Abstract
Objectives
Endosulfan is a lipophilic insecticide, which causes severe health issues due to its environmental stability, toxicity, and biological reservation in organisms. It is found in the atmosphere, soil, sediments, surface waters, rain, and food in almost equal proportions. The aim of this study was to isolate and identify endosulfan-degrading bacteria from the Kor River and evaluate the possibility of applying bioremediation in reducing environmental pollution in the desired region.
Methods
Samples of surface sediments and water were collected from three different stations in two seasons (summer and autumn), as these are areas with high agricultural activity. Isolated bacteria were identified by various biochemical tests and morphological characteristics. The amounts of degradation of endosulfan isomers and metabolites produced as a result of biodegradation were then analyzed using gas chromatography/mass spectrometry.
Results
In this study, the following five bacterial genera were able to degrade endosulfan: Klebsiella, Acinetobacter, Alcaligenes, Flavobacterium, and Bacillus. During biodegradation, metabolites of endosulfan diol, endosulfan lactone, and endosulfan ether were also produced, but these had lesser toxicity compared with the original compound (i.e., endosulfan).
Conclusion
The five genera isolated can be used as a biocatalyst for bioremediation of endosulfan.
Keywords: acinetobacter, endosulfan, gas chromatography/mass spectrometry, Klebsiella, Kor River
1. Introduction
During the past 50 years, pesticides have been the essential part of the agricultural world. Although the demand for production and distribution of pesticides to increase the quality and efficiency of the agricultural industry is evident, use of improper and unreasonable pesticides is likely [1]. Despite their benefits, pesticides are compounds that may have toxic side effects, causing potential environmental risk [2]. Endosulfan is an organochlorine pesticide, which belongs to the family of polycyclic chlorinated hydrocarbons. For more than 30 years, it has been used extensively in agriculture, horticulture, and forestry [3]. Endosulfan contains two stereoisomers [alfa- and beta-endosulfan (ratio, 3:7)] and has been registered under the trade names Thiodan, Cyclodan, Thimol, Thiofar, and Malix [4]. Endosulfan contamination and its persistence in soil and water environments cause it to accumulate in cells of phytoplankton products, zooplankton, fishes, and vegetables [5]. Endosulfan persists in soil and water for 3–6 months or longer [6]. Endosulfan attaches to gamma-aminobutyric acid receptors located on the membrane of neurons and reduces flow of chloride. Endosulfan poisoning causes seizures. All of these aforementioned problems encouraged the scientific community to develop biological methods to remove endosulfan instead of incineration and landfill methods [7].
Biodegradation is an efficient bioremediation technique in microorganisms that grow in different ecosystems and through symbiosis with xenobiotics, these microorganisms are able to survive even in incompatible conditions. Various studies have used endosulfan as a source of sulfur for microbial growth and as a carbon resource in bioremediation. Endosulfan is decomposed into endosulfan sulfate by oxidation pathway and into endosulfan diol by hydrolysis. Endosulfan sulfate is also toxic and stable as the major component (endosulfan). Endosulfan diol can be converted to endosulfan ether, endosulfan hydroxyl ether, endosulfan dialdehyde, and endosulfan lactone. However, these metabolites are less toxic [8]. There are many reports on the degradation of endosulfan by bacteria [9]. Klebsiella pneumoniae has the capability to biologically degrade (biodegradation) endosulfan. Pandoraea sp. degrades around 95–100% of alfa- and beta-endosulfan without producing endosulfan sulfate when incubated for 18 days. Klebsiella oxytoca was reported to degrade 145–260 mg of endosulfan in 6 days [6]. Some Gram-positive bacteria such as Bacillus are able to convert endosulfan to endosulfan sulfate [10]. Natural adsorbents such as sediments can control water quality indirectly by absorbing or releasing pesticides. Surface waters are good environments for degrading pesticides, particularly when microorganisms are capable of binding to the surface contour of the water, sediment, rocks, and plants. Many types of compounds decompose slowly in aerobic regions [11].
Kor River is one of the largest surface water resources in the Fars Province, with thousands of farmers depending on its water for agricultural use. In addition, the river provides a high percentage of drinking water to the regions of Shiraz, Marvdasht, and other villages along its way. The river also provides water to industries and factories located nearby. Urban, industrial, and agricultural activities are the main causes of pollution of this river. This water is mainly polluted by the Fars meat industrial complex, petrochemical industries, sugar mills, refineries, glazed tile factories, industrial towns, and wastewater of Marvdasht [12]. Urban and industrial wastewaters have adverse effects on fish breeding, environmental health, and particularly on drinking water of downstream residents. Thus, protecting the river is particularly important. The aim of this study was to isolate and identify endosulfan-degrading bacteria from the Kor River and also to evaluate the use of bioremediation techniques to improve the environmental status of this river.
2. Materials and methods
2.1. Sampling range
The study site is located in Fars Province in the southwestern region of Iran. Because of intense agricultural activities in the surrounding Kor River, three sampling stations were chosen.
2.2. Sampling method
Water and sediment samples were collected from areas with high agricultural activities at three different stations (3 times in each station) in two seasons [summer and autumn (fall)]. The samples were packaged in sterile plastic containers and flasks filled with ice and were transported to the laboratory within 4 hours [13].
2.3. Counting the bacterial colonies
Laboratory chemical manufactured by Merck Company (Darmstadt, Germany) were used in this study. After transporting the soil samples to the laboratory, bacteria were counted using the total viable plate count method. During the procedure, sediment and water samples were diluted with normal saline (from 10−1 to 10−9). Then 0.1 mL of each dilution was taken and surface cultured on two medium of nutrient agar: one containing the toxin and the other without toxin (control). The cultures were incubated for 48 hours at 37°C. Once the colonies appear, plates with identifiable and countable colonies were selected and the number of colonies was counted [14].
A mixture of different culture media, such as the mineral broth (1 g KH2PO4, 1 g K2HPO4, 1 g of NH4NO3, 0.2 g MgSO4, 0.02 g CaCl2, and 0.01 g FeSO4), deionized water, nutrient agar, and biochemical medium were used in the experiments [6].
2.4. Enrichment and isolation of endosulfan-degrading bacteria
To enrich endosulfan-degrading bacteria, 5 g of sediment of each station was added to an Erlenmeyer flask containing 50 mL of mineral medium and 50 μg/mL of endosulfan pesticide. The mixture was then incubated in a shaking incubator at 30°C at 150 rpm. After 10 days, 5 mL of the mixture from each flask was added to a fresh medium containing 50 mL of mineral medium and 50 μg/mL of endosulfan pesticide, and then incubated in a shaker incubator as described earlier. Then 0.1 mL of enrichment culture was cultured on agar medium. Pure colonies were obtained by streak cultures several times [6].
2.5. Identification of endosulfan-degrading bacteria
For identification of endosulfan-degrading bacteria, Gram staining, shape of the colony, and movement of bacteria were analyzed. In addition, various biochemical tests such as the production of acid from carbohydrates, urea breath test, gelatin hydrolysis, indole production, citrate consumption, methyl red, Voges Proskauer, oxidative-fermentative (OF), starch hydrolysis, reduction of nitrate to nitrite, catalase, and oxidase were used [6].
2.6. Biodegradation
The bacteria detected were grown in a mineral medium containing endosulfan (50 μg/mL) for 1 week. The endosulfan were then extracted by solid-phase extraction with methanol. The extracted samples were analyzed by gas chromatography/mass spectrometry (GC/MS), HP5840 series with a 30-m capillary column (DB 1-MS) using a mass-selective detector. The temperature settings were as follows: initial temperature, 120°C; rate, 20°C per minute; Step 2 temperature, 200°C; rate, 50°C per minute; final temperature, 270°C; injector temperature, 280°C; and interface temperature, 300°C.
The amount of biodegradation of alfa- and beta-endosulfan isomers was measured by GC/MS analysis. The metabolites produced during biodegradation were also identified and measured [2,15].
2.7. Statistical analysis
The obtained data were analyzed using analysis of variance and SPSS software version 15 (SPSS Inc., Chicago, IL, USA).
3. Results
3.1. Endosulfan-degrading bacteria
In this study, the following five bacterial genera were identified: Klebsiella, Acinetobacter, Alcaligenes, Bacillus, and Flavobacterium. Figure 1 shows the frequency of bacteria identified in summer and autumn.
Figure 1.
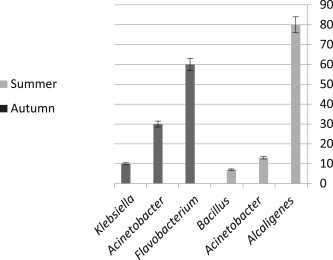
Average frequency of bacterial species isolated from different stations in summer and autumn.
3.2. Bacterial counts
There were differences between stations in terms of bacterial counts. The highest count of bacteria in the summer was found in Station 3 (8.082 CFU/mL; Figure 2). The lowest count of bacteria in the autumn was seen in Station 1 (6.033 CFU/mL; Figure 3). All of the stations showed significant difference at the 5% level. Based on the results, it was observed that there is significant difference between the station with and without endosulfan at the 5% level.
Figure 2.

Average of the logarithm of bacterial counts in three separate stations with and without endosulfan in summer.
Figure 3.
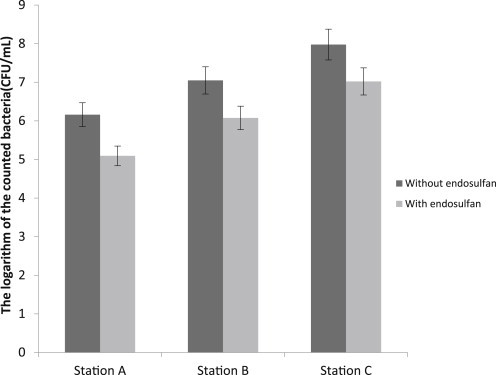
Average of the logarithm of bacterial counts in three separate stations with endosulfan and without endosulfan in autumn.
3.3. Biodegradation
Our experimental results show that alfa- and beta-endosulfan were degraded by bacteria at different levels. During the biodegradation process, metabolites of endosulfan diol, endosulfan ether, and endosulfan lactone have been produced, all of which have less toxicity than the original compound (i.e., endosulfan). The initial and remaining values (ppb) of the metabolites were measured. Figure 4 shows the GC/MS chromatogram of standard endosulfan.
Figure 4.
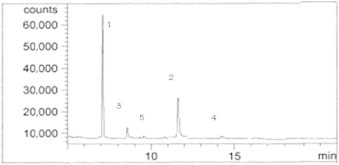
Gas chromatography/mass spectrometry chromatogram of standard endosulfan. Metabolite 1 = alfa-endosulfan; Metabolite 2 = beta-endosulfan; Metabolite 3 = endosulfan diol; Metabolite 4 = endosulfan lactone; Metabolite 5 = endosulfan ether.
According to the chromatogram obtained from the GC/MS system, the bacteria with the highest capacity to degrade endosulfan was Klebsiella. This bacterium brooks majority of endosulfan just after 1 week of incubation. It was observed that Klebsiella degrades 90% of alfa-endosulfan and 85% of beta-endosulfan. During the biodegradation process, 0.33 × 10−6 ppb (55%) endosulfan diol, 0.09 × 10−6 ppb (15%) endosulfan lactone, and 0.0 3 × 10−6 ppb (5%) endosulfan ether were produced (Figure 5).
Figure 5.
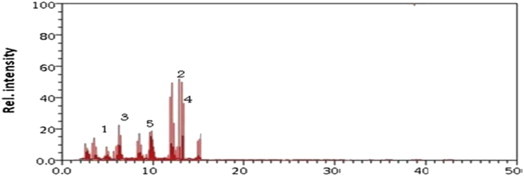
Gas chromatography/mass spectrometry chromatogram of Klebsiella. Metabolite 1 = alfa-endosulfan; Metabolite 2 = beta-endosulfan; Metabolite 3 = endosulfan diol; Metabolite 4 = endosulfan lactone; Metabolite 5 = endosulfan ether.
Acinetobacter bacterium also showed good degradation capability. This bacterium degraded 90% of the alfa-endosulfan and 90% of the beta-endosulfan. The metabolites produced contained 0.30 × 10−6 ppb (45%) endosulfan diol, 0.11 × 10−6 ppb (15%) endosulfan lactone, and 0.15 × 10−6 ppb (20%) endosulfan ether (Figure 6).
Figure 6.
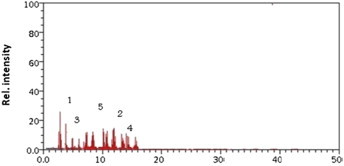
Gas chromatography/mass spectrometry chromatogram of the Acinetobacter. Metabolite 1 = alfa-endosulfan; Metabolite 2 = beta-endosulfan; Metabolite 3 = endosulfan diol; Metabolite 4 = endosulfan lactone; Metabolite 5 = endosulfan ether.
Alcaligenes was another good endosulfan-degrading bacterium, which was capable of degrading 88% of alfa-endosulfan and 87% of the beta-endosulfan, respectively. Upon degradation of endosulfan stereoisomers, this bacterium produces the metabolites of endosulfan diol [0.375 × 10−6 ppb (50%)], endosulfan lactone [0.06 × 10−6 ppb (8%)], and endosulfan ether [0.127 × 10−6 ppb (15%); Figure 7].
Figure 7.
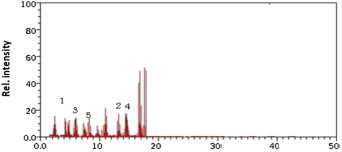
Gas chromatography/mass spectrometry chromatogram of the Alcaligenes. Metabolite 1 = alfa-endosulfan; Metabolite 2 = beta-endosulfan; Metabolite 3 = endosulfan diol; Metabolite 4 = endosulfan lactone; Metabolite 5 = endosulfan ether.
Flavobacterium degraded 75% of alfa-endosulfan and 95% of beta-endosulfan, respectively. This bacterium produced 0.4 × 10−6 ppb (40%) endosulfan diol, 0.15 × 10−6 ppb (15%) endosulfan lactone, and 0.15 × 10−6 ppb (15%) endosulfan ether (Figure 8).
Figure 8.
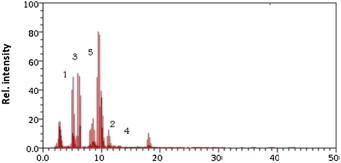
Gas chromatography/mass spectrometry chromatogram of Flavobacterium. Metabolite 1 = alfa-endosulfan; Metabolite 2 = beta-endosulfan; Metabolite 3 = endosulfan diol; Metabolite 4 = endosulfan lactone; Metabolite 5 = endosulfan ether.
Bacillus showed the minimum degrading capability and degrades 75% alfa-endosulfan and 80% beta-endosulfan. The metabolites produced contained 0.4 × 10−6 ppb (5%) endosulfan diol, 2.6 × 10−6 ppb (30%) endosulfan lactone, and 1.7 × 10−6 ppb endosulfan ether (Figure 9).
Figure 9.
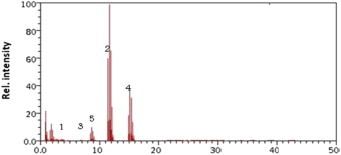
Gas chromatography/mass spectrometry chromatogram of the Bacillus.
4. Discussion
Biodegradation is a method of removing contaminants from water, and this is a natural process. Microorganisms survive by decomposing a xenobiotic or pesticide. Most of these microbes live in natural environments, but it is possible to change and strengthen them to decompose pesticides with greater speed. This ability of microbes can be used as a technology to remove contaminants. In addition, because using chemical methods to remove contaminants such as pesticide are very expensive, with advances in technology, the use of microorganisms for this purpose has been suggested. Different genera of bacteria have been isolated that could degrade the pesticide endosulfan. Because using microorganisms is an efficient and affordable method, many recent studies have focused on biodegradation of pesticides with microorganisms. The biodegradation process has also been observed in areas where these pesticides are used.
Many studies have been conducted around the world to identify pollutants in water/soil as well as to identify microorganisms that have the ability to degrade pesticides. These studies have evaluated the ability of various types of fungi and bacteria in biodegradation [16].
In a study conducted by Goswami and Singh [6], degradation of endosulfan and its metabolites was evaluated by GC/MS analysis. In this study, the main metabolite produced by Bordetella sp. B9 was endosulfan ether and endosulfan lactone, respectively. No endosulfan sulfate residual was detected. Endosulfan ether concentration after 6 days of incubation was 0.53% ± 0.2%, which declined to 0.41 ± 0.13 after the 18th day. Endosulfan lactone concentration after 6 days was 0.24 ± 0.09%, which increased to 0.35 ± 0.07% after 18 days. The study indicated that the Bordetella sp. degraded 80% alfa-endosulfan and 86% beta-endosulfan after 18 days of incubation [6].
Siddique et al [17] found that Pandoraea sp. is able to degrade 95–100% of the alfa- and beta-endosulfan after 18 days of incubation with the initial endosulfan concentration of 100 mg/mL, with no endosulfan sulfate produced during biodegradation.
Li et al [5] found that the Achromobacter xylosoxidans strain CS5 is able to use endosulfan as a source of carbon, sulfur, and energy. This study proves that CS5 can degrade > 24.8 mg/L of alfa-endosulfan and > 10.5 mg/L of beta-endosulfan in an aqueous environment after 8 days. Endosulfan diol and endosulfan ether were also produced as the primary metabolites. Their results suggested that the metabolism of endosulfan by the CS5 strain was accompanied by a significant reduction in the toxicity.
In another study, Kumar et al [18] examined degradation of endosulfan by a mixed bacterial culture containing Stenotrophomonas maltophilia and Rhodococcus erythropolis in soils contaminated with pesticides. After 2 weeks of incubation, the bacterial culture was able to degrade 73% and 81% of the alfa- and beta-endosulfan, respectively. Intermediate metabolites known as endodiol were produced during the biodegradation process. S. maltophilia demonstrated more degradation than R. erythropolis.
In 2006, a mixed culture of bacteria containing Staphylococcus, Bacillus circulance I, and B. circulance II was isolated from soil contaminated with endosulfan. After 3 weeks of incubation, the bacteria were able to decompose 71.58 ± 0.2% of alfa-endosulfan and 75.88 ± 0.2% beta-endosulfan under aerobic and anaerobic conditions. In addition, no intermediate metabolites were detected. The results showed that bacterial mixed cultures used can be applied to treat soil and water contaminated with endosulfan [19].
Bajaj et al [10] isolated and identified Pseudomonas sp. strain IITR01, which has the ability to degrade alfa-endosulfan. This strain quickly degrades endosulfan sulfate and converts it into the less toxic metabolites such as diol, ether, and lactone. In their study, the appropriate amount of lactone was prepared. The GC/MS analysis revealed degradation of alfa-endosulfan and endosulfan sulfate as well as production of endosulfan diol, endosulfan ether, and endosulfan lactone [10].
Xie et al [20] reported that bacteria can strengthen the production of endosulfan diol in a short period compared with fungi. It has also been reported that Pseudomonas aeruginosa degrades > 85% of the alfa- and beta-endosulfan after 16 days of incubation [16].
In a study by Kumar et al [21], endosulfan-degrading bacteria were isolated from soil contaminated with pesticides. These bacteria included Ochrobactrum, Burkholderia, Pseudomonas alcaligenes, Pseudomonas sp., and Arthrobacter sp. All cells of P. alcaligenes and Pseudomonas sp. absorbed 89% and 94% of alfa-endosulfan and 89% and 86% of beta-endosulfan, respectively. Endosulfan sulfate and a small amount of endosulfan diol were produced during biodegradation by Pseudomonas sp. By contrast, P. alcaligenes just generated endosulfan diol, indicating that there were no oxidation. Thus, in the case of P. alcaligenes, hydrolyzation is the only mechanism of endosulfan degradation. Whereas in the case of Pseudomonas sp., there is oxidation in addition to hydrolyzation. Based on these results, the bacteria can be used in various technologies for removing endosulfan or endosulfan sulfate from contaminated areas [21].
In 2007, in one study, 29 bacterial strains were isolated from soil contaminated with endosulfan, of which Pseudomonas spinosa, P. aeruginosa, and Burkholderia degraded bacteria faster and were able to degrade 90% of alfa- and beta-endosulfan [22].
In our study, which was carried out on water and sediments contaminated with the pesticide endosulfan, Klebsiella was the most powerful isolated strain, which could decrease the initial alfa-endosulfan concentration of 3.5 × 10−4 ppb to 0.06 × 10−6 ppb and the initial beta-endosulfan of 1.5 × 10−4 ppb to 0.09 × 10−6 ppb within a week, and therefore 90% of the alfa-endosulfan and 90% of the beta-endosulfan were degraded. In this study, endosulfan sulfate was not produced. The result of this study is parallel with results of Kwon et al [23] who found that Klebsiella sp. biologically degrades 81.72% of endosulfan after 10 days of incubation, and during this process endosulfan sulfate, which is a toxic metabolite of endosulfan, was not produced.
In this study, Alcaligenes, Acinetobacter, and Flavobacterium were reported to biodegrade the pesticide endosulfan in different regions of the Kor River, which was not previously reported.
According to Sutherland et al [24], degradation of endosulfan can be achieved by oxidation and hydrolysis pathways, and the toxic metabolite endosulfan sulfate along with other less toxic metabolites can be produced [24].
Bacterial culture causes rapid degradation of alfa- and beta-endosulfan isomers. The rate of degradation of isomers is comparable. Degradation of both isomers is associated with the production of metabolites of endosulfan sulfate, endosulfan diol, endosulfan lactone, endosulfan ether, and unknown metabolites [25].
In this study, endosulfan diol, endosulfan ether, and endosulfan lactone were metabolites obtained by degradation of alfa- and beta-endosulfan isomers, among which endosulfan diol was the greatest and most original metabolite. These are consistent with the results reported by Cotham and Bidleman [26] that endosulfan diol is produced by microorganisms in water and sediments [26]. Awasthi et al [25] also found that endosulfan diol is the major metabolite of endosulfan degradation.
Differences in the rate of degradation between alfa and beta isomers can be related to differences in rate and low solubility in the liquid medium. This is especially true because a concentration of 10 mg/L endosulfan is four times greater than its solubility in water. Degradation of endosulfan isomers is associated with the production of metabolites. Production of endosulfan ether has also been reported by other bioremediation studies. When oxidation occurs in a metabolic pathway, endosulfan sulfate is produced. In addition, by hydrolysis, both endosulfan diol and endosulfan ether are finally produced [8].
Furthermore, differences in the degradation of alfa and beta isomers may be due to stereo-isomerization because enzymes released from the bacterial system may correspond to only one of the stereoisomers [18]. As a result, endosulfan sulfate was not produced during the process of biodegradation in this study, with the predominant biodegradation route being hydrolysis.
Results of this study showed that endosulfan-degrading bacteria are widely distributed across various regions of the Kor River. A review of the previous studies and the results of this study showed that bioremediation can reduce endosulfan contamination in this river.
Conflicts of interest
The authors do not have any conflicts of interest.
Acknowledgments
The authors express their appreciation to the vice chancellor of Islamic Azad University, Jahrom Branch for the administrative support.
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- 1.Oliveira-Silva J.J., Alves S.R., Meyer A. Influence of socioeconomic factors on the pesticides poisoning, Brazil. Rev Saude Publica. 2001 Apr;35(2):130–135. doi: 10.1590/s0034-89102001000200005. [DOI] [PubMed] [Google Scholar]
- 2.Golfinopoulos S.K., Nikolaou A.D., Kostopoulou M.N. Organochlorine pesticides in the surface waters of Northern Greece. Chemosphere. 2003 Jan;50(4):507–516. doi: 10.1016/s0045-6535(02)00480-0. [DOI] [PubMed] [Google Scholar]
- 3.Daniel C.S., Agarwal S., Agarwal S.S. Human red blood cell membrane damage by endosulfan. Toxicol Lett. 1986 Jul–Aug;32(1–2):113–118. doi: 10.1016/0378-4274(86)90056-1. [DOI] [PubMed] [Google Scholar]
- 4.Peñuela G.A., Barceló D. Application of C18 disks followed by gas chromatography 18 techniques to degradation kinetics, stability and monitoring of endosulfan in water. J Chromatogr A. 1998 Jan;795(1):93–104. doi: 10.1016/s0021-9673(97)01068-6. [DOI] [PubMed] [Google Scholar]
- 5.Li W., Dai Y., Xue B. Biodegradation and detoxification of endosulfan in aqueous medium and soil by Achromobacter xylosoxidans strain CS5. J Hazard Mater. 2009 Aug;167(1–3):209–216. doi: 10.1016/j.jhazmat.2008.12.111. [DOI] [PubMed] [Google Scholar]
- 6.Goswami S., Singh D.K. Biodegradation of alpha and beta endosulfan in broth medium and soil microcosm by bacterial strain Bordetella sp. B9. Biodegradation. 2009 Apr;20(2):199–207. doi: 10.1007/s10532-008-9213-3. [DOI] [PubMed] [Google Scholar]
- 7.Soto A.M., Sonnenschein C., Chung K.L. The E-SCREEN assay as a tool to identify estrogens: an update on estrogenic environmental pollutants. Environ Health Perspect. 1995 Oct;103(Suppl. 7):113–122. doi: 10.1289/ehp.95103s7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castillo J.M., Casas J., Romero E. Isolation of an endosulfan-degrading bacterium from a coffee farm soil: persistence and inhibitory effect on its biological functions. Sci Total Environ. 2011 Dec;412–413:20–27. doi: 10.1016/j.scitotenv.2011.09.062. [DOI] [PubMed] [Google Scholar]
- 9.Kataoka R., Takagi K., Sakakibara F. A new endosulfan-degrading fungus, Mortierella species, isolated from a soil contaminated with organochlorine pesticides. J Pestic Sci. 2010 Sep;35(3):326–332. [Google Scholar]
- 10.Bajaj A., Pathak A., Mudiam M.R. Isolation and characterization of a Pseudomonas sp. strain IITR01 capable of degrading α-endosulfan and endosulfan sulfate. J Appl Microbiol. 2010 Dec;109(6):2135–2143. doi: 10.1111/j.1365-2672.2010.04845.x. [DOI] [PubMed] [Google Scholar]
- 11.Lal R., Saxena D.M. Accumulation, metabolism, and effects of organochlorine insecticides on microorganisms. Microbiol Rev. 1982 Mar;46(1):95–127. doi: 10.1128/mr.46.1.95-127.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mirzaei N., Kafilzadeh F., Kargar M. Isolation and identification of mercury resistant bacteria from Kor River. Iran J Biol Sci. 2008 July;8(5):935–939. [Google Scholar]
- 13.Regional Organization for the Protection of the Marine Environment (ROPME) 1999. Manual of oceanographic and observations pollutant analysis methods (MOOPAM) pp. 257–342. Safat, Kuwait: ROPME. [Google Scholar]
- 14.Udeani T.K.C., Obroh A.A., Okwuosa C.N. Isolation of bacteria from mechanic workshop soil environment contaminated with used engine oil. Afr J Biotechnol. 2009 Nov;8(22):6301–6303. [Google Scholar]
- 15.Kavoura O., Katsiris N., Zervas G. Proceedings of the 9th International Conference on Environmental Science and Technology. 2005 Sep. Determination of pesticides in drinking water using solid phase extraction GC/MS; pp. 1–3. Rhodes Island, Greece. [Google Scholar]
- 16.Singh D.K. Biodegradation and bioremediation of pesticide in soil: concept, method and recent developments. Indian J Microbiol. 2008 Mar;48(1):35–40. doi: 10.1007/s12088-008-0004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siddique T., Okeke B.C., Arshad M. Enrichment and isolation of endosulfan-degrading microorganisms. J Environ Qual. 2003 Jan–Feb;32(1):47–54. doi: 10.2134/jeq2003.4700. [DOI] [PubMed] [Google Scholar]
- 18.Kumar K., Devi S.S., Krishnamurthi K. Enrichment and isolation of endosulfan degrading and detoxifying bacteria. Chemosphere. 2007 Jun;68(2):317–322. doi: 10.1016/j.chemosphere.2006.12.076. [DOI] [PubMed] [Google Scholar]
- 19.Kumar M., Philip L. Adsorption and desorption characteristics of hydrophobic pesticide endosulfan in four Indian soils. Chemosphere. 2006 Feb;62(7):1064–1077. doi: 10.1016/j.chemosphere.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Xie H., Gao F., Tan W., Wang S.G. A short-term study on the interaction of bacteria, fungi and endosulfan in soil microcosm. Sci Total Environ. 2011 Dec;412–413:375–379. doi: 10.1016/j.scitotenv.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Kumar M., Lakshmi C.V., Khanna S. Biodegradation and bioremediation of endosulfan contaminated soil. Bioresour Technol. 2008 May;99(8):3116–3122. doi: 10.1016/j.biortech.2007.05.057. [DOI] [PubMed] [Google Scholar]
- 22.Jayashree R., Vasudevan N. Effect of Tween 80 added to the soil on the degradation of endosulfan by Pseudomonas aeruginosa. Int J Environ Sci Tech. 2007 Spring;4(2):203–210. [Google Scholar]
- 23.Kwon G.S., Sohn H.Y., Shin K.S. Biodegradation of the organochlorine insecticide, endosulfan, and the toxic metabolite, endosulfan sulfate, by Klebsiella oxytoca KE-8. Appl Microbiol Biotechnol. 2005 Jun;67(6):845–850. doi: 10.1007/s00253-004-1879-9. [DOI] [PubMed] [Google Scholar]
- 24.Sutherland T.D., Horne I., Lacey M.J. Enrichment of an endosulfan-degrading mixed bacterial culture. Appl Environ Microbiol. 2000 Jul;66(7):2822–2828. doi: 10.1128/aem.66.7.2822-2828.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Awasthi N., Singh A.K., Jain R.K. Degradation and detoxification of endosulfan isomers by a defined co-culture of two Bacillus strains. Appl Microbiol Biotechnol. 2003 Aug;62(2–3):279–283. doi: 10.1007/s00253-003-1241-7. [DOI] [PubMed] [Google Scholar]
- 26.Cotham W.E., Bidleman T.F. Degradation of malathion, endosulfan, and fenvalerate in seawater and seawater/sediment microcosms. J Agric Food Chem. 1989 May;37(3):824–828. [Google Scholar]


