Abstract
Osteoclastogenesis requires activation of RANK signaling as well as costimulatory signals from immunoreceptor tyrosine-based activation motif-containing (ITAM-containing) receptors/adaptors, predominantly tyrosine kinase–binding proteins DAP12 and FcRγ, in osteoclast precursors. It is not well understood how costimulatory signals are regulated and integrated with RANK signaling. Here, we found that osteopetrotic bone phenotypes in mice lacking DAP12 or DAP12 and FcRγ are mediated by the transcription factor RBP-J, as deletion of Rbpj in these mice substantially rescued the defects of bone remodeling. Using a TNF-α–induced model of inflammatory bone resorption, we determined that RBP-J deficiency enables TNF-α to induce osteoclast formation and bone resorption in DAP12-deficient animals. Thus, RBP-J imposes a requirement for ITAM-mediated costimulation of RANKL or TNF-α–induced osteoclastogenesis. Mechanistically, RBP-J suppressed induction of key osteoclastogenic factors NFATc1, BLIMP1, and c-FOS by inhibiting ITAM-mediated expression and function of PLCγ2 and activation of downstream calcium-CaMKK/PYK2 signaling. Moreover, RBP-J suppressed Plcg2 expression and downstream calcium oscillations indirectly by a TGF-β/PLCγ2/calcium axis. Together, our findings indicate that RBP-J suppresses ITAM-mediated costimulation, thereby limiting crosstalk between ITAM and RANK/TNFR signaling and allowing fine tuning of osteoclastogenesis during bone homeostasis and under inflammatory conditions. Furthermore, these data suggest that environmental cues that regulate RBP-J expression/function potentially modulate the requirement for costimulatory signaling for osteoclast differentiation and bone remodeling.
Introduction
Osteoclasts, derived from monocyte/macrophage precursors, are the exclusive bone-resorptive cells that play an important role in physiological skeleton development and homeostatic bone remodeling in concert with bone-forming osteoblasts/osteocytes (1–3). Abnormal generation and function of osteoclasts lead to either osteopetrosis or excessive bone resorption, such as occurs in osteoporosis and in inflammatory arthritis (4–6). In addition to the cellular survival factor macrophage colony-stimulating factor (M-CSF), osteoclastogenesis requires 2 essential signals induced by RANKL and costimulatory factors acting via their receptors RANK and immunoreceptor tyrosine-based activation motif–associated (ITAM-associated) receptors, respectively. Upon these stimulations, a broad range of signaling cascades is activated, such as canonical and noncanonical NF-κB pathways, MAPK pathways, calcium signaling, and the protein tyrosine kinases Bruton’s tyrosine kinase (BTK)/TEC. These signaling cascades mobilize a complex transcriptional network, inducing osteoclast-promoting factors such as NF-κB, AP-1, CREB, and BLIMP1 while downregulating repressors of osteoclast differentiation such as IRF8, MAFB, and BCL6, which function coordinately to induce the expression of “master regulator” nuclear factor of activated T cells c1 (NFATc1) and drive osteoclastogenesis (7–9). Although the requirement for ITAM-mediated costimulation for osteoclast differentiation is well established, it can be partially bypassed under certain conditions in vivo (10, 11). Thus, how costimulation is regulated and the mechanisms that determine the importance of costimulatory signaling remain unclear.
The main ITAM-containing adaptors expressed by myeloid osteoclast precursors are DNAX-activating protein 12 (DAP12) and Fc receptor common γ subunit (FcRγ). These adaptors associate with and mediate signaling by various receptors, including DAP12-associated triggering receptor expressed in myeloid cells 2 (TREM2) and signal-regulatory protein β 1 (SIRPβ1), FcRγ-associated osteoclast-associated receptor (OSCAR), paired immunoglobulin-like receptor-A (PIR-A), and FcRs. The ligands for many of these receptors are not known but appear to be constitutively expressed and mediate basal signaling. Ligation of ITAM-coupled receptors in myeloid cells leads to the phosphorylation of ITAM tyrosine residues by Src family kinases. The tyrosine kinase Syk is then recruited and activates signaling molecules, such as BTK/TEC, the scaffolding adaptor B cell linker (BLNK)/SLP76, and phospholipase-Cγ2 (PLCγ2). Phosphorylation of PLCγ2 increases its catalytic function and results in elevated intracellular calcium levels that activate calcium-mediated signaling pathways. Calcium-activated signaling molecules calcium/calmodulin-dependent protein kinase kinase (CaMKK), CaMK, protein tyrosine kinase 2 (PYK2; also known as PTK2B), and calcineurin further transmit calcium signaling to activate transcriptional factors, such as CREB, MEF2, and NFATc1, to induce various cellular activities (9, 12–17). Activation of PLCγ2 and downstream calcium-signaling pathways plays an essential role in NFATc1 induction during osteoclastogenesis (16, 18, 19).
Recent work has revealed an integration of RANK- and ITAM-signaling pathways by a multiprotein signaling complex comprising BTK/TEC/BLNK/SLP76/PLCγ2, which activates downstream calcium signaling and NFATc1 induction (19, 20). Deletion of PLCγ2 in mice results in an osteopetrotic phenotype due to a defect in osteoclastogenesis with impaired NFATc1 induction, demonstrating a key role for PLCγ2 in NFATc1 induction and osteoclast differentiation (19). Diminishment of ITAM-mediated costimulatory signaling in osteoclast precursors as occurs in Dap12–/–Fcrg–/– cells leads to attenuation of PLCγ2 activation followed by impaired NFATc1 induction and osteoclast differentiation even in the presence of RANKL (18, 19). Thus, ITAM-mediated costimulatory signals and PLCγ2 activity, which determine downstream calcium signaling, are required for osteoclast differentiation. RANK signaling induces a transient increase in PLCγ2 activity, but does not regulate its expression and is not required for basal PLCγ2 activity in osteoclast precursors. Instead, basal PLCγ2 activity, which has been observed in myeloid cells including osteoclast precursors (17, 21–24), appears to be maintained by tonic ITAM signaling mediated by engagement of ITAM-associated receptors by constitutively expressed ligands (13); consistently, addition of ligands/agonists for ITAM-associated receptors is not required for osteoclast differentiation in vitro. Importantly this basal level of costimulation and PLCγ2 activation together with RANKL is sufficient for NFATc1 induction and osteoclast differentiation (16, 18, 19). The ligands for FcRγ- and DAP12-associated receptors that mediate tonic signaling are suggested to be expressed by osteoblasts/stromal cells and osteoclast precursors, respectively (18, 24). Thus, cell-cell interaction could provide basal/“tonic” ITAM-mediated PLCγ2/calcium costimulatory signaling during osteoclastogenesis. There has been great interest in delineating the mechanisms by which ITAM signals crossregulate heterologous receptors, such as TLRs and cytokine receptors (13), but less is known about mechanisms/factors in the cells that fine tune ITAM-mediated PLCγ2/calcium signaling.
Recombinant recognition sequence binding protein at the Jκ site (RBP-J, also called RBP-Jκ, CSL, or CBF1) functions as a central transcription factor that receives input from several signaling pathways, such as the canonical NOTCH pathway, the WNT/β-catenin pathway, the NF-κB pathway, and the TLR- and TNF-signaling pathways (25–31), to mediate diverse cellular activities (25). We have recently identified RBP-J as a key inhibitor suppressing inflammatory osteoclastogenesis and bone resorption (29). To further dissect mechanisms by which RBP-J inhibits osteoclastogenesis, we tested whether RBP-J regulates ITAM-mediated costimulatory signaling during osteoclast differentiation and bone remodeling. We found that RBP-J deficiency substantially rescued impaired osteoclast formation and osteopetrotic bone phenotype in vivo in both Dap12–/– and Dap12–/–Fcrg–/– mice and enabled TNF-α to induce inflammatory bone resorption in Dap12–/– mice. Mechanistically, RBP-J inhibited induction of NFATc1, BLIMP1, and c-FOS by suppressing PLCγ2 expression and activity and downstream calcium-CaMKK-PYK2 signaling via the TGF-β pathway during osteoclastogenesis. Thus, RBP-J deficiency allows RANK or TNFR signaling to induce osteoclast differentiation independently of ITAM-mediated costimulation. These results indicate that RBP-J imposes a requirement for ITAM-mediated costimulation and limits the crosstalk between ITAM and RANK signaling, thereby restraining both pathways in osteoclastogenesis.
Results
RBP-J deficiency reverses the osteopetrotic bone phenotype of Dap12–/– and Dap12–/–Fcrg–/– mice.
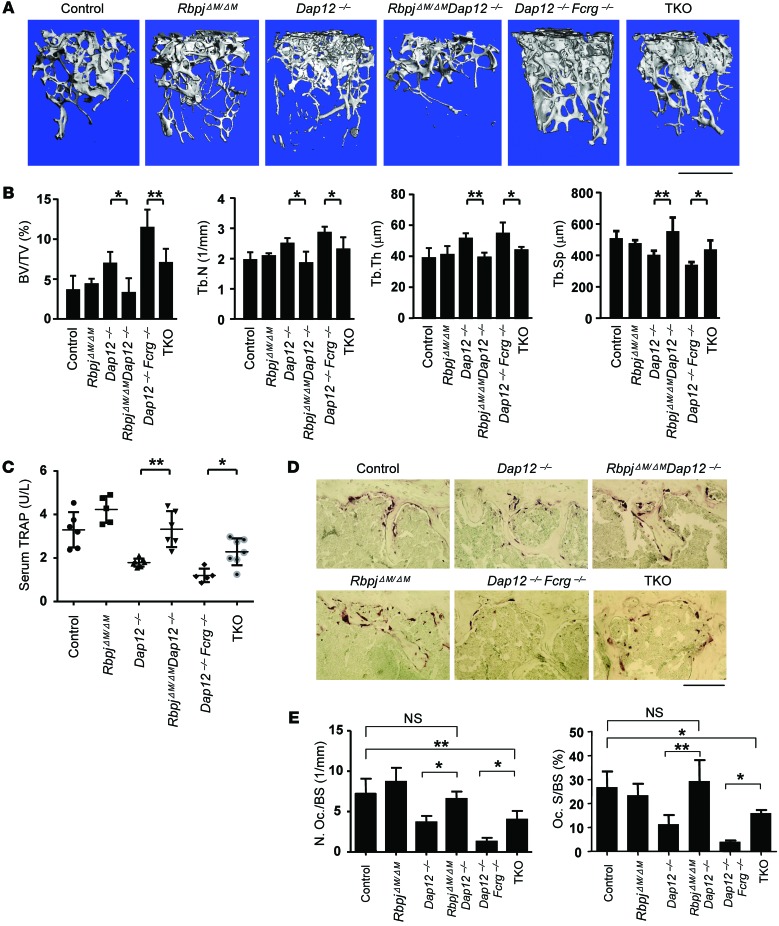
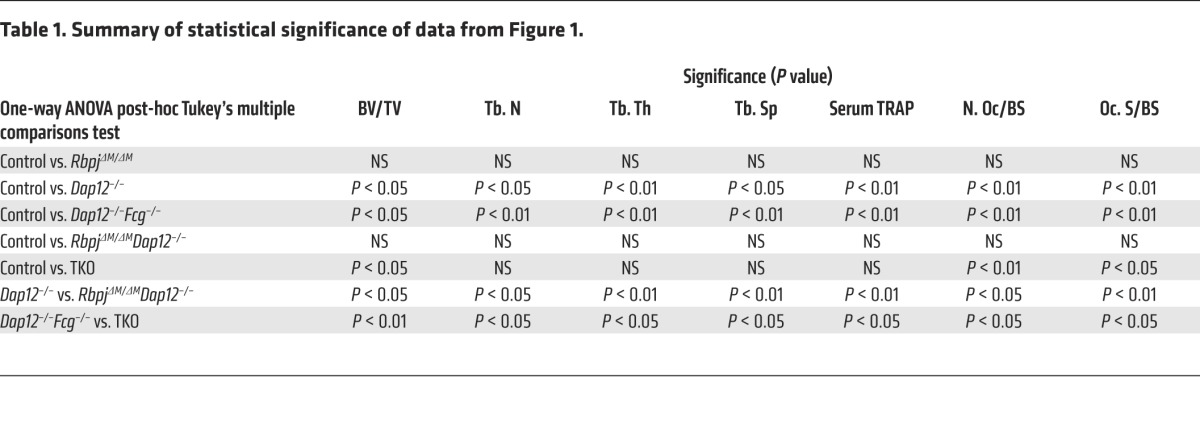
Loss of DAP12 or both DAP12 and FcRγ in mice leads to severe osteopetrosis due to impaired osteoclastogenesis and resorptive function (18, 24, 32, 33). To investigate the role of RBP-J in ITAM-mediated bone remodeling, we first deleted Rbpj in myeloid lineage osteoclast precursors in Dap12–/–, Fcrg–/–, and Dap12–/–Fcrg–/– mice by crossing these mice with RbpjΔM/ΔM (Rbpjflox/floxLysM-Cre+ mice as described in the literature; ref. 29) to generate RbpjΔM/ΔMDap12–/–, RbpjΔM/ΔMFcrg–/–, or RbpjΔM/ΔMDap12–/–Fcrg–/– (hereafter referred to as TKO) mice, respectively. Age- and sex-matched mice with a LysM-Cre+ genotype (hereafter referred to as control) were used as control mice. Consistent with the literature (18, 24), there were no significant bone defects in Fcrg–/– mice, and RBP-J deficiency in RbpjΔM/ΔMFcrg–/– mice did not affect osteoclast differentiation and bone phenotypes of Fcrg–/– mice (data not shown). Thus, in the present study, we focused on the effect of RBP-J on the Dap12–/– and Dap12–/–Fcrg–/– mice, which exhibit a severe osteopetrotic phenotype. In order to avoid potential effects of global deficiency of DAP12 or FcRγ, we generated bone marrow chimeric mice by transplanting the bone marrows of control, RbpjΔM/ΔM, Dap12–/–, Dap12–/–Fcrg–/–, RbpjΔM/ΔMDap12–/–, or TKO mice to age-matched male C57BL/6 recipient mice as described in Methods. Dap12–/– and Dap12–/–Fcrg–/– chimeric mice exhibited an osteopetrotic bone phenotype that was more severe in the double-knockout mice (Figure 1, A and B), which is consistent with previous literature (18, 24). In the presence of intact ITAMs, RbpjΔM/ΔM mice with 70% to 80% deletion did not exhibit obvious bone defects compared with control mice in homeostatic conditions (ref. 29, Figure 1, and Table 1). Strikingly, RBP-J deficiency in RbpjΔM/ΔMDap12–/– or TKO chimeric mice relieved the osteopetrotic phenotype caused by lack of DAP12 or both DAP12 and FcRγ, respectively. RbpjΔM/ΔMDap12–/– mice exhibited trabecular bone volume, number, thickness, and spacing similar to those of control mice (Figure 1, A and B, Table 1, and Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI71882DS1). Furthermore, the serum levels of tartrate-resistant acid phosphatase (TRAP), a biomarker reflecting the amount of osteoclasts and bone resorption in vivo, in RbpjΔM/ΔMDap12–/– or TKO chimeric mice were significantly enhanced relative to those of Dap12–/– or Dap12–/–Fcrg–/– chimeric mice, respectively (Figure 1C). Notably, serum TRAP levels in RbpjΔM/ΔMDap12–/– mice were comparable to those in control mice (Figure 1C and Table 1). These findings were further corroborated by markedly increased TRAP+ osteoclasts in RbpjΔM/ΔMDap12–/– or TKO chimeric mice compared with Dap12–/– or Dap12–/–Fcrg–/– chimeric mice, respectively (Figure 1, D and E). When compared with control mice, the bone defects in Dap12–/–Fcrg–/– mice were partially, while those in Dap12–/– mice were almost fully, compensated by RBP-J deficiency (Figure 1, A–E, and Table 1). In addition, RBP-J did not significantly affect cortical bone phenotype and osteoblastic serum marker alkaline phosphatase (ALP) in vivo (Supplemental Figure 2). These results indicate that myeloid-specific RBP-J deficiency can bypass the requirement for ITAM-mediated costimulation for osteoclast formation and homeostatic bone remodeling, suggesting that RBP-J constitutively plays an inhibitory role in this biological context.
Figure 1. RBP-J deficiency rescues the osteopetrotic bone phenotype in both Dap12–/– and Dap12–/–Fcrg–/– mice.
(A) Microcomputed tomography (μCT) images of trabecular bones of the distal femurs isolated from the indicated bone marrow chimeric mice. Top of images is toward the growth plate. Scale bar: 1 mm. (B) Bone morphometric analysis of femurs isolated from the chimeric mice. BV/TV, bone volume per tissue volume; Tb. N, trabecular number; Tb. Th, trabecular bone thickness; Tb. Sp, trabecular bone spacing. n = at least 5 per group. *P < 0.05; **P < 0.01. (C) Concentration of basal serum TRAP obtained from the chimeric mice. n = at least 5 per group. *P < 0.05; **P < 0.01. (D) TRAP staining and (E) Histomorphometric analysis of the histological sections obtained from the metaphysis region of distal femurs of the chimeric mice, N. Oc./BS, number of osteoclasts per bone surface; Oc. S/BS, osteoclast surface per bone surface. n = 5 per group. *P < 0.05; **P < 0.01. Scale bar: 100 μm.
Table 1.
Summary of statistical significance of data from Figure 1.
RBP-J deficiency bypasses the requirement for ITAM-mediated costimulation of RANKL-induced osteoclastogenesis.
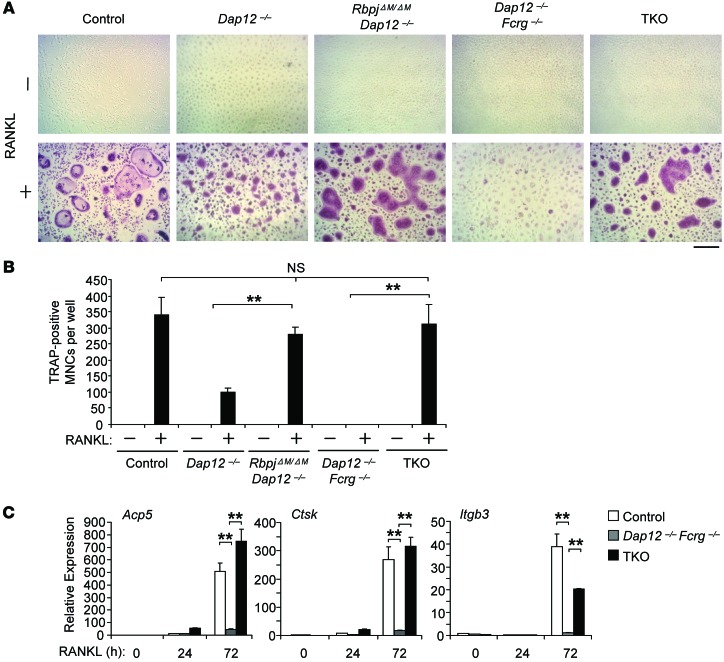
We then examined in vitro osteoclast differentiation using bone marrow–derived macrophages (BMMs) as osteoclast precursors. Dap12–/– BMMs, as described in previous literature (18, 24, 34), formed a low number of TRAP+ small “crenated” osteoclasts (Figure 2A), while there was no osteoclast formation in Dap12–/–Fcrg–/– cell cultures (Figure 2A) in response to RANKL stimulation compared with control cell cultures. RBP-J deficiency in RbpjΔM/ΔMDap12–/– cells significantly enhanced the number and size of the TRAP+ multinucleated cells (MNCs) to a level comparable to that seen in control cell cultures (Figure 2, A and B). Moreover, RBP-J deficiency in TKO cells substantially reversed the defects of osteoclastogenesis that resulted from DAP12 and FcRγ deficiency and resulted in formation of a large number of TRAP+ MNCs (Figure 2, A and B). In parallel with the increased generation of TRAP+ polykaryons, the expression of osteoclast marker genes Acp5 (encoding TRAP), Ctsk (encoding cathepsin K), and Itgb3 (encoding β3 integrin) was dramatically enhanced in RANKL-treated TKO cells relative to Dap12–/–Fcrg–/– cells, and the levels of Acp5 and CtsK were even comparable to the levels as induced by RANKL in control cell cultures (Figure 2C). Collectively, these results indicate that the defect in the osteoclast differentiation program in the absence of ITAM-mediated costimulation is almost completely rescued by RBP-J deficiency. This in vitro reversal of osteoclastogenesis defects by RBP-J deficiency further corroborates the reversal observed in vivo, supporting an important role of RBP-J in restraining ITAM-mediated osteoclast differentiation.
Figure 2. RBP-J imposes a requirement for ITAM-mediated costimulation in osteoclast differentiation.
BMMs derived from control, Dap12–/–, RbpjΔM/ΔMDap12–/–, Dap12–/–Fcrg–/–, and TKO mice were stimulated with RANKL for 5 days. TRAP staining was performed (A) and the number of TRAP-positive MNCs (≥ 3 nuclei per cell) per well was counted (B). TRAP-positive cells appear red in the photographs. Scale bar: 100 μm. Data are representative of at least 20 independent experiments. **P < 0.01. (C) Quantitative real-time PCR analysis of mRNA expression of Acp5 (encoding TRAP), Ctsk (encoding cathepsin K), and Itgb3 (encoding β3) in BMMs from control, Dap12–/–Fcrg–/–, and TKO mice treated with RANKL for the indicated times. Data are representative of at least 3 independent experiments. **P < 0.01.
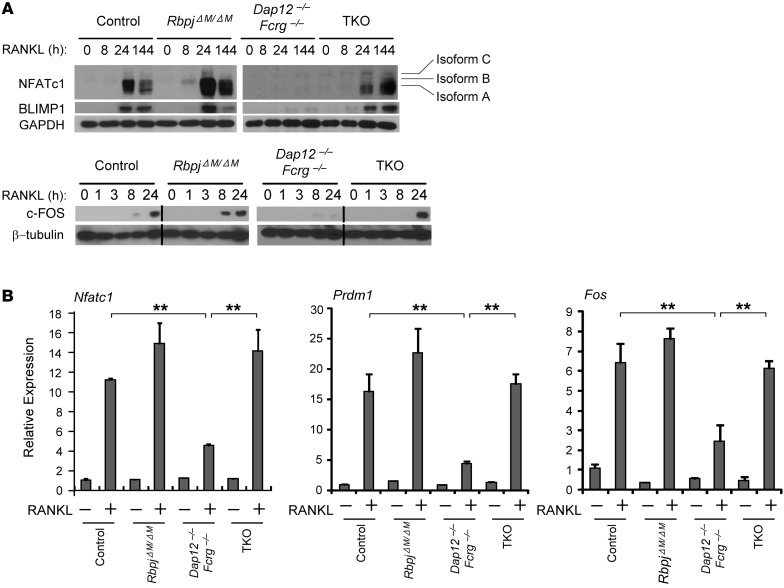
NFATc1 is well known as the “master regulator” of osteoclastogenesis. In addition to the regulation of Blimp1 by NFATc1 (35), genetic evidence obtained using Blimp1-KO cells or RNAi targeting of BLIMP1 shows that BLIMP1 conversely plays an important role in induction of NFATc1 (29, 36), indicating a critical reciprocal regulatory circuit between BLIMP1 and NFATc1 that coordinately controls the osteoclast differentiation program. Induction of NFATc1 required not only RANKL but also costimulatory signaling from ITAM-associated receptors (refs. 18, 19, and Figure 3A). In this study, we found that BLIMP1 induction also requires costimulatory signaling, as RANKL failed to induce BLIMP1 expression in Dap12–/–Fcrg–/– cells (Figure 3A). Consistent with previous literature (29), deletion of RBP-J enhanced the expression of NFATc1, BLIMP1, and c-FOS (Figure 3, A and B). Strikingly, RBP-J deficiency in TKO cells completely rescued the impaired induction of NFATc1, BLIMP1, and c-FOS by RANKL in Dap12–/–Fcrg–/– cells (Figure 3A). Although the kinetics of the induction of these transcription factors in TKO cells was slower than that in control cells, their expression levels were comparable to or even greater than control levels at late time points (Figure 3A). Furthermore, the diminished mRNA expression of Nfatc1, Blimp1, and Fos was also rescued in TKO cells (Figure 3B), suggesting that the regulation of the expression of Nfatc1, Prdm1 (which encodes Blimp1), and Fos (which encodes c-Fos) by RBP-J occurs at the transcriptional level. These results show that RBP-J deficiency bypassed the need for ITAM-mediated costimulation to induce NFATc1/BLIMP1/c-FOS expression and osteoclastogenesis. Thus, RBP-J imposes a requirement for costimulation of RANKL signaling.
Figure 3. RBP-J deficiency compensates for the defect of NFATc1, BLIMP1, and c-FOS induction in Dap12–/–Fcrg–/– cells by RANKL.
(A) Immunoblot analysis of NFATc1, BLIMP1, and c-FOS expression in whole-cell lysates obtained from control, RbpjΔM/ΔM, Dap12–/–Fcrg–/–, and TKO BMMs at the indicated time points after stimulation with RANKL. GAPDH or β-tubulin was measured as loading control. Isoform A of NFATc1 is the major isoform induced during osteoclastogenesis. Data are representative of at least 3 independent experiments. Lanes separated by a black line indicate that they were run on the same gel but were noncontiguous. (B) Quantitative real-time PCR analysis of mRNA expression of Nfatc1 (encoding NFATc1), Prdm1 (encoding BLIMP1), and Fos (encoding c-FOS) induced by RANKL for 24 hours in osteoclastogenic cell cultures from the indicated BMMs. Data are representative of at least 3 independent experiments. **P < 0.01.
RBP-J suppresses the expression and activity of basal PLCγ2 and downstream calcium-CaMKK-PYK2 signaling by regulating TGF-β signaling.
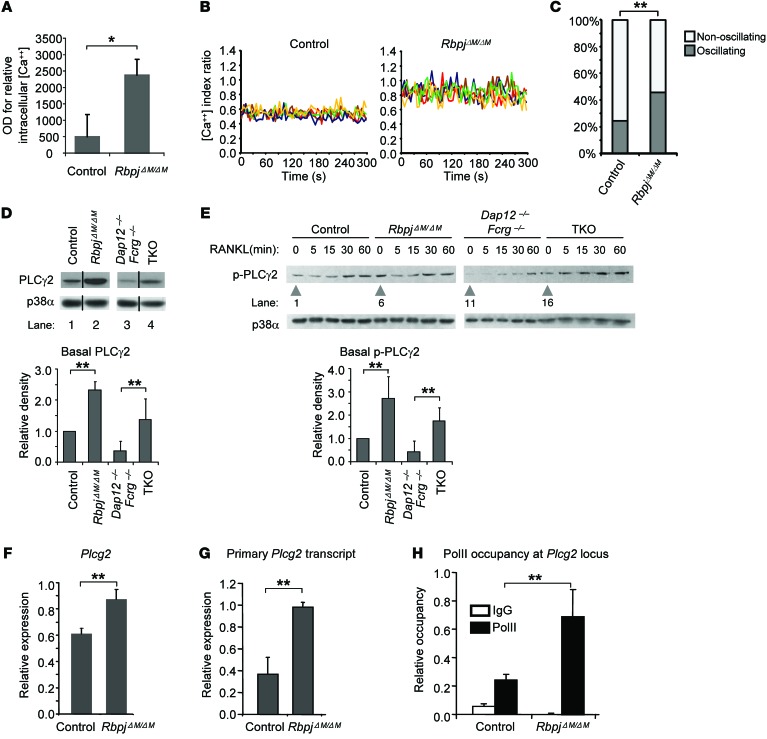
We then asked how RBP-J imposes a requirement for ITAM-mediated costimulation. To address this question, we first performed gene-expression profiling using high-throughput sequencing of RNA (RNAseq) with control and RbpjΔM/ΔM osteoclast precursors at baseline and after TNF-α stimulation to identify genes regulated by RBP-J during osteoclastogenesis. Gene-set enrichment analysis of the RBP-J–regulated genes revealed marked enrichment of calcium signaling pathway genes in RbpjΔM/ΔM cells (P = 7.72 × 10–5). Consistent with regulation of calcium signaling by RBP-J, we observed that RBP-J deficiency significantly increased intracellular calcium levels (Figure 4A). Moreover, we measured calcium oscillations including oscillatory amplitude in single cells (Figure 4B) and oscillating/nonoscillating cell proportion in each cell culture (Figure 4C). We found that calcium oscillations in RBP-J–deficient cells were dramatically increased (Figure 4, B and C). We thus hypothesized that RBP-J deficiency bypasses the need for ITAM-containing adaptors by enhancing the expression or function of calcium signaling components downstream of these adaptors. As PLCγ2 is an important mediator of calcium signaling, we examined the effects of RBP-J deficiency on the expression and activity of PLCγ2. Interestingly, we found that RBP-J deficiency significantly increased PLCγ2 expression (Figure 4D). In contrast, PLCγ2 expression was strongly diminished in Dap12–/–Fcrg–/– cells, and this decrease was reversed in TKO cells (Figure 4D). These data indicate that RBP-J suppresses while ITAM signaling maintains basal PLCγ2 expression. Next, we examined whether PLCγ2 activity was affected. As expected, RANKL increased phosphorylation of PLCγ2 over baseline (Figure 4E); strikingly, basal PLCγ2 phosphorylation was essentially absent in Dap12–/–Fcrg–/– cells (Figure 4E, lane 11), although RANKL was able to induce a modest increase, presumably due to the activation of BTC/TEC (20). However, such induction of PLCγ2 activity over a low baseline by RANKL is not sufficient to induce osteoclastogenesis, as shown in Figure 2A and previous literature (18, 19), as there is almost no osteoclast differentiation in Dap12–/–Fcrg–/– cells. The essentially nondetectable basal PLCγ2 phosphorylation in Dap12–/–Fcrg–/– cells (Figure 4E) indicates that tonic ITAM signaling makes a predominant contribution to maintaining basal activity of PLCγ2. Although the importance of basal PLCγ2 activity that is calibrated by ITAM pathways in response to environmental cues was not previously fully appreciated, our observations suggest that such basal PLCγ2 function acts additively with RANKL-induced PLCγ2 activity to surpass a threshold required to induce NFATc1 and osteoclast differentiation. Consistent with this notion, we found that RBP-J deficiency in RbpjΔM/ΔM or TKO cells, which resulted in increased osteoclastogenesis (ref. 29 and Figure 2), concomitantly, significantly, and consistently enhanced the basal level of phospho-PLCγ2 compared with control or Dap12–/–Fcrg–/– cells (Figure 4E, lane 6 vs. lane 1 and lane 16 vs. lane 11), but did not always affect the early transient induction of phospho-PLCγ2 by RANKL (Figure 4E).
Figure 4. RBP-J suppresses basal PLCγ2 expression and calcium levels during osteoclast differentiation.
(A) Basal intracellular calcium concentration in control and RbpjΔM/ΔM BMMs. *P < 0.05. (B) Calcium oscillations in control and RbpjΔM/ΔM BMMs. Representative traces of the [Ca++] index ratio in single cells for each condition were shown. Each color indicates a different cell in the same field. (C) Quantification of calcium oscillating cells in control and RbpjΔM/ΔM BMM cell cultures. Differences between conditions were statistically analyzed using the χ2 test. **P < 0.01. (D) Immunoblot analysis of basal total PLCγ2 and (E) phospho-PLCγ2 expression in control, RbpjΔM/ΔM, Dap12–/–Fcrg–/–, and TKO BMMs. p38α was measured as a loading control. Data are representative of 5 independent experiments. Samples (D) in lanes 1 and 2 were derived from noncontiguous lanes in the same gel; samples in lanes 3 and 4 were derived from noncontiguous lanes in a matched gel. The relative density of the immunoblot bands of total PLCγ2 (D, lower panel) or basal phospho-PLCγ2 (E, lower panel) versus those of loading control p38α were quantified by densitometry and normalized to the basal control condition. Results are shown as average values from 5 independent experiments. **P < 0.01. (F) mRNA expression of Plcg2 and (G) primary Plcg2 transcript expression examined by real-time PCR. (H) Pol II occupancy at Plcg2 locus using ChIP assay. **P < 0.01.
Previous data from our group and others show that NOTCH signaling inhibits osteoclastogenesis (29, 37). In the present study, we further found that inhibition of osteoclast differentiation by the NOTCH ligands delta-like1 and jagged1 is dependent on RBP-J (Supplemental Figure 3). Furthermore, activation of NOTCH signaling in osteoclast precursor cells suppressed phosphorylation of PLCγ2 and its total protein level (Supplemental Figure 4), indicating that NOTCH–RBP-J signaling negatively regulates PLCγ2 expression and further supporting the notion that the levels of PLCγ2 expression are important in determining its activity; the important adaptor functions of PLCγ2 (19) would also be compromised by its lower expression. We then investigated mechanisms that regulate PLCγ2 expression. We found that RBP-J deficiency increased Plcg2 mRNA amounts (Figure 4F). The elevation was modest, approximately a 30% increase compared with control cells, but was statistically significant (P < 0.01) and consistently similar to the elevation of PLCγ2 protein amounts in multiple experiments. RBP-J did not affect the mRNA stability of Plcg2 (Supplemental Figure 5), while the primary Plcg2 transcripts (unspliced transcripts of Plcg2) were significantly enhanced in the RBP-J–deficient cells (Figure 4G). These data indicate that RBP-J regulates PLCγ2 expression at the transcriptional level, which was further corroborated by enhanced RNA polymerase II (Pol II) occupancy at the Plcg2 locus in RBP-J–deficient cells (Figure 4H). To address whether RBP-J directly regulates Plcg2, we obtained the set of known RBP-J target genes from published databases (38–40) and overlapped these targets with genes that were regulated by RBP-J in our RNAseq experiments (Supplemental Figure 6), but did not find Plcg2 included. ChIPseq data (38) did not show any significant chromosomal DNA-binding peaks of RBP-J in a range of at least 100-kb upstream or downstream of the transcriptional start site of Plcg2. These suggest that RBP-J negatively regulates Plcg2 transcription in an indirect manner.
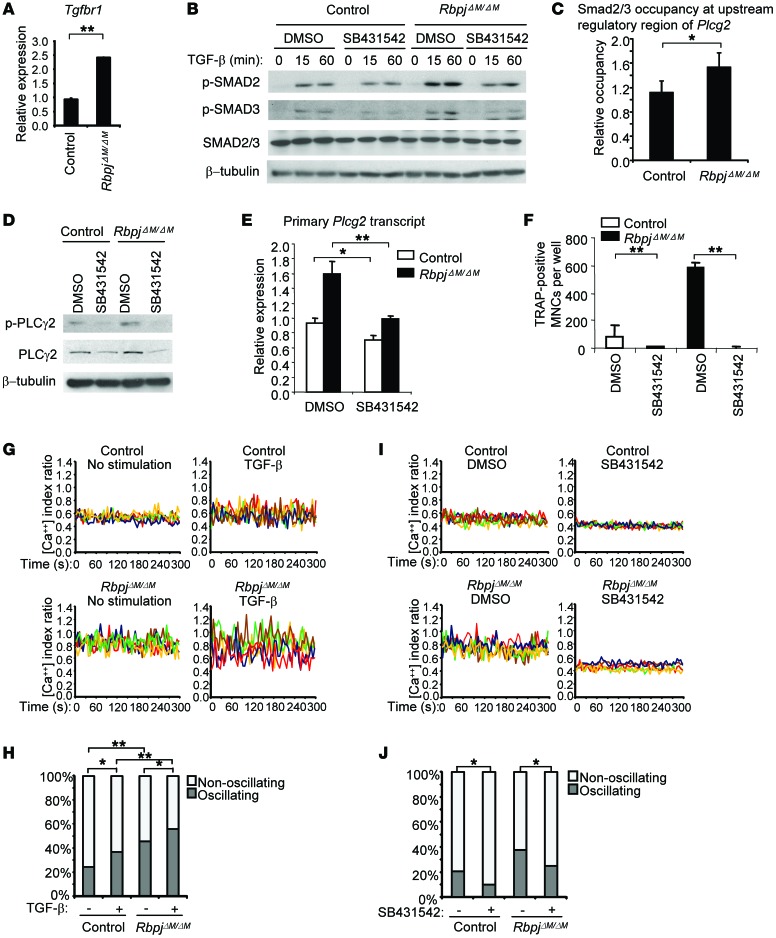
Insight into indirect mechanisms of regulation was provided by the striking finding that the direct RBP-J target gene Tgfbr1 (encoding TGF-β receptor I) was expressed at a rate approximately 2.5 times higher in RBP-J–deficient osteoclast precursors than in control cells (Figure 5A). This suggested that RBP-J deficiency could enhance the responsiveness of osteoclast precursors to TGF-β that is produced by these cells and is also present in serum, and is an important factor for osteoclastogenesis (41, 42). Previous literature showing crosstalk between TGF-β signaling and the Notch pathway in other settings (43–46) supports the possibility of regulation of TGF-β responses by RBP-J. Furthermore, TGF-β increases intracellular calcium concentration and activates downstream calcium signaling and PKC (47–49), suggesting that TGF-β signaling may regulate PLCγ2. Indeed, we found that RBP-J deficiency enhanced the expression of TGF-β–responsive genes, such as Id1, Id3, Cdkn1a, Cdkn2b, and Smad7 (Supplemental Figure 7), indicating that RBP-J deficiency potentiates cell responses to TGF-β. Corroboratively, phosphorylation of the TGF-β–activated key signaling components SMAD2 and SMAD3 was also enhanced in RBP-J–deficient cells (Figure 5B and Supplemental Figure 8). Previous ChIPseq data (50) showed that TGF-β–induced SMAD2/3 binding peaks in the upstream regulatory region of Plcg2. In our system, we found that RBP-J deficiency modestly, but significantly and consistently, elevated the occupancy of SMAD2/3 in response to TGF-β at the same upstream regulatory region of Plcg2 locus as shown in the literature (50) in multiple experiments (Figure 5C). This further supports the notion that RBP-J regulates Plcg2 expression via a TGF-β–SMAD2/3 signaling pathway. As TGF-β signaling primes osteoclast precursors and facilitates osteoclastogenesis (41, 42), we next examined whether the enhanced TGF-β signaling in RBP-J–deficient cells contributes to the enhanced PLCγ2 expression and osteoclastogenesis. We blocked the kinase activity of TGF-β receptor I and downstream signaling using the inhibitor SB431542 (Figure 5B and Supplemental Figure 8) and found that the basal expression and phosphorylation of PLCγ2 (Figure 5, D and E) as well as osteoclastogenesis were significantly suppressed (Figure 5F). Similar results were obtained when specifically knocking down Tgfbr1 (Supplemental Figure 9). These data suggest that repression of Tgfbr1 expression by RBP-J results in decreased responsiveness of osteoclast precursors to TGF-β, with a concomitant decrease in TGF-β–mediated PLCγ2 expression. Consistent with a role for TGF-β in calcium signaling, we, for what we believe is the first time, observed that TGF-β significantly induced calcium oscillations in control BMMs (Figure 5, G and H). RBP-J deficiency markedly increased calcium oscillations in both basal and TGF-β stimulatory conditions (Figure 5, G and H). The TGF-β signaling inhibitor SB431542 dampened basal calcium oscillation in both cell types and decreased the proportion of oscillating cells in RBP-J–deficient cells to a level comparable to that of control cells (Figure 5, I and J). In addition, inhibition of TGF-β signaling markedly decreased TNF-α–induced calcium oscillations in the RbpjΔM/ΔM cells (Supplemental Figure 10). This indicates a significant contribution of endogenous TGF-β signaling to calcium oscillations in control BMM cells as well as the enhanced oscillatory state in RBP-J–deficient cells. Collectively, the results show that RBP-J deficiency elevates the basal PLCγ2/calcium pathway via TGF-β–Smad2/3 signaling.
Figure 5. RBP-J suppresses PLCγ2 expression and calcium oscillations via a TGF-β–dependent pathway.
(A) Quantitative real-time PCR analysis of mRNA expression level of Tgfbr1 (encoding TGF-β receptor 1). Data are representative of at least 3 independent experiments. **P < 0.01. (B) Immunoblot analysis of phospho-SMAD2/3 in the cells pretreated with TGF-β receptor I inhibitor SB431542 (100 μM) or DMSO (vehicle) for 1 hour followed by stimulation with TGF-β (10 ng/ml) for the indicated times. β-Tubulin was measured as a loading control. (C) ChIP assay of SMAD2/3 occupancy at the upstream regulatory region of Plcg2 in BMM cells stimulated without or with TGF-β (10 ng/ml) for 1 hour. Relative occupancy of SMAD2/3 in response to TGF-β was normalized to each basal control condition. Results are shown as average values from 3 independent experiments. *P < 0.01. (D) Immunoblot analysis of phospho-PLCγ2 and total PLCγ2 and (E) primary Plcg2 transcript expression in BMMs treated with SB431542 (100 μM) or DMSO for 6 hours. (F) Quantification of TRAP-positive MNCs in the culture of BMMs (≥ 3 nuclei per cell) treated with SB431542 (10 μM) or DMSO for 4 days in the presence of TNF-α. *P < 0.05; **P < 0.01. (G) Representative calcium oscillatory traces of cells stimulated without or with TGF-β (10 ng/ml) for 24 hours; oscillating cells were quantified (H). Controls for basal calcium oscillation data without TGF-β stimulation were the same as shown in Figure 4B. (I) Representative calcium oscillatory traces of cells treated with SB431542 (100 μM) or DMSO for 24 hours; oscillating cells were quantified (J). *P < 0.05; **P < 0.01.
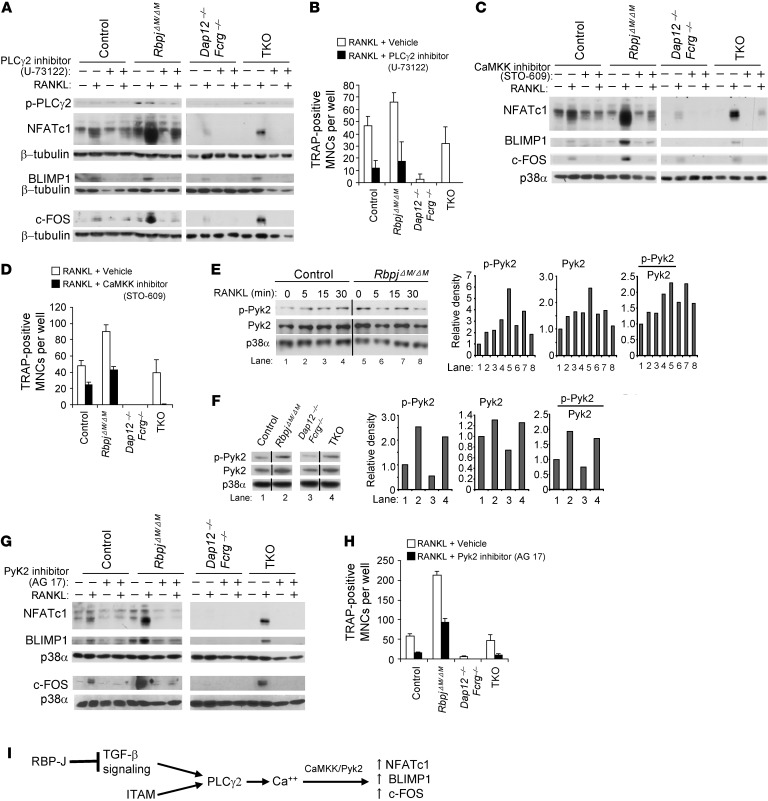
To directly test whether increased PLCγ2-calcium signaling in RBP-J–deficient cells contributes to increased expression of downstream NFATc1, BLIMP1, and c-FOS, we blocked PLCγ2 activity before RANKL treatment using the well-known PLCγ2 inhibitor U-73122. Indeed, the increased induction of NFATc1, BLIMP1, and c-FOS by RANKL in RbpjΔM/ΔM or TKO cells was attenuated to a level comparable to that in control or Dap12–/–Fcrg–/– cells, respectively, by the PLCγ2 inhibitor U-73122 (Figure 6A and Supplemental Figure 11A). The inhibition of NFATc1 and BLIMP1 expression by U-73122 lasted for at least 2 days (Supplemental Figure 12). Correspondingly, inhibition of PLCγ2 activity significantly reduced the enhanced RANKL-induced osteoclastogenesis in RBP-J–deficient cells; the inhibitory effect was most pronounced in TKO cells (Figure 6B). Thus, enhanced osteoclastogenesis in RBP-J–deficient cells is dependent on PLCγ2 signaling. PLCγ2/calcium signals are transmitted by CaMKK and PYK2 in other systems (12), where CaMKK functions downstream of PLCγ2 but upstream of PYK2. Thus, we first used CaMKK inhibitor STO-609 to block its activity in the osteoclastogenic system and found that the induction of NFATc1, BLIMP1, and c-FOS was suppressed in control cells, while the enhanced induction of NFATc1, BLIMP1, and c-FOS by RBP-J deficiency in RbpjΔM/ΔM or TKO cells was attenuated to the level seen in control or in Dap12–/–Fcrg–/– cells, respectively (Figure 6C and Supplemental Figure 11B). Osteoclastogenesis was also inhibited by CaMKK blockade, and inhibition was most pronounced in TKO cells (Figure 6D). Furthermore, we observed that RANKL induced phosphorylation of PYK2 (Figure 6E). RBP-J deficiency did not increase the RANKL-induced phosphorylation of PYK2, but significantly elevated its basal phosphorylation level and total amount in a CaMKK-dependent manner (Figure 6, E and F, and data not shown), which is likely due to increased upstream PLCγ2 signaling resulting from RBP-J deficiency. As such, the basal level of phosphorylation and total amounts of PYK2 were also higher in TKO cells than in Dap12–/–Fcrg–/– cells (Figure 6F). These results show that RBP-J regulates both PYK2 activity and total PYK2 amounts. Quantification data (Figure 6, E and F) showed that the active phospho-PYK2 relative to total PYK2 was still increased by RBP-J deficiency, indicating that the increased PYK2 activity by RBP-J is partially correlated with the increase in total amount of PYK2. Inhibition of PYK2 activity by its inhibitor AG 17 markedly suppressed the induction of NFATc1, BLIMP1,and c-FOS (Figure 6G) as well as osteoclastogenesis in control cells (Figure 6H), indicating that PYK2 plays an activating role in induction of NFATc1/BLIMP1/c-FOS and osteoclastogenesis. In RBP-J–deficient cells, inhibition of PYK2 activity by AG 17 dramatically decreased the enhanced expression level of NFATc1/BLIMP1/c-FOS (Figure 6G) and osteoclastogenesis, which was most pronounced in TKO cells (Figure 6H). The heightened suppression of osteoclastogenesis by PLCγ2, CaMKK, and Pyk2 inhibitors in TKO cells supports a critical role for augmented downstream PLCγ2 activity in compensating for missing upstream signals from DAP12 and FcRγ. Taken together, these findings indicate that RBP-J inhibits NFATc1/BLIMP1/c-FOS expression by keeping PLCγ2 in a basal suppressed state and thus downregulating downstream calcium-CaMKK-PYK2 signaling in osteoclastogenesis. These findings are consistent with a model whereby RBP-J deficiency rescues osteoclastogenesis by bypassing the need for upstream ITAM signaling by increasing TGF-β signaling to maintain downstream PLCγ2/calcium-CaMKK/PYK2 activity (Figure 6I).
Figure 6. RBP-J suppresses PLCγ2/calcium-CaMKK/Pyk2 signaling in osteoclast differentiation.
(A) Immunoblot analysis of phospho-PLCγ2, NFATc1, Blimp1, and c-Fos expression at basal levels or stimulated with RANKL for 24 hours in the BMMs. The cells were pretreated for 30 minutes with PLCγ2 inhibitor U-73122 (10 μM) or its vehicle ethanol; inhibitor or vehicle was completely washed away before addition of RANKL. (B) Quantitation of TRAP-positive MNCs (≥ 10 nuclei per cell) that were pretreated for 30 minutes with U-73122 or ethanol and then cultured with RANKL for 6 days. (C and G) Immunoblot analysis of NFATc1, Blimp1, and c-Fos expression at basal levels or after being stimulated with RANKL for 24 hours with CaMKK inhibitor STO-609 (5 μg/ml) (C) or PYK2 inhibitor AG 17 (3 μM) (G) or vehicle DMSO in the BMMs. (D and H) Quantitation of TRAP-positive MNCs (≥ 10 nuclei per cell) that were stimulated with RANKL with STO-609 (D), AG 17 (H), or DMSO for 6 days. (E and F) Immunoblot analysis of phospho-PYK2 and total PYK2 expression in the indicated conditions (E) or at basal levels (F). The lanes separated by a black line were run on the same gel but were noncontiguous. Quantification of the immunoblot bands by densitometry were shown in the right panels of E and F. β-Tubulin (A) or p38α (C and E–G) was measured as a loading control. (I) A model showing that RBP-J suppresses ITAM-mediated basal PLCγ2 expression/activity and downstream calcium-CaMKK-PYK2 signaling in induction of c-FOS, BLIMP1, and NFATc1 in a TGF-β dependent pathway.
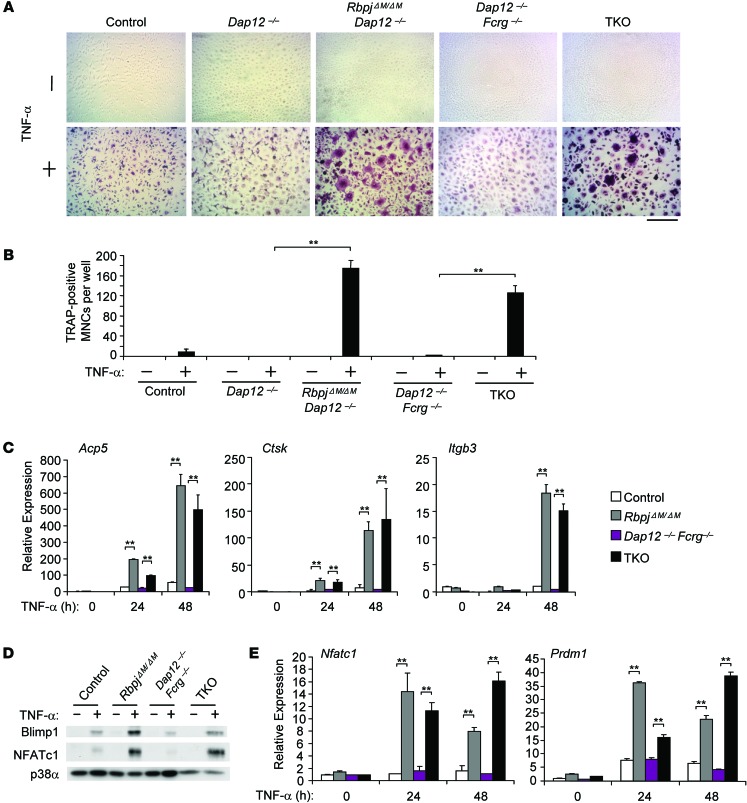
RBP-J deficiency enables TNF-α to induce NFATc1 and BLIMP1 expression and osteoclastogenesis in the absence of ITAM-mediated costimulatory signaling.
We reasoned that increased PLCγ2 activity in RBP-J–deficient cells would compensate for lesser induction of ITAM signaling by TNF-α than by RANKL (51) and thus contribute to increased TNF-α–mediated osteoclastogenesis. We tested this notion using a genetic approach to see whether TNF-α could induce osteoclastogenesis in RBP-J–deficient cells even in the absence of the ITAM-containing adaptors DAP12 and FcRγ. In Dap12–/– and Dap12–/–Fcrg–/– cells, as in control cells, TNF-α alone did not significantly induce a osteoclast differentiation program, including expression of osteoclast marker genes and NFATc1/BLIMP1 induction (Figure 7, A–E). However, RBP-J deficiency resulted in formation of TRAP+ MNCs in response to TNF-α in the absence of DAP12 or DAP12/FcRγ (Figure 7, A and B). Despite a “crenated” morphology, these TRAP+ MNCs in RbpjΔM/ΔMDap12–/– and TKO cells expressed osteoclast marker genes Acp5, Ctsk, and Itgb3 at levels similar to those induced in fully differentiated RbpjΔM/ΔM osteoclasts (Figure 7C and ref. 29). Furthermore, TNF-α induced expression levels of NFATc1 and BLIMP1 in RbpjΔM/ΔMDap12–/– and TKO cells comparable to that in RbpjΔM/ΔM cells (Figure 7, D and E, and data not shown). However, these crenated MNCs failed to fully spread on tissue culture plates and did not resorb dentin (data not shown). These results indicate that RBP-J deficiency that provides basal downstream PLCγ2-CaMKK-PYK2 signaling in the absence of upstream ITAMs enables TNF-α to fully induce the early osteoclast differentiation program in vitro in cultures of purified osteoclast precursors in the absence of stromal cells. However, ITAM-mediated costimulatory signaling is still required for osteoclast function, such as actin ring formation and resorptive activity, even in RBP-J–deficient cells (15, 32).
Figure 7. RBP-J deficiency allows TNF-α to induce osteoclast differentiation in Dap12–/– and Dap12–/–Fcrg–/– BMMs.
BMMs derived from control, Dap12–/–, RbpjΔM/ΔMDap12–/–, Dap12–/–Fcrg–/–, and TKO mice were stimulated with TNF-α for 5 days. TRAP staining was performed (A), and the number of TRAP-positive MNCs (≥ 3 nuclei per cell) per well was counted (B). TRAP-positive cells appear red in the photographs. Scale bar: 100 μm. Data are representative of at least 20 independent experiments. **P < 0.01. (C) Quantitative real-time PCR analysis of mRNA expression of Acp5 (encoding TRAP), Ctsk (encoding cathepsin K), and Itgb3 (encoding β3) in BMMs from control, RbpjΔM/ΔM, Dap12–/–Fcrg–/–, and TKO mice treated with TNF-α for the indicated times. Data are representative of at least 3 independent experiments. **P < 0.01. (D) Immunoblot analysis of NFATc1 and BLIMP1 expression in whole-cell lysates obtained from control, RbpjΔM/ΔM, Dap12–/–Fcrg–/–, and TKO BMMs after stimulation with TNF-α for 24 hours. p38α was measured as a loading control. Data are representative of at least 3 independent experiments. (E) Quantitative real-time PCR analysis of mRNA expression of Nfatc1 (encoding NFATc1) and Prdm1 (encoding Blimp1) induced by TNF-α in osteoclastogenic cell cultures from control, RbpjΔM/ΔM, Dap12–/–Fcrg–/–, and TKO BMMs. Data are representative of at least 3 independent experiments. **P < 0.01.
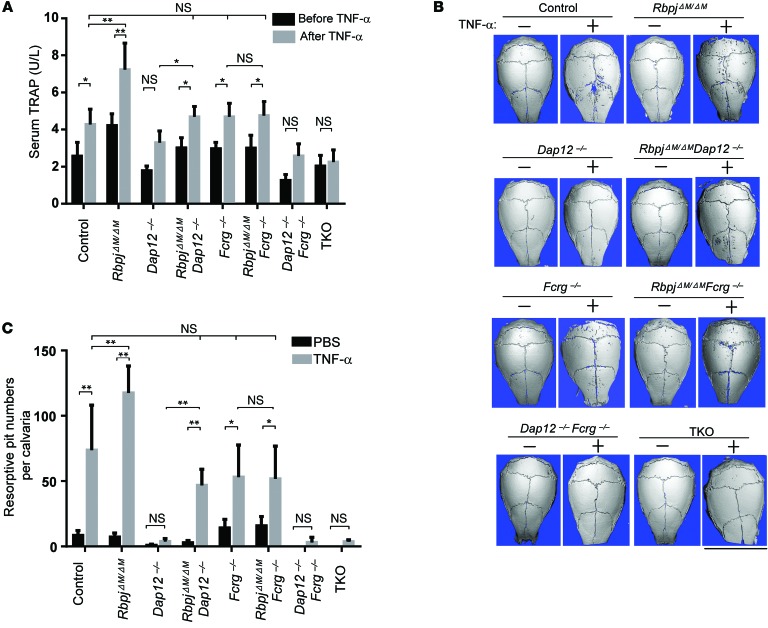
RBP-J deficiency compensates for DAP12-mediated costimulation in TNF-α–induced bone resorption in vivo.
We next examined whether RBP-J deficiency can compensate for the role of ITAM-mediated signaling in bone resorption in response to TNF-α stimulation in vivo. We used a well-established TNF-α–induced inflammatory bone resorption mouse model induced by injection of TNF-α into mouse calvarial periosteum (52). Administration of TNF-α to the calvarial periosteum resulted in more TRAP+ cells than PBS injection in all mouse types, but to a more dramatic extent in RbpjΔM/ΔM mice (ref. 29 and Supplemental Figure 13) and to a much lesser extent in Dap12–/–, Dap12–/–Fcrg–/–, and TKO mice (Supplemental Figure 13). Consistent with these results, a TNF-α challenge significantly increased TRAP levels in control, RbpjΔM/ΔM, RbpjΔM/ΔMDap12–/–, Fcrg–/–, and RbpjΔM/ΔMFcrg–/– mice, but there was no significant change in the TRAP level in TKO mice (Figure 8A). Although there was a trend toward increased serum TRAP levels in response to a TNF-α challenge in Dap12–/– and Dap12–/–Fcrg–/– mice, there was no statistically significant difference between these conditions (Figure 8A). RBP-J deficiency significantly compensated for the low TRAP levels in Dap12–/– mice, but did not affect TRAP levels of Fcrg–/– mice, which are comparable to control mice (Figure 8A). As ITAM signaling is also required for osteoclast resorptive function, regardless of multinucleated morphology, we performed μCT scanning of these calvarial bones and quantified resorptive pits to assess osteoclast function in vivo. The results showed that a local TNF-α challenge for a short period markedly induced resorptive pits in the calvarial bones of control, RbpjΔM/ΔM, RbpjΔM/ΔMDap12–/–, Fcrg–/–, and RbpjΔM/ΔMFcrg–/– mice with the most dramatic resorption level in RbpjΔM/ΔM mice (Figure 8, B and C), but not in Dap12–/–, Dap12–/–Fcrg–/–, and TKO mice, indicating that the TRAP+ cells induced by TNF-α in Dap12–/–, Dap12–/–Fcrg–/–, and TKO mice are functionally impaired. Dramatically, RBP-J deficiency almost completely compensated for the lack of bone resorption in Dap12–/– mice relative to control mice (Figure 8, B and C). In contrast, Fcrg–/– mice exhibit amounts of bone resorption similar to that of control mice in response to a local TNF-α challenge and RBP-J does not affect the level of bone resorption in Fcrg–/– mice (Figure 8, B and C). Interestingly, there was almost no bone resorption induced by TNF-α in TKO mice (Figure 8, B and C). This suggests that RBP-J deficiency compensates for the role of DAP12 in TNF-α–induced pathological bone resorption, probably in settings requiring additional signaling, such as that provided by FcRγ.
Figure 8. RBP-J deficiency enables TNF-α to induce inflammatory bone resorption in Dap12–/– mice.
(A) Concentration of serum TRAP obtained from the indicated mice before and after 5-day application of TNF-α to the calvarial periosteum. *P < 0.05; **P < 0.01. n = 5 in each group. (B) μCT images of the surface of whole calvaria obtained from the indicated mice. Scale bar: 1 cm. (C) Quantification of resorptive pit numbers of calvaria obtained from the indicated mice after 5-day application of PBS or TNF-α to the calvarial periosteum. n = 5 in PBS group. n = at least 5 in TNF-α group. *P < 0.05; **P < 0.01.
Discussion
Induction of osteoclastogenesis by RANKL requires calcium signaling to activate NFATc1, which is typically provided by costimulatory signals induced by ITAM-associated receptors to activate PLCγ2 and thus modulate intracellular calcium concentration. In this study, we identify RBP-J as a key transcription factor that controls calcium signaling downstream of ITAM-associated receptors by regulating PLCγ2 expression and function, thereby controlling NFATc1 and downstream osteoclastogenesis. Removal of RBP-J resulted in increased PLCγ2 expression and activity as well as basal calcium signaling that enabled NFATc1 activation independently of upstream signaling by the major ITAM-containing adaptors DAP12 and FcRγ. This newly described pathway was biologically important, as deletion of RBP-J resulted in ITAM adaptor-independent RANKL-induced osteoclastogenesis in vitro and in vivo under homeostatic and inflammatory conditions. These results identify a mechanism that regulates NFATc1 activation in osteoclast differentiation and is important for regulation of bone resorption in vivo.
RBP-J expression decreases over time after RANKL stimulation during osteoclastogenesis (29), and thus the effect of genetic knockout of RBP-J mimics and reflects its role under physiological conditions. The decreased RBP-J expression observed after RANKL stimulation likely contributes to the sustained PLCγ2/calcium signaling that has been described after RANKL stimulation (18), thereby facilitating the ability of upstream ITAM-generated signals to promote osteoclast differentiation. As the expression and function of RBP-J are controlled by various inputs, such as NOTCH, WNT/β-catenin, NF-κB, TLR, and TNF signaling pathways (25–30), our findings suggest that these various pathways can fine tune osteoclastogenesis and bone resorption by determining the strength of upstream ITAM signals required to provide adequate costimulation for NFATc1 activation. This model helps explain why, under certain conditions, such as high concentrations of M-CSF (10), ovariectomy, or low-calcium diet, ITAM-containing adaptors are not required for osteoclast formation and bone remodeling at specific skeletal sites (11). Allelic variants of RBPJ have been recently linked with increased disease susceptibility of rheumatoid arthritis (RA) (53–55). In parallel with these findings, we found that RBP-J expression level was significantly lower in RA patients’ synovial fluid macrophages, which can function as osteoclast precursors, than in monocyte-derived macrophages from healthy donors (Supplemental Figure 14), which supports the pathological relevance of RBP-J to RA. Thus, we speculate that the expression/function of RBP-J can be altered in various environmental cues and diminished RBP-J expression and/or function may contribute to the decreased requirement for ITAM adaptors under certain stress conditions in vivo.
We showed previously that in the presence of intact ITAM-mediated costimulatory signals (29), RBP-J more effectively suppresses TNF-α–induced than RANKL-induced osteoclast differentiation and bone resorption. Our current study helps explain this observation by showing that RBP-J decreases basal PLCγ2/calcium signaling, which imposes a requirement for a strong activating signal, such as that provided by RANKL. As TNF-α induces much weaker calcium signaling than RANKL (51), activation of NFATc1 by TNF-α would require a higher level of basal PLCγ2/calcium signaling than activation by RANKL. This suggests a model where the level of basal PLCγ2/calcium signaling, which is determined by the balance between ITAM-mediated induction and RBP-J–mediated suppression (Figure 6I), determines whether stimulation of osteoclast precursors with factors such as RANKL or TNF-α induces sufficient calcium signaling to cross the threshold required to effectively activate NFATc1.
NFATc1 activation requires sustained calcium signaling, which is manifested as increased intracellular calcium concentrations related to prolonged calcium oscillations (12) rather than an acute and transient calcium flux. Despite extensive study, how RANKL induces sustained calcium signaling is not understood, although induction of expression of ITAM-associated β3 integrins and OSCAR receptors that engage matrix components likely plays a role (56). Our results suggest that decreased RBP-J expression after RANKL stimulation facilitates RANKL-induced sustained calcium signaling, but also highlight an important role for basal PLCγ2 and downstream signaling in osteoclast precursors prior to RANKL stimulation. Basal PLCγ2 activity in osteoclast precursors is likely maintained primarily by constitutive ligation of ITAM-associated receptors, based on our results and previous reports showing that DAP12, FcRγ, and PLCγ2 are basally phosphorylated, that basal calcium concentrations are high, and that SYK and calcium-dependent PYK2 are basally active in myeloid cells (21, 22, 24). In addition to constitutive ITAM-mediated costimulation, we found in this study that endogenous TGF-β signaling also contributes to basal expression and activity of PLCγ2 and downstream basal calcium signaling. Our results show that RBP-J suppresses basal PLCγ2 expression and function, thereby lowering intracellular calcium concentrations and restraining calcium oscillations, thus making it more difficult to activate NFATc1 (12).
The correlation between PLCγ2 expression and phosphorylation in control, RbpjΔM/ΔM, Dap12–/–Fcrg–/–, and TKO cells (Figure 4, D and E) suggests that the expression levels of PLCγ2 may contribute to the regulation of its activity. Although the regulation of PLCγ2 activity has been extensively studied, the mechanism by which PLCγ2 expression is regulated has not been previously explored. Our data identify a new regulatory loop whereby RBP-J restrains expression of TGF-βR1 and thereby modulates cell responsiveness to TGF-β, which in turn regulates expression of PLCγ2 at the transcription level through binding of Smad2/3 to its upstream regulatory region. Suppression of TGF-β signaling by RBP-J in osteoclast precursors provides another piece of evidence of crossregulation of Notch pathway and TGF-β signaling (43–46). Although TGF-β was previously shown to increase intracellular calcium levels and activate downstream calcium signaling and PKC in other systems (47–49), our work, for what we believe is the first time, shows that in BMMs or osteoclast precursors, these effects are mediated by increased PLCγ2 expression and downstream calcium oscillations. Our results provide direct evidence that Plcg2 is a target of SMAD2/3. Thus, our data shows that, in addition to ITAMs and the RANK pathway in osteoclast precursors or BMMs, TGF-β–SMAD2/3 signaling also contributes to calcium signaling. Furthermore, the TGF-β–SMAD2/3–induced PLCγ2/calcium oscillation pathway is restrained by RBP-J. The complex convergence of RANK, ITAM, and TGF-β signaling and its interaction with NOTCH/RBP-J signaling may enable cells to fine tune downstream PLCγ2/calcium signaling in response to various environmental conditions. TGF-β–induced PLCγ2/calcium signaling may explain in part the positive priming effects of TGF-β on osteoclast precursors for differentiation (41, 42). Our single cell analysis of calcium oscillations shows that only a subset of cells in osteoclastogenic cell cultures oscillate at a given time. This heterogeneity of calcium oscillations in osteoclast precursors may help explain the distinct differentiation kinetics of individual cells and is consistent with previous reports that only a subset of cells strongly activate NFATc1 (57).
FcRγ does not significantly affect physiological bone remodeling (18, 24). Our results show that FcRγ-mediated costimulation is dispensable for TNF-α–induced local calvarial bone resorption, which differs from a previous report that used a TNF-α transgenic arthritis model (58). The different results in these 2 models may be explained by differences in the duration of TNF-α stimulation, different concentrations of TNF-α, or differences between effects of systemic and local expression of TNF-α (58). This supports a context-dependent role of FcRγ in regulating pathological bone resorption in various conditions, a notion that is further supported by our findings that a role for FcRγ in TNF-induced bone resorption emerged under conditions of DAP12 deficiency, as FcRγ was required for RBP-J to compensate for DAP12 deficiency in the TKO mice. Although the compensation of DAP12 function by RBP-J deficiency was almost complete in vivo, the multinucleated osteoclasts formed in vitro did not seem fully functional based on their crenated morphology and inefficient bone resorptive ability. This discrepancy between in vivo and in vitro osteoclastogenesis suggests that additional signals from the bony environment could promote osteoclast resorptive function in vivo (9, 18, 32). β3 Integrin plays an important role in cytoskeleton organization (32, 59). RBP-J deficiency significantly rescued β3 integrin expression in Dap12–/–Fcrg–/– cells in response to RANKL, but its level was less than that in control cells (Figure 2C), which may partially explain the cellular “crenated” morphology without obvious actin ring formation in these TKO cells.
RBP-J is a key regulator of osteoclastogenesis and bone resorption. Our previous work showed that RBP-J suppresses osteoclast differentiation by attenuating positive signals that lead to c-FOS/BLIMP1/NFATc1 activation while augmenting the expression of the transcriptional repressor IRF8, which suppresses Nfatc1 expression. RBP-J did not significantly affect the expression of several other negative factors for osteoclastogenesis, such as BCL6, MAFB, and noncanonical NF-κB signaling component p100 (Supplemental Figure 15). The current study reveals that the mechanism by which RBP-J suppresses positive signals that promote osteoclastogenesis is attenuation of costimulatory signaling by suppression of PLCγ2 and downstream calcium-dependent signaling pathways. In contrast, RBP-J deficiency downregulated the transcriptional repressor IRF8 (29) independently of its effects on PLCγ2 and calcium signaling (B. Zhao, unpublished data). Thus, RBP-J functions as a key central regulator that modulates both ITAM-dependent and independent RANK/TNFR signaling during osteoclastogenesis. Our findings establishing a key role for RBP-J in physiological and pathological bone remodeling suggest that modulating RBP-J expression/activity has therapeutic implications for abnormal osteoclastogenesis and associated bone resorption.
Methods
Mice and analysis of bone phenotype.
RbpjΔM/ΔM (Rbpjflox/floxLysM-Cre+) mice (29) and Dap12–/–, Fcrg–/–, and Dap12–/–Fcrg–/– mice (21) have been described previously. The extent of RBP-J deletion by LysM-Cre in vitro and in vivo was typically 70% to 80% (ref. 29 and Supplemental Figure 16). We deleted Rbpj in myeloid lineage osteoclast precursors in Dap12–/–, Fcrg–/–, or Dap12–/–Fcrg–/– mice by crossing these mice with RbpjΔM/ΔM to generate RbpjΔM/ΔMDap12–/–, RbpjΔM/ΔMFcrg–/–, or RbpjΔM/ΔM Dap12–/–Fcrg–/– mice (referred to as TKO), respectively. The mice with Dap12–/– or Dap12–/–Fcrg–/– genotypes also equivalently express LysM-Cre+. Age- and sex-matched mice with a LysM-Cre+ genotype (referred to as control) were used as controls. We generated bone marrow–chimeric mice by transplanting the bone marrows of control, Dap12–/–, Dap12–/–Fcrg–/–, RbpjΔM/ΔMDap12–/–, or TKO mice to 4-week-old male C57BL/6 mice that were obtained from The Jackson Laboratory and served as recipient mice. The method of bone marrow transplantation has been described before (60). We sacrificed chimeric mice 4 months after bone marrow transplantation. We used the protocol for in vivo analysis of TNF-α–induced inflammatory bone resorption as described before (29, 52). When the mice were sacrificed, serum was collected to determine TRAP concentration by using the Mouse TRAP Assay Kit (Immunodiagnostic Systems) according to the manufacturer’s instructions. Femur and calvarial bones were subjected to sectioning, TRAP staining, and histological analysis (60). Bioquant Osteo II software was used for bone histomorphometry.
Reagents.
Murine M-CSF, TNF-α, and sRANKL were purchased from Peprotech. Recombinant mouse TGF-β was from R&D systems. TGF-β receptor I inhibitor SB431542, PLCγ inhibitor U-73122, CaMKK inhibitor STO-609, and PYK2 inhibitor AG 17 were obtained from Millipore. Fluo-4 AM, Fura Red AM, and pluronic F127 were from Life Technologies.
Cell culture.
Murine bone marrow cells were harvested from tibiae and femora, and cultured overnight in petri dishes (BD Biosciences) in α-MEM medium (Invitrogen) with 10% FBS (Invitrogen). Except where stated, CMG14-12 supernatant, which contained the equivalent of 20 ng/ml of recombinant M-CSF, was used as a source of M-CSF, as described (10, 61), in experiments. Nonadherent cells were then replated into tissue-culture dishes and cultured in the same medium for 3 days to obtain BMMs, which are capable of differentiating to osteoclasts, and thus were used as osteoclast precursors. The attached BMMs were scraped, seeded at a density of 4.5 × 104/cm2, and cultured in α-MEM medium with 10% FBS and CMG14-12 supernatant for 1 day. Except where stated, the cells at this time point were used for the basal condition. The cells were then treated without or with RANKL (80 ng/ml) or TNF-α (40 ng/ml) for times indicated in the figure legends. Culture media were exchanged every 3 days. For osteoclast differentiation assays in 96-well plates, 4 wells per condition were used. TRAP staining was performed with an acid phosphatase leukocyte diagnostic kit (Sigma-Aldrich) in accordance with the manufacturer’s instructions.
In vitro gene silencing.
Antisense inhibition using locked nucleic acid (LNA) technology from Exiqon was applied to silence gene expression in vitro. LNA oligonucleotides specifically targeting Tgfbr1 and nontargeting control LNAs were from Exiqon and were transfected into murine BMMs using TransIT-TKO transfection reagent (Mirus) in accordance with the manufacturer’s instructions.
Intracellular calcium measurements.
BMMs were plated in 96-well culture plates with 200 μl culture medium/well. The cells were cultured for 24 hours and labeled with Fluo-4 NW using the Fluo-4 NW Calcium Assay Kit (Molecular Probes). The intracellular calcium concentration was measured according to the manufacturer’s instruction.
Calcium oscillation measurements.
Osteoclast precursors were cultured without or with exogenous TGF-β (10 ng/ml), or with 100 μM SB431542, an inhibitor blocking the kinase activity of TGF-β receptor I, in the presence of recombinant murine M-CSF (20 ng/ml) for 24 hours in glass-bottom culture dishes (MatTek Corporation) and subjected to calcium oscillation measurement, as previously described (18), with small modifications. Briefly, cells were loaded with 5 μM fluo-4 AM, 10 μM Fura Red AM, and 0.05% pluronic F127 and analyzed using confocal microscopy (Leica). Cells were excited at 488 nm, and emissions at 500 to 550 nm for fluo-4 and 600 to 680 nm for Fura Red were acquired simultaneously at 5-second intervals. The relative intracellular calcium levels in single cells were monitored for 300 seconds and indicated by the ratio of the fluorescence intensity of fluo-4 to that of Fura Red (referred to as [Ca++] index ratio in corresponding figure legends). Ionomycin (10 μM) was used to confirm cellular responsiveness to calcium influx for every experiment. At least 100 single cells in each condition were monitored, and each condition was repeated at least 3 times. The average amplitude of calcium oscillation in single cells was calculated using the TuneR and SeeWave packages from the R programming language (62, 63). Each cell trace was converted into a text file and used as input into a user-created function computer code, as follows: meanAmp <- function(x) {cell <- read.table(x); matrix <- as.matrix(cell); Wobj <- Wave(matrix, samp.rate = .2); object <- meanspec(Wobj.2, wl = n – 2, plot = FALSE); x <- object[,2]; return(mean(x[3: (wl/2)]))}, where n = number of time points in cell trace; wl must be an even number less than n.
Control BMM cells cultured with M-CSF alone had small amplitudes of oscillation. For each experiment, oscillating cells were defined as having oscillation amplitudes greater than 1.25 times the median oscillation amplitude of the control conditions. The proportions of oscillating/nonoscillating cells in each condition were calculated, and their differences were statistically analyzed using the χ2 test for independence (P < 0.05 and P < 0.01 were taken as statistically significant).
Reverse transcription and real-time PCR.
Reverse transcription and real-time PCR were performed as previously described (60). The primers for real-time PCR were as follows: Nfatc1: 5′-CCCGTCACATTCTGGTCCAT-3′ and 5′-CAAGTAACCGTGTAGCTCCACAA-3′; Prdm1: 5′-TTCTTGTGTGGTATTGTCGGGACTT-3′ and 5′-TTGGGGACACTCTTTGGGTAGAGTT-3′; Acp5: 5′-ACGGCTACTTGCGGTTTC-3′ and 5′-TCCTTGGGAGGCTGGTC-3′; Ctsk: 5′-AAGATATTGGTGGCTTTGG-3′ and 5′-ATCGCTGCGTCCCTCT-3′; Itgb3: 5′-CCGGGGGACTTAATGAGACCACTT-3′ and 5′-ACGCCCCAAATCCCACCCATACA-3′; Plcg2: 5′-AGACGAAGGCAGACAGCATTG-3′ and 5′-GCCCATTGAGCGAAAACAGC-3′; primary Plcg2: 5′-GCTCAACAACCAGCTCTTCC-3′ and 5′-TTGGAGGGGTTATGGGTACA-3′; Tgfbr1: 5′-TGCCATAACCGCACTGTCA-3′ and 5′- AATGAAAGGGCGATCTAGTGATG-3′; Id1: 5′-AGGTGAACGTCCTGCTCTACGA-3′ and 5′-CAGGATCTCCACCTTGCTCACT-3′; Id3: 5′-CACTTACCCTGAACTCAACGCC-3′ and 5′-CCCATTCTCGGAAAAGCCAG-3′; Cdkn1a: 5′-GCAGATCCACAGCGATATCC-3′ and 5′-CAACTGCTCACTGTCCACGG-3′; Cdkn2b: 5′-AGCTGGATCTGGTCCTTGAG-3′ and 5′-GATCCAAGAATTTCCCTTGC-3′; Smad7: 5′-TTCGGACAACAAGAGTCAGCTGGT-3′ and 5′-AGCCTTGATGGAGAAACCAGGGAA-3′; Gapdh: 5′-ATCAAGAAGGTGGTGAAGCA-3′ and 5′-AGACAACCTGGTCCTCAGTGT-3′.
Immunoblot analysis.
Total cell extracts were obtained using lysis buffer containing 20 mM HEPES (pH 7.0), 300 mM NaCl, 10 mM KCl, 1 mM MgCl2, 0.1% Triton X-100, 0.5 mM DTT, 20% glycerol, and 1× proteinase inhibitor cocktail (Roche). The cell membrane–permeable protease inhibitor Pefabloc (1 mM) was added immediately before harvesting cells. The protein concentration of extracts was quantified using the BCA Protein Assay Kit (Pierce). Cell lysates (10 μg/sample) were fractionated on 7.5% SDS-PAGE, transferred to Immobilon-P membranes (Millipore), and incubated with specific antibodies. Western Lightning plus-ECL (PerkinElmer) was used for detection. Densitometry was performed using ImageJ software (http://imagej.nih.gov/ij/). Phospho-PLCγ2, PLCγ2, phospho-PYK2, and phospho-SMAD2/3 antibodies were from Cell Signaling. c-FOS, BLIMP1, GAPDH, and p38α antibodies were from Santa Cruz Biotechnology Inc. NFATc1 and SMAD2/3 antibodies were from BD Biosciences, and β-tubulin antibody was from Abcam.
ChIP assay.
ChIP assays were performed as previously described (29). Briefly, 5 to 10 × 106 mouse BMMs (2.5 × 106 cells/100 mm dish) per condition were treated as indicated in figure legends, and 50% of chromatin from each condition was used per immunoprecipitation (IP). Cell fixation, lysis, and nuclear extraction were performed as described (64). A Branson Sonifier 250 with a microtip at a power output of 5 (equivalent to 12 w) was used. Samples were sonicated for 6 cycles at 20 seconds each, separated by 1 minute incubations on ice. Average DNA fragment size of 200 to 500 bp was confirmed by agarose gel electrophoresis. For IP, 4 μg per sample of antibodies against RNA polymerase II (Millipore), Smad2/3 (BD Biosciences), or corresponding control rabbit or mouse IgG (Santa Cruz Biotechnology) was used. Immunoprecipitated DNA was analyzed by quantitative real-time PCR and normalized relative to 28S rRNA-encoding gene segments that reflect amounts of nonspecific background DNA precipitation in each reaction. The primers used to detect Pol II occupancy at the Plcg2 locus were as follows: 5′-GCTCAACAACCAGCTCTTCC-3′ and 5′-TTGGAGGGGTTATGGGTACA-3′. The primers used to detect Smad2/3 binding to the upstream regulatory region of Plcg2 in BMMs were designed according to the Smad2/3-ChIPseq data (5 kb upstream of Plcg2) from literature (50); the primer sequences are as follows: 5′-ACTTCAGTCCCCCAAACTCC-3′ and 5′-TTGTCCTTTCTGCCAGTGAA-3′. The primers that amplify 28S rRNA–encoding gene segments are as follows: 5′-GATCCTTCGATGTCGGCTCTTCCTATC-3′ and 5′-AGGGTAAAACTAACCTGTCTCACG-3′.
RNA-seq.
Total RNA was extracted using RNeasy Mini Kit (QIAGEN) following the manufacturer’s instructions. RNA-seq libraries were constructed per the Illumina TrueSeq RNA Sample Preparation Kit. High-throughput sequencing was performed using the Illumina HiSeq 2500/1500. RNA-seq reads were aligned to the mouse genome (mm9, NCBI Build 37) using TopHat (65). Cufflinks (66) was subsequently used to assemble the aligned reads into transcripts and then estimate the transcript abundances as RPKM (reads per kilo base per million) values. Mapped reads are an average of 37 million per library. Gene Set Enrichment Analysis (GSEA program, Broad Institute) was performed according to the program’s instructions. All original RNA-seq data were deposited in the NCBI’s Gene Expression Omnibus (GEO GSE53218).
Statistics.
Statistical analysis was performed using Graphpad Prism® software. One-tailed Student’s t test was applied when there were only 2 groups of samples. In the case of more than 2 groups of samples with 1 condition, 1-way ANOVA followed by Tukey’s post hoc test was used to calculate differences between any groups of samples. In the case of more than 2 groups of samples with more than 1 condition/treatment, 2-way ANOVA followed by Sidak’s multiple comparisons was used. The χ2 test was used to calculate difference of proportions of oscillating/nonoscillating cells. P < 0.05 was taken as statistically significant; all data are presented as the mean ± SD.
Study approval.
All mouse experiments were approved by the Institutional Animal Care and Use Committee of the Hospital for Special Surgery.
Supplementary Material
Acknowledgments
We thank Yingli Shang for help with sectioning and Stephen Doty for help with analysis of the histological sections. We thank Takako Negishi-Koga and Hiroshi Takayanagi for technical support for calcium oscillation experiments. We are grateful to Steven Goldring, F. Patrick Ross, and Takenobu Katagiri for valuable discussion. This work was supported by grants from the NIH (to B. Zhao [AR062047] and L.B. Ivashkiv).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2014;124(11):5057–5073. doi:10.1172/JCI71882.
References
- 1.Novack DV, Teitelbaum SL. The osteoclast: friend or foe? Annu Rev Pathol. 2008;3:457–484. doi: 10.1146/annurev.pathmechdis.3.121806.151431. [DOI] [PubMed] [Google Scholar]
- 2.Nakashima T, Takayanagi H. New regulation mechanisms of osteoclast differentiation. Ann N Y Acad Sci. 2011;1240:E13–E18. doi: 10.1111/j.1749-6632.2011.06373.x. [DOI] [PubMed] [Google Scholar]
- 3.Boyce BF, et al. New roles for osteoclasts in bone. Ann N Y Acad Sci. 2007;1116:245–254. doi: 10.1196/annals.1402.084. [DOI] [PubMed] [Google Scholar]
- 4.Teitelbaum SL. Osteoclasts; culprits in inflammatory osteolysis. Arthritis Res Ther. 2006;8(1):201. doi: 10.1186/ar1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schett G, Gravallese E. Bone erosion in rheumatoid arthritis: mechanisms, diagnosis and treatment. Nat Rev Rheumatol. 2012;8(11):656–664. doi: 10.1038/nrrheum.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schett G, Teitelbaum SL. Osteoclasts and arthritis. J Bone Miner Res. 2009;24(7):1142–1146. doi: 10.1359/jbmr.090533. [DOI] [PubMed] [Google Scholar]
- 7.Asagiri M, Takayanagi H. The molecular understanding of osteoclast differentiation. Bone. 2007;40(2):251–264. doi: 10.1016/j.bone.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 8.Zhao B, Ivashkiv LB. Negative regulation of osteoclastogenesis and bone resorption by cytokines and transcriptional repressors. Arthritis Res Ther. 2011;13(4):234. doi: 10.1186/ar3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Humphrey MB, Lanier LL, Nakamura MC. Role of ITAM-containing adapter proteins and their receptors in the immune system and bone. Immunol Rev. 2005;208:50–65. doi: 10.1111/j.0105-2896.2005.00325.x. [DOI] [PubMed] [Google Scholar]
- 10.Faccio R, Takeshita S, Zallone A, Ross FP, Teitelbaum SL. c-Fms and the alphavbeta3 integrin collaborate during osteoclast differentiation. J Clin Invest. 2003;111(5):749–758. doi: 10.1172/JCI16924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Y, et al. Bone microenvironment specific roles of ITAM adapter signaling during bone remodeling induced by acute estrogen-deficiency. PLoS One. 2007;2(7):e586. doi: 10.1371/journal.pone.0000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivashkiv LB. A signal-switch hypothesis for cross-regulation of cytokine and TLR signalling pathways. Nat Rev Immunol. 2008;8(10):816–822. doi: 10.1038/nri2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivashkiv LB. Cross-regulation of signaling by ITAM-associated receptors. Nat Immunol. 2009;10(4):340–347. doi: 10.1038/ni.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bezbradica JS, Medzhitov R. Role of ITAM signaling module in signal integration. Curr Opin Immunol. 2012;24(1):58–66. doi: 10.1016/j.coi.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 15.Zou W, Teitelbaum SL. Integrins, growth factors, and the osteoclast cytoskeleton. Ann N Y Acad Sci. 2010;1192:27–31. doi: 10.1111/j.1749-6632.2009.05245.x. [DOI] [PubMed] [Google Scholar]
- 16.Negishi-Koga T, Takayanagi H. Ca2+-NFATc1 signaling is an essential axis of osteoclast differentiation. Immunol Rev. 2009;231(1):241–256. doi: 10.1111/j.1600-065X.2009.00821.x. [DOI] [PubMed] [Google Scholar]
- 17.Park-Min KH, et al. IL-10 suppresses calcium-mediated costimulation of receptor activator NF-κB signaling during human osteoclast differentiation by inhibiting TREM-2 expression. J Immunol. 2009;183(4):2444–2455. doi: 10.4049/jimmunol.0804165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koga T, et al. Costimulatory signals mediated by the ITAM motif cooperate with RANKL for bone homeostasis. Nature. 2004;428(6984):758–763. doi: 10.1038/nature02444. [DOI] [PubMed] [Google Scholar]
- 19.Mao D, Epple H, Uthgenannt B, Novack DV, Faccio R. PLCgamma2 regulates osteoclastogenesis via its interaction with ITAM proteins and GAB2. J Clin Invest. 2006;116(11):2869–2879. doi: 10.1172/JCI28775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shinohara M, et al. Tyrosine kinases Btk and Tec regulate osteoclast differentiation by linking RANK and ITAM signals. Cell. 2008;132(5):794–806. doi: 10.1016/j.cell.2007.12.037. [DOI] [PubMed] [Google Scholar]
- 21.Tassiulas I, et al. Amplification of IFN-α-induced STAT1 activation and inflammatory function by Syk and ITAM-containing adaptors. Nat Immunol. 2004;5(11):1181–1189. doi: 10.1038/ni1126. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, et al. ‘Tuning’ of type I interferon-induced Jak-STAT1 signaling by calcium-dependent kinases in macrophages. Nat Immunol. 2008;9(2):186–193. doi: 10.1038/ni1548. [DOI] [PubMed] [Google Scholar]
- 23.Kang YJ, et al. Calcineurin negatively regulates TLR-mediated activation pathways. J Immunol. 2007;179(7):4598–4607. doi: 10.4049/jimmunol.179.7.4598. [DOI] [PubMed] [Google Scholar]
- 24.Mocsai A, et al. The immunomodulatory adapter proteins DAP12 and Fc receptor gamma-chain (FcRγ) regulate development of functional osteoclasts through the Syk tyrosine kinase. Proc Natl Acad Sci U S A. 2004;101(16):6158–6163. doi: 10.1073/pnas.0401602101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu T, et al. Stabilized β-catenin functions through TCF/LEF proteins and the Notch/RBP-Jκ complex to promote proliferation and suppress differentiation of neural precursor cells. Mol Cell Biol. 2008;28(24):7427–7441. doi: 10.1128/MCB.01962-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izumiya Y, Izumiya C, Hsia D, Ellison TJ, Luciw PA, Kung HJ. NF-κB serves as a cellular sensor of Kaposi’s sarcoma-associated herpesvirus latency and negatively regulates K-Rta by antagonizing the RBP-Jκ coactivator. J Virol. 2009;83(9):4435–4446. doi: 10.1128/JVI.01999-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu X, et al. Integrated regulation of Toll-like receptor responses by Notch and interferon-γ pathways. Immunity. 2008;29(5):691–703. doi: 10.1016/j.immuni.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao B, Grimes SN, Li S, Hu X, Ivashkiv LB. TNF-induced osteoclastogenesis and inflammatory bone resorption are inhibited by transcription factor RBP-J. J Exp Med. 2012;209(2):319–334. doi: 10.1084/jem.20111566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu H, et al. Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nat Immunol. 2012;13(7):642–650. doi: 10.1038/ni.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engin F, Lee B. NOTCHing the bone: insights into multi-functionality. Bone. 2010;46(2):274–280. doi: 10.1016/j.bone.2009.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zou W, et al. Syk, c-Src, the alphavbeta3 integrin, and ITAM immunoreceptors, in concert, regulate osteoclastic bone resorption. J Cell Biol. 2007;176(6):877–888. doi: 10.1083/jcb.200611083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaifu T, et al. Osteopetrosis and thalamic hypomyelinosis with synaptic degeneration in DAP12-deficient mice. J Clin Invest. 2003;111(3):323–332. doi: 10.1172/JCI16923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou W, Reeve JL, Liu Y, Teitelbaum SL, Ross FP. DAP12 couples c-Fms activation to the osteoclast cytoskeleton by recruitment of Syk. Mol Cell. 2008;31(3):422–431. doi: 10.1016/j.molcel.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishikawa K, et al. Blimp1-mediated repression of negative regulators is required for osteoclast differentiation. Proc Natl Acad Sci U S A. 2010;107(7):3117–3122. doi: 10.1073/pnas.0912779107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyauchi Y, et al. The Blimp1-Bcl6 axis is critical to regulate osteoclast differentiation and bone homeostasis. J Exp Med. 2010;207(4):751–762. doi: 10.1084/jem.20091957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bai S, et al. NOTCH1 regulates osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblast lineage cells. J Biol Chem. 2008;283(10):6509–6518. doi: 10.1074/jbc.M707000200. [DOI] [PubMed] [Google Scholar]
- 38.Castel D, Mourikis P, Bartels SJ, Brinkman AB, Tajbakhsh S, Stunnenberg HG. Dynamic binding of RBPJ is determined by Notch signaling status. Genes Dev. 2013;27(9):1059–1071. doi: 10.1101/gad.211912.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Hibbs MA, Gard AL, Shylo NA, Yun K. Genome-wide analysis of N1ICD/RBPJ targets in vivo reveals direct transcriptional regulation of Wnt, SHH, and hippo pathway effectors by Notch1. Stem Cells. 2012;30(4):741–752. doi: 10.1002/stem.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamidi H, Gustafason D, Pellegrini M, Gasson J. Identification of novel targets of CSL-dependent Notch signaling in hematopoiesis. PLoS One. 2011;6(5):e20022. doi: 10.1371/journal.pone.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim N, et al. Osteoclast differentiation independent of the TRANCE-RANK-TRAF6 axis. J Exp Med. 2005;202(5):589–595. doi: 10.1084/jem.20050978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yasui T, et al. Regulation of RANKL-induced osteoclastogenesis by TGF-β through molecular interaction between Smad3 and Traf6. J Bone Miner Res. 2011;26(7):1447–1456. doi: 10.1002/jbmr.357. [DOI] [PubMed] [Google Scholar]
- 43.Zavadil J, Cermak L, Soto-Nieves N, Bottinger EP. Integration of TGF-β/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. EMBO J. 2004;23(5):1155–1165. doi: 10.1038/sj.emboj.7600069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niranjan T, et al. The Notch pathway in podocytes plays a role in the development of glomerular disease. Nat Med. 2008;14(3):290–298. doi: 10.1038/nm1731. [DOI] [PubMed] [Google Scholar]
- 45.Sassoli C, et al. Relaxin prevents cardiac fibroblast-myofibroblast transition via notch-1-mediated inhibition of TGF-β/Smad3 signaling. PLoS One. 2013;8(5):e63896. doi: 10.1371/journal.pone.0063896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Misra K, Luo H, Li S, Matise M, Xiang M. Asymmetric activation of Dll4-Notch signaling by Foxn4 and proneural factors activates BMP/TGFβ signaling to specify V2b interneurons in the spinal cord. Development. 2014;141(1):187–198. doi: 10.1242/dev.092536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mukherjee S, Kolb MR, Duan F, Janssen LJ. Transforming growth factor-β evokes Ca2+ waves and enhances gene expression in human pulmonary fibroblasts. Am J Respir Cell Mol Biol. 2012;46(6):757–764. doi: 10.1165/rcmb.2011-0223OC. [DOI] [PubMed] [Google Scholar]
- 48.Alevizopoulos A, Dusserre Y, Ruegg U, Mermod N. Regulation of the transforming growth factor β-responsive transcription factor CTF-1 by calcineurin and calcium/calmodulin-dependent protein kinase IV. J Biol Chem. 1997;272(38):23597–23605. doi: 10.1074/jbc.272.38.23597. [DOI] [PubMed] [Google Scholar]
- 49.Chow JY, Dong H, Quach KT, Van Nguyen PN, Chen K, Carethers JM. TGF-β mediates PTEN suppression and cell motility through calcium-dependent PKC-alpha activation in pancreatic cancer cells. Am J Physiol Gastrointest Liver Physiol. 2008;294(4):G899–G905. doi: 10.1152/ajpgi.00411.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mullen AC, et al. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell. 2011;147(3):565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yarilina A, Xu K, Chen J, Ivashkiv LB. TNF activates calcium-nuclear factor of activated T cells (NFAT)c1 signaling pathways in human macrophages. Proc Natl Acad Sci U S A. 2011;108(4):1573–1578. doi: 10.1073/pnas.1010030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kitaura H, Zhou P, Kim HJ, Novack DV, Ross FP, Teitelbaum SL. M-CSF mediates TNF-induced inflammatory osteolysis. J Clin Invest. 2005;115(12):3418–3427. doi: 10.1172/JCI26132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stahl EA, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42(6):508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eyre S, et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat Genet. 2012;44(12):1336–1340. doi: 10.1038/ng.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Orozco G, et al. Novel RA susceptibility locus at 22q12 identified in an extended UK genome wide association study. Arthritis Rheum. 2014;66(1):24–30. doi: 10.1002/art.38196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barrow AD, et al. OSCAR is a collagen receptor that costimulates osteoclastogenesis in DAP12-deficient humans and mice. J Clin Invest. 2011;121(9):3505–3516. doi: 10.1172/JCI45913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aliprantis AO, Glimcher LH. NFATc1 in inflammatory and musculoskeletal conditions. Adv Exp Med Biol. 2010;658:69–75. doi: 10.1007/978-1-4419-1050-9_8. [DOI] [PubMed] [Google Scholar]
- 58.Ochi S, et al. Pathological role of osteoclast costimulation in arthritis-induced bone loss. Proc Natl Acad Sci U S A. 2007;104(27):11394–11399. doi: 10.1073/pnas.0701971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feng X, et al. A Glanzmann’s mutation in β3 integrin specifically impairs osteoclast function. J Clin Invest. 2001;107(9):1137–1144. doi: 10.1172/JCI12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao B, et al. Interferon regulatory factor-8 regulates bone metabolism by suppressing osteoclastogenesis. Nat Med. 2009;15(9):1066–1071. doi: 10.1038/nm.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takeshita S, Kaji K, Kudo A. Identification and characterization of the new osteoclast progenitor with macrophage phenotypes being able to differentiate into mature osteoclasts. J Bone Miner Res. 2000;15(8):1477–1488. doi: 10.1359/jbmr.2000.15.8.1477. [DOI] [PubMed] [Google Scholar]
- 62.Dolmetsch RE, Lewis RS. Signaling between intracellular Ca2+ stores and depletion-activated Ca2+ channels generates [Ca2+]i oscillations in T lymphocytes. J Gen Physiol. 1994;103(3):365–388. doi: 10.1085/jgp.103.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. R Core Team. R: A Language And Environment For Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 64.Rogatsky I, Zarember KA, Yamamoto KR. Factor recruitment and TIF2/GRIP1 corepressor activity at a collagenase-3 response element that mediates regulation by phorbol esters and hormones. EMBO J. 2001;20(21):6071–6083. doi: 10.1093/emboj/20.21.6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.