Abstract
The pathogenesis of heart failure involves a complex interaction between genetic and environmental factors. Genetic factors may influence the susceptibility to the underlying etiology of heart failure, the rapidity of disease progression, or the response to pharmacologic therapy. The genetic contribution to heart failure is relatively minor in most multifactorial cases, but more direct and profound in the case of familial dilated cardiomyopathy. Early studies of genetic risk for heart failure focused on polymorphisms in genes integral to the adrenergic and renin-angiotensin-aldosterone system. Some of these variants were found to increase the risk of developing heart failure, and others appeared to affect the therapeutic response to neurohormonal antagonists. Regardless, each variant individually confers a relatively modest increase in risk and likely requires complex interaction with other variants and the environment for heart failure to develop.
Dilated cardiomyopathy frequently leads to heart failure, and a genetic etiology increasingly has been recognized in cases previously considered to be “idiopathic”. Up to 50% of dilated cardiomyopathy cases without other cause likely are due to a heritable genetic mutation. Such mutations typically are found in genes encoding sarcomeric proteins and are inherited in an autosomal dominant fashion. In recent years, rapid advances in sequencing technology have improved our ability to diagnose familial dilated cardiomyopathy and those diagnostic tests are available widely. Optimal care for the expanding population of patients with heritable heart failure involves counselors and physicians with specialized training in genetics, but numerous online genetics resources are available to practicing clinicians.
Keywords: Cardiomyopathy, dilated, genetic counseling, genetics, medical, genetic testing, heart failure, pharmacogenetics.
INTRODUCTION
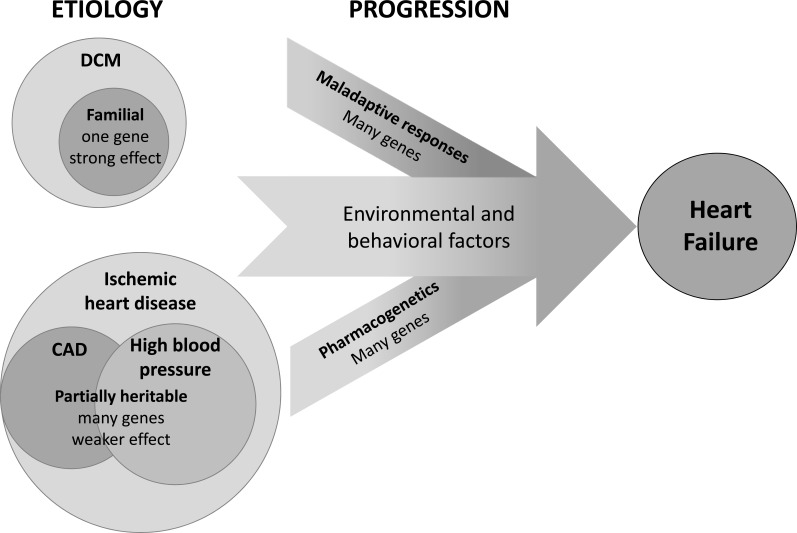
Heart failure (HF) is a clinical syndrome with many etiologies, initiated by an insult to the myocardium and perpetuated by maladaptive systemic responses. As with many common diseases, the pathogenesis of HF involves a complex multifactorial interaction between genetic predisposition, environmental effects and chance. Though the majority of cases develop sporadically, there is clear evidence demonstrating a heritable component of HF [1]. The genetic influence on HF can be understood by separate consideration of the determinants of the initial insult and the subsequent response to that insult (Fig. 1).
Fig. (1).
Heart failure arises from a complex interaction between genetic and environmental factors. Genetic factors may influence the susceptibility to the underlying etiology of heart failure, the progression of the disease, or the response to pharmacologic therapy. Dilated cardiomyopathy (DCM) can result directly from a mutation in a single gene, but the genetic component of heart failure typically consists of interacting variants in many genes with minor individual effects.
The most common index event in the development of HF remains myocardial infarction, and the most common comorbid illness in patients with HF is hypertension. To a certain extent, the genetic contribution to HF is defined by the predisposition to these diseases, both of which are complex and polygenic (i.e. influenced by many genes) in their own right. Though environmental factors play a strong role in determining the progression from underlying illness to clinically evident HF, genetic makeup also clearly affects the physiological response to injury. Thus, a similarly sized myocardial infarction or equivalent exposure to high blood pressure may lead to an effective compensatory response inone patient but overt HF in another. Response to HF medications also is variable, and recent investigations have elucidated potentially important genetic determinants of the efficacy of pharmacologic HF therapy. HF may also arise in the setting of a primary cardiomyopathy, most commonly dilated cardiomyopathy (DCM), which can be caused by single gene (monogenic) mutations that can be inherited in a classical Mendelian fashion. Distinguishing these cases from sporadic or “common” HF can have important implications for the patient and his/her family, and the rapid expansion of sequencing technology will facilitate diagnosis of familial DCM in the coming years.
In the following, we briefly review fundamental genetic principles and summarize our current understanding of the polygenic determinants of the etiologies, progression, and pharmacogenetics of common HF. Thereafter we consider the genetics of DCM, with a particular emphasis on providing clinicians with a practical guide to novel diagnostic modalities and resources to supplement the care of patients with heritable HF.
A GENETICS PRIMER FOR THE CARDIOLOGIST
The human genome contains 20,000–25,000 genes, most of which remain incompletely understood. Each individual genome contains several million genetic variants (differences from the standard “reference” genome), ranging from single nucleotide polymorphisms (SNPs) to larger (>1000 nucleotide) copy number variants [2]. Some specific genetic variants are present in the general population, and the frequency with which they occur in different populations reflects humanity’s genetic history [3]. Other genetic variants are rare or unique to a given individual or family, thus reflecting the continual addition of de novo mutations with each generation. In general, variants that occur with greater than 1% frequency are referred to as polymorphisms, whereas those that occur in less than 1% of the population are called mutations. A small number of variants are tied closely to specific disorders or measurable phenotypes (“quantitative traits”) with relevance to human health, but the overwhelming majority currently are neutral and considered part of the benign genetic variation between humans. Because of the vast number of genetic variants, there potentially are many different “alleles,” or versions of a gene, in the human population.
Two main categories typically are used when considering the genetic factors that contribute to disease: single gene disorders and multifactorial conditions. Though this dichotomy is somewhat artificial, the gene mutations implicated in Mendelian disorders generally are considered to be more deterministic, whereas the genetic variants involved in multifactorial conditions are considered probabilistic in nature.
SINGLE GENE (MENDELIAN) DISORDERS
Several thousand genes have been implicated in rare Mendelian disorders; elsewhere in this review we consider the Mendelian inheritance of familial DCM. A variety of different types of mutations, or genetic alterations, can cause single gene disorders. Some mutations result in a complete “loss of function” allele, or a protein with reduced or “hypomorphic” function. Other mutations result in toxic “gain of function” such as increased activity of an enzyme or cell surface receptor. Still other mutations have “dominant negative” effects that impair not only the mutant allele but also impair the function of the normal allele, thus completely eliminating functional protein. Adding to the complexity, different mutations in a given gene can cause different phenotypes, leading to specific “genotype-phenotype correlations”. In addition, the presence of purported disease-causing mutations in a presumed healthy population raises important questions about the veracity of assertions of pathogenicity and challenges our assumptions about the genetic causation of Mendelian forms of cardiomyopathy [4, 5].
The inheritance patterns observed in Mendelian disorders faithfully reflect the basic principle of random segregation of the two alleles of a given gene during gamete formation. These inheritance patterns subsequently allow us to determine which relatives are at risk for the familial disorder, enabling directed testing of other family members and possibly prevention of disease or mitigation of disease severity.
AUTOSOMAL DOMINANT INHERITANCE
A gene that is located on one of the 22 numbered chromosomes is considered to be “autosomal”, and most humans have two copies of each chromosome. A genetic disorder is said to be “dominant” when only one altered copy of a gene (allele) is required to manifest disease. Thus, in autosomal dominant disorders, the individual has one normally functioning copy of a gene and one copy that functions abnormally. Autosomal dominant inheritance is characterized by a 50% chance for an affected individual to transmit the mutant allele to each offspring, and the incidence of such disorders is equal in males and females. Many adult-onset disorders are characterized by autosomal dominant inheritance, including most heritable forms of cardiomyopathy. The clinical presentations of autosomal dominant disorders frequently are complicated by the confounding factors of “incomplete penetrance” and “variable expressivity”.
Penetrance: “Penetrance” reflects the chance that a person who harbors a genetic mutation will develop manifestations of disease. For many autosomal dominant disorders there is incomplete (sometimes called “reduced”) penetrance, which implies that some individuals can inherit a genetic mutation but never manifest disease.
Expressivity: A disorder is said to have variable “expressivity” when its signs and symptoms encompass a spectrum that differs between affected individuals. For example, DCM can be part of a broad phenotypic spectrum including neuropathy and accelerated aging seen with mutations of the LMNA gene, encoding the protein Lamin A/C. As with reduced penetrance, variable expressivity likely results from both genetic and environmental factors.
AUTOSOMAL RECESSIVE INHERITANCE
Recessive inheritance implies that a single working copy of a gene is sufficient for normal function but that two non-working copies of a gene result in disease. An individual with an autosomal recessive disorder transmits a disease-causing allele to each offspring. Since that person’s partner is extremely unlikely to be a carrier for the same mutation, their offspring most likely will be only carriers. Siblings of an affected individual have a 25% chance of being affected, a 50% chance of being a carrier, and a 25% chance of having two normal alleles.
X-LINKED INHERITANCE
Genetic disorders linked to genes on the X chromosome demonstrate unique inheritance characteristics. X-linked recessive disorders typically manifest only in males, whereas females are carriers. All daughters of an affected male are obligate carriers since they inherit his X chromosome, whereas all sons of an affected male are unaffected. Female carriers of some X-linked recessive disorders sometimes manifest milder or later onset of symptoms.
MITOCHONDRIAL DISORDERS
Tissues with a high energy requirement, such as the myocardium, rely heavily on mitochondria and therefore can be impacted by mitochondrial genetic disorders. Each cell may have 100’s to 1000’s of mitochondria, and each mitochondrion harbors several copies of a circular DNA molecule called mitochondrial DNA (mtDNA). The mtDNA consists of approximately 16,000 nucleotides and encodes 37 genes [6]. Since mitochondria are inherited only from the egg, disorders caused by mutations in the mtDNA are inherited in a matrilineal fashion: transmitted only from an affected female to each of her children. This unique inheritance pattern can often be recognized in a large pedigree, but may be confused with an autosomal dominant pattern in small pedigrees.
MULTIFACTORIAL DISORDERS
Most human health problems, including HF, have multifactorial etiologies influenced by environmental, lifestyle, and genetic factors. Multifactorial disorders are characterized by minor phenotypic contributions from many genes interacting with each other and with environmental factors, in contrast to much rarer disorders in which one gene has a very large effect on the phenotype. The degree to which the heritable components contribute to a disease phenotype varies from condition to condition; estimates of the heritability of HF will be discussed below.
Historically, the search for genetic variants that account for heritable risk has employed a “candidate gene” approach, only sequencing SNPs in genes within pathways known to influence the pathophysiology of a disease. However, advances in genotyping technology that allow simultaneous measurement of millions of SNP srecently have enabled unprecedented investigations into the genetic contribution to these conditions [7]. Genome-wide association studies (GWAS), comparing hundreds of thousands to millions of SNPs in affected and unaffected individuals, have identified numerous limited regions of the genome (loci) that are associated with different common disease or quantitative traits(such as cholesterol levels and susceptibility to atherosclerosis) [8]. Whereas GWAS undoubtedly have contributed to our understanding of common multifactorial conditions, they have had a less immediate effect on the practice of medicine. Importantly, these studies generally do not confirm the involvement of specific genes within the loci that are identified, but uncover the basic genetic pathways involved in disease and indirectly suggest potentially novel therapeutic approaches [9].
THE INFLUENCE OF GENETICS AND PHARMACOGENETICS ON HEART FAILURE
The clinical syndrome of HF is a final common pathway for many cardiovascular disorders including coronary artery disease (CAD), valvular heart disease, hypertension, and arrhythmias. To a certain extent, the risk of developing HF is influenced by the predisposition for each of these antecedent etiologies or insults. However, HF also appears to have a heritable component independent of identifiable comorbidities. After adjustment for comorbidities, analysis of the Framingham database indicates a 70% relative increase in the risk of HF for offspring of a parent diagnosed with HF. The authors of the analysis concluded that roughly 18% of the community HF burden was attributable to heritable factors [1]. Thus the majority of HF occurs sporadically, but the contribution of genetic factors is not insignificant.
Variants in multiple genes with individually minor effects likely account collectively for the heritability of multifactorial “common” HF. Genetic factors may influence the susceptibility to the initial insult, the progression of disease after the insult, and the response to therapy (Fig. 1). Though b-blockers and angiotensin converting enzyme (ACE) inhibitors conclusively confer benefit in broad populations of patients with HF and reduced ejection fraction, there is substantial inter-individual variation in response to these agents. Such variable response likely is influenced by genetic factors, and serves as the basis of the burgeoning field of pharmacogenetics. To date, the role of pharmacogenetic profiling in clinical practice remains uncertain, as no prospective study has demonstrated conclusive benefit to its use in HF therapy [10], though numerous variants with putative importance have been identified [11].
The search for genetic determinants of HF largely has employed a candidate gene approach, focusing on genes involved in the adrenergic and renin-angiotensin-aldosterone systems--pathways with known pathophysiological significance in HF [12, 13]. The variant that has received the greatest investigative attention is the Arg389Gly polymorphism in the gene encoding the beta-1-adrenergic receptor (b1-AR). The presence of two arginines (Arg) at amino acid 389 confers fully active b1-AR signaling, whereas two glycines (Gly) blunt the effects of b1-AR activation. As a result, mice that are homozygous for Arg389 display enhanced cardiac contractility in youth, but are predisposed to developing cardiac fibrosis and HF with aging [14].
The β1-AR Arg389Gly polymorphism has been studied extensively as a modulator of β-blocker response, but the findings of these studies are contradictory and inconclusive. Some find enhanced response to b-blockers in patients homozygous for the Arg389 allele, [14, 15] but others find no significant difference in efficacy [16, 17]. Controversy also exists regarding the effect of a polymorphism in the gene encoding the alpha-2C adrenergic receptor (a2C-AR), which interacts with the Arg389Gly β1-AR variant to either enhance [18] or diminish [19] the efficacy of b-blockers for the treatment of HF.
A rare variant (Ile164) in the gene encoding the β2-AR appears to confer significantly higher mortality in HF patients [20]. Studies in mice [21] and humans [22] suggest that this polymorphism blunts the effect of β2-AR activation by interfering with normal intracellular signaling cascades. Variable function of the proteins that inactivate β-ARs also has been associated with HF risk. G protein-coupled receptor kinases (GRKs) desensitize activated β1-ARs, somewhat analogous to “intracellular β-blockers”. Substitution of leucine for the glycine at amino acid 41 (Gln41Leu) in GRK5, leads to an increase in the function of the GRK5 protein, and thus a decrease in the cellular response to β-AR activation [23]. This polymorphism is much more common in individuals of African ancestry than Caucasians, and appears to be strongly protective against the development of HF with an effect similar in magnitude to the initiation of a β-blocker [24, 25].
Roughly 50% of the variation in circulating ACE levels can be attributed to genetic factors, chiefly the presence of an insertion (I) or deletion (D) in a non-coding portion of the ACE gene [26]. ACE activity is enhanced by a D allele and diminished by an I allele, and individuals with deletions in both of their ACE alleles (ACE DD) may have a higher risk of developing HF [27, 28], though a recent meta-analysis has called the previous findings into question [29]. The enhanced risk likely results from greater susceptibility of ACE DD carriers to pathological cardiac hypertrophy in the setting of hypertension [30, 31]. Thus, the ACE DD genotype may not contribute to the antecedent etiology of HF, but clearly appears to promote the progression of HF by enhancing maladaptive neurohormonal responses [32]. It also appears that the ACE insertion-deletion genotype predicts response to ACE inhibitors. Two retrospective analyses demonstrate that individuals with the ACE DD genotype are at greater risk for death from HF, and that treatment with ACE inhibitors powerfully mitigates that risk [33, 34].
Affordable high throughput sequencing technologies recently have expanded investigations into the genetic underpinnings of HF. Array-based profiling of 2000 candidate genes with putative importance to cardiovascular biology revealed variants in two genes that were associated with increased risk of HF [35]. The first published GWAS for incident HF identified two loci associated with elevated risk in individuals of either European or African ancestry [36], though these findings await replication in other populations.
GENETICS OF DILATED CARDIOMYOPATHY
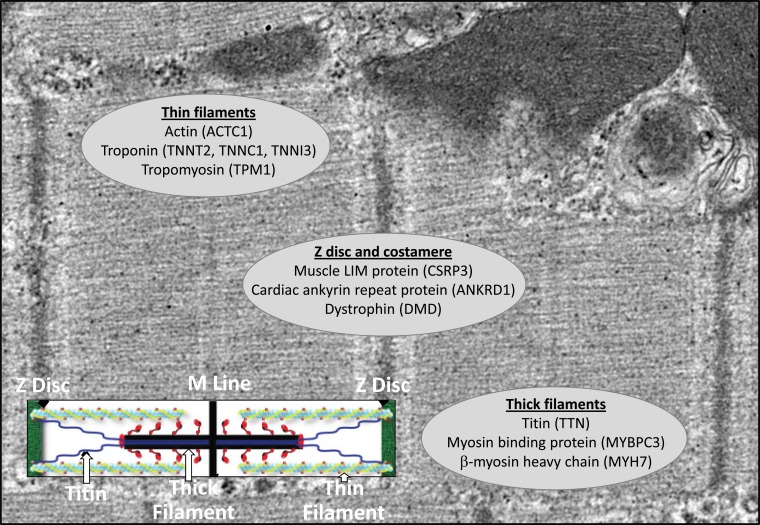
DCM is the second most common etiology of HF with reduced ejection fraction. It is a heterogeneous disorder with multiple etiologies, though it is estimated that 20 to 50% of DCM is caused by a genetic mutation inherited in a Mendelian fashion [37]. There are over 50 currently recognized genes associated with this condition, most of which encode proteins in the cardiomyocyte sarcomere (Fig. 2) [38]. Recent evidence suggests that the gene most commonly mutated in DCM is TTN, encoding titin, an element of the sarcomere and the largest protein in the human body [39]. Given the expanding evidence of heritability, a careful family history is essential in the initial evaluation of a patient who presents with DCM. At a minimum, a history of non-ischemic HF or sudden cardiac death should be solicited for all first and second-degree family members, though 3 to 4 generations ideally will be surveyed [40]. Given its prevalence in the general population, a family history of HF alone is not sufficient to indicate a diagnosis of familial DCM, though there are some strongly suggestive features (Table 1).
Fig. (2).
Selected sarcomeric gene mutations in familial dilated cardiomyopathy. Mutations in over 50 genes can cause familial dilated cardiomyopathy. Most of those mutations occur in genes encoding sarcomeric proteins, and a selected number of the most commonly affected genes are listed.
Table 1.
Determining heritability from the family history.
| Suggestive of Heritability (“Red Flag”) | Not Suggestive of Heritability |
|---|---|
| ➣“Heart attack” in young 1st degree relative (Males < 55 years, Females < 65 years) | ➣Congenital heart disease (“hole in the heart”) |
| ➣Sudden death, unexplained | ➣Murmur |
| ➣Syncope or near syncope, recurrent or unexplained | ➣Heart failure in older family members |
| ➣Heart failure in young 1st degree relative (<60 years) | ➣Rheumatic heart disease |
| ➣Heart transplantation in 1st degree relative | ➣Heart attack in older family members |
The pattern of inheritance in DCM is most frequently autosomal dominant, though penetrance is reduced such that all not all family members who harbor a mutation will develop DCM. There are X-linked, autosomal recessive and mitochondrial forms of heritable DCM as well. Expressivity also is variable, and the severity of the DCM phenotype may vary widely among affected family members. As a result, the identification of a particular mutation in a patient with DCM generally does not change the clinical management of that individual, though some authors recommend early ICD implantation for patients who carry an LMNA(Lamin A/C)mutation [41]. Regardless, current guidelines indicate that a new diagnosis of DCM without obvious precipitant should prompt a screening echocardiogram of all first degree relatives [42], though genetic testing can limit the need for ongoing screening in family members. Discovery of the familial mutation facilitates “cascade” screening in family members to determine those requiring surveillance for DCM, and those who do not [43].
A pedigree that includes a single family member with DCM onset prior to age 50, sudden unexplained death, cardiac transplant or two close relatives with DCM, warrants genetic evaluation and testing [42]. Given the broad implications of a positive result for the family members of a patient, such evaluation ideally should be undertaken with a multidisciplinary approach incorporating a clinical genetics professional (physician or genetic counselor) [42, 44, 45]. In the following we provide the practicing cardiologist with a practical introduction to genetic testing and counseling for DCM.
GENETIC COUNSELING AND TESTING
Genetic counseling “is the process of helping people understand and adapt to the medical, psychological and familial implications of genetic contributions to disease” [46]. It started formally in the 1970’s primarily in the prenatal setting and grew to include pediatric and adult populations. Genetic counselors educate the proband and his/her family about basic genetic concepts: what are genes, what genes do, how they are inherited and how can mistakes or mutations in genes be detected. They describe the pros, cons and limitations of genetic testing, and discuss the possible types of results of testing. Skilled genetic counselors also explore the family dynamics and provide resources for coping with challenging diagnoses. Though genetic counseling has been implemented more extensively in clinical oncology, its use is well established in cardiovascular medicine as well. As improved sequencing technologies and reduced costs facilitate further discovery of genetic contributions to heart disease, the role of cardiovascular genetic counselors likely will continue to expand.
Advances in genetic sequencing technology are rapidly altering the landscape of genetic testing for DCM [47]. Despite the growth in the field, most cardiologists do not have ready access to genetics professionals [48]. There are numerous online resources to assist a clinician in determining whether genetic testing is warranted, ordering tests, and interpreting results (Table 2). The NIH-sponsored website, www.genetests.org, offers comprehensive and unbiased information on testing for cardiovascular genetic disorders. Multiple companies offer genetic testing, though it is important for an ordering clinician to assure that the laboratory is approved by the Clinical Laboratory Improvement Amendments (CLIA) regulations that ensure quality of reported medical tests results.
Table 2.
Online resources for genetic testing information and support.
| Websites with General Information |
Websites with pre- and Post-test Decision Support | Laboratories Offering DCM Genetic Testing with Genetic Counselors on Staff |
|---|---|---|
| www.genetests.org | http://www.dnadirect.com/ http://informeddna.com/ |
http://www.genedx.com/ http://genetics.emory.edu/egl/ http://pcpgm.partners.org/lmm/tests/cardiomyopathy http://ambrygen.com/hereditary-cardiovascular-testing |
If genetic testing is indicated, the choice of which laboratory and which gene panel to use may depend on the type of insurance and financial means of the patient. Most insurance providers will cover the cost of testing if there is a compelling family history and most commercial laboratories offer comparable cost ($2500-$4000) and turn-around-time (6-10 weeks). The websites for the laboratories generally are easy to navigate and provide ready access to all required forms, including templates for letters of medical necessity often required by insurance companies. Blood can be drawn at any facility and should be shipped by two-day mail for processing.
The test result is sent to the ordering physician with an explanation of the potential significance of the findings. The results typically can be grouped into 3 categories:
A known deleterious mutation is detected: Such results are accompanied by an explanation of the DCM etiology, information for testing family members, and management advice.
No mutation is detected: None of the known mutations were detected. Importantly, this result does not exclude hereditary DCM, given that our knowledge of the causative mutations for DCM is incomplete. As such, management of the proband and recommendations for clinical screening of first-degree relatives remain unchanged by this result.
A variant or variants of uncertain significance (VUS) are detected: This result requires additional research, sometimes provided by the laboratory. Interpretation of VUS evolves as different laboratories collect more data. Unfortunately, those data are not always in the public domain, and there may be a long delay before the significance of VUS is determined. Such delays can cause distress for families until the VUS is reclassified as benign or harmful. Testing other affected family members can be helpful, and involving a genetics professional would be entirely appropriate.
If a causative mutation is identified, the type of inheritance pattern will determine the risk for family members.
Autosomal dominant (most common): Parents should be tested to determine which is a carrier or whether the mutation arose de novo. Siblings and children each have a 50% chance of carrying the familial mutation.
Autosomal recessive (rare): Siblings should be tested. Parents are obligate carriers of a pathogenic mutation; each sibling has a 25% chance of being affected with DCM, 25% chance of not being a carrier and 50% chance of being an unaffected carrier.
X-linked: In general only males are affected and their mothers are obligate carriers of a pathogenic mutation. Sons of affected males do not inherit the mutation, whereas daughters are obligate carriers.
Mitochondrial (rare): Only mothers can transmit the pathogenic mutation and they pass it on in variable degrees to all children. Affected males do not pass it onto their children.
CONCLUSION
Like most common diseases, HF develops as a result of complex interactions between genetic and environmental factors. Early studies identified variants in genes in the adrenergic and renin-angiotensin pathways that influence the likelihood of developing HF and response to evidence-based therapies. These polymorphisms confer relatively minor increases in risk and manifestation of clinically evident HF in patients who carry these alleles likely requires other genetic and environmental insults.
In contrast, mutations in single genes can cause DCM independently and typically are heritable in an autosomal dominant fashion. Diagnosis of familial DCM requires a careful family history and confirmatory genetic testing, and can have significant impact on the health of the patient’s family members. Though such testing ideally is undertaken in consultation with a genetics professional, numerous resources are available to guide the cardiologist through the process (Table 2). As sequencing technologies improve and cost decreases, our knowledge of the genetic contribution to cardiovascular diseases such as HF will expand [49] and a basic understanding of the principles of genetics will be essential to the contemporary practice of cardiology.
ACKNOWLEDGEMENTS
JSB received support from the NIH (U01HG006487); MSW received support from NIH (R01HL10412) and the Leducq Foundation; BCJ received support from the NIH (HL096836) and the UAI Research Fellowship.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- 1.Lee DS, Pencina MJ, Benjamin EJ , et al. Association of parental heart failure with risk of heart failure in offspring. N Engl J Med. 2006;355:138–47. doi: 10.1056/NEJMoa052948. [DOI] [PubMed] [Google Scholar]
- 2.Gonzaga-Jauregui C, Lupski Jr, Gibbs RA. Human genome sequencing in health and disease. Annu Rev Med. 2012;63:35–61. doi: 10.1146/annurev-med-051010-162644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conrad DF, Jakobsson M, Coop G , et al. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genet. 2006;38:1251–60. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- 4.Golbus Jr, Puckelwartz MJ, Fahrenbach JP , et al. Population-based variation in cardiomyopathy genes. Circ Cardiovasc Genet. 2012;5:391–9. doi: 10.1161/CIRCGENETICS.112.962928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Norton N, Robertson PD, Rieder MJ , et al. Evaluating pathogenicity of rare variants from dilated cardiomyopathy in the exome era. Circ Cardiovasc Genet. 2012;5:167–74. doi: 10.1161/CIRCGENETICS.111.961805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson S, Bankier AT, Barrell BG , et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290(5806):457–65. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 7.Musunuru K, Kathiresan S. HapMap and mapping genes for cardiovascular disease. Circ Cardiovasc Genet. 2008;1:66–71. doi: 10.1161/CIRCGENETICS.108.813675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manolio TA, Collins FS. The HapMap and genome-wide association studies in diagnosis and therapy. Annu Rev Med. 2009;60:443–56. doi: 10.1146/annurev.med.60.061907.093117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirschhorn JN, Gajdos ZK. Genome-wide association studies results from the first few years and potential implications for clinical medi-cine. Annu Rev Med. 2011;62:11–24. doi: 10.1146/annurev.med.091708.162036. [DOI] [PubMed] [Google Scholar]
- 10.Roden DM, Johnson JA, Kimmel SE , et al. Cardiovascular pharmacogenomics. Circ Res. 2011;109:807–20. doi: 10.1161/CIRCRESAHA.110.230995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Talameh JA, Lanfear DE. Pharmacogenetics in chronic heart failure new developments and current challenges. Curr Heart Fail Rep. 2012;9:23–32. doi: 10.1007/s11897-011-0076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cappola TP, Dorn GW. 2nd Clinical considerations of heritable factors in common heart failure. Circ Cardiovasc Genet. 2011:4701–9. doi: 10.1161/CIRCGENETICS.110.959379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorn GW. 2nd. The genomic architecture of sporadic heart failure. Circ Res. 2011;108:1270–83. doi: 10.1161/CIRCRESAHA.110.229260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mialet Perez J, Rathz DA, Petrashevskaya NN , et al. Beta 1-adrenergic receptor polymorphisms confer differential function and pre-disposition to heart failure. Nat Med. 2003;9:1300–5. doi: 10.1038/nm930. [DOI] [PubMed] [Google Scholar]
- 15.Liggett SB, Mialet-Perez J, Thaneemit-Chen S , et al. A polymorphism within a conserved beta(1)-adrenergic receptor motif alters cardiac function and beta-blocker response in human heart failure. Proc Natl Acad Sci USA. 2006;103:11288–93. doi: 10.1073/pnas.0509937103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cresci S, Kelly RJ, Cappola TP , et al. Clinical and genetic modifiers of long-term survival in heart failure. J Am Coll Cardiol. 2009;54:432–44. doi: 10.1016/j.jacc.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White HL, de Boer RA, Maqbool A , et al. An evaluation of the beta-1 adrenergic receptor Arg389Gly polymorphism in individuals with heart failure a MERIT-HF sub-study. Eur J Heart Fail. 2003;5:463–8. doi: 10.1016/s1388-9842(03)00044-8. [DOI] [PubMed] [Google Scholar]
- 18.Lobmeyer MT, Gong Y, Terra SG , et al. Synergistic polymorphisms of beta1 and alpha2C-adrenergic receptors and the influence on left ventricular ejection fraction response to beta-blocker therapy in heart failure. Pharmacogenet Genomics. 2007;17:277–82. doi: 10.1097/FPC.0b013e3280105245. [DOI] [PubMed] [Google Scholar]
- 19.Bristow MR, Murphy GA, Krause-Steinrauf H , et al. An alpha2C-adrenergic receptor polymorphism alters the norepinephrine-lowering effects and therapeutic response of the beta-blocker bucindolol in chronic heart failure. Circ Heart Fail. 2010;3:21–8. doi: 10.1161/CIRCHEARTFAILURE.109.885962. [DOI] [PubMed] [Google Scholar]
- 20.Liggett SB, Wagoner LE, Craft LL , et al. The Ile164 beta2-adrenergic receptor polymorphism adversely affects the outcome of con-gestive heart failure. J Clin Invest. 1998;102:1534–9. doi: 10.1172/JCI4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turki J, Lorenz JN, Green SA , et al. Myocardial signaling defects and impaired cardiac function of a human beta 2-adrenergic recep-tor polymorphism expressed in transgenic mice. Proc Natl Acad Sci USA. 1996;93:10483–8. doi: 10.1073/pnas.93.19.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brodde OE, Buscher R, Tellkamp R , et al. Blunted cardiac responses to receptor activation in subjects with Thr164Ile beta(2)-adrenoceptors. Circulation. 2001;103:1048–50. doi: 10.1161/01.cir.103.8.1048. [DOI] [PubMed] [Google Scholar]
- 23.Wang WC, Mihlbachler KA, Bleecker ER , et al. A polymorphism of G-protein coupled receptor kinase5 alters agonist-promoted desensitization of beta2-adrenergic receptors. Pharmacogenet Genomics. 2008;18:729–32. doi: 10.1097/FPC.0b013e32830967e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liggett SB, Cresci S, Kelly RJ , et al. A GRK5 polymorphism that inhibits beta-adrenergic receptor signaling is protective in heart failure. Nat Med. 2008;14:510–7. doi: 10.1038/nm1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lobmeyer MT, Wang L, Zineh I , et al. Polymorphisms in genes coding for GRK2 and GRK5 and response differences in antihyper-tensive-treated patients. Pharmacogenet Genomics. 2001;21:42–9. doi: 10.1097/FPC.0b013e328341e911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rigat B, Hubert C, Alhenc-Gelas F , et al. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene account-ing for half the variance of serum enzyme levels. J Clin Invest. 1990;86:1343–6. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang W, Xie C, Zhou H , et al. Association of the angiotensin-converting enzyme gene polymorphism with chronic heart failure in Chinese Han patients. Eur J Heart Fail. 2004;6:23–7. doi: 10.1016/j.ejheart.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 28.Palmer BR, Pilbrow AP, Yandle TG , et al. Angiotensin-converting enzyme gene polymorphism interacts with left ventricular ejection fraction and brain natriuretic peptide levels to predict mortality after myocardial infarction. J Am Coll Cardiol. 2003;41:729–36. doi: 10.1016/s0735-1097(02)02927-3. [DOI] [PubMed] [Google Scholar]
- 29.Bai Y, Wang L, Hu S, Wei Y. Association of angiotensin-converting enzyme I/D polymorphism with heart failure a meta-analysis. Mol Cell Biochem. 2012;361:297–304. doi: 10.1007/s11010-011-1115-8. [DOI] [PubMed] [Google Scholar]
- 30.Iwai N, Ohmichi N, Nakamura Y, Kinoshita M. DD genotype of the angiotensin-converting enzyme gene is a risk factor for left ventricular hypertrophy. Circulation. 1994;90:2622–8. doi: 10.1161/01.cir.90.6.2622. [DOI] [PubMed] [Google Scholar]
- 31.Schunkert H, Hense HW, Holmer SR , et al. Association between a deletion polymorphism of the angiotensin-converting-enzyme gene and left ventricular hypertrophy. N Engl J Med. 1994;330:1634–8. doi: 10.1056/NEJM199406093302302. [DOI] [PubMed] [Google Scholar]
- 32.Andersson B, Sylven C. The DD genotype of the angiotensin-converting enzyme gene is associated with increased mortality in idio-pathic heart failure. J Am Coll Cardiol. 1996;28:162–7. doi: 10.1016/0735-1097(96)00098-8. [DOI] [PubMed] [Google Scholar]
- 33.McNamara DM, Holubkov R, Postava L , et al. Pharmacogenetic interactions between angiotensin-converting enzyme inhibitor thera-py and the angiotensin-converting enzyme deletion polymorphism in patients with congestive heart failure. J Am Coll Cardiol. 2004;44:2019–26. doi: 10.1016/j.jacc.2004.08.048. [DOI] [PubMed] [Google Scholar]
- 34.Wu CK, Luo JL, Tsai CT , et al. Demonstrating the pharmacogenetic effects of angiotensin-converting enzyme inhibitors on long-term prognosis of diastolic heart failure. Pharmacogenomics J. 2010;10:46–53. doi: 10.1038/tpj.2009.39. [DOI] [PubMed] [Google Scholar]
- 35.Cappola TP, Li M, He J , et al. Common variants in HSPB7 and FRMD4B associated with advanced heart failure. Circ Cardiovasc Genet. 2010;3:147–54. doi: 10.1161/CIRCGENETICS.109.898395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith NL, Felix JF, Morrison AC , et al. Association of genome-wide variation with the risk of incident heart failure in adults of European and African ancestry a prospective meta-analysis from the cohorts for heart and aging research in genomic epidemiology (CHARGE) consortium. Circ Cardiovasc Genet. 2010;3:256–66. doi: 10.1161/CIRCGENETICS.109.895763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Callis TE, Jensen BC, Weck KE, Willis MS. Evolving molecular diagnostics for familial cardiomyopathies at the heart of it all. Expert Rev Mol Diagn. 2010;10:329–51. doi: 10.1586/erm.10.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McNally EM, Golbus Jr, Puckelwartz MJ. Genetic mutations and mechanisms in dilated cardiomyopathy. J Clin Invest. 2013;123:19–26. doi: 10.1172/JCI62862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herman DS, Lam L, Taylor MR , et al. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366:619–28. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hershberger RE, Siegfried JD. Update 2011: clinical and genetic issues in familial dilated cardiomyopathy. J Am Coll Cardiol. 2011;57:1641–9. doi: 10.1016/j.jacc.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Rijsingen IA, Arbustini E, Elliott PM , et al. Risk factors for malignant ventricular arrhythmias in lamin a/c mutation carriers a European cohort study. J Am Coll Cardiol. 2012;59:493–500. doi: 10.1016/j.jacc.2011.08.078. [DOI] [PubMed] [Google Scholar]
- 42.Hershberger RE, Lindenfeld J, Mestroni L , et al. Genetic evaluation of cardiomyopathy--a Heart Failure Society of America practice guideline. J Card Fail. 2009;15:83–97. doi: 10.1016/j.cardfail.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 43.Lakdawala NK, Funke BH, Baxter S , et al. Genetic testing for dilated cardiomyopathy in clinical practice. J Card Fail. 2012;18:296–303. doi: 10.1016/j.cardfail.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hershberger RE, Cowan J, Morales A, Siegfried JD. Progress with genetic cardiomyopathies screening, counseling, and testing in dilated, hypertrophic, and arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circ Heart Fail. 2009;2:253–61. doi: 10.1161/CIRCHEARTFAILURE.108.817346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Judge DP. Use of genetics in the clinical evaluation of cardiomyopathy. JAMA. 2009;302:2471–6. doi: 10.1001/jama.2009.1787. [DOI] [PubMed] [Google Scholar]
- 46.Task F, Resta R, Biesecker BB , et al. National Society of Genetic Counselors' Definition A new definition of Genetic Counseling National Society of Genetic Counselors' Task Force report. J Genet Couns. 2006;15:77–83. doi: 10.1007/s10897-005-9014-3. [DOI] [PubMed] [Google Scholar]
- 47.Teekakirikul P, Kelly MA, Rehm HL , et al. Inherited cardiomyopathies molecular genetics and clinical genetic testing in the post-genomic era. J Mol Diagn. 2013;15:158–70. doi: 10.1016/j.jmoldx.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 48.Patterson C. A good idea a physician's perspective on genetic counseling for hypertrophic cardiomyopathy. J Cardiovasc Transl Res. 2009;2:508–9. doi: 10.1007/s12265-009-9130-9. [DOI] [PubMed] [Google Scholar]
- 49.Velagaleti RS, O'Donnell CJ. Genomics of heart failure. Heart Fail Clin. 2010;6:115–24. doi: 10.1016/j.hfc.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]