Abstract
Polycystic ovary syndrome (PCOS) is the most common gynecological endocrine disorder. The genetic background is believed to play a crucial role in the pathogenesis of PCOS. In recent years, the role of insulin receptor (INSR) polymorphisms in PCOS predisposition has attracted much attention. We performed a meta-analysis to investigate the association between the single nucleotide polymorphisms (SNPs) of INSR and PCOS. Published literature from Pubmed, Embase, and Cochrane CENTRAL was retrieved up until 7 August 2014. A total of 20 case-control studies including 23,845 controls and 17,460 PCOS cases with an average Newcastle-Ottawa quality assessment scale (NOS) score of 6.75 were analyzed. Ninety-eight SNPs distributed in 23 exons and the flanking regions of INSR were investigated, among which 17 SNPs were found to be associated with PCOS. Three SNPs detected in more than three studies were selected for further analyses. Twelve studies including 1158 controls and 1264 PCOS cases entered the analysis of rs1799817, but no significant association was found for every genotype (p > 0.05). Further subgroup stratification by ethnicity and weight did not lead to discovery of significant correlation (p > 0.05). For rs2059806, four studies including 442 controls and 524 PCOS cases were qualified for meta-analysis, and no significant association with PCOS was found for any genotype (p > 0.05). Four studies including 12,830 controls and 11,683 PCOS cases investigated the correlation between rs2059807 and PCOS, and five of the six cohorts indicated a significant impact. Our current meta-analysis suggests no significant correlation between rs1799817/rs2059806 SNPs and susceptibility of PCOS, while rs2059807 could be a promising candidate SNP that might be involved in the susceptibility of PCOS.
Keywords: single nucleotide polymorphism (SNP), insulin receptor gene (INSR), polycystic ovary syndrome (PCOS), meta-analysis
1. Introduction
Polycystic ovary syndrome (PCOS) is the most common gynecological endocrine disorder that is characterized by ovarian dysfunction, hyperandrogenism, and polycystic ovary morphology [1]. It is associated with increased risks of infertility, impaired glucose tolerance, type 2 diabetes mellitus, and metabolic syndrome [2]. While 6%–17% of reproductive-age women worldwide suffer PCOS [3,4], some ethnicities, such as South Asian, have higher incidence rates [5]. The exact molecular mechanism and cellular pathways underlying this disorder are still unclear. Etiological studies demonstrated the significance of genetic susceptibility for the pathogenesis of PCOS [6]. It is believed that multiple, instead of single genes, such as fibrillin-3 (FBN3), fat and obesity associated gene (FTO), insulin (INS), insulin receptor (INSR), insulin receptor substrate 1 (ISR1), DENN/MADD domain containing 1A (DENND1A), thyroid adenoma associated protein (THADA), and luteinizing hormone receptor (LHR), may contribute to the development of PCOS [7,8]. For example, DENND1A may alter the aminopeptidase activity of endoplasmic reticulum and is involved in the development of PCOS [9].
INSR gene is located at the short arm of chromosome 19 and comprises 22 exons. The Insr knockout mice manifested extreme insulin resistance [10]. Insulin resistance may up-regulate LH secretion in pituitary, testosterone production in theca cells, and P450scc activity in granulosa cells, which may disturb follicular maturation and lead to PCOS [7]. Considering that accumulated data showed a strong association between INSR and insulin resistance [11], allelic polymorphism of INSR may impose a genetic predisposition for the development of PCOS. Indeed, a series of studies have been conducted to investigate the relationship between single nucleotide polymorphisms (SNPs) of INSR and PCOS. However, data from these studies appeared to be highly controversial [12,13]. Although a meta-analysis published in 2010 has reviewed then available data, and found no association between the SNPs of INSR and PCOS [14], two Genome-Wide Association Studies in 2011 [15] and 2012 [16] pointed to a positive association between the polymorphisms of INSR and PCOS. These findings ignited a renewed interest in the topic. Since then more studies have been performed, and both positive and negative results have been reported.
As there have been plenty of studies exploring the role of INSR polymorphisms in PCOS, and the results often contradict each other, we carried out an updated meta-analysis covering recent data. The findings will provide useful information for elucidating the relationship between the polymorphisms of INSR and the risk of PCOS.
2. Results
2.1. Selection of the Studies
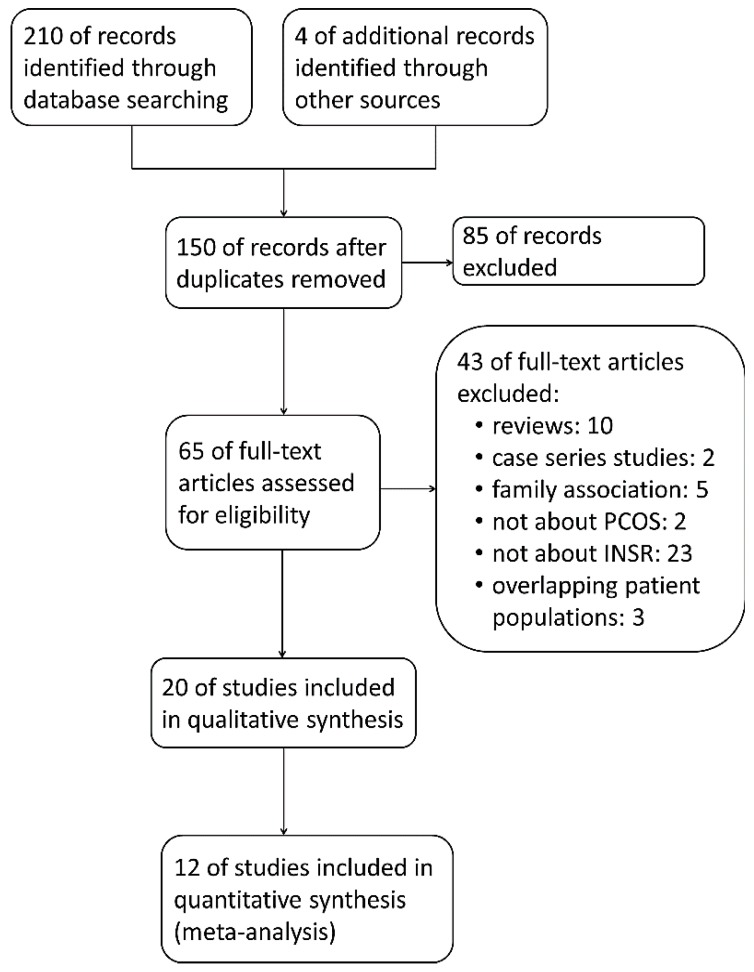
A total of 150 articles were recognized by database searching and reference reading (Figure 1), among which 85 studies were excluded based on information from titles and abstracts. Due to the exclusion reasons listed in the flow chart, including overlapping patient populations in three articles [17,18,19], 20 studies remained for the qualitative synthesis. Eight studies were removed because the involved SNPs were detected in fewer than three studies [20,21,22,23] or the failure to obtain original genotype data [15,16,24,25]. 12 studies were finally selected for meta-analysis.
Figure 1.
Flow chart of study selection for meta-analysis of insulin receptor (INSR) single nucleotide polymorphisms (SNPs) and risk of polycystic ovarian syndrome (PCOS).
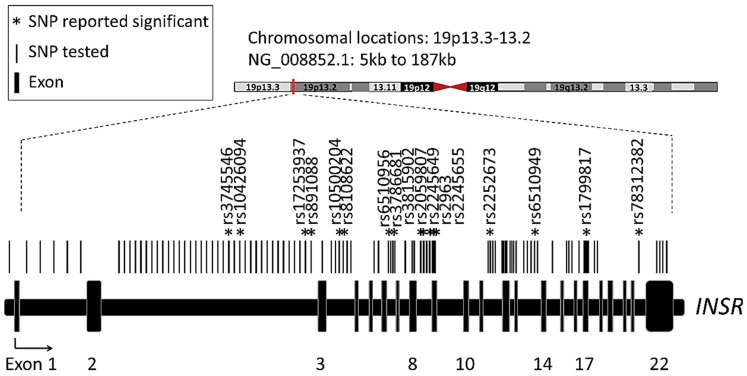
The initially included studies consist of 10 from Asia, seven from Europe, two from North America, and one from South America. These studies cover totally 17,460 cases and 23,845 controls (Table 1). All of these studies were case-control studies published between 1994 and 2013. All were published in English except for one in Chinese. Since the ethnicities in some countries such as Brazil are complicated, it is difficult to define the ethnicities in different studies. As shown in Figure 2 and Table 2, a total of 98 different SNPs were investigated in these studies, which distributed in 23 different exons and the flanking regions of INSR gene. We picked the SNPs that were examined in more than three studies for further meta-analysis. The PCOS patients and controls were genotyped for rs1799817 SNP in 12 studies, for rs2059806 in four studies, and for rs2059807 in four studies.
Table 1.
The characteristics of the studies included for qualitative analyses.
| Study | Year | Country | Ethnicity | Diagnosis Criteria | Number | Methodology | NOS Score | |
|---|---|---|---|---|---|---|---|---|
| Control | PCOS | |||||||
| Conway [26] | 1994 | UK | ND | ND | 8 | 22 | SSCP, sequencing | 6 |
| Talbot [27] | 1996 | UK | ND | Oligo/amenorrhea, hirsuitism, raised testosterone/androstenedione, polycystic ovaries | 5 | 24 | SSCP, sequencing | 6 |
| Siegel [12] | 2002 | USA | White | Oligomenorrhea, hyperandrogenism, polycystic ovaries | 136 | 99 | RFLP | 7 |
| Chen [28] | 2004 | China | Chinese | Oligo/amenorrhea, LH/FSH ≥ 2.5 or TT ≥ 1.56 nmol/L, polycystic ovaries, without endocrine disease or hypertension | 40 | 120 | RFLP | 8 |
| Jin [20] | 2006 | China | Chinese | Rotterdam criteria 2003 | 107 | 109 | SSCP, sequencing | 6 |
| Lee [29] | 2008 | Korea | Korean | Rotterdam criteria 2003 | 100 | 134 | sequencing | 7 |
| Mukherjee [30] | 2009 | India | Indian | Rotterdam criteria 2003 | 144 | 180 | sequencing | 7 |
| Unsal [13] | 2009 | Turkey | Caucasian | Rotterdam criteria 2003 | 50 | 44 | RFLP, sequencing | 8 |
| Hanzu [21] | 2010 | Romania | ND | Rotterdam criteria 2003 | 111 | 115 | sequencing | 7 |
| Goodarzi [22] | 2011 | USA | ND | Discover cohort: NIH criteria 1990; replication cohort: Rotterdam criteria 2003 | 3758 | 801 | Taqman assay, GWAS | 8 |
| Chen [15] | 2011 | China | Chinese | Rotterdam criteria 2003 | 6687 | 4082 | GWAS, LDR | 7 |
| Cirilo [31] | 2012 | Brazil | Brazian | Rotterdam criteria 2003 | 105 | 117 | RFLP | 6 |
| Ranjzad [32] | 2012 | Iran | ND | NIH criteria 1990 | 181 | 181 | RFLP, sequencing | 8 |
| Shi [16] | 2012 | China | Chinese | Rotterdam criteria 2003 | 9594 | 9736 | GWAS, LDR | 7 |
| Kashima [33] | 2013 | Japan | Japanese | Diagnostic criteria of the Japan Society of OBGY 2007 | 99 | 61 | Taqman assay | 7 |
| Skrgatic [34] | 2013 | Croatia | Crotian | Rotterdam criteria 2003 | 175 | 150 | Taqman assay | 5 |
| Ramezani [35] | 2013 | Iran | ND | NIH criteria | 156 | 186 | RFLP | 7 |
| Louwers [24] | 2013 | Netherlands | ND | Rotterdam criteria 2003 | 2164 | 703 | GWAS | 8 |
| Grigorescu [25] | 2013 | Romania | ND | ND | 177 | 500 | KASPar | 5 |
| Yin [23] | 2013 | China | Chinese | ND | 48 | 96 | ND | 5 |
ND: not determined; RFLP: restriction fragment length polymorphism analysis; SSCP: sensitive single strand conformation polymorphism analysis; LDR: ligation detection reaction method; GWAS: genome-wide association study; KASPar: fluorescence-based competitive allele-specific PCR.
Figure 2.
Map of investigated INSR SNPs in former studies. *: SNPs that were found to distribute differentially between PCOS and control.
Table 2.
Chromosome position and OR/RR of INSR SNPs detected in former studies.
| Study | SNP | Position | OR/RR (95% CI) | p Value |
|---|---|---|---|---|
| Conway [26] | rs1799817 | 7125286 | ND | ND |
| rs1799815 | 7125508 | ND | ND | |
| rs1799816 | 7125507 | ND | ND | |
| Talbot [27] | rs2860178 | 7170506 | ND | ND |
| rs2860177 | 7167940 | ND | ND | |
| rs2059806 | 7166365 | ND | ND | |
| rs2229429 | 7166377 | ND | ND | |
| rs41509747 | 7150480 | ND | ND | |
| rs2229431 | 7141764 | ND | ND | |
| rs41339753 | 7128812 | ND | ND | |
| rs1799815 | 7125508 | ND | ND | |
| rs1799817 | 7125286 | ND | ND | |
| Siegel [12] | rs1799817 * | 7125286 | RR: 2.1 in lean | 0.03 in lean, 0.32 in obese |
| Chen [28] | rs1799817 * | 7125286 | ND | <0.01 in total and lean, >0.05 in obese |
| Jin [20] | ND | 7125522 * | ND | <0.01 |
| Lee [29] | ND | 7184410 | ND | ND |
| rs6510959 | 7184227 | 1.39 (0.77–2.50) | 0.2726 | |
| rs2303672 | 7168394 | 0.82 (0.42–1.62) | 0.5707 | |
| rs2059806 | 7166365 | 1.31 (0.86–2.00) | 0.2134 | |
| rs2252673 | 7150407 | 0.90 (0.61–1.31) | 0.5688 | |
| rs2860175 | 7132070 | 1.46 (0.82–2.62) | 0.2010 | |
| rs1799817 | 7125286 | 0.83 (0.56–1.23) | 0.3541 | |
| ND | 7125064 | ND | ND | |
| rs78312382 * | 7117415 | 0.55 (0.32–0.96) | 0.0360 | |
| Mukherjee [30] | rs1799817 * | 7125286 | ND | 0.181 in total, 0.004 in lean |
| Unsal [13] | rs1799817 | 7125286 | ND | 0.437 |
| rs2229434 | 7142910 | ND | 1 | |
| rs2229430 | 7142832 | ND | 1 | |
| rs16994210 | 7142988 | ND | >0.05 | |
| rs35045353 | 7142927 | ND | >0.05 | |
| rs2162771 | 7142869 | ND | >0.05 | |
| rs1541806 | 7142844 | ND | >0.05 | |
| rs13306451 | 7142813 | ND | >0.05 | |
| Hanzu [21] | rs2245648 | 7163219 | ND | >0.05 |
| rs2962 | 7163054 | ND | >0.05 | |
| rs2245649 * | 7163203 | ND | 0.0086 | |
| rs2963 * | 7163143 | 2.99 (1.41–6.32) | 0.0025 | |
| rs2245655 * | 7163129 | ND | 0.0048 | |
| Goodarzi [22] | rs12459488 | 7206062 | 1.11 (0.92–1.34) | 0.3 |
| rs2971499 | 7214271 | 1.05 (0.86–1.28) | 0.067 | |
| rs2252673 * | 7150407 | 1.32 (1.08–1.60) | 0.006 | |
| rs10401628 | 7126207 | 1.00 (0.80–1.25) | 0.99 | |
| rs6510949 * | 7134391 | ND | 0.028 | |
| rs919275 | 7261430 | ND | 0.089 | |
| rs2042902 | 7204459 | ND | 0.099 | |
| rs4804404 | 7218371 | ND | 0.101 | |
| rs7254921 | 7278441 | ND | 0.111 | |
| rs7258382 | 7262558 | ND | 0.164 | |
| rs1549616 | 7132559 | ND | 0.166 | |
| rs7248939 | 7268427 | ND | 0.190 | |
| rs10408374 | 7127283 | ND | 0.249 | |
| rs8103483 | 7145363 | ND | 0.300 | |
| rs4804428 | 7235280 | ND | 0.360 | |
| rs3745550 | 7115562 | ND | 0.372 | |
| rs8112883 | 7179309 | ND | 0.404 | |
| rs10426094 | 7205229 | ND | 0.420 | |
| rs8110116 | 7143700 | ND | 0.511 | |
| rs2860172 | 7127364 | ND | 0.521 | |
| rs7245562 | 7218124 | ND | 0.522 | |
| rs890862 | 7233593 | ND | 0.536 | |
| rs10500204 | 7182952 | ND | 0.544 | |
| rs11668751 | 7251831 | ND | 0.586 | |
| rs3786680 | 7183540 | ND | 0.642 | |
| rs11667110 | 7136598 | ND | 0.644 | |
| rs6510960 | 7203721 | ND | 0.647 | |
| rs17254521 | 7238684 | ND | 0.689 | |
| rs4804195 | 7254933 | ND | 0.947 | |
| rs8111710 | 7292572 | ND | 0.962 | |
| rs12979424 | 7273481 | ND | 0.981 | |
| rs10408844 | 7244873 | ND | 0.992 | |
| rs11880337 | 7296441 | ND | ND | |
| rs6510975 | 7266867 | ND | ND | |
| rs2860183 | 7189364 | ND | ND | |
| Chen [15] | rs1864193 | 7114202 | 0.87 | 0.164 |
| rs11667110 | 7136598 | 0.92 | 0.343 | |
| rs16990074 | 7137340 | 1.14 | 0.494 | |
| rs4804304 | 7140514 | 1.18 | 0.387 | |
| rs2229431 | 7141764 | 1.03 | 0.832 | |
| rs2229430 | 7142832 | 1.09 | 0.672 | |
| rs2229434 | 7142910 | 1.05 | 0.814 | |
| rs2962 | 7163054 | 1.27 | 0.35 | |
| rs2059807 * | 7166098 | 1.34 | 3 × 10−4 | |
| rs3815902 * | 7166127 | 1.32 | 0.002 | |
| rs3786681 * | 7168922 | 1.34 | 9 × 10−4 | |
| rs6510956 * | 7169265 | 1.32 | 0.002 | |
| rs16994298 | 7170871 | 1.26 | 0.349 | |
| rs8109559 | 7171618 | 1.24 | 0.159 | |
| rs16994314 | 7176963 | 1.26 | 0.354 | |
| rs8108622 * | 7182742 | 1.5 | 7 × 10−5 | |
| rs10500204 * | 7182952 | 1.47 | 1 × 10−4 | |
| rs3786680 | 7183540 | 1.21 | 0.141 | |
| rs891088 * | 7184751 | 1.38 | 4 × 10−4 | |
| rs17253937 * | 7184790 | 1.43 | 0.001 | |
| rs7245757 | 7187617 | 0.94 | 0.476 | |
| rs1035939 | 7188968 | 0.93 | 0.337 | |
| rs4804368 | 7190279 | 0.96 | 0.602 | |
| rs7254358 | 7199330 | 0.91 | 0.204 | |
| rs1035942 | 7199792 | 0.93 | 0.354 | |
| rs8103883 | 7203411 | 1.24 | 0.075 | |
| rs2042901 | 7204383 | 0.91 | 0.241 | |
| rs10426094 * | 7205229 | 1.34 | 0.033 | |
| rs12459488 | 7206062 | 0.9 | 0.162 | |
| rs3745546 * | 7211805 | 1.25 | 0.036 | |
| rs3745545 | 7211830 | 1.07 | 0.584 | |
| rs7245562 | 7218124 | 0.93 | 0.364 | |
| rs7508679 | 7222821 | 0.99 | 0.879 | |
| rs4804415 | 7223808 | 1.05 | 0.636 | |
| rs4804416 | 7223837 | 0.97 | 0.677 | |
| rs7248104 | 7224420 | 0.97 | 0.643 | |
| rs4804424 | 7229666 | 1.06 | 0.553 | |
| rs10416429 | 7230427 | 1.03 | 0.758 | |
| rs890862 | 7233593 | 1.05 | 0.637 | |
| rs10424224 | 7240470 | 1.03 | 0.77 | |
| rs10404318 | 7247616 | 0.99 | 0.944 | |
| rs919275 | 7261430 | 1.09 | 0.283 | |
| rs8101064 | 7293108 | 1.15 | 0.273 | |
| Cirilo [31] | rs1799817 | 7125286 | ND | >0.05 |
| Ranjzad [32] | rs2059806 | 7166365 | ND | 0.519 |
| rs1799817 | 7125286 | ND | 0.630 | |
| Shi [16] | rs2059807 * | 7166098 | 1.14 | 1.09 × 10−8 |
| Kashima [33] | rs1799817 * | 7125286 | ND | 0.308 in total, 0.037 in lean, 0.644 in obese |
| Skrgatic [34] | rs1799817 | 7125286 | ND | 0.631 |
| Ramezani [35] | rs1799817 | 7125286 | ND | >0.05 |
| rs2059806 | 7166365 | ND | >0.05 | |
| Louwers [24] | rs2059807 | 7166098 | 0.93 | 0.27 |
| Grigorescu [25] | rs2059807 * | 7166098 | 3.1 (1.3–6.8) in non-insulin resistant | >0.05 in total, <0.006 in non-insulin resistant |
| Yin [23] | ND | 7125177 * | ND | 0.043 |
| ND | 7117386 * | ND | 0.055 in total, 0.042 in non-fat | |
| ND | 7117367 * | ND | 0.055 in total, 0.042 in non-fat |
ND: not determined; *: SNPs that distributed differentially between PCOS and control.
As shown in Table 1, three diagnosis criteria of PCOS were used in different studies. NIH criteria include hyperandrogenism and anovulation. The most widely used Rotterdam criteria require PCOS to be diagnosed by at least two of the following three items: oligo-anovulation, hyperandrogensim and PCO. Some studies all three items to be satisfied for the diagnosis of PCO. For rs1799817, five studies adopted Rotterdam criteria, four adopted the criteria including three items, two adopted NIH criteria, and one failed to document the criteria applied. For rs2059806, two studies adopted NIH criteria, one adopted Rotterdam criteria, and one adopted the three-item criteria. For rs2059807, three studies adopted Rotterdam criteria and one failed to document the criteria applied. BMI was not comparable between controls and PCOS cases in the majority of the studies.
2.2. Quality of the Included Studies
As shown in Table 1, the average score of Newcastle-Ottawa quality assessment scale (NOS) was 6.75 (range from 5 to 8). Two items, including the representativeness of the cases and the selection of controls, were the major sources of biases (80%).
2.3. Meta-Analysis Results
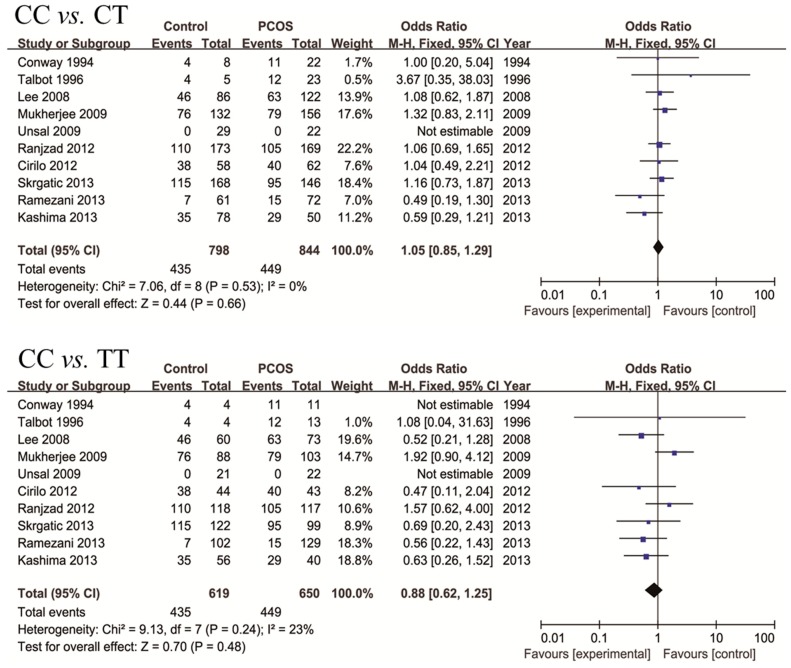
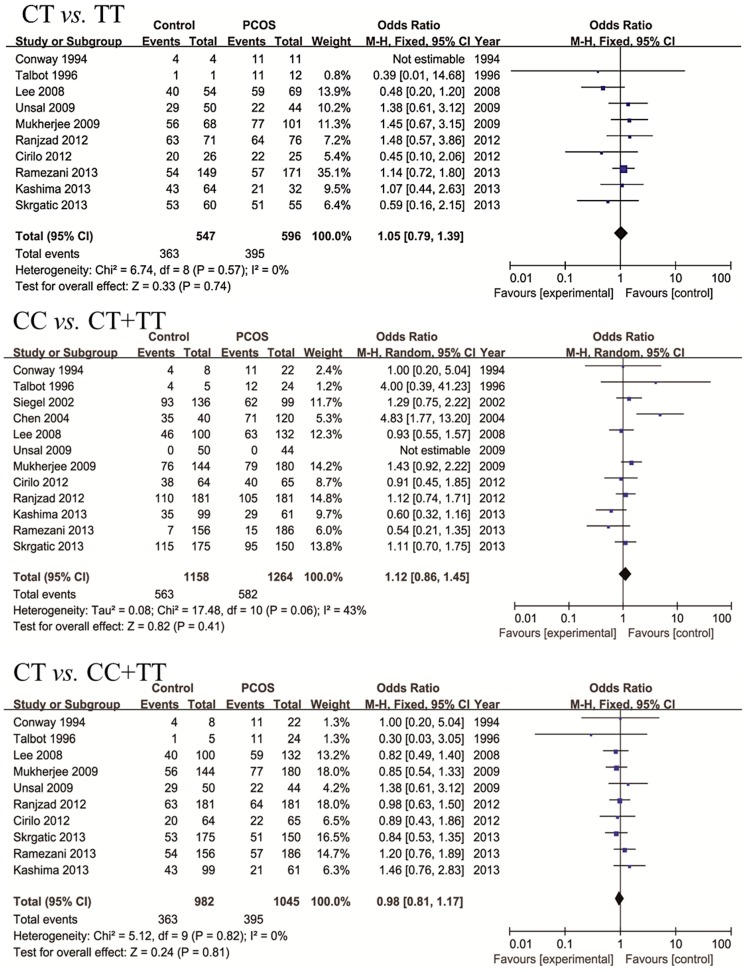
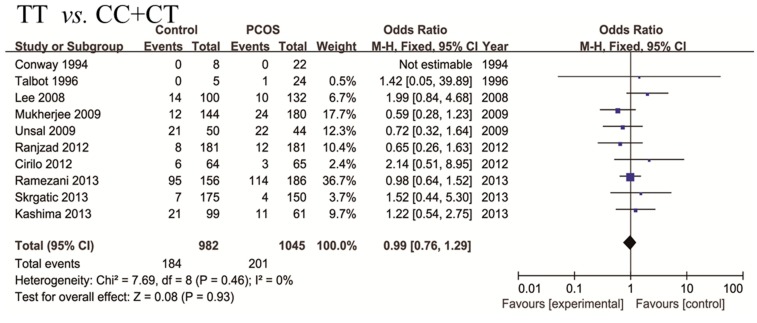
For rs1799817 (Table 3), a total of 12 studies were collected, including 1158 controls and 1264 PCOS cases. Except two studies in which Hardy-Weinberg Equilibrium (HWE) test could not be conducted due to a lack of information and one study that deviated from HWE [13], genotype distribution in the controls was consistent with HWE. In order to explore the potential correlation between genotypes of rs1799817 and PCOS, we compared every genotype (Figure 3). For CC vs. CT, CC vs. TT, CT vs. TT, CT vs. CC + TT, and TT vs. CC + CT, no significant association was found, with mild between-study heterogeneity. For CC vs. CT + TT, a random effect model was selected due to severe between-study heterogeneity (p = 0.06), and no significant association was found (p = 0.41).
Table 3.
Distribution of INSR rs1799817 genotype and the allele frequencies for controls and PCOS cases.
| Study | Year | Country | Age | BMI | Control | PCOS Cases | p-Value of HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | PCOS | Control | PCOS | Genotypes | Alleles | Cases | Genotypes | Alleles | Cases | ||||
| CC/CT/TT | C/T | CC/CT/TT | C/T | ||||||||||
| Conway [26] | 1994 | UK | ND | ND | 23.8 (21.0–24.7) | 32.1 (21.3–46.0) | 4/4/0 | 12/4 | 8 | 11/11/0 | 33/11 | 22 | 0.641 |
| Talbot [27] | 1996 | UK | 28 (25–33) | 24 (18–33) | 37.2 (30.5–45.8) | 36.6 (27.4–47) | 4/1/0 | 9/1 | 5 | 12/11/1 | 35/13 | 24 | 0.970 |
| Siegel [12] | 2002 | USA | age matched | 28 (16–52) | ND | ND | 93/43 | ND | 136 | 62/37 | ND | 99 | ND |
| Chen [28] | 2004 | China | 30 ± 4 | 29 ± 3 | 23 ± 3 | 25 ± 4 | 35/5 | ND | 40 | 71/49 | ND | 120 | ND |
| Lee [29] | 2008 | Korea | NDs | ND | 20.73 ± 2.36 | 23.22 ± 3.88 | 46/40/14 | 132/68 | 100 | 63/59/10 | 185/79 | 132 | 0.554 |
| Mukherjee [30] | 2009 | India | 24.94 ± 5.46 | 24.82 ± 5.26 | 22.16 ± 4.11 | 25.01 ± 5.63 | 76/56/12 | 208/80 | 144 | 79/77/24 | 235/125 | 180 | 0.934 |
| Unsal [13] | 2009 | Turkey | 14.0 ± 3.3 | 14.5 ± 1.3 | 20.7 ± 4.2 | 25.0 ± 5.5 | 0/29/21 | 29/71 | 50 | 0/22/22 | 22/66 | 44 | 0.015 * |
| Cirilo [31] | 2012 | Brazil | 32.53 ± 6.91 | 26.09 ± 6.90 | 25.61 ± 5.48 | 31.24 ± 7.15 | 38/20/6 | 96/32 | 64 | 40/22/3 | 102/28 | 65 | 0.411 |
| Ranjzad [32] | 2012 | Iran | 31.07 ± 5.84 | 27.13 ± 5.29 | 25.48 ± 4.21 | 26.80 ± 6.37 | 110/63/8 | 283/79 | 181 | 105/64/12 | 274/88 | 181 | 0.964 |
| Kashima [33] | 2013 | Japan | 34.6 ± 5.3 | 29.5 ± 3.7 | 20.8 ± 2.6 | 23.5 ± 5.2 | 35/43/21 | 113/85 | 99 | 29/21/11 | 79/43 | 61 | 0.528 |
| Skrgatic [34] | 2013 | Croatia | 29.1 ± 4.7 | 26.7 ± 5.9 | 22.4 ± 3.3 | 23.4 ± 4.2 | 115/53/7 | 283/67 | 175 | 95/51/4 | 241/59 | 150 | 0.960 |
| Ramezani [35] | 2013 | Iran | 30.8 ± 5.6 | 26.6 ± 5.6 | 25.5 ± 4.4 | 26.8 ± 6.4 | 7/54/95 | 68/244 | 156 | 15/57/114 | 87/285 | 186 | 0.982 |
* p < 0.05; ND: not determine; BMI: body mass index.
Figure 3.
Forest plots of meta-analyses on association between rs1799817 frequency and PCOS risk.
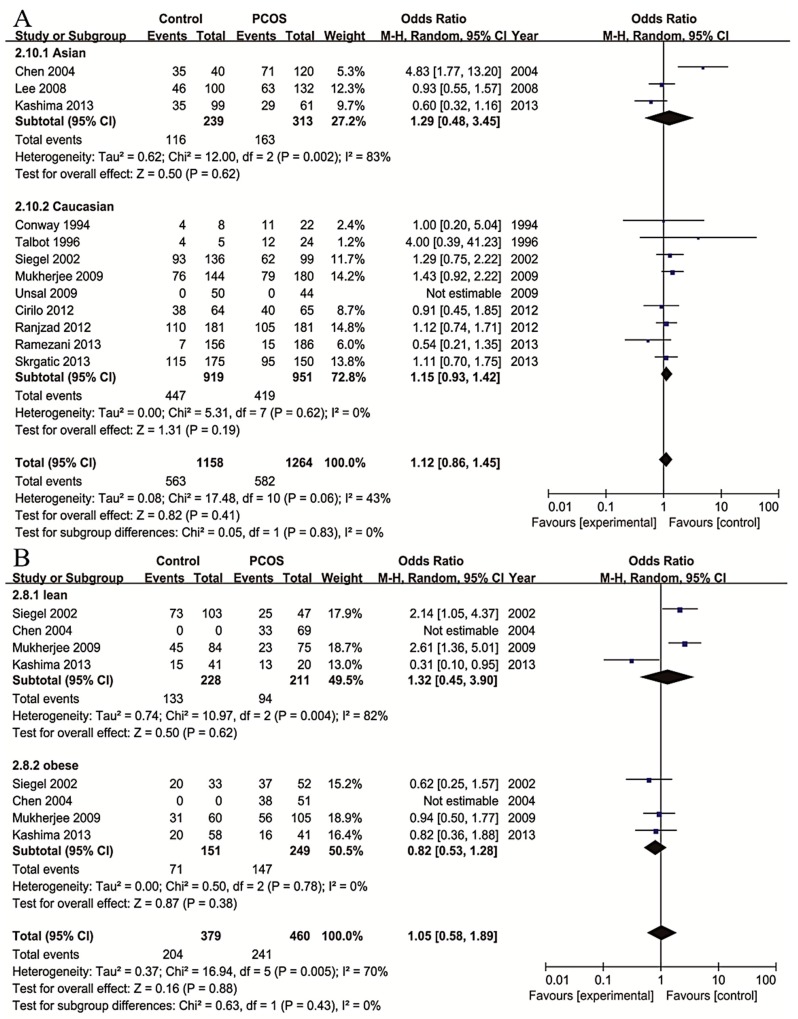
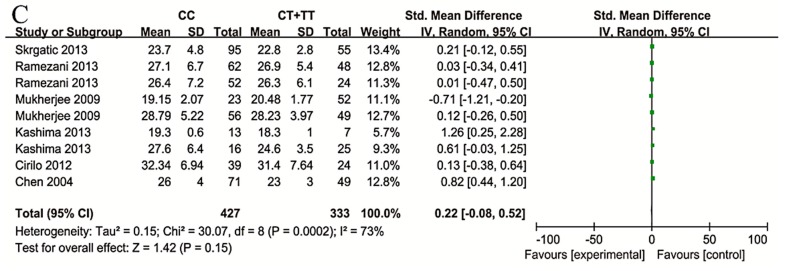
To explore the origin of between-study heterogeneity, subgroup analyses with stratification by ethnicity and weight were carried out (Figure 4). The heterogeneity in the studies from Caucasian population was mild (p = 0.62) while the heterogeneity from Asian population was still severe (p = 0.002). Both group indicated no significant association (p = 0.19 and p = 0.62, respectively). Subgroup analyses stratified by weight showed that the heterogeneity in obese population was mild (p = 0.78) while in lean population was still severe (p = 0.004). The random effect model was selected, and no significant correlation was found (p = 0.38 and p = 0.62, respectively). Only three studies carried sufficient data to be analyzed in the weight subgroups, and the data was limited to CC vs. CT + TT comparison. No correlation was found between rs1799817 and PCOS for CC vs. CT + TT, in both lean and obese subgroups. Further analyses indicated no significant difference of BMI between patients with different genotypes (p = 0.15). In addition, considering the divergence of diagnostic criteria, we performed the subgroup analysis according to the three diagnostic criteria. Consistent results were obtained from the analyses on groups diagnosed by the same criteria.
Figure 4.
Forest plots of association between genotype frequencies of SNP rs1799817 and PCOS risk. (A) Stratified by ethnicity; (B) stratified by BMI; (C) pooled standardized mean differences of BMI in PCOS patients with different genotypes.
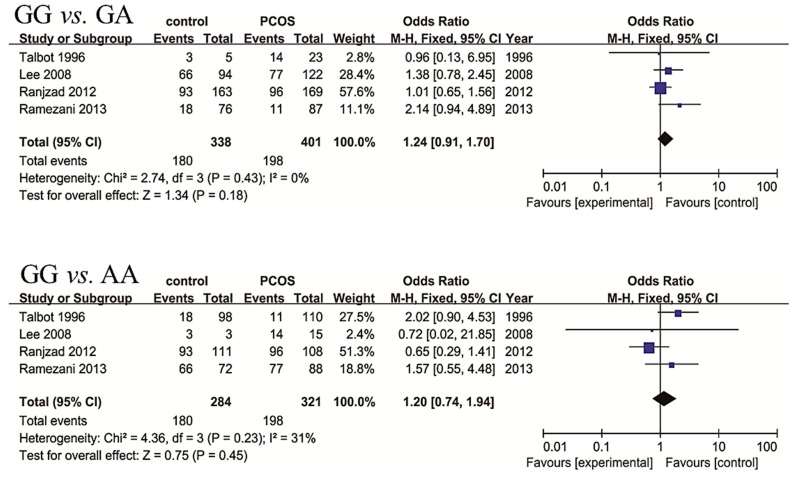
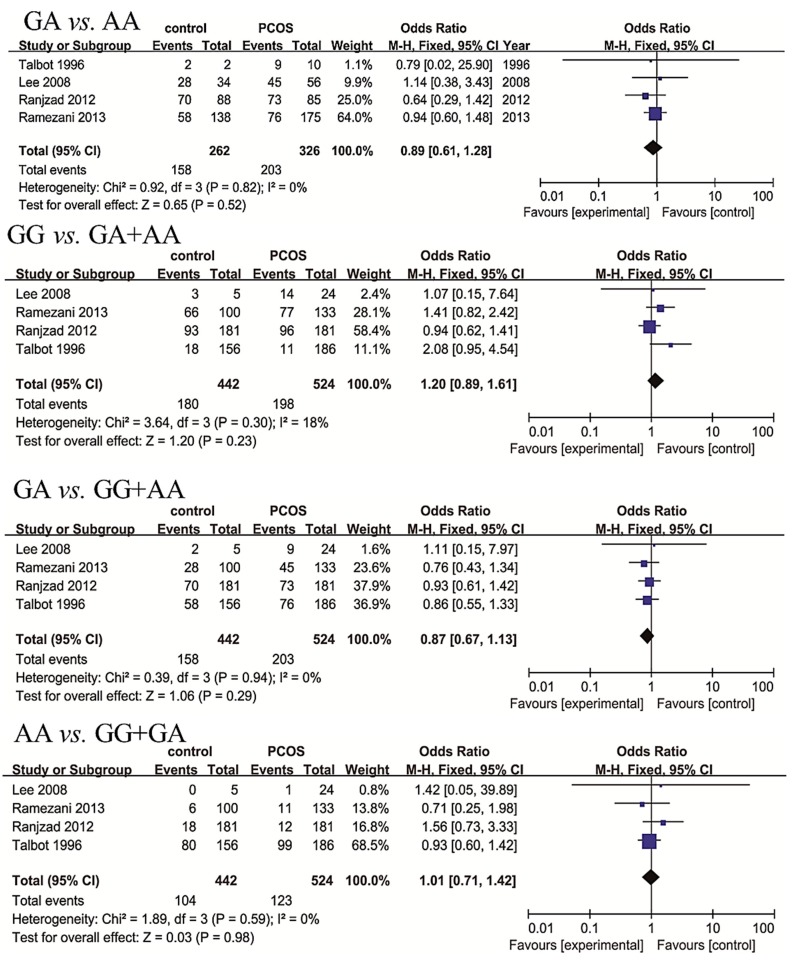
For rs2059806, four studies including 442 controls and 524 PCOS cases were selected for analyses, (Table 4). The between-study heterogeneity was mild and fixed effect model was applied. No significant association between genotype frequencies of rs2059806 and PCOS was found, including GG vs. GA, GG vs. AA, GA vs. AA, GG vs. GA + AA, GA vs. GG + AA, and AA vs. GG + GA (Figure 5).
Table 4.
Distribution of the INSR rs2059806 genotype and the allele frequencies for controls and PCOS cases.
| Study | Year | Country | Age | BMI | Control | PCOS | p-Value of HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | PCOS | Control | PCOS | Genotypes | Alleles | Cases | Genotypes | Alleles | Cases | ||||
| GG/GA/AA | G/A | GG/GA/AA | G/A | ||||||||||
| Talbot [27] | 1996 | UK | 28 (25–33) | 24 (18–33) | 37.2 (30.5–45.8) | 36.6 (27.4–47) | 3/2/0 | 8/2 | 5 | 14/9/1 | 37/11 | 24 | 0.855 |
| Lee [29] | 2008 | Korean | ND | ND | 20.73 ± 2.36 | 23.22 ± 3.88 | 66/28/6 | 160/40 | 100 | 77/45/11 | 199/67 | 133 | 0.458 |
| Ranjzad [32] | 2012 | Iran | 31.07 ± 5.84 | 27.13 ± 5.29 | 25.48 ± 4.21 | 26.80 ± 6.37 | 93/70/18 | 256/106 | 181 | 96/73/12 | 265/97 | 181 | 0.673 |
| Ramezani [35] | 2013 | Iran | 30.8 ± 5.6 | 26.6 ± 5.6 | 25.5 ± 4.4 | 26.8 ± 6.4 | 18/58/80 | 94/218 | 156 | 11/76/99 | 98/274 | 186 | 0.344 |
ND: not determined.
Figure 5.
Forest plots of association between genotype frequencies of SNP rs2059806 and PCOS risk.
Four studies covering 12,830 controls and 11,683 PCOS cases investigated the correlation between rs2059807 genotypes and PCOS. OR of allele C vs. allele T was calculated. As shown in Table 5, three studies detected significant correlations while one study found no association. All of these studies were performed in large population, and one of them investigated three cohorts [16]. However, pooled OR could not be calculated because the original information provided was insufficient.
Table 5.
Cohort characteristics and odds ratios (ORs) of the INSR rs2059807 (C/T).
| Study | Year | Country | Title of Cohort | Cases | Age | BMI | OR (95% CI) | p Value | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | PCOS | Control | PCOS | Control | PCOS | ||||||
| Chen [15] | 2011 | China | 895 | 744 | 30.68 ± 4.68 | 28.85 ± 3.62 | 22.68 ± 3.23 | 24.55 ± 3.99 | 1.34 (1.16–1.56) | 1.16 × 10−4 | |
| Shi [16] | 2012 | China | GWAS II | 2016 | 1510 | 53.95 ± 7.18 | 28.12 ± 2.75 | 24.93 ± 3.85 | 24.59 ± 3.17 | 1.19 (1.07–1.33) | 1.66 × 10−3 |
| REP I | 1913 | 1908 | 31.29 ± 5.00 | 28.01 ± 3.01 | 22.49 ± 2.17 | 24.23 ± 3.19 | 1.16 (1.05–1.28) | 4.40 × 10−3 | |||
| REP II | 5665 | 6318 | 29.52 ± 4.75 | 27.83 ± 3.11 | 21.95 ± 2.32 | 23.72 ± 3.18 | 1.09 (1.02–1.15) | 6.61 × 10−3 | |||
| Louwers [24] | 2013 | Dutch | 2164 | 703 | ND | 29.1 ± 5.73 | ND | 24.8 ± 5.6 | 0.93 | 0.27 | |
| Grigorescu [25] | 2013 | Romania | 177 | 500 | ND | ND | ND | 27.8 ± 0.35 | 3.1 (1.3-6.8) | <0.006 | |
ND: not determined.
2.4. Sensitivity Analysis
Sensitivity analysis was performed by removing one study at one time. For rs1799817, removing any study in the analysis of CC vs. CT, CC vs. TT, CT vs. TT, CC vs. CT + TT, CT vs. CC + TT, and TT vs. CC + CT did not impact the overall results. For rs2059806, when a study from Iran was removed [32], a significant association emerged in the analysis of GG vs. GA + AA, with OR 1.57 (1.02–2.42) and p = 0.04. No significant association was found from comparison of other genotypes, including GG vs. GA, GG vs. AA, GA vs. AA, GA vs. GG + AA, and AA vs. GG + GA, in the sensitivity analyses.
2.5. Publication Bias
Funnel plots were applied to evaluate the potential publication bias. Visual inspection of the funnel plots indicated no significant asymmetry in analyses for both rs1799817 (Table S1) and rs2059806 (Table S2). This result indicated the absence of severe publication bias.
3. Discussion
PCOS is associated with diversified genetic and environmental factors. Multiple candidate genes have been reported to increase the risk PCOS, but often the claims could not be confirmed in validation trials. In the past twenty years, a great number of studies were performed, and a dozen SNPs in different regions of INSR gene have been implicated in PCOS, but the results were divergent. For example, for rs2252673, a study by Lee et al. [29] indicated no association (p = 0.5688) while the study by Goodarzi et al. [22] reported a significant association (p = 0.006). One reason for the discrepancy could be the limited sample sizes and random errors. In this situation, a meta-analysis pooling together all the available data could effectively reduce the bias and achieve a more reliable conclusion.
Among the INSR polymorphisms related to PCOS, rs1799817 SNP is the most thoroughly investigated. INSR contains α and β subunits that are encoded by exons 1–11 and exons 12–22, respectively [36]. Exons 17–22 encode the tyrosine kinase domain, and mutations in this region can cause severe insulin resistance and hyperinsulinemia [37]. SNP rs1799817, located in exon 17, was considered to be involved in insulin resistance and PCOS. In the present study, however, we found no significant correlation between the genotypes of rs1799817 and PCOS. It is noteworthy that Ioannidis et al. [14] have performed a meta-analysis on rs1799817 and PCOS in 2010 and same result as current one were obtained in that study. A consistent negative result from previous and current studies support that this genetic variation is unlikely involved in the development of PCOS.
Considering that ethnic group possess diversified genetic and environmental backgrounds, and gene-disease associations may vary across these groups [38], we carried out a further subgroup analysis stratified by ethnicity. No significant difference was found in either Asian or Caucasian populations, indicating that the between-study heterogeneity was not mainly originated from population diversity. Again regarding stratification, previous studies indicated that significant association of genotypes with PCOS was not detected in total population, but observed in lean population. Therefore, we applied a subgroup analysis stratified by body mass index (BMI). Our analyses showed no significant correlation in the lean population. Furthermore, no significant difference was detected in the BMI between CC and CT + TT genotypes in PCOS patients. All together, the results suggested that in both lean and obese groups SNP rs1799817 may contribute little or no to the development of PCOS.
SNP rs2059806 located in the exon 8 of INSR gene was investigated in three Caucasian cohorts and one Asian cohort. In the present study, we pooled the results from previous studies and found no evidence that rs2039806 represents a predisposition of PCOS. The sensitivity analysis for GG vs. GA + AA showed that, when a study from Iran was removed, an association with PCOS arose with a marginal p value. It should be pointed out that the positive results are only based on three studies. Further investigation is needed to clarify the impact of this specific genotype on the risk of PCOS.
While many significant SNPs distribute in different exons and flanking regions of INSR gene, we noticed that five SNPs involved in the susceptibility of PCOS concentrate in exon 9 and 5' intron of exon 9, and rs2059807 is one of these SNP located in the “hot spot”. A haplotype constructed with four SNPs in this region was demonstrated to be associated with PCOS [21], indicating that this might be a PCOS susceptibility loci. Unfortunately, no pooled OR could be calculated due to the failure to obtain original data from the groups conducted the studies. Nevertheless, rs2059807 should be considered a candidate risk factor for genetic predisposition of PCOS.
This study has several limitations. First, as mentioned above, since we failed to connect with some authors to collect the original data, the power of the subgroup analysis of BMI was compromised, and the pooled OR of rs2059807 could not be calculated; Second, the ethnicity of some countries could not be clearly defined and we had to classify them as the majority. Similarly, cutoff values for lean and obese were somewhat different among different studies, and we accepted the various definitions by individual studies; Third, original studies used various control groups, including healthy women, infertile women, and elderly women, and various diagnostic criteria of PCOS, making it difficult to control the confounding factors.
In conclusion, our meta-analyses summarized the available data concerning the role of INSR polymorphisms for genetic predisposition of PCOS. Our results suggested the absence of significant correlation between rs1799817 or rs2059806 SNPs with the development of PCOS, even if ethnicity and BMI were taken into account. However, rs2059807 could be a promising candidate SNP that might be involved in the development of PCOS. Further investigation is required to clarify the role of rs2029807. Since metabolism abnormality is a major contributing factor for the pathogenesis of PCOS, subgrouped analyses according to BMI and/or insulin resistance status may much enhance the study power.
4. Experimental Section
This meta-analysis was developed according to the PRISM statement. A protocol was registered in PROSPERO, with registration number: CRD42014013145.
4.1. Literature Retrieval
Two authors independently searched the relevant studies published in all languages via 3 databases, including Pubmed, Embase, and Cochrane CENTRAL no later than 7 August 2014. The discrepancies were resolved by discussion. The following keywords were used: (PCOS OR “polycystic ovary syndrome” OR “polycystic ovarian syndrome”) AND (INSR OR CD220 OR HHF5 OR “insulin receptor”) AND (genotype OR “genetic predisposition” OR SNP OR polymorphism* OR variant* OR “genetic susceptibility” OR genetics OR allele). Additionally, we hand-searched the cited references to obtain more relevant studies.
Studies were included in this analysis if: (1) they evaluated the association between INSR SNPs and the risk of PCOS; (2) they were case-control or cohort studies; (3) sufficient data were provided to calculate odds ratio (OR) and 95% confidence interval (CI). Studies were excluded if: (1) they were family-association or case series studies; (2) the studies were from the same research group and the studied populations had overlaps. In this case, only the study with largest sample size was included for analysis and the others were excluded.
4.2. Data Extraction
The following data was extracted from every study by two reviewers independently, and the discrepancies were resolved by discussion: (1) name of the first author; (2) year of publication; (3) country; (4) ethnicity; (5) age; (6) BMI; (7) diagnosis criteria; (8) methods of genotype; (9) SNP detected; (10) OR; (11) p values; and (12) numbers of genotypes in cases and controls. We contacted investigators for additional information when extra information was required. When the SNPs were reported with different nomenclatures, we mapped them on the chromosome and unified them into reference SNP ID numbers.
4.3. Assessment of Study Quality
The NOS of case control studies was applied to evaluate the quality of studies. Each study was evaluated independently by two authors [39], and they discussed to resolve the disagreement. Three aspects including the selection of population, the comparability of two groups, and the exposure were assessed for eight items, with the highest score of nine.
4.4. Statistical Analysis
HWE was tested by the chi-square method. Review Manager 5.2 (Cochrane Collaboration, Oxford, UK) was used for the meta-analysis. The associations of INSR SNPs with PCOS were assessed by calculating the ORs and mean difference among the pooled data, and the statistical significance was calculated with Z test. Q test was employed to assess the between-study heterogeneity. A p value higher than 0.1 was considered to be of no serious heterogeneity, and a fixed-effect model (FEM) was subsequently applied to calculate the parameters of the data pool [40]. If the heterogeneity is serious, calculation was carried out based on a random-effect model (REM). To define where the severe heterogeneity come from, subgroup analyses on ethnicity and BMI were executed. Additionally, a sensitivity analysis was performed to verify the stability through removing one individual study a time. Publication bias was evaluated by examining the asymmetry of funnel plot. p < 0.05 was considered to be statistically significant.
Acknowledgments
The authors are grateful to Dinka Pavicic Baldani (Zagreb, Croatia), Mark O. Goodarzi (Los Angeles, CA, USA), Silvia Regina Rogatto (Sao Paulo, Brazil), and Yaron Tomer (New York, NY, USA) for their contribution in providing valuable information. This study was supported by the following research grants: National Basic Research Program of China (No. 2012CB944900); National Science Foundation of China (No. 81200446, No. 31171444 and 30973209); The National Science and Technology Support Program (No. 2012BAI32B01); National Science Foundation of Zhejiang Province (No. LY12H04010); The Georgia Research Alliance Scholarship; and research supports from Mercer University School of Medicine.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/16/02/2403/s1.
Author Contributions
Shi-Wen Jiang and Chun Feng designed the study and wrote the paper; Ping-Ping Lv and Tian-Tian Yu performed the experiments; Xue Wang, Min Jin, Jin-Ming Shen and Feng Zhou analyzed the data.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.The Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil. Steril. 2004;81:19–25. doi: 10.1016/j.fertnstert.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Moran L.J., Misso M.L., Wild R.A., Norman R.J. Impaired glucose tolerance, type 2 diabetes and metabolic syndrome in polycystic ovary syndrome: A systematic review and meta-analysis. Hum. Reprod. Update. 2010;16:347–363. doi: 10.1093/humupd/dmq001. [DOI] [PubMed] [Google Scholar]
- 3.March W.A., Moore V.M., Willson K.J., Phillips D.I., Norman R.J., Davies M.J. The prevalence of polycystic ovary syndrome in a community sample assessed under contrasting diagnostic criteria. Hum. Reprod. 2010;25:544–551. doi: 10.1093/humrep/dep399. [DOI] [PubMed] [Google Scholar]
- 4.Baptiste C.G., Battista M.C., Trottier A., Baillargeon J.P. Insulin and hyperandrogenism in women with polycystic ovary syndrome. J. Steroid Biochem. Mol. Biol. 2010;122:42–52. doi: 10.1016/j.jsbmb.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gustin S., Lee M., Westphal L. Ethnic Differences in Fertility and Assisted Reproduction. Springer; New York, NY, USA: 2013. Differences in fertility and assisted reproduction in South Asian women; pp. 105–113. [Google Scholar]
- 6.Vink J.M., Sadrzadeh S., Lambalk C.B., Boomsma D.I. Heritability of polycystic ovary syndrome in a Dutch twin-family study. J. Clin. Endocrinol. MeTable. 2006;91:2100–2104. doi: 10.1210/jc.2005-1494. [DOI] [PubMed] [Google Scholar]
- 7.Diamanti-Kandarakis E., Papavassiliou A.G. Molecular mechanisms of insulin resistance in polycystic ovary syndrome. Trends Mol. Med. 2006;12:324–332. doi: 10.1016/j.molmed.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 8.Ben-Shlomo I., Younis J.S. Basic research in PCOS: Are we reaching new frontiers. Reprod. Biomed. Online. 2014;28:669–683. doi: 10.1016/j.rbmo.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 9.Goodarzi M.O., Jones M.R., Li X., Chua A.K., Garcia O.A., Chen Y.D., Krauss R.M., Rotter J.I., Ankener W., Legro R.S., et al. Replication of association of DENND1A and THADA variants with polycystic ovary syndrome in European cohorts. J. Med. Genet. 2012;49:90–95. doi: 10.1136/jmedgenet-2011-100427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Accili D., Drago J., Lee E.J., Johnson M.D., Cool M.H., Salvatore P., Asico L.D., Jose P.A., Taylor S.I., Westphal H. Early neonatal death in mice homozygous for a null allele of the insulin receptor gene. Nat. Genet. 1996;12:106–109. doi: 10.1038/ng0196-106. [DOI] [PubMed] [Google Scholar]
- 11.Chu Y.L., Gong Y.D., Su Z.H., Yu H.N., Cui Q., Jiang H.Y., Qu H.M. Relationship between tyrosine phosphorylation and protein expression of insulin receptor and insulin resistance in gestational diabetes mellitus. J. Huazhong Univ. Sci. Technol. Med. Sci. 2014;34:393–397. doi: 10.1007/s11596-014-1289-x. [DOI] [PubMed] [Google Scholar]
- 12.Siegel S., Futterweit W., Davies T.F., Concepcion E.S., Greenberg D.A., Villanueva R., Tomer Y. A C/T single nucleotide polymorphism at the tyrosine kinase domain of the insulin receptor gene is associated with polycystic ovary syndrome. Fertil. Steril. 2002;78:1240–1243. doi: 10.1016/S0015-0282(02)04241-3. [DOI] [PubMed] [Google Scholar]
- 13.Unsal T., Konac E., Yesilkaya E., Yilmaz A., Bideci A., Ilke O.H., Cinaz P., Menevse A. Genetic polymorphisms of FSHR, CYP17, CYP1A1, CAPN10, INSR, SERPINE1 genes in adolescent girls with polycystic ovary syndrome. J. Assist. Reprod. Genet. 2009;26:205–216. doi: 10.1007/s10815-009-9308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ioannidis A., Ikonomi E., Dimou N.L., Douma L., Bagos P.G. Polymorphisms of the insulin receptor and the insulin receptor substrates genes in polycystic ovary syndrome: A Mendelian randomization meta-analysis. Mol. Genet. Metable. 2010;99:174–183. doi: 10.1016/j.ymgme.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 15.Chen Z.J., Zhao H., He L., Shi Y., Qin Y., Shi Y., Li Z., You L., Zhao J., Liu J., et al. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat. Genet. 2011;43:55–59. doi: 10.1038/ng.732. [DOI] [PubMed] [Google Scholar]
- 16.Shi Y., Zhao H., Shi Y., Cao Y., Yang D., Li Z., Zhang B., Liang X., Li T., Chen J., et al. Genome-wide association study identifies eight new risk loci for polycystic ovary syndrome. Nat. Genet. 2012;44:1020–1025. doi: 10.1038/ng.2384. [DOI] [PubMed] [Google Scholar]
- 17.Jin L. Polymorphism in insulin receptor gene exon 17 in women with polycystic ovary syndrome. Zhonghua Fu Chan Ke Za Zhi. 2005;40:323–326. (in Chinese) [PubMed] [Google Scholar]
- 18.Lee E.J., Yoo K.J., Kim S.J., Lee S.H., Cha K.Y., Baek K.H. Single nucleotide polymorphism in exon 17 of the insulin receptor gene is not associated with polycystic ovary syndrome in a Korean population. Fertil. Steril. 2006;86:380–384. doi: 10.1016/j.fertnstert.2005.12.073. [DOI] [PubMed] [Google Scholar]
- 19.Louwers Y.V. Insulin receptor gene polymorphisms are associated with polycystic ovary syndrome (PCOS) Hum. Reprod. 2010;25 doi: 10.1093/humrep/de.25.s1.438. [DOI] [Google Scholar]
- 20.Jin L., Zhu X.M., Luo Q., Qian Y., Jin F., Huang H.F. A novel SNP at exon 17 of INSR is associated with decreased insulin sensitivity in Chinese women with PCOS. Mol. Hum. Reprod. 2006;12:151–155. doi: 10.1093/molehr/gal022. [DOI] [PubMed] [Google Scholar]
- 21.Hanzu F.A., Radian S., Attaoua R., Ait-El-Mkadem S., Fica S., Gheorghiu M., Coculescu M., Grigorescu F. Association of insulin receptor genetic variants with polycystic ovary syndrome in a population of women from Central Europe. Fertil. Steril. 2010;94:2389–2392. doi: 10.1016/j.fertnstert.2010.04.029. [DOI] [PubMed] [Google Scholar]
- 22.Goodarzi M.O., Louwers Y.V., Taylor K.D., Jones M.R., Cui J., Kwon S., Chen Y.D., Guo X., Stolk L., Uitterlinden A.G., et al. Replication of association of a novel insulin receptor gene polymorphism with polycystic ovary syndrome. Fertil. Steril. 2011;95:1736–1741. doi: 10.1016/j.fertnstert.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin C. Natural Cycle IVF and Mild Stimulation. J. Assist. Reprod. Genet. 2013;30:172–173. [Google Scholar]
- 24.Louwers Y.V., Stolk L., Uitterlinden A.G., Laven J.S. Cross-ethnic meta-analysis of genetic variants for polycystic ovary syndrome. J. Clin. Endocrinol. Metable. 2013;98:E2006–E2012. doi: 10.1210/jc.2013-2495. [DOI] [PubMed] [Google Scholar]
- 25.Grigorescu F. Sub-classification of polycystic ovary syndrome by rotterdam criteria reveals stronger association of the insulin receptor (INSR) and FTO genes in central European population. Diabetes. 2013;62:A716–A717. [Google Scholar]
- 26.Conway G.S., Avey C., Rumsby G. The tyrosine kinase domain of the insulin receptor gene is normal in women with hyperinsulinaemia and polycystic ovary syndrome. Hum. Reprod. 1994;9:1681–1683. doi: 10.1093/oxfordjournals.humrep.a138773. [DOI] [PubMed] [Google Scholar]
- 27.Talbot J.A., Bicknell E.J., Rajkhowa M., Krook A., OʼRahilly S., Clayton R.N. Molecular scanning of the insulin receptor gene in women with polycystic ovarian syndrome. J. Clin. Endocrinol. MeTable. 1996;81:1979–1983. doi: 10.1210/jcem.81.5.8626868. [DOI] [PubMed] [Google Scholar]
- 28.Chen Z.J., Shi Y.H., Zhao Y.R., Li Y., Tang R., Zhao L.X., Chang Z.H. Correlation between single nucleotide polymorphism of insulin receptor gene with polycystic ovary syndrome. Zhonghua Fu Chan Ke Za Zhi. 2004;39:582–585. [PubMed] [Google Scholar]
- 29.Lee E.J., Oh B., Lee J.Y., Kimm K., Lee S.H., Baek K.H. A novel single nucleotide polymorphism of INSR gene for polycystic ovary syndrome. Fertil. Steril. 2008;89:1213–1220. doi: 10.1016/j.fertnstert.2007.05.026. [DOI] [PubMed] [Google Scholar]
- 30.Mukherjee S., Shaikh N., Khavale S., Shinde G., Meherji P., Shah N., Maitra A. Genetic variation in exon 17 of INSR is associated with insulin resistance and hyperandrogenemia among lean Indian women with polycystic ovary syndrome. Eur. J. Endocrinol. 2009;160:855–862. doi: 10.1530/EJE-08-0932. [DOI] [PubMed] [Google Scholar]
- 31.Ramos C.P.D., Rosa F.E., Moreira F.M.F., Rainho C.A., Pontes A., Rogatto S.R. Genetic polymorphisms associated with steroids metabolism and insulin action in polycystic ovary syndrome. Gynecol. Endocrinol. 2012;28:190–194. doi: 10.3109/09513590.2011.593661. [DOI] [PubMed] [Google Scholar]
- 32.Ranjzad F., Mahmoudi T., Irani S.A., Mahban A., Nikzamir A., Vahedi M., Ashrafi M., Gourabi H. A common variant in the adiponectin gene and polycystic ovary syndrome risk. Mol. Biol. Rep. 2012;39:2313–2319. doi: 10.1007/s11033-011-0981-1. [DOI] [PubMed] [Google Scholar]
- 33.Kashima K., Yahata T., Fujita K., Tanaka K. Polycystic ovary syndrome: Association of a C/T single nucleotide polymorphism at tyrosine kinase domain of insulin receptor gene with pathogenesis among lean Japanese women. J. Reprod. Med. 2013;58:491–496. [PubMed] [Google Scholar]
- 34.Skrgatic L., Baldani D.P., Gersak K., Cerne J.Z., Ferk P., Coric M. Genetic polymorphisms of INS, INSR and IRS-1 genes are not associated with polycystic ovary syndrome in Croatian women. Coll. Antropol. 2013;37:141–146. [PubMed] [Google Scholar]
- 35.Ramezani T.F., Daneshpour M., Hashemi S., Zarkesh M., Azizi F. Relationship between polymorphism of insulin receptor gene, and adiponectin gene with PCOS. Iran. J. Reprod. Med. 2013;11:185–194. [PMC free article] [PubMed] [Google Scholar]
- 36.Seino S., Seino M., Nishi S., Bell G.I. Structure of the human insulin receptor gene and characterization of its promoter. Proc. Natl. Acad. Sci. USA. 1989;86:114–118. doi: 10.1073/pnas.86.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moller D.E., Yokota A., White M.F., Pazianos A.G., Flier J.S. A naturally occurring mutation sof insulin receptor alanine 1134 impairs tyrosine kinase function and is associated with dominantly inherited insulin resistance. J. Biol. Chem. 1990;265:14979–14985. [PubMed] [Google Scholar]
- 38.Ioannidis J.P., Ntzani E.E., Trikalinos T.A. “Racial” differences in genetic effects for complex diseases. Nat. Genet. 2004;36:1312–1318. doi: 10.1038/ng1474. [DOI] [PubMed] [Google Scholar]
- 39.Wells G.A., Shea B., OʼConnell D., Peterson J., Welch V., Losos M., Tugwell P. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta analyses. [(accessed on 1 July 2014)]. Available online: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp.
- 40.Lau J., Ioannidis J.P., Schmid C.H. Quantitative synthesis in systematic reviews. Ann. Intern. Med. 1997;127:820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]