Abstract
In stratified epithelial tissues, homeostasis relies on the self-renewing capacity of stem cells located within the innermost basal layer1. As basal cells become suprabasal, they lose proliferative potential and embark on a terminal differentiation programme2,3. Here, we show that microRNA-203 is induced in the skin concomitantly with stratification and differentiation. By altering miR-203’s spatiotemporal expression in vivo, we show that miR-203 promotes epidermal differentiation by restricting proliferative potential and inducing cell-cycle exit. We identify p63 as one of the conserved targets of miR-203 across vertebrates. Notably, p63 is an essential regulator of stem-cell maintenance in stratified epithelial tissues4–9. We show that miR-203 directly represses the expression of p63: it fails to switch off suprabasally when either Dicer1 or miR-203 is absent and it becomes repressed basally when miR-203 is prematurely expressed. Our findings suggest that miR-203 defines a molecular boundary between proliferative basal progenitors and terminally differentiating suprabasal cells, ensuring proper identity of neighbouring layers.
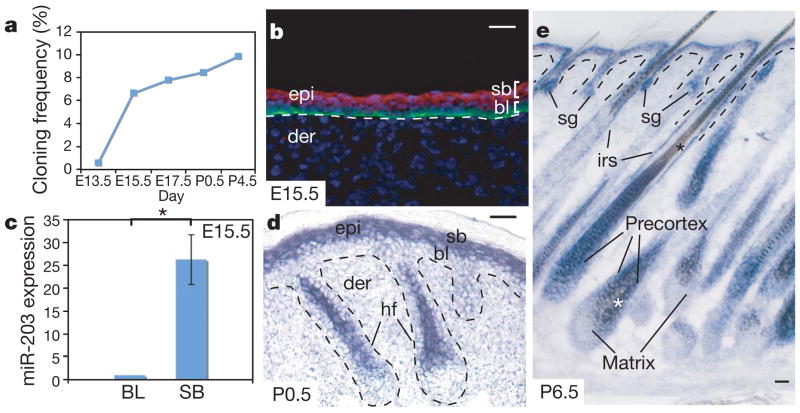
MicroRNAs are small, non-coding RNAs that regulate gene expression post-transcriptionally by directly targeting RNA-induced silencing complex (RISC) to cognate messenger RNA targets10. When miRNAs are globally ablated in skin epithelium by conditionally targeting the gene that encodes the miRNA-processing enzyme Dicer1, hair follicles fail to invaginate. This distorts epidermal morphology, compromising the barrier and underscoring the functional importance of these small RNAs in skin development11,12. To gain further insight into the possible significance of different skin miRNAs, we constructed epidermal miRNA libraries using total RNAs isolated from pure epidermis starting from embryonic day 13.5 (E13.5), when it is still a single-layered epithelium, to postnatal day 4.5 (P4.5), when it is fully stratified. Among more than 100 epidermal miRNAs cloned, miR-203 barely surfaced in the pool of E13.5 clones but emerged as one of the most abundant epidermal miRNAs from E15.5 onwards (Fig. 1a).
Figure 1. Spatiotemporal expression of miR-203 during skin development.
a, Temporal induction of miR-203 as measured by cloning frequency. b, d, e, In situ hybridization reveals restriction of miR-203 to suprabasal, differentiating layers of skin. sb, suprabasal; bl, basal layer; epi, epidermis; der, dermis; sg, sebaceous gland; irs, inner root sheath; hf, hair follicle. Asterisks in e represent brown melanin pigment that is not a hybridization signal. In b, anti-β4-integrin co-labelling is in green and miR-203 in situ pseudocoloured signal is in red. c, qRT–PCR of FACS-purified cells from E15.5 epidermis reveals 25-fold more miR-203 in suprabasal (SB) versus basal layer (BL) cells (*P < 0.002). Error bars (s.d.) are derived from three experiments with basal layer level set as 1. Scale bars are 30 μm.
The significant upregulation of miR-203 between E13.5 and E15.5 was suggestive that this miRNA may be absent in multipotent progenitors of single-layered epidermis, but is induced upon stratification and differentiation. By in situ hybridization13, miR-203 was largely confined to suprabasal cells of epithelial tissues, and was especially prominent in skin (Fig. 1b, d, e and Supplementary Fig. 1). The specificity of hybridization was confirmed by analysing conditionally ablated Dicer1 skin, in which expression of miR-203 and other mature miRNAs was abolished11 (Supplementary Fig. 1a). Quantification of its differential expression by quantitative PCR with reverse transcription (qRT–PCR) revealed ~25-fold more miR-203 in E15.5 suprabasal cells than in their basal counterparts (Fig. 1c). Similarly, miR-203 was rapidly upregulated when primary mouse keratinocytes were induced by calcium to differentiate in vitro (Supplementary Fig. 1b).
Epidermal development precedes that of its appendages. However, at early stages of hair follicle development, miR-203 was not detected. By E17.5, faint miR-203 hybridization was detected within the emerging suprabasal cells of developing hair follicles and expression was also seen in stratified layers of developing tongue epithelia (Supplementary Fig. 1f, g). As development advanced, miR-203 expression intensified in differentiating cells of epidermis, hair follicles and sebaceous glands (Fig. 1d, e). Present throughout transcriptionally active, terminally differentiating cells of skin epithelium, miR-203 was conspicuously absent in its proliferating progenitor compartments. Interestingly, miR-203’s sequence and expression pattern seemed to be conserved among vertebrates, in which the epidermis is stratified, but not in eukaryotes that have a single-layered epidermis (Supplementary Fig. 2a, b).
If miR-203 functions in the switch between proliferative and terminally differentiating compartments in vertebrate skin, it may be expected to repress its basal targets once basal cells become suprabasal and enter the terminal differentiation programme. To test this hypothesis, we generated transgenic mice expressing miR-203 under the control of the keratin 14 (K14) promoter, active by E15 in basal progenitors of stratified epithelia14. Most K14-miR-203 mice died shortly after birth owing to apparent dehydration and/or malnutrition. By E18.5, the level of transgenic basal miR-203 was comparable to endogenous suprabasal miR-203 (Fig. 2a). Moreover, transgenic miR-203 expression did not interfere with endogenous miRNA processing15 (Supplementary Fig. 3).
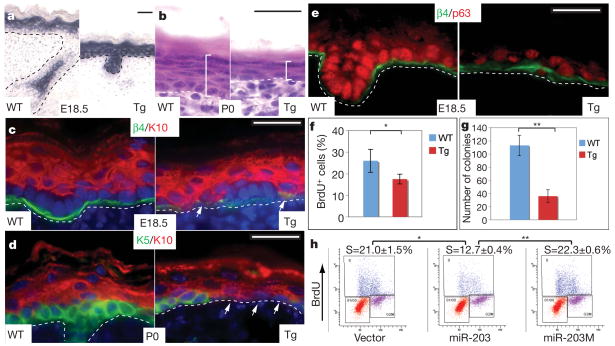
Figure 2. Premature activation of miR-203 in epidermis restricts its proliferative potential.
a, In situ hybridization detects precociously expressed miR-203 in basal epidermis and hair germs of K14-miR-203 Tg skin. WT, wild type. b, By P0, Tg epidermis is thinner than the wild type. c–f, Signs of basal cell depletion in Tg skin. Arrows denote keratin-10 (K10)-positive, keratin-5 (K5)-negative, β4-integrin-low cells aberrantly juxtaposed to basement membrane. Note marked reduction of p63 in e. In f, quantifications reflect *P < 0.003, n = 8. g, Marked reduction in Tg versus wild-type colony-forming efficiency in vitro (**P < 0.0001, n = 3). In f and g, error bars (s.d.) are derived from eight or three experiments as indicated. h, Transduction of primary mouse keratinocytes with miR-203, but not empty vector or mutant miR-203M, results in a marked decline in S-phase cells that incorporate BrdU (*P < 0.001, **P < 0.0001). Values represent mean ± s.d. from three experiments. Scale bars are 30 μm.
At E18.5, K14-miR-203 back skin epidermis was noticeably thinner than that of its wild-type littermates (Supplementary Fig. 4a). By the time of birth (P0), transgenic epidermis consisted of a layer of flattened basal cells and one layer of suprabasal cells (Fig. 2b). Hence, a thin epidermis can reflect either defective differentiation, as in mice conditionally targeted by K14-Cre for Notch effector protein RBPj16, or impaired stem cells, as in p63 null epidermis4,6,9. To distinguish between these possibilities, we first examined keratin 10, an early differentiation marker downstream of canonical Notch signalling16. At E18.5, many keratin-10-positive cells within K14-miR-203 epidermis were aberrantly juxtaposed to the basement membrane (Fig. 2c). As development progressed, basal progenitor/stem cells marked by keratin 5 were missing over increasingly larger epidermal stretches that were replaced by flat cells expressing keratin 10 (Fig. 2d). No discernible apoptotic cells were detected as judged by TdT-mediated dUTP nick end labelling (TUNEL) assay and active caspase-3 (data not shown).
The depletion of basal stem cells in K14-miR-203 epidermis bore a resemblance to p63 null epidermis6,9. Interestingly, anti-p63 immunolocalization revealed only sporadic, weak expression in E18.5 K14-miR-203 basal cells (Fig. 2e). Quantification of 5-bromodeoxyuridine (BrdU)-positive cells after a 2-h pulse-labelling revealed a significant reduction in the proliferative pool of E18.5 transgenic progenitors (Fig. 2f).
To address whether miR-203 restricts the proliferative potential of epidermal stem cells, we compared the clonogenic capacity of primary mouse keratinocytes cultured from E18.5 transgenic with wild-type littermates. As expected, wild-type keratinocytes formed typical holoclones composed of small, undifferentiated cells capable of long-term passage17,18. By contrast, transgenic keratinocytes produced mostly paraclones, composed of large, flattened cells17,18 (Supplementary Fig. 4b). Moreover, approximately fourfold fewer, and significantly smaller, colonies formed from transgenic compared with wild-type keratinocytes (Fig. 2g and Supplementary Fig. 4c). On passage, only wild-type clones gave rise to colonies. Consistent with a requirement for p63 in maintaining proliferative potential of epidermal stem cells9, p63 was markedly diminished in transgenic keratinocytes.
To test whether these dramatic effects of transgene expression are specific to miR-203, we transduced primary mouse keratinocytes with retroviral vectors expressing either miR-203 or miR-203M, a miR-203 mutant harbouring mutations within the 5′ seed (Supplementary Fig. 4d), or empty vector alone. Only miR-203 impaired keratinocyte proliferation and colony formation (Fig. 2h and Supplementary Fig. 4e).
To identify early consequences of losing miR-203 expression, we first examined p63 and cell-cycle status in E18.5 Dicer1 null epidermis. In contrast to the wild type2,5,9, Dicer1 null epidermis showed frequent p63-positive, BrdU-positive and phospho-histone-H3-positive mitotic suprabasal cells (Supplementary Fig. 5a–c). Fluorescence-activated cell sorting (FACS) quantification revealed an approximately threefold increase in the number of G2/M phase suprabasal cells (Supplementary Fig. 5d).
To test specifically whether miR-203 represses proliferative potential of suprabasal cells in vivo, we adapted the antagomir method19 to repress miR-203 expression in dorsal skin of neonatal mice, in which basal-layer proliferation is still high. We first used a dye-conjugated antagomir to document efficient incorporation in epidermis within 24 h following a single subcutaneous injection (Supplementary Fig. 6a). One day after three dorsal injections, antagomir-203, but not mm-antagomir-203 (a 4-nucleotide mismatched control) or PBS markedly and specifically repressed miR-203 expression in surrounding skin (Fig. 3a and Supplementary Fig. 6b, c).
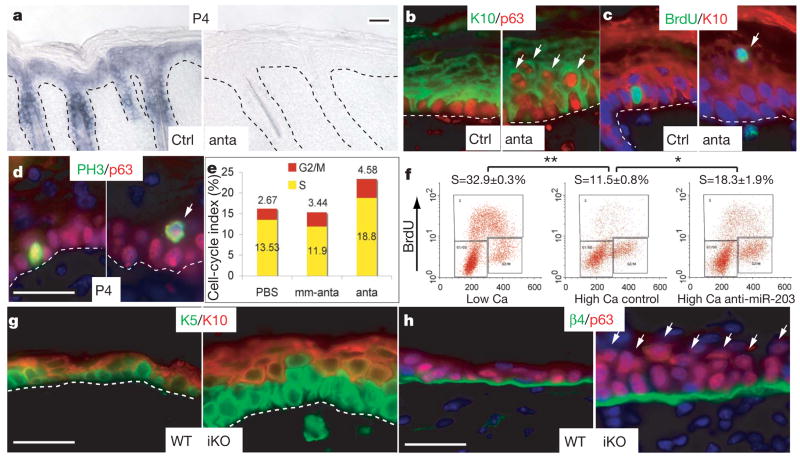
Figure 3. Inhibition of miR-203 results in increased epidermal proliferation.
a–d, P4 skins analysed after treatment of mice with mm-antagomir-203 (Ctrl) or antagomir-203 (anta). a, miR-203 in situ hybridizations. b–d, Immunofluorescence microscopy. Arrows denote basal-like features in suprabasal cells of antagomir-203-treated epidermis. PH3, phospho-histone H3. e, Elevated epidermal proliferation determined by BrdU injections at 4-h intervals and FACS quantification. f, The reduction in cycling cells that typically occurs upon calcium-induced differentiation of primary mouse keratinocytes in vitro is partially abrogated by anti-miR-203 oligonucleotides. Values represent mean ± s.d. from three experiments (*P < 0.005, **P < 0.0001). g, h, Epidermal defects that arise 30 days after tamoxifen-induced Dicer1 ablation (iKO). Arrows denote suprabasal p63. Scale bars are 30 μm.
Similar to E18.5 Dicer1 null skin, antagomir-treated P4 epidermis showed atypical expansion of p63 expression and BrdU-positive suprabasal cells (Fig. 3b, c). This was further substantiated by the presence of suprabasal cells positive for p63 and the mitotic marker phospho-histone H3 (Fig. 3d). Finally, quantification of epidermal cells harvested 4 h after BrdU injections revealed that proliferative cells were more numerous in antagomir-203-treated dorsal epidermis than either PBS- or mm-antagomir-203-treated skin (Fig. 3e).
Because these abnormalities occurred early after miR-203 repression, it was unlikely that they arose secondarily from physical perturbations to the skin or mouse. Moreover, the alterations were recapitulated in vitro with an antisense oligonucleotide20 that specifically blocked calcium-induced expression of endogenous miR-203 (Supplementary Fig. 6d). BrdU labelling and FACS analysis revealed that anti-miR-203 in vitro partially blocked the ability of keratinocytes to exit the cell cycle during calcium-induced differentiation (Fig. 3f).
Having established early consequences of inhibiting miR-203 expression, we examined longer-term effects in vivo. Although tail-vein injections of antagomirs afford efficient delivery for some adult tissues19, it was only partially effective in targeting miR-203 knockdown to skin (Supplementary Fig. 7). Similarly, sustained subcutaneous antagomir injections were not effective once skin matured. To ablate miR-203 efficiently in adult skin, we engineered K14-CreTm/Dicer1 inducible conditional knockout (iKO) mice and examined long-term consequences of conditionally inducing Dicer1 ablation. By 30 days, the epidermis had thickened markedly and suprabasal cells were more numerous (Fig. 3g). Atypical p63-positive suprabasal cells were now prevalent in iKO epidermis (Fig. 3h). These results suggested that miR-203 may be required to effectively repress supra-basal p63 and in turn restrict cell proliferation in differentiating epidermis. Importantly, by targeting Dicer1 after follicles had matured and anchored within the dermis, we could attribute these defects specifically to alterations originating in the epidermis rather than secondarily arising from perturbed hair follicle morphogenesis.
Whereas miR-203 markedly impaired proliferative potential, it had minimal effects on the induction of terminal differentiation markers whether in vivo (Fig. 2c, d) or in vitro (Supplementary Fig. 8). Thus, of the two key features involved in terminal differentiation, suprabasal miR-203 seemed to act predominantly by restricting proliferative potential of progenitors as they transitioned from basal to suprabasal layers. Although loss of miR-203 resulted in increased suprabasal proliferation, neither p63 overexpression nor miR-203 reduction are associated with psoriasis21,22, suggesting that neither p63 nor the miRNA are obligatory features of the hyperproliferative state.
Bioinformatics suggest that miRNAs and their predicted targets tend to be mutually exclusive in neighbouring tissues23,24. On the basis of the inverse correlation that we observed between miR-203 and p63 in expression and function, p63 mRNA surfaced as a possible miR-203 target. Consistent with in vivo results (Fig. 2e), p63 was markedly diminished in K14-miR-203 transgenic primary mouse keratinocytes that were cultured in low calcium (Fig. 4a). Conversely, knocking down endogenous miR-203 in calcium-treated wild-type primary mouse keratinocyte cultures strongly impaired the rapid down-regulation of p63 protein upon terminal differentiation (Fig. 4b), even though p63 mRNA levels remained appreciable (Supplementary Fig. 9). Conversely, transduction of miR-203 in wild-type primary mouse keratinocytes cultured in low calcium strongly inhibited p63 protein expression (Fig. 4b), while only slightly decreasing p63 mRNA (Fig. 4b right-hand graph). Thus, miR-203 seemed to regulate p63 primarily through translational repression.
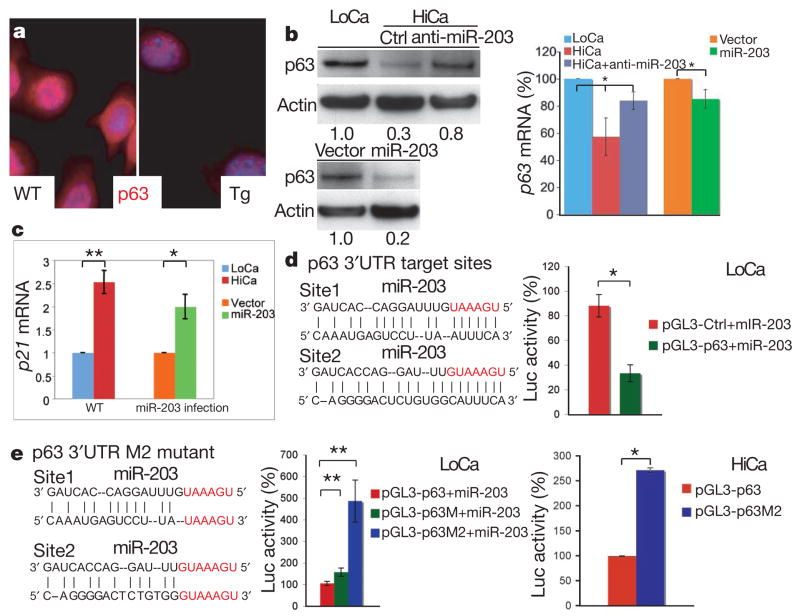
Figure 4. MiR-203 targets p63 mRNA at 3′UTR.
a, p63 protein is markedly diminished in Tg versus wild-type primary mouse keratinocytes.
b, Immunoblot and qRT–PCR quantification of p63 expression. Top panel, calcium-induced (HiCa) p63 downregulation is largely relieved by anti-miR-203 oligonucleotides. Bottom panel, miR-203 transduction is sufficient to repress p63 protein expression in low calcium (LoCa). Right-hand graph, anti-miR-203 treatment slightly recovers p63 mRNA whereas miR-203 transduction marginally reduces p63 mRNA (*P < 0.01, n = 3). c, miR-203 transduction in LoCa primary mouse keratinocytes elevates p21 mRNA level (**P < 0.005, *P < 0.002, n = 3). d, Insertion of two miR-203 target sequences within mouse p63 3′UTR leads to diminished luciferase (Luc) reporter activity in the presence of miR-203 (*P < 0.002, n = 3). pGL3 is a luciferase-expressing plasmid from Promega. e, Mutation of both target sites (p63M2) abolishes miR-203-mediated repression of luciferase activity; a single-site mutation (p63M) has marginal effects (**P < 0.0001, n = 6). In HiCa, wild-type but not mutant p63 3′UTR represses luciferase activity (*P < 0.002, n = 3). For all graphs, error bars (s.d.) are derived from the number of experiments as indicated.
To further define the parallels, we examined the status of p21, a well-established p63 target that functions to restrict proliferative potential of basal epidermal cells25 and is normally repressed in proliferative keratinocytes26–28. Upon miR-203 transduction, however, p21 was upregulated in undifferentiated wild-type primary mouse keratinocytes to an extent that was nearly comparable to its levels in differentiated primary mouse keratinocytes (Fig. 4c). This finding extends the correlation between miR-203 and p63 to p63’s downstream targets, and provides mechanistic insight into how miR-203 may inhibit proliferative potential of epidermal stem cells.
DeltaNp63α is the main isoform expressed in epidermis. Within the 3′UTR of ΔNp63α mRNA, a hexamer and heptamer match perfectly to miR-203’s 5′seed sequence (Fig. 4d). When introduced into the 3′UTR of a luciferase reporter gene, a 599-base-pair mouse ΔNp63α 3′UTR fragment encompassing these two putative target sites caused a threefold reduction in activity in K14-miR-203-expressing keratinocytes cultured in low calcium (Fig. 4d). Mutations within these two sites together abolished miR-203-mediated repression under the same conditions (Fig. 4e, LoCa). When these assays were performed in high-calcium primary mouse keratinocytes in which endogenous miR-203 was induced (Supplementary Fig. 1b), exogenous miR-203 was not needed to elicit the wild-type p63 3′UTR-specific reduction in luciferase activity (Fig. 4e, HiCa). Together, these results indicate that miR-203’s effect on p63 is direct and is mediated through these 3′UTR target sites.
MiR-203 and its suprabasal expression seem to have emerged in vertebrate evolution concomitant with a stratified epidermis. Similarly, the basal expression and function of p63 in stratified epithelia also seems to be conserved4–7,9. Interestingly, the putative miR-203 recognition sequence exists four times in the p63 3′UTR of zebrafish and twice in mouse and human. Moreover, when similarly tested, the p63 3′UTR fragments flanking the hexamer motifs from either zebrafish or human reduced luciferase reporter activity comparably to that from mouse (Supplementary Fig. 10). These data suggest that this mechanism for regulating p63 by miR-203 may be conserved across vertebrates.
Strictly on the basis of bioinformatic studies, the mRNA encoding a zinc-finger protein, Zfp281, is the top candidate of miR-203 targets, as it harbours four perfectly conserved miR-203 consensus sites in its 3′UTR29 (Supplementary Fig. 11a). Similarly to p63, Zfp281 is also enriched in basal epidermal stem cells (Supplementary Fig. 11b). In both K14-miR-203 transgenic mice and miR-203-transduced primary mouse keratinocytes, Zfp281 mRNA was markedly reduced (Supplementary Fig. 11c). Using a luciferase reporter assay, we showed that the Zfp281 3′UTR, but not the mutant form with mutations at four target sites, was specifically regulated by co-transfected and endogenous miR-203 (Supplementary Fig. 11a). Intriguingly, even though Zfp281 has not been previously studied in skin development, it has been implicated in maintaining the proliferative potential of embryonic stem cells30, and in this regard its function resembles that ascribed to p63 in keratinocytes.
Taken together, our studies provide evidence that miR-203 acts at least in part by targeting and negatively regulating suprabasal expression of basal genes, thereby acting as a switch between proliferation and differentiation. Although the mechanisms underlying this balance are complex and likely to be dominated by transcriptional controls, the mutually exclusive expression patterns, opposite functions and evolutionarily conserved regulation offer strong support for the intimate relation between miR-203 and its targets in refining the boundary between these two stages in epidermis.
METHODS SUMMARY
Antagomir synthesis and injection
Antagomir-203 and mm-antagomir-203 were designed and synthesized (Dharmacon) as described19. The antagomir-203 sequence was 5′-UpsCps UAGUGGUCCUAAACAUUpsUpsCpsAps-Chol-3′and mm-antagomir-203 was 5′-UpsCpsUcGUGuUCaUAAACAcUpsUpsCpsAps-Chol-3′. Lower-case letters indicate the mismatched nucleotide. Subcutaneous injection was performed on newborn CD-1 mice at a dosage of 80 mg kg−1, according to an established protocol approved by LARC animal facility at the Rockefeller University. Tail-vein injections were performed as described19. After three initial injections, a booster injection was performed every 4 days. Total treatment was 15 days.
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Acknowledgments
We thank D. O’Carroll and A. Tarakhovsky for Dicer1fl/fl mice; A. Schaefer and P. Greengard for dye-conjugated antagomir-124; Z. Zhang and F. Dietrich for miRNA library sequencing; B. Liu for bioinformatics assistance; A. Giraldez for zebrafish cDNA; N. Stokes and L. Polak for assistance with animals; S. Mazel and X. Fan for assistance in the flow cytometry core facility; J. Racelis for assistance with in situ hybridization; and D. Wang and E. Fuchs laboratory members for discussions. R.Y. is supported by the Pathway to Independence Award from the NIH. M.N.P. is supported by the Ruth L. Kirschstein NRSA Fellowship from the NIH. E.F is an investigator of the Howard Hughes Medical Institute. This work was supported by the HHMI and the NIH.
Footnotes
Author Contributions R.Y. and E.F. designed the research. R.Y. performed the experiments. M.N.P. and M.S. designed and contributed to the antagomir experiments. R.Y. and E.F. analysed the data and wrote the paper.
Author Information Reprints and permissions information is available at www.nature.com/reprints. The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature.
References
- 1.Fuchs E. Scratching the surface of skin development. Nature. 2007;445:834–842. doi: 10.1038/nature05659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lechler T, Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005;437:275–280. doi: 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clayton E, et al. A single type of progenitor cell maintains normal epidermis. Nature. 2007;446:185–189. doi: 10.1038/nature05574. [DOI] [PubMed] [Google Scholar]
- 4.Mills AA, et al. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 5.Parsa R, Yang A, McKeon F, Green H. Association of p63 with proliferative potential in normal and neoplastic human keratinocytes. J Invest Dermatol. 1999;113:1099–1105. doi: 10.1046/j.1523-1747.1999.00780.x. [DOI] [PubMed] [Google Scholar]
- 6.Yang A, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 7.Lee H, Kimelman D. A dominant-negative form of p63 is required for epidermal proliferation in zebrafish. Dev Cell. 2002;2:607–616. doi: 10.1016/s1534-5807(02)00166-1. [DOI] [PubMed] [Google Scholar]
- 8.Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari P. A p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Senoo M, Pinto F, Crum CP, McKeon F. p63 is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Yi R, et al. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nature Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- 12.Andl T, et al. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol. 2006;16:1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wienholds E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 14.Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci USA. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimm D, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 16.Blanpain C, Lowry WE, Pasolli HA, Fuchs E. Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 2006;20:3022–3035. doi: 10.1101/gad.1477606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrandon Y, Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci USA. 1987;84:2302–2306. doi: 10.1073/pnas.84.8.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 19.Krutzfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 20.Davis S, Lollo B, Freier S, Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34:2294–2304. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haider AS, et al. Genomic analysis defines a cancer-specific gene expression signature for human squamous cell carcinoma and distinguishes malignant hyperproliferation from benign hyperplasia. J Invest Dermatol. 2006;126:869–881. doi: 10.1038/sj.jid.5700157. [DOI] [PubMed] [Google Scholar]
- 22.Sonkoly E, et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS ONE. 2007;2:e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farh KK, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 24.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 25.Topley GI, Okuyama R, Gonzales JG, Conti C, Dotto G. P p21(WAF1/Cip1) functions as a suppressor of malignant skin tumor formation and a determinant of keratinocyte stem-cell potential. Proc Natl Acad Sci USA. 1999;96:9089–9094. doi: 10.1073/pnas.96.16.9089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okuyama R, et al. p53 homologue, p51/p63, maintains the immaturity of keratinocyte stem cells by inhibiting Notch1 activity. Oncogene. 2007;26:4478–4488. doi: 10.1038/sj.onc.1210235. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen BC, et al. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev. 2006;20:1028–1042. doi: 10.1101/gad.1406006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westfall MD, Mays DJ, Sniezek JC, Pietenpol JA. The Delta Np63 alpha phosphoprotein binds the p21 and 14–3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Mol Cell Biol. 2003;23:2264–2276. doi: 10.1128/MCB.23.7.2264-2276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.