Abstract
Silent information regulator 2 (SIRT2) is a member of the sirtuin family of class III NAD (nicotinamide adenine dinucleotide)-dependent protein deacetylases and may regulate senescence, metabolism and apoptosis. The aims of this study were to investigate whether the SIRT2 gene could be used as a candidate gene in the breeding of Qinchuan cattle. Real-time polymerase chain reaction (RT-PCR) results showed that among all types of tissue that were analyzed, the highest mRNA expression levels of the gene were found in subcutaneous fat. DNA sequencing of 468 individual Qinchuan cattle identified two novel, single nucleotide polymorphisms (g.19501 C > T and g.19518 C > T) in the 3' untranslated region (3'UTR) of the SIRT2 gene. The frequencies of SNP g.19501 C > T and g.19518 C > T were in Hardy-Weinberg disequilibrium in all the samples (chi-square test, χ2 < χ0.052). An association analysis showed that the two loci were significantly correlated with some body size traits and the H2H2 (-CT-CT-) diplotypes performed better than other combinations. These results indicated that the variations in the SIRT2 gene and their corresponding genotypes may be considered as molecular markers for economic traits in cattle breeding.
Keywords: SIRT2 gene, expression study, body size traits, Qinchuan cattle
1. Introduction
In mammals, the yeast silent information regulator factor 2 (SIR2) are called sirtuins; they belong to the family of class III nicotinamide adenine dinucleotide (NAD)-dependent histone deacetylase [1]. Seven members of the sirtuin family have been identified to date, and designated as SIRT1-SIRT7. They share a conserved central deacetylase domain but have different N- and C-termini. These proteins play complex and important roles in aging-related pathological conditions, such as cancer and the deregulation of metabolism [2]. Among them, SIRT2 is a cytoplasmic protein that may be either beneficial or detrimental for cell survival, depending on the conditions [3]. Studies of the central nervous system of mice have suggested that the SIRT2 gene is ubiquitously expressed as NAD+-dependent protein deacetylases [4] and it appears to be involved in mediating genomic silencing, DNA repair, and longevity [5,6].
SIRT2 is a NAD+-dependent protein deacetylase of several substrate proteins in the cytoplasm. Particularly, SIRT2 has been implicated in adipocyte differentiation through modulation of acetylation/phosphorylation of forkhead box O1 (FOXO1) [7]. Overexpression of SIRT2 in 3T3-L1 cells causes FoxO1 binding to the promoter of peroxisome proliferator-activated receptor-gamma (PPARγ) and subsequently represses its activity, which leads to abnormal mitochondrial morphology and inhibits adipogenesis [8]. Transcriptional repression of SIRT2 results in an inhibition of fatty acid oxidation (FAO) and energetic uncoupling via deacetylate hypoxia-inducible factor 1α (HIF1α) in vivo and in vitro [9]. Cell-based studies have suggested that SIRT2 deacetylates and stabilizes phosphoenolpyruvate carboxykinase1 (PEPCK1), thereby modulating the cellular response to glucose [10]. Combined, all of these findings support the hypothesis that SIRT2 is a potential candidate gene for the selection of growth-related traits in livestock.
Few SIRT2 variants have been reported to date for cattle, as most research has focused on rodents and human models [11]. The Qinchuan breed of cattle (Bos taurus) used in our research is one of the outstanding indigenous breeds in China, known for its excellent adaptability and physical features. However, it has obvious weaknesses compared to imported commercial beef cattle breeds, such as slow growth and underdeveloped hind hip [12]. Therefore, this study aimed to predict SIRT2 gene function in this cattle breed using bioinformatics information, in order to analyze tissue expression pattern using Real-time PCR, and to identify associated quantitative traits for the benefit of cattle breeding and genetics.
2. Results and Discussion
2.1. Molecular Cloning and Sequence of the Bovine SIRT2 Gene
Based on sequencing of a PCR product that encodes the SIRT2 cDNA of Qinchuan cattle, comparison of the SIRT2 amino acid sequence that we obtained with seven other animal species from GenBank revealed the following similarities (Table 1). Sus scrofa (93.30%), Homo sapiens (88.92%), Rattus norvegicus (88.00%), Mus musculus (85.27%), Gallus gallus (68.63%), Danio rerio (61.93%) and Drosophila melanogaster (49.86%). The relatively high amino acid similarity observed among mammalians (85.27%–93.30%) suggested that the SIRT2 gene was more conserved within this group.
Table 1.
Comparative analysis of SIRT2 gene amino acid sequence of different animals.
| Species | GenBank Accession | Similarity |
|---|---|---|
| Sus scrofa | NP_001107743.1 | 93.30% |
| Homo sapiens | NP_036369.2 | 88.92% |
| Mus musculus | NP_001116237.1 | 85.27% |
| Rattus norvegicus | NP_001008369.1 | 88.00% |
| Gallus gallus | NP_001088636.1 | 68.63% |
| Drosophila melanogaster | NP_001287422.1 | 49.86% |
| Danio rerio | NP_955890.1 | 61.93% |
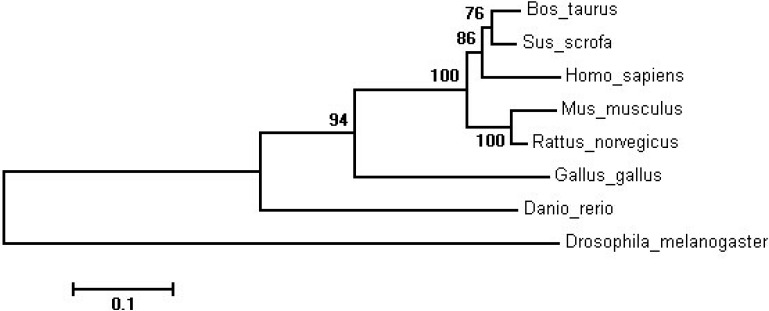
To better understand the relationship between bovine SIRT2 and the potential evolutional process, we constructed the phylogenetic tree based on amino acid sequence of SIRT2 (Figure 1). It showed that the bovine SIRT2 is phylogenetically closest to pig SIRT2 and then to human, with the mouse and rat forming a separate group, while the non-mammalian species formed an even more distant group.
Figure 1.
Phylogenetic tree of the SIRT2 gene in different species.
2.2. Ontogenic Expression of SIRT2 in Qinchuan Cattle
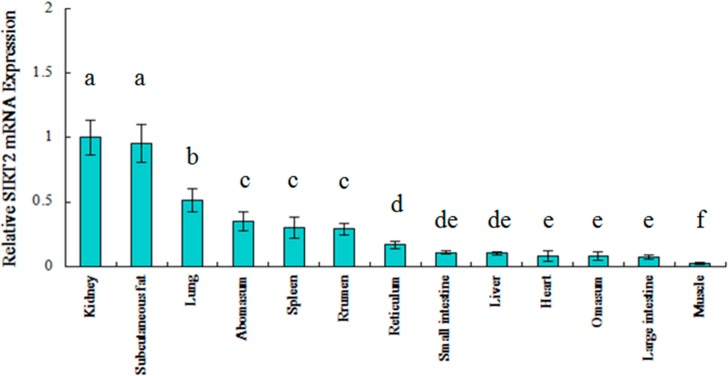
Tissue distribution analysis in pigs has indicated that sirtuin genes are expressed ubiquitously and with the highest abundance in brain, spinal cord and genital tissue [13]. In a calorie-restriction model for rats, SIRT2 was expressed predominantly in white adipose and kidney tissue [14]. Human SIRT2 is expressed in a variety of tissues, with high levels of expression in skeletal muscle and brain tissue [15]. To date, there have been no studies of the expression pattern of bovine SIRT2. We performed RT-PCR to determine the expression of SIRT2 in different tissues. The relative expression results were obtained using the 2−ΔΔCt method, which was first normalized to the geometric mean of β-actin, RPS9 and GAPDH. As shown in Figure 2, SIRT2 was widely expressed in Qinchuan cattle. Moreover, the comparison of SIRT2 gene expression in diverse tissues demonstrated that the bovine SIRT2 gene was highly expressed in kidney, subcutaneous fat, and lung tissue, moderately expressed in rumen, spleen and abomasum tissue, and only slightly expressed in muscle, large and small intestine, heart, omasum, liver and reticulum tissue. There seem to be clear connections (directly or indirectly) between body size traits and modulation by SIRT2 genes in Qinchuan cattle, which fits to what we know about the important functions of SIRT2 genes in metabolism, especially in gluconeogenesis and lipid oxidation [16].
Figure 2.
Tissue expression analysis of SIRT2 mRNA in Qinchuan cattle. Different lowercase letters above the bars (a–f) indicate significant difference between tissues (p < 0.05).
2.3. Genetic Polymorphism of Qinchuan Cattle SIRT2 and χ2 Test
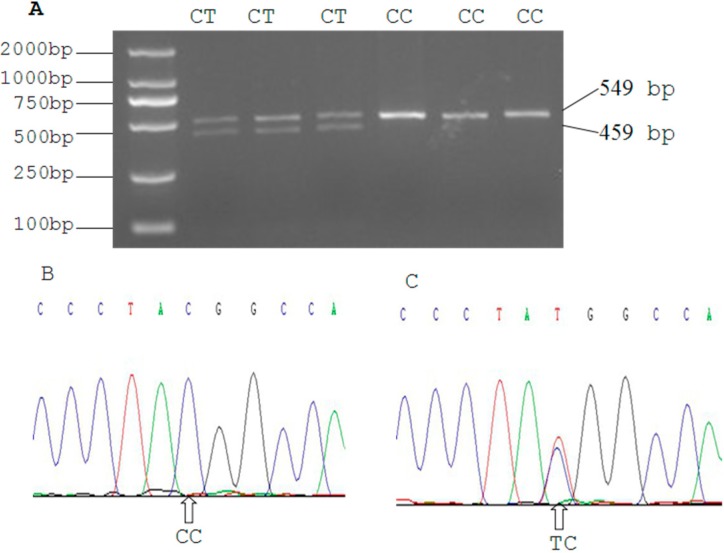
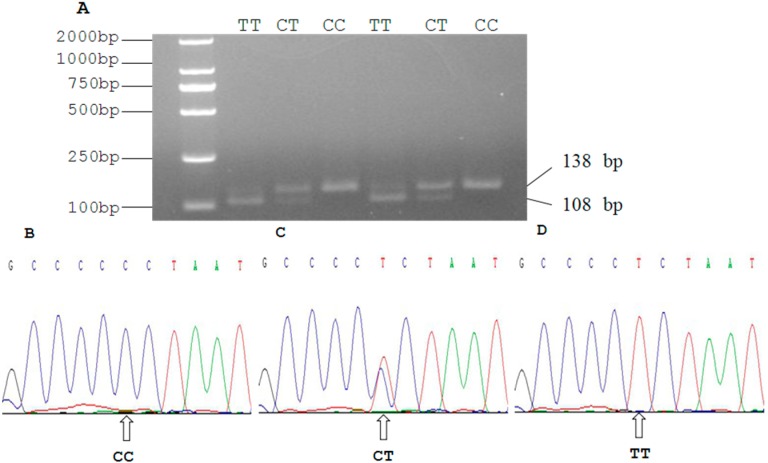
Sequence analysis of the SIRT2 gene revealed two C > T mutations in the 3'UTR region at 19,501 bp (Figure 3) and 19,518 bp (Figure 4). At the g.19501 C > T locus digestion of the 549 bp PCR fragment of SIRT2 3'UTR with BstX1 resulted in fragment lengths of 549, 459, and 90 bp for genotype CT, and 549 bp for genotype CC. The frequency of allele C was dominant in Qinchuan cattle, and genotype CC was more frequent than CT. At the g.19518 C > T locus, digestion of the 138 bp PCR fragment of SIRT2 3'UTR with Xba1 resulted in fragment lengths of 138 bp for genotype CC; 138, 108, and 30 bp for CT, and 108, 30 bp for TT. The frequency of allele C was dominant in Qinchuan cattle and genotype CT was more frequent than other genotypes.
Figure 3.
(A) PCR-RFLP detection results of SIRT2 gene PCR product (19,501 bp locus); (B,C) The sequencing maps of the novel SNP of SIRT2 gene (19,501 bp locus).
Figure 4.
(A) PCR-RFLP detection results of SIRT2 gene PCR product (19,518 bp locus); (B–D) The sequencing maps of the novel SNP of SIRT2 gene (19,518 bp locus).
We found that the g.19501 C > T locus had 2 genotypes, and the genotype TT was not observed in the sampled animals. The absence of that genotype in this population of Qinchuan cattle might mean that it does not exist in the population, or that the size of the experimental population was too small to capture its full genetic variation.
Based on analysis of genotype and allele frequencies (Table 2), we found that for the g.19501 C > T mutation, the CT genotype (20.09%) was less frequent than the wild allele CC (79.91%). Allele frequencies, gene heterozygosity (He), effective allele numbers (Ne) and polymorphism information content (PIC) at the current locus were 0.8996 (C), 0.1004 (T), 0.1807, 1.2205 and 0.1644, respectively. For the g.19518 C > T mutation, the CT genotype was the most prevalent (43.16%) followed by CC (41.03%) and TT (15.81%). The values of He, Ne, and PIC at the current locus were 0.6261 (C), 0.3739 (T), 0.4682, 1.8805 and 0.3586, respectively.
Table 2.
Genotype frequencies (%) of the SIRT2 gene for the SNPs in the Qinchuan cattle populations. Note: HWE, Hardy-Weinberg equilibrium; χ0.052 = 5.991, χ0.012 = 9.21.
| Locus | Genotypic Frequencies (N) | Total | Allelic Frequencies | χ2 (HWE) | PIC | He | Ne | |||
|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | C | T | ||||||
| g.19501 C > T | 0.7991 | 0.2009 | 0 | 468 | 0.8996 | 0.1004 | 5.8328 | 0.1644 | 0.1807 | 1.2205 |
| g.19518 C > T | 0.4103 | 0.4316 | 0.1581 | 468 | 0.6261 | 0.3739 | 2.8581 | 0.3586 | 0.4682 | 1.8805 |
According to the conventions for PIC classification (PIC value <0.25 is considered low polymorphism, 0.25–0.50 is intermediate polymorphism, and >0.50 is high polymorphism), our data showed that g.19501 C > T had an intermediate level of polymorphism, while g.19518 C > T locus had low polymorphism. The genotypic distributions of these two mutations were in Hardy-Weinberg disequilibrium (chi-square test, χ2 < χ0.052), indicating that the genotypic frequencies had been affected by selection, mutation or migration.
2.4. Effects of Single Marker on Body Size Traits
Analysis of the SIRT2 gene in five main breeds of Chinese cattle demonstrated that the polymorphism of g.4140A > G was significantly related to body weight in Nanyang cattle [17]. Here, two novel SNPs (g.19501 C > T and g.19518 C > T) were found in the SIRT2 gene. Association results of single markers with nine economic traits in the Qinchuan population are shown in Table 3. The g.19501 C > T was significantly associated with body length. Individuals with genotype CC had significantly greater body length than those with genotype CT (p = 0.0022). For g.19518 C > T, individuals with genotype TT had significantly greater rump length compared with genotype CC (p = 0.0025), indicating that allele T might be associated with an increase in rump length in Qinchuan cattle. Such associations remained significant even after Bonferroni correction (p < 2.78 × 10−3).
Table 3.
Association of different genotypes of single nucleotide polymorphisms (SNPs) in SIRT2 with body size traits in Qinchuan cattle. Note: Values are shown as the least squares means ± standard error. a,b Means with different superscripts are significantly different (p < 2.78 × 10−3) after Bonferroni correction. Body length (BL), withers height (WH), hip height (HH), rump length (RL), hip width (HW), chest depth (CD), chest circumference (CC), and pin bone width (PBW), fat thickness (BF) and ultrasound loin muscle area (ULA).
| Locus | Genotypes | BL (cm) | WH (cm) | RL (cm) | HW (cm) | CD (cm) | CC (cm) | PBW (cm) | BF (cm) | ULA (cm2) |
|---|---|---|---|---|---|---|---|---|---|---|
| g.19501 C > T | CC (374) | 132.608 ± 0.478 a | 119.222 ± 0.481 | 41.664 ± 0.201 | 38.286 ± 0.258 | 58.345 ± 0.315 | 161.993 ± 0.734 | 18.409 ± 0.142 | 0.873 ± 0.014 | 45.188 ± 0.673 |
| CT (94) | 129.210 ± 0.721 b | 117.329 ± 0.770 | 41.287 ± 0.351 | 36.968 ± 0.324 | 57.159 ± 0.447 | 158.596 ± 0.959 | 17.776 ± 0283 | 0.860 ± 0.023 | 42.343 ± 0.962 | |
| P-value | 0.0022 | 0.0593 | 0.3071 | 0.0204 | 0.1051 | 0.0193 | 0.0387 | 0.3685 | 0.0367 | |
| g.19518 C > T | CC (192) | 130.844 ± 0.672 | 118.612 ± 0.671 | 41.099 ± 0.278 b | 37.177 ± 0.359 | 57.419 ± 0.440 | 159.276 ± 1.023 | 17.849 ± 0.197 | 0.850 ± 0.019 | 44.600 ± 2.162 |
| CT (202) | 132.810 ± 0.655 | 118.381 ± 0.654 | 41.683 ± 0.271 | 38.649 ± 0.350 | 58.369 ± 0.429 | 162.552 ± 0.998 | 18.629 ± 0.193 | 0.883 ± 0.021 | 45.017 ± 2.246 | |
| TT (74) | 133.682 ± 1.082 | 120.696 ± 0.882 | 42.608 ± 0.447 a | 38.500 ± 0.578 | 59.176 ± 0.522 | 163.203 ± 1.648 | 18.459 ± 0.318 | 0.890 ± 0.031 | 43.566 ± 1.554 | |
| P-value | 0.0175 | 0.1004 | 0.0025 | 0.0031 | 0.0267 | 0.0127 | 0.0380 | 0.2007 | 0.2068 |
Sequence alignment demonstrated that these two SNPs located in the 3'UTR did not change the structure of their encoded proteins. However, recent research has provided evidence that mutation in the 3'UTR could affect protein expression and phenotype by altering the stability of mRNA [18]. In Texel sheep, Clop et al. (2006) [19] reported a significant association between a G/A polymorphism in the 3'UTR of the MSTN gene and the phenotype of muscular hypertrophy. Ren et al. (2010) [20] identified a HeaⅢ polymorphism in the 3'UTR of the LHX4 gene that was associated with body mass and length in Nanyang cattle. Therefore, we hypothesized that 3'UTR mutation g.19501C > T and g.19518 C > T of SIRT2 may play significant roles in modifying gene expression patterns, which would therefore affect body size traits in cattle.
2.5. Linkage Disequilibrium (LD) and Haplotype Analysis
In regression analysis, r2 values above 0.33 might imply LD that is strong enough to be used for mapping [21]. In our study, the pair-wise D' and r2 values of the SNPs were 0.918 and 0.158, respectively. The r2 values in SIRT2 were lower than 0.33, indicating that those SNPs had little LD. The rate of recombination may therefore be high, and LD would hence be low in genovariation-dense regions [22].
In order to perform haplotype-based association analysis, three different haplotypes were constructed in SIRT2. The three most frequent haplotypes had a summed probability of 0.995, and those of frequency less than 0.05 were ignored (Table 4). Hap1 (-CC-) had the highest haplotype frequencies (62.10%), followed by Hap2 (-CT-), and Hap3 (-TT-). The haplotypes with high frequency have probably been present in the population for a long time, which may be directly or indirectly regulated by different rearing environments [23].
Table 4.
Haplotypes of SIRT2 gene and their frequencies in Qinchuan cattle.
| Haplotype | g.19501 C > T | g.19518 C > T | Frequency |
|---|---|---|---|
| Hap1 | C | C | 0.621 |
| Hap2 | C | T | 0.279 |
| Hap3 | T | T | 0.095 |
| Hap4 | T | C | 0.005 |
2.6. Effects of Haplotype Combinations on Body Size Traits
We inferred that the effects of variation of a gene could be demonstrated more readily by integrating analysis of the haplotype combinations with the single locus effects. Then the effects of the combinations of the two SNPs were evaluated, and a total of five haplotype combinations were identified for further analysis. Compared with the combination results, individuals with H2H2 diplotypes performed better in terms of their body traits (Table 5). Specifically, the H2H2 diplotypes had significantly greater body length (p = 0.0004), withers height (p = 0.0005), hip width (p = 0.0024), chest depth (p = 0.0023) and pin bone width (p = 0.0019), than H1H3 diplotypes; such associations remained significant even after Bonferroni correction for multiple testing (p < 2.78 × 10−3). Based on our findings, we infer that the H2H2 diplotypes could be used as a molecular marker of combined genotypes for future selection of body size traits in Qinchuan cattle.
Table 5.
Associations of haplotypes with body size traits in Qinchuan cattle. Note: Values are shown as the least squares means ± standard error. a–c Means with different superscripts are significantly different (p < 2.78 × 10−3) after Bonferroni correction. Body length (BL), withers height (WH), hip height (HH), rump length (RL), hip width (HW), chest depth (CD), chest circumference (CC), and pin bone width (PBW), fat thickness (BF) and ultrasound loin muscle area (ULA).
| Combined Genotypes | Body Measurement | Meat Quality Trait | |||||||
|---|---|---|---|---|---|---|---|---|---|
| BL (cm) | WH (cm) | RL (cm) | HW (cm) | CD (cm) | CC (cm) | PBW (cm) | BF (cm) | ULA (cm2) | |
| Hap1/1 (188) | 131.016 ± 0.658 b | 118.332 ± 0.674 | 41.309 ± 0.281 | 37.319 ± 0.357 | 57.582 ± 0.440 | 159.297 ± 1.016 | 17.899 ± 0.197 b | 0.851 ± 0.020 | 44.054 ± 1.152 |
| Hap1/2 (168) | 133.717 ± 0.696 a,b | 119.884 ± 0.713 a | 41.851 ± 0.297 | 39.089 ± 0.377 a | 58.919 ± 0.465 | 164.354 ± 1.075 a | 18.863 ± 0.209 | 0.891 ± 0.021 | 46.341 ± 1.073 |
| Hap1/3 (34) | 125.359 ± 1.520 c | 113.779 ± 1.584 b | 40.294 ± 0.660 | 35.735 ± 0.839 b | 55.588 ± 0.881 b | 153.970 ± 2.389 b | 17.088 ± 0.464 b | 0.811 ± 0.046 | 40.579 ± 1.741 |
| Hap2/2 (18) | 138.868 ± 1.776 a | 122.333 ± 1.399 a | 43.667 ± 0.907 | 40.726 ± 0.559 a | 60.944 ± 0.824 a | 168.121 ± 2.283 a | 19.500 ± 0.638 a | 0.927 ± 0.063 | 46.266 ± 1.271 |
| Hap2/3 (56) | 132.009 ± 1.204 a,b | 119.438 ± 1.234 | 41.857 ± 0.514 | 37.788 ± 0.653 | 58.161 ± 0.806 | 161.464 ± 1.861 | 18.161 ± 0.362 | 0.898 ± 0.036 | 43.347 ± 2.271 |
| P-value | 0.0004 | 0.0005 | 0.0028 | 0.0024 | 0.0023 | 0.0084 | 0.0019 | 0.1142 | 0.0192 |
Several studies have reported that marker-assisted selection (MAS), which selects particularly for beneficial traits that have low heritability, can accelerate genetic gains dramatically compared to conventional breeding [24]. The use of MAS technology has demonstrated that many genes are related to growth [25], production [26] and meat quality traits [27] in livestock. Our previous study revealed significant associations between polymorphisms in SIRT1 and body size in Qinchuan cattle [28]. Functional studies of SIRT1 and SIRT2 showed that they share a conserved central deacetylase domain [29] and that both proteins inhibit proliferation and differentiation in adipocytes [8,30]. Those similar genetic effects can be attributed to their similar function in metabolism. These findings suggest that the SIRT2 gene may have an important influence on animal body size traits, thus making it useful in MAS in cattle.
3. Experimental Section
3.1. Bioinformatic Study
Sequence similarity between bovine SIRT2 protein and its homologue were obtained from Genbank using a search of the BLAST protein databases (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Phylogenetic and molecular evolutionary analysis was conducted using ClustalX software, and the results were further analysed with Mega5.1 software. Numbers at each branch indicated the percentage of times a node was supported in 1000 bootstrap pseudoreplications, based on neighbor joining.
3.2. Collection of RNA Samples
Three purebred two-year-old bulls of the Qinchuan breed were purchased from the Experiment farm of the National Beef Cattle Improvement Center (Yangling, China). The animals were euthanized with a captive bolt gun and then exsanguinated. Samples were collected from 13 types of tissue: Heart, liver, spleen, lung, kidney, muscle, subcutaneous fat, rumen, reticulum, omasum, abomasum, and small and large intestine. They were frozen immediately in liquid nitrogen, and kept at −80 °C until analysis.
3.3. RNA Purification and cDNA Synthesis
Total RNA was isolated from each tissue sample using an RNA simple Total RNA kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. The RNA was quantified using the NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE, USA) at 260 nm. The ratios of OD260 and OD280 for all RNA isolates ranged from 1.8 to 2.0. The integrity of the RNA samples was verified by electrophoresis in a 0.8% agarose gel stained with 0.5 μg/mL ethidium bromide. Then a reverse transcription reaction was performed for the first-strand cDNA synthesis (TaKaRa, Dalian, China). Firstly, to destabilize the secondary structure of RNA, total RNA for each individual was incubated at 65 °C for 5 min; secondly, 10 μL of total RNA was combined with 5× PrimeScript Buffer, 0.5 μL RNase Inhibitor, 0.5 μL PrimeScript RTase, and RNase water, with a total volume 20 μL. The mixture was put on a thermal cycler, and the first-strand cDNA synthesis was accomplished using the program of 37 °C 10 min, 42 °C for 30 min, and for 70 °C 10 min.
3.4. SYBR Green RT-PCR Analysis of Expression Patterns
For normalization of the expression analyses, three reference genes were evaluated: β-actin (AY141970.1), RPS9 (NM_001101152.2), and GAPDH (NM_001034034). Each sample for Real-time PCR was conducted in triplicate and SIRT2 levels were quantified relative to the geometric mean of β-actin, RPS9 and GAPDH via the 2−ΔΔCt method. Primers were designed for the SIRT2 gene and reference genes (Table 6), and gene expression profiles were than analyzed by Real-time PCR using the SYBR Premix Ex TaqII (TaKaRa, Dalian, China). The program consisted of a pre-incubation cycle at 95 °C for 30 s, 40 quantification cycles of a 5 s denaturation at 95 °C, and 45 s annealing at 60 °C. PCRs were conducted in triplicate and then SIRT2 levels were then quantitated using 7500 System SDS Software V 1.4.0 (Applied Biosystems, Waltham, MA, USA).
Table 6.
Primers used in these experiments.
| Name | Function | Primer Sequence (5' to 3') | Tm (°C) | Product Length | Amplified Region |
|---|---|---|---|---|---|
| SIRT2 | RT-PCR | CAACCTGGAGAAATACCGTCTT | 61.0 | 166 bp | 400–565 |
| CAGTCCTTTTTCCTTCAGCAG | |||||
| β-actin | Reference | CACCAACTGGGACGACAT | 61.0 | 202 bp | 320–521 |
| ATACAGGGACAGCACAGC | |||||
| RPS9 | Reference | CCTCGACCAAGAGCTGAAG | 61.0 | 64 bp | 128–191 |
| CCTCCAGACCTCACGTTTGTTC | |||||
| GAPDH | Reference | CCAACGTGTCTGTTGTGGAT | 61.0 | 80 bp | 778–857 |
| CTGCTTCACCACCTTCTTGA | |||||
| Primer A | 3'UTR region amplification | ACCCCTGACCTCACCAAAT | 56.5 | 549 bp | 19411–19959 |
| GCACCTTTCAGCACTCTTC | |||||
| Primer B | 3'UTR region amplification | ACCCCTGACCTCACCAAAT | 62.3 | 138 bp | 19411–19548 |
| CCTGTGGCCCCCTGAGCAGTTAGAGTCTAG |
3.5. Animal Source, Data Collection and Genomic DNA Isolation
In total, 468 adult cows (that were female, 18–24 months old, and unrelated for at least three generations) were randomly selected from National Beef Cattle Improvement Center’s experiment farm (Yangling, China). Body measurement traits were measured as described previously [31], including body length, withers height, rump length, chest depth, hip width, chest circumference and pin bone width. Ultrasound was used to measure some additional traits on carcasses, namely fat thickness and ultrasound loin muscle area.
DNA was extracted from blood samples collected from the jugular vein prior to euthanasia and stored at 80 °C according to the standard phenol chloroform protocol [32]. DNA content was estimated by spectrophotometer. Genomic DNA was diluted to 50 ng/μL. All DNA samples were stored at 20 °C until subsequent analysis.
3.6. PCR Amplification and Sequencing
Based upon the sequence of the bovine SIRT2 gene (GenBank accession No. NM_001113531.1), two pairs of primers were designed to amplify it with PCR using primer premier 5 (Table 6). They produced two sequences, of 549 bp (Primer A) and 138 bp (Primer B) respectively. The PCR amplification product was a mixture of 20 μL composed of 50 ng DNA, 10 pM of each primer, 0.2 mM dNTP, 2.5 mM MgCl2 and 0.5 U Taq DNA polymerase (TaKaRa, Dalian, China). The cycling protocol consisted of denaturation for 5 min at 95 °C, 35 cycles of 94 °C for 30 s, annealing for 30 s, primer extension at 72 °C for 30 s, and a final extension performed at 72 °C for 10 min.
3.7. DNA Pooling, PCR-RFLP and DNA Sequencing
Thirty random individual DNA samples were mixed to form a DNA pool that was used to detect mutations in the SIRT2 gene. DNA sequencing was then applied to screen variations within the amplified regions of the DNA pool, and the products amplified from genomic DNA were imported directly into the BioXM software version 2.6.
Two SNPs (g.19501 C> T and g.19518 C > T) were identified within the 3'UTR of the SIRT2 gene based on DNA sequencing. Further analysis with Primer Premier 5.0 software revealed that the g.19501 C > T and g.19518 C > T were located in the BstX and Xba restriction sites, respectively. Therefore, genotyping of the two SNPs was performed using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). Aliquots of 10 μL of PCR products were digested with 10 U BstX (g.19501 C > T) and Xba (g.19518 C > T) for 8 h at 37 °C, respectively. The digested products were detected by electrophoresis on a 2.5% agarose gel stained with ethidium bromide. Finally, PCR products with different electrophoresis patterns were sequenced from both directions using the DNA sequencer (Applied Biosystems 3730 Genetic Analyzer, USA), and the results were analyzed by DNAMAN software version 5.2.2 (Madison, WI, USA).
3.8. Statistical Analysis
Several parameters including allelic frequencies, genotypic frequencies, He, Ne, Hardy-Weinberg equilibriums and PIC were analyzed statistically according to Nei’s methods [33,34]. LD and haplotype distributions of the SNPs were analyzed using the expectation maximization algorithm with Haploview software [35].
The association between single SNPs and body size traits were analyzed using the general linear models (GLM) procedure in SPSS (version 13.0). The linear model was used:
| Yijklm = u + Gi + Aj + Ak + Sl + Sm + Eijklm | (1) |
where Yijklm were the traits measured on each individual cow, μ was the overall population mean for the traits, Gi was the fixed effect associated with the genotype, Ai was the fixed effect due to the age, Ak was the fixed effect due to the age of dam, Sl was the fixed effect due to the season of sampling (spring vs. fall), Sm was the fixed effect due to the sire, and Eijklm was the standard error.
We tested for an association between combined genotypes for different haplotypes with ultrasound carcass and body measurement traits, to explore any possible interaction between the different haplotypes. The following statistical linear model was used:
| Yijklm = u + Gi + Aj + Ak + Sl + Sm + Eijklm | (2) |
where Yijklm, µ, Aj, Ak, Sl, Sm and Eijklm were the same as for model 1, and Gi was the fixed effect associated with combined genotype.
To avoid Type I errors derived from multiple statistical tests, we calculated the Bonferroni correction, which uses a modified criterion for significance (a/k, where a = 0.05, and k is the overall number of independent statistical tests conducted on the given data). In this study, we analyzed nine traits for two different SNPs, resulting in an adjusted p-value of 2.78 × 10−3 for the 5% significance threshold.
4. Conclusions
In summary, we showed that mRNA expression levels of the SIRT2 gene were highest in subcutaneous fat and kidney tissue of Qinchuan cattle. Two novel SNPs (g.19501 C >T and g.19518 C > T) in the 3'UTR of the gene were identified. Association analysis of the two SNPs with nine economic traits revealed that cattle with the C allele of g.19501 C > T and the T allele of g.19501 C > T loci had better body measurements. When in combination, the H2H2 diplotypes had better body size traits. Our findings suggest that the two SNPs could be used as molecular markers for eliminating or selecting preferred individuals in MAS breeding. This could facilitate the breeding of desired production traits in Qinchuan cattle.
Acknowledgments
The research was supported by the National 863 Program of China (#2013 AA102505 & #2011AA100307-02), National Natural Science Foundation (#31272411), National Beef and Yak Industrial Technology System (#CARS-38), National Science-technology Support Plan Projects (#2012BAD28B04-03) and Technical Innovation Engineering Project of Shaanxi Province (#2014 KTZB02-02-01).
Author Contributions
Lin-Sen Zan conceived and designed the experiments. Lin-Sheng Gui and Ya-Ran Zhang performed the experiments. Gui-Yao Liu and Lin-Sheng Gui analyzed the data. Lin-Sen Zan and Lin-Sheng Gui wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Neugebauer R.C., Sippl W., Jung M. Inhibitors of NAD+ dependent histone deacetylases (sirtuins) Curr. Pharm. Des. 2008;14:562–573. doi: 10.2174/138161208783885380. [DOI] [PubMed] [Google Scholar]
- 2.Vassilopoulos A., Fritz K.S., Petersen D.R., Gius D. The human sirtuin family: Evolutionary divergences and functions. Hum. Genomics. 2011;5:485–496. doi: 10.1186/1479-7364-5-5-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peck B., Chen C.Y., Ho K.K., di Fruscia P., Myatt S.S., Coombes R.C., Fuchter M.J., Hsiao C.D., Lam E.W. SIRT inhibitors induce cell death and p53 acetylation through targeting both SIRT1 and SIRT2. Mol. Cancer Ther. 2010;9:844–855. doi: 10.1158/1535-7163.MCT-09-0971. [DOI] [PubMed] [Google Scholar]
- 4.Inoue T., Hiratsuka M., Osaki M., Oshimura M. The molecular biology of mammalian SIRT proteins: SIRT2 in cell cycle regulation. Cell Cycle. 2007;9:1011–1018. doi: 10.4161/cc.6.9.4219. [DOI] [PubMed] [Google Scholar]
- 5.Blander G., Guarente L. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 6.Marmorstein R. Structure and chemistry of the Sir2 family of NAD+-dependent histone/protein deactylases. Biochem. Soc. Trans. 2004;32:904–909. doi: 10.1042/BST0320904. [DOI] [PubMed] [Google Scholar]
- 7.Jing E., Gesta S., Kahn C.R. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab. 2007;6:105–114. doi: 10.1016/j.cmet.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang F., Tong Q. SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1’s repressive interaction with PPARγ. Mol. Biol. Cell. 2009;20:801–808. doi: 10.1091/mbc.E08-06-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krishnan J., Danzer C., Simka T., Ukropec J., Walter K.M., Kumpf S., Mirtschink P., Ukropcova B., Gasperikova D., Pedrazzini T., et al. Dietary obesity-associated Hif1α activation in adipocytes restricts fatty acid oxidation and energy expenditure via suppression of the Sirt2-NAD+ system. Genes Dev. 2012;26:259–270. doi: 10.1101/gad.180406.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang W., Wang S., Xiao M., Lin Y., Zhou L., Lei Q., Xiong Y., Guan K.L., Zhao S. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol. Cell. 2011;43:33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghinis-Hozum Y., Antaramian A., Villarroya F., Pina E., Mora O. Potential role of sirtuins in livestock production. Animal. 2013;7:101–108. doi: 10.1017/S1751731112001115. [DOI] [PubMed] [Google Scholar]
- 12.Adoligbe C., Zan L., Farougou S., Wang H., Ujjan J.A. Bovine GDF10 gene polymorphism analysis and its association with body measurement traits in Chinese indigenous cattle. Mol. Biol. Rep. 2012;39:4067–4075. doi: 10.1007/s11033-011-1188-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin D., Tan H.J., Lei T., Gan L., Chen X.D., Long Q.Q., Feng B., Yang Z.Q. Molecular cloning and characterization of porcine sirtuin genes. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2009;153:348–358. doi: 10.1016/j.cbpb.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Wang F., Nguyen M., Qin F.X., Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell. 2007;6:505–514. doi: 10.1111/j.1474-9726.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- 15.Frye R.A. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 16.Houtkooper R.H., Pirinen E., Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M., Sun X., Jiang J., Sun Y.J., Lan X.Y., Lei C.Z., Zhang C.L., Chen H. Tetra-primer ARMS-PCR is an efficient SNP genotyping method: An example from SIRT2. Anal. Methods. 2014;6:1835–1840. doi: 10.1039/c3ay41370e. [DOI] [Google Scholar]
- 18.Yie S.M., Li L.H., Xiao R., Librach C.L. A single base-pair mutation in the 3'-untranslated region of HLA-G mRNA is associated with pre-eclampsia. Mol. Hum. Reprod. 2008;14:649–653. doi: 10.1093/molehr/gan059. [DOI] [PubMed] [Google Scholar]
- 19.Clop A., Marcq F., Takeda H., Pirottin D., Tordoir X., Bibe B., Bouix J., Caiment F., Elsen J.M., Eychenne F., et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat. Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 20.Ren G., Chen H., Zhang L.Z., Lan X.Y., Wei T.B., Li M.J., Jing Y.J., Lei C.Z., Wang J.Q. A coding SNP of LHX4 gene is associated with body weight and body length in bovine. Mol. Biol. Rep. 2010;37:417–422. doi: 10.1007/s11033-009-9486-6. [DOI] [PubMed] [Google Scholar]
- 21.Ardlie K.G., Kruglyak L., Seielstad M. Patterns of linkage disequilibrium in the human genome. Nat. Rev. Genet. 2002;3:299–309. doi: 10.1038/nrg777. [DOI] [PubMed] [Google Scholar]
- 22.Huang Y.Z., Wang K.Y., He H., Shen Q.W., Lei C.Z., Lan X.Y., Zhang C.L., Chen H. Haplotype distribution in the GLI3 gene and their associations with growth traits in cattle. Gene. 2013;513:141–146. doi: 10.1016/j.gene.2012.10.052. [DOI] [PubMed] [Google Scholar]
- 23.Huang Y.Z., He H., Sun J.J., Wang J., Li Z.J., Lan X.Y., Lei C.Z., Zhang C.L., Zhang E.P., Wang J.Q., et al. Haplotype combination of SREBP-1c gene sequence variants is associated with growth traits in cattle. Genome. 2011;45:507–516. doi: 10.1139/g11-016. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen M.T., Andrew C.B., Peter B.M., Yutao L., Russell E.L. Single nucleotide polymorphisms in the actin and crustacean hyperglycemic hormone genes and their correlation with individual growth performance in giant freshwater prawn Macrobrachium rosenbergii. Aquaculture. 2010;301:7–15. doi: 10.1016/j.aquaculture.2010.02.001. [DOI] [Google Scholar]
- 25.Camargo G.M., Cardoso D.F., Gil F.M., Fonseca P.D., Zetouni L., Braz C.U., de Freitas A.C., de Souza F.R., Aspilcueta-Borquis R.R., Baldi F., et al. First polymorphisms in JY-1 gene in cattle (Bos taurus indicus) and their association with sexual precocity and growth traits. Mol. Biol. Rep. 2012;39:10105–10109. doi: 10.1007/s11033-012-1884-5. [DOI] [PubMed] [Google Scholar]
- 26.Alim M.A., Fan Y.P., Wu X.P., Xie Y., Zhang Y., Zhang S.L., Sun D.X., Zhang Y., Zhang Q., Liu L., et al. Genetic effects of stearoyl-coenzyme A desaturase (SCD) polymorphism on milk production traits in the Chinese dairy population. Mol. Biol. Rep. 2012;39:8733–8740. doi: 10.1007/s11033-012-1733-6. [DOI] [PubMed] [Google Scholar]
- 27.Cinar M.U., Kayan A., Uddin M.J., Jonas E., Tesfaye D., Phatsara C., Ponsuksili S., Wimmers K., Tholen E., Looft C., et al. Association and expression quantitative trait loci (eQTL) analysis of porcine AMBP, GC and PPP1R3B genes with meat quality traits. Mol. Biol. Rep. 2012;39:4809–4821. doi: 10.1007/s11033-011-1274-4. [DOI] [PubMed] [Google Scholar]
- 28.Gui L., Wang H., Wei S., Zan L. Molecular characterization, expression profiles, and analysis of Qinchuan cattle SIRT1 gene association with meat quality and body measurement traits (Bos taurus) Mol. Biol. Rep. 2014;41:5237–5246. doi: 10.1007/s11033-014-3393-1. [DOI] [PubMed] [Google Scholar]
- 29.Greiss S., Gartner A. Sirtuin/Sir2 phylogeny, evolutionary considerations and structural conservation. Mol. Cells. 2009;28:407–415. doi: 10.1007/s10059-009-0169-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiao L., Shao J. SIRT1 regulates adiponectin gene expression through Foxo1-C/enhancer-binding protein α transcriptional complex. J. Biol. Chem. 2006;281:39915–39924. doi: 10.1074/jbc.M607215200. [DOI] [PubMed] [Google Scholar]
- 31.Sambrock J., Russell D.W. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; New York, NY, USA: 2001. [Google Scholar]
- 32.Gilbert R.P., Bailey D.R.C., Shannon N.H. Linear body measurements of cattle before and after 20 years of selection for postweaning gain when fed two different diets. J. Anim. Sci. 1993;71:1712–1720. doi: 10.2527/1993.7171712x. [DOI] [PubMed] [Google Scholar]
- 33.Nei M., Roychoudhury A.K. Sampling variance of heterozygosity and genetic distance. Genetics. 1974;76:379–390. doi: 10.1093/genetics/76.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nei M., Li W.H. Mathematic model for studying genetic variation in terms of restriction endonucULAses. Proc. Natl. Acad. Sci. USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]