Abstract
Objectives
Klebsiella pneumoniae is a gram-negative rod bacterium, a known cause of community-acquired bacterial pneumonia and is an important hospital-acquired pathogen that causes severe morbidity and mortality. The aim of this study was to identify the TEM and SHV genes in K. pneumoniae isolated from cockroaches obtained from hospitals.
Methods
In this study, 250 cockroaches were collected from different hospitals in the province of Chaharmahal Va Bakhtiari, which is located in southwest Iran. The samples were examined for the presence of K. pneumoniae by plating onto a combination of culture media, and the antimicrobial susceptibility patterns of isolated K. pneumoniae from samples were evaluated using the disk diffusion test. In addition, from the culture, genomic bacterial DNA was extracted, and sequence-specific targets (TEM and SHV genes) were amplified using the polymerase chain reaction (PCR) method.
Results
Out of 250 cockroach samples collected from various hospitals, 179 samples (71.60%) were positive for K. pneumoniae. PCR reaction was performed using specific oligonucleotide primers (TEM-F, TEM-R and SHV-F, SHV-R) for the amplification of each gene, and amplified products were visualized on 1% agarose gel electrophoresis. Of all the specimens amplified by PCR in this research, 32 samples (17.87%) were positive for TEM and 15 samples (8.37%) were positive for SHV.
Conclusion
Detection of TEM and SHV genes using molecular methods and their pattern of antimicrobial resistance can provide useful information about the epidemiology of and risk factors associated with K. pneumoniae infection.
Keywords: cockroaches, Klebsiella pneumoniae, PCR, SHV, TEM
1. Introduction
Studies done in the 19th century have proven that insects have an important role in the transmission of diseases to humans, one of which are cockroaches [1]. Most cockroaches are infected with pathogenic bacteria causing leprosy, dysentery, bubonic plague, pimples, abscesses, and food poisoning. Cockroaches also have a symbiotic relationship with more than 100 bacteria species, 60 yeast species, 90 protozoa species, and 45 parasite worm species [2]. Important carriers of bacteria include the German cockroach (Blattella germanica) and the American cockroach (Periplaneta americana). German cockroaches, one of the most commonplace pests that carry pathogenic bacteria, have been reported many years ago [3]. The bacteria contaminant could be from air, water, food, or contact with vectors harboring the pathogens. Cockroaches stay in filthy environments in households, shops, and even hospitals where both clinical and environmental samples are kept [4,5].
Moreover, the medical importance of cockroaches is rather substantial. They are among the medically important pests in urban environments that cause serious public health problems, and they have been especially associated with an outbreak of dysentery [6]. Most pathogenic bacteria have been recently isolated from cockroaches, such as Salmonella spp., Campylobacter spp., Shigella spp., Pseudomonas aeruginosa, and K. pneumoniae [7,8].
Klebsiella was first discovered by Karl Friedlander at 1882 [9]. Klebsiella consists of seven types, of which K. oxytoca and K. rhinoscleromatis have been demonstrated in human clinical specimens. K. pneumoniae is a gram-negative, rod-shaped, lactose-fermenting, nonmotile, and encapsulated bacillus of the Enterobacteriaceae family [10,11]. It is an opportunistic pathogen found in water, soil, and plants; it also exists as a normal flora in mucosal surfaces such as the intestines, pharynx, mouth, and skin in mammals. This bacterium was also recognized as a community-acquired pulmonary pathogen, mainly among patients with a history of chronic alcoholism [10,12,13]. In humans, K. pneumoniae can colonize the gastrointestinal tract, bowel, bladder, skin, or pharynx, which may cause various clinical outcomes, including pneumonia, thrombophlebitis, bacteremia, and urinary tract infection [13]. Almost 160 variants of the TEM-1 and TEM-2 penicillinases have been described, many of which exhibit activity against extended-spectrum cephalosporins [14]. Class A enzymes are mainly plasmid encoded, and the first to be described at amino-acid sequence level were the enzymes TEM-1 and TEM-2 [15].
The SHV family has been derived from Klebsiella spp. SHV-1 is universally found in K. pneumonia [16], evolved as a chromosomal gene in Klebsiella spp., and was later incorporated into a plasmid, which has spread to other enterobacterial species [17]. A total of 40 types of SHV have already been reported [18].
In this research, we identified the presence of TEM and SHV genes in K. pneumoniae isolated from cockroaches from Shahrekord hospitals using the polymerase chain reaction (PCR) technique and their antibiotic resistance pattern.
2. Material and methods
2.1. Sampling
Cockroaches were randomly collected for 6 months (January–June 2013), from six hospitals located in Chaharmahal Va Bakhtiari province (Kashani Shahrekord, Imam Ali Farrokhshahr, Shohada Farsan, Imam Reza Lordegan, and Valiasr Boroujen). A total of 250 cockroaches were collected from different parts of the hospitals by handlers who wore sterile gloves. Live cockroach specimens were immediately carried to the Biotechnology Research Center of the Islamic Azad University and killed using chloroform-soaked cotton. The tubes containing the samples were filled with 70% ethanol for 5 minutes to decontaminate their external surface and then allowed to air dry. Next, cockroaches were washed with sterile normal saline to remove the residue ethanol. Finally, their viscera were removed with sterile forceps under a dissecting microscope, and the instruments were sterilized after every dissection. Cockroach gut was kept in 2 mL sterile normal saline for 5 minutes to produce a homogenate specimen.
2.2. Bacterial isolation
After the samples were isolated and placed in test tubes containing normal saline, the gut homogenate was kept in buffered peptone water, after which the samples were inoculated on blood agar and Mac-Conkey agar (MCA), then incubated for 24 hours at 37°C. For the growth of gram-negative bacteria such as K. pneumonia, BPW was used that was inoculated in seven primary media (Sheep Blood Agar, Chocolate Agar, Mac-Conkey, Deoxycholate Citrate Agar, SS agar, Mannitol Salt Agar, and Xylose Lysine Deoxycholate). In addition, identification of gram-negative bacteria was achieved by use of standard methods (API System; bioMerieux, Marcy-l'Étoile, France). The biochemical reagents and test used to identify K. pneumoniae included triple sugar iron agar, simmons citrate, indole, urease, motility, and H2S. The biochemical characteristics of K. pneumoniae identified were as follows: positive citrate utilization test, negative methyl red test, negative indole test, positive urease test, positive Voges–Proskauer test, sucrose, acid and abundant gas production from glucose, lactose, mannitol sugar fermentation tests, and maltose.
2.3. Picking up the cultivation of susceptibility
Antimicrobial susceptibility profiles were determined using the dilution method on Mueller–Hinton agar, according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI). The antimicrobial agents tested included ceftazidime, nalidixic acid, cefixime, gentamicin, amikacin, cephalexin, and imipenem. The CLSI breakpoints were used for the interpretation of minimum inhibitory concentrations (CLSI 2012). The results were interpreted after 24 hours of incubation at 37°C, as sensitive, intermediately sensitive, and resistant according to the zone diameter around each antibiotic disk.
2.4. DNA extraction and PCR
K. pneumoniae genomic DNA was extracted using the DNA extraction kit (DNP Kit; CinnaGen, Tehran, Iran) according to the manufacturer's recommendation. The extracted DNA was quantified with spectrophotometric measurement at a wavelength of 260 nm according to the method described by Sambrook and Russell [19].
To detect the TEM and SHV genes, PCR reactions were performed in a total volume of 25 μL containing 2 μL of the DNA sample, 1 μM of each primers, 2 mM Magnesium chloride (MgCl2), 5 μL of 10 × PCR buffer AMS, 200μM dNTPs, and 1 unit of Taq DNA polymerase (CinnaGen Co., Tehran, Iran). The PCR assay was performed at 95°C for 5 minutes and then for 32 cycles of 94°C for 1 minute, 58°C for 40 seconds, 72°C for 40 seconds, and a final extension at 72°C for 5 minutes, with a final hold at 10°C in a thermal cycler (Mastercycler gradient; Eppendorf, Hamburg, Germany). For SHV amplifications, conditions for thermal cycling remained the same except for the annealing temperature (55°C). The primer sequences for TEM and SHV are shown in Table 1.
Table 1.
Primers for identification of Klebsiella pneumoniae TEM and SHV genes.
| Gene target | Primer name | Sequence | Annealing temperature | Amplicon size (bp) |
|---|---|---|---|---|
| TEM | TEM-F TEM-R |
5′-TCCGCTCATGAGACAATAACC-3′ 3′-ATAATACCGCACCACATAGCAG-5′ |
58 | 296 |
| SHV | SHV-F SHV-R |
5′-TACCATGAGCGATAACAGCG-3′ 3′-GATTTGCTGATTTCGCTCGG-5′ |
60 | 450 |
The amplified products were run on 1% agarose gel and staining with ethidium bromide (0.5 mg/ml) in a dark room. The electrode buffer used was Tris-borate-EDTA (TBE), which consists of Tris-base 10.8 g 89 mM, boric acid 5.5 g 2 mM, EDTA (pH 8.0) 4 mL of 0.5 M EDTA (pH 8.0) (all components were combined in sufficient H2O and stirred to dissolve). A 100-bp ladder molecular weight marker (Roche, New Jersey, USA) was used to measure the molecular weight of the amplified products. Aliquots (14 μL) of PCR products were applied to the gel. A constant voltage of 84 V for 20 minutes was used for product separation. The images of ethidium bromide stained DNA bands were digitized using a UVItec documentation system (UVItec, Paisley, UK).
2.5. Statistical analysis
All data were analyzed by using MS Excel 2007 and SPSS software (Version 17, SPSS Inc., Chicago, USA), and the p value was calculated using Chi-square and Fisher's exact tests to find any significant relationship. A p value of <0.05 was considered statistically significant.
3. Results
The quality of extracted DNA from samples was examined by electrophoretic analysis through a 1% agarose gel. Out of 250 cockroach samples collected from various Iranian hospitals, 179 samples (71.60%) were infected with K. pneumoniae. The TEM and SHV genes were successfully amplified with the TEM-F and TEM-R and SHV-F and SHV-R primers. The agarose gel electrophoresis of the PCR-amplified products for TEM and SHV genes is shown in Figures 1 and 2, respectively. From the samples that were assayed by PCR in this research, only 32 samples (17.87%) were positive for the TEM gene and 15 samples (8.37%) were positive for the SHV gene (Table 2). Moreover, Table 3 shows the antimicrobial susceptibility pattern of K. pneumoniae TEM and SHV genes. The overall susceptibility of isolated K. pneumoniae strains to antimicrobial agents was 64.3% for nalidixic acid, 65.4% for cephalexin, 69.8% for cefixime, 73.50% for gentamicin, 83.2% for ceftazidime, 85.1% for amikacin, and 100% for imipenem. According to these results, imipenem, amikacin, and ceftazidime were the most effective agents against isolated K. pneumoniae.
Figure 1.
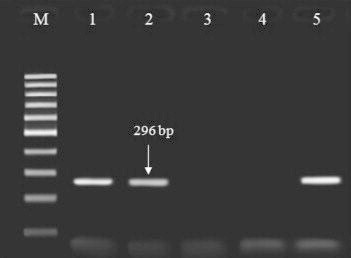
Agarose gel electrophoresis of the products amplified with polymerase chain reaction (PCR) using the specific primers for Klebsiella pneumoniae TEM gene samples. Lane M: 100-bp DNA ladder (Fermentase, Leon-Rot, Germany); lanes 1, 2, and 5: PCR products of the positive samples; lanes 3 and 4: negative samples.
Figure 2.
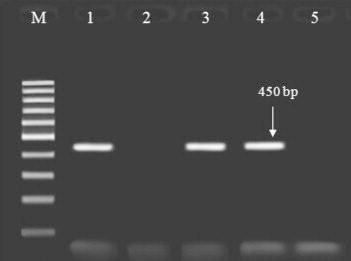
Agarose gel electrophoresis of the products amplified with polymerase chain reaction (PCR) using the specific primers for K. pneumoniae SHV gene samples. Lane M: 100-bp DNA ladder (Fermentase, Leon-Rot, Germany); lanes 1, 3 and 4: PCR products of the positive samples; lanes 2 and 5: negative samples.
Table 2.
Number of K. pneumoniae TEM and SHV genes.
| Hospitals | Number of samples | Positive for Klebsiella | Positive for TEM gene | Positive for SHV gene |
|---|---|---|---|---|
| Imam Reza Lordegan | 60 | 39 | 9 | 0 |
| Valiasr Boroujen | 60 | 52 | 11 | 7 |
| Shohada Farsan | 40 | 19 | 0 | 2 |
| Imam Ali Farokhshahr | 30 | 21 | 3 | 1 |
| Kashani Shahrekord | 60 | 48 | 9 | 5 |
| Total | 250 | 179 | 32 | 15 |
Table 3.
Antibiotic resistance patterns of K. pneumoniae TEM and SHV genes.
| Antimicrobial agent | Sensitive | Half-resistant | Resistant | Total |
|---|---|---|---|---|
| Nalidixic acid | 64.30 | 0 | 35.70 | 100 |
| Cephalexin | 65.40 | 2.9 | 31.70 | 100 |
| Cefixime | 69.80 | 0 | 30.20 | 100 |
| Gentamicin | 73.50 | 0 | 26.50 | 100 |
| Ceftazidime | 83.20 | 1.80 | 15 | 100 |
| Amikacin | 85.10 | 6.30 | 8.60 | 100 |
| Imipenem | 100 | 0 | 0 | 100 |
4. Discussion
Klebsiella consists of seven types—K. pneumoniae, K. planticola, K. terrigena, K. rhinoscleromatis, K. ozaenae, K. ornithinolytica, and K. oxytoca—and two of these species, K. oxytoca and K. rhinoscleromatis, have been demonstrated in human clinical specimens [10,11]. K. pneumoniae, one of the most common causes of gram-negative sepsis, usually inhabits the human and animal intestinal tract [10,20]. Some gram-negative bacilli such as K. pneumoniae and Escherichia coli strains of particular types produce β-lactamases. The Enterobacteriaceae family produces β-lactamases, which are encoded by plasmids. Among the most important β-lactamases are SHV and TEM. For the first time, TEM-1 from a blood culture (Temonera) in Greece was isolated from E. coli [21,22]. One of the first reported β-lactamases is TEM-1, which is encoded by plasmids; however, other bacteria such as Vibrio cholerae and Haemophilus strains of P. aeruginosa and Neisseria also have the ability to produce it. Today, reports indicate the prevalence of TEM-1 β-lactamase in certain parts of the world, suggesting that this type of β-lactamase is a global problem [23,24]. According to a 2011 study by Shebani et al [24], out of 70 samples of E. coli, 27 (38.58%) were ESBL (extended spectrum beta-lactamase) positive and 43 samples (61.42%) were ESBL negative, and that there were only 10 samples (37.04%) with TEM-1 β-lactamase gene and 17 samples (62.96%) were free of the TEM-1 β-lactamase gene [24]. TEM has been responsible for several unrelated outbreaks in the United States [25] and a recently reported outbreak in Europe with the same frequency [26]. A study from Korea revealed that SHV is the most common ESBL found in Korea [27]. SHV (especially SHV-5) is commonly encountered and reported worldwide [27,28]. In another study in India, the reported frequency of ESBL-producing Klebsiella spp. was between 6% and 87% [29–31].
In conclusion, our results suggest that the PCR method can be used for the specific, rapid, simple, and highly sensitive detection of K. pneumoniae in samples. Furthermore, isolated K. pneumoniae from cockroaches collected from various hospitals has more resistance to nalidixic acid. Imipenem seems to be the only antimicrobial agent that showed 100% sensitivity and may be used as the drug of choice for this type of infection.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
This research was supported by the Research Deputy of Islamic Azad University, Shahrekord. Thanks all the staff members of the Biotechnology Research Center of Islamic Azad University of Shahrekord Branch for their support in this research.
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- 1.Heidari Soureshjani E., Doosti A. Examination methicillin-resistant Staphylococcus aureus (MRSA) prevalence in cockroaches from hospital in Chaharmahal-va-Bakhtiari province, Iran by polymerase chain reaction (PCR) Acad J. 2013 July;5(7):324–329. [Google Scholar]
- 2.Nejati J., Keyhani A., Moosa-Kazemi S.H. Cockroaches' bacterial infections in wards of hospitals, Hamedan city, west of Iran. Asian Pac J Trop Dis. 2012 Oct;2(5):381–384. [Google Scholar]
- 3.Birkneh T., Bogale W., Erdaw T. High load of multi-drug resistant nosocomial neonatal pathogens carried by cockroaches in a neonatal intensive care unit at Tikur Anbessa specialized hospital, Addis Ababa, Ethiopia. Antimicrob Resist Infect Control. 2012 Mar;1(12):1–7. doi: 10.1186/2047-2994-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fortedor R., Shriniwas U.B., Verma A. Cockroaches (Blattella germanica) as carriers of microorganisms of medical importance in hospitals. Epidemiol Infect. 1991 Aug;107(1):181–187. doi: 10.1017/s0950268800048809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bala A.Y., Sule H. Vectorial potential of cockroaches in transmitting parasites of medical importance in Arkilla, Sokoto, Nigeria. Nig J Basic Appl Sci. 2012 Jun;20(2):111–115. [Google Scholar]
- 6.Tachbele E., Erku W., Gebre-Michael T. Cockroach-associated food-borne bacterial pathogens from some hospitals and restaurants in Addis Ababa, Ethiopia: distribution and antibiograms. J Rural Trop Public Health. 2006;5:34–41. [Google Scholar]
- 7.Salehzadeh A., Tavacol P., Mahjub H. Bacterial, fungal and parasitic contamination of cockroaches in public hospitals of Hamadan. Iran J Vector Borne Dis. 2007 Jun;44(2):105–110. [PubMed] [Google Scholar]
- 8.Cotton M.F., Wasserman E., Pieper C.H. Invasive disease due to extended spectrum beta-lactamase producing Klebsiella pneumoniae in a neonatal unit: the possible role of cockroaches. J Hosp Infect. 2000 Jan;44(1):13–17. doi: 10.1053/jhin.1999.0650. [DOI] [PubMed] [Google Scholar]
- 9.Lawlor M.S., Hsu J., Rick P.D. Identification of Klebsiella pneumoniae virulence determinants using an intranasal infection model. Mol Microbiol. 2005 Nov;58(4):1054–1073. doi: 10.1111/j.1365-2958.2005.04918.x. [DOI] [PubMed] [Google Scholar]
- 10.Mun Teng W., Chee Sian K., Siang Ling C. Structural modeling and biochemical characterization of recombinant KPN_02809, a zinc-dependent metalloprotease from Klebsiella pneumoniae MGH 78578. Int J Mol Sci. 2012 Nov;13:901–917. doi: 10.3390/ijms13010901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mungloo-Rujubali S.K., Issack M.I., Jaufeerally-Fakim Y. Study of Klebsiella pneumoniae isolates with ESBL activity, from ICU and nurseries, on the island of Mauritius. J Cell Mol Biol. 2013 Aug;10(1):39–51. [Google Scholar]
- 12.Wilksch J.J., Yang Ji, Clements A. MrkH, a novel c-di GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLOS Pathog. 2012 Aug;7(8):1–22. doi: 10.1371/journal.ppat.1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao F., Bai J., Zhou M. Sequencing and genetic variation of multidrug resistance plasmids in Klebsiella pneumoniae. PLoS ONE. 2010 Apr;5(4):1–9. doi: 10.1371/journal.pone.0010141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mulvey M.R., Boyd D.A. TEM-168, a heretofore laboratory-derived TEM β-lactamase variant found in an Escherichia coli clinical isolate. Antimicrob Agents Chemother. 2009 Nov;53(11):4955–4956. doi: 10.1128/AAC.00547-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Black M.T., Stachyra T., Coleman K. Mechanistic studies of the inactivation of TEM-1 and P99 by NXL104, a novel non-β-lactam β-lactamase inhibitor. Antimicrob Agents Chemother. 2010 Dec;54(12):5132–5180. doi: 10.1128/AAC.00568-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sturenburg E., Mack D. Extended spectrum beta-lactamases: implications for the clinical microbiology laboratory, therapy, and infection control. J Infect. 2003 Nov;47(4):273–295. doi: 10.1016/s0163-4453(03)00096-3. [DOI] [PubMed] [Google Scholar]
- 17.Livermore D.M. β-lactamase in laboratory and clinical resistance. Clin Microbiol Rev. 1995 Oct;8(4):557–584. doi: 10.1128/cmr.8.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain A., Mondal R. TEM & SHV genes in extended spectrum β-lactamase producing Klebsiella species & their antimicrobial resistance pattern. Indian J Med Res. 2008 Dec;128(6):759–764. [PubMed] [Google Scholar]
- 19.Sambrook J., Russell D.W. 3rd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, New York; 2001. Molecular cloning: a laboratory manual; p. 517. [Google Scholar]
- 20.Achouit A., Vogl T., Florquin S. Myeloid-related protein-14 contributes to protective immunity in gram-negative pneumonia derived sepsis. PLOS Pathog. 2012;8(10):1–14. doi: 10.1371/journal.ppat.1002987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medeiros A.A. Beta-lactamases. Br Med Bull. 1984 Jan;40(1):18–27. doi: 10.1093/oxfordjournals.bmb.a071942. [DOI] [PubMed] [Google Scholar]
- 22.Shojapour M., Shariati L., Karimi A. Prevalence of TEM-1 type beta-lactmase genes in Pseudomonas aeruginosa strains isolated from burn infections using duplex PCR in Shahrekord 2008. Arak Med Univ J. 2011;14(54):55–61. [Google Scholar]
- 23.Jajić-Bencic I., Bedenić B., Mikoc A. Characterization of extended-spectrum beta-lactamases in Enterobacteriaceae causing nosocomial infections in a Zagreb University Hospital. J Chemother. 2009 Jun;21(3):282–289. doi: 10.1179/joc.2009.21.3.282. [DOI] [PubMed] [Google Scholar]
- 24.Shebani AA, Ebrahimi Varkiyani M, Aghaei SS, et al. Prevalence of genes TEM-1 strains of E. coli isolated from clinical specimens city of Damghan. J Microb Biotechnol Res Islamic Azad Univ 1390;3(11):15–22.
- 25.Bradford P.A. Extended spectrum beta-lactamases in 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001;14(4):933–951. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barroso H., Viera H.A., Lito L.M. Survey of K. pneumonia producing extended spectrum beta-lactamase at a Portuguese hospital: TEM-10 as the endemic enzyme. J Antimicrob Chemother. 2000 May;45(5):611–616. doi: 10.1093/jac/45.5.611. [DOI] [PubMed] [Google Scholar]
- 27.Kim J., Kwon H., Pai J. Survey of K. pneumoniae strains producing extended spectrum betalactamase: prevalence of SHV-12 and SHV-2 in Korea. J Clin Microbiol. 1998;36:1446–1449. doi: 10.1128/jcm.36.5.1446-1449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jain A., Mondal R. Prevalence and antimicrobial resistant pattern of extended spectrum β-lactamase producing Klebsiella spp. isolated from cases of neonatal septisaemia. Indian J Med Res. 2007 Jan;125(1):89–94. [PubMed] [Google Scholar]
- 29.Jain A., Roy I., Gupta M.K. Prevalence of extended spectrum beta-lactamase producing Gram negative bacteria in septicemic neonates in a tertiary care hospital. J Med Microbiol. 2003 May;52(Pt5):421–425. doi: 10.1099/jmm.0.04966-0. [DOI] [PubMed] [Google Scholar]
- 30.Mathur P., Kapil A., Das B. Prevalence of extended spectrum beta-lactamase producing Gram-negative bacteria in a tertiary care hospital. Indian J Med Res. 2005 Apr;115:153–157. [PubMed] [Google Scholar]
- 31.Hansotia J.B., Agarwal V., Pathak A.A. Extended spectrum beta-lactamase mediate resistance to third generation cephalosporins in Klebsiella pneumoniae in Nagpur, central India. Indian J Med Res. 1997 Apr;105:158–161. [PubMed] [Google Scholar]


