Abstract
Cholestenoic acids are formed as intermediates in metabolism of cholesterol to bile acids, and the biosynthetic enzymes that generate cholestenoic acids are expressed in the mammalian CNS. Here, we evaluated the cholestenoic acid profile of mammalian cerebrospinal fluid (CSF) and determined that specific cholestenoic acids activate the liver X receptors (LXRs), enhance islet-1 expression in zebrafish, and increase the number of oculomotor neurons in the developing mouse in vitro and in vivo. While 3β,7α-dihydroxycholest-5-en-26-oic acid (3β,7α-diHCA) promoted motor neuron survival in an LXR-dependent manner, 3β-hydroxy-7-oxocholest-5-en-26-oic acid (3βH,7O-CA) promoted maturation of precursors into islet-1+ cells. Unlike 3β,7α-diHCA and 3βH,7O-CA, 3β-hydroxycholest-5-en-26-oic acid (3β-HCA) caused motor neuron cell loss in mice. Mutations in CYP7B1 or CYP27A1, which encode enzymes involved in cholestenoic acid metabolism, result in different neurological diseases, hereditary spastic paresis type 5 (SPG5) and cerebrotendinous xanthomatosis (CTX), respectively. SPG5 is characterized by spastic paresis, and similar symptoms may occur in CTX. Analysis of CSF and plasma from patients with SPG5 revealed an excess of the toxic LXR ligand, 3β-HCA, while patients with CTX and SPG5 exhibited low levels of the survival-promoting LXR ligand 3β,7α-diHCA. Moreover, 3β,7α-diHCA prevented the loss of motor neurons induced by 3β-HCA in the developing mouse midbrain in vivo.Our results indicate that specific cholestenoic acids selectively work on motor neurons, via LXR, to regulate the balance between survival and death.
Introduction
The vertebrate CNS is composed of a wide variety of neurons that are generated following tightly regulated developmental programs. Characterization of the function and specificity of molecules working on distinct neuronal populations is thus essential in order to enhance our understanding of how such complexity is achieved in the developing brain and how it is maintained in the adult brain. One means of developmental and adult regulation is via nuclear receptors. Examples of nuclear receptors expressed in embryonic and adult brain having both a developmental role (1) and a function in the adult brain (2) are the liver X receptors (LXRα and LXRβ), which are activated by oxysterols (3, 4). Analysis of double-knockout Lxra–/–Lxrb–/– mice revealed that LXRs are required for neurogenesis during ventral midbrain (VM) development (1). Moreover, adult male Lxrb–/– show progressive accumulation of lipids in the brain and loss of spinal cord motor neurons, suggestive of a neuroprotective role of LXRs and their ligands on adult motor neurons (5). Similarly, the number of islet-1+ oculomotor neurons is lower in the developing midbrain of Lxra–/–Lxrb–/– mice, indicating a role of LXRs not only in the maintenance of adult motor neurons, but also in their development (1). In agreement with these findings, enzymes involved in the synthesis of cholesterol and oxysterols, such as 2,3-oxidosqualene-lanosterol cyclase, are localized in islet-1+ oculomotor neurons in the mouse VM at E11.5 (1). In addition to the above, we recently found that oxysterols and other endogenous brain LXR ligands are sufficient to regulate neurogenesis in the developing VM (1, 6). While endogenous brain LXR ligands have been identified and found to regulate the development of midbrain dopamine neurons and red nucleus neurons (6), to date, no endogenous ligand capable of regulating the survival of motor neurons in vivo has been identified. In a recent study, we found that cholesterol metabolites that had the capacity to activate LXRs can be identified in human cerebrospinal fluid (CSF) (7). In order to identify novel LXR ligands that regulate motor neuron function, we delved deeper into the human CSF sterolome and examined plasma of patients with 2 different human diseases associated with upper motor neuron degeneration, hereditary spastic paresis type 5 (SPG5) and cerebrotendinous xanthomatosis (CTX). These diseases result from mutations in the cytochrome P450 (CYP) genes CYP7B1 and CYP27A1, respectively (8–17). The enzymes coded by these genes are responsible for 7α-hydroxylation of oxysterols and (25R),26-hydroxylation of sterols, respectively, reactions that generate further oxysterols and ultimately cholestenoic acids (Figure 1A and ref. 18). Note that we have adopted the sterol nomenclature recommended by the lipid maps consortium: 26-hydroxycholesterol (26-HC) refers to cholest-(25R)-5-en-3β,26-diol; similarly, carboxylic acids that introduce 25R stereochemistry to the side-chain are at C-26 (19). We found that specific cholestenoic acids with a 3β-hydroxy-5-ene, but not a 3-oxo-4-ene, structure activated LXRα and LXRβ in neuronal cells, increased expression of islet-1, a transcription factor required for the development of motor neurons (20–22), and promoted the survival of islet-1+ oculomotor neurons. Moreover, these effects were abolished by knockdown or knockout of the LXR-encoding genes in zebrafish or in rodent models, respectively. In addition, we showed that patients with CTX and SPG5 are unable to synthesize normal amounts of the LXR ligand 3β,7α-dihydroxycholest-5-en-26-oic acid (3β,7α-diHCA), a cholestenoic acid that we found to promote neuronal survival. This is of interest in relation to the fact that patients with SPG5 present with motor neuron degeneration and spastic paraplegia. Patients with CTX may sometimes also present with spasticity, possibly due to upper motor neuron degeneration. Our findings may have important implications for neurological diseases leading to motor neuron degeneration, since LXR ligands, as well as inhibitors of specific biosynthetic enzymes in the cholestenoic acid biosynthetic and metabolic pathways, may be useful pharmaceuticals for the treatment of motor neuron disorders.
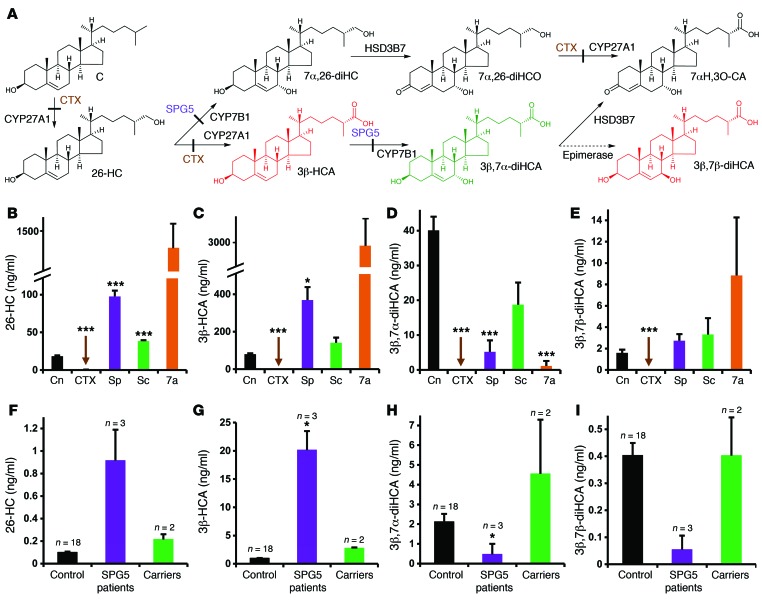
Figure 1. Biosynthesis of cholestenoic acids in brain and levels of cholestenoic acids in the circulation and CSF.
(A) Suggested metabolic pathway for the biosynthesis of 3β-HCA, 3β,7α-diHCA, 3β,7β-diHCA, and 7αH,3O-CA. Sterols are abbreviated according to cholesterol (C), cholest-4-en-3-one (CO), and cholesten-26-oic acid (CA) structures; numbers (with Greek letters) indicate the location of hydroxy (H) and oxo (O) groups. The enzymes in the pathway are indicated. With the exception of the epimerase, each enzyme is known to be expressed in brain. Enzyme defects in CTX and SPG5 are depicted by solid bars across arrows. Metabolites that are toxic toward neurons (red) or neuroprotective (green) are indicated. (B–E) Levels of (B) 26-HC, (C) 3β-HCA, (D) 3β,7α-diHCA, and (E) 3β,7β-diHCA in plasma/serum from control subjects (Cn; n = 50), CTX patients (n = 4), SPG5 patients (Sp; n = 9), SPG5 carriers (Sc; n = 3) and infants with O7AHD (7a; n = 3). (F–I) Levels of (F) 26-HC, (G) 3β-HCA, (H) 3β,7α-diHCA, and (I) 3β,7β-diHCA in CSF from control subjects (n = 18), SPG5 patients (n = 3), and SPG5 carriers (n = 2). Measurements were made by LC-ESI-MS (see Supplemental Tables 1 and 2). *P < 0.05, **P < 0.01, ***P < 0.001 vs. control, Dunnett T3 test.
Results
Specific cholestenoic acids are abundant in human CSF.
We have previously shown that cholestenoic acids are abundant in human plasma (23, 24) and that human CSF also contains specific cholestenoic acids (7). Surprisingly, in CSF, the levels of cholestenoic acids were higher than those of oxysterols. In this study, we delved deeper into the CSF sterolome, particularly into the cholestenoic acid portion, using liquid chromatography–electrospray ionization–mass spectrometry (LC-ESI-MS) analysis. We hereby report the exact identity of 16 oxysterols and downstream metabolites, including cholestenoic acids, found in human CSF (Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI68506DS1). The most abundant of these metabolites (19.48–0.40 ng/ml; Supplemental Figure 1) were 7α-hydroxy-3-oxocholest-4-en-26-oic acid (7αH,3O-CA), 3β-hydroxycholest-5-en-26-oic acid (3β-HCA), and 2 newly identified metabolites in CSF, 3β,7α-diHCA and 3β,7β-dihydroxycholest-5-en-26-oic acid (3β,7β-diHCA). Precursors of these acids, including 26-HC and newly identified 7α,26-dihydroxycholesterol (7α,26-diHC; cholest-5-ene-3β,7α,26-triol) and 7α,26-dihydroxycholest-4-en-3-one (7α,26-diHCO), were also found, but at lower levels (0.15–0.03 ng/ml). Our results thus identified 4 novel oxysterol metabolites in human CSF that were downstream of 26-HC (Figure 1A). 26-HC is metabolized via 7α,26-diHC and 7α,26-diHCO, or via 3β-HCA and 3β,7α-diHCA, to 7αH,3O-CA. While 26-HC can cross the blood-brain barrier (BBB) and enter the brain from the circulation (25), 7αH,3O-CA traverses the BBB and is exported from the brain (26). Very low levels of 24S-hydroxycholesterol (24S-HC; cholest-5-ene-3β,24S-diol), 25-hydroxycholesterol (25-HC; cholest-5-ene-3β,25-diol), and newly identified 7α,25-dihydroxycholesterol (7α,25-diHC; cholest-5-ene-3β,7α,25-triol) and 7α,25-dihydroxycholest-4-en-3-one (7α,25-diHCO) were also found in CSF (0.08–0.03 ng/ml).
Reduced levels of 7α-hydroxylated cholestenoic acids in CSF and plasma/serum of human patients with SPG5.
SPG5 presents with upper motor neuron signs and results from mutations in CYP7B1, encoding the oxysterol 7α-hydroxylase responsible for 7α-hydroxylation of side-chain oxidized sterols that is required for extrahepatic synthesis of 7αH,3O-CA and its precursor, 3β,7α-diHCA (Figure 1A and ref. 18). In order to examine the pathogenic role of such mutations, we sought to identify alterations in oxysterol and cholestenoic acid profiles in CSF and plasma from these patients and then examine the biological activities of the altered metabolites. We first studied the CSF from 3 patients with SPG5 (see Supplemental Table 3 for patient clinical data). Elevated levels of the CYP7B1 substrates 25-HC (P < 0.10), 26-HC (P < 0.07), and 3β-HCA (P < 0.02), as well as reduced levels of its products, 3β,7α-diHCA (P < 0.03) and 7αH,3O-CA (P < 0.001), were found compared with 18 individual control subjects; similar results were found when the SPG5 patients were compared with 2 healthy carrier heterozygotes with a single mutation in CYP7B1 (Figure 1, F–I, and Supplemental Table 1). When plasma was analyzed from 9 SPG5 patients (8, 10, 14), significantly elevated 25-HC (P < 0.03), 26-HC (P < 0.001), and 3β-HCA (P < 0.02) and reduced 3β,7α-diHCA (P < 0.001) and 7αH,3O-CA (P < 0.02) were found compared with control subjects (Figure 1, B–E, and Supplemental Table 2). Similar differences were found between patient samples and 3 healthy carriers. This indicates that for these metabolites, plasma represents a good surrogate for CSF. However, while 3β,7α-diHCA and 7αH,3O-CA in the CNS is normally derived from 26-HC, that found in the circulation can be derived via either the 26-HC or the 7α-hydroxycholesterol (7α-HC; cholest-5-ene-3β,7α-diol) pathway (acidic and neutral, respectively) of bile acid biosynthesis (18). Thus, in SPG5 patients (CYP7B1 mutation), the liver-specific 7α-hydroxylase CYP7A1 (neutral pathway) accounts for the residual content of 3β,7α-diHCA and 7αH,3O-CA found in the circulation. We previously investigated the plasma oxysterol and cholestenoic acid profile of 3 infants with mutations in CYP7B1 (Supplemental Table 2) resulting in oxysterol 7α-hydroxylase deficiency (O7AHD) and neonatal liver disease (27–30), as well as SPG5 in adults (31). The first identification of CYP7B1 mutations were found in a child with severe cholestasis (32), defining a new inborn error of bile acid biosynthesis. As expected by the absence of functional CYP7B1 in these patients, we found very low plasma levels of 3β,7α-diHCA (P < 0.001; Figure 1D and Supplemental Table 2), as described above for SPG5. These patients also had considerably elevated plasma levels of 24S-HC, 25-HC, and 26-HC and high levels of hepatotoxic 3β-hydroxychol-5-en-24-oic acid (3βH-Δ5-BA) compared with SPG5 patients and controls. These findings suggest that additional factors, including increased levels of toxic 3β-hydroxy-5-ene acids, may contribute to the progressive liver disease in these patients at an early age.
CTX is a second human disease that may present with signs of motor neuron loss. It is characterized by mutations in CYP27A1. We found that the plasma of patients with CTX was essentially devoid of 26-HC and the downstream cholestenoic acids (Figure 1, B–E, and Supplemental Table 2). As reported previously (33), elevated levels of 7α-HC plus 7α-hydroxycholest-4-en-3-one (7α-HCO) and/or 7α,12α-dihydroxycholesterol (7α,12α-diHC; cholest-5-ene-3β,7α,12α-triol) plus 7α,12α-dihydroxycholest-4-en-3-one (7α,12α-diHCO) were observed (Supplemental Table 2). The absence of cholestenoic acids in plasma indicates an inability to biosynthesize C27 acids in extrahepatic steroidogenic tissue, including the CNS.
In summary, our data showed specific changes in 7α-hydroxylated cholestenoic acids in plasma of CTX and SPG5 patients; we thus decided to examine their effect on neural function.
Specific cholestenoic acids are ligands to LXRα and LXRβ in neural cells.
In order to gain insights into the mechanism by which alterations in cholesterol metabolism causes neurological disease, we studied whether any of the cholestenoic acids present at high levels in control human CSF and deregulated in CSF or plasma of SPG5 or CTX patients work as LXR ligands. We thus focused on 26-HC, 3β-HCA, 3β,7α-diHCA, its isomer 3β,7β-diHCA, and 7αH,3O-CA (Figure 1A) and tested their capacity to activate LXRα and LXRβ in neural cells. We previously found that 3β-HCA activates LXR (7) and here observed that 3β,7α-diHCA, its isomer 3β,7β-diHCA, and the necessary intermediate for the interconversion of the isomers, 3β-hydroxy-7-oxocholest-5-en-26-oic acid (3βH,7O-CA), had the ability to activate both LXRs and therefore act as LXR ligands in neural cells (Figure 2A and Supplemental Figure 2, A and B). 3β,7α-diHCA was the most potent cholestenoic acid, exhibiting 61% activity/efficacy toward LXRβ activation compared with the known LXR ligand 22R-hydroxycholesterol (22R-HC; cholest-5-ene-3β,22R-diol), followed by 3β-HCA and 3β,7β-diHCA (Supplemental Figure 2B). However 3β,7β-diHCA had the lowest 50% effective concentration (EC50), followed by 3β,7α-diHCA and 3β-HCA. In addition, we confirmed in our system the capacity of 24S-HC and 25-HC to activate LXR and observed that 26-HC had no significant effect. We also tested the activational capacity of 7αH,3O-CA, its 7β-isomer, 7β-hydroxy-3-oxocholest-4-en-26-oic acid (7βH,3O-CA), and the precursors 7α,26-diHC and 7α,26-diHCO, none of which showed significant activity (Figure 2A and Supplemental Figure 2A). Moreover, in order to examine whether the identified acidic ligands exert their effect by binding to LXR, we used the LXR antagonist geranylgeranyl pyrophosphate (GGPP; ref. 34), which blocked their activity (Figure 2C), indicating that the acids are indeed LXR ligands. The activity of the known LXR ligands 22R-HC and 25-HC was similarly blocked. In order to confirm that the cholestenoic acids are indeed agonists to LXR, we tested the LXRβ activational capacity of 3β,7α-diHCA, 3β,7β-diHCA, or 3β-HCA in combination with 22R-HC. No additive effect was observed (Supplemental Figure 3A), which indicates that these cholestenoic acids work, as does 22R-HC, via LXR. To further examine the specificity of the cholestenoic acids described above, farnesoid X receptor (FXR) luciferase reporter assays were performed (Figure 2B). As previously described (35), chenodeoxycholic acid (CDCA; 3α,7α-dihydroxy-5β-cholan-24-oic acid) activated FXR. Interestingly, 3βH-Δ5-BA (36), a cholesterol metabolite identified in plasma (Supplemental Table 2), also activated FXR, but was less potent than CDCA. However, none of the other compounds tested activated the FXR luciferase reporter or modulated the activity of CDCA in neural cells (Figure 2B and Supplemental Figure 3B). Thus, the cholestenoic acids do not exert their effect via modulating the activity of the FXR nuclear receptor or its ligands. Similarly, cholestenoic acids did not activate luciferase reporter assays under the control of a DR5 element (activated by Nur-related factor 1/retinoid X receptor [NURR1/RXR] heterodimers), whereas 9-cis-retinoic acid (9-cis-RA) activated this reporter (Supplemental Figure 2D). Furthermore, while lithocholic acid (LCA; 3α-hydroxy-5β-cholan-24-oic acid) activated a vitamin D receptor (VDR) luciferase reporter, none of the cholestenoic acids tested showed any significant effect on VDR activation in neural cells (Supplemental Figure 2C). Thus, our results indicate that 3β-HCA (7), 3β,7α-diHCA, and 3β,7β-diHCA are specific LXR ligands in neural cells.
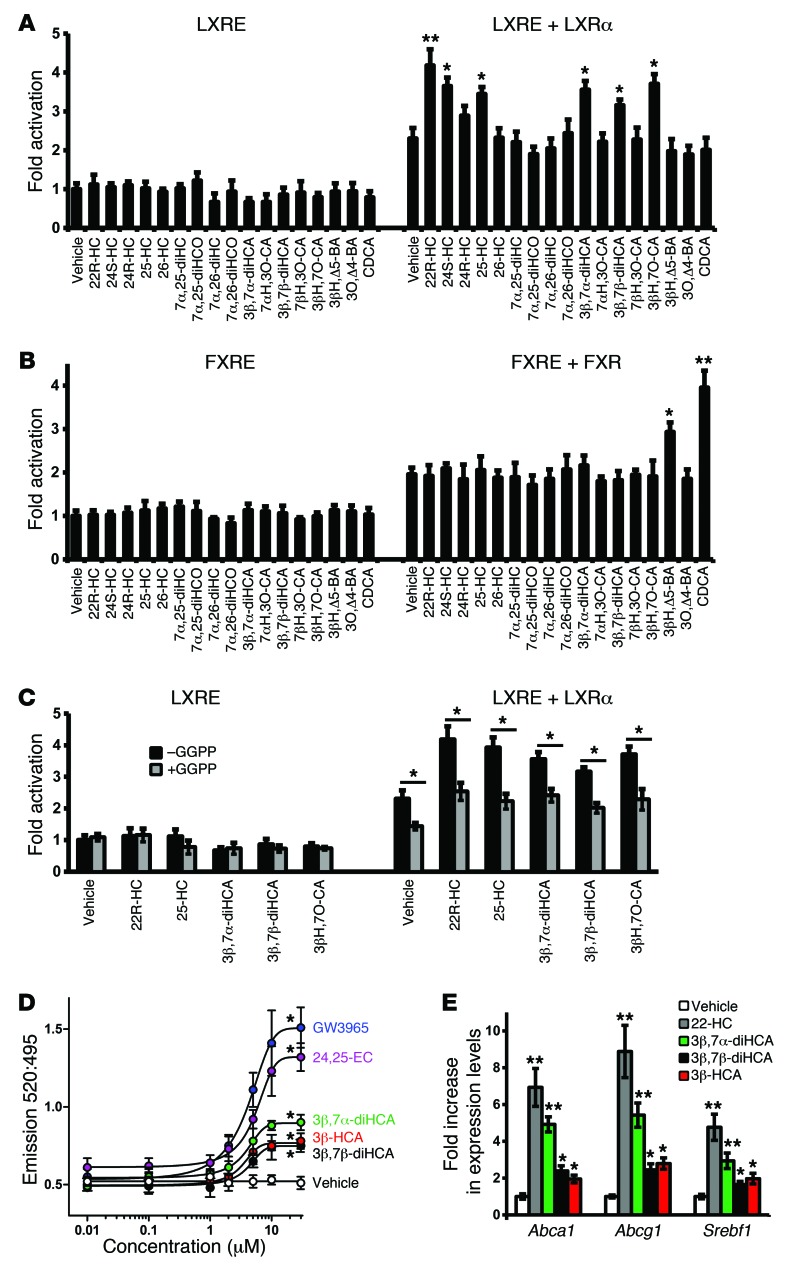
Figure 2. Analysis of the nuclear receptor activational capacity of oxysterols and cholestenoic acids.
(A) Luciferase activity in SN4741 neural cells transfected with an LXR-responsive luciferase reporter construct (LXRE) with or without LXRα and stimulated for 24 hours with 22R-HC (10 μM), a known LXRα ligand (3, 4), or the compounds indicated. (B) Similar assay in cells transfected with an FXR-responsive luciferase reporter construct (FXRE) with or without FXR and stimulated for 24 hours with CDCA, a known FXR ligand, or the compounds indicated. (C) Additional luciferase assays performed as in A with or without addition of the LXR antagonist GGPP (10 μM) along with the indicated cholesterol metabolites (10 μM each). Values in A–C are fold activation over the basal LXRE or FXRE luciferase activity (set to 1) (n = 3). *P < 0.05, **P < 0.01 vs. vehicle or as indicated by brackets, Mann-Whitney test. (D) TR-FRET LXRβ coactivator assay, used to determine the binding affinity of cholestenoic acids as well as the known LXR ligands GW3965 and 24,25-EC toward the LXRβ-LBD (n = 3). The 520:495 TR-FRET ratio was determined as described in Methods. *P < 0.05 vs. vehicle for ≥10 μM 3β,7α-diHCA, ≥10 μM 3β,7β-diHCA, ≥10 μM 3β-HCA, ≥5 μM 24,25-EC, and ≥5 μM GW3965, Mann-Whitney test. (E) 3β,7α-diHCA, 3β,7β-diHCA and 3β-HCA induced significant increases in Abca1, Abcg1, and Srebf1 in SN4741 cells (n = 3). *P < 0.05, **P < 0.01 vs. vehicle, Mann-Whitney test.
Specific cholestenoic acids bind and activate LXR.
In order to show that specific cholestenoic acids directly interact with LXR, we performed a binding and activation fluorescence resonance energy transfer (FRET) assay, in which ligand binding to the ligand binding domain (LBD) of LXRβ (referred to herein as LXRβ-LBD) recruits a fluorescent coactivator. 3β,7α-diHCA induced FRET, in a dose-dependent manner, to a greater extent than the other acids, but to a lesser extent than the known LXR ligands GW3965 and 24S,25-epoxycholesterol (24,25-EC; 3β-hydroxycholest-5-en-24S,25-epoxide) (Figure 2D). The respective efficacy (GW3965-normalized) and EC50 values were 1 and 4.04 μM for GW3965, 0.79 and 4.94 μM for 24,25-EC, 0.41 and 4.16 μM for 3β,7α-diHCA, 0.25 and 4.51 μM for 3β,7β-diHCA, and 0.27 and 3.83 μM for 3β-HCA. To provide further evidence that cholestenoic acids are LXR ligands in neural cells, we treated neural cells with the individual acids for 3 hours and measured Abca1, Abcg1, and Srebf1 transcript levels. 3β,7α-diHCA increased transcripts levels to a similar degree as 22R-HC, while 3β,7β-diHCA and 3β-HCA induced transcription, but to a lesser extent (Figure 2E). These results provide further proof that 3β-HCA, 3β,7α-diHCA, and 3β,7β-diHCA are specific LXR ligands in neural cells.
3β,7α-diHCA and 3βH,7O-CA increase islet-1 expression in islet-1–GFP zebrafish embryos.
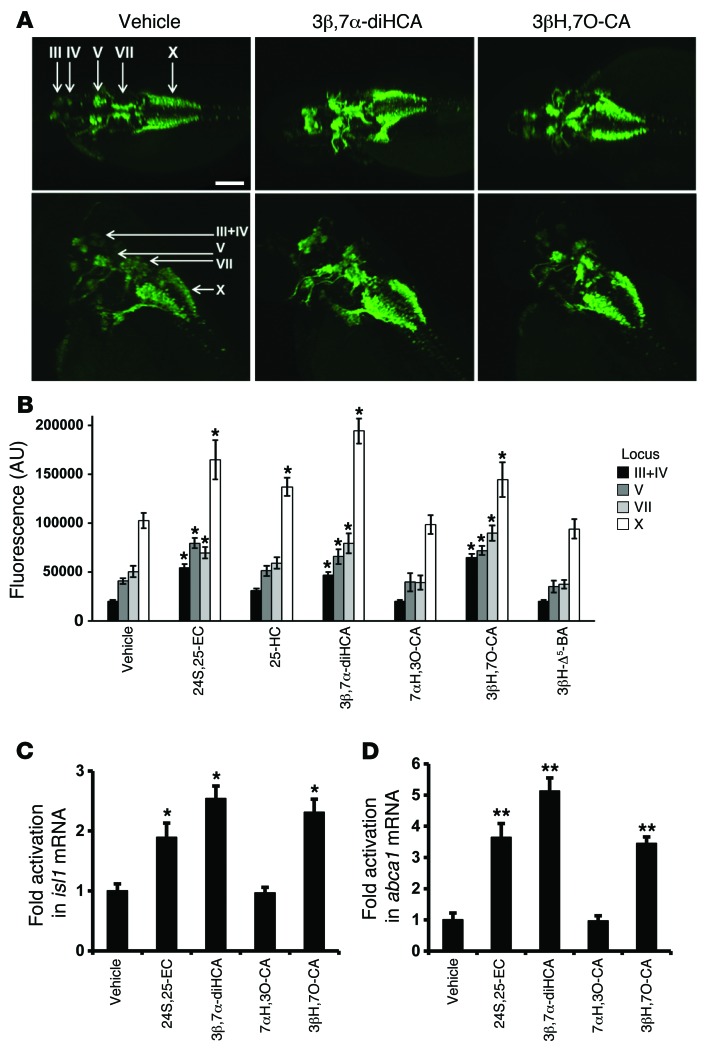
Having established that cholestenoic acids that are altered in CTX or SPG5 can activate LXRs in vitro, we next sought to identify their effect in vivo. In particular, we focused on the expression of islet-1, a transcription factor expressed in all postmitotic motor neurons (20, 37) and required for multiple aspects of motor neuron development, including motor neuron specification, motor column formation, axonal growth, and maintenance of spinal motor neuron identity (38). We used transgenic zebrafish embryos expressing GFP driven by the isl1 gene promoter/enhancer sequences (Tg[isl1:GFP]) (39, 40) to screen for biologically active compounds in vivo. Previous studies have indicated that islet-1 protein is required for the formation of zebrafish primary motor neurons and is conserved throughout vertebrate evolution (21). Treatment of Tg[isl1:GFP] embryos with the weak LXR ligands 3β-HCA and 3β,7β-diHCA, as well as CDCA, the most potent FXR ligand, had a deleterious toxic effect that impaired survival and precluded further in vivo analysis. Interestingly, we found that 2 LXR agonists, 3β,7α-diHCA and 3βH,7O-CA, increased islet-1–GFP expression, but had no significant effect on the number of islet-1+ cells, in the different cranial nerves examined (loci III, IV, V, VII, and X; Figure 3, A and B). These effects were specific, as 7αH,3O-CA, which is not an LXR ligand, failed to regulate islet-1–GFP expression (Figure 3B). Notably, the increase in islet-1–GFP expression observed in response to the specific acidic compounds was evident in all cranial nerves studied (Figure 3B). Importantly, the increased islet-1–GFP expression in response to 3β,7α-diHCA and 3βH,7O-CA was also accompanied by an increased level of isl1 mRNA, as assessed by quantitative PCR (Figure 3C). To further verify our results, we examined whether these cholestenoic acids regulated the in vivo expression of endogenous LXR target genes, such as abca1. Our results showed enhanced expression of abca1 by both 3β,7α-diHCA and 3βH,7O-CA, but not by 7αH,3O-CA (Figure 3D). Finally, in order to determine whether these effects are actually mediated by LXRs, we performed lxr morpholino (MO) injections in Tg[isl1:GFP] zebrafish. Interestingly, we found that lxr MO injections abolished the in vivo increase in islet-1–GFP levels by 3β,7α-diHCA and 3βH,7O-CA, compared with control scrambled MO (Supplemental Figure 4). Thus, our data showed that cholestenoic acids are capable of activating endogenous LXR target genes in vivo and regulate the in vivo expression of islet-1 in brain motor neurons via LXR.
Figure 3. Effects of cholestenoic acids on zebrafish motor neurons.
Tg[isl1:GFP] embryos were incubated with 10 μM test compound or vehicle added to medium, and the medium was replaced every 12 hours with fresh solution (containing test compound or vehicle). Immunocytochemistry was performed using an anti-GFP antibody at 48 hpf. (A) Dorsal (top) and dorsolateral (bottom) views of the head/upper back region of embryos treated with vehicle, 3β,7α-diHCA, or 3βH,7O-CA. Arrows indicate loci III, IV, V, VII, and X (37), which are evolutionarily homologous to the cranial nerves in humans (40). Locus III contains the oculomotor neurons, locus IV the trochlear neurons, locus V the trigeminal motor neurons, locus VII the facial motor neurons, and locus X the cell bodies of the vagus nerve. Scale bar: 50 μm. (B) Quantification of islet-1–GFP signal intensity in the different loci (n = 4). 24,25-EC was used as a positive control. (C and D) mRNA levels of (C) isl1 and (D) abca1 after treatment with the indicated compounds. Data are mean ± SEM (n = 3). *P < 0.05, **P < 0.01 vs. respective vehicle, Mann-Whitney test.
3β,7α-diHCA and 3βH,7O-CA increase, whereas 3β,7β-diHCA and 3β-HCA decrease, the number of rodent oculomotor neurons in vitro.
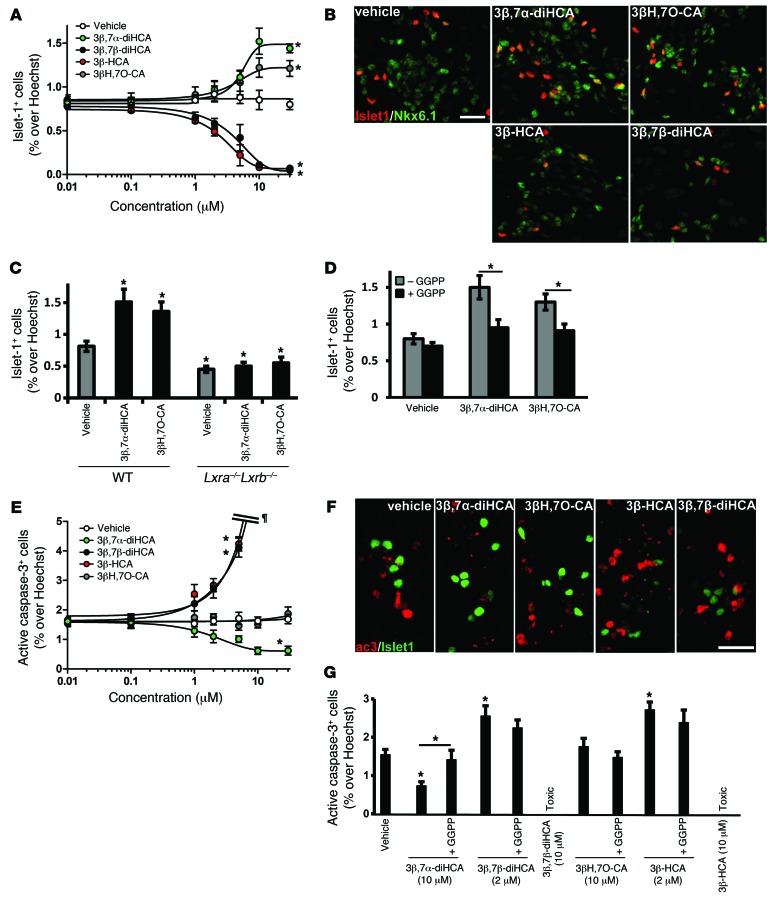
In order to determine the functional relevance of our findings in mammalian cells, we examined whether any of the cholestenoic acids present in human CSF and implicated in motor neuron loss are also capable of regulating islet-1 expression in mouse primary brain progenitor cultures. We first performed dose-response analysis of the cholestenoic acids of interest, using a wide range of concentrations; whereas 3β,7β-diHCA and 3β-HCA caused loss of islet-1+ cells in the cultures, equimolar concentrations of 3β,7α-diHCA and 3βH,7O-CA increased islet-1+ oculomotor cell numbers (Figure 4A). EC50 values were 5.21 μM for 3β,7α-diHCA and 3.58 μM for 3βH,7O-CA, and IC50 values were 4.44 μM for 3β,7β-diHCA and 2.63 μM for 3β-HCA. Moreover, islet-1+ cells coexpressed the transcription factor NKX6.1 as well as choline acetyltransferase (ChAT) (Figure 4B and data not shown), which indicates that they are bona fide oculomotor neurons. Time-course analysis confirmed the stimulatory effect of 3β,7α-diHCA and 3βH,7O-CA, as well as the toxic effect of 3β,7β-diHCA and 3β-HCA, over a range of incubation times (Supplemental Figure 5A). The number of oculomotor neurons peaked after 2 days in culture and leveled thereafter. These results confirmed in rodent cells the dual effects of cholestenoic acids first observed in zebrafish, where 3β,7β-diHCA and 3β-HCA caused death, whereas 3β,7α-diHCA and 3βH,7O-CA regulated islet-1 expression. Thus, our data provide further evidence of the differential effect of cholestenoic acids on islet-1+ oculomotor neurons. Interestingly, the effects of 3β,7α-diHCA and 3βH,7O-CA were specific, as they did not affect the number of other midbrain neurons, such as tyrosine hydroxylase–positive (TH+) dopamine neurons, GABAergic neurons, or red nucleus neurons in the cultures (data not shown). In order to examine whether the effects of 3β,7α-diHCA and 3βH,7O-CA were specifically mediated by LXR receptors in the rodent brain, we performed progenitor brain cultures from Lxra–/–Lxrb–/– mice. Remarkably, the effects of 3β,7α-diHCA and 3βH,7O-CA (at their most efficient concentration) on islet-1+ neurons were eliminated (Figure 4C), confirming that they regulate the number of islet-1+ cells in the rodent brain through LXRs. Moreover, the effects of these acids were blocked by GGPP (Figure 4D), further suggesting that the observed effects were mediated by LXR receptors.
Figure 4. Specific cholestenoic acids increase the number of islet-1+ oculomotor neurons, promote neuronal survival, or are toxic in mouse E11.5 brain primary cultures.
(A) Dose-response curves for quantification of islet-1+ cells in mouse E11.5 brain primary cultures from WT embryos treated with the indicated compounds. *P < 0.05 vs. vehicle for ≥5 μM 3β,7α-diHCA, ≥10 μM 3βH,7O-CA, ≥2 μM 3β,7β-diHCA, and ≥2 μM 3β-HCA, Mann-Whitney test. (B) Representative islet-1– and NKX6.1-stained cell nuclei treated with 3β,7α-diHCA (10 μM), 3βH,7O-CA (10 μM), 3β,7β-diHCA (2 μM), or 3β-HCA (2 μM). (C) Quantification of islet-1+ cells in primary cultures from WT or Lxra–/–Lxrb–/– embryos treated with 3β,7α-diHCA or 3βH,7O-CA (10 μM). *P < 0.05 vs. vehicle, Mann-Whitney test. (D) Effect of 10 μM GGPP on treatment in the WT group. *P < 0.05, Mann-Whitney test. (E) Dose-response curves for quantification of active caspase-3+ cells in mouse E11.5 brain primary cultures. Very high cell death in the cultures is denoted by ¶. *P < 0.05 vs. vehicle for ≥10 μM 3β,7α-diHCA, ≥2 μM 3β,7β-diHCA, and ≥2 μM 3β-HCA, Mann-Whitney test. (F) Representative active caspase-3– and islet-1–stained cell nuclei treated as in B. (G) Quantification of active caspase-3+ cells in primary cultures (WT group) treated as indicated, with or without 10 μM GGPP. *P < 0.05 vs. vehicle or as indicated by brackets, Mann-Whitney test. All data are mean ± SEM (n = 3). Scale bars: 20 μm.
Interestingly, 26-HC, a precursor of cholestenoic acids in the acidic pathway of bile acid biosynthesis (18) that had no LXR activational effect in luciferase assays in neural cells (Figure 2 and ref. 6), also exhibited no effect on the number of islet-1+ cells (data not shown). Furthermore, the effects of 3β,7α-diHCA, 3β,7β-diHCA, and 3β-HCA on islet-1+ cells were not altered by the known LXR ligand 22R-HC (Supplemental Figure 3C), which does not show an effect toward islet-1+ cells (6). These findings are very exciting, as none of the previously known endogenous brain LXR ligands regulates the function of motor neurons. We recently reported that brain endogenous LXR ligands selectively regulate neurogenesis and/or survival of other cell populations in the developing midbrain (6). We thus sought to examine the mechanism by which 3β,7α-diHCA and 3βH,7O-CA increase the number of islet-1+ cells and studied their role in neurogenesis, proliferation, and motor neuron survival in the developing brain.
3β,7α-diHCA promotes rodent oculomotor neuron survival in vitro, whereas 3β,7β-diHCA and 3β-HCA are toxic.
Neurogenesis was examined in BrdU pulse-chase experiments, in which neuronal progenitors in primary cultures are labeled with a pulse of BrdU at the beginning of the experiment and then are examined for their differentiation into motor neurons, as assessed by the acquisition of islet-1 expression, a marker for motor neuron fate (41). Surprisingly, none of the cholestenoic acids studied affected the number of double-positive BrdU+islet-1+ cells (Supplemental Figure 6A), which indicates that they do not promote motor neuron neurogenesis. Similarly, none of the cholestenoic acids affected the total number of BrdU+ cells in the cultures (Supplemental Figure 6B), which suggests that they do not modulate proliferation. Finally, we tested whether cholestenoic acids regulate neuronal survival, as assessed by active caspase-3 staining to detect the number of cells undergoing apoptosis in the cultures. Interestingly, treatment with 3β,7α-diHCA over a wide range of concentrations decreased the number of active caspase-3+ cells, whereas 3βH,7O-CA had no effect (Figure 4, E and F). Since proliferation, neurogenesis, and cell death were not affected by 3βH,7O-CA, we attribute the increase in islet-1+ cells to an increase in differentiation, given the known role of islet-1 in this process (20). Cholestenoic acids that reduced the number of islet-1+ cells were also examined for their capacity to induce cell death in oculomotor neuron cultures. Interestingly, low concentrations of either 3β,7β-diHCA or 3β-HCA increased the number of active caspase-3+ cells (Figure 4, E and F), with toxicity starting at 1 μM. No surviving cells were detected when cultures were treated with concentrations of 3β,7β-diHCA or 3β-HCA higher than 5 μM. Time-course experiments showed that toxicity was evident after 2 days of incubation (Supplemental Figure 5B).
The survival-promoting effect of 3β,7α-diHCA was completely blocked by coincubation with GGPP (Figure 4G), which indicated that this effect was mediated by LXR. Conversely, the cell death effects of 3β,7β-diHCA or 3β-HCA (at 2 μM) were not blocked by GGPP (Figure 4G), which suggested that the survival-promoting effects of 3β,7α-diHCA, but not the toxic effects of 3β,7β-diHCA or 3β-HCA, were mediated by LXR.
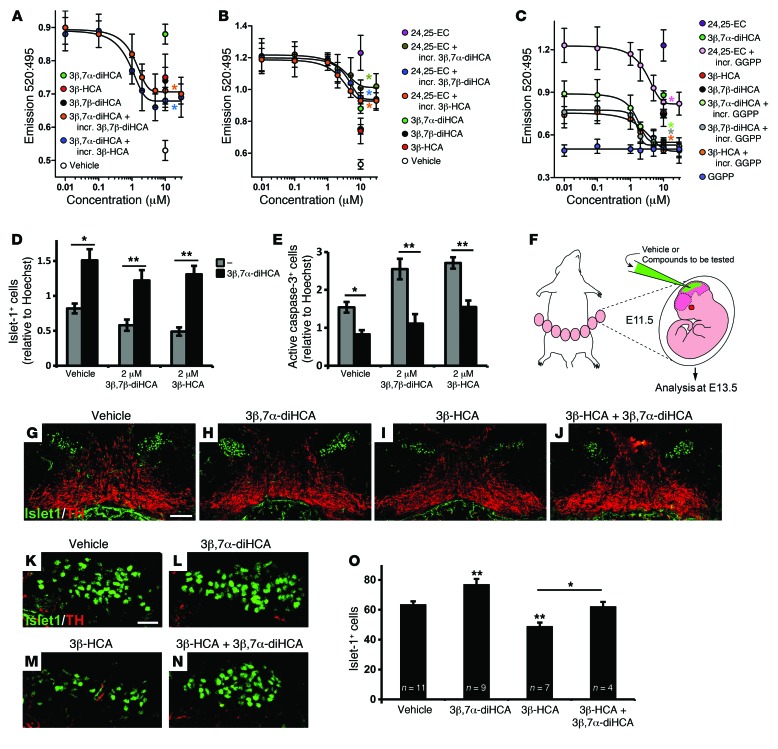
We next examined whether the toxic effects of 3β,7β-diHCA or 3β-HCA could be mediated by receptor blockade or competition. We first performed a binding and activation FRET assay using 3β,7α-diHCA at its most effective concentration and increasing concentrations of 3β,7β-diHCA or 3β-HCA. We found that 3β,7β-diHCA and 3β-HCA reduced the binding of 3β,7α-diHCA in a dose-dependent manner (Figure 5A), which indicates that these toxic cholestenoic acids compete with the neuroprotective 3β,7α-diHCA for binding to the LXRβ-LBD. Ki values were 0.91 μM for 3β-HCA and 1.22 μM for 3β,7β-diHCA.
Figure 5. Competition between cholestenoic acid effects, and 3β,7α-diHCA promotes motor neuron survival in vivo.
(A–C) TR-FRET LXRβ coactivator assays. (A) The effect of 10 μM 3β,7α-diHCA was dose-dependently competed by increasing concentrations (incr.) of 3β,7β-diHCA and 3β-HCA (both *P < 0.05 for ≥2 μM). Similarly, (B) the effect of 10 μM 24,25-EC was dose-dependently competed by 3β,7α-diHCA (*P < 0.05 for ≥10 μM), 3β,7β-diHCA (*P < 0.05 for ≥10 μM), and 3β-HCA (*P < 0.05 for ≥5 μM), and (C) the effects of 3β,7α-diHCA, 3β-HCA, 3β,7β-diHCA, and 24,25-EC (all 10 μM) were dose-dependently competed by GGPP (all *P < 0.05 vs. respective no-GGPP group). Data were analyzed by Mann-Whitney test. (D and E) 10 μM 3β,7α-diHCA rescued the toxic effect of 2 μM 3β,7β-diHCA or 3β-HCA on islet-1+ cells and reduced neuronal cell death induced by these acids, as indicated by the number of active caspase-3+ cells, in mouse E11.5 brain primary cultures. *P < 0.05, **P < 0.01, Mann-Whitney test. (F) Mouse embryos were injected with vehicle or cholestenoic acid into the mesencephalic ventricle in utero at E11.5 and collected at E13.5. (G–N) Midbrain coronal sections showing islet-1+ motor neurons (green) and neighboring TH+ neurons (red) (G–J), and higher magnification of islet-1+ cells (K–N), for embryos injected with vehicle (G and K), 5 μM 3β,7α-diHCA (H and L), 1 μM 3β-HCA (I and M), and 1 μM 3β-HCA plus 5 μM 3β,7α-diHCA (J and N). (O) Quantification of islet-1+ cell number in each group. *P < 0.05, **P < 0.01, 1-way ANOVA with LSD post-hoc test. Scale bars: 100 μm (G–J); 20 μm (K–N). All data are mean ± SEM (n = 3).
Furthermore, we performed competition experiments using the known LXR ligand 24,25-EC (at its most effective concentration) and increasing concentrations of each of the cholestenoic acids studied. 3β,7α-diHCA, 3β-HCA, and 3β,7β-diHCA each reduced the binding of 24,25-EC to the LXRβ-LBD in a dose-dependent manner (Figure 5B), which suggests that each of these cholestenoic acids competes with 24,25-EC for binding to the LXRβ-LBD. 3β-HCA had the lowest Ki value (3β,7α-diHCA, 3.56 μM; 3β,7β-diHCA, 3.45 μM; 3β-HCA, 2.43 μM).
In order to further prove that 3β,7α-diHCA, 3β-HCA, and 3β,7β-diHCA are specific LXR ligands in neural cells, we incubated the acids with GGPP (6). This antagonized and inhibited the ligand-dependent induction of FRET by 3β,7α-diHCA, 3β-HCA, and 3β,7β-diHCA as well as by the known LXR ligand 24,25-EC (Figure 5C), which indicates that 3β,7α-diHCA, 3β-HCA, and 3β,7β-diHCA bind to LXRs. The Ki values for the compounds of interest when incubated with GGPP were 3.96 μM for 24,25-EC, 1.95 μM for 3β,7α-diHCA, 2.51 μM for 3β,7β-diHCA, and 2.02 μM for 3β-HCA.
We next tested whether 3β,7α-diHCA could also compete and reduce the toxic effect of 3β,7β-diHCA or 3β-HCA. Treatment with 10 μM 3β,7α-diHCA reversed the loss of islet-1+ cells and the increased number of activated caspase-3+ cells induced by 2 μM of 3β,7β-diHCA or 3β-HCA (Figure 5, D and E). These observations reinforce the notion of balance between the survival- and death-inducing effects of cholestenoic acids and suggest a new avenue for therapeutic intervention.
3β,7α-diHCA promotes survival of rodent motor neurons in vivo and prevents toxicity by 3β-HCA.
In order to examine whether the observed effects of cholestenoic acids on motor neuron survival could also be detected in the mammalian brain in vivo, we performed cholestenoic acid injections into the cerebral aqueduct of E11.5 mice in utero and analyzed brain sections at the midbrain level at E13.5 (Figure 5F). While only occasional double-positive islet-1+activate caspase-3+ cells remained at the end of the experiment (Supplemental Figure 7), reliable cell counts could be performed for islet-1 and TH (Figure 5, G–O). 3β,7α-diHCA did not affect the number of TH+ neurons, but increased the number of islet-1+ oculomotor neurons, indicative of a selective survival effect of 3β,7α-diHCA on oculomotor neurons in vivo. Upon injection of 3β-HCA, the number of islet-1+ oculomotor neurons was reduced, showing the toxic effect of this metabolite on motor neurons in vivo. Finally, coinjection of 3β-HCA and 3β,7α-diHCA reversed the loss of oculomotor neurons induced by 3β-HCA, indicating that 3β,7α-diHCA protects oculomotor neurons against cholestenoic acids such as 3β-HCA in vivo. These data further confirm the specificity of the neuronal survival and toxic effects of these cholestenoic acids on islet-1+ neurons, but not TH+ neurons, as well as the competitive interaction between prosurvival and toxic cholestenoic acids on motor neurons in vivo.
In summary, our results indicate that cholestenoic acids regulate the number of islet-1+ motor neurons by controlling islet-1 expression (3βH,7O-CA) and competitively regulating neuronal survival in a positive (3β,7α-diHCA) or negative (3β,7β-diHCA and 3β-HCA) manner.
Discussion
The results presented herein show that cholestenoic acids are not mere intermediate metabolites of bile acid biosynthesis, but rather represent a diverse family of bioactive compounds capable of regulating nuclear receptor function. Cholestenoic acids were found to specifically activate LXR and elicit an exquisite array of functions, ranging from the regulation of islet-1 expression to the positive and negative regulation of motor neuron survival both in vitro and in vivo. Moreover, our findings identified cholestenoic acids in human CSF to be deregulated in plasma of CTX and SPG5 patients. Importantly, an absence of neuroprotective cholestenoic acids was found in CTX patient plasma, while a combination of decreased neuroprotective and increased toxic cholestenoic acids was detected in SPG5 plasma. These results are consistent with cholestenoic acids being key regulators of motor neuron function in development and disease.
Cholesterol is present at high levels in the CNS of vertebrates and is metabolized in brain predominantly to 24S-HC (42), which accounts for about two-thirds of brain cholesterol metabolism (43). Low levels of 26-HC have been found in human and mouse brain (Supplemental Results and Supplemental Table 4) and in human CSF, where it may be imported from the blood (25, 44–46). Conversely, 7αH,3O-CA, a metabolic product of 26-HC, is exported from brain to blood in humans (26). In the current study, we found 4 intermediates in the biosynthesis of 7αH,3O-CA from 26-HC via CYP27A1 and CYP7B1 to be present in normal CSF: 3β-HCA, 3β,7α-diHCA, 7α,26-diHC, and 7α,26-diHCO (Figure 1A). While 3β-HCA and 7α,26-diHC were previously found in human neural tissue (46, 47), 3β,7α-diHCA and 7α,26-diHCO have not been previously found in neural tissue or CSF. Importantly, the identification of these intermediates in the biosynthesis of 7αH,3O-CA lends further support to the hypothesis that 7αH,3O-CA is biosynthesized in the human brain (26). This pathway is also conserved in rodents, in which 26-HC is the precursor for the synthesis of 3β-HCA in fetal neurons and of 3β,7α-diHCA and 7αH,3O-CA in fetal astrocytes (Figure 1A and ref. 48). There have been few studies of specific brain regions for cholestenoic acids; however, human retina has been found to contain 3β-HCA at a level of up to 130 pmol/mg protein, about 10% of the level of 24S-HC in brain gray matter (47), whereas in chronic subdural hematoma, 7αH,3O-CA has been found at a level of 1.5 μM (49). Future studies will be directed at profiling cholestenoic acids in distinct brain regions. Surprisingly, Cyp7b1–/– and Cyp27a1–/– mice, unlike human SPG5 patients and some CTX patients, do not suffer from motor neuron disease, despite an absence of 3β,7α-diHCA in brain and plasma (Supplemental Table 4). These findings suggest that other LXR ligands may compensate for the accumulative effect of altered levels of cholestenoic acids in Cyp7b1–/– and Cyp27a1–/– mice.
Classic studies by Lehmann et al. (4) and Janowski et al. (3) defined the general structural requirements of steroidal LXR ligands to be a 3β-hydroxy-5-ene function in the ring system and a hydroxy, oxo, or epoxide function on the C-17 side-chain (3, 4). The side-chain functions have more recently been extended to include a carboxylic acid group (7, 50), a functional group that is also present in the synthetic nonsteroidal LXR ligand GW3965. In the current study, we confirmed the LXR-activational capacity of cholestenoic acids with a 3β-hydroxy-5-ene structure in neural cells and showed that despite the introduction of a 7α-hydroxy, 7β-hydroxy, or 7-oxo group, LXR activity was maintained. On the contrary, the 3-oxo-4-ene equivalents of these acids were not LXR ligands. Thus, 3β-HCA, 3β,7α-diHCA, its 7β-isomer (3β,7β-diHCA), and the necessary 7-oxo intermediate in the epimerization reaction (3βH,7O-CA) are all LXR ligands effective at micromolar concentrations. Moreover, none of these acids were found to activate FXR, VDR, or NURR1 in neural cells, thereby confirming the specificity of their effect on LXR.
Numerous studies in recent years have linked LXR to neuronal degeneration (2, 5, 51–53). These studies have used Lxrb–/– and Lxra–/–Lxrb–/– mice. Indeed, both LXR isoforms are expressed in brain (1, 54), and the knockout mice show progressive lipid accumulation in brain, abnormal BBB, increased reactive microglia, astrogliosis, and degeneration of adult spinal cord motor neurons (2, 5). Interestingly, a decrease in the number of oculomotor neurons was also detected during development in Lxra–/–Lxrb–/– mice at E11.5 (1). However, the identity of endogenous brain LXR ligands that regulate motor neuron function was unknown. Here we used zebrafish to study the in vivo function of acidic LXR ligands newly identified in human CSF (i.e., 3β,7α-diHCA and 3β,7β-diHCA), and the 7-oxo intermediate in their epimerization, to study the effect of these molecules on islet-1+ cranial motor neurons. While metabolites that did not activate LXR, such as 7αH,3O-CA, did not regulate islet-1 expression, the LXR ligands 3β,7α-diHCA and 3βH,7O-CA enhanced isl1 transcript expression in zebrafish embryos (Figure 3), effects that were abolished by injection of lxr MO (Supplemental Figure 4). These effects were confirmed in rodent primary oculomotor neuron cultures, which showed that only 3β,7α-diHCA and 3βH,7O-CA increased the number of islet-1+ neurons (Figure 4, A–D). Importantly, their activity was specific to and mediated by LXR, as their biological activities were eliminated in cultures from Lxra–/–Lxrb–/– mice or by the LXR inhibitor GGPP. The effect of 3β,7α-diHCA on WT cultures was accompanied by a decrease in the number of active caspase-3+ cells (Figure 4E), but no change in neurogenesis or proliferation was detected (Supplemental Figure 6), which indicates that the mechanism by which it increased the number of motor neurons was by promoting neuronal survival. In vivo experiments performed on embryonic brain in utero confirmed this neuroprotective effect of 3β,7α-diHCA on oculomotor neurons (Figure 5, G–O). In contrast, 3βH,7O-CA had no effect on cell death, neurogenesis, or survival in WT cultures, but increased the expression of islet-1, which suggested that it promoted the maturation of precursor cells into islet-1+ neurons. Analysis of the function of 3β,7β-diHCA and 3β-HCA revealed a toxic effect, manifested by an increase in the number of active caspase-3+ cells at low concentrations (Figure 4E). Thus, our results indicated that only some of the cholestenoic acids capable of activating LXR promote motor neuron development. Intriguingly, our findings also suggested that cholestenoic acids regulate motor neuron number by distinct mechanisms regulating differentiation and survival. Moreover, we found that the regulation of motor neuron survival could be either positive, as shown by the neuroprotective effect of 3β,7α-diHCA, or negative, as shown by the toxic effect of 3β,7β-diHCA and 3β-HCA (Figures 4 and 5). These results were quite unexpected, but correlated very well with the cholestenoic acid profiles in patients with the human diseases CTX and SPG5 (characterized by mutations in CYP27A1 and CYP7B1, respectively) that may present with signs of motor neuron loss. Indeed, we found that 3β-HCA, one of the LXR ligands identified to have a toxic effect, was present at higher levels in SPG5 patients in both plasma and CSF (Figure 1, C and G, and Supplemental Tables 1 and 2). At the same time, the LXR ligand found to be neuroprotective, 3β,7α-diHCA, was present at lower levels in SPG5 patients and absent in CTX patients (Figure 1, D and H). These observations suggest a double-hit model for SPG5, in which loss of motor neurons is effected by both mechanisms, whereas for CTX we propose that the predominant mechanism would be loss of neuroprotection. Moreover, our finding that the neuroprotective cholestenoic acid 3β,7α-diHCA prevented the loss of oculomotor neurons induced by 3β-HCA, both in vitro and in vivo, suggests a possible application of 3β,7α-diHCA in the treatment of both SPG5 and CTX. The fact that deletion of LXRs results in early developmental motor neuron loss (1), while the onset of disease in SPG5 or CTX is usually in adolescent or adults, indicates that additional endogenous LXR ligands may partially compensate for the loss of 3β,7α-diHCA in SPG5 and CTX patients. Future studies should examine whether 3β,7α-diHCA and other yet-unidentified LXR ligands represent useful therapeutic tools to treat these and perhaps other devastating diseases affecting motor neurons.
In summary, we identified 3β,7α-diHCA and 3β,7β-diHCA as LXR ligands present in human CSF. Of these, 3β,7β-diHCA and the previously identified LXR ligand 3β-HCA were found to cause cell death, and the latter was present at high levels in patients with SPG5. Instead, 3β,7α-diHCA, which was found at low levels in SPG5 and absent in CTX patients, promoted motor neuron survival, while 3βH,7O-CA regulated islet-1 expression levels. Thus, our results uncovered several novel functions of cholestenoic acids and identified them as LXR ligands and key regulators of motor neuron function in development and disease. We suggest that factors with neuroprotective function, such as 3β,7α-diHCA, may thus find a therapeutic application to prevent motor neuron loss. This suggestion is supported by our finding that 3β,7α-diHCA, a cholestenoic acid that decreased in SPG5, prevented cell loss by 3β-HCA, a cholestenoic acid that accumulates in SPG5.
Methods
Extraction of sterols.
Sterols were extracted from CSF, plasma, or mouse brain into ethanol and fractionated by reversed phase solid phase extraction (SPE) to give a cholestenoic acid– and oxysterol-rich fraction devoid of cholesterol (7, 55, 56).
Charge-tagging of sterols.
The sterols were charge-tagged with the GP-hydrazine as described previously (7, 55, 56). This greatly enhances their response when analyzed by LC-ESI-MS and tandem mass spectrometry (MSn).
Reagents.
HPLC-grade water and solvents were from Fisher Scientific or Sigma-Aldrich. Authentic sterols, steroids, cholestenoic acids, bile acids, and their precursors were from Avanti Polar Lipids, Steraloids, Sigma-Aldrich, or from previous studies in our laboratories. Girard P (GP) reagent [1-(carboxymethyl)pyridinium chloride hydrazide] was from TCI Europe or synthesized in earlier studies, and cholesterol oxidase from Streptomyces sp. was from Sigma-Aldrich. Certified Sep-Pak C18 200 mg SPE cartridges were from Waters. Luer-lock syringes were from BD Biosciences.
LC-ESI-MSn on the LTQ-Orbitrap.
LC-ESI-MS and LC-ESI-MSn were performed using an Ultimate 3000 HPLC system (Dionex) linked to the ESI source of a LTQ-Orbitrap XL or LTQ-Orbitrap Velos (Thermo Fisher) mass spectrometer as described previously (7, 55, 56).
Luciferase reporter assay.
The ability of oxysterols and their acidic metabolites to activate several nuclear receptors (i.e., LXRα, LXRβ, FXR, VDR, and NURR1) was tested in luciferase assays. Transient transfection studies were performed in the mouse neuronal cell line SN4741. This cell line was selected because the oxysterols and acidic metabolites tested were initially identified in CSF. Cells were plated in 24-well plates (5 × 105 cells/well) 24 hours before transfection and transfected with 1 μg plasmid DNA/well complexed with 2 μl Lipofectamine 2000 (Invitrogen). Cells were transfected with 400 ng of a LXR-, FXR-, VDR-, or NURR1-responsive luciferase reporter construct and 200 ng LXRα, LXRβ, FXR, VDR, or NURR1. A reporter gene expressing the Renilla luciferase (pRL-TK; Promega) was cotransfected in all experiments as an internal control for normalization of transfection efficiency. After a 12-hour incubation, the lipid/DNA mix was replaced with fresh 2.5% serum medium containing vehicle or appropriate ligand (10 μM), as specified in each experiment. The ability of cholestenoic acids to activate LXR was confirmed in experiments with or without the LXR inhibitor GGPP (10 μM) also added to the medium. Luciferase activities were assayed 24 hours later using the Dual-Luciferase Reporter Assay System (Promega), following the manufacturer’s protocol.
LXRβ ligand binding assay.
For LXRβ ligand binding activity measurement, we used the Lanthascreen TR-FRET LXRβ Coactivator Assay (Invitrogen). The assay uses a terbium-labeled anti-GST antibody, a fluorescein-labeled coactivator peptide, and the LXRβ-LBD tagged with glutathione-S-transferase (GST). Binding of the agonist/ligand to the LXRβ-LBD causes a conformational change that increases the affinity of LXRβ for the coactivator peptide. Close proximity of the fluorescently labeled coactivator peptide to the terbium-labeled antibody increases the TR-FRET signal intensity. The 520:495 TR-FRET ratio was calculated using a Victor multilabel reader with an excitation wavelength of 340 nm and emission wavelengths of 520 nm and 495 nm. The activational capacity of potential ligands was tested in a 382-well polypropylene plate following the manufacturer’s protocol.
Zebrafish experiments.
Zebrafish were raised on a 14-hour day/10-hour night cycle and kept at 28.5°C. Tg[isl1:GFP] embryos were obtained via natural spawning and staged in hours postfertilization (hpf) or days post fertilization, according to Kimmel et al. (57). Embryos older than 24 hpf were treated with 0.03% phenylthiourea (PTU) to inhibit pigmentation. MO injections were performed using splice site–specific zebrafish lxr MO, as previously reported (58). MO-injected embryos were immediately dechorionated, transferred to a 96-well plate, and exposed to DMSO- or ligand-treated medium. Tg[isl1:GFP] embryos were collected by natural mating, immediately dechorionated (at the 1-cell stage), and transferred to a 96-well plate. Each compound tested was obtained as 10 mM stock and diluted in embryo medium to a final concentration of 10 μM, and 200 μl was added to each well. DMSO- or propan-2-ol–treated embryo medium was used as control. Ligand solutions were replaced every 12 hours with fresh ligand solution prepared in PTU-treated embryo medium. Embryos were collected at 48 hpf, fixed for 4 hours at room temperature with 4% paraformaldehyde (PFA), and then washed and kept in PBST. Immunocytochemistry was performed using an anti-GFP antibody, and fluorescence was viewed and photographed using a Zeiss Axioplan compound microscope and a Zeiss Axiocam digital camera.
Quantitative PCR.
Total RNA was extracted from SN4741 cells and from zebrafish treated with the compounds of interest using the RNeasy Mini Kit (Qiagen); 1 μg was treated with RQ1 RNase-free DNase (Promega) and reverse transcribed using SuperScript II Reverse Transcriptase (Invitrogen) and random primers (Invitrogen) (RT+ reaction). Parallel reactions without reverse transcriptase enzyme were performed as a control (RT– reaction), and Sybergreen real-time quantitative PCR assays were carried out. Expression levels were obtained by normalization to the value of the housekeeping gene encoding actin or 18S, obtained for every sample in parallel assays.
Mice.
Mice were housed, bred, and treated according to the guidelines of the European Communities Council (directive 86/609/EEC) and the Society for Neuroscience. Lxra–/–Lxrb–/– mouse cell cultures were from the colony at the Department of Biosciences and Nutrition at Novum, Karolinska Institutet. Male and female WT and Lxra–/–Lxrb–/– mice were generated as previously described (59, 60). Mice were backcrossed onto a C57BL/6 background for 10 generations. Male Cyp7b1–/– mouse brain and plasma were from animals generated at the University of Edinburgh. Male mice homozygous for targeted disruption of the Cyp7b1 gene (61) congenic on the C57BL/6 genetic background (>15 generations backcrossed to C57BL/6) and WT littermate controls were generated from Cyp7b1–/+ crosses. Male Cyp27a1–/– mouse tissue and plasma was purchased from The Jackson Laboratory (strain B6.129-Cyp27a1tm1Elt/J; ref. 62). The Cyp27a1–/– colony was backcrossed to C57BL/6J inbred mice for approximately 12 generations by the donating investigator (62) prior to sending to The Jackson Laboratory Repository. Upon arrival, mice were bred to C57BL/6J inbred mice for at least 1 generation to establish the colony. WT animals from the colony were used as controls. Tissue and blood sampling from these mice was performed under the aegis of the UK Scientific Procedures (Animals) Act, 1986.
Primary midbrain cultures.
Brains from E11.5 mice were obtained, and the midbrain region was dissected, mechanically dissociated, plated on poly-d-lysine (150,000 cells/cm2), and grown in serum-free N2 media consisting of F12/DMEM (1:1 mixture) with 10 ng/ml insulin, 100 μg/ml apo-transferrin, 100 μM putrescine, 20 nM progesterone, 30 nM selenium, 6 mg/ml glucose, and 1 mg/ml BSA. Cells were treated for 3 days in vitro with the compounds of interest, fixed with 4% PFA, and processed for staining using appropriate antibodies.
For BrdU analysis, cells were treated with BrdU 1 hour after plating, and media was replaced with fresh medium after 16 hours. After a further 2 days in culture, cells were treated for 30 minutes with 2N HCl, and immunocytochemistry was performed to evaluate the number of double-positive BrdU+islet-1+ cells (a measure of motor neuron neurogenesis). Hoechst staining was performed by permeabilizing cells with a 0.3% Triton-X 100/PBS solution for 5 minutes followed by incubation with Hoechst 33258 (Sigma-Aldrich) for 10 minutes.
Immunocytochemistry.
Cells were fixed in 4% PFA, washed in PBS, and blocked in 5% normal goat serum/PBS for 1 hour at room temperature. Primary antibodies were diluted in PBS (pH 7.4), 0.3% Triton X-100, and 1% BSA, and incubations were carried out overnight at 4°C or at room temperature for 2 hours. The antibodies used were anti-BrdU (diluted 1:400; Abcam), anti–islet-1 (1:100; Developmental Studies Hybridoma Bank), anti–cleaved caspase-3 (Asp175) (1:100; Cell Signaling Technology), anti-TH (1:1,000; Pel-Freeze), anti-GABA (1:1,000; Sigma-Aldrich), anti-BRN3A (1:250; Millipore), anti-NKX6.1 (1:200; Novus Biologicals), anti-ChAT (1:500; Millipore), and appropriate secondary antibodies (Jackson ImmunoResearch or Alexa). Cells positive for the corresponding marker were counted directly at the microscope at ×20 magnification. Cells were counted in every well — in 8 consecutive fields (from one side of the well to the other, passing through the center) — in 3 different wells per experiment and 3 different experiments per condition. Random images of the wells were taken for every condition to document the result, and representative images were subsequently selected to represent the quantitative data. Photos were acquired with a Zeiss Axioplan microscope and a Hamamatsu camera C4742-95 using Openlab software.
Intraventricular injections in utero.
Female WT CD-1 mice (25–35 g; Charles River Breeding Laboratories) were used for these experiments. For embryo analyses, WT CD-1 mice were mated overnight, and noon of the day the plug was considered E0.5. E11.5 pregnant females were deeply anesthetized using isofluorane (IsoFlo; Abbott Labs), and the uterine horns were accessed through an abdominal incision. 1 μl 3β,7α-diHCA (5 μM) or 3β-HCA (1 μM) or vehicle solution (isopropanol; 50% v/v) was injected into the cerebral aqueduct. The uterine horns were replaced into the abdominal cavity, which was then closed with sutures. Embryos were analyzed 48 hours later.
Immunohistochemical analysis of rodent embryos.
Embryos were dissected out of the uterine horns in ice-cold PBS, fixed in 4% PFA for 4 hours to overnight, cryoprotected in 15%–30% sucrose, frozen in Tissue-Tek OCT compound (Sakura Finetek) on dry ice, and stored at –80°C until use. Serial coronal 14-μm sections of the brain were obtained on a cryostat. 14-μm serial coronal sections through the midbrain region were cut on a cryostat and placed serially on 10 slides. Slides 1 and 6 were subjected to immunohistochemistry. Sections were preincubated for 1 hour in blocking solution followed by incubation at 4°C overnight with the following primary antibodies: anti-TH (1:750, Pel-Freeze), anti–islet-1 (1:100; Developmental Studies Hybridoma Bank), and anti–cleaved caspase-3 (Asp175) (1:100; Cell Signaling Technology). After washing, slides were incubated for 1–2 hours at room temperature with the appropriate fluorophore-conjugated (Cy2, Cy3, and Cy5, 1:300, Jackson Laboratories; Alexa Fluor 488, Alexa Fluor 555, and Alexa Fluor 647, 1:1,000, Invitrogen) secondary antibodies. Confocal images were taken with a Zeiss LSM5 Exciter or LSM700 microscope.
Statistics.
Statistical analyses (Mann-Whitney test, Dunnett T3 test, and 1-way ANOVA with LSD post-hoc test) were performed using Prism 4 (GraphPad Software). All data represent mean ± SEM. A P value less than 0.05 was considered significant.
Study approval.
Human samples, collected according to the principles of the Declaration of Helsinki, were provided to University Hospital Basel, Barts Health NHS Trust, St. Mary’s Hospital, Institute of Child Health, Conegliano Research Centre, Federico II University, University of Tubingen, Kurume University School of Medicine, and University Hospital “Attikon” with written informed consent and IRB and ethical approval. Ethical approval was granted by Stockholm Norra Djurförsöksetisks Nämnd for zebrafish experimentation (nos. N293/09 and N338/10), mouse experimentation (nos. N154/06, N145/09, N370/09, and N273/11), and in utero experimentation (no. N486/12).
Supplementary Material
Acknowledgments
Work in Swansea was supported by funding from the UK Research Councils BBSRC (BBC5157712, BBC5113561, BBI0017351 to W.J. Griffiths, BBH0010181 to Y. Wang, studentship to A. Meljon) and EPSRC (studentship to M. Ogundare). The EPSRC National Mass Spectrometry Service Centre is acknowledged for allowing access to the LTQ-Orbitrap XL mass spectrometer. Work in Karolinska Institutet was supported by funding from the Swedish Research Council (DBRM program; VR2011-3116 and VR2011-3318), the European Union (DDPD program), the Swedish Foundation for Strategic Research (DBRM and SRL program), and the Hjärnfonden and Karolinska Institutet (SFO Thematic Center in Stem cells and Regenerative Medicine). S. Theofilopoulos was supported by the Swedish MRC and the Onassis Foundation. P.T. Clayton is funded by Great Ormond Street Hospital Children’s Charity. M.T. Bassi was supported by Italian Ministry of Health funds (grant no. RC2011-2012). Work in Houston was supported by the Robert A. Welch Foundation (grant no. E-0004, to J.-Å. Gustafsson). Work in Edinburgh was supported by the Wellcome Trust (WT73429) and a Carter Fellowship to J.L. Yau from Alzheimer’s Research UK. Work in Tubingen was supported by an EU grant to NEUROMICS (grant no. 2012-305121). Members of the European Network for Oxysterol Research are thanked for informative discussions.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2014;124(11):4829–4842. doi:10.1172/JCI68506.
References
- 1.Sacchetti P, et al. Liver X receptors and oxysterols promote ventral midbrain neurogenesis in vivo and in human embryonic stem cells. Cell Stem Cell. 2009;5(4):409–419. doi: 10.1016/j.stem.2009.08.019. [DOI] [PubMed] [Google Scholar]
- 2.Wang L, et al. Liver X receptors in the central nervous system: from lipid homeostasis to neuronal degeneration. Proc Natl Acad Sci U S A. 2002;99(21):13878–13883. doi: 10.1073/pnas.172510899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janowski BA, et al. Structural requirements of ligands for the oxysterol liver X receptors LXRα and LXRβ. Proc Natl Acad Sci U S A. 1999;96(1):266–271. doi: 10.1073/pnas.96.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lehmann JM, et al. Activation of the nuclear receptor LXR by oxysterols defines a new hormone response pathway. J Biol Chem. 1997;272(6):3137–3140. doi: 10.1074/jbc.272.6.3137. [DOI] [PubMed] [Google Scholar]
- 5.Andersson S, Gustafsson N, Warner M, Gustafsson JA. Inactivation of liver X receptor β leads to adult-onset motor neuron degeneration in male mice. Proc Natl Acad Sci U S A. 2005;102(10):3857–3862. doi: 10.1073/pnas.0500634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Theofilopoulos S, et al. Brain endogenous liver X receptor ligands selectively promote midbrain neurogenesis. Nat Chem Biol. 2012;9(2):126–133. doi: 10.1038/nchembio.1156. [DOI] [PubMed] [Google Scholar]
- 7.Ogundare M, et al. Cerebrospinal fluid steroidomics: are bioactive bile acids present in brain? J Biol Chem. 2010;285(7):4666–4679. doi: 10.1074/jbc.M109.086678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnoldi A, et al. Clinical phenotype variability in patients with hereditary spastic paraplegia type 5 associated with CYP7B1 mutations. Clin Genet. 2012;81(2):150–157. doi: 10.1111/j.1399-0004.2011.01624.x. [DOI] [PubMed] [Google Scholar]
- 9.Björkhem I. Cerebrotendinous xanthomatosis. Curr Opin Lipidol. 2013;24(4):283–287. doi: 10.1097/MOL.0b013e328362df13. [DOI] [PubMed] [Google Scholar]
- 10.Criscuolo C, et al. Two novel CYP7B1 mutations in Italian families with SPG5: a clinical and genetic study. J Neurol. 2009;256(8):1252–1257. doi: 10.1007/s00415-009-5109-3. [DOI] [PubMed] [Google Scholar]
- 11.Diekstra FP, et al. Mapping of gene expression reveals CYP27A1 as a susceptibility gene for sporadic ALS. PLoS One. 2012;7(4):e35333. doi: 10.1371/journal.pone.0035333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manganelli F, et al. Electrophysiological characterisation in hereditary spastic paraplegia type 5. Clin Neurophysiol. 2011;122(4):819–822. doi: 10.1016/j.clinph.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 13.Mondelli M, Rossi A, Scarpini C, Dotti MT, Federico A. Evoked potentials in cerebrotendinous xanthomatosis and effect induced by chenodeoxycholic acid. Arch Neurol. 1992;49(5):469–475. doi: 10.1001/archneur.1992.00530290051011. [DOI] [PubMed] [Google Scholar]
- 14.Schüle R, et al. Marked accumulation of 27-hydroxycholesterol in SPG5 patients with hereditary spastic paresis. J Lipid Res. 2010;51(4):819–823. doi: 10.1194/jlr.M002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugama S, et al. Frontal lobe dementia with abnormal cholesterol metabolism and heterozygous mutation in sterol 27-hydroxylase gene (CYP27). J Inherit Metab Dis. 2001;24(3):379–392. doi: 10.1023/A:1010564920930. [DOI] [PubMed] [Google Scholar]
- 16.Verrips A, et al. Spinal xanthomatosis: a variant of cerebrotendinous xanthomatosis. Brain. 1999;122(pt 8):1589–1595. doi: 10.1093/brain/122.8.1589. [DOI] [PubMed] [Google Scholar]
- 17.Verrips A, et al. Clinical and molecular genetic characteristics of patients with cerebrotendinous xanthomatosis. Brain. 2000;123(pt 5):908–919. doi: 10.1093/brain/123.5.908. [DOI] [PubMed] [Google Scholar]
- 18.Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- 19.Fahy E, et al. A comprehensive classification system for lipids. J Lipid Res. 2005;46(5):839–861. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 20.Ericson J, Thor S, Edlund T, Jessell TM, Yamada T. Early stages of motor neuron differentiation revealed by expression of homeobox gene Islet-1. Science. 1992;256(5063):1555–1560. doi: 10.1126/science.1350865. [DOI] [PubMed] [Google Scholar]
- 21.Hutchinson SA, Eisen JS. Islet1 and Islet2 have equivalent abilities to promote motoneuron formation and to specify motoneuron subtype identity. Development. 2006;133(11):2137–2147. doi: 10.1242/dev.02355. [DOI] [PubMed] [Google Scholar]
- 22.Song MR, et al. Islet-to-LMO stoichiometries control the function of transcription complexes that specify motor neuron and V2a interneuron identity. Development. 2009;136(17):2923–2932. doi: 10.1242/dev.037986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Axelson M, Mörk B, Sjövall J. Occurrence of 3 β-hydroxy-5-cholestenoic acid, 3 β,7 α-dihydroxy-5-cholestenoic acid, and 7 α-hydroxy-3-oxo-4-cholestenoic acid as normal constituents in human blood. J Lipid Res. 1988;29(5):629–641. [PubMed] [Google Scholar]
- 24.Griffiths WJ, et al. Analytical strategies for characterization of oxysterol lipidomes: liver X receptor ligands in plasma. Free Radic Biol Med. 2013;59:69–84. doi: 10.1016/j.freeradbiomed.2012.07.027. [DOI] [PubMed] [Google Scholar]
- 25.Heverin M, et al. Crossing the barrier: net flux of 27-hydroxycholesterol into the human brain. J Lipid Res. 2005;46(5):1047–1052. doi: 10.1194/jlr.M500024-JLR200. [DOI] [PubMed] [Google Scholar]
- 26.Meaney S, et al. Novel route for elimination of brain oxysterols across the blood-brain barrier: conversion into 7α-hydroxy-3-oxo-4-cholestenoic acid. J Lipid Res. 2007;48(4):944–951. doi: 10.1194/jlr.M600529-JLR200. [DOI] [PubMed] [Google Scholar]
- 27. Chong CP, Mills PB, McClean P, Clayton PT. Response to chenodeoxycholic acid therapy in an infant with oxysterol 7α-hydroxylase deficiency. In: Abstracts of the Annual Symposium of the Society for the Study of Inborn Errors of Metabolism; Aug 31–Sep 6, 2010; Istanbul, Turkey. Abstract S382. [Google Scholar]
- 28.Dai D, et al. Liver disease in infancy caused by oxysterol 7α-hydroxylase deficiency: successful treatment with chenodeoxycholic acid. J Inherit Metab Dis. doi: 10.1007/s10545-014-9695-6. [published online ahead of print]. doi: 10.1007/s10545-014-9695-6. [DOI] [PubMed] [Google Scholar]
- 29.Mizuochi T, et al. Successful heterozygous living donor liver transplantation for an oxysterol 7α-hydroxylase deficiency in a Japanese patient. Liver Transpl. 2011;17(9):1059–1065. doi: 10.1002/lt.22331. [DOI] [PubMed] [Google Scholar]
- 30.Ueki I, et al. Neonatal cholestatic liver disease in an Asian patient with a homozygous mutation in the oxysterol 7α-hydroxylase gene. J Pediatr Gastroenterol Nutr. 2008;46(4):465–469. doi: 10.1097/MPG.0b013e31815a9911. [DOI] [PubMed] [Google Scholar]
- 31.Goizet C, et al. CYP7B1 mutations in pure and complex forms of hereditary spastic paraplegia type 5. Brain. 2009;132(pt 6):1589–1600. doi: 10.1093/brain/awp073. [DOI] [PubMed] [Google Scholar]
- 32.Setchell KD, et al. Identification of a new inborn error in bile acid synthesis: mutation of the oxysterol 7alpha-hydroxylase gene causes severe neonatal liver disease. J Clin Invest. 1998;102(9):1690–1703. doi: 10.1172/JCI2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Björkhem I, Hansson M. Cerebrotendinous xanthomatosis: an inborn error in bile acid synthesis with defined mutations but still a challenge. Biochem Biophys Res Commun. 2010;396(1):46–49. doi: 10.1016/j.bbrc.2010.02.140. [DOI] [PubMed] [Google Scholar]
- 34.Forman BM, Ruan B, Chen J, Schroepfer GJ, Jr, Evans RM. The orphan nuclear receptor LXRα is positively and negatively regulated by distinct products of mevalonate metabolism. Proc Natl Acad Sci U S A. 1997;94(20):10588–10593. doi: 10.1073/pnas.94.20.10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiang JY, Kimmel R, Weinberger C, Stroup D. Farnesoid X receptor responds to bile acids and represses cholesterol 7α-hydroxylase gene (CYP7A1) transcription. J Biol Chem. 2000;275(15):10918–10924. doi: 10.1074/jbc.275.15.10918. [DOI] [PubMed] [Google Scholar]
- 36.Nishimaki-Mogami T, et al. Identification of intermediates in the bile acid synthetic pathway as ligands for the farnesoid X receptor. J Lipid Res. 2004;45(8):1538–1545. doi: 10.1194/jlr.M400102-JLR200. [DOI] [PubMed] [Google Scholar]
- 37.Higashijima S, Hotta Y, Okamoto H. Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J Neurosci. 2000;20(1):206–218. doi: 10.1523/JNEUROSCI.20-01-00206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang X, et al. Isl1 Is required for multiple aspects of motor neuron development. Mol Cell Neurosci. 2011;47(3):215–222. doi: 10.1016/j.mcn.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Higashijima S. Transgenic zebrafish expressing fluorescent proteins in central nervous system neurons. Dev Growth Differ. 2008;50(6):407–413. doi: 10.1111/j.1440-169X.2008.01023.x. [DOI] [PubMed] [Google Scholar]
- 40.Uemura O, et al. Comparative functional genomics revealed conservation and diversification of three enhancers of the isl1 gene for motor and sensory neuron-specific expression. Dev Biol. 2005;278(2):587–606. doi: 10.1016/j.ydbio.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 41.Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 1996;84(2):309–320. doi: 10.1016/S0092-8674(00)80985-X. [DOI] [PubMed] [Google Scholar]
- 42.Lütjohann D, et al. Cholesterol homeostasis in human brain: evidence for an age-dependent flux of 24S-hydroxycholesterol from the brain into the circulation. Proc Natl Acad Sci U S A. 1996;93(18):9799–9804. doi: 10.1073/pnas.93.18.9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lund EG, et al. Knockout of the cholesterol 24-hydroxylase gene in mice reveals a brain-specific mechanism of cholesterol turnover. J Biol Chem. 2003;278(25):22980–22988. doi: 10.1074/jbc.M303415200. [DOI] [PubMed] [Google Scholar]
- 44.Heverin M, et al. Changes in the levels of cerebral and extracerebral sterols in the brain of patients with Alzheimer’s disease. J Lipid Res. 2004;45(1):186–193. doi: 10.1194/jlr.M300320-JLR200. [DOI] [PubMed] [Google Scholar]
- 45.Leoni V, et al. Side chain oxidized oxysterols in cerebrospinal fluid and the integrity of blood-brain and blood-cerebrospinal fluid barriers. J Lipid Res. 2003;44(4):793–799. doi: 10.1194/jlr.M200434-JLR200. [DOI] [PubMed] [Google Scholar]
- 46.Shafaati M, et al. Marked accumulation of 27-hydroxycholesterol in the brains of Alzheimer’s patients with the Swedish APP 670/671 mutation. J Lipid Res. 2011;52(5):1004–1010. doi: 10.1194/jlr.M014548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liao WL, et al. Quantification of cholesterol-metabolizing P450s CYP27A1 and CYP46A1 in neural tissues reveals a lack of enzyme-product correlations in human retina but not human brain. J Proteome Res. 2011;10(1):241–248. doi: 10.1021/pr1008898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J, Akwa Y, el-Etr M, Baulieu EE, Sjövall J. Metabolism of 27-, 25- and 24-hydroxycholesterol in rat glial cells and neurons. Biochem J. 1997;322(pt 1):175–184. doi: 10.1042/bj3220175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagata K, et al. Identification of 7 alpha-hydroxy-3-oxo-4-cholestenoic acid in chronic subdural hematoma. Biochim Biophys Acta. 1992;1126(2):229–236. doi: 10.1016/0005-2760(92)90295-7. [DOI] [PubMed] [Google Scholar]
- 50.Song C, Liao S. Cholestenoic acid is a naturally occurring ligand for liver X receptor alpha. Endocrinology. 2000;141(11):4180–4184. doi: 10.1210/endo.141.11.7772. [DOI] [PubMed] [Google Scholar]
- 51.Bigini P, et al. Neuropathologic and biochemical changes during disease progression in liver X receptor β–/– mice, a model of adult neuron disease. J Neuropathol Exp Neurol. 2010;69(6):593–605. doi: 10.1097/NEN.0b013e3181df20e1. [DOI] [PubMed] [Google Scholar]
- 52.Dai YB, Tan XJ, Wu WF, Warner M, Gustafsson JA. Liver X receptor beta protects dopaminergic neurons in a mouse model of Parkinson disease. Proc Natl Acad Sci U S A. 2012;109(32):13112–13117. doi: 10.1073/pnas.1210833109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim HJ, et al. Liver X receptor β (LXRβ): a link between beta-sitosterol and amyotrophic lateral sclerosis-Parkinson’s dementia. Proc Natl Acad Sci U S A. 2008;105(6):2094–2099. doi: 10.1073/pnas.0711599105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whitney KD, et al. Regulation of cholesterol homeostasis by the liver X receptors in the central nervous system. Mol Endocrinol. 2002;16(6):1378–1385. doi: 10.1210/mend.16.6.0835. [DOI] [PubMed] [Google Scholar]
- 55.Griffiths WJ, Wang Y. Analysis of oxysterol metabolomes. Biochim Biophys Acta. 2011;1811(11):784–799. doi: 10.1016/j.bbalip.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 56.Meljon A, et al. Analysis of bioactive oxysterols in newborn mouse brain by LC/MS. J Lipid Res. 2012;53(11):2469–2483. doi: 10.1194/jlr.D028233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 58.Archer A, et al. The Liver X-receptor (Lxr) governs lipid homeostasis in zebrafish during development. Open J Endocrine Metab Dis. 2012;2(4):74–81. doi: 10.4236/ojemd.2012.24012. [DOI] [Google Scholar]
- 59.Alberti S, et al. Hepatic cholesterol metabolism and resistance to dietary cholesterol in LXRβ-deficient mice. J Clin Invest. 2001;107(5):565–573. doi: 10.1172/JCI9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Steffensen KR, Robertson K, Gustafsson JA, Andersen CY. Reduced fertility and inability of oocytes to resume meiosis in mice deficient of the Lxr genes. Mol Cell Endocrinol. 2006;256(1–2):9–16. doi: 10.1016/j.mce.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 61.Rose K, et al. Neurosteroid hydroxylase CYP7B: vivid reporter activity in dentate gyrus of gene-targeted mice and abolition of a widespread pathway of steroid and oxysterol hydroxylation. J Biol Chem. 2001;276(26):23937–23944. doi: 10.1074/jbc.M011564200. [DOI] [PubMed] [Google Scholar]
- 62.Rosen H, et al. Markedly reduced bile acid synthesis but maintained levels of cholesterol and vitamin D metabolites in mice with disrupted sterol 27-hydroxylase gene. J Biol Chem. 1998;273(24):14805–14812. doi: 10.1074/jbc.273.24.14805. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.