Abstract
The demonstrated ability to differentiate both human embryonic stem cells (hESCs) and patient-derived induced pluripotent stem cells (hiPSCs) into hepatocyte-like cells (HLCs) holds great promise for both regenerative medicine and liver disease research. Here, we determined that, despite an immature phenotype, differentiated HLCs are permissive to hepatitis C virus (HCV) infection and mount an interferon response to HCV infection in vitro. HLCs differentiated from hESCs and hiPSCs could be engrafted in the liver parenchyma of immune-deficient transgenic mice carrying the urokinase-type plasminogen activator gene driven by the major urinary protein promoter. The HLCs were maintained for more than 3 months in the livers of chimeric mice, in which they underwent further maturation and proliferation. These engrafted and expanded human HLCs were permissive to in vivo infection with HCV-positive sera and supported long-term infection of multiple HCV genotypes. Our study demonstrates efficient engraftment and in vivo HCV infection of human stem cell–derived hepatocytes and provides a model to study chronic HCV infection in patient-derived hepatocytes, action of antiviral therapies, and the biology of HCV infection.
Introduction
The technology to developmentally program human embryonic stem cells (hESCs) to various cell lineages offers great promise for in vitro study and cell therapy of various diseases (1, 2). Recent advances in generating human induced pluripotent stem cells (hiPSCs) from somatic cells by forced expression of reprogramming factors provide unique opportunities to generate patient-specific cell types of all lineages (3–5). Several protocols have been described to differentiate both hESCs and hiPSCs into hepatocyte-like cells (HLCs) (6–11). The differentiation process is based on the induction of definitive endoderm, followed by hepatic specification to generate α-fetoprotein–positive (AFP-positive) HLCs, and finally hepatic maturation, leading to HLCs capable of producing high levels of albumin. These HLCs reproduce key features of primary human hepatocytes (PHHs). However, the HLCs exhibit a phenotype of incompletely matured hepatocytes, compared with in vitro culture of PHHs (9). The in vitro monolayer differentiation culture does not reproduce the complex, 3D, multicellular environment of the native liver, which could be important for the proper maturation of hepatocytes during in vitro differentiation. In this context, the engraftment and maintenance of HLCs in a native liver parenchyma constitutes a relevant approach to promote further maturation and long-term residence of engrafted cells. Engraftment of human stem cell–derived HLCs has been described in different models of immunodeficient mice with transgene-induced (9, 10, 12) or chemically induced (9, 13–16) liver toxicity with low efficiencies (12, 14). Furthermore, not much is known about the effect of in situ engraftment on the level of maturation and permissiveness of these HLCs to infection by hepatitis viruses.
Hepatitis C virus (HCV) infection is a serious public health problem in the world; 80% of HCV-infected patients developing chronic infection, which can progress to liver cirrhosis and hepatocellular carcinoma (17). Hepatitis C is difficult to treat, with only half of the patients responding to the standard treatment based on PEG-IFN-α and ribavirin. Recent development and licensure of direct-acting antivirals offer promise in improving the treatment response (18). Research on HCV infection has been hampered by the lack of relevant in vitro and in vivo models. The most widely used cell culture model is based on human hepatocellular carcinoma cell line Huh7 and a unique strain of HCV of genotype 2a (JFH1). Infected Huh7 cells produce infectious progeny viruses (called HCVcc for cell culture) and thus recapitulate the entire life cycle of the virus (19–21). However, these transformed, rapidly dividing Huh7 cells are not a relevant biological model for the natural host of the virus: the PHHs. PHHs are permissive to infection with HCVcc and HCV+ sera both in vitro (22–24) and in vivo after engraftment (25, 26). However, their general use is hampered by their limited availability, variable quality, cost, and short-term use. In this context, HLCs derived from patient-specific hiPSCs are a valuable model to study HCV infection in vitro (27, 28), but their permissiveness after engraftment has not been investigated yet.
Here, we demonstrate that HLCs, generated from both hESCs and patient-specific hiPSCs, can be engrafted into the livers of transgenic mice carrying the uPA gene driven by the major urinary protein promoter onto a SCID/beige background (referred to as MUP-uPA/SCID/Bg mice) (26, 29), in which they undergo further maturation and can be persistently infected in vivo by HCV of different genotypes for more than 3 months. This approach represents a unique and highly relevant model to study HCV infection both in vitro and in vivo, in the context of patient-specific genetic background.
Results
Hepatic differentiation of human pluripotent stem cells.
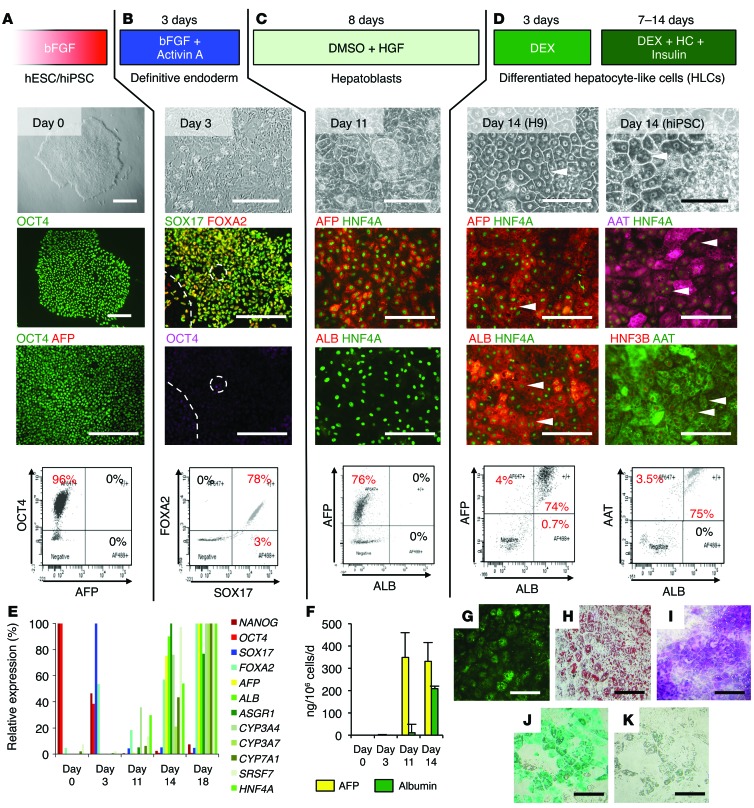
hiPSCs were generated from skin fibroblasts using STEMCCA lentiviral vectors (ref. 30 and Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI75456DS1). The hiPSCs share the same characteristics as the hESCs (Supplemental Figure 1B). hESCs remain the gold standard for in vitro differentiation. In this context, we used HSF-6 and H9 hESCs as positive controls for our experiments. Both hiPSCs and hESCs were subjected to a 3-step differentiation protocol (Figure 1). At day 0, hESC and hiPSC colonies were positive for OCT4, a pluripotency marker, and were negative for definitive endoderm and hepatic markers (Figure 1A, day 0). They were first treated for 3 days with 100 ng/ml activin A and basic fibroblast growth factor to generate definitive endoderm exhibiting a more spiky appearance. At day 3, more than 80% of the cells lost expression of OCT4 and became strongly positive for SOX17 and FOXA2 (also known as HNF3β), 2 markers of definitive endoderm (Figure 1B, day 3).
Figure 1. Hepatic differentiation of human pluripotent stem cells.
(A–D) Protocol, phase-contrast microscopy, immunofluorescent assay, and FACS analysis of the cells at different stages. (A) hESCs and hiPSCs. (B) Stem cells were treated for 3 days with 100 ng/ml activin A and basic fibroblast growth factor (bFGF) to generate definitive endoderm. (C) Hepatic specification was induced by maintaining the cells in presence of 100 ng/ml hepatocyte growth factor (HGF) and 0.1% DMSO for 8 days. (D) Finally, hepatic maturation was achieved by treating the hepatoblasts for 3 days with 10–7 M dexamethasone. Differentiated HLCs were maintained for up to 2 weeks in culture in the presence of dexamethasone (DEX), hydrocortisone (HC), and insulin. Arrowheads indicate binucleate cells. Scale bars: 200 μm. (E) Expression of differentiation markers assessed by RTqPCR along the differentiation process (day 0: stem cells, day 3: definitive endoderm, day 11: hepatoblasts, days 14 and 18: differentiated hepatocytes). Results are expressed as relative expression. (F) Secretion of hepatic proteins AFP and albumin assessed by ELISA. Data represent mean ± SEM. (G–K) Functional characterization of HLCs at day 14 of differentiation. (G) Lipoprotein uptake assessed by incubation with Alexa 488–conjugated LDL. (H) Lipid storage demonstrated by Oil Red O staining of the lipid droplets. (I) Glycogen storage demonstrated by periodic acid–Schiff staining. (J) Cells were examined for uptake of indocyanine green. (K) Six hours later, internalized indocyanine green was released. Scale bars: 100 μm.
Confluent definitive endoderm cells were passaged at a ratio of 1:3 on growth factor–reduced Matrigel and cultured for 8 days in medium containing 1% DMSO and 100 ng/ml hepatocyte growth factor for hepatic specification. At day 11, around 80% of the cells were positive for AFP, a marker of hepatoblasts, but remained negative for albumin, a marker of mature hepatocytes. Hepatoblasts were also positive for the transcription factor HNF4α, a master regulator of hepatic differentiation (Figure 1C, day 11).
Finally, cells were maturated for 3 more days in medium containing 10–7 M dexamethasone. At the end of this protocol (day 14), differentiated cells exhibited characteristic hepatocyte morphology: cuboidal shape, distinctive round small nuclei, and compact cell-cell contacts (Figure 1D, day 14). On day 14 and under the best conditions, 80% of the cells were positive for albumin, consistent with hepatic maturation. Treatment with oncostatin M did not show an additional effect on hepatic differentiation and maturation (data not shown). The differentiated HLCs were also positive for α-1-antitrypsin (AAT) and the hepatic transcription factors HNF3B, also known as FOXA2, and HNF4α (Figure 1D and Supplemental Figure 2A). However, most of the cells remained AFP positive, suggesting that they maintained a more fetal phenotype than PHHs (Supplemental Figure 3A).
Staining of HLCs for different isoforms of CYP450, a marker for mature hepatocytes, remained mainly negative (CYP2D6) or weakly positive in few cells (CYP3A4), confirming that the ALB-positive cells differentiated in vitro did not fully mature (Supplemental Figure 2B), as seen in PHHs (Supplemental Figure 3A). Some cells maintained expression of CK7 (cholangiocyte marker) (Supplemental Figure 2C), suggesting that our cell population consists of maturated HLCs, AFP-positive hepatoblasts, and CK7-positive hepatic biprogenitor cells. The differentiated cells were then maintained in hepatocyte culture medium containing 1 μM insulin, 10 μM hydrocortisone, and 10–7 M dexamethasone for up to 1 week. Cell preparations consisting of more than 60% polygonal cells were considered good enough for further experimentation.
Quantitative RT-PCR (RTqPCR) (Figure 1E) and ELISA assays (Figure 1F) illustrated the transition from pluripotent NANOG- and OCT4-positive stem cells to SOX17- and FOXA2-positive definitive endoderm cells, followed by hepatic specification toward AFP-secreting hepatoblasts (day 11) and further maturation into albumin-secreting HLCs (day 14 and 18). HLCs exhibit a less mature phenotype than adult hepatocytes when compared with PHHs (Supplemental Figure 3, B and C).
Finally, we demonstrated other hepatocyte-specific functions of these differentiated cells, including LDL uptake (Figure 1G), lipid storage using Oil Red O staining (Figure 1H), glycogen storage using periodic acid–Schiff staining (Figure 1I), and uptake and excretion of indocyanine green, an organic anion exclusively eliminated by mature hepatocytes through the liver-specific organic anion transporter 1 (Figure 1, J and K). These results indicate that this protocol is highly efficient for generating differentiated hepatocytes from human pluripotent stem cells.
Expression of cellular factors important for HCV infection.
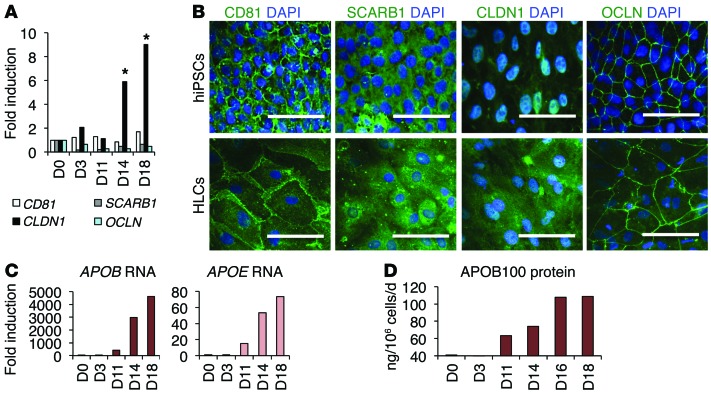
We confirmed that these cells express the cellular factors necessary for productive HCV infection. Interestingly, the stem cells expressed detectable amounts of 4 HCV entry factors: CD81, SCARB1, CLDN1, and OCLN (Figure 2A). During the differentiation process, expression of CD81, SCARB1, and OCLN was maintained at a stable level, while CLDN1 mRNA expression was significantly increased at the hepatic maturation stage. This result was confirmed by visualization of these factors by immunofluorescence of pluripotent stem cells (day 0) and HLCs at day 14 (Figure 2B). Of note, the levels of expression of HCV entry factors were lower in HLCs than in PHHs (Supplemental Figure 3D).
Figure 2. Cellular factors associated with HCV infection.
(A and B) Expression of the 4 HCV entry factors during hepatic differentiation (day 0 [D0]) by (A) RTqPCR and (B) IFA. *P < 0.05. Scale bars: 50 μm. (C and D) Expression of apolipoproteins APOB and APOE during hepatic differentiation by (C) RTqPCR and (D) ELISA. For comparison to PHHs, see Supplemental Figure 3, C and D.
HCV assembly and secretion have been associated with the lipoprotein secretion pathway and particularly with pathways mediated by APOB and APOE (31–33). Here, we show that the expression of APOB and APOE increased during hepatic differentiation, as demonstrated by RTqPCR (Figure 2C). The hepatic-specific APOB100 was not expressed or secreted by stem cells and definitive endoderm cells. However, after hepatic specification and during hepatic maturation, APOB was highly overexpressed, and increasing levels of APOB100-containing lipoproteins were detected by ELISA in the supernatant of the hepatoblasts and differentiated hepatocytes (Figure 2D), but at a lower level than the supernatants of PHHs (Supplemental Figure 3, D and E). Taken together, these results show that the differentiated HLCs express several of the main cellular cofactors important for the various steps of the HCV life cycle.
We had previously performed a genome-wide siRNA screen for host factors required for productive HCV infection (34), leading to the identification of 262 host factors involved in different steps of the HCV life cycle in Huh7-derived cell lines. We compared this list, updated with all other host factors described in the literature (a total of 323 genes) (35), with a list of 374 genes shown by microarray as significantly induced (i.e., >4-fold) during one or several steps of in vitro hepatic differentiation of hiPSCs (36). From this comparison, we identified 8 genes that are important in HCV replication and significantly induced during hepatic differentiation. We confirmed the expression profile of these 8 genes in our differentiation protocol (Supplemental Table 1). Among these factors are the CLDN1 and APOB, as described above (Figure 2, A and C). In addition, microsomal triglyceride transfer protein (MTTP), another gene implicated in the lipoprotein secretion pathway and the assembly and maturation of the viral particles (31, 33), was also significantly induced during hepatic specification and maturation. The functions of 5 other identified genes (FER1L3, [also known as MYOF], FOXA2, SMAD6, TWIST1, HIST1H2BK) in the HCV infection cycle are unknown and under investigation.
In vitro infection of HLCs with HCV.
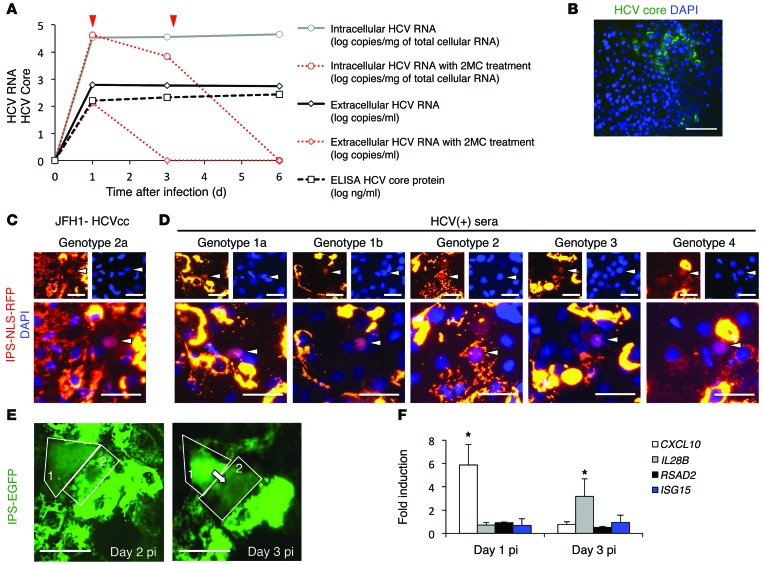
At day 14, differentiated cells were inoculated overnight with JFH1-HCVcc at a multiplicity of infection of 0.5. Intracellular HCV RNA was detected by RTqPCR on day 1, 3, and 6 postinfection (pi), indicative of HCV replication (Figure 3A). The levels of intracellular HCV RNA were similar among HCV-infected UCO6-, H9-, and hiPSC-derived HLCs, suggesting similar hepatic maturation among these cells suitable for HCV infection (data not shown). Treatment of the infected cells with 2′-C-methylcytidine, an HCV polymerase inhibitor, inhibited 90% of HCV replication after 2 days of treatment, confirming authentic HCV replication (Figure 3A, red lines). HCV RNA and core antigen were detected in the supernatant by RTqPCR and ELISA, respectively, indicating the production of progeny virus (Figure 3A). More importantly, infectivity assays, consisting of incubation of Huh7.5.1 cells with supernatant from infected HLCs, followed by immunostaining for HCV core, confirmed that these progeny virions were infectious (Figure 3B). The infectious titers in the medium were relatively low (~2 log focus-forming units/ml).
Figure 3. In vitro HCV infection of HLCs.
(A) Quantification of intracellular and extracellular HCV RNA by RTqPCR, and quantification of HCV core antigen in the supernatant by ELISA, after infection with JFH1-HCVcc. Inhibition of JFH1-HCV replication by 100 mM 2′-C-methylcytidine (2MC) is indicated in red (red arrows indicate when the 2MC was added). (B) Inoculation of Huh7.5.1 with supernatant of JFH1-HCVcc–infected HLCs, followed by immunofluorescence detection of HCV core protein, indicating production of infectious virus. Scale bar: 100 μm. (C and D) HLCs were transduced to express the IPS-NLS-RFP vector. One day later, cells were inoculated with (C) JFH1-HCVcc– or (D) HCV-positive clinical isolates of different genotypes. Two days pi, cells were observed for relocalization events, confirming infection of the cells by HCVcc and authentic HCV particles from patients’ sera. Scale bars: 50 μm. (E) Time-dependent visualization of HDFR, suggesting cell-to-cell transmission of HCV infection. Scale bars: 50 μm. (F) Induction of an antiviral innate immunity in response to JFH1-HCVcc infection of HLCs, as assessed by RTqPCR. *P < 0.05. Data are expressed as mean ± SEM.
HCV protein expression in infected HLCs, like in PHHs, was not detectable by regular immunofluorescence, probably due to a low level of replication. To circumvent this limitation, we used a recently described approach to visualize low-level infection: the hepatitis C–dependent relocalization (HDFR) assay (37, 38). HLCs were transduced with EGFP-IPS or RFP-NLS-IPS reporters and then infected with HCV. Cells exhibiting relocalization of fluorescence (i.e., cells in which the viral protease NS3-4A cleaved the reporter) could be observed as early as day 2 pi, confirming the infection of these cells by HCVcc (Figure 3C). Three days after inoculation with JFH1-HCVcc at a multiplicity of 0.5, less than 1% of the HLCs showed relocalization events, suggesting that HLCs are much less permissive to HCVcc infection than Huh7 cell lines. Immunostaining for ALB on infected cells expressing the HDFR vector confirmed that only differentiated cells (ALB positive) were permissive to HCV infection (Supplemental Figure 4). Relocalization events were also detectable after in vitro inoculation with HCV-positive sera of different genotypes (1a, 1b, 2, 3, 4) (Figure 3D and Supplemental Table 2), confirming that HLCs are permissive to infection with authentic HCV particles of different genotypes. Moreover, we observed propagation of HCV infection from infected “relocalized” cells to adjacent cells (Figure 3E), suggesting possible direct cell-to-cell transmission of HCV, as has been described previously in Huh7 cells (37, 38).
We also demonstrated that the HLCs responded to HCV infection by mounting an interferon response, as demonstrated by an early induction of CXCL10 (also known as IP-10), followed by IL-28B (also known as IFNL3) (Figure 3F), as observed in PHHs upon HCV infection (24). However, the percentage of infected cells was probably too low to detect IFN-α, IFN-β, or IL-28 proteins by ELISA in the supernatant. Small amount of CXCL10 protein could be detected in the supernatant of infected cells (data not shown).
Engraftment of HLCs in MUP-uPA/SCID/Bg mice.
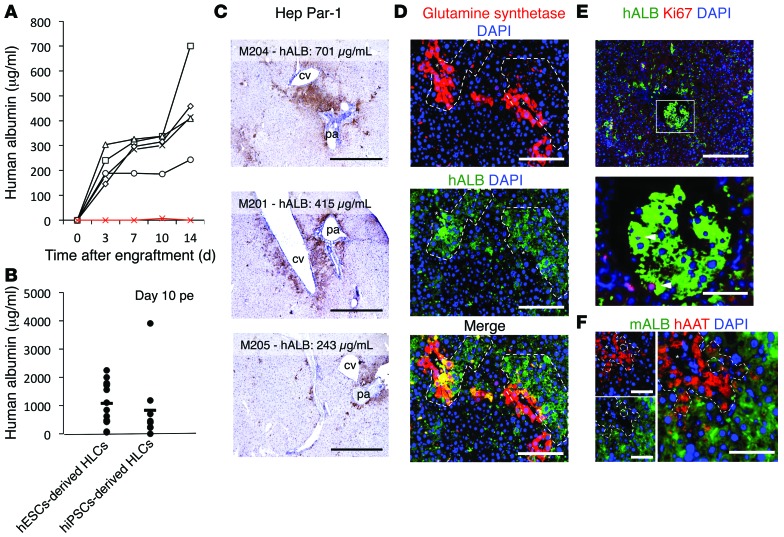
Four million hiPSC-derived HLCs were injected into the spleens of MUP-uPA/SCID/Bg mice (26, 29). Human albumin (hALB) was then monitored by ELISA in the sera of 5 engrafted mice (black lines) and 1 control nonengrafted mouse (red line) for 2 weeks post-engraftment (pe) (Figure 4A). hALB was detectable in every injected mouse as early as day 4 pe and increased during the 2 weeks pe. A total of 25 more mice were engrafted with 4 million hESC- or hiPSC-derived HLCs. hALB was detectable in every injected mouse by day 10 pe, confirming engraftment of HLCs. It is noteworthy that the concentration at day 10 pe varied from 50 to 3,900 μg/ml, but no statistical difference could be seen between hESC- or hiPSC-derived cells (Figure 4B).
Figure 4. Engraftment of HLCs in the liver parenchyma of engrafted MUP-uPA/SCID/Bg mice.
(A) hALB quantification in the sera of engrafted mice within 2 weeks after intrasplenic injection of 4 million hiPSC-derived HLCs. (B) ELISA for hALB at day 10 pe did not reveal a significant difference between hESC- and hiPSC-derived HLCs. Horizontal bars indicate the mean. (C) Visualization of engrafted HLCs 14 days pe by IHC for Hep Par-1. cv, central vein; pa, portal area. Scale bars: 1 mm. (D) IFA for hALB and glutamine synthetase confirms engraftment around central veins. Scale bars: 200 μm. (E) Staining for Ki67 in the nuclei of cells positive for hALB confirms proliferation of engrafted HLCs (arrowheads). Scale bar: 200 μm (top); 50 μm (bottom). (F) Staining for hAAT and mouse ALB in distinct cells confirms the absence of cell fusion between human HLCs and mouse hepatocytes. Scale bars: 100 μm. Asterisks indicate central veins.
At day 14 pe, clusters of engrafted cells could be visualized by immunohistochemistry (IHC) for human Hep Par-1 (Figure 4C) and immunofluorescence assay (IFA) for hALB (Figure 4, D and E). At this time, the percentage of liver repopulation is between 1% and 7% of the liver parenchyma and correlates with serum hALB titers. Tissue morphology after hematoxylin staining (Figure 4C) and immunostaining for glutamine synthetase (Figure 4D) showed that engraftment mainly occurred around central veins and to a lesser extent around portal areas. Nuclear staining for the proliferation marker Ki67 in hALB-positive HLCs confirmed that the engrafted HLCs proliferate in the mouse parenchyma (Figure 4E). Importantly, costaining for human AAT (hAAT) and mouse ALB visualized in different cells confirmed the absence of fusion of the engrafted HLCs with mouse hepatocytes (Figure 4F).
Maintenance of engrafted HLCs.
Importantly, a detectable level of hALB in the sera of engrafted mice was maintained for more than 100 days pe, confirming long-term engraftment of HLCs (Figure 5A). To better examine the kinetics of hALB production in these engrafted mice, we showed the time course of serum hALB in individual mice (Supplemental Figure 5A). hALB was never detectable in the sera of control nonengrafted mice.
Figure 5. Maintenance of HLCs in the liver parenchyma of engrafted mice.
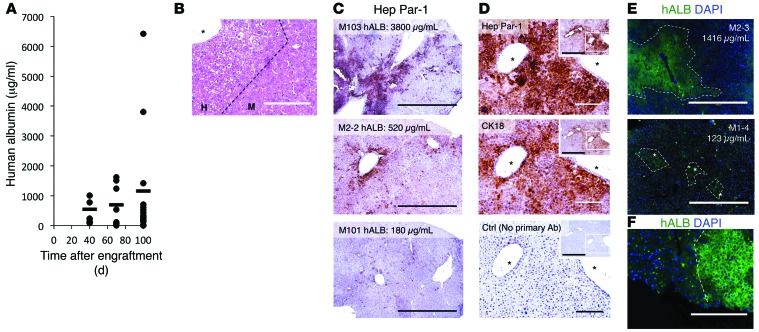
(A) ELISA for hALB 40, 70, and 100 days pe. For individual hALB ELISAs, see Supplemental Figure 5A. Horizontal bars indicate the mean. (B) Liver sections of MUP-uPA/SCID/Bg mice, 100 days pe, analyzed by H&E staining, showing white cells believed to be human engrafted cells. Scale bar: 200 μm. (C) Three representative IHC stainings for Hep Par-1 in mice showing high, average, and low concentrations of hALB 100 days pe. Scale bar: 1 mm. (D) IHC with antibodies anti–human hepatocyte-specific antigen (Hep Par-1) and CK18 and control without primary antibody. Scale bar: 200 μm; 1,000 μm (insets). (E) Immunostaining for hALB visualized using an inverted fluorescence microscope on sections of mouse liver 100 days pe. Scale bar: 1,000 μm. (F) Visualization of the difference in cell density and size between human and mouse cells. Scale bar: 200 μm. Asterisks indicate central veins.
One hundred days pe, H&E staining of sections of engrafted liver revealed large areas of hypoeosinophilic cells, believed to be engrafted human cells (Figure 5B). IHC assays with the Hep Par-1 antibody showed engrafted human HLCs in all engrafted mice. The percentage of repopulation varied from mouse to mouse, from scattered single cells (<1%) to large areas of positive cells constituting up to 15% to 20% of the liver parenchyma (Figure 5C and Supplemental Figure 5B for pictures of whole sections). Importantly, the percentage of engraftment correlated with the concentration of hALB in the sera of the mice 100 days pe (Figure 5, A and C). Hep Par-1–positive–engrafted human hepatocytes could also be detected with anti–cytokeratin 18 (anti-CK18) (Figure 5D). Human cells could be visualized by IFA for hALB (Figure 5E), from small colonies to large areas of the liver parenchyma, consistent with IHC, mainly around central veins (Supplemental Figure 5C). Interestingly, hALB-positive areas often exhibited a distinct histological morphology, with smaller polygonal cells, smaller nuclei, and higher cell density (Figure 5F), similar to what was observed after H&E staining (Figure 5B). On day 100 pe, some of the hALB-positive cells could be stained for proliferation marker Ki67, indicating that HLCs continued to proliferate in the mouse liver parenchyma 100 days pe (Supplemental Figure 5D, arrows and inlay). Sections of nonengrafted control liver did not show areas of lighter-stained cells after H&E staining and were negative for human Hep Par-1, CK18, and hALB (Supplemental Figure 5E).
In situ maturation of engrafted HLCs.
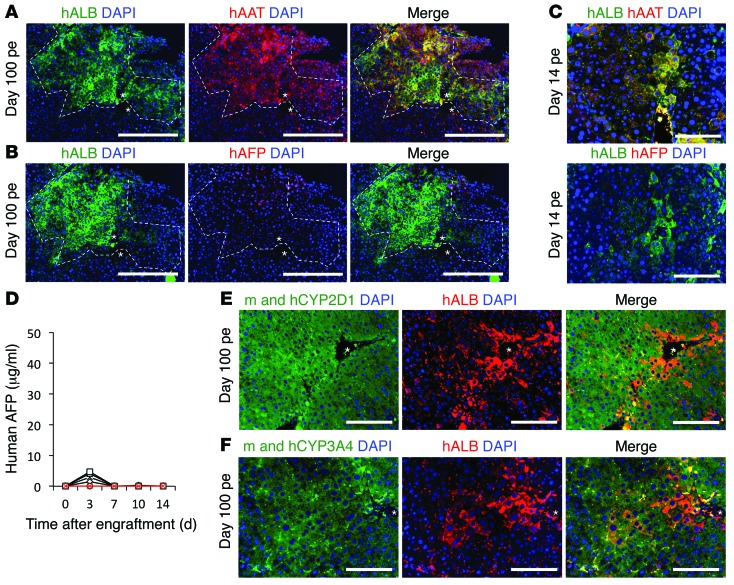
We performed coimmunostaining on sections of engrafted liver at day 14 and 100 pe and compared them to the HLCs used for the engraftment. HLCs at the time of engraftment were positive for human AFP (hAFP), hAAT, and hALB (Figure 1D and Supplemental Figure 2A). However, after engraftment, all engrafted hALB-positive cells were positive for hAAT expression (Figure 6A and Supplemental Figure 6A) but negative for hAFP expression (Figure 6B and Supplemental Figure 6A), similar to engrafted PHHs (Supplemental Figure 6B) (see Supplemental Figure 6C for positive control of AFP staining on section of human fetal liver). In situ IFA (Figure 6C) and hAFP ELISA assay (Figure 6D) during the first 2 weeks pe showed that hAFP expression was lost during the first week pe. Nonengrafted control liver tissues and sera were negative for hALB, hAAT, and hAFP (data not shown).
Figure 6. In situ maturation of engrafted HLCs.
(A and B) Coimmunostaining for hALB, hAAT, and hAFP on liver sections of engrafted mice 100 days pe, visualized by confocal microscopy. Asterisks indicate central veins. Original magnification, ×10; scale bar: 400 μm. Images at ×20 magnification are shown in Supplemental Figure 6. (C) IFA for hALB, hAFP, and hAAT 14 days pe. Scale bar: 200 μm. (D) ELISA for hAFP in sera of engrafted mice during the first 2 weeks pe (black lines) and control nonengrafted mice (red line). (E and F) Coimmunostaining for hALB and human and mouse CYP450 isoforms, confirming expression of mature hepatic markers in engrafted HLCs. Scale bars: 200 μm. Asterisks indicate central veins.
In vitro HLCs were largely negative for isoforms of CYP450 (Supplemental Figure 2B). Here, we show that engrafted hALB-positive HLCs can be stained for different cytochrome P450 isoforms, which are markers of mature hepatocytes (Figure 6, E and F). The CYP antibodies recognized both human and mouse isoforms and were expressed only in the liver parenchyma cells. Interestingly, engrafted HLCs on day 14 pe were already positive for CYPs (Supplemental Figure 7A), like engrafted PHHs (Supplemental Figure 7B), confirming that the maturation of the human cells occurred within 2 weeks pe in the liver parenchyma. Together, these results demonstrate that engrafted HLCs underwent maturation in situ.
HCV infection of engrafted HLCs in vivo.
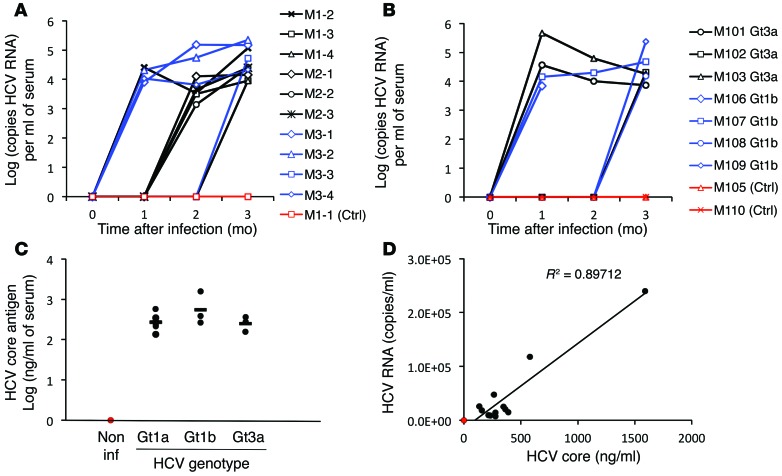
We investigated the permissiveness of the engrafted HLCs to HCV infection in vivo. HLC-engrafted mice were inoculated by tail vein injection with 100 μl diluted plasma from chimpanzees chronically infected with HCV genotype 1a (strain HC-TN) (26). At 1, 2, and 3 months pi, 100 μl sera were harvested and assessed for HCV RNA titer by RTqPCR (Figure 7A). When inoculated with a low dose of HCV (30 chimpanzee infectious dose 50 [CID50]), only 1 of 6 mice exhibited detectable HCV RNA 1 month pi. On the other hand, inoculation with high titer of HCV (1,000 CID50, experiment M3, blue line) led to detectable HCV RNA 1 month pi in 75% of the inoculated mice (Figure 7A). Interestingly, HCV RNA could not be detected 2 weeks after infection, no matter what infectious dose was inoculated (data not shown). However, HCV RNA was readily detectable in the blood of all infected engrafted mice 3 months pi, with viral titers varying from 3.95 to 5.17 log (copies HCV RNA/ml of sera), consistent with long-term establishment of HCV infection. As expected, HCV RNA was not detected in the sera of control noninfected engrafted mice.
Figure 7. HCV infection of engrafted HLCs.
Two weeks pe, chimeric mice were inoculated with HCV+ plasma of genotype 1a, 1b, or 3a. (A and B) HCV RNA quantified by RTqPCR, 1, 2, and 3 months after inoculation, in the sera of mice inoculated with (A) diluted chimpanzee sera containing 30 (black) or 1,000 (blue) CID50 HCV of genotype 1a (strain HC-TN) or with (B) chimpanzee sera of HCV of genotype 1b (strain CG1B) (blue) or genotype 3a (strain S52) (black) (100 CID50). Control noninoculated engrafted mice are depicted in red. Individual lines represent individual mice. (C) HCV core antigen assessed by ELISA in the sera of the mice 3 months after inoculation. Horizontal bars indicate the mean. Non inf, noninfected. (D) Correlation between HCV RNA titers and HCV core antigen concentrations in the sera of mice 3 months pe.
Engrafted HLCs were also permissive to infection with sera from chimpanzees infected with genotype 1b (strain CG1B) or 3a (strain S52) (100 CID50) (Figure 7B). More than half the mice showed detectable HCV RNA as early as 1 month pi, and all of them were positive at 3 months pi, with titers between 3.86 and 5.38 log (copies HCV RNA/ml). At 3 months pi, HCV core antigen was detectable in the sera of all infected mice by ELISA (Figure 7C) and correlated with HCV RNA titers pi (Figure 7D). Only mice with hALB over 450 μg/ml at the time of inoculation showed detectable HCV RNA 1 month pi, but in those mice with positive HCV RNA 1 month pi, there was no statistically significant correlation between hALB levels before inoculation and HCV RNA titers (Supplemental Figure 8A). There was also no correlation between HCV RNA and hALB 3 months pi (Supplemental Figure 8B). Attempts to visualize HCV proteins core, E2, and NS5A by immunostaining on sections of infected liver were not conclusive, consistent with previous observations of a low in situ level of HCV replication and a high level of autofluorescence of the liver parenchyma (39). These results demonstrate that HLCs, efficiently engrafted in the liver parenchyma of MUP-uPA/SCID/Bg mice, can be persistently infected in vivo with HCV of different genotypes.
Discussion
The natural history of HCV infection and response to antiviral treatment varies from patient to patient, and host genetic factors may play an important role in these differences. The most widely used cell culture model of HCV infection is based on subclones of the Huh7 cell line (19), which is derived from one individual and thereby is not useful for investigating the influence of patient-specific genetic background on the HCV infection. Productive in vitro HCV infection has also been achieved in PHHs (23), but several limitations (availability, invasive procedure, short window of time for cell culture) make them difficult to use for routine research purposes. It has been shown that human somatic cells can be reprogrammed into a stem cell–like state (hiPSC) and differentiated into mature HLCs (6–11). This approach may hold the key to more relevant models to investigate the link between patient genetic background and HCV infection, pathology, and treatment. Here, we demonstrate that both hESCs and fibroblast-derived hiPSCs can be efficiently differentiated into functional HLCs with many hepatocyte-specific functions. These HLCs do maintain some characteristics of immature fetal hepatocytes, such as expression of AFP and CYP3A7. However, despite these limitations, the differentiated cells express various CYP450 isoforms and reproduce key features of mature hepatocytes, such as glycogen and lipid storage, lipoprotein internalization, and indocyanine green metabolism. More importantly, the HLCs exhibit a level of hepatic differentiation high enough to be infected by HCV, a hepatotropic virus notoriously difficult to culture in vitro (40, 41). Consistent with previous publications (27, 28), we demonstrate that HLCs can be infected with HCVcc in vitro at a low level.
The reason for this low level of viral replication in HLCs may be several-fold. Initially, the virus most widely used for research (JFH1-HCVcc) (19) is produced and highly adapted to the Huh7 cell line, is limited to the genotype 2a (JFH1) replicating machinery, and poorly reproduces the characteristic of the natural virus found in the blood of HCV-infected patients in terms of specific infectivity and buoyant density (23, 42, 43). Using sera from HCV-infected patients, we demonstrated that the HLCs are also permissive to infection with authentic HCV of genotype 1a, 1b, 2, 3, and 4. HCV infection also triggers an antiviral innate immune response. In highly permissive Huh7 subclones, the high level of replication is a result of defects in the RIG-1 and TLR pathways, two key actors of the innate immunity pathways (44, 45). These pathways are expected to be functional in our HLCs. HCV-infected PHHs can mount an effective intrinsic innate immunity, with production of interferon and induction of antiviral genes, thereby limiting HCV replication and spread (24). Upon infection of HLCs with HCV, we detected an increased expression of CXCL10 and IL-28B. Thus, this model is a valuable tool to study the innate immune response to HCV infection.
Finally, the low replication of the virus in HLCs may be linked to the less matured phenotype of the HLCs, as compared with that of primary adult hepatocytes (PHHs). We confirmed that proviral host factors are expressed in our HLCs but at lower levels than those of PHHs. Interestingly, the infectious titers in the supernatant of infected HLCs were low compared with the intracellular HCV RNA level, suggesting a relative deficiency in assembly and/or secretion of infectious progeny viruses by the HLCs. Assembly and secretion of progeny virions involve the lipoproteins secretion pathway. While we confirmed that our HLCs expressed high levels of APOB and APOE, the secretion of APOB100-containing lipoprotein was lower than that described for PHHs.
Different strategies have been used to improve the level of differentiation of the HLCs in vitro, such as the use of special matrix, 3D culture, spheroids formation, or coculture with stromal cells (46–52). However, none of them so far allowed maturation of the HLCs at a level similar to that of PHHs. We therefore hypothesized that engraftment of HLCs in the native liver niche, a complex 3D multicellular environment, may allow further maturation of the cells.
Several articles reported engraftment of stem cell–derived HLCs in the livers of various models of transgenic mice (9, 12–16). While one study did not report any engraftment using stem cell–derived hepatocytes in Alb-uPA Rag2–/– γc–/– mice (12), other studies showed low levels of repopulation in induced hepatotoxicity models (9, 10, 13) and in Alb-uPA SCID mice (9). In contrast, 2 studies showed 8%–15% (14) and up to 24% repopulation (15) after transplantation of hESCs or hiPSC-derived HLCs in 2 mouse models of induced hepatotoxicity. Recently, coculture of hiPSC-derived hepatocytes with human umbilical vein endothelial cells and human mesenchymal stem cells has been described to result in formation of 3D “liver buds” in vitro that could be efficiently engrafted and vascularized (53). Engraftment of spheroid-forming HLCs into CCl4-injured mice has also been described (54). Finally, two studies showed that human induced hepatocytes, transdifferentiated from human fibroblasts, could be engrafted in the liver parenchyma of transgenic mice and repopulate up to 0.3% to 4.2% of the liver parenchyma of Fah–/– Rag2–/– mice (55) and up to 30% in Tet-uPA Rag2–/– γc–/– mice (56).
Here, we describe engraftment of hESC- and hiPSC-derived HLCs in the liver parenchyma of the MUP-uPA/SCID/Bg mouse model. The MUP-uPA/SCID/Bg model is relatively easy to use for human hepatocyte engraftment (26, 29). Healthy mice are used in this model, which allows a longer window for engraftment compared with the more commonly used ALB-uPA/SCID model. In this model, the differentiated cells were maintained for more than 100 days in the liver parenchyma. The efficiency of repopulation at day 100 pe varied from about 1% to a maximum of 15% to 20% of the liver parenchyma. Concentrations of hALB in the sera of engrafted mice also varied from 100 to 6,415 μg/ml but correlated with percentages of repopulation. The percentage of repopulation is similar to that in some of the published results using human stem cell–derived HLCs (14, 15); however, the hALB titers in our MUP-uPA/SCID/Bg mice are surprisingly high. Only 2 studies, using hiPSC-derived HLCs in a chemically induced hepatotoxicity model (13) or transdifferentiated human induced hepatocytes in a transgene-induced toxicity model (56), reported concentrations of hALB at levels close to ours. The reasons for these differences are not clear and may be explained by different protocols and models used.
One difference in our protocol is the use of the MUP-uPA/SCID/Bg mouse model. Previous studies showed that this model allows high repopulation by PHHs — an optimal number of 4 million PHHs could lead to an average of 40% repopulation and to an average hALB titer of 1,750 μg/ml after intrasplenic injection in mice aged 5–8 months (26). In comparison, HLCs are somewhat less efficient than PHHs for engraftment in the MUP-uPA/SCID/Bg model.
Importantly, our HLC population at day 14 of differentiation was heterogeneous, mainly composed of hALB+hAAT+hAFP+ hepatocytes, but also contained hALB–hAAT+hAFP+ hepatoblasts and hAFP+hAAT+hCK7+ biprogenitor cells. It is not clear whether these heterogeneous cells have a different ability to engraft or be maintained in the liver parenchyma, but this approach differs from that of previous studies engrafting purified, homogeneous HLC populations (9, 15). Interestingly, a previous study demonstrated that hiPSC-derived cells at a different stage of hepatic differentiation have different abilities to repopulate the liver parenchyma of NSG mice, with more matured cells repopulating less efficiently (14). In our experiments, we did not see a correlation between global hepatic maturation (assessed by ELISA for hAFP and hALB at day 14 of differentiation) and efficiency of engraftment (assessed by ELISA for hALB 10 days pe) or long-term maintenance (hALB ELISA at day 100 pe) in the mouse liver parenchyma (data not shown). At this point, we can only hypothesize that the level or nature of maturation of our cells plays a role in the efficiency of engraftment or maintenance in vivo. The absence of quantitative and defined criteria to assess hepatic maturation and cellular homogeneity makes it difficult to compare quality of differentiated cells from study to study.
Increasing hALB concentration during the 14 days pi, higher level of albumin in some mice 100 days pe, and immunostaining for Ki67 at day 14 and 100 suggest that HLCs are capable of proliferation once engrafted in the liver parenchyma, but this ability varies from experiment to experiment.
The individual profiles of secretion of hALB show variable engraftment kinetics — some mice exhibited a high level of serum hALB at early time points followed by gradual decrease. Other mice, exhibiting a lower concentration of hALB on day 10 pe, showed a gradual increase during the next 3 months, similar to the kinetics of PHH-engrafted mice. However, all the mice but one, engrafted with hESC- or hiPSC-derived HLCS, exhibited detectable hALB titer for more than 100 days pe, confirming that long-term engraftment of HLCs is feasible. The reason for this variability is not totally clear. Decrease of some of the hALB titers suggests death of engrafted HLCs. Analyzing the environmental and cellular factors influencing the survival or death of engrafted cells would be of particular interest to understand and achieve long-term engraftment of transplanted human hepatocytes in the liver parenchyma.
Our data suggest that the quality of the cell preparation may be important for the engraftment kinetics. Four mice (mice 1-1 to 1-4; see Supplemental Figure 5A) were engrafted with the same lot of HLCs and showed decreasing hALB titers at late time points. However, cell quality was not the only reason, as cells from a same preparation achieved different fates after engraftment in mice of a same litter (for example, M101/102 compared with M103/105; see Supplemental Figure 5A). To be noted, we didn’t see correlation between repopulation efficiency and sex or age of the mice at time of the engraftment (data not shown).
More importantly, we demonstrated that engrafted stem cell–derived HLCs undergo further maturation once in the hepatic niche, as described recently with transdifferentiated hepatocytes (56) and iMPC-Heps (57). While in vitro HLCs always expressed high levels of both fetal (hAFP) and mature (hALB, hAAT) hepatocyte markers, all engrafted cells lost expression of hAFP. ELISA assay at early time points revealed loss of AFP within the first week pe. After engraftment HLCs became positive for CYP450 isoforms, markers of mature hepatocytes, confirming that the cells acquired a more mature phenotype in the liver parenchyma within 2 weeks pe. Importantly, the in situ maturation of the cells could be associated with a higher expression and production of hALB by the cells, which could be partly accountable for the rapid increase of serum hALB titers after engraftment.
Because mice were injected with a heterogeneous cell population, we looked for appearance of teratomas at the time of sacrifice. No teratomas were detected in mice sacrificed 100 days pe. Recently, a study showed that hepatocytes generated from human fibroblasts, functionally similar to HLCs, could be safely engrafted in the livers of FAG mice (57) by directly differentiating the fibroblast-derived hiPSCs toward definitive endoderm, thus bypassing the pluripotent stage.
We demonstrated here for the first time that engrafted human HLCs are permissive to HCV infection in vivo. Interestingly, after inoculation with HCV-positive sera of different genotypes, none of the mice exhibited a detectable level of HCV RNA in the serum at 2 weeks pi, and only about half of them exhibited a detectable level at 1 month pi. This is in contrast with what has been described after inoculation of chimeric mice engrafted with PHHs (25, 26, 58, 59), where all the engrafted mice inoculated with HCV exhibit detectable viral titers at early time points. Our data show that only mice with hALB titers over 450 μg/ml at the time of HCV inoculation, equivalent to a liver repopulation around 5%, had detectable HCV RNA 1 month pi. This observation is consistent with what has been observed in the HCV-infected, PHH-engrafted Fah–/– Rag2–/– Il2rg–/– mouse (58).
Three months pi, all the inoculated HLC-engrafted mice showed high serum titers of HCV RNA (4–6 log [copies HCV RNA/ml]). The reason for this delayed appearance of detectable HCV RNA in some of the HLC-engrafted mice is not clear but may be linked to an expansion of the engrafted HLCs or to a higher level of maturation. However, at day 100 pe, HCV RNA did not correlate with the concentration of hALB, suggesting that the level of repopulation is not a limiting factor to HCV spreading and that a higher level of repopulation is not enough to attain high viral titer.
This in vivo model of HCV infection shows long-term infection. The viral titers after in vitro infection tend to drop after 2 weeks (data not shown), associated with a dedifferentiation of the HLCs with long-term cell culture. It is similar to what has been described with PHHs. Here, the engrafted HLCs maintained a high level of maturation and detectable level of HCV RNA for more that 3 months. We believe this novel in vivo system constitutes a unique model for studying long-term HCV infection of HLCs.
Genome-wide association studies have demonstrated that genetic polymorphisms near the IL28B gene are highly associated with spontaneous virus clearance (60) and response to antiviral treatment (61) in patients infected with HCV. However, in vitro studies of these mechanisms are limited by the lack of a relevant in vitro model to study HCV in the context of the patient’s genetic background. Here, for the first time, we demonstrate that hepatocytes differentiated from patient-specific hiPSCs can be infected both in vitro and in vivo by HCV. Our approach represents a pertinent and valuable model to study HCV infection and subsequent innate immune response in the context of patient-specific genetic background.
Methods
Hepatic differentiation, characterization of differentiated cells, and in vitro HCV infection of HLCs.
Detailed protocols for hepatic differentiation of hESCs and hiPSCs, characterization of HLCs, and in vitro HCV infection and monitoring are provided in the Supplemental Methods and Supplemental Tables 3–5.
Engraftment of HLCs in MUP-uPA/SCID/Bg mice.
HLCs were incubated for 10 minutes at 37°C in a solution of 0.25% Trypsin and 2.21 mM EDTA in HBSS (Cellgro), further dissociated by pipetting, and washed twice in PBS (centrifugation 50 g) for 10 minutes at 4°C). Resuspended cell pellet was filtered through a 40-μm nylon mesh to remove nondissociated cells. Trypan blue exclusion test was performed: only cell preparations with viability over 60% were used for engraftment. After one more wash, cells were resuspended in HBSS (Vedco) at a final concentration of 20 million living cells per ml.
Engraftment was performed following the procedure based on our experience with PHHs (26). Only heterozygotes MUP-uPA/SCID/Bg mice between 5 and 8 months old were used, as this has been described as the best age to ensure the most optimal engraftment with PHHs. Briefly, after anesthesia with isoflurane inhalation, a short incision (<1 cm) was performed in the upper left quadrant of the abdomen of the recipient mouse. The dark spleen could be visualized through the intact peritoneum. 4 million HLCs (the optimal number of cells when injecting PHHs; ref. 26), in a maximum volume of 200 μl HBSS, were injected with a 27-gauge needle directly into the spleen. Control nonengrafted mice were injected with 200 μl HBSS. The skin was finally sutured. Details about mice, cell type, and the differentiation protocol used for the engraftments are provided in Supplemental Table 5.
At different time points after engraftment, mice were bled from the tail veins, and sera was collected after centrifugation. 30 μl sera were used to measure the concentration of hALB (Bethyl Laboratories) and hAFP (Calbiotech) by ELISA according to the manufacturers instructions.
Mice were sacrificed at day 14 or 100 pe, and liver tissues were collected. Liver tissues were fixed with 4% phosphate-buffered formalin overnight, processed, and embedded in paraffin. Sections were routinely stained with Eosin and Hematoxylin (both from Sigma-Aldrich) to monitor the liver parenchyma architecture and visualize the human cells (appearing white after eosin staining). Detailed protocols and antibody information for in situ IHC and IFAs are provided in the Supplemental Methods.
HCV in vivo infection of chimeric mice.
The chimeric mice were inoculated intravenously with 100 μl diluted plasma from chimpanzees infected with genotype 1a (strain H), 1b (strain CG1b) (62), and 3a (strain S52) (30 or 1,000 CID50 per ml of genotype 1a, 100 CID50 per ml for genotype 1b and 3a). The infectivity of genotype 1a and 3a has been previously demonstrated in MUP-uPA/SCID/Bg mice engrafted with PHHs (26).
Serum samples were obtained by tail bleeding at day 30, 60, and 90 after inoculation. Viral RNA was isolated using the QIAamp UltraSens Virus Kit (QIAGEN) and assessed for HCV RNA by RTqPCR as described previously (21, 63, 64). HCV core antigen was assessed using the QuickTiter HCV Core Antigen ELISA Kit (Cell Biolabs) according to the manufacturer’s instructions.
Statistics.
In vitro data are expressed as average of representative experiment, and 2-tailed Student’s t test was performed to assess statistical significance. The results from in vivo experiments are presented for each individual animal. A P value of less than 0.05 was considered significant.
Study approval.
All engraftment and animal procedures were performed according to the NIH guidelines for animal care and were approved by the Office of Animal Care and Use, Bethesda, Maryland, USA (protocol NIDDK-K090-LDB-13) and the Center for Biologics Evaluation and Research/FDA Institutional Animal Care and Use Committee, Bethesda, Maryland, USA (animal study protocol no. 2001-42).
Supplementary Material
Acknowledgments
We wish to thank Ronda Sapp, Frank Qisheng Li, Zhensheng Zhang, Zongyi Hu, Emmanuel Thomas, and Chen Du (all from NIDDK); Marian E. Major (FDA); Stephen Strom (Karolinska Institute); Hengli Tang (University of Florida); and the Flow Cytometry Core of National Heart, Lung, and Blood Institute for technical assistance and helpful discussions. This work was supported by the Intramural Research Program of the NIDDK (Z01 DK54504 and DK54505) and the NIH Center for Regenerative Medicine. PHHs were provided by the NIH-funded Liver Tissue Procurement and Cell Distribution System (N01-DK-7-0004/HHSN26700700004C).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2014;124(11):4953–4964. doi:10.1172/JCI75456.
References
- 1.Gearhart J. New potential for human embryonic stem cells. Science. 1998;282(5391):1061–1062. doi: 10.1126/science.282.5391.1061. [DOI] [PubMed] [Google Scholar]
- 2.Irion S, Nostro MC, Kattman SJ, Keller GM. Directed differentiation of pluripotent stem cells: from developmental biology to therapeutic applications. Cold Spring Harb Symp Quant Biol. 2008;73:101–110. doi: 10.1101/sqb.2008.73.065. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Lowry WE, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A. 2008;105(8):2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 6.Cai J, et al. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology. 2007;45(5):1229–1239. doi: 10.1002/hep.21582. [DOI] [PubMed] [Google Scholar]
- 7.Agarwal S, Holton KL, Lanza R. Efficient differentiation of functional hepatocytes from human embryonic stem cells. Stem Cells. 2008;26(5):1117–1127. doi: 10.1634/stemcells.2007-1102. [DOI] [PubMed] [Google Scholar]
- 8.Hay DC, et al. Highly efficient differentiation of hESCs to functional hepatic endoderm requires Activin a and wnt3a signaling. Proc Natl Acad Sci U S A. 2008;34(105):12301–12306. doi: 10.1073/pnas.0806522105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basma H, et al. Differentiation and transplantation of human embryonic stem cell-derived hepatocytes. Gastroenterology. 2009;136(3):990–999. doi: 10.1053/j.gastro.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Si-Tayeb K, et al. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51(1):297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sullivan GJ, et al. Generation of functional human hepatic endoderm from human induced pluripotent stem cells. Hepatology. 2010;51(1):329–335. doi: 10.1002/hep.23335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haridass D, et al. Repopulation efficiencies of adult hepatocytes, fetal liver progenitor cells, and embryonic stem cell-derived hepatic cells in albumin-promoter-enhancer urokinase-type plasminogen activator mice. Am J Pathol. 2009;175(4):1483–1492. doi: 10.2353/ajpath.2009.090117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asgari S, Moslem M, Bagheri-Lankarani K, Pournasr B, Miryounesi M, Baharvand H. Differentiation and transplantation of human induced pluripotent stem cell-derived hepatocyte-like cells. Stem Cell Rev. 2013;9(4):493–504. doi: 10.1007/s12015-011-9330-y. [DOI] [PubMed] [Google Scholar]
- 14.Liu H, Kim Y, Sharkis S, Marchionni L, Jang YY. In vivo liver regeneration potential of human induced pluripotent stem cells from diverse origins. Sci Transl Med. 2011;3(82):82ra39. doi: 10.1126/scitranslmed.3002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woo DH, et al. Direct and indirect contribution of human embryonic stem cell-derived hepatocyte-like cells to liver repair in mice. Gastroenterology. 2012;142(3):602–611. doi: 10.1053/j.gastro.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 16.Li HY, et al. Reprogramming induced pluripotent stem cells in the absence of c-Myc for differentiation into hepatocyte-like cells. Biomaterials. 2011;32(26):5994–6005. doi: 10.1016/j.biomaterials.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Ioannou GN, Boyko EJ, Lee SP. The prevalence and predictors of elevated serum aminotransferase activity in the United States in 1999-2002. Am J Gastroenterol. 2006;101(1):76–82. doi: 10.1111/j.1572-0241.2005.00341.x. [DOI] [PubMed] [Google Scholar]
- 18.Liang TJ, Ghany MG. Current and future therapies for hepatitis C virus infection. N Engl J Med. 2013;368(20):1907–1917. doi: 10.1056/NEJMra1213651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wakita T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11(7):791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heller T, et al. An in vitro model of hepatitis C virion production. Proc Natl Acad Sci U S A. 2005;102(7):2579–2583. doi: 10.1073/pnas.0409666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato T, et al. Production of infectious hepatitis C virus of various genotypes in cell cultures. J Virol. 2007;81(9):4405–4411. doi: 10.1128/JVI.02334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ploss A, et al. Persistent hepatitis C virus infection in microscale primary human hepatocyte cultures. Proc Natl Acad Sci U S A. 2010;107(7):3141–3145. doi: 10.1073/pnas.0915130107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Podevin P, et al. Production of infectious hepatitis C virus in primary cultures of human adult hepatocytes. Gastroenterology. 2010;139(4):1355–1364. doi: 10.1053/j.gastro.2010.06.058. [DOI] [PubMed] [Google Scholar]
- 24.Thomas E, et al. HCV infection induces a unique hepatic innate immune response associated with robust production of type III interferons. Gastroenterology. 2012;142(4):978–988. doi: 10.1053/j.gastro.2011.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mercer DF, et al. Hepatitis C virus replication in mice with chimeric human livers. Nat Med. 2001;7(8):927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- 26.Tesfaye A, et al. Chimeric mouse model for the infection of hepatitis B and C viruses. PLoS One. 2013;8(10):e77298. doi: 10.1371/journal.pone.0077298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu X, et al. Productive hepatitis C virus infection of stem cell-derived hepatocytes reveals a critical transition to viral permissiveness during differentiation. PLoS Pathog. 2012;8(4):e1002617. doi: 10.1371/journal.ppat.1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz RE, et al. Modeling hepatitis C virus infection using human induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012;109(7):2544–2548. doi: 10.1073/pnas.1121400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weglarz TC, Degen JL, Sandgren EP. Hepatocyte transplantation into diseased mouse liver. Kinetics of parenchymal repopulation and identification of the proliferative capacity of tetraploid and octaploid hepatocytes. Am J Pathol. 2000;157(6):1963–1974. doi: 10.1016/S0002-9440(10)64835-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sommer CA, et al. Excision of reprogramming transgenes improves the differentiation potential of iPS cells generated with a single excisable vector. Stem Cells. 2010;28(1):64–74. doi: 10.1002/stem.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang H, et al. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc Natl Acad Sci U S A. 2007;104(14):5848–5853. doi: 10.1073/pnas.0700760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang KS, Jiang J, Cai Z, Luo G. Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J Virol. 2007;81(24):13783–13793. doi: 10.1128/JVI.01091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gastaminza P, et al. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J Virol. 2008;82(5):2120–2129. doi: 10.1128/JVI.02053-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Q, et al. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci U S A. 2009;106(38):16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Q, et al. Integrative functional genomics of hepatitis C virus infection identifies host dependencies in complete viral replication cycle. PLoS Pathog. 2014;10(5):e1004163. doi: 10.1371/journal.ppat.1004163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeLaForest A, et al. HNF4A is essential for specification of hepatic progenitors from human pluripotent stem cells. Development. 2011;138(19):4143–4153. doi: 10.1242/dev.062547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones CT, et al. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotech. 2010;28(2):167–171. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brimacombe CL, et al. Neutralizing antibody-resistant hepatitis C virus cell-to-cell transmission. J Virol. 2011;85(1):596–605. doi: 10.1128/JVI.01592-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang Y, et al. Visualizing hepatitis C virus infections in human liver by two-photon microscopy. Gastroenterology. 2009;137(4):1448–1458. doi: 10.1053/j.gastro.2009.07.050. [DOI] [PubMed] [Google Scholar]
- 40.von Hahn T, McKeating JA. In vitro veritas? The challenges of studying hepatitis C virus infectivity in a test tube. J Hepatol. 2007;46(3):355–358. doi: 10.1016/j.jhep.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 41.Gottwein JM, Bukh J. Cutting the gordian knot-development and biological relevance of hepatitis C virus cell culture systems. Adv Virus Res. 2008;71:51–133. doi: 10.1016/S0065-3527(08)00002-X. [DOI] [PubMed] [Google Scholar]
- 42.Andre P, et al. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J Virol. 2002;76(14):6919–6928. doi: 10.1128/JVI.76.14.6919-6928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindenbach BD, et al. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc Natl Acad Sci U S A. 2006;103(10):3805–3809. doi: 10.1073/pnas.0511218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sumpter R., Jr et al. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79(5):2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foy E, et al. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci U S A. 2005;102(8):2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farzaneh Z, Pournasr B, Ebrahimi M, Aghdami N, Baharvand H. Enhanced functions of human embryonic stem cell-derived hepatocyte-like cells on three-dimensional nanofibrillar surfaces. Stem Cell Rev. 2010;6(4):601–610. doi: 10.1007/s12015-010-9179-5. [DOI] [PubMed] [Google Scholar]
- 47.Yu YD, et al. Hepatic differentiation from human embryonic stem cells using stromal cells. J Surg Res. 2011;170(2):e253–e261. doi: 10.1016/j.jss.2011.06.032. [DOI] [PubMed] [Google Scholar]
- 48.Ramasamy TS, Yu JS, Selden C, Hodgson H, Cui W. Application of three-dimensional culture conditions to human embryonic stem cell-derived definitive endoderm cells enhances hepatocyte differentiation and functionality. Tissue Eng Part A. 2013;19(3–4):360–367. doi: 10.1089/ten.tea.2012.0190. [DOI] [PubMed] [Google Scholar]
- 49.Nagamoto Y, et al. The promotion of hepatic maturation of human pluripotent stem cells in 3D co-culture using type I collagen and Swiss 3T3 cell sheets. Biomaterials. 2012;33(18):4526–4534. doi: 10.1016/j.biomaterials.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 50.Sivertsson L, Synnergren J, Jensen J, Bjorquist P, Ingelman-Sundberg M. Hepatic differentiation and maturation of human embryonic stem cells cultured in a perfused three-dimensional bioreactor. Stem Cell Dev. 2013;22(4):581–594. doi: 10.1089/scd.2012.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takayama K, et al. 3D spheroid culture of hESC/hiPSC-derived hepatocyte-like cells for drug toxicity testing. Biomaterials. 2013;34(7):1781–1789. doi: 10.1016/j.biomaterials.2012.11.029. [DOI] [PubMed] [Google Scholar]
- 52.Subramanian K, et al. Spheroid culture for enhanced differentiation of human embryonic stem cells to hepatocyte-like cells. Stem Cell Dev. 2014;23(2):121–131. doi: 10.1089/scd.2013.0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takebe T, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499(7459):481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 54.Kim SE, et al. Engraftment potential of spheroid-forming hepatic endoderm derived from human embryonic stem cells. Stem Cell Dev. 2013;22(12):1818–1829. doi: 10.1089/scd.2012.0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang P, et al. Direct reprogramming of human fibroblasts to functional and expandable hepatocytes. Cell Stem Cell. 2014;14(3):370–384. doi: 10.1016/j.stem.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 56.Du Y, et al. Human hepatocytes with drug metabolic function induced from fibroblasts by lineage reprogramming. Cell Stem Cell. 2014;14(3):394–403. doi: 10.1016/j.stem.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 57.Zhu S, et al. Mouse liver repopulation with hepatocytes generated from human fibroblasts. Nature. 2014;508(7494):93–97. doi: 10.1038/nature13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bissig KD, et al. Human liver chimeric mice provide a model for hepatitis C and B virus infection and treatment. J Clin Invest. 2010;120(3):924–930. doi: 10.1172/JCI40094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vanwolleghem T, et al. Factors determining successful engraftment of hepatocytes and susceptibility to hepatitis B and C virus infection in uPA-SCID mice. J Hepatol. 2010;53(3):468–476. doi: 10.1016/j.jhep.2010.03.024. [DOI] [PubMed] [Google Scholar]
- 60.Thomas DL, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461(7265):798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ge D, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461(7262):399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 62.Thomson M, et al. Emergence of a distinct pattern of viral mutations in chimpanzees infected with a homogeneous inoculum of hepatitis C virus. Gastroenterology. 2001;121(5):1226–1233. doi: 10.1053/gast.2001.28669. [DOI] [PubMed] [Google Scholar]
- 63.Thomas E, et al. Ribavirin potentiates interferon action by augmenting interferon-stimulated gene induction in hepatitis C virus cell culture models. Hepatology. 2011;53(1):32–41. doi: 10.1002/hep.23985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takeuchi T, et al. Real-time detection system for quantification of hepatitis C virus genome. Gastroenterology. 1999;116(3):636–642. doi: 10.1016/S0016-5085(99)70185-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.