Abstract
Inflammatory pathways are central mechanisms in diabetic kidney disease (DKD). Serum amyloid A (SAA) is increased by chronic inflammation, but SAA has not been previously evaluated as a potential DKD mediator. The aims of this study were to determine whether SAA is increased in human DKD and corresponding mouse models and to assess effects of SAA on podocyte inflammatory responses. SAA was increased in the plasma of people with DKD characterized by overt proteinuria and inversely correlated with estimated glomerular filtration rate (creatinine-based CKD-EPI). SAA was also elevated in plasma of diabetic mouse models including type 1 diabetes (streptozotocin/C57BL/6) and type 2 diabetes (BTBR-ob/ob). SAA mRNA (Nephromine) was increased in human DKD compared with non-diabetic and/or glomerular disease controls (glomerular fold change 1.5, P=0.017; tubulointerstitium fold change 1.4, P=0.021). The kidneys of both diabetic mouse models also demonstrated increased SAA mRNA (quantitative real-time PCR) expression compared with non-diabetic controls (type 1 diabetes fold change 2.9; type 2 diabetes fold change 42.5, P=0.009; interaction by model P=0.57). Humans with DKD and the diabetic mouse models exhibited extensive SAA protein deposition in the glomeruli and tubulointerstitium in similar patterns by immunohistochemistry. SAA localized within podocytes of diabetic mice. Podocytes exposed to advanced glycation end products, metabolic mediators of inflammation in diabetes, increased expression of SAA mRNA (fold change 15.3, P=0.004) and protein (fold change 38.4, P=0.014). Podocytes exposed to exogenous SAA increased NF-κB activity, and pathway array analysis revealed upregulation of mRNA for NF-κB-dependent targets comprising numerous inflammatory mediators, including SAA itself (fold change 17.0, P=0.006). Inhibition of NF-κB reduced these pro-inflammatory responses. In conclusion, SAA is increased in the blood and produced in the kidneys of people with DKD and corresponding diabetic mouse models. Podocytes are likely to be key responder cells to SAA-induced inflammation in the diabetic kidney. SAA is a compelling candidate for DKD therapeutic and biomarker discovery.
Diabetic kidney disease (DKD) is one of the most serious complications of diabetes because it is a leading cause of death, end-stage renal disease, and cardiovascular disease.1, 2, 3 There is urgent need for targeted therapies to improve clinical outcomes and for informative biomarkers to better identify patients at high risk for DKD progression. Inflammation is a central mechanistic pathway for DKD.4, 5 Yet, there is a wide gap in knowledge about specific mediators promoting these responses. In DKD, inflammation is well recognized to be triggered by advanced glycation end products (AGE), aberrant metabolic by-products of oxidative stress produced by high levels of glucose, amino acids, and/or lipids.6, 7 AGE bind the receptor for AGE (RAGE) to activate expression signals for numerous inflammatory genes.8, 9, 10
Serum amyloid A (SAA), a potent pro-inflammatory protein, increases by an AGE–RAGE interaction in podocytes, suggesting that it may promote glomerular inflammation.10 SAA is produced as distinct acute-phase isoforms classified as SAA1, 2 and 3.11, 12 Isoforms 1 and 2 have high sequence similarity in both mice (91% protein identity) and humans (93% protein identity). In mice, SAA is primarily expressed in tissues including the kidney as SAA3, while SAA1, 2 expression is limited in amount and distribution.13 To the contrary, SAA is predominantly co-expressed as isoforms 1 and 2 in human tissues, whereas the SAA3 gene is non-expressed.11, 12, 14, 15 Human SAA1, 2 is comparable to mouse SAA3 in structure, function, and distribution.11, 16, 17 Despite genetic similarities of SAA1, 2 between species, the amino acid compositions of human SAA1, 2 protein has greater identity to mouse SAA3 (73%) protein than mouse SAA1, 2 (68%) protein.11, 18 In addition to dissemination by the systemic circulation, human and mouse SAA isoforms are locally produced at inflammatory sites. For example, SAA mRNA expression is found in mouse adipocytes (SAA3) and in adipocytes of obese people, as well as in synovial fibroblasts of individuals with rheumatoid arthritis (SAA1, 2).19, 20 In human atherosclerosis, SAA1, 2 is expressed in the vascular wall, where it promotes inflammation and disrupts endothelial function.21, 22
The contribution of SAA to DKD has not been previously studied, even though diabetes is well known as a state of chronic inflammation. We recently discovered that AGE induces SAA production in cultured podocytes.10 As SAA is another ligand for RAGE, once it is upregulated SAA could perpetuate inflammation.23, 24, 25 The aims of this study were to determine whether SAA is increased in human DKD and corresponding mouse models and to assess effects of SAA on podocyte inflammatory responses.
MATERIALS AND METHODS
Human Studies of DKD
Plasma SAA protein levels
Plasma samples were obtained from participants in a clinical study of DKD in people with type 2 diabetes who provided written informed consent for use of their samples for future research. The parent study was reviewed and approved by the Institutional Review Board, Spokane, and its primary findings have been published.26 In brief, DKD was defined by the presence of overt proteinuria, characterized by urinary excretion of total protein >500 mg/day. Diabetic controls had urinary excretion of total protein <100 mg/day and normal controls had neither diabetes nor increased urinary excretion of total protein. The study entry criteria required that participants were free of other systemic diseases or chronic inflammatory conditions. Plasma samples were stored at −70 °C until assayed. SAA protein expression was measured by an ELISA kit for SAA1 (www.hycultbiotech.com).26 The estimated glomerular filtration rate (eGFR) based on serum creatinine was computed by the CKD-EPI equation.
Nephromine analysis of SAA mRNA expression in kidney tissue samples
SAA1, 2 mRNA expression (probe set 214456_x_at from Affymetrix array U133A) was evaluated in two human data sets, the Schmid and Woroniecka diabetes studies.27, 28 Total RNA was isolated from human kidney biopsies of glomerular and tubulointerstitial compartments in these studies. Standard histological measurements were used to define DKD.27, 28 Samples from individuals with DKD were compared with control samples, including tissue from healthy and deceased donors for kidney transplantation and biopsies from those with other proteinuric glomerular diseases (for example, minimal change disease). Gene expression profiles were generated using array analysis and have been made publically available by Nephromine (Compendia Bioscience, part of Life Technologies, Ann Arbor, MI). As the probe set designed for the SAA2 gene cross-reacts with SAA1, it provides a measure SAA 1, 2 mRNA (http://xavierlab2.mgh.harvard.edu/EnrichmentProfiler/primary/Expression/214456_x_at.html).
Kidney histology and SAA protein detection by immunohistochemistry
Kidney biopsy samples were obtained from banked tissue at the University of Washington. Samples of human kidneys were graded according to a recently published classification system for DKD.29 Samples from glomerular class IIa (type 1 diabetes) and glomerular class III–IV (type 2 diabetes) DKD and controls were studied (n=4–6 per diabetic group; n=6 normal controls: n=2 non-diabetic glomerular disease–monocolonal immunoglobulin deposition disease, MIDD). Class IIa included samples with mild-to-moderate mesangial expansion. Class III–IV exhibited extensive and nodular mesangial expansion.
Kidney tissue was immersion fixed in 10% buffered formalin, embedded in paraffin, sectioned, and stained with periodic acid–Schiff (PAS) using standard histologic techniques.30 Human kidney sections (4 μm) were subjected to antigen retrieval by heating in citrate-based unmasking solution followed by immunohistochemistry (IHC) procedures using a mouse monoclonal antibody to human amyloid A protein diluted 1:20 (MC1, Thermo Fisher Scientific, www.thermoscientific.com).31 Due to the high degree of identity between SAA isoforms 1 and 2 (93%), both isotypes are recognized by this antibody. Negative controls included substitution of the primary antibody with an irrelevant isotype-matched immunoglobulin and phosphate-buffered saline (PBS) alone. After overnight incubation, the primary antibody was detected using Impress HRP Reagent (Vector laboratories, www.vectorlabs.com), visualized with 3,3′ diaminobenzidine (DAB). Sections were counterstained with hematoxylin.
Studies in Mouse Models of Diabetes
Plasma SAA protein levels
The Animal Care Committee at the University of Washington approved experimental protocols used in the mouse studies. The models were of mild DKD in type 1 diabetes (C57BL/6 strain, 20 weeks after streptozotocin, and non-diabetic controls) and severe DKD in type 2 diabetes (BTBR-ob/ob strain and BTBR wild-type controls, 24 weeks old). The mild DKD in type 1 diabetes model developed abnormalities including reduced number of podocytes, mesangial expansion with no-to-minimal tubulointerstitial fibrosis, and moderately increased albuminuria.10 In contrast, the severe DKD in type 2 diabetes model showed extensive mesangial matrix expansion and mesangiolysis with moderate-to-severe tubulointerstitial fibrosis and severely increased albuminuria.30 Both models have also been shown to exhibit glomerular basement membrane thickening consistent with DKD.30, 32 Stored samples of plasma were obtained from mice in these studies.10, 30 Plasma was stored at −70 °C until assayed by ELISA for mouse SAA3 (Mouse Serum Amyloid A-3 kit, EZMSAA3–12 K, EMD Millipore, www.millipore.com).
SAA mRNA expression by quantitative real-time PCR in kidney tissue samples
Stored kidney tissues were procured from the same groups of mice.10, 30 Whole-kidney cortex was obtained from control and diabetic mice of both mouse models. Kidney cortex total RNA was isolated by the RiboPure kit (Ambion, www.lifetechnologies.com) and quantified using the Quant-iT RiboGreen RNA Reagent and kit (Invitrogen, www.lifetechnologies.com). Equal amounts of RNA were then DNAse treated using amplification-grade DNase I (Invitrogen). cDNA was synthesized using Superscript III (Invitrogen). Expression of the mouse SAA3 transcript was measured by quantitative real-time PCR (qRT-PCR) on an Applied Biosystems (www.lifetechnologies.com) 7900HT Fast RT-PCR System using SA Biosciences SYBR Green reagent (Qiagen, www.qiagen.com). Gene amplification results were quantified with SDS v2.4 software (Applied Biosystems) and normalized to mouse TATA-box binding protein, a housekeeping gene.
Kidney histology and SAA protein deposition by IHC
Collected kidneys from both mouse studies were perfused with PBS, immersion fixed in 10% neutral-buffered formalin, and embedded in paraffin. Sections were stained with silver methenamine and PAS for routine histology.10, 30
Rabbit anti-mouse SAA3 antibody was developed against a glutathione-S-transferase (GST)-C-terminal SAA3 fragment fusion protein.33 Anti-GST antibodies were removed by affinity chromatography with agarose-coupled GST (Thermo Fisher Scientific). SAA3 negative control antiserum was obtained by affinity absorption of SAA3 antibodies with agarose-coupled GST-C-terminal SAA3. Immunostaining for mouse SAA3 (1:2000 dilution) was performed on kidney sections after antigen retrieval.31 SAA3-absorbed antiserum recovered from the affinity column was diluted to match the protein concentration of anti-SAA3 antiserum. After overnight incubation, the primary antibody was detected using Impress HRP Reagent (Vector laboratories) and visualized with DAB. Sections were counterstained with hematoxylin.
SAA3 immunostaining abundance and intensity were assessed by a blinded observer on five sections per group and 10 randomly selected areas per section. Scoring of glomerular and tubulointerstitial compartments was based on staining of the area (0, 25, 50, 75, or 100%) and intensity (0—none; 1—light; 2—medium; and 3—dark). The immunostaining score was a product of area and intensity scores, averaged over the 10 selected areas for each section.
Double labeling for mouse SAA3 and Wilm's tumor 1 (WT-1), a marker of podocytes, was performed using the rabbit anti-mouse SAA3 antibody and a mouse monoclonal anti-mouse WT-1 antibody (Novus Biologicals, www.novusbio.com). Secondary antibodies were DyLight 594 Anti-Rabbit IgG and DyLight488 Anti-Mouse IgG (Vector laboratories) for fluorescence detection of bound primary antibodies. IHC images were captured on an Olympus fluorescence microscope equipped with a DP-70 digital camera.
Cell Culture Studies
Podocytes
Mouse podocytes (Dr Stuart Shankland, University of Washington) are a conditionally immortalized cell line.34 Podocytes were grown on Collagen I (BD Biosciences, www.bdbiosciences.com)-coated Primaria plates (VWR, www.vwrsp.com) in RPMI 1640 medium (Sigma Chemical, www.sigmaaldrich.com) containing 10% heat-inactivated fetal bovine serum (FBS, HyClone, Thermo Fisher, www.hyclone.com) with interferon-γ (50 U/ml) at 33 °C. Cell differentiation was promoted by incubation at 37 °C without interferon-γ for 10–12 days. Podocytes were placed on RPMI with reduced FBS (0.5%) for 1 day before exposure to experimental conditions in the same media. Control samples were podocytes from the same isolates and passages as the experimental groups without exposure to SAA or AGE. Recombinant human SAA1 protein (rSAA; #300-53, PeproTech, www.peprotech.com) was used at 10 μg/ml unless otherwise specified. rSAA was tested for endotoxin activity and found to be free of a detectable endotoxin-related inflammatory response (Supplementary Figure S1). AGE-bovine serum albumin (BSA) was produced by incubating fatty acid-free fraction IV BSA (100 mg/ml, Sigma-Aldrich, www.sigmaaldrich.com) with 0.5 M glucose for 45 days at 37 °C.10 The resulting AGE solution was dialyzed with PBS (pH 7.4) and sterile filtered. Endotoxin was not detected (<0.01 EU, E-toxate, Sigma-Aldrich). Control BSA did not produce cellular responses (not shown). The final AGE-BSA concentration used in experiments was 300 μg/ml. The NF-κB inhibitor ammonium pyrrolidine-di-thiocarbamate (PDTC, Sigma-Aldrich) was used in experiments with rSAA at concentrations of 5 and 20 μM. PDTC was added to cells 30 min before addition of rSAA and remained present for the duration of the experiment.
NF-κB activity assay
NF-κB activity was measured in podocytes exposed to rSAA or AGE for 6 and 24 h. Whole-cell lysates were collected and NF-κB activity was measured using the Pierce NF-κB p65 Transcription Factor Assay Kit (Thermo Fisher Scientific, www.thermoscientific.com). The NF-κB activity readout of chemiluminescence was captured using a Chemidoc XRS system and Quantity One version 4.5.2 software (Bio-Rad Laboratories, www.biorad.com). Activity was normalized to cellular protein (DC protein Assay, Bio-Rad Laboratories) and displayed relative to control.
mRNA measurements by qRT-PCR and RT2 Profiler Array analysis
Podocyte total RNA was isolated by the RiboPure kit (Ambion) and quantified using the Quant-iT RiboGreen RNA Reagent and kit (Invitrogen). Equal amounts of RNA were DNAse treated using amplification-grade DNase I (Invitrogen). cDNA was synthesized using Superscript III (Invitrogen). Podocyte transcripts were then quantified by qRT-PCR as described for kidney tissue mRNA.
To prepare RNA for pathway array analysis, differentiated podocytes were exposed to rSAA for 20 h. Total RNA from each of four separate experiments was isolated and quantified. Equal amounts of RNA from control and the rSAA condition of the experiments were pooled and DNAse treated, followed by cDNA synthesis with Superscript III (Invitrogen). Gene expression from cDNA was quantitated using SA Biosciences RT2 Profiler PCR Arrays (Qiagen) featuring genes associated with NF-κB Signaling (PAMM-025A) and Inflammatory Response and Autoimmunity (PAMM-077Z). Array data were analyzed via the RT2 Profiler PCR Array Data Analysis Template v 4.0 (SA Biosciences). Profiler PCR Array ‘housekeeping' gene expression between control and experimental samples had <10% inter-array variance with array amplifications repeated 6–8 times. Genes with at least a threefold increase in expression and P-value <0.02 were considered significantly upregulated. A subset of genes from the cDNA used for array analysis was validated by amplification with separately designed primers by qRT-PCR. Another validation was conducted with subsequent experiments to corroborate expression of representative inflammatory genes (MCP-1, RANTES, ENA-78, and iNOS) by qRT-PCR.
SAA3 protein measurements
Media and whole-cell lysates were collected and clarified of debris by centrifugation at 12 000 g for 5 min. Media SAA3 was measured by ELISA (EZMSAA3–12 K, EMD Millipore). The mouse anti-SAA3 antibody was verified not to cross-react with rSAA and was confirmed by lack of reaction in the SAA3 ELISA (EMD Millipore). SAA3 from media was expressed relative to protein (DC Protein Assay) from cell layers collected in RIPA buffer.
Statistics
Continuous data are expressed as mean±s.d. for normally distributed data or median and interquartile range for non-normally distributed data. Data were logarithmically transformed or coded into tertiles for statistical analyses in the case of skewness (for example, human urinary albumin-to-creatinine ratio; human and mouse kidney tissue SAA mRNA). For the human studies, one-way analysis of variance (ANOVA) was used to analyze study participant characteristics. Analysis of covariance was used to assess differences in plasma SAA levels between normal controls, diabetic controls, and the DKD group with covariates of age, gender, body mass index (BMI), and eGFR. The relationship between eGFR and plasma SAA in humans was determined by Pearson's correlation coefficient. Mouse plasma SAA protein and SAA mRNA expression in human kidney tissue (Nephromine analysis, version 4.0) were assessed by two sample student's t-tests. Generalized estimating equations (GEE) were used to detect between-group differences for SAA mRNA in the mouse models. Measures of immunostaining in mouse kidney tissue were analyzed by two-way ANOVA. In the GEE and ANOVA analyses of mouse models, main effects and interactions were tested. For studies of podocytes, two sample student's t-tests or two-way ANOVA analyses were used to evaluate differences between groups, doses, and/or times. Statistical analyses were conducted using SPSS version 17. The RT2 Profiler PCR Arrays data analysis was performed by DATA Analysis Template v.4.0 software supplied with the Arrays (SA Biosciences).
RESULTS
Plasma SAA in Humans and Mice with DKD
In the human study participants with DKD and diabetic controls, mean (±s.d.) duration of diabetes was 16±7 vs 8±8 years (P=0.06); eGFR was 61±21 vs 74±16 ml/min per 1.73 m2 (P=0.016); median (interquartile range) urine albumin-to-creatinine ratio was 1127 (228–4307) and 16 (14–28) mg/g (P<0.001), respectively.26 Normal controls had an eGFR of 86±18 ml/min per 1.73 m2 and urine albumin-to-creatinine ratio of 11 (7–14) mg/g (P<0.001 vs DKD). BMI was 26±4 kg/m2 in normal controls, 35±6 kg/m2 in diabetic controls, and 31±6 kg/m2 in DKD (P=0.011 normal vs diabetic control).
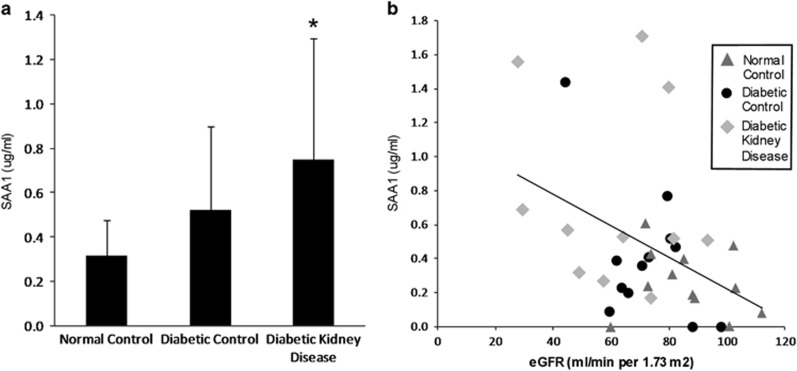
Plasma SAA1 protein was higher in a graded manner from normal controls to diabetic controls to those with DKD independent of age, sex, BMI, and eGFR (Figure 1a). Plasma SAA1 inversely correlated with eGFR across these groups (Figure 1b).
Figure 1.
SAA1 in human plasma. (a) Plasma levels of human SAA1 in normal controls (n=11), diabetic controls (n=10), and people with DKD defined by overt proteinuria (n=11); P=0.026 for linear trend by analysis of covariance controlled for covariates of age, sex, BMI, and eGFR; *P=0.048 for DKD vs normal controls. Data are shown as mean±s.d. (b) Plasma SAA1 levels inversely correlated with eGFR across groups, r=−0.38, P=0.029. BMI, body mass index; eGFR, estimated glomerular filtration rate; DKD, diabetic kidney disease; SAA, serum amyloid A.
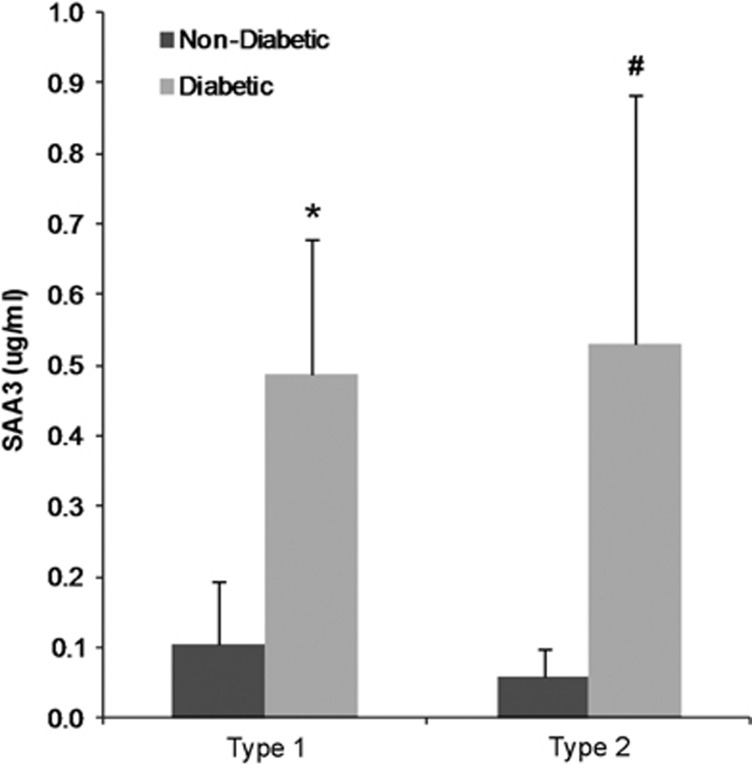
Findings were similar in mice, with SAA3 protein markedly increased (approximately fivefold higher) in the plasma of mild DKD, type 1 diabetes mice and severe DKD, type 2 diabetes mice compared with non-diabetic controls in the mouse models (Figure 2).
Figure 2.
SAA3 in mouse plasma. Plasma levels of mouse SAA3 in type I diabetic mouse model (streptozotocin-treated C57BL/6), *P=0.004 vs non-diabetic mice (n=7); and type 2 diabetic mouse model (BTBR-ob/ob), #P=0.009 vs non-diabetic mice (n=5–6). Data are shown as mean±s.d. SAA, serum amyloid A.
SAA mRNA in the Kidneys of Humans and Mice with DKD
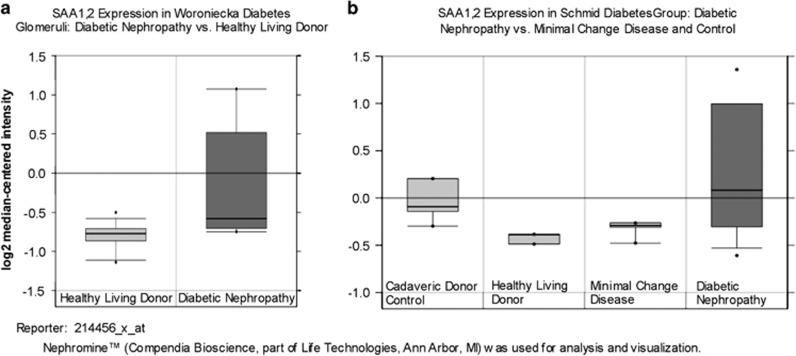
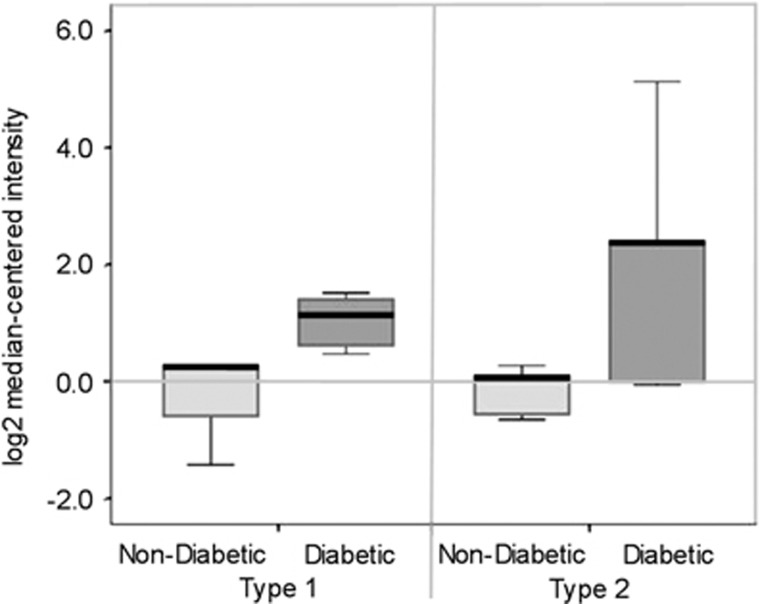
SAA 1, 2 mRNA was increased in human DKD compared with non-diabetic and/or glomerular disease controls (Figure 3). The glomerular fold change was 1.5 (P=0.017), whereas the tubulointerstitium fold change was 1.4 (P=0.021). Concordant findings in the kidneys of both diabetic mouse models also demonstrated increased SAA3 mRNA expression compared with non-diabetic controls (Figure 4). In the mild DKD, type 1 diabetes model, the fold change was 2.9 and in the severe DKD, type 2 diabetes model the fold change was 42.5 (P=0.009; interaction by model P=0.57)
Figure 3.
Human SAA1, 2 mRNA levels in Nephromine data sets from humans with DKD. (a) Woroniecka study of glomerular samples: DKD (diabetic nephropathy) samples vs healthy living donor controls, P=0.017 (control n=13, DKD n=9). (b) Schmid study of tubulointerstitial compartment: DKD (diabetic nephropathy) samples vs controls, P=0.021 (n=11 for controls, n=11 for DKD). Data are displayed as median and interquartile range with 95% confidence intervals for log2-transformed data. DKD, diabetic kidney disease; SAA, serum amyloid A.
Figure 4.
Expression of SAA3 mRNA in kidney cortex of diabetic mice. Diabetic vs non-diabetic mice (type 1: streptozotocin-treated C57BL/6 model; type 2: BTBR-ob/ob model), P=0.009, P=0.57 for interaction by model (n=9–10 per mouse model). Data are displayed as median and interquartile range with 95% confidence intervals for log2-transformed data. SAA, serum amyloid A.
SAA Protein in the Kidneys of Humans and Mice with DKD
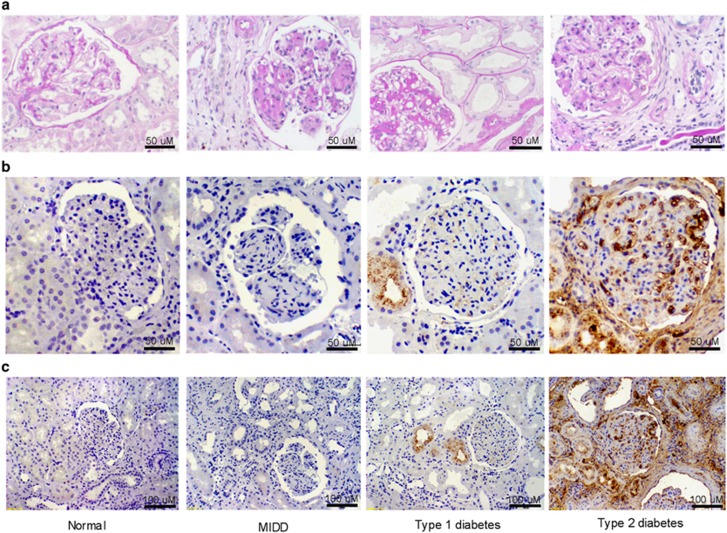
Human kidneys demonstrated expanded glomerular matrix in mild (class IIa), severe (class III–IV) DKD, and MIDD, a non-diabetic glomerular disease control (Figure 5a). A representative section from a sample of class III–IV DKD, type 2 diabetes (glomerulosclerosis and tubulointerstitial fibrosis) demonstrates extensive glomerular and tubulointerstitial human SAA1, 2 protein deposition by IHC. A section from a person with class IIa DKD, type 1 diabetes (mild glomerulopathy and tubulointerstitial changes) showed a lesser amount of SAA1, 2 staining primarily in the proximal tubular epithelium with faint glomerular staining (Figures 5b and c). SAA1, 2 protein was exclusively detected in kidneys of humans with DKD (n=4 of 10 samples). Human SAA1, 2 was undetectable in any normal control or non-diabetic glomerular disease control kidneys (n=0 of 8 samples).
Figure 5.
Human kidney histology and immunostaining for SAA1, 2. Representative sections from kidney biopsies of normal and non-DKD (MIDD) controls, type 1 diabetes with class IIa DKD, and type 2 diabetes with class III-IV DKD. (a) PAS staining; (b, c) human SAA1, 2 immunostaining (brown). Magnification bars are shown in the lower right corner of each image. DKD, diabetic kidney disease; MIDD, monocolonal immunoglobulin deposition disease; PAS, periodic acid–Schiff; SAA, serum amyloid A.
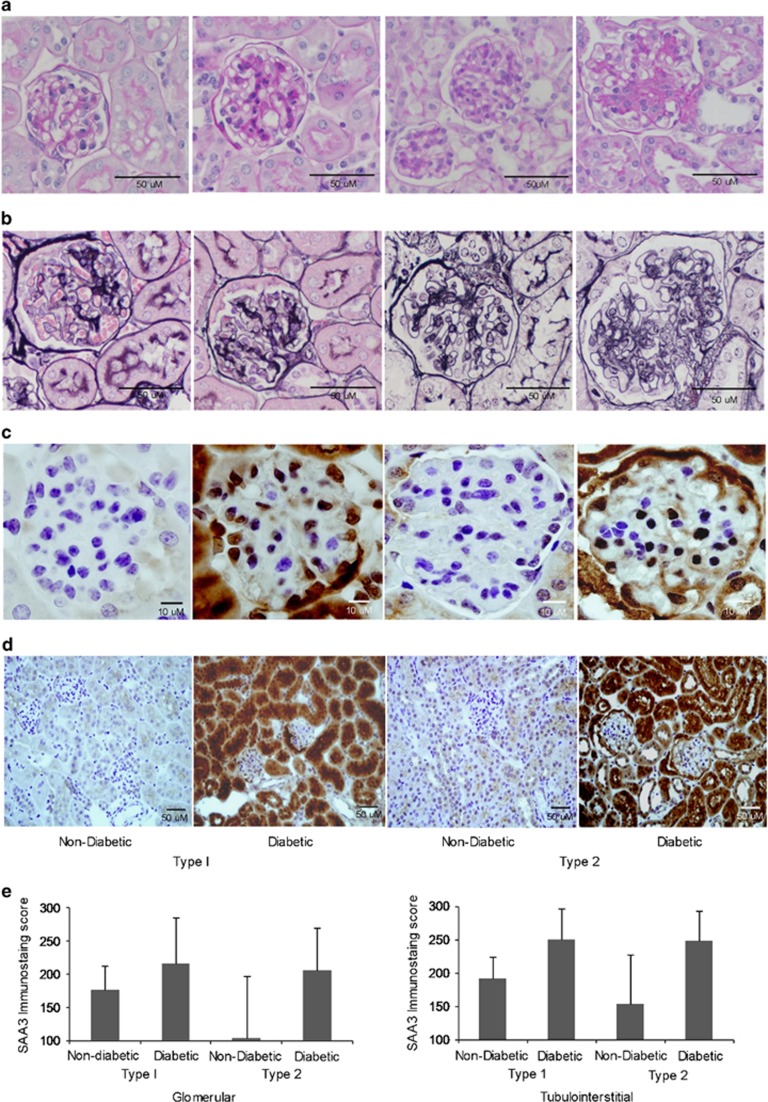
Comparable SAA3 patterns of expression were found in diabetic mouse models. In the model of mild DKD, type 1 diabetes mice exhibited mildly increased mesangial matrix, whereas the model of severe DKD, type 2 diabetes displayed markedly expanded mesangial matrix expansion and mesangiolysis (Figures 6a and b). Mouse SAA3 protein was observed in the glomeruli of both diabetic mouse models (Figure 6c). Extensive staining of tubular cells was readily evident in the diabetic mice compared with faint staining in control mice (Figure 6d). No IHC staining was detected with anti-SAA3 antiserum after antigen absorption of anti-SAA3 antibodies, indicating specific reactivity (Supplementary Figure S2). SAA3 immunostaining was increased in diabetic kidneys compared with controls in glomeruli (Figure 6e; fold change 1.2 in type 1 diabetes and 2.0 in type 2 diabetes, P=0.036, interaction by model P=0.33) and the tubulointerstitium (fold change 1.3 and 1.6, respectively, P=0.004, interaction by model P=0.47).
Figure 6.
Mouse kidney histology and immunostaining for SAA3. Representative sections of non-diabetic and diabetic mouse kidneys (type 1: streptozotocin-treated C57BL/6 model; type 2: BTBR-ob/ob model). (a) PAS; (b) silver methenamine; (c) immunostaining of SAA3 in glomerulus; and (d) tubulointerstitium. Magnification bars are shown in the lower right corner of each image. (e) SAA3 quantification by immunostaining scores. Glomerular immunostaining was increased in diabetic vs non-diabetic mice (P=0.036, interaction by model P=0.331, n=10). Tubulointerstitial immunostaining was increased in diabetic vs non-diabetic mice (P=0.004, interaction by model, P=0.465, n=10). Data are shown as mean±s.d. PAS, periodic acid–Schiff; SAA, serum amyloid A.
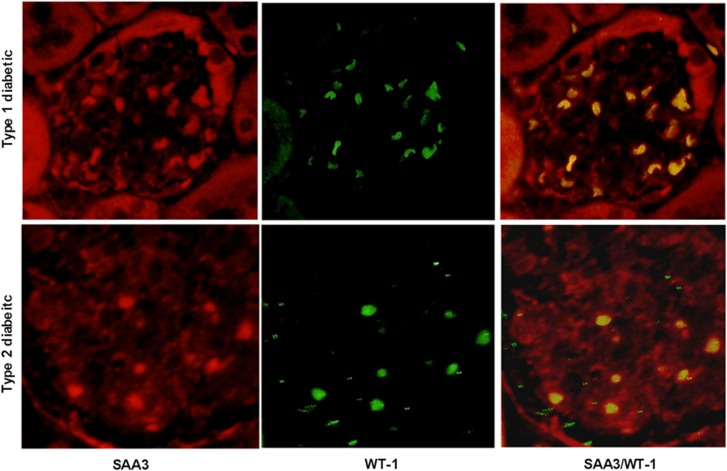
SAA3 protein was present in glomerular podocytes from both mouse models of diabetes (Figure 7). Double immunofluorescence staining of SAA3 (red) and podocyte marker WT-1 (green) shows SAA3 localization within podocytes in the merged image (yellow).
Figure 7.
Colocalization of mouse SAA3 and podocyte WT-1 by immunofluorescence. Representative glomeruli of diabetic mice (type 1: streptozotocin-treated C57BL/6 model; type 2: BTBR-ob/ob model), immunostained with anti-mouse SAA3 antibody (red), the podocyte-specific marker, WT-1 (green) and merged photos showing colocalization of SAA3 and WT-1 in podocytes (yellow). SAA, serum amyloid A.
Podocyte Studies
Effect of AGE exposure on expression of SAA in podocytes
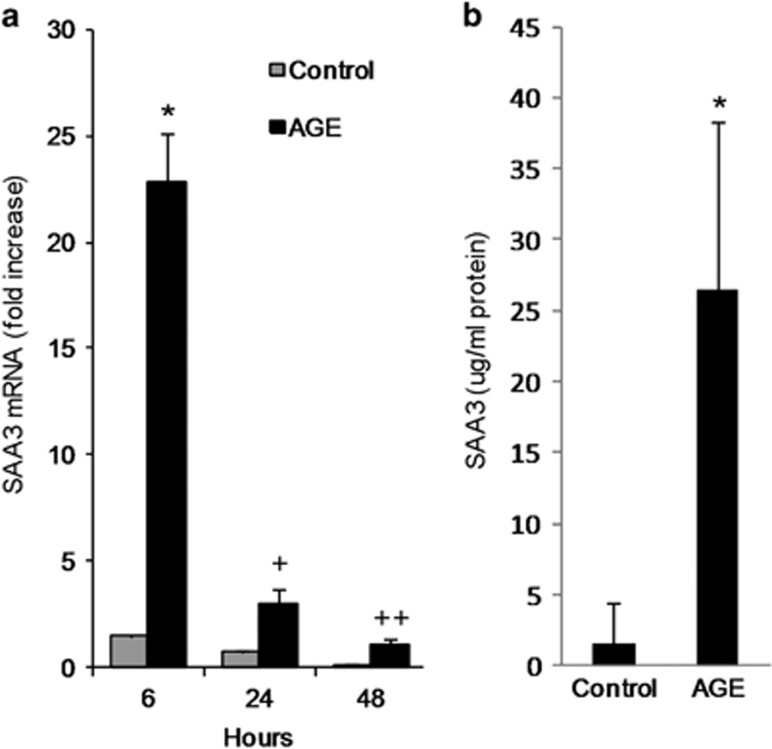
Mouse podocytes produced increased amounts of SAA3 mRNA when exposed to AGE over a time course from 6 to 48 h (Figure 8a). The corresponding SAA3 protein was also secreted and increased in conditioned media after AGE exposure (Figure 8b).
Figure 8.
SAA3 expression in mouse podocytes exposed to AGE. (a) SAA3 mRNA after AGE exposure for 6, 24, and 48 h; *P=0.004, +P=0.018, ++P=0.039 vs control (n=3). (b) SAA3 protein after exposure to AGE for 20 h; *P=0.014 vs control (n=6). Data are shown as mean±s.d. AGE, advanced glycation end products; SAA, serum amyloid A.
Podocyte inflammatory responses to AGE and SAA
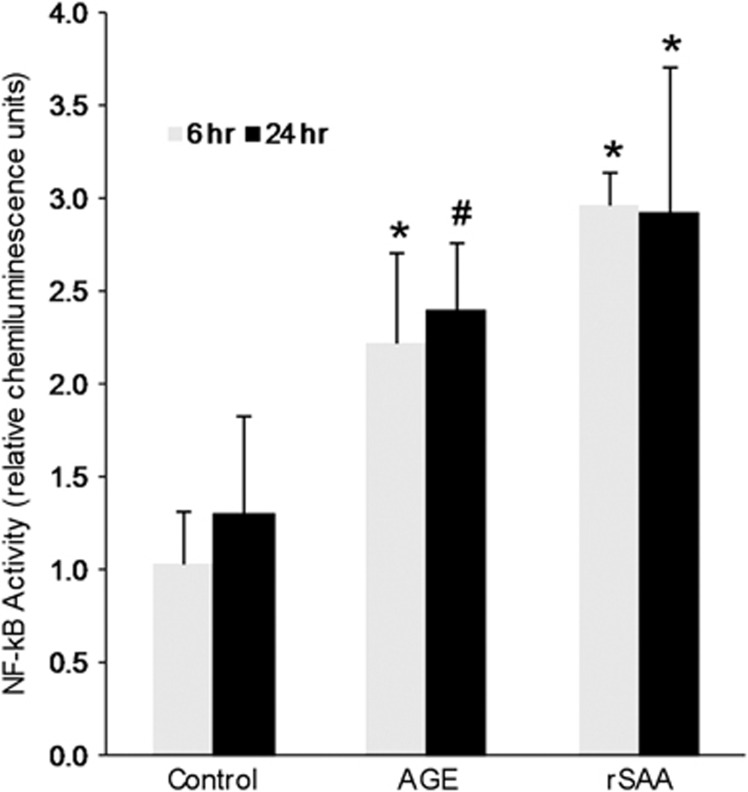
NF-κB activity was increased in podocytes exposed to rSAA at both 6 and 20 h, comparable to the effect of AGE exposure (Figure 9). An rSAA-induced podocyte inflammatory gene transcription profile was generated by array analysis (Table 1). A large number of genes associated with NF-κB activation and inflammation were upregulated. Clusters of related chemokine and cytokine genes were upregulated. CC (beta) chemokine ligands included: Ccl2 (MCP-1), Ccl5 (RANTES), Ccl7 (MARC or MCP-3), Ccl8 (MCP-2), and Ccl20 (LARC). Another induced chemokine family was the CXC (alpha) chemokine ligands: Cxcl1 (GRO1 or NAP-3), Cxcl2 (GRO2 or MIP-2a), and Cxcl3 (GRO3 or MIP2b). Cxcl5 (ENA-78) revealed marked induction with a 166-fold increase. Other genes showing robust induction were C3 (complement 3), G-CSF 2 and 3, IL-6, Kng1, and NOS2. Validation of pathway array results using independently designed primers and qRT-PCR on the pooled cDNA samples confirmed these results (Table 1). Subsequent experiments of podocytes exposed to rSAA further validated results obtained from the pathway array analyses of the pooled samples (Table 1).
Figure 9.
Effect of AGE and rSAA on podocyte NF-κB activity. NF-κB activity after exposure to AGE or rSAA for 6 or 24 h; *P<0.001 and #P=0.004 vs control (n=4–5). Data are shown as mean±s.d. AGE, advanced glycation end products; rSAA, recombinant human SAA1 protein.
Table 1. Inflammatory genes expressed in podocytes exposed to rSAA.
| Discovery data sets |
Validation data sets |
||||
|---|---|---|---|---|---|
| Gene symbol | Gene name and/or description | P-value | RT2 Profiler Array | RT2 Profiler Array experiments, independent primers | Subsequent experiments |
| C3 | Complement component 3 | <0.001 | 24.4 | 25.4 | |
| Ccl2 | MCP-1, monocyte chemoattractant protein-1, chemokine CC motif 2 | <0.001 | 18.2 | 17.9 | 6.4 |
| Ccl5 | RANTES, regulated on activation, normal T-cell expressed and secreted, chemokine CC motif 5 | <0.001 | 36.6 | 25.21 | 101.0 |
| Ccl7 | MCP-3, monocyte chemotactic protein-3, chemokine CC motif 7 | <0.001 | 16.8 | 17.6 | |
| Ccl8 | MCP-2, monocyte chemotactic protein-2, chemokine CC motif 8 | 0.002 | 22.6 | 21.0 | |
| Ccl17 | TARC, thymus and activation-regulated chemokine CC motif 17 | <0.001 | 3.4 | ||
| Ccl19 | MIP-3, macrophage inflammatory protein-3-beta, chemokine CC motif 19 | 0.013 | −5.6 | ||
| Ccl20 | MIP-3, macrophage inflammatory protein-3-alpha, chemokine CC motif 20 | <0.001 | 37.6 | ||
| Cxcl1 | GRO1, growth-related oncogene alpha chemokine CXC motif 1 | <0.001 | 30.0 | 25.7 | |
| Cxcl2 | GRO2, growth-related oncogene beta chemokine CXC motif 2 | <0.001 | 28.3 | 16.0 | |
| Cxcl3 | GRO3, growth-related oncogene gamma chemokine CXC motif 3 | <0.001 | 8.5 | ||
| Cxcl5 | ENA-78, epithelial-derived neutrophil-activating protein 78, chemokine CXC motif 5 | <0.001 | 166.1 | 52.5 | 18.5 |
| Cxcl11 | I-TAC, interferon-inducible T-cell alpha chemoattractant, chemokine CXC motif 11 | 0.003 | 3.6 | ||
| Csf2 | G-CSF2, granulocyte colony stimulating factor 2 | <0.001 | 72.2 | ||
| Csf3 | G-CSF3, granulocyte colony stimulating factor 3 | <0.001 | 34.2 | ||
| Il1a | Interleukin-1 | 0.018 | 3.0 | ||
| Il1rn | IL-1ra, interleukin-1 receptor antagonist | <0.001 | 5.3 | ||
| Il-6 | Interleukin 6 | 0.007 | 27.2 | ||
| Itgb2 | Cd18, integrin beta 2 | <0.001 | 3.6 | ||
| Kng1 | Kng, kininogen1 | 0.003 | 10.0 | ||
| Nos2 | iNOS, nitric oxide synthase 2 | <0.001 | 19.9 | 56.8 | 18.9 |
| Ptgs2 | Cox-2, prostaglandin-endoperoxide synthase 2 | <0.001 | 3.3 | ||
| Tlr5 | Toll-like receptor 5 | 0.006 | −7.4 | ||
Data are represented as fold change vs control.
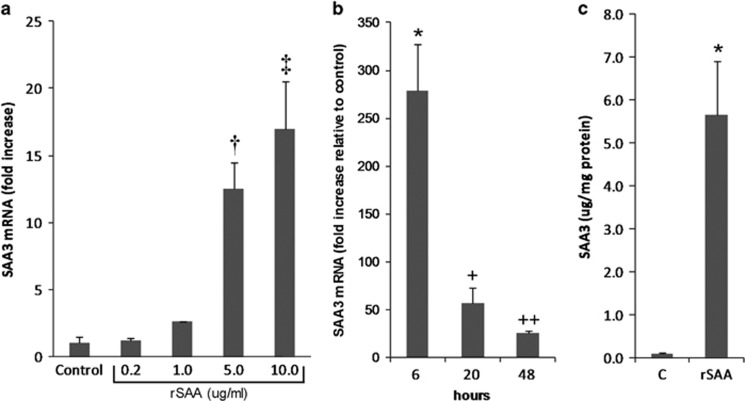
Effect of exogenous SAA exposure on endogenous SAA3 expression
SAA3 mRNA increased in response to doses of rSAA ranging from 1 to 10 μg/ml (Figure 10a). SAA3 expression peaked at 6 h and remained elevated at 20 and 48 h (Figure 10b). SAA3 protein also increased (>50-fold) in podocyte media after exposure to rSAA for 20 h (Figure 10c).
Figure 10.
Effects of exogenous SAA on endogenous SAA3 expression by podocytes. (a) SAA3 mRNA dose response to rSAA for 20 h; †P=0.019, ‡P=0.006 vs control (n=3). (b) SAA3 mRNA time course for response to rSAA (10 μg/ml); *P=0.010, +P=0.024, ++P=0.001 vs control (n=3). (c) SAA3 protein after exposure to rSAA for 20 h; *P<0.001 vs control (n=7). Data are expressed as mean±s.d. rSAA, recombinant human SAA1 protein; SAA, serum amyloid A.
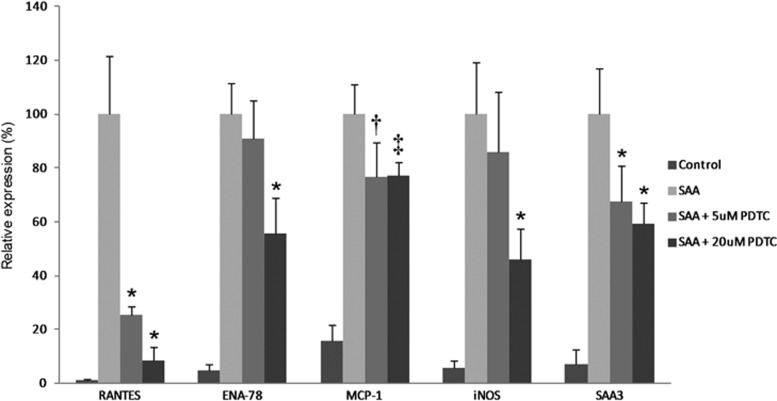
Effect of NF-κB inhibition on SAA-induced gene expression
Increased expression of representative inflammatory genes (RANTES, ENA-78, MCP-1, iNOS, SAA3) was reduced in a dose-dependent manner by NF-κB inhibition (Figure 11).
Figure 11.
Effects of NF-κB inhibition on SAA-induced inflammatory gene expression in podocytes. Levels of mRNA after 20 h of exposure to rSAA with two dose levels of PDTC, an NF-κB inhibitor. Expression levels are shown relative to no inhibition; *P<0.001, †P=0.005, ‡P=0.002 vs rSAA without PDTC (n=5–6). Data are expressed as mean±s.d. PDTC, pyrrolidine-di-thiocarbamate; rSAA, recombinant human SAA1 protein.
DISCUSSION
The present data reveal increased production and deposition of SAA in the kidneys of human DKD as well as in corresponding diabetic mouse models. Notably, SAA was elevated in the plasma of both people and mice, suggesting its candidacy as a DKD biomarker. Although SAA was expressed throughout the kidney, it specifically localized within glomerular podocytes in two different diabetic mouse models. Exposure of mouse podocytes to exogenous SAA increased endogenous SAA expression and a broad pro-inflammatory response manifest by NF-κB dependent upregulation of multiple genes.
Previous studies have reported elevated levels of plasma SAA in small numbers of people with type 2 diabetes and increased albuminuria/proteinuria as part of inflammatory biomarker panels.35, 36 The present data considerably expand earlier human observations by showing direct association between plasma SAA and DKD defined by overt proteinuria along with an inverse relationship between plasma SAA and eGFR. Furthermore, increased plasma SAA levels in DKD were independent of eGFR and BMI, denoting that its accumulation is not simply due to reduced renal clearance or obesity. Nephromine provided two additional human data sets that showed upregulation of SAA1, 2 mRNA in the kidneys of people with DKD. These data indicate that SAA deposition in the glomeruli and the tubulointerstitium may result from local production in addition to systemic inflammation.
Mouse models of types 1 and 2 diabetes produced results with striking similarities to human DKD: increased plasma levels of SAA, increased transcription of SAA mRNA in the kidney cortex, and increased glomerular and tubulointerstitial SAA protein deposition. Notably, both diabetic mouse models showed colocalization of SAA with WT-1, revealing the presence of SAA in glomerular podocytes. These observations of increased amounts of circulating and local SAA in the kidneys of diabetic mouse models are highly concordant with human DKD and novel to the best of our knowledge.
Mouse podocytes cultured with AGE, metabolic mediators of inflammation, increased SAA3 expression indicating that it can be locally produced in the glomerulus under diabetic-like conditions. Cellular SAA production has potential for autocrine and/or paracrine inflammatory actions in the kidney. For example, mouse podocytes cultured with exogenous SAA (rSAA) increased expression of endogenous SAA3 consistent with autocrine activation of transcription.37 If SAA accumulation in resident glomerular podocytes produces similar effects as it does in in vitro, it could amplify inflammation through self-upregulation. An SAA–RAGE interaction in the podocyte could provide a mechanistic pathway for such an autocrine amplification loop. The concept of autocrine activation adds to a growing body of evidence implicating the podocyte as an ongoing source of inflammatory mediators.38, 39 We have previously demonstrated that RAGE inhibition blocks AGE-induced SAA3 gene transcription and apoptosis in mouse podocytes.10 Thus, we propose that RAGE may be a central receptor for SAA-induced podocyte responses including inflammation.
As RAGE activation signals through NF-κB, the effect of SAA exposure on NF-κB activity was studied in mouse podocytes. Exposure to exogenous SAA upregulated many NF-κB-dependent inflammatory genes. Inflammation is now clearly recognized as a mechanism of kidney injury in diabetes.40 We propose that SAA may be an important regulator of inflammatory gene transcription in podocytes. Pro-inflammatory genes that were upregulated by exogenous SAA exposure in podocytes were also differentially expressed in human DKD in the Nephromine data sets (for example, C3, Ccl2, Ccl5, Itgb2, Il1rn, Cxcl1).27, 28 The podocytes could promote inflammatory responses by production of chemokines and cytokines via SAA-induced NF-κB activation. Indeed, many of the SAA-induced genes expressed by podocytes are involved in DKD.5, 41, 42 For instance, CXC or α-chemokines stimulate chemotaxis and neutrophil activation and accumulation.43, 44 In particular, Cxcl5 showed a very large increase in response to SAA exposure and has been detected in the urine of people with DKD.45 Another group of CC or β-chemokines that recruit inflammatory cells to the kidney was upregulated by SAA in podocytes.46, 47 Ccl2, also known as MCP-1, a well-known member of this group, is increased by RAGE activation and promotes inflammatory injury by monocyte and macrophage recruitment in DKD.42, 48, 49 SAA3 has been shown to recruit monocytes and macrophages, which accumulate in the glomeruli of STZ-diabetic C57BL/6 and BTBR-ob/ob mice, and thus may contribute to local inflammation.17, 30, 48, 50 SAA exposure in podocytes also produced marked increases in expression of C3, IL-6, and iNOS, each of which has been detected in diabetic kidneys.28, 51, 52 Finally, podocyte-produced cytokines such as G-CSF 2 and 3 stimulate production, differentiation, and activation of granulocytes and macrophages. Although their role in DKD is undetermined, G-CSF 2 and 3 levels are elevated in the blood of diabetic mice.53 Overall, SAA exposure triggered upregulation of myriad inflammatory genes via NF-κB, implicating the podocyte as a key responder cell for SAA-mediated inflammatory responses.
This study has several limitations to note. First, the human studies included several data sets and samples. Scarcity of well-characterized human samples for kidney disease research is a major challenge.54 Kidney tissue, blood, urine, and information about clinical phenotypes are very limited in availability for human DKD. As such, different sources must be pieced together to create an over-arching picture. Nevertheless, increased amounts of SAA in the blood and kidneys of DKD were reproducible between murine and human species, different groups of people, and two mouse models of diabetes. We submit that this general concordance strengthens generalizability of the findings. Comprehensive collection of human data and samples is needed to more fully characterize SAA and other candidate mediators and biomarkers that will identify patients at high risk for progressive DKD. Second, SAA has been implicated in producing AA amyloidosis involving the kidney. Inflammatory conditions producing AA-type amyloidosis with unique tissue deposition of amyloid fibrils are typically marked by periods of abrupt high-level inflammation and markedly elevated circulating levels of SAA. However, in diabetes, sustained lower-level SAA exposure with smoldering inflammation may instead contribute to inflammatory mechanisms in DKD. Third, in vitro studies may not fully translate to human disease conditions. For example, treating cultured mouse podocytes with exogenous SAA (rSAA) may not produce the same effects as endogenous SAA in vivo. However, both rSAA and rabbit SAA3 protein have been shown to have similar capacity to stimulate matrix metalloproteinase production in mouse and human chondrocytes, indicating similar function between SAA isoforms and species.20 In addition, the present data indicate that rSAA elicits a cytokine-inducing response in mouse podocytes that is similar to the effect of purified human or mouse SAA in mouse monocytes and phagocytes.23, 55 We have also tested custom-made recombinant mouse SAA3 and found it to have similar capacity to rSAA for inducing SAA3 mRNA and related inflammatory cytokines (Supplementary Figure S3). Thus, it is reasonable to propose that podocytes treated with exogenous SAA provide disease-relevant discoveries.
In conclusion, SAA was elevated at the protein and/or mRNA levels in the blood and kidneys of people with DKD. Mouse models of both mild and severe DKD in !types 1 and 2 diabetes were concordant with these findings. SAA was widely distributed in the diabetic kidney, including specific glomerular localization in podocytes of mice. Exposure to exogenous SAA directly elicited a broad pro-inflammatory response in podocytes with NF-κB-dependent induction of many chemokines and cytokines including endogenous SAA itself, indicating potential for autocrine upregulation. Podocytes appear to be key responder cells to SAA-induced inflammation. Together, these data make SAA a compelling candidate for DKD therapeutic and biomarker discovery.
Acknowledgments
We thank Dr Robert Short for assistance with statistical analysis and Dr Maria Bertagnolli, Dr Carolyn Johnson, Dr Brad Dieter, and Mr Thomas Cooney for their editorial commentaries on this manuscript. We also thank the Applied Systems Biology Core of the O'Brien Renal Center at the University of Michigan and Compendia Bioscience, part of Life Technologies, for making the Nephromine analysis engine freely available to us and other researchers.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Laboratory Investigation website (http://www.laboratoryinvestigation.org)
Supplementary Material
References
- Adler AI, Stevens RJ, Manley SE, et al. Development and progression of nephropathy in type 2 diabetes: the United Kingdom Prospective Diabetes Study (UKPDS 64) Kidney Int. 2003;63:225–232. doi: 10.1046/j.1523-1755.2003.00712.x. [DOI] [PubMed] [Google Scholar]
- Afkarian M, Sachs MC, Kestenbaum B, et al. Kidney disease and increased mortality risk in type 2 diabetes. J Am Soc Nephrol. 2013;24:302–308. doi: 10.1681/ASN.2012070718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danaei G, Finucane MM, Lu Y, et al. National, regional, and global trends in fasting plasma glucose and diabetes prevalence since 1980: systematic analysis of health examination surveys and epidemiological studies with 370 country-years and 2.7 million participants. Lancet. 2011;378:31–40. doi: 10.1016/S0140-6736(11)60679-X. [DOI] [PubMed] [Google Scholar]
- Tuttle KR. Linking metabolism and immunology: diabetic nephropathy is an inflammatory disease. J Am Soc Nephrol. 2005;16:1537–1538. doi: 10.1681/ASN.2005040393. [DOI] [PubMed] [Google Scholar]
- Navarro-Gonzalez JF, Mora-Fernandez C, Muros de Fuentes M, et al. Inflammatory molecules and pathways in the pathogenesis of diabetic nephropathy. Nat Rev Nephrol. 2011;7:327–340. doi: 10.1038/nrneph.2011.51. [DOI] [PubMed] [Google Scholar]
- Kalousova M, Zima T, Tesar V, et al. Advanced glycation end products in clinical nephrology. Kidney Blood Press Res. 2004;27:18–28. doi: 10.1159/000075533. [DOI] [PubMed] [Google Scholar]
- Goldin A, Beckman JA, Schmidt AM, et al. Advanced glycation end products: sparking the development of diabetic vascular injury. Circulation. 2006;114:597–605. doi: 10.1161/CIRCULATIONAHA.106.621854. [DOI] [PubMed] [Google Scholar]
- Basta G, Schmidt AM, De Caterina R. Advanced glycation end products and vascular inflammation: implications for accelerated atherosclerosis in diabetes. Cardiovasc Res. 2004;63:582–592. doi: 10.1016/j.cardiores.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Thallas-Bonke V, Coughlan MT, Tan AL, et al. Targeting the AGE-RAGE axis improves renal function in the context of a healthy diet low in advanced glycation end-product content. Nephrology (Carlton) 2013;18:47–56. doi: 10.1111/j.1440-1797.2012.01665.x. [DOI] [PubMed] [Google Scholar]
- Meek RL, LeBoeuf RC, Saha SA, et al. Glomerular cell death and inflammation with high-protein diet and diabetes. Nephrol Dial Transplant. 2013;28:1711–1720. doi: 10.1093/ndt/gfs579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlar CM, Whitehead AS. Serum amyloid A, the major vertebrate acute-phase reactant. Eur J Biochem. 1999;265:501–523. doi: 10.1046/j.1432-1327.1999.00657.x. [DOI] [PubMed] [Google Scholar]
- Sipe J. Revised nomenclature for serum amyloid A (SAA). Nomenclature Committee of the International Society of Amyloidosis. Part 2. Amyloid. 1999;6:67–70. doi: 10.3109/13506129908993291. [DOI] [PubMed] [Google Scholar]
- Meek RL, Benditt EP. Amyloid A gene family expression in different mouse tissues. J Exp Med. 1986;164:2006–2017. doi: 10.1084/jem.164.6.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluve-Beckerman B, Drumm ML, Benson MD. Nonexpression of the human serum amyloid A three (SAA3) gene. DNA Cell Biol. 1991;10:651–661. doi: 10.1089/dna.1991.10.651. [DOI] [PubMed] [Google Scholar]
- Upragarin N, Landman WJ, Gaastra W, et al. Extrahepatic production of acute phase serum amyloid A. Histol Histopathol. 2005;20:1295–1307. doi: 10.14670/HH-20.1295. [DOI] [PubMed] [Google Scholar]
- Reigstad CS, Backhed F. Microbial regulation of SAA3 expression in mouse colon and adipose tissue. Gut Microbes. 2010;1:55–57. doi: 10.4161/gmic.1.1.10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hartigh LJ, Wang S, Goodspeed L, et al. Deletion of serum amyloid a3 improves high fat high sucrose diet-induced adipose tissue inflammation and hyperlipidemia in female mice. PLoS One. 2014;9:e108564. doi: 10.1371/journal.pone.0108564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Migita S. Complete primary structures of two major murine serum amyloid A proteins deduced from cDNA sequences. Proc Natl Acad Sci USA. 1985;82:2915–2919. doi: 10.1073/pnas.82.9.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly M, Marrelli A, Blades M, et al. Acute serum amyloid A induces migration, angiogenesis, and inflammation in synovial cells in vitro and in a human rheumatoid arthritis/SCID mouse chimera model. J Immunol. 2010;184:6427–6437. doi: 10.4049/jimmunol.0902941. [DOI] [PubMed] [Google Scholar]
- Vallon R, Freuler F, Desta-Tsedu N, et al. Serum amyloid A (apoSAA) expression is up-regulated in rheumatoid arthritis and induces transcription of matrix metalloproteinases. J Immunol. 2001;166:2801–2807. doi: 10.4049/jimmunol.166.4.2801. [DOI] [PubMed] [Google Scholar]
- Meek RL, Urieli-Shoval S, Benditt EP. Expression of apolipoprotein serum amyloid A mRNA in human atherosclerotic lesions and cultured vascular cells: implications for serum amyloid A function. Proc Natl Acad Sci USA. 1994;91:3186–3190. doi: 10.1073/pnas.91.8.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua S, Song C, Geczy CL, et al. A role for acute-phase serum amyloid A and high-density lipoprotein in oxidative stress, endothelial dysfunction and atherosclerosis. Redox Rep. 2009;14:187–196. doi: 10.1179/135100009X12525712409490. [DOI] [PubMed] [Google Scholar]
- Yan SD, Zhu H, Zhu A, et al. Receptor-dependent cell stress and amyloid accumulation in systemic amyloidosis. Nat Med. 2000;6:643–651. doi: 10.1038/76216. [DOI] [PubMed] [Google Scholar]
- Okamoto H, Katagiri Y, Kiire A, et al. Serum amyloid A activates nuclear factor-kappaB in rheumatoid synovial fibroblasts through binding to receptor of advanced glycation end-products. J Rheumatol. 2008;35:752–756. [PubMed] [Google Scholar]
- Cai H, Song C, Endoh I, et al. Serum amyloid A induces monocyte tissue factor. J Immunol. 2007;178:1852–1860. doi: 10.4049/jimmunol.178.3.1852. [DOI] [PubMed] [Google Scholar]
- Saha SA, LaSalle BK, Clifton GD, et al. Modulation of advanced glycation end products by candesartan in patients with diabetic kidney disease—a dose-response relationship study. Am J Ther. 2010;17:553–558. doi: 10.1097/MJT.0b013e3181b96c27. [DOI] [PubMed] [Google Scholar]
- Schmid H, Boucherot A, Yasuda Y, et al. Modular activation of nuclear factor-kappaB transcriptional programs in human diabetic nephropathy. Diabetes. 2006;55:2993–3003. doi: 10.2337/db06-0477. [DOI] [PubMed] [Google Scholar]
- Woroniecka KI, Park AS, Mohtat D, et al. Transcriptome analysis of human diabetic kidney disease. Diabetes. 2011;60:2354–2369. doi: 10.2337/db10-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tervaert TW, Mooyaart AL, Amann K, et al. Pathologic classification of diabetic nephropathy. J Am Soc Nephrol. 2010;21:556–563. doi: 10.1681/ASN.2010010010. [DOI] [PubMed] [Google Scholar]
- Hudkins KL, Pichaiwong W, Wietecha T, et al. BTBR Ob/Ob mutant mice model progressive diabetic nephropathy. J Am Soc Nephrol. 2010;21:1533–1542. doi: 10.1681/ASN.2009121290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpers CE, Hudkins KL, Segerer S, et al. Localization of SPARC in developing, mature, and chronically injured human. Kidney Int. 2002;62:2073–2086. doi: 10.1046/j.1523-1755.2002.00680.x. [DOI] [PubMed] [Google Scholar]
- Wang A, Ziyadeh FN, Lee EY, et al. Interference with TGF-beta signaling by Smad3-knockout in mice limits diabetic glomerulosclerosis without affecting albuminuria. Am J Physiol Renal Physiol. 2007;293:F1657–F1665. doi: 10.1152/ajprenal.00274.2007. [DOI] [PubMed] [Google Scholar]
- Meek RL, Eriksen N, Benditt EP. Murine serum amyloid A3 is a high density apolipoprotein and is secreted by macrophages. Proc Natl Acad Sci USA. 1992;89:7949–7952. doi: 10.1073/pnas.89.17.7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankland SJ, Pippin JW, Reiser J, et al. Podocytes in culture: past, present, and future. Kidney Int. 2007;72:26–36. doi: 10.1038/sj.ki.5002291. [DOI] [PubMed] [Google Scholar]
- Kumon Y, Suehiro T, Itahara T, et al. Serum amyloid A protein in patients with non-insulin-dependent diabetes mellitus. Clin Biochem. 1994;27:469–473. doi: 10.1016/0009-9120(94)00044-v. [DOI] [PubMed] [Google Scholar]
- Dalla Vestra M, Mussap M, Gallina P, et al. Acute-phase markers of inflammation and glomerular structure in patients with type 2 diabetes. J Am Soc Nephrol. 2005;16 (Suppl 1:S78–S82. doi: 10.1681/asn.2004110961. [DOI] [PubMed] [Google Scholar]
- Shvartsman SY, Hagan MP, Yacoub A, et al. Autocrine loops with positive feedback enable context-dependent cell signaling. Am J Physiol Cell Physiol. 2002;282:C545–C559. doi: 10.1152/ajpcell.00260.2001. [DOI] [PubMed] [Google Scholar]
- Banas MC, Banas B, Hudkins KL, et al. TLR4 links podocytes with the innate immune system to mediate glomerular injury. J Am Soc Nephrol. 2008;19:704–713. doi: 10.1681/ASN.2007040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahler S, Ising C, Hagmann H, et al. Intrinsic proinflammatory signaling in podocytes contributes to podocyte damage and prolonged proteinuria. Am J Physiol Renal Physiol. 2012;303:F1473–F1485. doi: 10.1152/ajprenal.00031.2012. [DOI] [PubMed] [Google Scholar]
- Galkina E, Ley K. Leukocyte recruitment and vascular injury in diabetic nephropathy. J Am Soc Nephrol. 2006;17:368–377. doi: 10.1681/ASN.2005080859. [DOI] [PubMed] [Google Scholar]
- Ruster C, Wolf G. The role of chemokines and chemokine receptors in diabetic nephropathy. Front Biosci. 2008;13:944–955. doi: 10.2741/2734. [DOI] [PubMed] [Google Scholar]
- Mezzano S, Aros C, Droguett A, et al. NF-kappaB activation and overexpression of regulated genes in human diabetic nephropathy. Nephrol Dial Transplant. 2004;19:2505–2512. doi: 10.1093/ndt/gfh207. [DOI] [PubMed] [Google Scholar]
- Geiser T, Dewald B, Ehrengruber MU, et al. The interleukin-8-related chemotactic cytokines GRO alpha, GRO beta, and GRO gamma activate human neutrophil and basophil leukocytes. J Biol Chem. 1993;268:15419–15424. [PubMed] [Google Scholar]
- Vieira S, Lemos H, Grespan R, et al. A crucial role for TNF-α in mediating neutrophil influx induced by endogenously generated or exogenous chemokines, KC/CXCL1 and LIX/CXCL5. Br J Pharmacol. 2009;158:779–789. doi: 10.1111/j.1476-5381.2009.00367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higurashi M, Ohya Y, Joh K, et al. Increased urinary levels of CXCL5, CXCL8 and CXCL9 in patients with Type 2 diabetic nephropathy. J Diabetes Complications. 2009;23:178–184. doi: 10.1016/j.jdiacomp.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Van Damme J, Proost P, Lenaerts JP, et al. Structural and functional identification of two human, tumor-derived monocyte chemotactic proteins (MCP-2 and MCP-3) belonging to the chemokine family. J Exp Med. 1992;176:59–65. doi: 10.1084/jem.176.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schall TJ, Bacon K, Toy KJ, et al. Selective attraction of monocytes and T lymphocytes of the memory phenotype by cytokine RANTES. Nature. 1990;347:669–671. doi: 10.1038/347669a0. [DOI] [PubMed] [Google Scholar]
- Chow FY, Nikolic-Paterson DJ, Ozols E, et al. Monocyte chemoattractant protein-1 promotes the development of diabetic renal injury in streptozotocin-treated mice. Kidney Int. 2006;69:73–80. doi: 10.1038/sj.ki.5000014. [DOI] [PubMed] [Google Scholar]
- Gu L, Hagiwara S, Fan Q, et al. Role of receptor for advanced glycation end-products and signalling events in advanced glycation end-product-induced monocyte chemoattractant protein-1 expression in differentiated mouse podocytes. Nephrol Dial Transplant. 2006;21:299–313. doi: 10.1093/ndt/gfi210. [DOI] [PubMed] [Google Scholar]
- Han CY, Subramanian S, Chan CK, et al. Adipocyte-derived serum amyloid A3 and hyaluronan play a role in monocyte recruitment and adhesion. Diabetes. 2007;56:2260–2273. doi: 10.2337/db07-0218. [DOI] [PubMed] [Google Scholar]
- Trachtman H, Futterweit S, Pine E, et al. Chronic diabetic nephropathy: role of inducible nitric oxide synthase. Pediatr Nephrol. 2002;17:20–29. doi: 10.1007/s004670200004. [DOI] [PubMed] [Google Scholar]
- Moriwaki Y, Yamamoto T, Shibutani Y, et al. Elevated levels of interleukin-18 and tumor necrosis factor-[alpha] in serum of patients with type 2 diabetes mellitus: relationship with diabetic nephropathy. Metabolism. 2003;52:605–608. doi: 10.1053/meta.2003.50096. [DOI] [PubMed] [Google Scholar]
- Starkey JM, Haidacher SJ, LeJeune WS, et al. Diabetes-induced activation of canonical and noncanonical nuclear factor-κB pathways in renal cortex. Diabetes. 2006;55:1252–1259. doi: 10.2337/db05-1554. [DOI] [PubMed] [Google Scholar]
- Breyer MD, Coffman TM, Flessner MF, et al. Diabetic nephropathy: a national dialogue. Clin J Am Soc Nephrol. 2013;8:1603–1605. doi: 10.2215/CJN.03640413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MH, de Beer MC, Wroblewski JM, et al. SAA does not induce cytokine production in physiological conditions. Cytokine. 2013;61:506–512. doi: 10.1016/j.cyto.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.