Abstract
Three-dimensional (3D) cell culture is an important tool that facilitates biological discoveries by bridging the divide between standard two-dimensional cell culture and the complex, high-cell-density in vivo environment. Typically, the internal structures of 3D tissue-engineered samples are visualized through an involved process of physical sectioning, immunostaining, imaging, and computational reconstruction. However, recent progress in tissue-clearing methods has improved optical-imaging-depth capabilities in whole embryos and brains by reducing tissue opacity and light scattering, thus decreasing the need for physical sectioning. In this study, we assessed the application of the recently published clearing techniques ClearT2, Scale, and SeeDB to tissue-engineered neural spheres. We found that scaffold-free self-assembled adult hippocampal neural stem cell spheres of 100-μm diameter could be optically cleared and imaged using either ClearT2 or Scale, while SeeDB only marginally improved imaging depth. The ClearT2 protocol maintained sphere size, while Scale led to sample expansion, and SeeDB led to sample shrinkage. Additionally, using ClearT2 we cleared and successfully imaged spheres of C6 glioma cells and spheres of primary cortical neurons. We conclude that ClearT2 is the most effective protocol of those tested at clearing neural spheres of various cell types and could be applied to better understand neural cell interactions in 3D tissue-engineered samples.
Introduction
In recent years, adult neural stem cells (NSCs) have been identified as important players in the brain milieu during health, aging, injury, and disease, and therefore as potential therapeutic targets.1–8 However, the differentiation pathways and injury responses of NSCs remain inadequately understood. Increased understanding can be gained by studying NSCs in three-dimensional (3D) in vitro environments. Three-dimensional cell culture has been shown to maintain natural cell polarity and gene expression, increase cell–cell contacts, and slow cellular proliferation, relative to traditional two-dimensional (2D) culture.9,10 Historically, spherical 3D tissues have been used as tumor models, and current 3D culture systems, which include scaffold constructs and scaffold-free cell aggregates, are also leading to widespread insights in areas such as drug screening, development, and stem cell dynamics and differentiation.11–14
While 3D cell culture has many benefits, it also introduces new challenges, as typical imaging techniques and 2D assays must be adapted for 3D applications. The standard method for visualizing the internal structures of 3D tissues has been serial sectioning, subsequent histology or fluorescent staining, imaging, and software reconstruction. This process is notoriously labor intensive and time consuming, it permanently disrupts the 3D sample structure, and slice reconstruction can introduce structural artifacts.15 Algorithms have been investigated to reconstruct serial histological sections, aiming to correct for sectioning artifacts.16,17 Alternatively, improvements in confocal and two-photon microscopes have allowed researchers to image deeper into thick samples.18 However, large, opaque 3D tissues remain difficult to image.
Refractive index mismatches between cellular components cause light scattering, which makes fluorescent signal collection challenging in dense tissues. To address this problem, optical clearing agents have been developed to reduce scattering by better matching refractive indices within tissues or by removing scatter-inducing cellular components.18–20 One of the earliest chemical clearing protocols used a mixture of organic solvents, one part benzyl alcohol and two parts benzyl benzoate (BABB), which renders tissues transparent but unfortunately quenches green fluorescence.21–23 In 2011, a water-based clearing reagent, Scale, was first used in whole mouse brains and embryos.24 In 2012, the 3D Imaging of Solvent-Cleared Organs, or 3DISCO, protocol was published, providing a BABB-inspired, green-fluorescence-friendly, organic solvent protocol.25 In 2013, three new clearing reagent protocols were published: ClearT2, Clarity, and SeeDB.26–28 ClearT2 uses a detergent- and solvent-free mixture of formamide and poly-ethylene glycol. Clarity uses hydrogel embedding and electrophoresis to yield a lipid-free polyacrylamide-gel-embedded sample. Finally, SeeDB is a protocol that involves moving samples through an increasing fructose gradient. All of these clearing protocols have been developed for use on whole mouse embryos or organs.
To our knowledge, the application of optical clearing techniques to 3D tissue-engineered neural structures has not been explored. Herein we aimed to optically clear scaffold-free self-assembled spheres, with microscale diameters ranging from 100 to 300 μm. Although these spheres have a smaller size range than a mouse embryo or organ, their cellular density and optical opacity often exceeds that of in vivo tissues because there is no initial matrix material. The ideal clearing protocol would allow the user to image into the sphere center, lead to negligible sample changes, be minimally complex, and be compatible with immunostaining fluorophores and a standard confocal microscope. To this end we tested the clearing techniques ClearT2, Scale, and SeeDB on 3D self-assembled NSC spheres with the goal of determining an appropriate protocol. We compared imaging of whole uncleared spheres with control cryosectioned spheres, and assessed the effects of clearing methods on sample transparency, size, and visualization of fluorescently labeled cellular components.
Materials and Methods
Cell culture
Adult hippocampal NSCs
Rat adult hippocampal NSCs (Millipore) were expanded on tissue culture flasks coated with 10 μg/mL poly-L-ornithine (Sigma) in water and 5 μg/mL laminin (Life Technologies) in phosphate-buffered saline (PBS). Cell medium was composed of Neurobasal-A (Life Technologies) with 1% penicillin/streptomycin, 2 mM l-glutamine, 1X B27 growth supplement without vitamin A (Life Technologies), and 20 ng/mL basic fibroblast growth factor (bFGF; Millipore). Cells were passaged with Accutase (Millipore) for detachment and used before passage 5. All centrifugations were performed at 300 g for 3 min.
C6 glioma cells
Rat C6 glioma cells (American Type Culture Collection) were maintained in Dulbecco's modified Eagle's medium (11995-065 Life Technologies) with 1% penicillin/streptomycin and 10% fetal bovine serum. Cells were passaged with 0.25% trypsin for detachment. All centrifugations were performed at 1360 g for 6 min.
Cortical neurons
Primary cortical neurons were dissected from postnatal day 1 CD rats (Charles River Laboratories). Isolation protocol was adapted from the BrainBits Primary Neuronal Cell Culture Protocol (BrainBits, LLC). Harvested cells were seeded directly into 3D microtissue gels (see “Formation of 3D self-assembled microtissues” subheading). Cortical neurons were maintained in Neurobasal-A medium with 1% penicillin/streptomycin, 0.5 mM Glutamax (Gibco), and 1X B27 growth supplement (Life Technologies).
Formation of 3D self-assembled microtissues
Scaffold-free microtissue spheres were fabricated using agarose gels with spherical microwells to guide self-assembly.29 Briefly, agarose gels of 2% wt/vol agarose and 0.9% wt/vol sodium chloride were formed from molds with 400-μm-diameter rounded pegs (Microtissues, Inc.). NSCs, C6 cells, and cortical neurons were seeded into gels at a density of 50, 200, and 2000 cells/sphere, respectively. Cell medium was changed every 2 days and phase images were taken daily on a Nikon Eclipse TS100 microscope with a 10× objective.
Whole-sphere fixation and immunostaining
NSC, C6, and cortical neuron spheres were fixed in 4% vol/vol paraformaldehyde and 8% wt/vol sucrose in PBS overnight on days 3, 1, and 7, respectively, followed by three 1-h washes in PBS. Spheres were then removed from agarose gels. All subsequent steps were performed at room temperature on a VWR DS-500 shaker plate at 90 rpm. Spheres were permeabilized and nonspecific staining was blocked by incubation in 1% Triton X-100, 10% normal goat serum, and 4% bovine serum albumin in PBS (B-PBT) for 2 h. Spheres were incubated in primary antibody diluted in B-PBT overnight (see Table 1 for antibodies used). Next, spheres underwent two 2-h washes in 0.2% Triton X-100 in PBS (PBT), followed by one 2-h wash in B-PBT. Spheres were then incubated in secondary antibody diluted in B-PBT overnight in the dark (Table 1). Next, spheres underwent two 2-h washes in PBT, followed by incubation in a 1 μg/mL 4′,6-diamidino-2-phenylindole (DAPI) PBT solution for 1 h. Spheres were then returned to PBS.
Table 1.
Primary and Secondary Antibodies
| Primary antibody | Corresponding secondary antibody | ||||
|---|---|---|---|---|---|
| Antibody | Dilution | Company catalog No. | Antibody | Dilution | Company catalog No. |
| Mouse monoclonal | 1:200 | Millipore | Goat Cy3 | 1:500 | Jackson |
| anti-nestin | MAB353 | anti-mouse | 115-165-068 | ||
| Rabbit polyclonal | 1:100 | BTI | Goat dylight 488 | 1:500 | Jackson |
| anti-laminin | BT-594 | anti-rabbit | 111-485-144 | ||
| Mouse monoclonal | 1:50 | Abcam | Goat Cy3 | 1:200 | Jackson |
| anti-pan cadherin | Ab6528 | anti-mouse | 115-165-068 | ||
| Rabbit polyclonal | 1:200 | Dako | Goat Cy3 | 1:500 | Jackson |
| anti-S100 | Z0311 | anti-rabbit | 111-165-144 | ||
| Mouse monoclonal | 1:50 | Covance | Goat Cy3 | 1:50 | Jackson |
| anti-neuronal class III β-tubulin | MMS-435P | anti-mouse | 115-165-068 | ||
Clearing protocols
Clearing protocols were performed at room temperature on a shaker plate at 90 rpm. Control, uncleared spheres were kept in PBS solution. Control spheres for evaluation of autofluorescence, treated with secondary antibody and no primary antibody, were tested for each clearing protocol. Whole spheres were kept in control PBS or final clearing solution and transferred to a glass-bottom confocal dish for imaging.
ClearT2
Following the ClearT2 method,26 stock solutions of 20% and 40% wt/vol 8000-MW poly-ethylene glycol (PEG; Sigma) were made in water, while a 50% vol/vol formamide (Sigma) stock solution was made in PBS. The protocol for sections between 200 and 1000 μm was followed (Table 2).
Table 2.
Summary of Clearing Protocols
| Protocol | Reagents | Protocol summary | Length | Notes | Reference |
|---|---|---|---|---|---|
| ClearT2 | FormamidePEG | 1. 10 min in 25% formamide/10% PEG | 1.5 h | Kuwajima et al.26 | |
| 2. 5 min in 50% formamide/20% PEG | |||||
| 3. 60 min in 50% formamide/20% PEG | |||||
| Scale | Urea | 1. 24 h in Scale | 3 days | Samples become mechanically fragile | Hama et al.24 |
| Triton X-100 | 2. 24 h in fresh Scale | ||||
| Glycerol | 3. 24 h in fresh Scale | ||||
| SeeDB | Fructose α-Thioglycerol | 1. 8 h in 20% fructose | 3 days | Solutions become increasingly viscous and difficult to manipulate | Ke et al.28 |
| 2. 8 h in 40% fructose | |||||
| 3. 8 h in 60% fructose | |||||
| 4. 12 h in 80% fructose | |||||
| 5. 12 h in 100% fructose | |||||
| 6. 24 h in 115% fructose |
PEG, poly-ethylene glycol.
Scale
Following the Scale method variant, ScaleA2,24 clearing solution consisted of 4 M urea, 0.2% wt/vol Triton X-100, and 10% wt/wt glycerol in water. The published ScaleA2 solution contained 0.1% wt/vol Triton X-100, which was doubled to improve clearing. See Table 2 for Scale protocol.
SeeDB
Following the SeeDB method variation for minimal sample size change, SeeDBp,28 solutions of 20%, 40%, 60%, and 80% wt/vol fructose in PBS, and 100% and 115% wt/vol fructose in water were made. All solutions contained 0.5% vol/vol α-thioglycerol. See Table 2 for SeeDB protocol.
Sphere embedding and cryosectioning
Day-3 NSC spheres were fixed and washed as earlier and then taken through a gradient of 15% and 30% wt/vol sucrose in PBS over 6 h. Whole agarose gels containing spheres were removed from sucrose, blotted dry, and embedded in Tissue-Tek CRYO-OCT Compound (Andwin Scientific). Blocks were stored at −80°C, sectioned on a Leica CM3050 S cryostat into 8-μm-thick sections, and placed on SuperFrost Plus slides (Fisher Scientific).
Cryosection immunostaining
After air-drying for 15 min, sections were postfixed in 4% paraformaldehyde in PBS at room temperature for 10 min. Sections were then placed in two 5-min washes in PBS, and permeabilized for 15 min in PBT, followed by two 5-min PBS washes. Nonspecific staining was blocked by incubation in B-PBT for 1 h at room temperature. After blocking, primary antibody solution was placed on sections overnight at 4°C in a humidified chamber (Table 1). Sections then underwent two 30-min PBS washes, followed by a 30-min PBT wash. Secondary antibody solution was put on sections for 1 h at room temperature (Table 1), followed by two 10-min PBT washes. Samples were counterstained with 1 μg/mL DAPI for 10 min. Sections underwent a final PBS wash before mounting with Fluoromount-G (Southern Biotech).
Imaging
All confocal imaging was performed on a Zeiss LSM 510 Meta Confocal Laser Scanning Microscope built on an Axiovert 200M inverted microscope with ZEN 2009 software. Objectives used included a 20×air objective (Plan Apochromat, NA=0.8, working distance=0.55 mm) and a 40×water objective (C Apochromat, NA=1.2, working distance=0.28 mm). The size of the confocal aperture was 1 Airy disk and for 20×z-stacks, the optimal slice thickness of 1.035 μm was used. For z-stacks, master gain was adjusted for differences in signal intensity along structure thickness using the Auto Z Brightness Correction function, which performs spline interpolation between inputted z-depth/gain pairs. Laser power was kept constant during image acquisition. DAPI was visualized using a 405-nm diode laser, Dylight 488 was visualized with a 488-nm Argon laser, and Cy3 was visualized with a 561-nm diode laser. Images were pseudocolored to optimize visualization of fine cellular details.
Transmission images and statistical analysis
Transmission images of whole spheres were taken on a Leica Z00M 2000 dissection scope. Diameter measurements were performed in ImageJ. Statistical significance between live sphere diameters prior to fixation and sphere diameters after immunostaining was tested with a two-tailed t-test. Statistical significance between PBS control and cleared spheres was tested with a Kruskal–Wallis one-way analysis of variance followed by Dunn's method for multiple comparisons versus a control group using SigmaPlot 12.3 (Systat Software, Inc.).
Results
We prepared rat adult hippocampal NSC spheres by self-assembling cells in nonadhesive agarose gels with spherical microwells of 400-μm diameter. After 3 days in culture, NSC spheres seeded at a density of 50 cells per sphere were 104.64±22.01 μm in diameter.
Optical sectioning of whole spheres preserved cellular morphology but did not allow for inner-tissue imaging
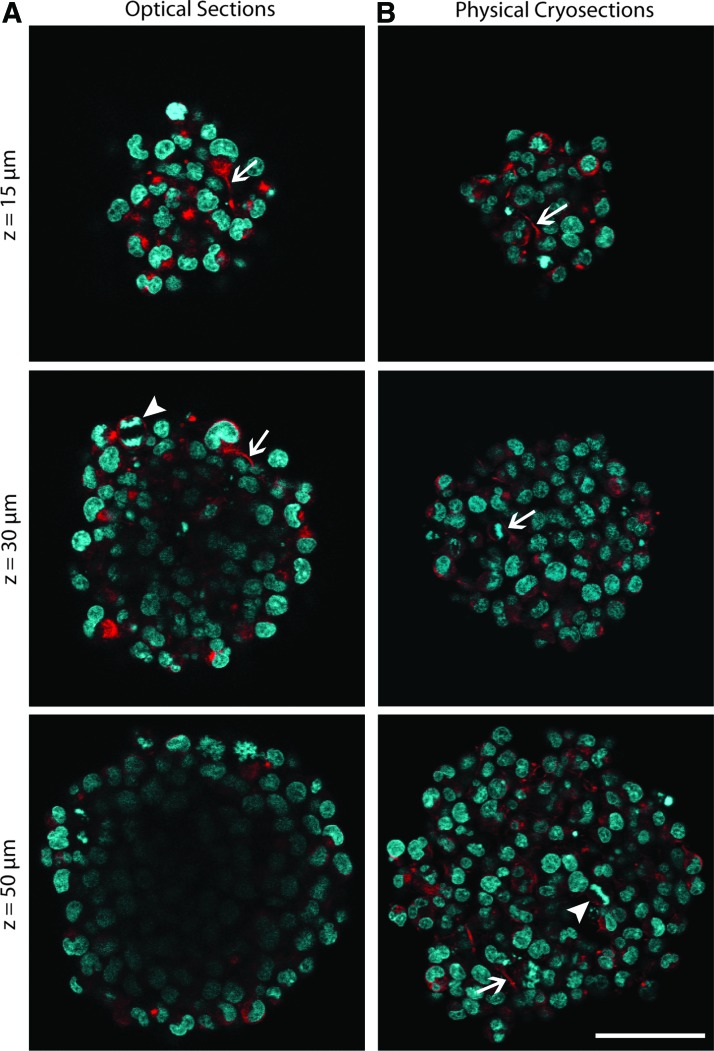
Using a confocal microscope, we imaged whole NSC spheres that had been fixed after 3 days in culture. Optical sectioning allowed for visualization of fine NSC structures, including nestin-labeled extensions and DAPI-counterstained dividing cells (Fig. 1A). However, fluorescent signal losses began at a z-depth of 20–30 μm into the sphere and soon prevented further imaging.
FIG. 1.
Comparison of optical and physical sections of neural stem cell (NSC) spheres shows signal losses in center of optical sections. NSC spheres fixed after 3 days in culture were 104.64±22.01 μm in diameter. (A) Optical sections of immunostained whole spheres in phosphate-buffered saline (PBS), at depths of 15, 30, and 50 μm. (B) Cryosections of 8-μm thickness were immunostained and images show sections of corresponding sizes to optical sections. Immunostaining was performed for nestin (red), and counterstained for DAPI (cyan). Arrows highlight cellular extensions; arrowheads highlight dividing cells. Scale bar indicates 50 μm. Color images available online at www.liebertpub.com/tec
Physical cryosectioning preserved cellular morphology and allowed for imaging of sections of inner tissue
We assessed the use of physical sectioning to view the inner structures of NSC spheres. To allow for comparisons between physical and optical sections at corresponding depths, physical sections were selected whose diameters matched those of previously obtained optical sections. Eight-micron-thick cryosections retained fine NSC features, including nestin-labeled stem cell processes and cells in different stages of division (Fig. 1B).
Optical clearing agents affected light transmission and sample size
To achieve inner-tissue imaging while preserving cellular morphology and 3D organization, we tested optical clearing agents on whole NSC spheres. Potential optical clearing agents in the literature were identified and tested, including ClearT2, Scale, and SeeDB.24,26,28
The ClearT2 protocol took ∼1.5 h. The published ClearT2 protocol variant for thicker samples was tested and did not further improve clearing in tested samples. Both the Scale and the SeeDB protocols were 72 h in duration. Using the published Scale protocol length of 21 days did not further improve clearing in tested samples. Scale treatment resulted in spheres that were more fragile and susceptible to tearing than PBS controls. SeeDB solutions of 100% and 115% fructose were highly viscous, which made sphere manipulation difficult, and resulted in the loss of many samples. Protocols are summarized in Table 2.
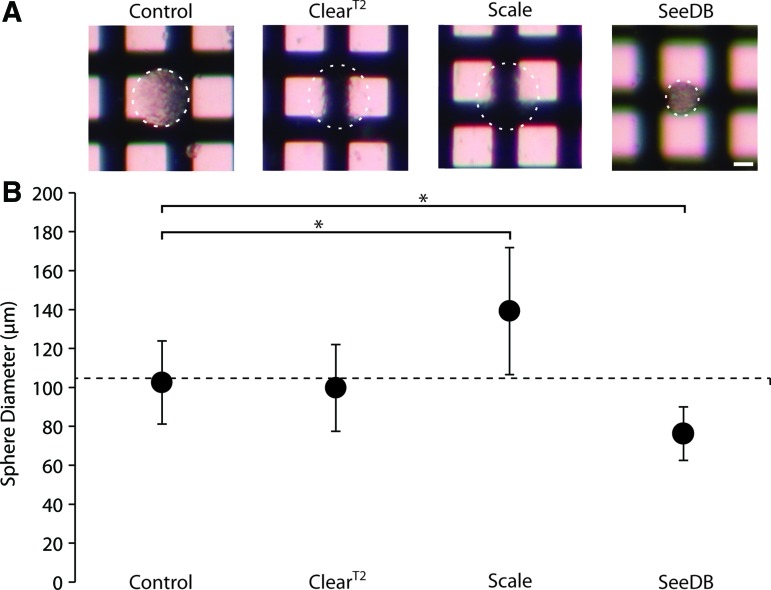
All clearing protocols resulted in more optically transparent samples, compared with PBS control spheres (Fig. 2A). Scale and ClearT2 allowed the most light transmission, while SeeDB was slightly more transparent than a control sample. It should be noted that the viscosity of 115% SeeDB solution made the acquisition of an in-focus image containing both a sample sphere and background challenging.
FIG. 2.
Changes in transparency and size were observed in 3-day-old NSC spheres after clearing with different methods. (A) Light transmission images of control and cleared spheres imaged on crosshatched backgrounds to show relative differences in transparency. White dotted lines outline spheres. Scale bar indicates 50 μm. (B) Graph of NSC sphere diameters after fixation, immunostaining, and incubation in either control PBS or clearing solutions. Horizontal dotted line indicates average sphere diameter prior to fixation. *p<0.05. Color images available online at www.liebertpub.com/tec
Because clearing protocols can cause tissue swelling or shrinkage, we measured the diameter of spheres at day 3 in culture, after the immunostaining protocol, and after each clearing protocol (Fig. 2B). On day 3 in culture, live NSC spheres had an average diameter of 104.64±22.01 μm (n=72 spheres, dotted line Fig. 2B). After fixation and immunostaining, spheres in PBS had an average diameter of 102.53±21.40 μm (n=73), which was not significantly different than before fixation (p=0.561). ClearT2-treated spheres had a diameter of 99.77±22.3 (n=47), which was not significantly different from PBS control spheres. Scale protocol treatment resulted in sphere expansion (p<0.05) to an average diameter of 139.2±32.64 (n=58). SeeDB protocol treatment resulted in sphere shrinkage (p<0.05) to an average diameter of 76.2±13.77 (n=17).
Tissue clearing with ClearT2 and Scale allowed confocal imaging to a depth of 100 μm
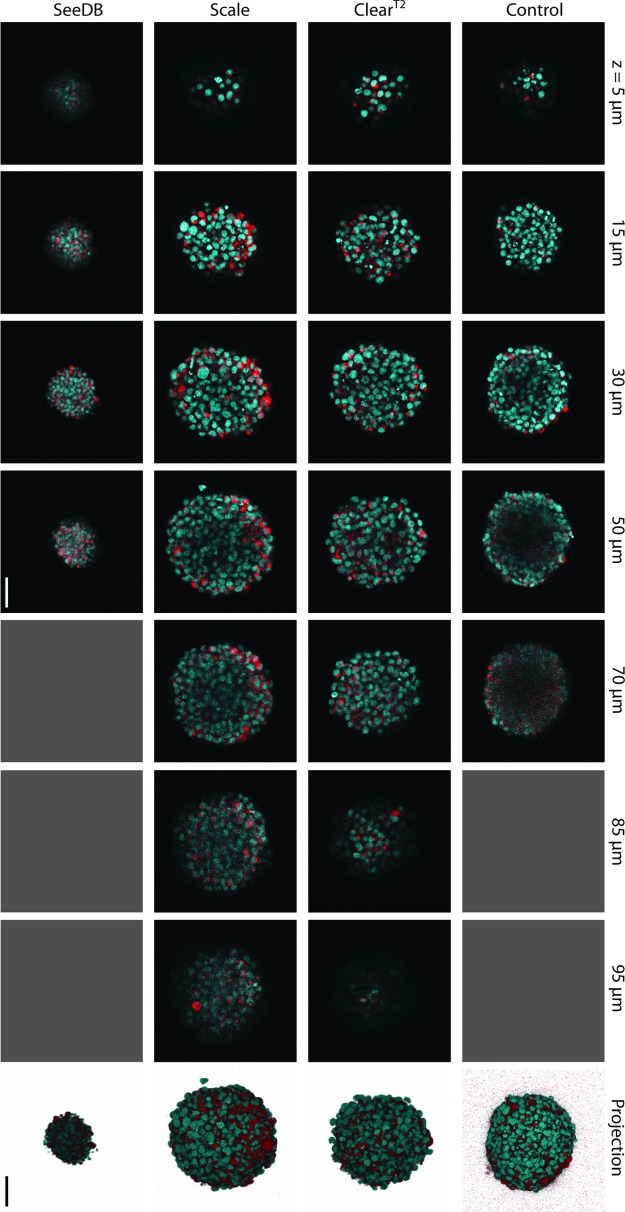
To visualize the effects of clearing protocols on imaging ability, we imaged immunofluorescently labeled samples on a confocal microscope (Fig. 3). As expected in control spheres, fine features were visible at z-depths of up to 30 μm, while at larger z-depths there was a low signal-to-noise ratio, which resulted in difficulties distinguishing cellular features. ClearT2- and Scale-treated spheres were both imageable up to 100 μm, the full diameter of the ClearT2-cleared sphere. In ClearT2-treated spheres, fine features within the center of the sphere were visible. Scale-treated spheres had more light scattering than ClearT2-treated spheres, as can be visualized by a decrease in signal from the center of the 50- and 70-μm Scale z-stack slices. Spheres in SeeDB had a low signal-to-noise ratio and DAPI-labeled nuclei were difficult to distinguish. Comparing projection images of control to ClearT2- and Scale-treated spheres demonstrated that the outer structures of spheres remained intact with these methods. While ClearT2 and Scale both allowed imaging 100 μm into NSC spheres, the sample expansion seen with Scale treatment (Fig. 2B) led us to choose ClearT2 for further study.
FIG. 3.
Confocal slices at denoted z-depths and corresponding projections of 3-day-old NSC spheres show changes in fluorescent nestin (red) and DAPI (cyan) signal visualization due to the application of different clearing protocols. Gray boxes represent depths at which an image could not be acquired. Scale bar indicates 50 μm. Color images available online at www.liebertpub.com/tec
ClearT2 allowed visualization of extracellular matrix and transmembrane proteins in NSC spheres
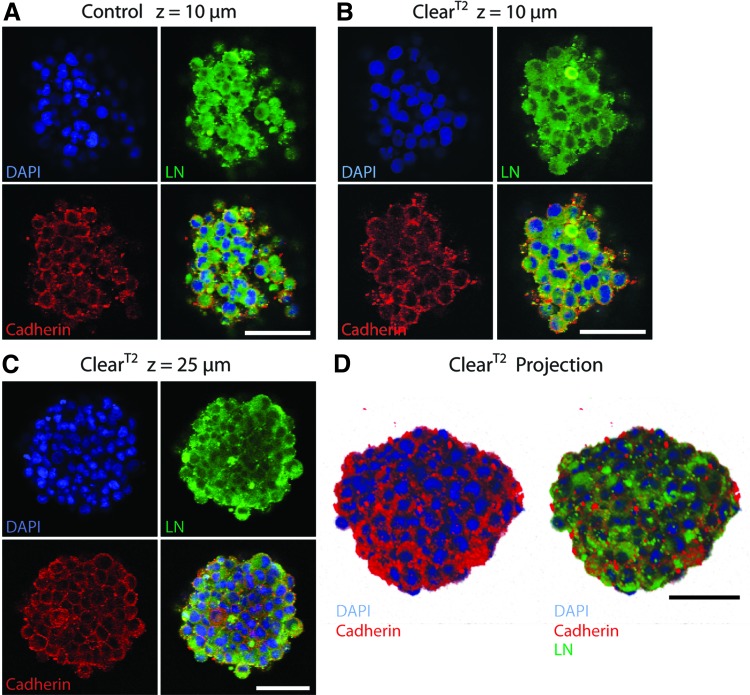
Although self-assembled microtissues are initially formed without matrix material, we have observed that maturing neural spheres express extracellular matrix proteins (Yu-Ting Liu Dingle, unpublished data). To see whether NSC extracellular and transmembrane proteins were preserved during the clearing process, spheres were stained for the extracellular matrix molecule laminin (LN) and the cell–cell adhesion protein cadherin (Fig. 4). In 3-day-old NSC spheres, LN was observed localized to cells and in the extracellular space, while cadherin was present at sites of cell–cell contact. Imaging at comparable depths (z=10 μm) in control and ClearT2-treated spheres confirmed that antigens were preserved after clearing (Fig. 4A, B). LN and cadherin expression was visible throughout the ClearT2 NSC sphere (Fig. 4C, D).
FIG. 4.
Extracellular laminin and transmembrane cadherin were preserved after the ClearT2 protocol. NSC spheres were fixed after 3 days in culture, immunostained, DAPI counterstained (blue), and cleared. Comparing superficial optical sections of control (A) and ClearT2-treated (B) spheres demonstrates that laminin (green) and cadherin (red) antigens were preserved after clearing (z=10 μm). (C) Laminin and cadherin staining was visible within the ClearT2-treated sphere (z=25 μm). (D) Z-stack projection of an entire sphere (diameter=76 μm) shows cadherin located at cell–cell junctions and laminin present extracellularly. All scale bars indicate 50 μm. Color images available online at www.liebertpub.com/tec
ClearT2 was successfully applied to self-assembled spheres of other cell types
We explored whether ClearT2 could be applied to spheres composed of different neural cell types. C6 glioma cells were selected for testing because spheres are commonly used as tumor models, and cortical neurons were chosen to assess ClearT2 compatibility with primary neuronal cell spheres.
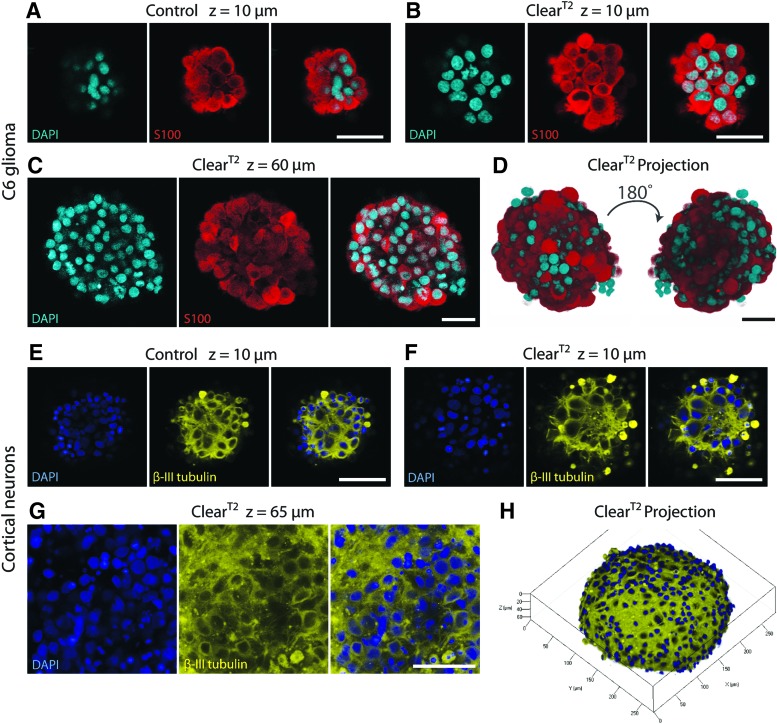
In 1-day-old C6 spheres, fine detail in the calcium-binding protein S100 could be seen in both control (Fig. 5A) and ClearT2-treated (Fig. 5B) samples, which demonstrated that the S100 antigen was preserved after clearing. An inner optical slice (Fig. 5C) and the reversal of a projection image (Fig. 5D) illustrated that cleared spheres could be imaged at all z-depths.
FIG. 5.
ClearT2 protocol was successfully applied to spheres composed of various neural cell types. (A–D) C6 glioma spheres were fixed after 1 day in culture, immunostained, DAPI counterstained (cyan), and cleared. Comparing superficial optical sections of control (A) and ClearT2-treated (B) spheres illustrates that S100 (red) antigen was preserved after clearing (z=10 μm). (C) ClearT2-treated sphere shows S100 staining halfway through the sphere (z=60 μm). (D) 180° rotation of 110-μm z-stack projection demonstrates effective clearing throughout the entire C6 sphere. (E–H) Primary cortical neuron spheres were fixed after 7 days in culture, immunostained, DAPI counterstained (blue), and cleared. Comparing superficial optical sections of control (E) and ClearT2-treated (F) spheres confirms that β-III tubulin (yellow) antigen was preserved after clearing (z=10 μm). (G) ClearT2-treated sphere shows β-III tubulin staining halfway through the sphere (z=65 μm). (H) Z-stack projection of half of a cortical sphere (depth=67 μm) displays β-III tubulin network formation in sphere periphery. All scale bars indicate 50 μm. Color images available online at www.liebertpub.com/tec
In 7-day-old primary cortical neuron spheres, a comparison of β-III tubulin-labeled processes between control (Fig. 5E) and ClearT2-treated (Fig. 5F) samples displayed that clearing preserved β-III tubulin antigens. In ClearT2-treated spheres, β-III tubulin could be observed even in the middle of central z-slices (Fig. 5G). A projection image of a z-stack taken through half of the sphere thickness allowed for visualization of the dense β-III tubulin-labeled neurite network within these spheres (Fig. 5H).
Discussion
Significant effort has been focused on improving optical imaging of thick native tissues through clearing agents, with an emphasis on maximizing fluorescent signal collection by reducing light scattering, and maintaining the physical characteristics of samples. Herein we assessed the efficacy of existing optical clearing protocols on self-assembled, scaffold-free spheres composed of different neural cell types. Specifically, we tested ClearT2, Scale, and SeeDB on 3-day-old rat adult hippocampal NSC spheres of ∼100 μm in diameter, and found that ClearT2 was the most effective in clearing these samples. Additionally, we investigated the application of ClearT2 to spheres composed of C6 glioma cells or primary cortical neurons, and found that ClearT2 was also able to clear these structures. These clearing techniques were developed for whole native tissues and had not been previously tested on tissue-engineered neural spheres; this work provides the first evidence of such an application.
ClearT2 was developed with the goals of creating a method that could be performed in an abbreviated time-span, and preserved sample volume and fluorescent signal. It was originally applied to whole mouse embryos and postnatal mouse brains in conjunction with lipophilic dyes, fluorescent tracers, endogenous fluorescence, and fluorescent antibody immunostaining.26 In our application of ClearT2 to neural spheres, we found that sample volume was preserved and fluorescent signal was detectable at all depths within spheres of 100-μm diameter. Both counterstain DAPI excitation fluorescence of 405 nm and secondary antibodies with excitation wavelengths of 488 and 550 nm were preserved after clearing. No changes in physical structure of, for example, β-III tubulin-labeled processes, were observed after treatment. Extracellular, transmembrane, and intracellular proteins were preserved following ClearT2 treatment.
The development of Scale was focused on preserving endogenous fluorescent signal and clearing deep into mouse embryos and mouse brains of all development stages. With Scale treatment, the authors observed sample expansion, and quantified this expansion as 1.25-fold linear after 5 days in Scale.24 Atsushi Kaida et al. applied the Scale clearing protocol to self-assembled spheres composed of cancerous HeLa cells expressing the fluorescent ubiquitination-based cell-cycle indicator (Fucci).30 In that study, Scale clearing of 10–13-day-old, 500-μm-diameter spheres allowed for an increase of imageable z-distance from 60 μm in live spheres, to 200 μm in fixed, cleared spheres. However, sample expansion was observed and the 1.25-fold linear expansion reported in the Scale publication was assumed, suggesting that the imaging depth was actually 160 μm (not 200 μm) of the original sphere. Based on the diameter expansion reported in the present study of 1.36-fold after only 3 days, we suggest that expansion with Scale may be dependent on sample, including cell type, matrix components, and maturity, and should be calculated in a sample-dependent manner. Spheres presented herein were relatively immature and likely had little connective matrix material, potentially making them more permissive to expansion than the matured and highly connected adult mouse brain.
In the original Scale publication, the authors restored the original optical opacity of cleared samples using PBS, and subsequently performed immunostaining to ensure that expansion was not permanently disrupting or distorting physical structures. Restored, immunostained samples were seen to have identical fluorescent cell structures and patterning to uncleared samples.24 Our protocol differs as immunostaining is performed prior to Scale clearing; we investigate whether Scale will allow for the subsequent visualization of immunofluorescently labeled cell structures. Of note, we demonstrate that antibody binding and fluorescence are compatible with 3 days of Scale treatment in NSC spheres.
SeeDB was developed as a method to clear adult mouse brain and olfactory bulb while preserving the fluorescence of lipophilic dextran tracers and endogenous proteins, and maintaining sample volume.28 The refractive indices of SeeDB and Scale are 1.49 and 1.38, respectively. While the refractive index of the ClearT2 solution was not reported,26 we can estimate the refractive index to be between 100% formamide and water, which have indices of 1.45 and 1.33, respectively.31 Differences in refractive indices may make particular clearing protocols suitable to clear different tissue types.20 The refractive index of SeeDB is around that of lipids and, as shown in the SeeDB publication, SeeDB was more successful than Scale at clearing lipid-rich white matter in brains, suggesting that refractive index matching was optimized with SeeDB. The NSC samples tested herein were maintained in a stem-like state with bFGF, so differentiation into myelinating cells was minimal. This suggests that the refractive index of our NSC spheres was lower than white matter, and the refractive index of SeeDB was not matched to our samples, which resulted in poor clearing.
While sample volume changes were not observed in the original SeeDB publication,28 we observed a 0.74-fold sphere diameter reduction after SeeDB treatment. We hypothesize that differences in maturity between our 3-day-old spheres, and developing or mature mouse brains, may have made our samples more susceptible to compression by the saturated fructose solution. This finding also highlights the need to investigate the effects of different clearing protocols on a sample-dependent basis.
The original Scale and SeeDB publications included the development of microscope objectives corrected for the specific refractive indices of the clearing solutions. We did not use these specialized microscope objectives even though they may have increased imaging depths in the current study; our goal was to investigate clearing protocols that were compatible with accessible and affordable confocal microscope objectives. Additionally, while it has been observed that refractive index mismatches between the immersion medium and tissue can lead to measurement errors, these mismatches can be simply accounted for by using correction factors to rescale image dimensions.32,33
There are clearing protocols that did not fall within the scope of this study. The 3DISCO protocol involves tissue dehydration with tetrahydrofuran and subsequent clearing with dibenzyl ether.25 3DISCO demonstrated excellent clearing of whole brain and spinal cord. However, it is established that the dehydration steps required for organic solvent use can cause sample shrinkage, and our goal was to identify a protocol that did not induce sample size changes.25,34
The clearing protocol Clarity has received much attention because of its success in clearing whole mouse brains and human brain tissue, and its compatibility with and enhancement of typical immunostaining methods.27 This protocol involves the perfusion of tissues with polyacrylamide monomers and paraformaldehyde, polymerization of the polyacrylamide-tissue complex, and removal of tissue lipids through an ionic extraction technique. While Clarity is useful and perhaps necessary for whole-organ tissues, this protocol may be overly complex and cost prohibitive for samples on the order of hundreds of microns. For these reasons the Clarity protocol was not tested in this study; however, future research may find that it can be adapted for small microtissues.
There are many innovative image and data acquisition techniques that can be applied in conjunction with optical clearing. For example, Calle et al. recently investigated the use of the clearing technique BABB in conjunction with two-photon-induced tissue autofluorescence and second-harmonic generation to visualize extracellular matrix molecules in thick samples.35 Chen et al. have applied the commercially available clearing reagent FocusClear to 3D intestinal crypt organoids and biopsied tissues to supplement conventional 2D slice histopathology.36 Combining acquisition techniques with clearing protocols will allow researchers to maximize the information obtained from tissue-engineered samples and increase the throughput of imaging and analysis of intact 3D tissues.
Conclusions
Through the application of optical clearing protocols to scaffold-free self-assembled spheres we highlight that clearing protocols can have diverse effects on sample size and opacity, and should be chosen in a sample-dependent fashion. We show that ClearT2 maintains the size of scaffold-free self-assembled spheres, is compatible with typical immunostaining techniques, and allows imaging through 100-μm-diameter spheres. Thus we conclude that ClearT2 is a valuable tool for studying tissue-engineered 3D structures of various neural cell types.
Acknowledgments
Funding was provided by the National Science Foundation (NSF) GRFP to M.B., NSF CBET 1134166, National Institutes of Health (NIH) 5R01HL110791 subcontract, NIH R21HL113918 subcontract, and Brown Institute for Brain Science Pilot Award to D.H.K. The authors thank Geoff Williams of the Leduc Bioimaging Facility for technical assistance with confocal microscopy. The authors thank Melinda Golde and Paula Weston of the Molecular Pathology Core at Brown University, and Maggie Vantangoli for cryosectioning assistance. The authors thank Sean Curran for the preculturing of C6 glioma cells and Michael Kader for Scale pilot studies. The authors additionally thank Jeffrey Morgan for critical reading of the article.
Disclosure Statement
No competing financial interests exist.
References
- 1.Nakatomi H., Kuriu T., Okabe S., Yamamoto S., Hatano O., Kawahara N., Tamura A., Kirino T., and Nakafuku M.Regeneration of hippocampal pyramidal neurons after ischemic brain injury by recruitment of endogenous neural progenitors. Cell 110,429, 2002 [DOI] [PubMed] [Google Scholar]
- 2.Salman H., Ghosh P., and Kernie S.G.Subventricular zone neural stem cells remodel the brain following traumatic injury in adult mice. J Neurotrauma 21,283, 2004 [DOI] [PubMed] [Google Scholar]
- 3.Doetsch F.A niche for adult neural stem cells. Curr Opin Genet Dev 13,543, 2003 [DOI] [PubMed] [Google Scholar]
- 4.Temple S.The development of neural stem cells. Nature 414,112, 2001 [DOI] [PubMed] [Google Scholar]
- 5.Ming G.-L., and Song H.Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron 70,687, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanai N., Tramontin A.D., Quinones-Hinojosa A., Barbaro N.M., Gupta N., Sandeep K., Lawton M.T., McDermott M.W., Parsa A.T., Verdugo J.M.-G., Berger M.S., and Alvarez-Buylla A.Unique astrocyte ribbon in adult human brain contains neural stem cells but lacks chain migration. Nature 427,740, 2004 [DOI] [PubMed] [Google Scholar]
- 7.Yoshimura S., Teramoto T., Whalen M.J., Irizarry M.C., Takagi Y., Qiu J., Harada J., Waeber C., Breakefield X.O., and Moskowitz M.A.FGF-2 regulates neurogenesis and degeneration in the dentate gyrus after traumatic brain injury in mice. J Clin Invest 112,1202, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshimura S., Takagi Y., Harada J., Teramoto T., Thomas S.S., Waeber C., Bakowska J.C., Breakefield X.O., and Moskowitz M.A.FGF-2 regulation of neurogenesis in adult hippocampus after brain injury. Proc Natl Acad Sci USA 98,5874, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birgersdotter A., Sandberg R., and Ernberg I.Gene expression perturbation in vitro-a growing case for three-dimensional (3D) culture systems. Semin Cancer Biol 15,405, 2005 [DOI] [PubMed] [Google Scholar]
- 10.Pampaloni F., Reynaud E.G., and Stelzer E.H.K.The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol 8,839, 2007 [DOI] [PubMed] [Google Scholar]
- 11.Kim J.B.Three-dimensional tissue culture models in cancer biology. Semin Cancer Biol 15,365, 2005 [DOI] [PubMed] [Google Scholar]
- 12.Achilli T.-M., McCalla S., Meyer J., Tripathi A., and Morgan J.R.Multilayer spheroids to quantify drug uptake and diffusion in 3D. Mol Pharm 11,2071, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lancaster M.A., Renner M., Martin C.-A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., and Knoblich J.A.Cerebral organoids model human brain development and microcephaly. Nature 501,373, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baraniak P.R., and McDevitt T.C.Scaffold-free culture of mesenchymal stem cell spheroids in suspension preserves multilineage potential. Cell Tissue Res 347,701, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ju T., Warren J., Carson J., Bello M., Kakadiaris I., Chiu W., Thaller C., and Eichele G.3D volume reconstruction of a mouse brain from histological sections using warp filtering. J Neurosci Methods 156,84, 2006 [DOI] [PubMed] [Google Scholar]
- 16.Skoglund T.S., Hammar I., Olsson C., and Berthold C.H.A method for 2D reconstruction of intracellularly labeled neurons from sequential sections. J Neurosci Methods 53,199, 1994 [DOI] [PubMed] [Google Scholar]
- 17.Ourselin S., Roche A., Subsol G., Pennec X., and Ayache N.Reconstructing a 3D structure from serial histological sections. Image Vis Comput 19,25, 2000 [Google Scholar]
- 18.Kim S.-Y., Chung K., and Deisseroth K.Light microscopy mapping of connections in the intact brain. Trends Cogn Sci 17,596, 2013 [DOI] [PubMed] [Google Scholar]
- 19.Tuchin V. V.Optical clearing of tissues and blood using the immersion method. J Phys D Appl Phys 38,2497, 2005 [Google Scholar]
- 20.Zhu D., Larin K. V., Luo Q., and Tuchin V.V.Recent progress in tissue optical clearing. Laser Photon Rev 7,732, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spalteholz W.Über das Durchsichtigmachen von menschlichen und tierischen Präparaten und seine theoretischen Bedingungen. Hirzel S., ed. Leipzig: Druck von Breitkopf & Härtel, 1914 [Google Scholar]
- 22.Dent J.A., Polson A.G., and Klymkowsky M.W.A whole-mount immunocytochemical analysis of the expression of the intermediate filament protein vimentin in Xenopus. Development 105,61, 1989 [DOI] [PubMed] [Google Scholar]
- 23.Dodt H.-U., Leischner U., Schierloh A., Jahrling N., Mauch C.P., Deininger K., Deussing J.M., Eder M., Zieglgansberger W., and Becker K.Ultramicroscopy : three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods 4,331, 2007 [DOI] [PubMed] [Google Scholar]
- 24.Hama H., Kurokawa H., Kawano H., Ando R., Shimogori T., Noda H., Fukami K., Sakaue-Sawano A., and Miyawaki A.Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci 14,1481, 2011 [DOI] [PubMed] [Google Scholar]
- 25.Ertürk A., Becker K., Jährling N., Mauch C.P., Hojer C.D., Egen J.G., Hellal F., Bradke F., Sheng M., and Dodt H.-U.Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat Protoc 7,1983, 2012 [DOI] [PubMed] [Google Scholar]
- 26.Kuwajima T., Sitko A.A., Bhansali P., Jurgens C., Guido W., and Mason C.ClearT: a detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development 140,1364, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung K., Wallace J., Kim S.-Y., Kalyanasundaram S., Andalman A.S., Davidson T.J., Mirzabekov J.J., Zalocusky K.A., Mattis J., Denisin A.K., Pak S., Bernstein H., Ramakrishnan C., Grosenick L., Gradinaru V., and Deisseroth K.Structural and molecular interrogation of intact biological systems. Nature 497,332, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ke M.-T., Fujimoto S., and Imai T.SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci 16,1154, 2013 [DOI] [PubMed] [Google Scholar]
- 29.Napolitano A.P., Chai P., Dean D.M., and Morgan J.R.Dynamics of the self-assembly of complex cellular aggregates on micromolded nonadhesive hydrogels. Tissue Eng 13,2087, 2007 [DOI] [PubMed] [Google Scholar]
- 30.Kaida A., and Miura M.Visualizing the effect of tumor microenvironments on radiation-induced cell kinetics in multicellular spheroids consisting of HeLa cells. Biochem Biophys Res Commun 439,453, 2013 [DOI] [PubMed] [Google Scholar]
- 31.Cases A.M., Marigliano A.C.G., Bonatti C.M., and Solimo H.N.Density, viscosity, and refractive index of formamide, three carboxylic acids, and formamide+ carboxylic acid binary mixtures. J Chem Eng Data 46,712, 2001 [Google Scholar]
- 32.Hell S., Reiner G., Cremer C., and Stelzer E.H.K.Aberrations in confocal fluorescence microscopy induced by mismatches in refractive index. J Microsc 169,391, 1993 [Google Scholar]
- 33.Bucher D., Scholz M., Stetter M., Obermayer K., and Pflüger H.J.Correction methods for three-dimensional reconstructions from confocal images: I. Tissue shrinking and axial scaling. J Neurosci Methods 100,135, 2000 [DOI] [PubMed] [Google Scholar]
- 34.Ertürk A., Mauch C.P., Hellal F., Förstner F., Keck T., Becker K., Jährling N., Steffens H., Richter M., Hübener M., Kramer E., Kirchhoff F., Dodt H.U., and Bradke F.Three-dimensional imaging of the unsectioned adult spinal cord to assess axon regeneration and glial responses after injury. Nat Med 18,166, 2012 [DOI] [PubMed] [Google Scholar]
- 35.Calle E.A., Vesuna S., Dimitrievska S., Zhou K., Huang A., Zhao L., Niklason L.E., and Levene M.J.The use of optical clearing and multiphoton microscopy for investigation of three-dimensional tissue-engineered constructs. Tissue Eng Part C 00,1, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y., Tsai Y.H., Liu Y.A., Lee S.H., Tseng S.H., and Tang S.C.Application of three-dimensional imaging to the intestinal crypt organoids and biopsied intestinal tissues. Sci World J 2013,2013 [DOI] [PMC free article] [PubMed] [Google Scholar]