Abstract
The suprachiasmatic nucleus (SCN) in the mammalian hypothalamus functions as an endogenous pacemaker that generates and maintains circadian rhythms throughout the body. Next to this central clock, peripheral oscillators exist in almost all mammalian tissues. Whereas the SCN is mainly entrained to the environment by light, peripheral clocks are entrained by various factors, of which feeding/fasting is the most important. Desynchronization between the central and peripheral clocks by, for instance, altered timing of food intake can lead to uncoupling of peripheral clocks from the central pacemaker and is, in humans, related to the development of metabolic disorders, including obesity and Type 2 diabetes. Diets high in fat or sugar have been shown to alter circadian clock function. This review discusses the recent findings concerning the influence of nutrients, in particular fatty acids and glucose, on behavioral and molecular circadian rhythms and will summarize critical studies describing putative mechanisms by which these nutrients are able to alter normal circadian rhythmicity, in the SCN, in non-SCN brain areas, as well as in peripheral organs. As the effects of fat and sugar on the clock could be through alterations in energy status, the role of specific nutrient sensors will be outlined, as well as the molecular studies linking these components to metabolism. Understanding the impact of specific macronutrients on the circadian clock will allow for guidance toward the composition and timing of meals optimal for physiological health, as well as putative therapeutic targets to regulate the molecular clock.
Keywords: clock genes, metabolism, saturated fatty acid, glucose, nutrient sensors, hypothalamus
because of the rotation of the earth around its axis, all organisms are subjected to a daily exposure to sunlight. Consequently, in a broad range of species, cellular clock mechanisms have evolved that are sensitive to light and permit anticipation of and entrainment to the changing environmental light conditions of many circadian rhythms, including activity (5, 40). The biological clock, located within the suprachiasmatic nucleus (SCN) in the anterior hypothalamus, is responsible for the generation and maintenance of circadian rhythms throughout the body (40). The most obvious circadian rhythm in humans regulated by the SCN is the sleep-wake cycle, but many other physiological, biochemical, and behavioral variables are under SCN control as well, including core body temperature, blood pressure, and the secretion of numerous hormones, like melatonin, cortisol, prolactin, and growth hormone (40). In mammals, the SCN functions as a master clock that entrains to the light-dark schedule, enabling proper physiology and behavior for the time of the day, which is evolutionarily advantageous and important for survival of the individual (40, 114). For instance, the SCN generates a diurnal rhythm in cortisol secretion that prepares the cardiovascular system to the upcoming active period. A compromised cardiovascular preparation could lead to increased risk of cardiovascular incidents in the early morning (114).
The SCN generates an approximate (“circa”) 24 h (“diem”) rhythm by means of a transcriptional-translational feedback loop (5). The core of this mechanism is formed by the basic helix-loop-helix (bHLH) Per-Arnt-Sim (PAS) domain containing transcription factors circadian locomotor output cycles kaput (CLOCK) and brain and muscle ARNT-like 1 (BMAL1), which drive the expression of three Period (Per 1–3) and two Cryptochrome (Cry 1–2) genes by binding to their E-box (5′-CACGTG-3′) promoter elements. In the cytoplasm, PER and CRY proteins rhythmically accumulate, heterodimerize, and translocate to the nucleus to suppress their own transcription by interaction with the CLOCK:BMAL1 complex. CLOCK:BMAL1 heterodimers also activate the transcription of the DNA-binding orphan nuclear receptor reverse erythroblastosis virus-α and -β (REV-ERBα, -β), retinoid-related orphan receptor-α, -β, and -γ (ROR-α, -β -γ), peroxisome proliferator-activated receptor-α (PPARα), and PPAR-γ coactivator 1-α (PGC1-α). The transcription factors PPARα, RORs, and PGC1-α activate, whereas REV-ERBs repress the transcription of Bmal1 (13, 75, 105). Recently, it was found that there is also a robust association between Cry1 and ROR-α and -γ (68). This additional loop, in which RORs and REV-ERBs modulate Bmal1 transcription, is believed to make the clock more robust (68). Posttranslational events like protein-protein interactions, intracellular translocation, and protein modification delay the PER-CRY feedback on CLOCK-BMAL1 activity, thereby creating a feedback rhythm with an approximate 24-h period. In this way, the core CLOCK-BMAL1-Per/Cry loop regulates the rhythmic expression of hundreds of clock-controlled genes that define the physiological state of the cell. Posttranslational modifications in the mammalian clock system include phosphorylation (all clock genes), acetylation (BMAL1 and PER2), SUMOylation (BMAL1), ubiquitination (probably most clock proteins), poly(ADP-ribosylation) (8), and O-GlcNAcylation (BMAL1, PER, CLOCK, CRY) (52, 74).
Entrainment
To synchronize with the outside world, the approximate 24-h rhythm generated by the SCN is set to exactly 24 h by environmental stimuli of which light is the strongest entraining signal (Zeitgeber) in mammals (105). Other Zeitgebers include temperature, fasting/eating, rest/activity, and social cues (10, 78), although compared with light these Zeitgebers are much weaker (23). Photoentrainment, the process of adjusting circadian time by light, occurs via retinal photoreceptors, which project to the SCN via the retinohypothalamic tract (RHT) (28, 97, 99). Retinal light stimulation results in secretion of the neurotransmitters glutamate and pituitary adenylate cyclase-activating peptide (PACAP) from the RHT terminal, stimulating their receptors in the SCN neurons and leading to the upregulation of some immediate early genes and Per1 and 2 (105). In addition to the photic input via the RHT, nonphotic inputs reach the SCN via afferent neuropeptide Y (NPY)-containing projections from the intergeniculate leaflet and serotonergic inputs from the midbrain raphe (39, 91). In the mammalian brain, the SCN mainly projects to the paraventricular nucleus of the hypothalamus (PVN), the subparaventricular zone (sPVZ), the dorsomedial nucleus of the hypothalamus (DMH), the paraventricular nucleus of the thalamus (PVT), and the intergeniculate leaflet (IGL) (89) utilizing classic neurotransmitters, such as glutamate and γ-aminobutyric acid (GABA), and neuropeptides, including vasoactive intestinal peptide (VIP), vasopressin (VP), and gastrin-releasing peptide (GRP) (114).
Peripheral Clocks
Until the 1990s, it was believed that time keeping was solely designated to specific pacemaker cells in the SCN. Since then, independent clocks have been found in most cells outside of the SCN in a variety of species, and it is well accepted that all mammalian peripheral tissues contain the molecular machinery (Clock, Per) necessary for circadian oscillation (reviewed in Refs. 9 and 105). These so-called peripheral oscillators receive phase resetting signals from the SCN, but their phase can also be reset by a number of other entraining factors, of which the feeding/fasting cycle is one of the most important factors, at least for some tissues (23, 43). These factors are able to stimulate signal transduction pathways that influence the molecular oscillator in peripheral cells (reviewed in Ref. 16). The SCN coordinates peripheral clocks via behavioral, neuroendocrine, and autonomic pathways (16, 17). First, the SCN can inhibit or activate (extra-) hypothalamic centers that facilitate behavior. Second, the SCN can influence neuroendocrine hypothalamic centers that are responsible for hormone secretion. Finally, the SCN can influence preautonomic hypothalamic neurons that affect parasympathetic and sympathetic autonomic centers in the brain stem and spinal cord [dorsal motor nucleus of the vagus (DMV) and intermediolateral column of the spinal cord (IML)], respectively (17).
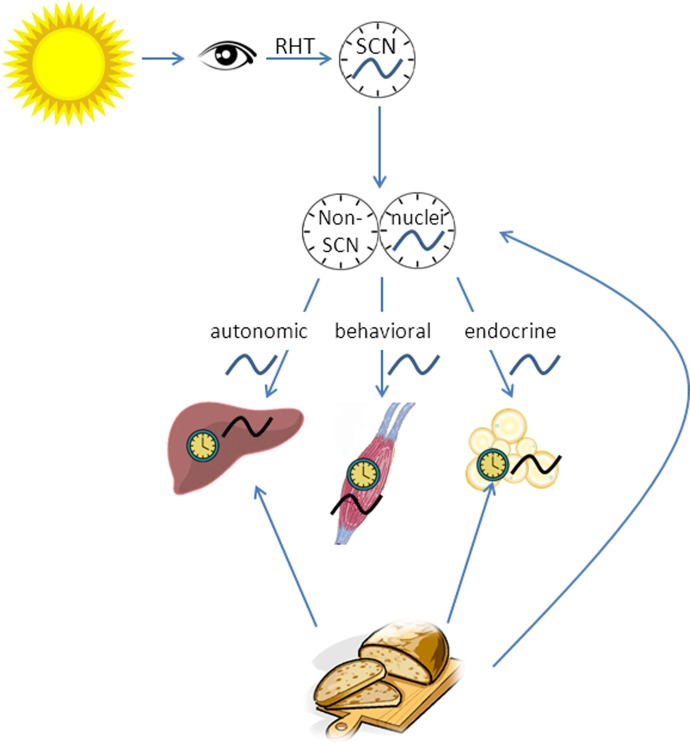
From peripheral organs, metabolic information feeds back to the hypothalamus through hormones and/or through axons from the nucleus of the solitary tract (NTS) and the parabrachial nucleus. Together, the central and peripheral clocks provide the hypothalamus with information to adjust and maintain energy homeostasis (16). Peripheral clocks within the brain, so-called non-SCN brain clocks, have been found in many brain nuclei, including the retina, olfactory bulb, and mediobasal hypothalamus (MBH). These non-SCN clocks are in synchrony with the SCN, but can be uncoupled from SCN control by, for instance, restricted feeding (127). The mediobasal hypothalamus, which comprises the DMH, ventromedial hypothalamus (VMH), PVN, and arcuate nucleus, is an important brain structure in regulating feeding and energy metabolism. Thus, for optimal energy homeostasis, it seems important that also non-SCN clocks within the brain are in synchrony with the SCN (Fig. 1).
Fig. 1.
Suprachiasmatic nucleus (SCN) and its output to non-SCN brain clocks and peripheral clocks. The SCN generates an approximate 24-h rhythm, which is adjusted to exact 24 h by the light-dark cycle. SCN receives photic information through the retinohypothalamic tract and transmits its information to other hypothalamic nuclei, mostly to the paraventricular nucleus of the hypothalamus (PVN). From here, information is translated into hormonal and autonomic signals, which will reach peripheral organs. Non-SCN brain areas and peripheral organs contain endogenous clocks as well, and these clocks are synchronized by the SCN, but also by external signals, including the feeding/fasting cycle.
Thus the SCN generates a circadian rhythm that is transmitted, through other brain nuclei, to the rest of the body. DNA microarrays demonstrate that ∼8–10% of the transcriptome in any tissue is under SCN control (2, 78), indicating the importance of the circadian clock in many processes in the body.
Circadian Clock and Metabolism
There is an intricate link between the circadian clock and metabolism. The SCN is important for the circadian rhythm of food intake, because SCN-lesioned animals show complete abolishment of daily rhythms in food intake (92). An intact circadian system has also been shown to be important for circadian variation in energy metabolism and insulin sensitivity. The SCN contributes to circadian variations in glucose uptake and insulin release (66), although peripheral clocks also play an important role in these processes independent of the SCN (70, 79, 109). Furthermore, many genes involved in glucose and lipid metabolism cycle with a circadian period (98). Peripheral circadian oscillators in the liver regulate the temporal switch between oxidizing short- and medium-chain fatty acids during nocturnal feeding and the storage of very-long-chain fatty acids during the daytime resting period. In addition to the circadian system influencing metabolism, elements of the molecular clock are regulated by metabolic transcription factors at the cellular level. For instance, Bmal1 transcription is inhibited by REV-ERBα, a transcription factor regulated by adipogenesis (20, 103), whereas it is activated by RORα, a nuclear receptor involved in lipid homeostasis (71). Cryptochromes (CRYs) are directly regulated by the nutrient sensor AMPK, suggesting a close relationship between metabolism and circadian rhythms (69).
An intact molecular clock is essential for balanced clock output and healthy metabolism. A dampening in SCN rhythmicity, induced, for instance, by exposing mice to constant light, resulted in a strong reduction of the circadian rhythm in energy metabolism and insulin sensitivity, possibly contributing to obesity and diabetes development (22). Disruptions in individual core clock genes can have profound biological and metabolic consequences, as reviewed previously (24, 54, 59). For instance, cryptochromes, repressors that feedback on CLOCK-BMAL, can affect glucose homeostasis by directly repressing glucocorticoid receptor-mediated induction of gluconeogenic enzymes in the liver, thereby limiting glucocorticoid-induced hyperglycemia (68). Molecular studies have revealed a critical role for the clock genes Bmal1 and Clock in regulating lipid metabolism (95, 116, 124). Inactivation of Bmal1 suppresses the 24-h variation in plasma glucose and triglyceride concentrations and can lead to insulin resistance and glucose intolerance (107). Clock mutant mice on a C57Bl6/6J background, lacking one of the core elements of the molecular circadian clock, rapidly gained weight compared with wild-type mice and showed signs of metabolic disease, including hyperleptinemia, hyperglycemia, and hypoinsulinemia (124), whereas a Clock mutation in ICR mice attenuated obesity due to disrupted dietary fat absorption (62). Furthermore, a liver-specific deletion of Bmal1 resulted in hypoglycemia and loss of rhythmic expression of hepatic glucose regulatory genes (70). A pancreas-specific deletion of Bmal1 resulted in elevated glucose levels throughout the day and impaired glucose tolerance and decreased insulin secretion (79, 109), showing the importance of peripheral clocks in glucose homeostasis.
Thus the existence of clocks in peripheral organs, the knowledge that their timing is set by metabolic cues, and the fact that many genes are under circadian transcriptional control, makes it attractive to speculate that the clock is involved in adapting the organism to changing metabolic needs during the day (68). Furthermore, as peripheral clocks are mainly entrained by feeding and fasting signals, alterations in timing, amount, or composition of food intake can affect, and possibly disrupt, normal clock function.
Metabolic effects of desynchronization between central and peripheral clocks.
In the most optimal situation, central and peripheral oscillations are synchronized. However, the lifestyle and timing of the current Western 24/7 society does not always allow these rhythms to act in concert. Because of the different input signals to the central and peripheral clocks, changes in, for instance, timing of feeding can lead to an uncoupling of peripheral oscillators from the central pacemaker. Studies in rodents have linked disruptions in the timing of the daily rhythm of food intake with weight gain (4, 23, 41, 86, 87). Also in humans, conflicting environmental signals to the circadian system, such as altered sleep-wake patterns, meal schedules, and lighting regimes can disturb the balanced clock network (40). Indeed, epidemiological studies have shown that shift workers are at higher risk for developing metabolic disorders, including obesity and Type 2 diabetes, which is most likely the result of internal desynchronization (58, 77). In the 1970s, it was already reported that in rats the rhythm of food intake was more important than the light-dark cycle for the production of a circadian rhythm of the gluconeogenic enzyme phosphoenolpyruvate carboxykinase (PEPCK) in the liver and kidney (93, 119). During prolonged fasting, >80% of the 2,997 rhythmically cycling transcripts in the ad libitum-fed mouse liver is blunted, indicating that food intake is indeed an important contributor to rhythmic expression of hepatic transcripts (126). Rhythmic hepatic transcripts are products of both direct circadian control (i.e., the intrinsic circadian clock) and an indirect homeostatic response (i.e., feeding) (2). In the liver, 14% of rhythmic transcripts are dependent on systemic signals, whereas 86% are dependent on local oscillators (61), which can be influenced by, for instance, feeding. The central clock, i.e., the SCN, can affect hepatic gene expression either by regulating immediate early genes in the liver, which in their turn transmit the signal to core clock genes of hepatocyte oscillators, or by regulating genes that are directly participating in rhythmic liver physiology and metabolism (8). Under normal conditions, i.e., spontaneous and ad libitum feeding, the liver oscillator is synchronized by signals from the SCN, and these signals are enhanced by appropriately timed feeding activities (23). However, when feeding is changed from night to day in rodents, conflicting signals can reach the liver clock, resulting in uncoupling from the central pacemaker; in other words, the normal phase relation between circadian gene expression in the liver and circadian gene expression in the SCN is changed. This uncoupling could be facilitated by changes in body temperature rhythms that accompany the altered feeding behavior (23). A desynchronization between peripheral clocks and the central clock may furthermore result in a dissociation of the rhythm of metabolic genes (e.g., Pparα) from clock genes within an organ, which was suggested to form a molecular basis for metabolic disruption due to circadian misalignment (112). To support this, in Wistar rats it was found that both forced activity and restricted feeding during the normal rest phase resulted in uncoupling between the SCN and the liver, as well as in uncoupling of clock genes Bmal1, Per1, Clock from metabolic genes, e.g., Pparα, within the liver, which may contribute to the development of metabolic disorders like obesity (112).
Effect of Fat and Sugar on the Circadian Clock
Thus many studies have shown intricate relationships between timing of food intake, energy metabolism, and the circadian clock mechanism. In addition to the timing of food intake, the composition of the food could be an important contributor to disruptions of the normal clock output. In other words, the feeding-dependent resetting of the peripheral and non-SCN brain circadian network may be a result of periodic availability of circulating macronutrients. Glucose, for instance, can initiate circadian gene expression of Per1, Per2, albumin D site-binding protein (Dbp), and Bmal1 in cultured fibroblasts (46). This may suggest a role for glucose in synchronizing (peripheral and perhaps central) clocks. Alterations in the temporal availability of glucose may possibly lead to disruptions in clock gene expression, or desynchronization of peripheral clocks from SCN control, or desynchronization between clock genes and metabolic genes.
In keeping with the increasing fat and sugar consumption in the Western society, concomitantly with an increasing incidence of obesity and obesity-related metabolic symptoms, this review will focus on the effects of fat and sugar on the molecular circadian clock as well as on metabolism and how these factors influence each other. Although it has been shown that many elements in the human diet, including amino acids, caffeine, and alcohol can directly or indirectly affect the clock, it is beyond the scope of this review to discuss them in detail and they are clearly presented in another recent review (29).
Fat intake and energy homeostasis.
Mice with ad libitum access to a high-fat diet increase their diurnal intake, resulting in a longer feeding period and shorter fasting period, as well as a longer free-running period, compared with animals with ad libitum access to a low-fat diet (41, 60, 84, 102). In contrast, however, Yanagihara et al. reported that C57BL/6J female mice fed a high-fat diet (40%) for 6 wk did not show increased diurnal food intake, but they did develop obesity, hyperlipidemia, and hyperglycemia (128). Disruption of the circadian clock by exposure to constant light, as well as a high-fat diet, were shown to have both independent and additive effects on body weight gain. This indicates that the combination of a disruption in the molecular circadian clock and a hypercaloric diet is more detrimental to body weight homeostasis than either of these factors alone (22). Timing appears important for the effects of a high-fat diet on energy homeostasis. Bray et al. (15) found that feeding mice a high-fat diet toward the end of the active period resulted in increased adiposity, compared with mice fed a similar high-fat diet in the beginning of the active period, likely because the temporal regulation of β-oxidation was disturbed (15). Restricting high-fat diet to the dark period as opposed to ad libitum access to high-fat diet, resulted in lower body weight and restored glucose tolerance, diurnal rhythms in metabolic regulators, and prevented liver disease (41), whereas mice fed a high-fat diet during the light period gained significantly more body weight than mice fed the same diet with the same amount of calories during the dark period (4). Interestingly, and in contrast to the latter, Sherman et al. (115) showed that restricted high-fat feeding during 4 h in the light period improved insulin sensitivity and lipid oxidation and resulted in a lower body weight and decreased fat profile compared with mice on an ad libitum high-fat diet and comparable to mice on an ad libitum low-fat diet. It needs to be mentioned that the total caloric intake in the ad libitum high-fat group was higher than in the timed high-fat group (115). It thus seems that restricted feeding or perhaps the prolonged fasting period, per se, is beneficial for metabolism, even when it takes place in the normally inactive period.
To summarize, as described above, feeding mice a high-fat diet could lead to disruptions in normal circadian patterns of behavior and lipid and glucose metabolism, suggesting that a high-fat diet could influence both the central and peripheral clocks. Perhaps, fat intake can be a synchronizing factor for the molecular clock. Only a few studies have focused on the effects of fatty acids on the molecular circadian clock, which will be described below.
EFFECT ON CENTRAL CLOCK.
Despite important studies showing that the SCN is not phase shifted by restricted food intake during the light period (23, 118), recent studies suggest that the phase of the central clock might be sensitive to nutrients. For instance, Kohsaka and colleagues (60) observed that a high-fat diet altered the free-running period in male C57BL6/CJ mice under unentrained (dark-dark) conditions, whereas clock gene expression in the mediobasal hypothalamus was not affected by a high-fat diet, suggesting that alterations in metabolic cycles can lead to alterations in diurnal cycles, i.e., the central clock, although the precise mechanism needs to be elucidated.
The effect of high-fat diet on the central clock might be through a direct feedback to the clock. The SCN receives metabolic information from the peripheral organs through different pathways. AgRP/NPY neurons in the arcuate nucleus (ARC) are activated by ghrelin from the stomach during fasting and project to the SCN (132). Recently, it was found that not only the ARC, but also the IGL, transmits metabolic signals to the SCN, probably through sensory neurons in the brain stem that transmit metabolic signals from organs to the IGL (110). NPY and GABA were found to have an important role in transmitting metabolic information from the IGL to the SCN (110). The effect of high-fat diet on the SCN was shown by Mendoza et al. In mice fed a high-fat diet, the induction of the immediate early gene c-fos by light was reduced in the SCN. This reduction was most pronounced in the ventral area of the SCN, which expresses VIP, suggesting that VIP may be a specific target for the chronobiotic effect of high-fat feeding (84). Recently, it was found that VIP in the mouse SCN is coexpressed with the NPY6-receptor (Npy6r), which is activated by pancreatic polypeptide, released from the pancreas in response to food intake. VIP expression was reduced in the absence of Npy6r signaling (134). This is in accordance with the recent finding that NPY is an important neurotransmitter in signaling to the SCN (110). As VIP synchronizes SCN neurons, alterations in VIP due to feeding may be a mechanism via which food intake can alter the output of the central clock, at least in the mouse. Furthermore, the adipokine leptin was shown in vitro to dose dependently cause a forward shift in the peak time of SCN activity, i.e., phase advanced the SCN, suggesting that the rhythmic pattern of endogenous leptin production may affect the central clock (104). In vivo it was shown that leptin plays an important role in the modulation of the SCN light response (83).
The effects of fatty acids on the central clock could also be indirect, through alterations in peripheral clocks that feed back to the central clock. For instance, Paschos et al. reported that in adipocyte-specific Bmal1 knockout mice, the adipocyte clock was disturbed. Consequently, there was a disruption in the normal daily rhythm of circulating polyunsaturated fatty acids. Concomitantly, an alteration in feeding rhythms was noticed. The authors suggest that there seems to be a bidirectional relationship between the peripheral clocks and the SCN, which may explain the altered feeding behavior (central effect) due to a disruption of a peripheral (pancreas) clock (100).
EFFECT ON PERIPHERAL CLOCKS.
In mice fed a high-fat diet, alterations in diurnal patterns of circulating metabolic markers, including leptin and insulin, clock-regulating nuclear factors, including RORα and PPARα, hypothalamic neuropeptides, including AgRP and NPY, and factors involved in lipid metabolism, including sterol regulatory element-binding protein 1c (SREBP1c) and acetyl-coenzyme A carboxylase (ACC), were found. Furthermore, the amplitude of the circadian expression profiles of Clock, Bmal1, and Per2 in fat tissue and liver was reduced. Interestingly, in these mice, there was a desynchronization between factors cycling in the liver and in adipose tissue (60). Barnea et al. found that in mice fed a high-fat diet for 7 wk, rhythmic expression of hepatic clock genes as well as components of the adiponectin signaling pathway, including Pparα and Ampk, were disrupted. As Pparα and Ampk promote β-oxidation, disruption of their rhythmic expression may lead to the impairment of lipid metabolism in the liver. The authors propose a vicious cycle in which the diet-induced obesity leads to disrupted circadian rhythms, which, in turn, can lead to enhanced obesity (12). By using a high-throughput profiling of the mouse liver metabolome and transcriptome, Eckel-Mahan et al. (25) showed that a high-fat diet could alter normal cycling of circadian genes in the liver. This included a loss of oscillations of normally rhythmically expressed transcripts and metabolites (e.g., NAD+ and Nampt), as well as gain of oscillation in otherwise arrhythmic transcripts (e.g., the histone methyltransferase Ehmt2), and a phase advance of metabolites and transcript oscillations (e.g., Dbp). This induction was rapid and did not require the development of obesity, and it appeared to be reversible on a normocaloric diet. The alterations rely on two mechanisms: first, the lack of proper BMAL1 and CLOCK chromatin recruitment at the level of target promoters of previously oscillating genes; and second, the genesis of novel rhythmicity of a great number of genes was found to rely on changes in the presence and pattern of oscillation and chromatin recruitment of PPARγ (25). This study illustrates that the (peripheral) circadian system is dynamic and can change under high-fat conditions.
EFFECT ON NON-SCN BRAIN CLOCKS.
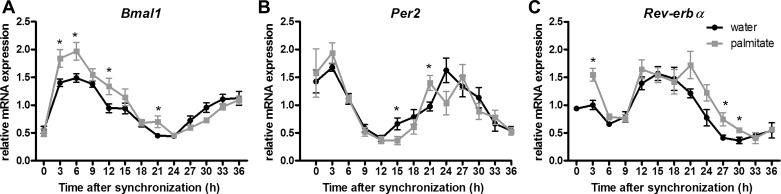
While many studies have focused on the effects of high-fat diet on clock gene expression in peripheral tissues, limited analysis of individual neuronal mechanisms is available. In the immortalized, hypothalamic murine neuronal cell line mHypoE-44, it was shown that high levels of the saturated fatty acid palmitate (300 μM) caused alterations in the circadian expression profiles of Bmal1, Rev-erbα, and Per2 (27). Recently, in another murine immortalized hypothalamic cell line mHypoE-37, it was shown that physiologically elevated levels of palmitate (25 μM) caused an upregulation of Bmal1 mRNA and a phase shift of Bmal1 and Rev-erbα mRNA (Fig. 2; 32), indicating that palmitate can directly affect clock genes in a model of hypothalamic neurons. Interestingly, in the presence of the unsaturated fatty acid docosahexaenoic acid (DHA) the palmitate-induced changes in Bmal1 transcript levels were less pronounced. Although the exact mechanism by which this occurs remains to be delineated, activation of PPARs by palmitate provides a putative mechanism for the changes seen in the expression profile of Bmal1. The phosphorylation levels of AMPK and GSK were not affected, suggesting that these pathways are not affected by palmitate in this cell model (32).
Fig. 2.
Effects of saturated fatty acids on the expression profile Bmal1, Per2, and Rev-erbα in mHypoE-37 neuronal cells. Relative mRNA transcript levels of Bmal1, Per2, and Rev-erbα, respectively, are shown. Bmal1 transcript levels are upregulated in the palmitate-treated cells compared with controls. *P < 0.05 between palmitate and control group at the indicated time point as determined by two-way ANOVA with post hoc t-test. Values are plotted as mean values ± SE. From Greco JA et al. (32).
The studies in cell lines indicate that the effects of fatty acids on the clock are direct, i.e., independent of obesity. This is supported in vivo by the study of Eckel-Mahan et al.(25), in which the alterations in the mouse liver clock were already present after 3 days of high-fat diet, suggesting that the high-fat diet rather than the development of obesity initiates the reprogramming of the circadian clock. Similarly, Kohsaka et al. (60) did not find a correlation between body mass and clock gene disruption in male C57BL/6J mice. In contrast, Yanagihara et al. (128) report that female C57BL/6J mice on a high-fat diet became obese but did not develop alterations in clock gene expression in visceral adipose tissue and liver. The authors discuss that the relatively mild obesity might explain the lack of alterations in clock genes (128). Alternatively, there could be a sex-specific effect, as Yanagihara and colleagues used female mice, as opposed to male mice used in the studies of Eckel-Mahan and Kohsaka (25, 60).
To summarize, there is evidence that on a molecular level, fatty acids can alter the circadian molecular clock, both centrally and peripherally. However, the downstream pathways through which alterations in the molecular clock eventually lead to alterations in metabolism and body weight gain have yet to be elucidated. Indeed, rodent models with global or tissue-specific deletions of clock elements have shown the effects on clock disturbances on physiology (e.g., 62, 70, 79, 109), but the effects of changes in circadian clock gene expression due to a high-fat diet may be less pronounced and mechanisms through which this alters physiology are yet unknown.
Sugar intake and energy homeostasis.
Similar to the timing of fat intake, the timing of glucose intake could be important for normal energy homeostasis, as Morris et al. (90) found that mice with restricted access to fructose only during the light period gained more body weight and showed a greater increase in white adipose tissue as well as increased levels of insulin and leptin compared with mice with nighttime-restricted access to fructose (90). Rats with diurnal access to liquid sugar, on top of a high-fat diet, gained more body weight compared with rats exposed to an isocaloric diet with only nocturnal access to sugar, indicating that the timing of sugar is important for body weight homeostasis (67).
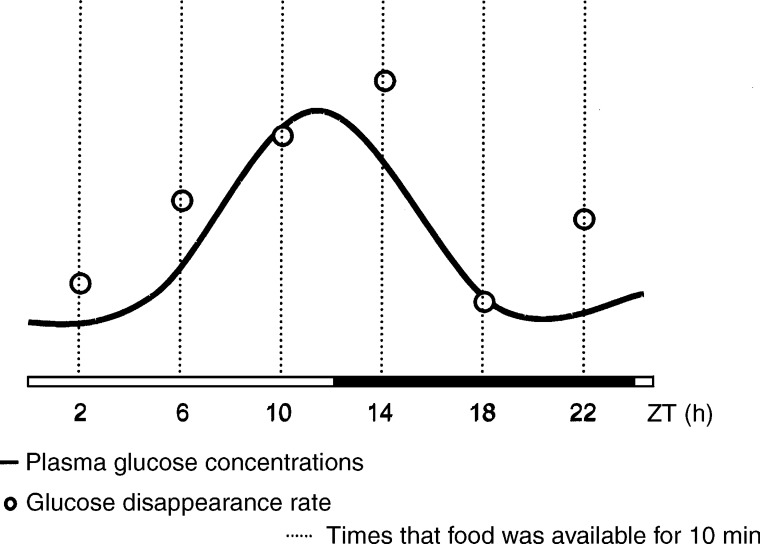
The brain derives its metabolic energy mostly from glucose and therefore it is crucial that circulating glucose levels do not fall below ∼5 mM. To ensure this, glucose-sensing systems are located in the periphery, as well as in the central nervous system, to control glucose homeostasis, feeding behavior, and energy storage (81). The central clock is important in the control of glucose homeostasis. Plasma glucose levels show circadian variation, which is independent from food intake, and surgical ablation of the SCN impairs the control of glucose homeostasis (65). Furthermore, it is widely accepted that glucose uptake shows diurnal variation (Fig. 3; see references in 55, 64). Because of the sensitivity of the brain for glucose, and since the SCN in the hypothalamus is important for glucose homeostasis, it is interesting to identify whether alterations in glucose level due to feeding are able to alter the molecular circadian clock, either centrally in the SCN, in non-SCN brain nuclei, or peripherally.
Fig. 3.
Temporal relationship between plasma glucose concentrations and glucose disappearance rate. Both plasma glucose concentrations and glucose disappearance rate rise before the onset of the activity period, independently from food intake. From la Fleur SE (64).
EFFECTS ON THE CENTRAL CLOCK AND NON-SCN BRAIN CLOCKS.
Parenteral infusion of glucose during the light phase in male Wistar rats shifted mRNA expression levels in the SCN and in the liver in opposite directions (50). This indicates that glucose may be a strong entraining signal for the SCN, but it also indicates that glucose is able to differentially affect central and peripheral clocks. In a recent study using the immortalized hypothalamic cell line mHypoE-37 (14), it was found that treatment with 5.5 mM glucose, as opposed to 0.5 mM glucose, shortened the period and advanced the acrophase of rhythmic Per2 mRNA levels (Oosterman and DD Belsham, unpublished observations), indicating that glucose can affect the molecular clock in non-SCN brain nuclei.
EFFECTS ON PERIPHERAL CLOCKS.
Glucose is one of the factors that is able to induce cellular circadian rhythms (46). Glucose induced a downregulation of Per1 and Per2 gene expression in rat cultured fibroblasts in a metabolism-dependent manner. Transforming growth factor β-inducible early gene 1 (Tieg1) and vitamin D3 upregulated protein 1 (Vdup1) were identified as glucose-responsive immediate early genes (44). Physiologically, alterations in serum glucose levels are accompanied by alterations in other hormones, including insulin, glucagon, and somatostatin, which in their turn may affect the clock. For instance, several studies showed effects of insulin on the clock and clock gene expression. In vitro studies in rat fibroblasts have shown that insulin upregulates mRNA expression of Per1 and Per2 (11), whereas in vivo insulin-induced hypoglycemia reduces photic resetting (19). Tahara et al. (120) showed alterations in hepatic expression of Per2 and Rev-erbα after refeeding, which was not observed in streptozotocin (STZ)-induced insulin-deficient mice, suggesting that that insulin can affect the liver clock. In another study it was shown that nonobese, STZ-induced insulin-resistance in C57BL/6 mice resulted in alterations in hepatic clock gene expression, suggesting that the molecular clock is sensitive to mild elevations in glucose and insulin levels, as seen in insulin resistance (129), although the underlying mechanism is unclear. Interestingly, treatment with the insulin sensitizer and PPARα agonist pioglitazone reverted the changes in clock-gene expression, suggesting that enhancing insulin sensitivity may strengthen the circadian molecular system and improve the adverse effects of metabolic syndrome (129), but it also suggests that the nuclear receptor PPARα might be involved in the effects of insulin on the hepatic clock. Insulin receptor signaling pathways have been extensively examined and shown to regulate several kinases, including glycogen synthase kinase 3 (GSK3) and mitogen-activated protein kinase (MAPK), which play important roles in the regulation of the circadian clock. For instance, MAPK downregulates the activity of BMAL1 (113) and GSK3 can phosphorylate CRY2, leading to its degradation (35).
The above-mentioned studies demonstrate that it is difficult to predict the effects of sugar on the expression of circadian clock genes in vivo, as increased glucose levels will be accompanied by alterations in hormones, including an increase in insulin levels. Both glucose and insulin can affect the molecular clock, although not many studies have been published on this topic.
Influence of Energy Status
Thus far this review has focused on the effect of fatty acids and sugar on the molecular clock. It has been shown that both of these macronutrients are able to affect the clock: both centrally and peripherally, within and outside of the brain. Although different pathways have been proposed through which fatty acids and sugar can exert their effect on the molecular clock, exact mechanisms are still unknown. It was, for instance, suggested that the effects of high-fat diet on the clock could be just the result of rhythmically driven nutrient-sensing activities (76). In our search to find evidence for differential effects of fat and sugar on the molecular clock, we noticed a paucity of studies in which nutrient content was compared with energy content. Therefore, it remains unclear whether the specific metabolites or the caloric content are causal for the effects observed. Especially the described high-fat diets seem to be used as a model for a high-caloric diet, rather than as a model to study the effects of specific nutrients on the circadian clock. It thus remains a possibility that effects of fat or sugar on the clock are through alterations in energy status, rather than through specific effects of the metabolites of fatty acids and sugar. Recent studies suggest that energetic state can indeed affect the central clock, despite previous evidence showing that food intake has no effect on the output of the SCN (23, 118). For instance, when glucose availability was reduced by fasting, or by administration of the competitive inhibitor of glucose metabolism 2-deoxy-d-glucose (2-DG), light-induced phase delays of the circadian clock were attenuated (19). Furthermore, a hypocaloric diet was shown to speed up reentrainment to a shifted light-dark cycle (105a), whereas high-fat feeding slowed it down (84), indicating that the amount of calories is possibly more important for photic resetting than the actual metabolites of a high-fat diet. A timed hypocaloric diet led to alterations in the diurnal expression patterns of Per2 and Cry1 in the SCN, as well as phase advances in locomotor activity, melatonin, and vasopressin mRNA levels. This indicates that hypocaloric feeding can both alter central clock gene expression as well as the photic response of the circadian system (82). Feillet et al. (26) showed that hypocaloric feeding resulted in an upregulation of Cry and a downregulation of Bmal1 mRNA in the SCN. Thus, under certain food-restricted conditions, phase shifts and alterations in clock gene expression can occur, suggesting that clock genes could play an important role in feeding back information about the nutrient status to the SCN (26). These cues could involve glucose metabolism and circulating levels of AMP (136), indicating that either energy status or metabolites of glucose can influence the clock.
Although the above-described studies suggest that energy status, rather than specific metabolites of the diet, is important for the effects of a high-fat or high-sugar diet on the molecular clock, comparative studies are lacking. Until specific studies are performed that directly compare energy status with nutrient content, it remains a possibility that fatty acids and sugar may directly affect the clock, independently of energy status.
Nutrient Sensors
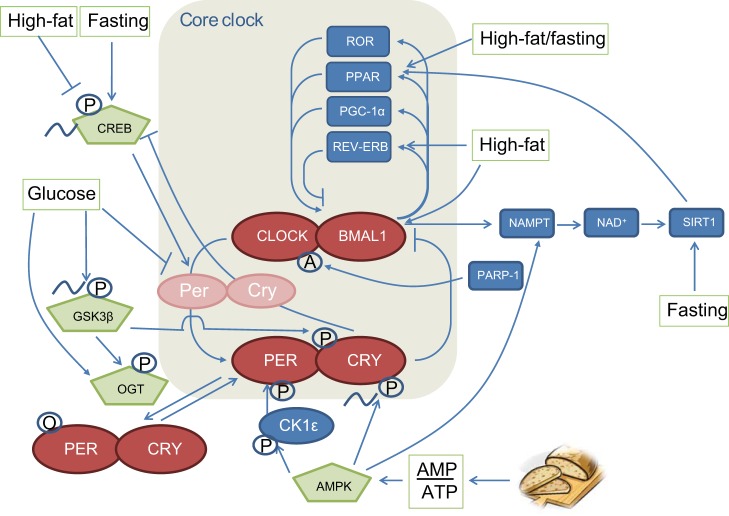
From an evolutionary point of view, it is beneficial to anticipate the transitions in nutrient availability. For instance, the body clock helps to adjust the energy needs of our body to the sleep/wake cycle. As the central pacemaker in the suprachiasmatic nucleus prepares the body for environmental changes, it is only logical that nutrient sensing and the clock would be intimately related. To assess molecular links between the circadian clock and energy balance, multiple putative clock-related components have been studied, including AMP-activated protein kinase (AMPK), cAMP-responsive element binding protein (CREB), glycogen synthase kinase 3β (GSK3β), sirtuins (SIRTs), and PPARs. In this section, these will be discussed in more detail (Fig. 4). While there are certainly many other putative signaling components that can be connected to the molecular circadian clock, especially given ∼10% of genes are under circadian control, at present these are the ones of particular relevance to metabolic control.
Fig. 4.
Overview of the molecular circadian clock mechanism in relation to metabolism. The clock regulates many clock output genes involved in metabolism. Conversely, the clock receives input from transcription factors and nutrient sensors with important roles in metabolic processes. Feeding and fasting can influence the core clock in a number of ways, and glucose and fat intake can also directly or indirectly influence the clock, as depicted in this figure. It becomes clear that energy status is important in regulating the clock, whereas there is a paucity of studies showing the specific effects of fat and sugar. O, O-glcNAcylation; P, phosphorylation; A, poly-ADP-ribosylation.
AMP-activated protein kinase.
AMPK AS ENERGY SENSOR.
AMPK is a critical nutrient sensor, which is present in every tissue, including neurons. Nutrient levels in the hypothalamus, often dictated by lipid metabolites and intracellular fuel, are monitored by AMP kinase (37). AMPK is activated by a decrease in intracellular ATP levels, with a concomitant increase in the amounts of ADP and AMP. Once activated, AMPK acts to restore intracellular ATP by inhibiting biosynthetic processes and inducing FFA catabolic pathways (36). Hypothalamic AMPK is important in regulating energy balance by altering metabolism and food intake in response to nutritional and endocrine food signals. Modulation of hypothalamic AMPK activity alters food intake and body weight (85). Hypothalamic AMPK is stimulated by orexigenic agents, including ghrelin and agouti-related protein (AgRP), and is suppressed by anorexigenic agents including leptin, insulin, and glucose (85). Activated hypothalamic AMPK leads to increased food intake and body weight, in concert with increased expression of the orexigenic neuropeptides NPY and AgRP in the arcuate nucleus (131). An abundance of nutrients (for instance, high levels of glucose) reduces AMPK activity, leading to decreased phosphorylation of acetyl-CoA carboxylase (ACC). This result in turn leads to increased amounts of malonyl-CoA, thereby inhibiting carnitine palmitoyltransferatse 1 (CPT-1), resulting in decreased β-oxidation and reduced food intake (37, 94). Recently, in GT1–7 and mHypoE-46 hypothalamic cell lines, it was found that glucose inhibits long chain fatty acid oxidation in hypothalamic neurons via AMPK (121). This indicates an important role for hypothalamic AMPK in the regulation of energy homeostasis. In peripheral tissues, activated AMPK switches on catabolic pathways, including fatty acid oxidation in the liver, and switches off anabolic pathways, such as lipogenesis or gluconeogenesis (37).
AMPK AND THE CIRCADIAN CLOCK.
It has been proposed that AMPK replaced light as a clock-resetting signal in organisms in which light could not penetrate into all cells, as the presence of AMPK-dependent phosphorylation sites seems associated with increased body size (51). Thus, from an evolutionary point of view, AMPK may have evolved as important time-giving nutrient sensor. Indeed, AMPK exerts multiple actions on the circadian clock (reviewed in Ref. 69). For instance, AMPK contributes to the circadian phosphorylation of CRY1, thereby reducing its association with PER2 and instead increasing its binding to F-box and Leu-rich repeat protein 3 (FBXL3), a ubiquitin ligase that promotes CRY1 ubiquitination and degradation. This leads to subsequent derepression of the PER-CRY dimer on CLOCK-BMAL, thus lengthening the circadian rhythm (69). Furthermore, AMPK can phosphorylate (and activate) casein kinase 1ϵ (CK1ϵ), which leads to phosphorylation and degradation of PER (69). Levels of the mRNA encoding AMPK2β subunit were found to oscillate in a circadian manner, concomitantly with a circadian rhythm in nuclear translocation of AMPK. Loss of AMPK signaling in the mouse liver in vivo resulted in disrupted circadian rhythms in hepatic clock genes (69). In synchronized mouse embryo fibroblasts, treatment with low glucose or the AMPK activator AICAR reduced the amplitude and increased the period of the circadian rhythm of a luciferase reporter gene driven by the BMAL1-CLOCK promoter; these effects were lost in cells lacking AMPK (69).
Thus the presence or absence of glucose and fatty acids can regulate the activity of AMPK, both centrally in the hypothalamus, as in peripheral tissues. Because of its tight connections to the circadian clock, this may be a putative mechanism through which fat and sugar can alter the molecular circadian clock.
cAMP-response element binding protein.
CREB AS NUTRIENT SENSOR.
cAMP-response element (CRE) binding protein (CREB) is a cellular transcription factor that stimulates target gene expression at promoters that contain CREs. In the hypothalamus, CREB is involved in appetite regulation, whereas in adipose tissue, activated CREB increased insulin resistance (3). In the liver, CREB controls gluconeogenesis by activating or repressing the transcription of gluconeogenic genes and regulates fatty acid oxidation (3, 125). CREB in peripheral organs is phosphorylated in response to several signaling and hormonal stimuli, including catecholamines that increase intracellular cAMP production, Ca2+, and the ERK-MAPK pathway (3, 123). In the liver, the circadian phosphorylation of CREB is dependent on feeding rhythms. Fasting increases, whereas feeding decreases, CREB phosphorylation and activity (126). During short-term fasting, increasing levels of glucagon increase CREB activity, which stimulates transcription of gluconeogenic genes by binding to the CRE in their promoters. Activated CREB increases the activity of PGC1α and NR4A1, which elevate the transcription of gluconeogenic genes during prolonged fasting (3).
CREB AND THE CIRCADIAN CLOCK.
CREB is closely linked to the circadian clock. Within the SCN, CREB is phosphorylated in response to photic stimuli and hence shows a circadian rhythm in activity. A light stimulus during the subjective night demonstrated an increased phosphorylation of CREB in the hamster and rat SCN (31, 123). The increased phosphorylation of CREB coincided with increased transcription of c-fos in the SCN, suggesting that phosphorylation of CREB regulates gene expression in the SCN through c-fos, and CREB may thus be important for the entrainment of the SCN (31). Furthermore, the Per gene contains CRE binding sites and phosphorylated CREB has been shown to activate Per transcription in rat SCN explants (123). In the mouse liver, activated CREB can activate Per transcription (126), and CRY can repress phosphorylation of CREB through inhibition of glucagon-stimulated cAMP production, leading to repressed transcription of gluconeogenic genes, including phosphoenolpyruvate carboxykinase-1 (Pck1) and glucose-6-phosphatase (G6pc) (135).
Thus the feeding-fasting cycle regulates the activation of CREB in peripheral organs, and activated CREB can affect the molecular clock by activating Per transcription. A high-fat diet in mice resulted in downregulation of CREB protein levels in the liver (49). Given the importance of CREB for entrainment of the SCN, it could be speculated that feeding-induced alterations in CREB only takes place in peripheral organs and not in the SCN. This might be a possible mechanism through which food intake can lead to desynchronization between peripheral clocks and the central clock. Further analysis of the role of CREB in clock-controlled gene expression is warranted.
Glycogen synthase kinase 3β.
GSK3β AS A NUTRIENT SENSOR.
GSK3β is a ubiquitous kinase that regulates a number of cellular functions, including glucose homeostasis, and it has been implicated in the development of insulin resistance, based on its role in regulation of glycogen synthesis. GSK is constitutively active and is inactivated upon phosphorylation (72). High concentrations of glucose (30 mM) and insulin (1 nM) increased the phosphorylation of GSK3β in murine kidney proximal tubular epithelial cells (80). Furthermore, glucose addition increased the levels of phosphorylated GSK3β in the immortalized hypothalamic cell line mHypoE-37 (JE Oosterman and DD Belsham, unpublished observations). In contrast, 12-h treatment with palmitate decreased phosphorylation of insulin-stimulated pGSK3 in rat hepatocytes (106).
GSK AND THE CIRCADIAN CLOCK.
The kinase activity of GSK3β is circadian, both in the SCN and in peripheral clocks, including liver and fibroblasts, although with inverted patterns between SCN and liver (47). Various studies have shown that phosphorylation of GSK3β can alter circadian period length but with opposing results. A high-throughput approach demonstrated that small molecule inhibitors of GSK3β and small interfering RNA (siRNA)-mediated knockdown of GSK3β leads to shortening of the period in cultured human osteosarcoma U2OS cells (45), whereas the GSK3β inhibitor lithium has been shown to increase free-running period length in cultured murine SCN cells (1). In another study, both lithium and GSK3 knockdown caused lengthening of the period of PER2 mRNA transcription rhythms in synchronized mouse fibroblasts (53). In mammals, GSK3β has been reported to phosphorylate PER2, CRY2, and REV-ERBα with differential effects: GSK3β-mediated phosphorylation induces the degradation of CRY2 in the mouse liver (35), causes phosphorylation of BMAL, and primes it for ubiquitination, followed by proteasomal degradation in cultured peripheral cells (111), whereas it stabilizes Rev-erbα in cultured peripheral cells (133). As Cry2 and Rev-erbα knockout mice show long- and short-period phenotypes, respectively, it is possible that the period shortening by GSK3β inhibition is partly mediated by stabilization of CRY2 and degradation of REV-ERBα (45). Thus the effect of GSK3β on the circadian clock takes place both in the central clock in the SCN, and in peripheral tissues.
Recently a novel mechanism was described by which sugar can affect the molecular clock. O-GlcNAcylation is the covalent attachment and cycling of N-acetylglucosamine (GlcNAc) on serine or threonine residues of nucleocytoplasmic proteins. Intracellular concentrations of UDP-GlcNAc, the donor for O-GlcNAcylation, are controlled by flux through several major metabolic pathways, including metabolism of glucose, amino acids, and fatty acids (38). Because O-GlcNAcylation is dependent on UDP-GlcNAc concentrations, O-GlcNAc serves as an important nutrient sensor. O-GlcNAcylation competitively inhibits phosphorylation of PER and CRY proteins and can control the turnover of CLOCK and BMAL proteins by inhibiting ubiquitin-mediated degradation, thereby regulating circadian period length. Because O-GlcNAc transferase (OGT), the enzyme responsible for O-GlcNAcylation, is activated through phosphorylation by GSK3β, whose activity cycles with the circadian clock (52, 74), this could potentially be an important mechanism by which nutrient intake can affect circadian control mechanisms at the molecular level.
The above-described studies indicate that the activity of GSK3β and OGT can be regulated by alterations in nutrient levels. Furthermore, posttranslational O-GlcNAcylation by OGT is highly dependent on nutrients. Because of their intricate links to the circadian clock, nutrients may affect the clock through regulating GSK3β activity. It would be of interest to study whether fat and sugar can differentially affect the clock through this mechanism. Furthermore, since GSK3β and OGT exert their actions both in the periphery and in the SCN, it would also be of interest to study whether nutrients differentially alter these factors in the brain and in peripheral tissues, which could potentially lead to alterations in the respective clocks, ultimately leading to disruptions of rhythms controlling normal energy metabolism.
Redox state.
SIRTs are a class of enzymes with NAD+-dependent histone deacetylase activity. The activity of SIRT1 is modulated via the redox state of the cell, i.e., by the NAD(P)+-to-NAD(P)H ratio (6). SIRTs regulate many metabolic processes, including gluconeogenesis and insulin sensitivity. Many of the transcription factors regulated by SIRT are involved in the cellular response to stress and nutrient flux, including peroxisome proliferator-activated receptor-γ coactivator (PGC-1α), sterol regulatory element-binding protein 1 (SREBP-1c), and signal transducer and activator of transcription-3 (STAT3) (34). SIRT1 was identified as a regulator of circadian gene expression, and its protein level was found to be expressed in a circadian manner in mouse liver, mouse hepatocytes and cultured fibroblasts. Furthermore, SIRT1 binds to CLOCK-BMAL and PER2 in a circadian fashion and SIRT1 was found to deacetylate PER2 in vivo, leading to PER2 degradation (6).
REDOX STATE AND THE CIRCADIAN CLOCK.
The circadian control of NAD+ metabolism possibly couples internal energetic cycles with oscillations in the external nutrient environment (reviewed in Ref. 101). In a normal feeding-fasting cycle, NAD+ cycles, with peaks during the fasting phase and troughs during the feeding phase. As SIRT1 is activated by NAD+, clock-driven oscillations of NAD+ contribute to daily rhythms in many metabolic pathways regulated by SIRT1. However, under conditions of a high-fat diet, there are constantly elevated and nonoscillatory levels of glucose, and hence there is no real fasting phase, resulting in loss of circadian oscillation in NAD+ (25). This may lead to altered activity of SIRT and hence alterations in transcription factors involved in metabolism. Mendoza et al. (84) speculate that if high-fat feeding, through increased glucose levels, modifies the redox state of SCN cells, daily changes in binding activity of CLOCK-BMAL1 to E-boxes of clock genes may result in slower circadian oscillations and lengthening of the endogenous period.
In vitro DNA-binding activity of CLOCK-BMAL1 is modulated by the redox states of NAD(H) or NADP(H). The oxidized form NADP (NAD+) was found to inhibit, while the reduced form NAD(P)H was found to enhance DNA binding of the CLOCK-BMAL heterodimer. Hence, fluctuations in cellular redox state can directly entrain the molecular clock (108). Recently, another feeding-dependent regulator of the circadian clock was discovered, poly (ADP-ribose) polymerase 1 (PARP-1), that is also regulated by NAD+. PARP-1 is a nuclear protein that possesses ADP-ribosylation activity. PARP-1 ADP-ribosylation activity cycles in a circadian fashion and is regulated by feeding. PARP-1 poly-ADP-ribosylates CLOCK in a circadian way and can thereby affect CLOCK function. Interestingly, mice lacking the Parp-1 gene have a longer circadian period, suggesting that PARP-1 may feedback to and alter the central clock (7). Although the exact mechanism through which PARP-1 cycles remains elusive, it seems to play an important role in the link between metabolism and circadian rhythms (63).
Thus alterations in circulating nutrient levels can alter the molecular circadian clock via alterations in redox state. This might happen in the SCN, in non-SCN brain areas, as well as in the periphery. Further studies need to be undertaken to dissect possible differential effects of nutrient intake on the central and peripheral clock and the subsequent alterations in metabolism.
Peroxisome proliferator-activated receptors.
PPARS AS NUTRIENT SENSORS.
PPARs are a group of nuclear hormone receptors that function as transcription factors involved in many cellular processes including lipid, carbohydrate, and protein metabolism. Three types of PPARs have been identified: α, β/δ, and γ. PPARα is expressed most abundantly in the liver, PPARβ/δ i esxpressed in many organs, whereas PPARγ is mainly expressed in adipose tissue and to a lesser extent in other tissues (48, 49). PPARα controls numerous genes that are directly or indirectly involved in lipid metabolism and energy homeostasis, in response to activation by lipophilic compounds, including fatty acids (56). PPARγ, which exhibits a circadian expression pattern (130), is involved in adipocyte differentiation and plays an important role in lipid and glucose metabolism (57, 122). PPARγ in the liver is upregulated upon a high-fat diet (49), whereas fasting causes an upregulation of PPARα (56). PPARα and PPARγ directly interact with clock genes, suggesting their role in linking energy metabolism to the peripheral molecular clock (reviewed in Ref. 21). For instance, PER2 interacts with PPARγ and suppresses its pro-adipogenic transcriptional activity, specifically in white adipose tissue. Indeed, lack of PER2 results in increased adipogenesis in fibroblasts, although in vivo, PER2 deficiency in mice did not result in increased adiposity but resulted in a lean phenotype (33). Furthermore, a recent study by Eckel-Mahan and coworkers (25) revealed a critical role for PPARγ in driving de novo gene expression under high-fat diet conditions by binding to the HFD-specific gene promoters, while rhythmic expression was lost in the presence of a PPARγ antagonist. Although PPARs are expressed mostly in tissues with a high rate of fatty oxidation, they have also been found in the brain (88).
PPARS AND THE CIRCADIAN CLOCK.
PPARs have been proposed to integrate the circadian clock with energy metabolism, yet how exactly they link the two together is unknown (21). Next to its function in metabolism, PPARs are directly linked to the molecular circadian clock. Ppar transcription is driven by BMAL1, and PPARs in their turn activate the transcription of Bmal1. PPARs are rhythmically expressed in several peripheral tissues, including liver and adipose tissue, and to a lesser extent in the SCN (PPARα). Fibrates, a group of PPARα agonists, are able to alter the rhythmic expression of Bmal1, Per1, Per2, Per3, and Rev-erbα in cultured hepatocytes and multiple mouse peripheral tissues including the liver (18, 21, 30, 96, 117). The PPARα ligand bezafibrate advanced locomotor activity in wild-type and SCN-lesioned mice, indicating that PPARα is involved in circadian clock control but independently of the SCN (117). This might be explained by the fact that PPARα, in contrast to other PPARs, was expressed at low levels in the rat and mouse hypothalamus (73, 88, 117). This is in accordance with a study by Canaple et al. (18) who reported that PPARα−/− mice exhibited normal entrainment in constant darkness, indicating that PPARα is not essential for circadian rhythm generation in the SCN, although it was found to be important in photoentrainment of the clock (96). The circadian expression of PPARα in the liver was reset by feeding (18). Thus PPARα may play an important role in peripheral, but not central, circadian control. Because of the low expression of PPARα in the SCN, alterations in rhythmic expression of PPARα, due to food intake, may potentially lead to a desynchronization between the peripheral and central clock. As PPARs regulates Bmal1 transcription, it can be speculated that feeding can influence the clock through PPAR, which might ultimately lead to alterations in energy metabolism. Alternatively, and as shown by Salgado-Delgado et al. (112), food intake may alter the rhythmic expression of Pparα without altering the rhythmic expression of the clock genes, leading to desynchronization of clock genes from metabolic genes, which was found to be associated with adiposity and liver steatosis. Further analysis should be undertaken to understand the role of PPARs in feeding-dependent clock alterations.
Interrelation between nutrient sensors.
In the previous section, nutrient sensors were described that may be involved in linking nutrient intake to the molecular circadian clock. Although discussed separately, these factors do not all function independently, but form a highly interactive metabolic network. For instance, AMPK enhances SIRT1 activity by increasing cellular NAD+ levels, and activated SIRT1, in turn, can phosphorylate AMPK via deacetylation-dependent activation of the AMPK-activating kinase liver kinase B1 (LKB1) (reviewed in Ref. 51). Similarly, AMPK can regulate PARP-1 activity through regulating NAD+ levels (63), and AMPK can inhibit CREB activity (125) and PPARγ. Furthermore, it was proposed that CREB controls hepatic lipid metabolism through PPARγ (42). In the liver, SIRT1 promotes fatty acid oxidation and gluconeogenesis via PGC-1α and PPARα, and in white adipose tissue, SIRT1 decreases fat storage by repressing PPARγ (34). Determining the interrelationship between the different factors in response to nutrients will be important in determining how circadian and metabolic rhythms are linked.
Conclusion and Recommendations for Further Studies
This review has focused on the effects of fatty acids and sugar on the circadian clock. Studies have shown that high-fat and high-sugar intake can lead to alterations in clock gene expression in peripheral tissues, in non-SCN brain nuclei, as well as in the SCN. Putative mechanisms through which this can occur were discussed with an important role for signaling components that link nutrient status to the molecular clock mechanism. Indeed, it was shown that alterations in circulating levels of fatty acids and glucose can affect various nutrient sensing mechanisms, which can affect the molecular clock, thereby linking fat and/or sugar intake to the circadian clock. Furthermore, many studies have focused on how altered energetic status can affect the clock. Current evidence supports the idea that the amount of nutrients may be more important for resetting the molecular clock than the source of nutrients, but it remains elusive whether the specific metabolites of fatty acids and sugar can contribute to alterations in the circadian clock and whether there are differential mechanisms through which fatty acids and sugar regulate the clock.
Perspectives and Significance
From an evolutionary point of view, it is advantageous to adapt the clock to nutrient status, so that activity is synchronized to periods in which food is available. In our search for evidence for differential effects of fat and sugar on the molecular clock, we noticed a paucity of studies in which nutrient content was compared with energy content. To assess if, and which, specific macronutrient components in (human) diet can alter normal circadian rhythmicity, further analysis should be undertaken to compare the effects of fatty acids and sugar on the circadian clock and clock output rhythms.
Another outstanding question is how nutrients can differentially affect the central clock versus the peripheral clocks. For instance, high-fat intake affects both the central and peripheral clocks. Conversely, feeding-dependent alterations in the rhythmic expression of PPARα only takes place in peripheral organs and not in the central clock, although individual nuclei and specific neuropeptide-expressing neurons were not individually studied. If alterations in PPARα lead to alterations in the molecular clock, and subsequently in clock output genes, this might be a mechanism that promotes circadian desynchronization between the central and peripheral clocks. Furthermore, it would be of interest to study whether nutrients themselves can differentially alter the central and peripheral clocks, and whether this leads to desynchrony in clock output rhythms and altered rhythms of downstream metabolic pathways.
Given the reciprocal relationship between the molecular circadian clock and energy homeostasis, these are important questions that directly affect physiological homeostasis. Understanding the impact of specific macronutrients on the circadian clock will allow for guidance toward the composition and timing of meals optimal for physiological health, as well as putative therapeutic targets to regulate the molecular clock. Further studies will be necessary to explore this intriguing and complex field of research.
GRANTS
We acknowledge funding from the Natural Sciences and Engineering Research Council (NSERC), Canadian Institutes for Health Research (CIHR), Canada Foundation for Innovation and Canada Research Chairs Program (to D. D. Belsham), and Technology Foundation STW grant Feeding OnTime (no. 12195, to A. Kalsbeek). J. E. Oosterman was supported by an AMC PhD Scholarship.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: J.E.O. prepared figures; J.E.O. drafted manuscript; J.E.O., A.K., S.E.l.F., and D.D.B. edited and revised manuscript; J.E.O., A.K., S.E.l.F., and D.D.B. approved final version of manuscript.
REFERENCES
- 1.Abe M, Herzog ED, Block G. Lithium lengthens the circadian period 3261–3264 of individual suprachiasmatic nucleus neurons. Neuroreport 11: 3261–3264, 2000. [DOI] [PubMed] [Google Scholar]
- 2.Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray is driven by the suprachiasmatic nucleus. Curr Biol 12: 540–550, 2002. [DOI] [PubMed] [Google Scholar]
- 3.Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol 12: 141–51, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arble DM, Bass J, Laposky AD, Vitaterna MH, Turek FW. Circadian timing of food intake contributes to weight gain. Obesity (Silver Spring) 17: 2100–2102, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aschoff J. Circadian timing. Ann NY Acad Sci 423: 442–68, 1984. [DOI] [PubMed] [Google Scholar]
- 6.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134: 317–328, 2008. [DOI] [PubMed] [Google Scholar]
- 7.Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell 142: 943–953, 2010. [DOI] [PubMed] [Google Scholar]
- 8.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab 13: 125–137, 2011. [DOI] [PubMed] [Google Scholar]
- 9.Balsalobre A. Clock genes in mammalian peripheral tissues. Cell Tissue Res 309: 193–199, 2002. [DOI] [PubMed] [Google Scholar]
- 10.Balsalobre A. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289: 2344–7, 2000. [DOI] [PubMed] [Google Scholar]
- 11.Balsalobre A, Marcacci L, Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr Biol 10: 1291–1294, 2000. [DOI] [PubMed] [Google Scholar]
- 12.Barnea M, Madar Z, Froy O. High-fat diet delays and fasting advances the circadian expression of adiponectin signaling components in mouse liver. Endocrinology 150: 161–168, 2009. [DOI] [PubMed] [Google Scholar]
- 13.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science 330: 1349–1354, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belsham DD, Cai F, Cui H, Smukler SR, Salapatek AM, Shkreta L. Generation of a phenotypic array of hypothalamic neuronal cell models to study complex neuroendocrine disorders. Endocrinology 145: 393–400, 2004. [DOI] [PubMed] [Google Scholar]
- 15.Bray MS, Young ME. Regulation of fatty acid metabolism by cell autonomous circadian clocks: time to fatten up on information? J Biol Chem 286: 11883–11889, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buijs RM, Kalsbeek A. Hypothalamic integration of central and peripheral clocks. Nat Rev Neurosci 2: 521–526, 2001. [DOI] [PubMed] [Google Scholar]
- 17.Buijs RM, la Fleur SE, Wortel J, Van Heyningen C, Zuiddam L, Mettenleiter TC, Kalsbeek A, Nagai K, Niijima A. The suprachiasmatic nucleus balances sympathetic and parasympathetic output to peripheral organs through separate preautonomic neurons. J Comp Neurol 464: 36–48, 2003. [DOI] [PubMed] [Google Scholar]
- 18.Canaple L, Rambaud J, Dkhissi-Benyahya O, Rayet B, Tan NS, Michalik L, Delaunay F, Wahli W, Laudet V. Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor alpha defines a novel positive feedback loop in the rodent liver circadian clock. Mol Endocrinol 20: 1715–1727, 2006. [DOI] [PubMed] [Google Scholar]
- 19.Challet E, Losee-Olson S, Turek FW. Reduced glucose availability attenuates circadian responses to light in mice. Am J Physiol Regul Integr Comp Physiol 276: R1063–R1070, 1999. [DOI] [PubMed] [Google Scholar]
- 20.Chawla A, Lazar MA. Induction of Rev-ErbAα, an orphan receptor encoded on the opposite strand of the a-thyroid hormone receptor gene, during adipocyte differentiation. J Biol Chem 268: 16265–16269, 1993. [PubMed] [Google Scholar]
- 21.Chen L, Yang G. PPARs integrate the mammalian clock and energy metabolism. PPAR Res 2014: 653017, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coomans CP, van den Berg SA, Houben T, van Klinken JB, van den Berg R, Pronk AC, Havekes LM, Romijn JA, van Dijk KW, Biermasz NR, Meijer JH. Detrimental effects of constant light exposure and high-fat diet on circadian energy metabolism and insulin sensitivity. FASEB J 27: 1721–1732, 2013. [DOI] [PubMed] [Google Scholar]
- 23.Damiola F. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 14: 2950–2961, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eckel-Mahan K, Sassone-Corsi P. Metabolism and the circadian clock converge. Physiol Rev 93: 107–135, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eckel-Mahan KL, Patel VR, de Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, Dilag-Penilla SA, Dyar KA, Baldi P, Sassone-Corsi P. Reprogramming of the circadian clock by nutritional challenge. Cell 155: 1464–1478, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feillet CA, Ripperger JA, Magnone MC, Dulloo A, Albrecht U, Challet E. Lack of food anticipation in Per2 mutant mice. Curr Biol 16: 2016–2022, 2006. [DOI] [PubMed] [Google Scholar]
- 27.Fick LJ, Fick GH, Belsham DD. Palmitate alters the rhythmic expression of molecular clock genes and orexigenic neuropeptide Y mRNA levels within immortalized, hypothalamic neurons. Biochem Biophys Res Commun 413: 414–419, 2011. [DOI] [PubMed] [Google Scholar]
- 28.Foster RG, Hankins MW. Circadian vision. Curr Biol 17: R746–R51, 2007. [DOI] [PubMed] [Google Scholar]
- 29.Froy O. The relationship between nutrition and circadian rhythms in mammals. Front Neuroendocrinol 28: 61–71, 2007. [DOI] [PubMed] [Google Scholar]
- 30.Gervois P, Chopin-Delannoy S, Fadel A, Dubois G, Kosykh V, Fruchart JC, Najïb J, Laudet V, Staels B. Fibrates increase Human REV-ERB expression in liver via a novel peroxisome proliferator-activated receptor response element. Mol Endocrinol 13: 400–409, 1999. [DOI] [PubMed] [Google Scholar]
- 31.Ginty DD, Kornhauser JM, Thompson MA, Bading H, Mayo KE, Takahashi JS, Greenberg ME. Regulation of CREB phosphorylation in the suprachiasmatic nucleus by light and a circadian clock. Science 260: 238–241, 1993. [DOI] [PubMed] [Google Scholar]
- 32.Greco JA, Oosterman JE, Belsham DD. Differential effects of omega-3 fatty acid DHA and palmitate on the circadian transcriptional profile of clock genes in immortalized hypothalamic neurons. Am J Physiol Regul Integr Comp Physiol (August 20, 2014). doi: 10.1152/ajpregu.00100.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grimaldi B, Bellet MM, Katada S, Astarita G, Hirayama J, Amin RH, Granneman JG, Piomelli D, Leff T, Sassone-Corsi P. PER2 controls lipid metabolism by direct regulation of PPARgamma. Cell Metab 12: 509–520, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol 5: 253–295, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harada Y, Sakai M, Kurabayashi N, Hirota T, Fukada Y. Ser-557-phosphorylated mCRY2 is degraded upon synergistic phosphorylation by glycogen synthase kinase-3 beta. J Biol Chem 280: 31714–31721, 2005. [DOI] [PubMed] [Google Scholar]
- 36.Hardie DG. AMPK: a key regulator of energy balance in the single cell and the whole organism. Int J Obes (Lond) 32, Suppl 4: S7–S12, 2008. [DOI] [PubMed] [Google Scholar]
- 37.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol 13: 251–262, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem 80: 825–858, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hastings MH. Entrainment of the circadian system of mammals by nonphotic cues. Chronobiol Int 15: 425–445, 1998. [DOI] [PubMed] [Google Scholar]
- 40.Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci 4: 649–661, 2003. [DOI] [PubMed] [Google Scholar]
- 41.Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, Ellisman MH, Panda S. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab 15: 848–860, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herzig S, Hedrick S, Morantte I, Koo SH, Galimi F, Montminy M. CREB controls hepatic lipid metabolism through nuclear hormone receptor PPAR-γ. Nature 426: 190–193, 2003. [DOI] [PubMed] [Google Scholar]
- 43.Hirota T, Fukada Y. Resetting mechanism of central and peripheral circadian clocks in mammals. Zool Sci 21: 359–368, 2004. [DOI] [PubMed] [Google Scholar]
- 44.Hirota T, Kon N, Itagaki T, Hoshina N, Okano T, Fukada Y. Transcriptional repressor TIEG1 regulates Bmal1 gene through GC box and controls circadian clockwork. Genes Cells 15: 111–121, 2010. [DOI] [PubMed] [Google Scholar]
- 45.Hirota T, Lewis WG, Liu AC, Lee JW, Schultz PG, Kay SA. A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3β. Proc Natl Acad Sci USA 105: 20746–207451, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirota T, Okano T, Kokame K, Shirotani-Ikejima H, Miyata T, Fukada Y. Glucose down-regulates Per1 and Per2 mRNA levels and induces circadian gene expression in cultured Rat-1 fibroblasts. J Biol Chem 277: 44244–44251, 2002. [DOI] [PubMed] [Google Scholar]
- 47.Iitaka C, Miyazaki K, Akaike T, Ishida N. A role for glycogen synthase kinase-3beta in the mammalian circadian clock. J Biol Chem 280: 29397–29402, 2005. [DOI] [PubMed] [Google Scholar]
- 48.Inoue I, Shinoda Y, Ikeda M, Nomura M, Matsunaga T, Awata T, Komoda T, Hayashi K, Xu H, Kawai S, Katayama S. CLOCK/BMAL1 is involved in lipid metabolism via transactivation of the peroxisome proliferator-activated receptor (PPAR) response element. J Atheroscler Thromb 12: 169–174, 2005. [DOI] [PubMed] [Google Scholar]
- 49.Inoue M, Ohtake T, Motomura W, Takahashi N, Hosoki Y, Miyoshi S, Suzuki Y, Saito H, Kohgo Y, Okumura T. Increased expression of PPARgamma in high fat diet-induced liver steatosis in mice. Biochem Biophys Res Commun 336: 215–222, 2005. [DOI] [PubMed] [Google Scholar]
- 50.Iwanaga H, Yano M, Miki H, Okada K, Azama T, Takiguchi S, Fujiwara Y, Yasuda T, Nakayama M, Kobayashi M, Oishi K, Ishida N, Nagai K, Monden M. Per2 gene expressions in the suprachiasmatic nucleus and liver differentially respond to nutrition factors in rats. J Parenter Enteral Nutr 29: 157–161, 2005. [DOI] [PubMed] [Google Scholar]
- 51.Jordan SD, Lamia KA. AMPK at the crossroads of circadian clocks and metabolism. Mol Cell Endocrinol 366: 163–169, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaasik K, Kivimae S, Allen JJ, Chalkley RJ, Huang Y, Baer K, Kissel H, Burlingame AL, Shokat KM, Ptacek LJ, Fu YH. Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab 17: 291–302, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaladchibachi SA, Doble B, Anthopoulos N, Woodgett JR, Manoukian AS. Glycogen synthase kinase 3, circadian rhythms, and bipolar disorder: a molecular link in the therapeutic action of lithium. J Circadian Rhythms 5: 3, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalsbeek A, la Fleur S, Fliers E. Circadian control of glucose metabolism. Mol Metab 3: 372–383, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalsbeek A, Ruiter M, La Fleur SE, Cailotto C, Kreier F, Buijs RM. The hypothalamic clock and its control of glucose homeostasis. Prog Brain Res 153: 283–307, 2006. [DOI] [PubMed] [Google Scholar]
- 56.Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor α mediates the adaptive response to fasting. J Clin Invest 103: 1489–1498, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim H, Ahn Y. Role of peroxisome proliferator-activated receptor-γ in the glucose-sensing apparatus of liver and β-cells. Diabetes 53S60–S65, 2004. [DOI] [PubMed] [Google Scholar]
- 58.Knutsson A. Health disorders of shift workers. Occup Med 53: 103–108, 2003. [DOI] [PubMed] [Google Scholar]
- 59.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet 15 Spec No 2: R271–R277, 2006. [DOI] [PubMed] [Google Scholar]
- 60.Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab 6: 414–421, 2007. [DOI] [PubMed] [Google Scholar]
- 61.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol 5: e34, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kudo T, Tamagawa T, Kawashima M, Mito N, Shibata S. Attenuating effect of clock mutation on triglyceride contents in the ICR mouse liver under a high-fat diet. J Biol Rhythms 22: 312–323, 2007. [DOI] [PubMed] [Google Scholar]
- 63.Kumar V, Takahashi JS. PARP around the clock. Cell 142: 841–843, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.la Fleur SE. Daily rhythms in glucose metabolism: suprachiasmatic nucleus output to peripheral tissue. J Neuroendocrinol 15: 315–322, 2003. [DOI] [PubMed] [Google Scholar]
- 65.la Fleur SE, Kalsbeek A, Wortel J, Buijs R. A Suprachiasmatic nucleus generated rhythm in basal glucose concentrations. J Neuroendocrinol 11: 643–652, 1999. [DOI] [PubMed] [Google Scholar]
- 66.la Fleur SE, Kalsbeek A, Wortel J, Fekkes ML, Buijs R. A daily rhythm in glucose tolerance: a role for the suprachiasmatic nucleus. Diabetes 50: 1237–1243, 2001. [DOI] [PubMed] [Google Scholar]
- 67.la Fleur SE, Luijendijk MC, van der Zwaal EM, Brans MA, Adan RA. The snacking rat as model of human obesity: effects of a free-choice high-fat high-sugar diet on meal patterns. Int J Obes (Lond) 38: 643–649, 2014. [DOI] [PubMed] [Google Scholar]
- 68.Lamia KA, Papp SJ, Yu RT, Barish GD, Uhlenhaut NH, Jonker JW, Downes M, Evans RM. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 480: 552–526, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, Thompson CB, Evans RM. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326: 437–440, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA 105: 15172–15127, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lau P, Nixon SJ, Parton RG, Muscat GE. RORalpha regulates the expression of genes involved in lipid homeostasis in skeletal muscle cells: caveolin-3 and CPT-1 are direct targets of ROR. J Biol Chem 279: 36828–36840, 2004. [DOI] [PubMed] [Google Scholar]
- 72.Lee J, Kim MS. The role of GSK3 in glucose homeostasis and the development of insulin resistance. Diabetes Res Clin Pract 77, Suppl 1: S49–S57, 2007. [DOI] [PubMed] [Google Scholar]
- 73.Lemberger T, Braissant O, Juge-Aubry C, Keller H, Saladin R, Staels B, Auwerx J, Burger AG, Meier CA, Wahli W. PPAR tissue distribution and interactions with other hormone-signaling pathways. Ann NY Acad Sci 804: 231–251, 1996. [DOI] [PubMed] [Google Scholar]
- 74.Li MD, Ruan HB, Hughes ME, Lee JS, Singh JP, Jones SP, Nitabach MN, Yang X. O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell Metab 17: 303–310, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet 4: e1000023, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu S, Evans RM. Metabolism: Tick, tock, a high-fat clock. Nat Rev Endocrinol 10: 191–192, 2014. [DOI] [PubMed] [Google Scholar]
- 77.Lowden A, Moreno C, Holmbäck U, Lennernäs M, Tucker P. Eating and shift work-effects on habits, metabolism and performance. Scand J Work Environ Health 36: 150–162, 2010. [DOI] [PubMed] [Google Scholar]
- 78.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet 5: 407–441, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, Lopez JP, Philipson LH, Bradfield CA, Crosby SD, JeBailey L, Wang X, Takahashi JS, Bass J. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 466: 627–631, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mariappan MM, Shetty M, Sataranatarajan K, Choudhury GG, Kasinath BS. Glycogen synthase kinase 3beta is a novel regulator of high glucose- and high insulin-induced extracellular matrix protein synthesis in renal proximal tubular epithelial cells. J Biol Chem 283: 30566–30575, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marty N, Dallaporta M, Thorens B. Brain glucose sensing, counterregulation, and energy homeostasis. Physiology (Bethesda) 22: 241–251, 2007. [DOI] [PubMed] [Google Scholar]
- 82.Mendoza J, Graff C, Dardente H, Pevet P, Challet E. Feeding cues alter clock gene oscillations and photic responses in the suprachiasmatic nuclei of mice exposed to a light/dark cycle. J Neurosci 25: 1514–1522, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mendoza J, Lopez-Lopez C, Revel FG, Jeanneau K, Delerue F, Prinssen E, Challet E, Moreau JL, Grundschober C. Dimorphic effects of leptin on the circadian and hypocretinergic systems of mice. J Neuroendocrinol 23: 28–38, 2011. [DOI] [PubMed] [Google Scholar]
- 84.Mendoza J, Pevet P, Challet E. High-fat feeding alters the clock synchronization to light. J Physiol 586: 5901–5910, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Minokoshi Y, Alquier T, Furukawa N, Kim YB, Lee A, Xue B, Mu J, Foufelle F, Ferre P, Birnbaum MJ, Stuck BJ, Kahn BB. AMP-kinase regulates food intake by responding to hormonal and nutrient signals in the hypothalamus. Nature 428: 569–574, 2004. [DOI] [PubMed] [Google Scholar]
- 86.Mistlberger RE, Lukman H, Nadeau BG. Circadian rhythms in the Zucker obese rat: assessment and intervention. Appetite 30: 255–267, 1998. [DOI] [PubMed] [Google Scholar]
- 87.Morales L, Del Olmo N, Valladolid-Acebes I, Fole A, Cano V, Merino B, Stucchi P, Ruggieri D, Lopez L, Alguacil LF, Ruiz-Gayo M. Shift of circadian feeding pattern by high-fat diets is coincident with reward deficits in obese mice. PLos One 7: e36139, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moreno S, Farioli-Vecchioli S, Ceru MP. Immunolocalization of peroxisome proliferator-activated receptors and retinoid X receptors in the adult rat CNS. Neuroscience 123: 131–145, 2004. [DOI] [PubMed] [Google Scholar]
- 89.Morin LP, Goodless-Sanchez N, Smale L, Moore RY. Projections of the suprachiasmatic nuclei, subparaventricular zone and retrochiasmatic area in the golden hamster. Neuroscience 61: 391–410, 1994. [DOI] [PubMed] [Google Scholar]
- 90.Morris M, Araujo IC, Pohlman RL, Marques MC, Rodwan NS, Farah VM. Timing of fructose intake: an important regulator of adiposity. Clin Exp Pharmacol Physiol 39: 57–62, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mrosovsky N. Locomotor activity and non-photic influences on circadian clocks. Biol Rev 71: 343–372, 1996. [DOI] [PubMed] [Google Scholar]
- 92.Nagai K, Nishio T, Nakagawa H, Nakamura S, Fukuda Y. Effect of bilateral lesions of the suprachiasmatic nuclei on the circadian rhythm of food-intake. Brain Res 142: 384–389, 1978. [DOI] [PubMed] [Google Scholar]
- 93.Nagai K, Suda M, Yamagishi O, Toyama Y, Nakagawa H. Studies on the circadian rhythm of phosphoenolpyruvate carboxykinase. III. Circadian rhythm in the kidney. J Biochem 77: 1249–1254, 1975. [PubMed] [Google Scholar]
- 94.Obici S, Feng Z, Arduini A, Conti R, Rossetti L. Inhibition of hypothalamic carnitine palmitoyltransferase-1 decreases food intake and glucose production. Nat Med 9: 756–761, 2003. [DOI] [PubMed] [Google Scholar]
- 95.Oishi K, Atsumi G, Sugiyama S, Kodomari I, Kasamatsu M, Machida K, Ishida N. Disrupted fat absorption attenuates obesity induced by a high-fat diet in Clock mutant mice. FEBS Lett 580: 127–130, 2006. [DOI] [PubMed] [Google Scholar]
- 96.Oishi K, Shirai H, Ishida N. PPARa is involved in photoentrainment of the circadian clock. Neuroreport 19: 487–489, 2008. [DOI] [PubMed] [Google Scholar]
- 97.Panda S. Multiple photopigments entrain the mammalian circadian oscillator. Neuron 53: 619–621, 2007. [DOI] [PubMed] [Google Scholar]
- 98.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109: 307–320, 2002. [DOI] [PubMed] [Google Scholar]
- 99.Panda S, Sato TK, Castrucci AM, Rollag MD, DeGrip WJ, Hogenesch JB, Provencio I, Kay SA. Melanopsin (Opn4) requirement for normal light-induced circadian phase shifting. Science 298: 2213–2216, 2002. [DOI] [PubMed] [Google Scholar]
- 100.Paschos GK, Ibrahim S, Song WL, Kunieda T, Grant G, Reyes TM, Bradfield CA, Vaughan CH, Eiden M, Masoodi M, Griffin JL, Wang F, Lawson JA, Fitzgerald GA. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med 18: 1768–1777, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Peek CB, Ramsey KM, Marcheva B, Bass J. Nutrient sensing and the circadian clock. Trends Endocrinol Metab 23: 312–318, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pendergast JS, Branecky KL, Yang W, Ellacott KL, Niswender KD, Yamazaki S. High-fat diet acutely affects circadian organisation and eating behavior. Eur J Neurosci 37: 1350–1356, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Preitner F, Damiola F, Molina LL, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110: 251–260, 2002. [DOI] [PubMed] [Google Scholar]
- 104.Prosser RA, Bergeron HE. Leptin phase-advances the rat suprachiasmatic circadian clock in vitro. Neurosci Lett 336: 139–142, 2003. [DOI] [PubMed] [Google Scholar]
- 105.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature 418: 935–941, 2002. [DOI] [PubMed] [Google Scholar]
- 105a.Resuehr D, Olcese J. Caloric restriction and melatonin substitution: effects on murine circadian parameters. Brain Res 1048: 146–52, 2005. [DOI] [PubMed] [Google Scholar]
- 106.Ruddock MW, Stein A, Landaker E, Park J, Cooksey RC, McClain D, Patti ME. Saturated fatty acids inhibit hepatic insulin action by modulating insulin receptor expression and post-receptor signalling. J Biochem 144: 599–607, 2008. [DOI] [PubMed] [Google Scholar]
- 107.Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol 2: e377, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science 293: 510–514, 2001. [DOI] [PubMed] [Google Scholar]
- 109.Sadacca LA, Lamia KA, deLemos AS, Blum B, Weitz CJ. An intrinsic circadian clock of the pancreas is required for normal insulin release and glucose homeostasis in mice. Diabetologia 54: 120–124, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Saderi N, Cazarez-Marquez F, Buijs FN, Salgado-Delgado RC, Guzman-Ruiz MA, del Carmen Basualdo M, Escobar C, Buijs RM. The NPY intergeniculate leaflet projections to the suprachiasmatic nucleus transmit metabolic conditions. Neuroscience 246: 291–300, 2013. [DOI] [PubMed] [Google Scholar]
- 111.Sahar S, Zocchi L, Kinoshita C, Borrelli E, Sassone-Corsi P. Regulation of BMAL1 protein stability and circadian function by GSK3beta-mediated phosphorylation. PLos One 5: e8561, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Salgado-Delgado RC, Saderi N, Basualdo Mdel C, Guerrero-Vargas NN, Escobar C, Buijs RM. Shift work or food intake during the rest phase promotes metabolic disruption and desynchrony of liver genes in male rats. PLos One 8: e60052, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sanada K, Okano T, Fukada Y. Mitogen-activated protein kinase phosphorylates and negatively regulates basic helix-loop-helix-PAS transcription factor BMAL1. J Biol Chem 277: 267–271, 2002. [DOI] [PubMed] [Google Scholar]
- 114.Scheer FA, Kalsbeek A, Buijs R. Cardiovascular control by the suprachiasmatic nucleus: neural and neuroendocrine mechanisms in human and rat. Biol Chem 384: 697–709, 2003. [DOI] [PubMed] [Google Scholar]
- 115.Sherman H, Genzer Y, Cohen R, Chapnik N, Madar Z, Froy O. Timed high-fat diet resets circadian metabolism and prevents obesity. FASEB J 26: 3493–502, 2012. [DOI] [PubMed] [Google Scholar]
- 116.Shimba S, Ishii N, Ohta Y, Ohno T, Watabe Y, Hayashi M, Wada T, Aoyagi T, Tezuka M. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc Natl Acad Sci USA 102: 12071–12076, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shirai H, Oishi K, Kudo T, Shibata S, Ishida N. PPARalpha is a potential therapeutic target of drugs to treat circadian rhythm sleep disorders. Biochem Biophys Res Commun 357: 679–662, 2007. [DOI] [PubMed] [Google Scholar]
- 118.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science 291: 490–493, 2001. [DOI] [PubMed] [Google Scholar]
- 119.Suda M, Nagai K, Nakagawa H. Studies on the circadian rhythm of phosphoenolpyruvate carboxykinase activity in rats. I. Mechanism of circadian increase in liver enzyme with special reference to hormonal and dietary effects. J Biochem 73: 727–738, 1973. [DOI] [PubMed] [Google Scholar]
- 120.Tahara Y, Otsuka M, Fuse Y, Hirao A, Shibata S. Refeeding after fasting elicits insulin-dependent regulation of Per2 and Rev-erbalpha with shifts in the liver clock. J Biol Rhythms 26: 230–240, 2011. [DOI] [PubMed] [Google Scholar]
- 121.Taib B, Bouyakdan K, Hryhorczuk C, Rodaros D, Fulton S, Alquier T. Glucose regulates hypothalamic long-chain fatty acid metabolism via AMP-activated kinase (AMPK) in neurons and astrocytes. J Biol Chem 288: 37216–37229, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem 77: 289–312, 2008. [DOI] [PubMed] [Google Scholar]
- 123.Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci USA 99: 7728–7733, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, Bass J. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308: 1043–1045, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vila-Brau A, De Sousa-Coelho AL, Goncalves JF, Haro D, Marrero PF. Fsp27/CIDEC is a CREB target gene induced during early fasting in liver and regulated by FA oxidation rate. J Lipid Res 54: 592–601, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci USA 106: 21453–21458, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Waddington Lamont E, Harbour VL, Barry-Shaw J, Renteria Diaz L, Robinson B, Stewart J, Amir S. Restricted access to food, but not sucrose, saccharine, or salt, synchronizes the expression of Period2 protein in the limbic forebrain. Neuroscience 144: 402–411, 2007. [DOI] [PubMed] [Google Scholar]
- 128.Yanagihara H, Ando H, Hayashi Y, Obi Y, Fujimura A. High-fat feeding exerts minimal effects on rhythmic mRNA expression of clock genes in mouse peripheral tissues. Chronobiol Int 23: 905–914, 2006. [DOI] [PubMed] [Google Scholar]
- 129.Yang SC, Tseng HL, Shieh KR. Circadian-clock system in mouse liver affected by insulin resistance. Chronobiol Int 30: 796–810, 2013. [DOI] [PubMed] [Google Scholar]
- 130.Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell 126: 801–810, 2006. [DOI] [PubMed] [Google Scholar]
- 131.Yang Y, Atasoy D, Su HH, Sternson SM. Hunger states switch a flip-flop memory circuit via a synaptic AMPK-dependent positive feedback loop. Cell 146: 992–1003, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yi CX, van der Vliet J, Dai J, Yin G, Ru L, Buijs RM. Ventromedial arcuate nucleus communicates peripheral metabolic information to the suprachiasmatic nucleus. Endocrinology 147: 283–294, 2006. [DOI] [PubMed] [Google Scholar]
- 133.Yin L, Wang J, Klein PS, Lazar MA. Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science 311: 1002–1005, 2006. [DOI] [PubMed] [Google Scholar]
- 134.Yulyaningsih E, Loh K, Lin S, Lau J, Zhang L, Shi Y, Berning BA, Enriquez R, Driessler F, Macia L, Khor EC, Qi Y, Baldock P, Sainsbury A, Herzog H. Pancreatic polypeptide controls energy homeostasis via Npy6r signaling in the suprachiasmatic nucleus in mice. Cell Metab 19: 58–72, 2014. [DOI] [PubMed] [Google Scholar]
- 135.Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, Hirota T, Nusinow DA, Sun X, Landais S, Kodama Y, Brenner DA, Montminy M, Kay SA. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med 16: 1152–1156, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang J, Kaasik K, Blackburn MR, Lee CC. Constant darkness is a circadian metabolic signal in mammals. Nature 439: 340–343, 2006. [DOI] [PubMed] [Google Scholar]