Abstract
Objectives
Coxsackievirus A group 16 strain (CVA16) is one of the predominant causative agents of hand, foot, and mouth disease (HFMD).
Methods
Using a specimen from a male patient with HFMD, we isolated and performed sequencing of the Korean CVA16 strain and compared it with a G10 reference strain. Also, we were investigated the effects of medicinal plant extract on the cytopathic effects (CPE) by CPE reduction assay against Korean CVA16.
Results
Phylogenetic analysis showed that the Korean CVA16 isolate belonged to cluster B-1 and was closely related to the strain PM-15765-00 isolated in Malaysia in 2000. The Korean CVA16 isolate showed 73.2% nucleotide identity to the G10 prototype strain and 98.7% nucleotide identity to PM-15765-00. Next, we assessed whether the Korean CVA16 isolate could be used for in vitro screening of antiviral agents to treat HFMD infection. Vero cells infected with the Korean CVA16 isolate showed a cytopathic effect 2 days after the infection, and the treatment of cells with Cornus officinalis, Acer triflorum, Pulsatilla koreana, and Clematis heracleifolia var. davidiana Hemsl extracts exhibited strong antiviral activity against CVA16.
Conclusion
Collectively, our work provides potential candidates for the development of vaccine and novel drugs to treat the CVA16 strain isolated from a Korean patient.
Keywords: coxsackievirus A16, antiviral drug, HFMD, medicinal plant
1. Introduction
Coxsackievirus A16 (CVA16) is a serotype of the genus Enterovirus belonging to the family Picornaviridae, and one of the causative agents of hand, foot, and mouth disease (HFMD) in humans. In rare instances, it may cause neurological diseases, including aseptic meningitis, exanthems, and herpangina. HFMD is a self-limiting exanthematous eruption characterized by vesicles in the oral cavity and peripherally distributed cutaneous lesions on the hands and feet [1,2]. Recently, large outbreaks of HFMD by CVA16 and enterovirus 71 (EV71) were reported, especially in the Asia–Pacific area [3,4]. Outbreaks of HFMD by CVA16 occurred in Taiwan (1996–2006) and in Singapore (2001–2007), as well as in England (1994) [1,5]. In Korea, outbreaks of HFMD caused by human enterovirus (HEV) infections have been consistently reported since 2009 [6–8]. HFMD has become a serious public health problem in South-East Asia, with periodic large epidemics occurring in recent decades. Most of therapeutic and vaccine strategies against HFMD have focused on EV71 [9,10]. In China, Phase III trials of antiviral vaccines against EV71 were recently successfully completed [11]. In addition, no HEV-specific antiviral drugs are yet available for clinical use. Many synthetic anti-HEV compounds have been described in vitro, but only a few of them are effective in vivo [12,13]. However, no vaccines are currently available to prevent CVA16 infections. Thus, research and development of vaccines and antiviral drugs to prevent CVA16 infections and treat HFMD must be spurred on.
A suitable alternative to antiviral drugs may be traditional medicinal plants. They have multiple targets, minimal side effects, low potential to cause resistance, and are less expensive. Screening of medicinal plants has led to the discovery of potent in vitro inhibitors of viral replication [14,15].
In this study, the molecular biological characteristics and genetic diversity of a Korean isolate of CVA16 were analyzed through complete nucleotide sequencing and phylogenetic analysis, respectively. Finally, Korean medicinal plants were screened for their potential anti-CVA16 effects.
2. Materials and methods
2.1. Virus isolation and identification
The Korean CVA16 strain was isolated from the stool sample of a male patient with HFMD admitted to the Department of Pediatrics at the Soonchunhyang University Cheonan Hospital, Cheonan, Korea, in June 2008. This study was conducted in accordance with the ethical principles adopted by the World Medical Association Declaration of Helsinki and approved by the Institutional Review Board (IRB No. 2012-48) of the ethical committee of Soonchunhyang University Cheonan Hospital. The pretreated sample was inoculated into Vero cells and incubated at 37°C in an atmosphere of 5% CO2 until the appearance of cytopathic effects (CPE). Basic Local Alignment Search Tool (BLAST) search of VP1 sequences verified the identity of the Korean isolate. The VP1 sequences of the Korean isolate had the maximum nucleotide similarity with CVA16 serotype strains [16].
2.2. Nucleotide sequencing and sequence analysis
The complete nucleotide sequence of the Korean CVA16 strain was determined using a primer walking strategy; the sequences of the genome termini were determined by random amplification of cDNA ends system (Invitrogen, Carlsbad, CA, USA). Polymerase chain reaction (PCR) products were purified using a QIAquick PCR Purification Kit (Qiagen, Hamburg, Germany). The purified DNA was added to a reaction mixture containing 2 μL of BigDye terminator reaction mix (ABI Prism BigDye Terminator Cycle Sequencing Kit; Applied Biosystems, Foster, CA, USA) and 2 pmol of each primer. Sequencing reactions were subjected to an initial denaturation at 96°C for 1 minute and 25 cycles consisting of 96°C for 10 seconds, 50°C for 5 seconds, and 60°C for 4 minutes in a Gene Amp PCR system 2700 (Applied Biosystems). The products were purified by precipitation with 100% cold ethanol and 3M sodium acetate (pH 5.8), and then loaded on an automated 3100 Genetic Analyzer (Applied Biosystems). Nucleotide sequences of CVA 16 strains were constructed to contig and compared with the reference G10 strain (accession no. U05876). G10 strain, obtained from Genbank databases, was isolated in South Africa in 1954 and subsequently sequenced in 1994 [17,18]. The complete sequences of the Korean CVA16 isolate are deposited in the GenBank sequence database under the accession number JX839965. Complete nucleotide sequences of CVA16 isolate were compared with the reference strains by using CLUSTAL W (version 1.81) [19]. The phylogenetic relationships of each virus isolate were inferred using free MEGA software version 5.05 (Available from www.megasoft.net). The maximum composite likelihood method was used as the substitution method, while the neighbor-joining method was used to reconstruct the phylogenetic tree [20]. The reliability of the phylogenetic tree was determined by bootstrap resampling of 1000 replicates.
2.3. High-throughput screening of medicinal plant extracts for antiviral activity against CVA16
A medicinal plant extract library was supplied by the Ginseng Research Division, National Institute of Horticultural and Herbal Science, Eumseong-gun, Chungcheongbuk-do, Korea. To screen 492 medicinal plant extracts, Vero cells were seeded in 96-well plates. After CVA16 infection, the plates were incubated with individual plant extract at 10 μg/mL for 48 hours. The antiviral activity was measured using a sulforhodamine B (SRB) assay.
2.4. Antiviral activity and cytotoxicity assays
Antiviral activity and cytotoxicity were evaluated by the SRB method using CPE reduction recently reported [21]. The effect of Cornus officinalis, Acer triflorum, Pulsatilla koreana, and Clematis heracleifolia var. davidiana extracts on CVA16-induced CPE was observed. Briefly, Vero cells were seeded onto a 96-well culture plate at a concentration of 2 × 104 cells/well. Next day, the medium was removed and washed with phosphate-buffered saline. Then, 0.09 mL of diluted virus suspension and 0.01 mL of the medium supplemented with FBS containing 0.4 μg/mL, 2 μg/mL, 10 μg/mL, and 50 μg/mL C. officinalis, A. triflorum, P. koreana, or C. heracleifolia var. davidiana Hemsl were added. After incubation at 37°C in 5% CO2 for 2 days, cell morphology was microscopically observed at 4 × 10 magnification (Axiovert10, Zeiss, Germany), and images were recorded. Cytotoxicity was measured by the antiviral activity assay as described above. To calculate the concentration required to reduce cell growth by 50% (CC50) values, the data were expressed as percentages relative to controls, and CC50 values were obtained from the resulting dose-response curves.
2.5. Real-time reverse transcription-PCR
Total RNA was isolated from Vero cells. Reverse transcription was performed using SuperScript II Reverse Transcriptase (Invitrogen, Grand Island, NY, USA) according to the manufacturer's instructions. For real-time PCR analysis, the cDNA was serially diluted 10-fold and amplified using the 7500 Real Time PCR System (Applied Biosystems, Grand Island, NY, USA) with Power SYBR Green PCR Master Mix (Applied Biosystems). After the first denaturation step (95°C for 10 minutes), amplification was performed for 40 cycles at 95°C for 15 seconds, 55°C for 15 seconds, and 72°C for 40 seconds. The final cycle was followed by the dissociation stage. The following primers were used: 5′ NCR gene, 5′-CCG GCC CCT GAA TGC GG-3′ and 5′-ATT CTT TAA TTG TCA CCA TAA GCA GCC A-3′ and β-actin gene, 5′-CAA TCA TGA AGT GTG ACG TGG-3′ and 5′-GTC CGC CTA GAA GCA TTT GCG-3′.
2.6. Statistical analyses
Differences across more than three groups are analyzed using one-way Analysis of Variance (ANOVA) (Graphpad P, version 5.01). All results were expressed as means ± standard deviation. Significant differences in direct comparisons were determined using Tukey's post hoc test. Differences with p < 0.05 were considered statistically significant.
3. Results
3.1. Analysis of nucleotide sequence of Korean CVA 16
The complete sequences of the Korean CVA16 isolate are deposited in the GenBank sequence database under the accession number JX839965. The genome is 7,411 nt in length, excluding the poly(A) tail. The 5′NCR contains 745 nt, followed by an ORF that encodes a viral polyprotein consisting of 2,194 codons, between a start codon (AUG) at position 746 and a stop codon (UGA) at position 7,327. The 3′NCR is 84 nt in length. These nucleotide sequences were used to construct a phylogenetic tree with 39 reference strains of the same serotype extracted from GenBank database. The CVA16 strains including Korean CVA16 isolate were segregated into four distinct genetic groups, which were supported by high bootstrap values (Figure 1). In phylogenetic relationships, the Korean CVA16 isolate belonged to cluster B-1 and was closely related to strain PM-15765-00 isolated in Malaysia in 2000. The Korean CVA16 isolate showed 73.2% nucleotide identity to the G10 prototype strain and 98.7% nucleotide identity to the PM-15765-00 strain.
Figure 1.
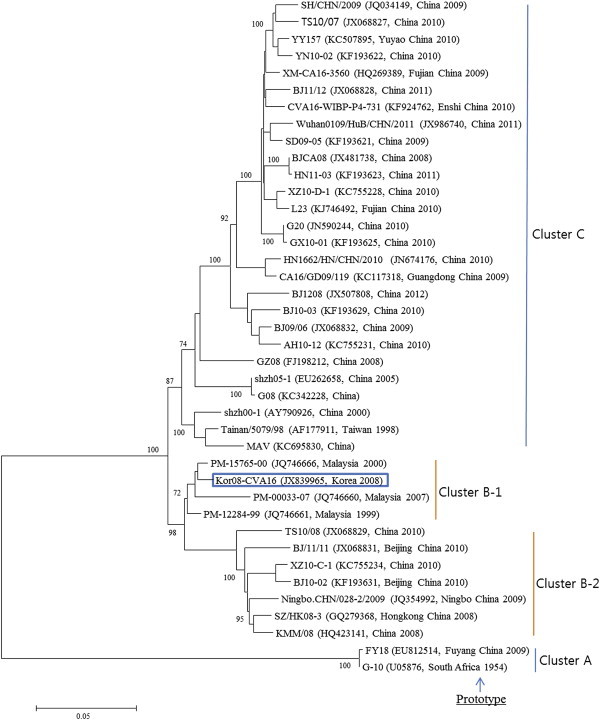
Phylogenetic analysis based on full-length genomic sequences of a CVA16 isolate from a patient with hand, foot, and mouth disease. Nucleotide sequences were analyzed by the neighbor-joining method. The numbers at the branches indicate the bootstrap values for 1000.
3.2. Identification of antiviral activity of four medicinal plant extract against CVA16
We sought to identify the antiviral activity of medicinal plant extracts against CVA16. Four extracts were identified from 492 medicinal plant extracts screened that showed significant cell viability of > 50%, indicating antiviral activity against CVA16 (data not shown). The four extracts are C. officinalis, A. triflorum, P. koreana, and C. heracleifolia var. davidiana Hemsl.
3.3. Antiviral activity of four medicinal plant extracts against CVA16
To determine drug potency, CVA16-infected Vero cells were treated with various doses of the four medicinal plant extracts. The antiviral assays demonstrated that P. koreana and C. heracleifolia var. davidiana Hemsl possessed strong antiviral activity of > 60% against CVA16 at concentrations of 2 μg/mL, 10 μg/mL, and 50 μg/mL. C. officinalis and A. triflorum showed 80% and 70% antiviral activity, respectively, against CVA16 at the concentration of 50 μg/mL (Figure 2A). The values of concentration required to inhibit virus-induced cytopathic effects by 50% were 32.9 μg/mL, 32.3 μg/mL, 1.51 μg/mL, and 2.55 μg/mL for C. officinalis, A. triflorum, P. koreana, and C. heracleifolia var. davidiana Hemsl extract, respectively. The CC50 values of C. officinalis, A. triflorum, P. koreana, and C. heracleifolia var. davidiana Hemsl extracts were superior to 50 μg/mL (Table 1). In addition, we confirmed the antiviral activity of the four medicinal plant extracts against CVA16 by real-time PCR analysis. The highest inhibition of CVA16 replication was observed at 50 μg/mL for the C. officinalis and A. triflorum extracts, while the P. koreana and C. heracleifolia var. davidiana Hemsl extracts inhibited CVA16 replication only marginally at all concentrations (Figure 2B).
Figure 2.
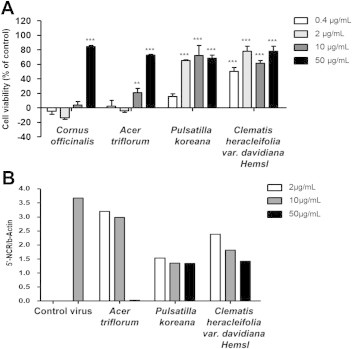
Antiviral activity of medicinal plant extracts against CVA16 in Vero cells. (A) Vero cells were infected with CVA16 and treated with the indicated concentrations (0.4–50 μg/mL) of medicinal plant extracts for 48 hours. Antiviral activity was assessed using a cytopathic effects reduction assay. Results are presented as the mean percentage values obtained from three independent experiments carried out in triplicate ± standard deviation. (B) Vero cells were infected with CVA16 and treated with various concentrations of Cornus officinalis, Acer triflorum, Pulsatilla koreana, and Clematis heracleifolia var. davidiana Hemsl for 48 hours. CVA16 5′NCR genes were quantitatively measured using real-time reverse transcription polymerase chain reaction.
Table 1.
Antiviral activity of four medicinal plant extracts against CVA16.
| Medicinal plant extract | Coxsackievirus A16 |
||
|---|---|---|---|
| CC50 | IC50 | TI | |
| Cornus officinalis | > 50 | 32.9 ± 1.30 | 1.52 |
| Acer triflorum | > 50 | 32.3 ± 1.67 | 1.55 |
| Pulsatilla koreana | > 50 | 1.51 ± 0.05 | 33.11 |
| Clematis heracleifolia var. davidiana Hemsl | > 50 | 2.55 ± 1.06 | 19.61 |
Results are presented as the mean IC50 values ± standard deviation obtained from three independent experiments carried out in triplicate.
CC50 = concentration required to reduce cell growth by 50% (μg/mL); IC50 = concentration required to inhibit virus-induced cytopathic effects by 50% (μg/mL); TI = therapeutic index = CC50/IC50.
3.4. The effect of four medicinal plant extracts on CVA16 virus-induced CPE
After Day 2 post infection with CVA16, there was no difference between mock or Vero cells treated with 50 μg/mL of C. officinalis, A. triflorum, P. koreana, or C. heracleifolia var. davidiana Hemsl, in terms of typical spread-out shapes and normal morphology. Infection of Vero cells with CVA16 resulted in a severe CPE, whereas CVA16-infected Vero cells treated with the medicinal plant extracts exhibited noticeably reduced CPE compared with untreated CVA16-infected cells (Figure 3). These results indicate that the CPE of CVA16 infection is prevented by C. officinalis, A. triflorum, P. koreana, or C. heracleifolia var. davidiana Hemsl.
Figure 3.
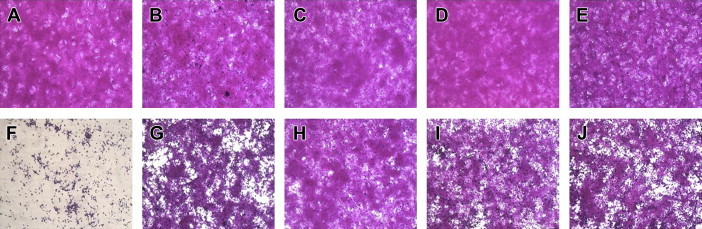
Morphological assessment of CVA16-infected Vero cells following treatment with four medicinal plant extracts. CVA16-infected and uninfected Vero cells were treated with 50 μg/mL medicinal plant extracts. After staining of viable cells with sulforhodamine B, cell morphology was assessed by microscopy. (A) Untreated, (B) uninfected cells treated with Cornus officinalis, (C) Acer triflorum, (D) Pulsatilla koreana, (E) Clematis heracleifolia var. davidiana Hemsl, (F) untreated CVA16-infected cells, (G) CVA16-infected cells treated with Cornus officinalis, (H) CVa16-infected cells treated with Acer triflorum, (I) CVA16-infected cells treated with Pulsatilla koreana, and (J) CVA16-infected cells treated with Clematis heracleifolia var. davidiana Hemsl.
4. Discussion
CVA16 and EV71 infections are both responsible for widespread HFMD and present serious public health problems in the Asia–Pacific region [22,23]. However, there are no available therapeutics to treat CVA16 or EV71 infections effectively. Most of antiviral and vaccine studies have been focusing on EV71, since it was reported to be associated with severe complications involving neuropathological complications and significant mortality [24,25]. However, evidence has shown that CVA16 infection also causes severe neurological complications and death [26,27], and HFMD caused by CVA16 is now considered as a serious public health threat in young children, especially in the Asia–Pacific region. In the current study, the first complete nucleotide sequence of a CVA16 isolate from a HFMD patient in Korea was described and its genetic diversity explored by phylogenetic analysis against 39 reference strains. In addition, a Korean medicinal plant library of 492 extracts was screened to find candidates with antiviral properties against the Korean CVA16 isolate.
CVA16 usually has a high mutation rate during viral replication due to the deficiency of proofreading activity [28], and there was > 20% genetic difference between G10 prototype strain and the current widespread strain. Therefore, it would be difficult to develop a therapeutic antiviral drug against the current epidemic strain [29]. In the current study, we isolated a CVA16 from Korean HFMD patients, and confirmed that the CVA16 isolate could be used for development and screening of antiviral drugs in vitro.
Many plant-derived natural products have been used in traditional medicine for the treatment of various diseases, including viral infection. In the current study, we found that CVA16 infection in Vero cells could be prevented by extract of C. officinalis, A. triflorum, P. koreana, or C. heracleifolia var. davidiana Hemsl. Among them, C. officinalis is a species of dogwood and a known edible plant. Particularly, the fruit of C. officinalis is well known for its chemotherapeutic benefits. Recently, cornuside, isolated from the fruit of C. officinalis, was reported to have anti-inflammatory activity by inhibiting tumor necrosis factor-α production [30]. Cornuside has also been reported to inhibit lipopolysaccharide-induced inflammation by inhibiting nuclear factor-κB pathway [31]. Next, P. koreana is a perennial plant found in South Korea, and its roots have been used as traditional medicine for the treatment of dysentery, malaria, chills, and fever [32]. In addition, previous studies suggested that it has antifungal and antibiotic properties and antitumor effects and that it can lower blood pressure [33,34]. Unlike C. officinalis and P. koreana, there are few reports regarding the biological activities or medical uses of A. triflorum and C. heracleifolia var. davidiana Hemsl. More interestingly, there are no reports suggesting an antiviral activity of the C. officinalis, A. triflorum, P. koreana, or C. heracleifolia var. davidiana Hemsl extracts, and thus, further studies to elucidate the antiviral constituents of those plants are needed.
In this study, the CC50 values of all extracts were > 50 μg/mL, suggesting that the extracts of those plants are nontoxic to Vero cells even in high concentrations. In addition, when their antiviral activities were assessed after CVA16 infection, all of the extracts showed significant antiviral activity against the Korean CVA16 isolate. In particular, C. officinalis and A. triflorum revealed high suppression potency against viral gene replication as assessed by real-time PCR, which correlates with their antiviral activity tested by SRB assay. Although the extracts of P. koreana and C. heracleifolia var. davidiana Hemsl also showed high antiviral activity, their suppressive ability on viral replication was moderate.
In conclusion, this study is the first report of the complete nucleotide sequence of the Korean CVA16, and shows the possibility of using the virus for the screening of antiviral drug candidates against HFDM infection in Korea. Antiviral activity screening of 492 medicinal plant extracts showed that C. officinalis, A. triflorum, P. koreana, and C. heracleifolia var. davidiana Hemsl possessed strong antiviral activity against this Korean CVA16 isolate. The data would be useful in preventing future outbreaks of CVA16 and in treating patients infected with the strain. Prospectively, identification of antiviral constituents included in the medicinal plant extracts and study of the mechanisms underlying their antiviral activity is necessary for the development of antiviral therapeutics to treat Korean HFDM patients infected with CVA16.
Conflicts of interest
All contributing authors declare no conflicts of interest.
Acknowledgments
This paper was supported by Daejeon Health Sciences College (grant No. 2013015) in 2013.
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Contributor Information
Kyungah Yoon, Email: artemis3993@gmail.com.
Hyun-Jeong Ko, Email: hjko@kangwon.ac.kr.
References
- 1.Bendig J.W., Fleming D.M. Epidemiological, virological, and clinical features of an epidemic of hand, foot, and mouth disease in England and Wales. Commun Dis Rep CDR Rev. 1996 May 24;6(6):R81–R86. [PubMed] [Google Scholar]
- 2.Chang L.Y., Huang Y.C., Lin T.Y. Fulminant neurogenic pulmonary oedema with hand, foot, and mouth disease. Lancet. 1998 Aug 1;352(9125):367–368. doi: 10.1016/S0140-6736(98)24031-1. [DOI] [PubMed] [Google Scholar]
- 3.Iwai M., Masaki A., Hasegawa S. Genetic changes of coxsackievirus A16 and enterovirus 71 isolated from hand, foot, and mouth disease patients in Toyama, Japan between 1981 and 2007. Jpn J Infect Dis. 2009 Jul;62(4):254–259. [PubMed] [Google Scholar]
- 4.Rabenau H.F., Richter M., Doerr H.W. Hand, foot and mouth disease: seroprevalence of Coxsackie A16 and Enterovirus 71 in Germany. Med Microbiol Immunol. 2010 Feb;199(1):45–51. doi: 10.1007/s00430-009-0133-6. [DOI] [PubMed] [Google Scholar]
- 5.Ang L.W., Koh B.K., Chan K.P. Epidemiology and control of hand, foot and mouth disease in Singapore, 2001-2007. Ann Acad Med Singapore. 2009 Feb;38(2):106–112. [PubMed] [Google Scholar]
- 6.Choi C.S., Choi Y.J., Choi U.Y. Clinical manifestations of CNS infections caused by enterovirus type 71. Korean J Pediatr. 2011 Jan;54(1):11–16. doi: 10.3345/kjp.2011.54.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park K., Lee B., Baek K. Enteroviruses isolated from herpangina and hand-foot-and-mouth disease in Korean children. Virol J. 2012 Sep 17;9:205. doi: 10.1186/1743-422X-9-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bawany F.I., Khan M.S., Khan A. Circulating endothelial cells: a new predictor of myocardial infarction. J Pak Med Assoc. 2014 Jul;64(7):855. [PubMed] [Google Scholar]
- 9.Chen C.W., Lee Y.P., Wang Y.F. Formaldehyde-inactivated human enterovirus 71 vaccine is compatible for co-immunization with a commercial pentavalent vaccine. Vaccine. 2011 Mar 24;29(15):2772–2776. doi: 10.1016/j.vaccine.2011.01.094. [DOI] [PubMed] [Google Scholar]
- 10.Chou A.H., Liu C.C., Chang J.Y. Formalin-inactivated EV71 vaccine candidate induced cross-neutralizing antibody against subgenotypes B1, B4, B5 and C4A in adult volunteers. PLoS One. 2013 Nov 21;8(11):e79783. doi: 10.1371/journal.pone.0079783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.JX1 Li, Mao Q.Y., Liang Z.L. Development of enterovirus 71 vaccines: from the lab bench to Phase III clinical trials. Expert Rev Vaccines. 2014 May;13(5):609–618. doi: 10.1586/14760584.2014.897617. [DOI] [PubMed] [Google Scholar]
- 12.Carrasco L. Picornavirus inhibitors. Pharmacol Ther. 1994;64(2):215–290. doi: 10.1016/0163-7258(94)90040-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rotbart H.A. Treatment of picornavirus infections. Antiviral Res. 2002 Feb;53(2):83–98. doi: 10.1016/s0166-3542(01)00206-6. [DOI] [PubMed] [Google Scholar]
- 14.Briskin D.P. Medicinal plants and phytomedicines. Linking plant biochemistry and physiology to human health. Plant Physiol. 2000 Oct;124(2):507–514. doi: 10.1104/pp.124.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jassim S.A., Naji M.A. Novel antiviral agents: a medicinal plant perspective. J Appl Microbiol. 2003;95(3):412–427. doi: 10.1046/j.1365-2672.2003.02026.x. [DOI] [PubMed] [Google Scholar]
- 16.Oberste M.S., Maher K., Kilpatrick D.R. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol. 1999 Mar;73(3):1941–1948. doi: 10.1128/jvi.73.3.1941-1948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sickles G.M., Mutterer M., Feorino P. Recently classified types of Coxsackie virus, group A; behavior in tissue culture. Proc Soc Exp Biol Med. 1955 Nov;90(2):529–531. doi: 10.3181/00379727-90-22088. [DOI] [PubMed] [Google Scholar]
- 18.Pöyry T., Hyypiä T., Horsnell C. Molecular analysis of coxsackievirus A16 reveals a new genetic group of enteroviruses. Virology. 1994 Aug 1;202(2):982–987. doi: 10.1006/viro.1994.1423. [DOI] [PubMed] [Google Scholar]
- 19.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994 Nov 11;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura K., Dudley J., Nei M. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007 Aug;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 21.Song J., Yeo S.G., Hong E.H. Antiviral activity of Hederasaponin B from Hedera helix against enterovirus 71 subgenotypes C3 and C4a. Biomol Ther (Seoul) 2014 Jan;22(1):41–46. doi: 10.4062/biomolther.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho M., Chen E.R., Hsu K.H. An epidemic of enterovirus 71 infection in Taiwan. Taiwan Enterovirus Epidemic Working Group. N Engl J Med. 1999 Sep 23;341(13):929–935. doi: 10.1056/NEJM199909233411301. [DOI] [PubMed] [Google Scholar]
- 23.Yang F., Zhang T., Hu Y. Survey of enterovirus infections from hand, foot and mouth disease outbreak in China, 2009. Virol J. 2011 Nov;6(8):508. doi: 10.1186/1743-422X-8-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S.C., Chang H.L., Yan T.R. An eight-year study of epidemiologic features of enterovirus 71 infection in Taiwan. Am J Trop Med Hyg. 2007 Jul;77(1):188–191. [PubMed] [Google Scholar]
- 25.Qiu J. Enterovirus 71 infection: a new threat to global public health? Lancet Neurol. 2008 Oct;7(10):868–869. doi: 10.1016/S1474-4422(08)70207-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C.Y., Li Lu F., Wu M.H. Fatal coxsackievirus A16 infection. Pediatr Infect Dis J. 2004 Mar;23(3):275–276. doi: 10.1097/01.inf.0000115950.63906.78. [DOI] [PubMed] [Google Scholar]
- 27.Yen F.B., Chang L.Y., Kao C.L. Coxsackieviruses infection in northern Taiwan—epidemiology and clinical characteristics. J Microbiol Immunol Infect. 2009 Feb;42(1):38–46. [PubMed] [Google Scholar]
- 28.Drake J.W. Rates of spontaneous mutation among RNA viruses. Proc Natl Acad Sci U S A. 1993 May 1;90(9):4171–4175. doi: 10.1073/pnas.90.9.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park K., Song J., Baek K. Genetic diversity of a Korean echovirus 5 isolate and response of the strain to five antiviral drugs. Virol J. 2011 Feb 24;8:79. doi: 10.1186/1743-422X-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang D.G., Moon M.K., Lee A.S. Cornuside suppresses cytokine-induced proinflammatory and adhesion molecules in the human umbilical vein endothelial cells. Biol Pharm Bull. 2007 Sep;30(9):1796–1799. doi: 10.1248/bpb.30.1796. [DOI] [PubMed] [Google Scholar]
- 31.Choi Y.H., Jin G.Y., Li G.Z. Cornuside suppresses lipopolysaccharide-induced inflammatory mediators by inhibiting nuclear factor-kappa B activation in RAW 264.7 macrophages. Biol Pharm Bull. 2011;34(7):959–966. doi: 10.1248/bpb.34.959. [DOI] [PubMed] [Google Scholar]
- 32.Kim Y., Bang S.C., Lee J.H. Pulsatilla saponin D: the antitumor principle from Pulsatilla koreana. Arch Pharm Res. 2004 Sep;27(9):915–918. doi: 10.1007/BF02975843. [DOI] [PubMed] [Google Scholar]
- 33.Martín M.L., San Román L., Domínguez A. In vitro activity of protoanemonin, an antifungal agent. Planta Med. 1990 Feb;56(1):66–69. doi: 10.1055/s-2006-960886. [DOI] [PubMed] [Google Scholar]
- 34.Kim Y., Kim S.B., You Y.J. Deoxypodophyllotoxin; the cytotoxic and antiangiogenic component from Pulsatilla koreana. Planta Med. 2002 Mar;68(3):271–274. doi: 10.1055/s-2002-23140. [DOI] [PubMed] [Google Scholar]


