 10. Sena G, Wang X, Liu HY, Hofhuis H, Birnbaum KD (2009) Organ regeneration does not require a functional stem cell niche in plants. Nature 457: 1150–1153. 10.1038/nature07597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, et al. (2004) Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16: 1365–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Argyros RD, Mathews DE, Chiang YH, Palmer CM, Thibault DM, et al. (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116. 10.1105/tpc.108.059584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tian Q, Uhlir NJ, Reed JW (2002) Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14: 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, et al. (2008) A Genetic Framework for the Control of Cell Division and Differentiation in the Root Meristem. Science 322: 1380–1384. 10.1126/science.1164147 [DOI] [PubMed] [Google Scholar]
- 15. Moubayidin L, Di Mambro R, Sozzani R, Pacifici E, Salvi E, et al. (2013) Spatial coordination between stem cell activity and cell differentiation in the root meristem. Dev Cell 26: 405–415. 10.1016/j.devcel.2013.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Byrne ME (2006) Shoot meristem function and leaf polarity: the role of class III HD-ZIP genes. PLoS Genet 2: e89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou J, Sebastian J, Lee JY (2011) Signaling and gene regulatory programs in plant vascular stem cells. Genesis 49: 885–904. 10.1002/dvg.20795 [DOI] [PubMed] [Google Scholar]
- 18. Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, et al. (2005) Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17: 61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, et al. (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13: 1768–1774. [DOI] [PubMed] [Google Scholar]
- 20. Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, et al. (2004) MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5' region. EMBO J 23: 3356–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grigg SP, Galinha C, Kornet N, Canales C, Scheres B, et al. (2009) Repression of apical homeobox genes is required for embryonic root development in Arabidopsis. Curr Biol 19: 1485–1490. 10.1016/j.cub.2009.06.070 [DOI] [PubMed] [Google Scholar]
- 22. Smith ZR, Long JA (2010) Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature 464: 423–426. 10.1038/nature08843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Iwata Y, Takahashi M, Fedoroff NV, Hamdan SM (2013) Dissecting the interactions of SERRATE with RNA and DICER-LIKE 1 in Arabidopsis microRNA precursor processing. Nucleic Acids Res 41: 9129–9140. 10.1093/nar/gkt667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, et al. (2010) Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321. 10.1038/nature08977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Perilli S, Sabatini S (2010) Analysis of root meristem size development. Methods in Mol Biol 655: 177–187. 10.1007/978-1-60761-765-5_12 [DOI] [PubMed] [Google Scholar]
- 26. Ingouff M, Jullien PE, Berger F (2006) The female gametophyte and the endosperm control cell proliferation and differentiation of the seed coat in Arabidopsis. Plant Cell 18: 3491–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sabatini S, Heidstra R, Wildwater M, Scheres B (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17: 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, et al. (2007) PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057. [DOI] [PubMed] [Google Scholar]
- 29. Nawy T, Lee J-Y, Colinas J, Wang JY, Thongrod SC, et al. (2005) Transcriptional Profile of the Arabidopsis Root Quiescent Center. Plant Cell 17: 1908–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee J-Y, Colinas J, Wang JY, Mace D, Ohler U, et al. (2006) Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc Natl Acad Sci USA 103: 6055–6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mahonen AP, Bonke M, Kauppinen L, Riikonen M, Benfey PN, et al. (2000) A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev 14: 2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Birnbaum K, Jung JW, Wang JY, Lambert GM, Hirst JA, et al. (2005) Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nature Methods 2: 615–619. [DOI] [PubMed] [Google Scholar]
- 33. Heyer LJ, Kruglyak S, Yooseph S (1999) Exploring Expression Data: Identification and Analysis of Coexpressed Genes. Genome Res 9: 1106–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, et al. (2003) A Gene Expression Map of the Arabidopsis Root. Science 302: 1956–1960. [DOI] [PubMed] [Google Scholar]
- 35. Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, et al. (2006) Whole-Genome Analysis of the SHORT-ROOT Developmental Pathway in Arabidopsis. PLoS Biol 4: e143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brady SM, Orlando DA, Lee J-Y, Wang JY, Koch J, et al. (2007) A High-Resolution Root Spatiotemporal Map Reveals Dominant Expression Patterns. Science 318: 801–806. [DOI] [PubMed] [Google Scholar]
- 37. Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, et al. (2009) AmiGO: online access to ontology and annotation data. Bioinformatics 25: 288–289. 10.1093/bioinformatics/btn615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moubayidin L, Di Mambro R, Sabatini S (2009) Cytokinin-auxin crosstalk. Trends Plant Sci 14: 557–562. 10.1016/j.tplants.2009.06.010 [DOI] [PubMed] [Google Scholar]
- 40. Lucas M, Swarup R, Paponov IA, Swarup K, Casimiro I, et al. (2011) Short-Root regulates primary, lateral, and adventitious root development in Arabidopsis. Plant Physiol 155: 384–398. 10.1104/pp.110.165126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nordström A, Tarkowski P, Tarkowska D, Norbaek R, Åstot C, et al. (2004) Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: A factor of potential importance for auxin-cytokinin-regulated development. Proc Natl Acad Sci USA 101: 8039–8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jones B, Gunnerås SA, Petersson SV, Tarkowski P, Graham N, et al. (2010) Cytokinin Regulation of Auxin Synthesis in Arabidopsis Involves a Homeostatic Feedback Loop Regulated via Auxin and Cytokinin Signal Transduction. Plant Cell 22: 2956–2969. 10.1105/tpc.110.074856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marhavy P, Bielach A, Abas L, Abuzeineh A, Duclercq J, et al. (2011) Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Dev Cell 21: 796–804. 10.1016/j.devcel.2011.08.014 [DOI] [PubMed] [Google Scholar]
- 44. Cui H, Hao Y, Kovtun M, Stolc V, Deng XW, et al. (2011) Genome-wide direct target analysis reveals a role for SHORT-ROOT in root vascular patterning through cytokinin homeostasis. Plant Physiol 157: 1221–1231. 10.1104/pp.111.183178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Miyawaki K, Tarkowski P, Matsumoto-Kitano M, Kato T, Sato S, et al. (2006) Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc Natl Acad Sci USA 103: 16598–16603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rashotte AM, Carson SD, To JP, Kieber JJ (2003) Expression profiling of cytokinin action in Arabidopsis. Plant Physiol 132: 1998–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Muller B, Sheen J (2008) Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097. 10.1038/nature06943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang W, To JP, Cheng CY, Eric Schaller G, Kieber JJ (2011) Type-A response regulators are required for proper root apical meristem function through post-transcriptional regulation of PIN auxin efflux carriers. Plant J 68: 1–10. 10.1111/j.1365-313X.2011.04668.x [DOI] [PubMed] [Google Scholar]
- 49. Mahonen AP, Bishopp A, Higuchi M, Nieminen KM, Kinoshita K, et al. (2006) Cytokinin Signaling and Its Inhibitor AHP6 Regulate Cell Fate During Vascular Development. Science 311: 94–98. [DOI] [PubMed] [Google Scholar]
- 50. Eshed Y, Baum SF, Perea JV, Bowman JL (2001) Establishment of polarity in lateral organs of plants. Curr Biol 11: 1251–1260. [DOI] [PubMed] [Google Scholar]
- 51. Miyashima S, Koi S, Hashimoto T, Nakajima K (2011) Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138: 2303–2313. 10.1242/dev.060491 [DOI] [PubMed] [Google Scholar]
- 52. Hsu YC, Li L, Fuchs E (2014) Transit-amplifying cells orchestrate stem cell activity and tissue regeneration. Cell 157: 935–949. 10.1016/j.cell.2014.02.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kalaitzidis D, Scadden DT (2014) Tic-TACs: refreshing hair growth. Cell 157: 769–770. 10.1016/j.cell.2014.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lander AD, Kimble J, Clevers H, Fuchs E, Montarras D, et al. (2012) What does the concept of the stem cell niche really mean today? BMC Biol 10: 19 10.1186/1741-7007-10-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Barker N (2014) Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol 15: 19–33. 10.1038/nrm3721 [DOI] [PubMed] [Google Scholar]
- 56. Buczacki SJ, Zecchini HI, Nicholson AM, Russell R, Vermeulen L, et al. (2013) Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature 495: 65–69. 10.1038/nature11965 [DOI] [PubMed] [Google Scholar]
- 57. Rost TL (2011) The organization of roots of dicotyledonous plants and the positions of control points. Ann Bot 107: 1213–1222. 10.1093/aob/mcq229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Baum SF, Dubrovsky JG, Rost TL (2002) Apical organization and maturation of the cortex and vascular cylinder inArabidopsis thaliana (Brassicaceae) roots. Am J Bot 89: 908–920. 10.3732/ajb.89.6.908 [DOI] [PubMed] [Google Scholar]
- 59. Shishkova S, Rost TL, Dubrovsky JG (2008) Determinate root growth and meristem maintenance in angiosperms. Ann Bot 101: 319–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, et al. (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815. [DOI] [PubMed] [Google Scholar]
- 61. Gallagher KL, Benfey PN (2009) Both the conserved GRAS domain and nuclear localization are required for SHORT-ROOT movement. Plant J 57: 785–797. 10.1111/j.1365-313X.2008.03735.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dello Ioio R, Linhares FS, Sabatini S (2008) Emerging role of cytokinin as a regulator of cellular differentiation. Curr Opin Plant Biol 11: 23–27. [DOI] [PubMed] [Google Scholar]
- 63. Dello Ioio R, Galinha C, Fletcher AG, Grigg SP, Molnar A, et al. (2012) A PHABULOSA/Cytokinin Feedback Loop Controls Root Growth in Arabidopsis. Curr Biol 22: 1699–1704. 10.1016/j.cub.2012.07.005 [DOI] [PubMed] [Google Scholar]
- 64. Jander G, Baerson SR, Hudak JA, Gonzalez KA, Gruys KJ, et al. (2003) Ethylmethanesulfonate saturation mutagenesis in Arabidopsis to determine frequency of herbicide resistance. Plant Physiol 131: 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ingram P, Dettmer J, Helariutta Y, Malamy JE (2011) Arabidopsis Lateral Root Development 3 is essential for early phloem development and function, and hence for normal root system development. Plant J 68: 455–467. 10.1111/j.1365-313X.2011.04700.x [DOI] [PubMed] [Google Scholar]
- 66. Sundaresan V, Springer P, Volpe T, Haward S, Jones JD, et al. (1995) Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev 9: 1797–1810. [DOI] [PubMed] [Google Scholar]
- 67. van den Berg C, Willemsen V, Hage W, Weisbeek P, Scheres B (1995) Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature 378: 62–65. [DOI] [PubMed] [Google Scholar]
- 68. Willemsen V, Wolkenfelt H, de Vrieze G, Weisbeek P, Scheres B (1998) The HOBBIT gene is required for formation of the root meristem in the Arabidopsis embryo. Development 125: 521–531. [DOI] [PubMed] [Google Scholar]
- 69. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 70. Hejatko J, Blilou I, Brewer PB, Friml J, Scheres B, et al. (2006) In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nature Protoc 1: 1939–1946. [DOI] [PubMed] [Google Scholar]
- 71. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, et al. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. [DOI] [PubMed] [Google Scholar]
- 72. Naouar N, Vandepoele K, Lammens T, Casneuf T, Zeller G, et al. (2009) Quantitative RNA expression analysis with Affymetrix Tiling 1.0R arrays identifies new E2F target genes. Plant J 57: 184–194. 10.1111/j.1365-313X.2008.03662.x [DOI] [PubMed] [Google Scholar]
- 73. Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3. [DOI] [PubMed] [Google Scholar]
- 74. Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, et al. (2006) TM4 microarray software suite. Methods Enzymol 411: 134–193. [DOI] [PubMed] [Google Scholar]
- 75. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 76. Novák O, Tarkowski P, Tarkowská D, Doležal K, Lenobel R, et al. (2003) Quantitative analysis of cytokinins in plants by liquid chromatography-single-quadrupole mass spectrometry. Anal Chim Acta 480: 207–218. [Google Scholar]
- 77. Bieleski RL (1964) The problem of halting enzyme action when extracting plant tissues. Anal Biochem 9: 431–442. [DOI] [PubMed] [Google Scholar]
- 78. Faiss M, Zalubilova J, Strnad M, Schmulling T (1997) Conditional transgenic expression of the ipt gene indicates a function for cytokinins in paracrine signaling in whole tobacco plants. Plant J 12: 401–415. [DOI] [PubMed] [Google Scholar]
- 79. Novak O, Hauserova E, Amakorova P, Dolezal K, Strnad M (2008) Cytokinin profiling in plant tissues using ultra-performance liquid chromatography-electrospray tandem mass spectrometry. Phytochemistry 69: 2214–2224. 10.1016/j.phytochem.2008.04.022 [DOI] [PubMed] [Google Scholar]
- 80. Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- 81. Cho YH, Yoo SD (2010) Expression of epitope-tagged proteins in Arabidopsis leaf mesophyll protoplasts. Methods Mol Biol 657: 33–42. 10.1007/978-1-60761-783-9_3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Restoration in root length in shr phb double mutants. New phb alleles; phb-15 & 16 also resulted in a significant recovery in the shr root growth. (B) Root meristem size in wild-type, shr-2, phb-6, shr-2 phb-6, scr-4, and scr-4 phb-6 plants (5 DAG). (C) Comparable levels of pCycB1.2:GUS expression in the proximal meristem cells in wild-type and shr phb roots (15 DAG). (D) Quantification of pCycB1.2:GUS expression is shown in the panel (C). White arrowheads marks the end of the meristem. Error bars represent the standard error (n = 15–30 plants). Scale bars: B, 50 μm; C, 20 μm.
(TIF)
(A) Aniline blue staining of sieve plates in phloem sieve elements in wild-type, shr-2, and shr-2 phb-6 roots. Two sieve plates are organized in parallel in the wild-type root, whereas only one is found in the shr-2 and shr-2 phb-6 roots. (B) Expression patterns of known QC marker genes in wild-type, shr-2 and shr-2 phb-6 roots. Red arrowheads mark position of sieve plates and white arrowheads mark QC. Scale bars represent 25 μm.
(TIF)
(A) Similar levels of pWOX5:erGFP expression in wild-type, shr-2, and shr-2 phb-6 embryos at the late heart stage. (B) A significant decrease in pWOX5:erGFP expression in shr-2 and shr-2 phb-6 embryos in contrast to the wild type at the bent-cotyledon stage. (C) Lugol’s staining shows precocious differentiation of columella stem cells in the shr-2 phb-6 roots (5 DAG). Black and red arrowheads indicate QC and columella stem cells respectively. Scale bars represent A and B, 25 μm; C, 20 μm.
(TIF)
(A) Four synonymous mutations introduced into the miRNA 165/6 binding site of PHB via site-directed mutagenesis. Mutated bases are shown in purple. (B) A comparison of PHB-GFP expression domains in pPHB:PHB:GFP, pPHB:PHB-m:GFP and pPHB:PHB:GFP shr-2 roots (5 DAG). Scale bars represent 20 μm. (C) A comparison of root meristem size in wild-type, pWOL:PHB-m:GFP NLS and pWOL:PHB-em:GFP NLS plants (upper panel: 7 DAG, lower panel: 5 DAG). Scale bars represent 20 μm (upper panel) and 10 μm (lower panel). White arrowheads mark where meristem ends. Asterisks indicate cortex cells. (D, E) Expression of pWOX5:YFP (D) and pQC25:GUS (E) in 15 day-old pWOL:PHB-em:GFP NLS roots. (F) Aberrant cell divisions in the QC position of pWOL:PHB-em:GFP NLS roots (10 DAG). (G) WOX5 in situ suggests decline in its expression levels in the QC region of pWOL:PHB-em:GFP NLS roots. White arrows indicate QC cells undergoing abnormal cell divisions; Scale bars represent 25 μm for D-G.
(TIF)
Expression pattern of pWOL:erGFP in the wild-type, shr-2, and shr-2 phb-6 roots. Scale bars represent 25 μm.
(TIF)
(A) Correlation coefficients (R2) between cell sorting/tiling array data. Biological replicates are highlighted in yellow. (B, C) Expression dynamics of differentially expressed genes between the shr and wild-type plants, summarized with PCA (B) and QT clustering (C). shr-2, SHR1 and 2; wild type, WT1, 2 and 3; shr-2 phb-6, DM1 and 2.
(TIF)
Expression patterns are classified by Hierarchical Clustering.
(TIF)
(A) pDR5:YFPvenus shows a reduced signal in shr-2 mutants in comparison to wild-type and shr-2 phb-6 plants (7 DAG). (B, C) Reduced expression of pPIN1:PIN1:GFP in 5- (B) and 10- (C) day-old shr-2 roots in comparison to the wild type. Expression is partially restored in shr-2 phb-6. Scale bars represent 25 μm.
(TIF)
(A) Cytokinin content in the wild-type, shr-2 and shr-2 phb-6 roots. tZ, Transzeatin; tZR, tZ riboside; cZ, cis-zeatin; cZR, cZ riboside; iP, N6-(Δ2-isopentenyl) adenine; iPR, iP riboside; DHZ, Dihydrozeatin; DHZR, Dihydrozeatin riboside; DHZ9G, DHZ-9-glucoside; iP9G, iP-9-glucoside; tZ9G, tZ-9-glucoside; cZ9G, cZ-9-glucoside. (B) Heat map showing expression levels of A-ARRs in the wild-type (WT1, 2 and 3), shr (SHR1 and 2), and shr phb (DM1 and 2) root stele. (C) Relative mRNA levels of ARR5, ARR7 and ARR15 in the shr-2 and shr-2 phb-6 roots. Data are normalized to the wild-type plants. (D) Root growth in shr-2 phb-6 arr7 mutant. (E) Expression pattern of pTCS:erGFP in phb-7d roots. (F) pS32:erGFP expression in the wild-type, shr-2 and shr-2 phb-6 roots. Scale bars represent 25 μm. SD: standard deviation (n = 3 biological replicates).
(TIF)
(A) Quantitative comparison of TCS-GFP levels in the stele and root cap expressing the wild type (WT) and transgenic line expressing PHB-em fused to the glucocorticoid receptor (GR) under the PHB promoter. (B) Expression patterns of pTCS:erGFP in WT and transgenic line crossed with pPHB:PHBem:GR. All seedlings grew under the same conditions. After 5 DAG, seedlings were transferred to new media including no DEX or 10 uM DEX and were incubated for 3 h. GFP expression is reduced following treatment with 10 uM DEX in transgenic lines.
(TIF)
(A) A high dosage of PHB (p35S:PHB-em) suppressed ARR10 activities under the high cytokinin. In contrast, PHB expressed by p35S:PHB was not effective on ARR10 activities. (B) PHB expression is analyzed by qRT-PCR using 100 nM BAP-treated protoplasts. The error bar represents standard deviation. BAP, 6-benzylaminopurine.
(TIF)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLSX)