Abstract
Patients with BRAFV600E/K-driven melanoma respond to the BRAF inhibitor vemurafenib due to subsequent deactivation of the proliferative RAS/RAF/MEK/ERK pathway. In BRAF WT cells and those with mutations that activate or result in high levels of the BRAF activator RAS, BRAF inhibition can lead to ERK activation, resulting in tumorigenic transformation. We describe a patient with malignant melanoma who developed chronic lymphocytic leukemia (CLL) in the absence of RAS mutations during vemurafenib treatment. BRAF inhibition promoted patient CLL proliferation in culture and in murine xenografts and activated MEK/ERK in primary CLL cells from additional patients. BRAF inhibitor–driven ERK activity and CLL proliferation required B cell antigen receptor (BCR) activation, as inhibition of the BCR-proximal spleen tyrosine kinase (SYK) reversed ERK hyperactivation and proliferation of CLL cells from multiple patients, while inhibition of the BCR-distal Bruton tyrosine kinase had no effect. Additionally, the RAS-GTP/RAS ratio in primary CLL cells exposed to vemurafenib was reduced upon SYK inhibition. BRAF inhibition increased mortality and CLL expansion in mice harboring CLL xenografts; however, SYK or MEK inhibition prevented CLL proliferation and increased animal survival. Together, these results suggest that BRAF inhibitors promote B cell malignancies in the absence of obvious mutations in RAS or other receptor tyrosine kinases and provide a rationale for combined BRAF/MEK or BRAF/SYK inhibition.
Introduction
BRAF kinase inhibition has revolutionized the treatment of melanoma with somatic V600E or V600K mutations and led to improved overall survival (1). However, in tumors and normal cells with WT RAF, drug-bound BRAF cooperates with GTP-loaded, activated RAS proteins in eliciting paradoxical activation of the MEK/ERK pathway by stimulating drug-free RAF molecules via dimerization, in particular with the RAF1 isoform (2–5). This paradoxical ERK activation underlies the occurrence of keratoacanthomas, squamous cell carcinomas, and even de novo melanomas in the context of RAF inhibitor treatment (6, 7). Compatible with the concept that increased RAS signaling mediates paradoxical ERK activation under BRAF inhibition, activating mutations in RAS genes were found in the majority of cutaneous squamous lesions (8), as secondary events in previously vemurafenib-responsive BRAF mutant melanoma (9), in a chronic myelomonocytic leukemia (CMML) (10), and in a pancreatic carcinoma (11) progressing under BRAF inhibition. Overexpression, mutation, and microenvironment-mediated hyperactivation of RTKs were identified as drug-resistance mechanisms in melanoma. In any scenario, RTK hyperactivity is very likely to increase RAS activity and thereby could contribute to paradoxical ERK activation as well (12, 13).
To our knowledge, no previous reports have documented progression of a lymphoid malignancy driven by BRAF inhibition in the absence of a RAS mutation. Instead, this malignancy was driven by spleen tyrosine kinase activity (SYK) that is likely the result of chronic B cell antigen receptor (BCR) signaling. Here, we present a patient in whom chronic lymphocytic leukemia (CLL) with WT RAS developed shortly after the initiation of vemurafenib therapy for metastatic BRAF mutant melanoma. Upon discontinuation of vemurafenib, CLL disease burden diminished. The observed phenomenon was not restricted to individual patients, but was reproducible in CLL cells from multiple patients. We were able to model dependence of the CLL clone on BRAF inhibition in vivo in multiple patient-derived CLL samples and provide evidence for the biochemical mechanism responsible for RAS-independent promotion of CLL cells by vemurafenib.
Results
Exacerbation of CLL during vemurafenib treatment.
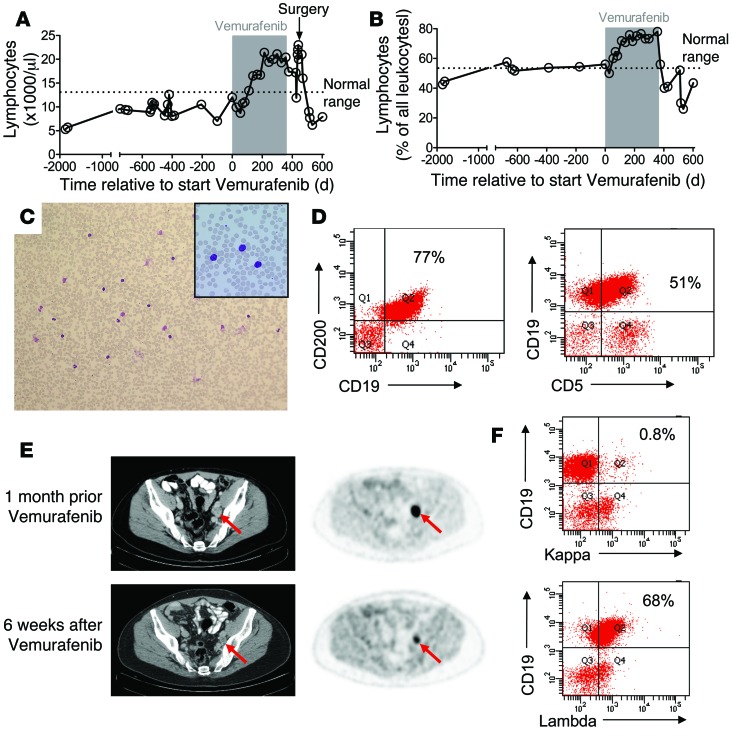
A 49-year-old patient with stage IV (pT2bpN3pM1a, AJCC classification 2009; ref. 14) BRAF V600 mutant melanoma presented to our dermatology outpatient clinic. Six years earlier, the patient had been diagnosed with melanoma located on the left lower extremity (tumor thickness 1.2 mm according to Breslow with ulceration, Clark’s level of invasion IV, sentinel node biopsy inguinal left without evidence of metastasis). Following surgical resection, the patient received adjuvant immunotherapy with IFN-α–2a 3 times, 3 million IU per week, subcutaneously for 18 months. Relapse with a subcutaneous metastasis of the left lower extremity and in the inguinal and iliacal lymph nodes (LNs) had been documented 4 years after primary diagnosis, and the tumor manifestation was surgically removed twice and irradiated (60 Gy) at the site of the subcutaneous metastasis due to R1-resection status. Ten months later, new LN metastases occurred at the same locations, and surgery and radiotherapy could not adequately control disease. To treat progression of inoperable inguinal, iliacal, and paraaortal LN metastases, the patient received 960 mg of vemurafenib twice a day (study ID number MO25515; ClinicalTrials.gov identifier: NCT01307397; Hoffmann-La Roche). At the time vemurafenib was initiated, his white cell and lymphocyte counts were in the normal and upper normal ranges, respectively. The patient developed significant leukocytosis and lymphocytosis during vemurafenib treatment (Figure 1, A and B).
Figure 1. Clinical course of a melanoma patient with CLL progressing during treatment with vemurafenib.
Displayed are the white-cell count (A) and the lymphocyte count (B) at multiple time points prior to and after vemurafenib treatment (gray area). (C) A representative blood smear of the patient during vemurafenib treatment is shown. Original magnification, ×100; ×200 (inset). The dominant population has a mature lymphocyte phenotype. (D) Immunophenotyping of the white blood cells during vemurafenib treatment revealed a CD19+CD200+ population that could also be seen as CD19+CD5+ cells. (E) Combined CT and FDG PET scans obtained 1 month before the patient started taking vemurafenib and 6 weeks after vemurafenib was discontinued showed a partial response to treatment. The maximal standardized uptake value (SUVmax; semi-quantitative measure of tumor glucose metabolism) decreased from 9.5 to 4.6 g/ml (–52%) in the parailiacal LN (red arrow) and from 6.6 g/ml to 1.9 g/ml (–71%) in the right inguinal LN (not shown). Arrows indicate melanoma metastasis. (F) Lambda light-chain restriction in CD19+ B lymphocytes on day 732 relative to treatment initiation is shown.
Investigation of the peripheral blood smear revealed lymphocytes with a mature phenotype (Figure 1C). A BM biopsy specimen and aspirate were obtained, which revealed increased numbers of mature lymphocytes (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI76539DS1). Immunophenotyping of BM aspirate (Supplemental Figure 1B) and peripheral blood (Figure 1D) with the use of flow cytometry identified a lymphocyte population characterized by expression of CD19, CD5, CD23, and CD200, which was also CD10 and FMC7 negative. Together, the clinical and pathological findings were diagnostic of CLL. Fluorescence in situ hybridization revealed del13q14 and gain on 2p23 (Supplemental Figure 1C), which was previously described in CLL (15), and PCR-based immunoglobulin VH (IgVH) gene analysis revealed an unmutated IgVH status, which is associated with an adverse prognosis (16) and chronic BCR signaling (17, 18).
Although the manifestation of the melanoma in inguinal and parailiacal LN responded well to vemurafenib treatment, as documented by PET-CT scan 6 weeks after vemurafenib had been discontinued (Figure 1E), the drug was discontinued based on the previous report on its growth-promoting effects in CMML (10). Three weeks after vemurafenib was discontinued, the patient’s white cell and lymphocyte counts had decreased to normal ranges (Figure 1, A and B), but a monoclonal B cell population still persisted on day 732 after discontinuation of vemurafenib (Figure 1F). Since the iliacal melanoma metastasis had responded well and because vemurafenib was not considered a treatment option over the next months, the iliacal tumor manifestation was surgically removed. After surgery, the patient’s white cell count increased and then dropped again into the normal range, while the lymphocyte counts remained normal (Figure 1, A and B). To determine whether BRAF inhibition can trigger the development of lymphatic malignancies, we screened 7 patients with melanoma treated with BRAF inhibition, but found no other patients with CLL under BRAF inhibitor treatment (Supplemental Table 1). These data indicate that the CLL was most likely preexisting in our patient, possibly in the form of monoclonal B cell lymphocytosis (MBL). At present, the patient has been off vemurafenib for 12 months, and no melanoma progression has occurred.
BRAF inhibition leads to primary CLL expansion in vivo, which can be antagonized by SYK inhibition.
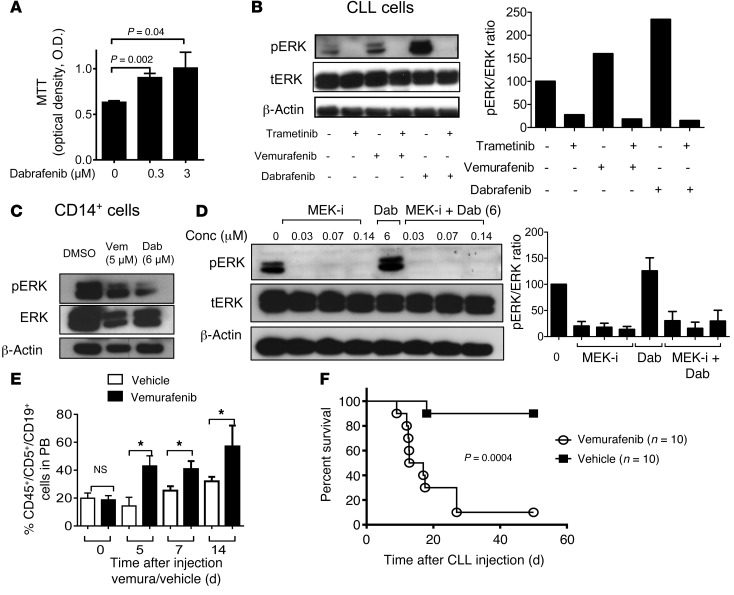
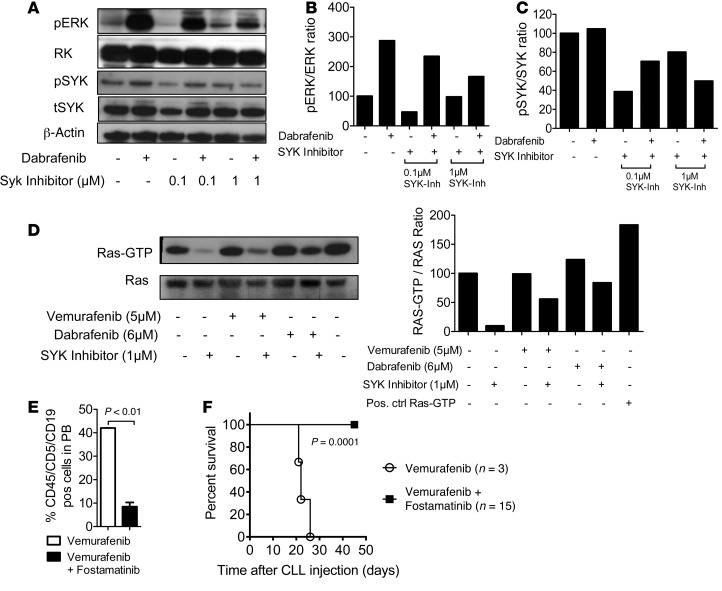
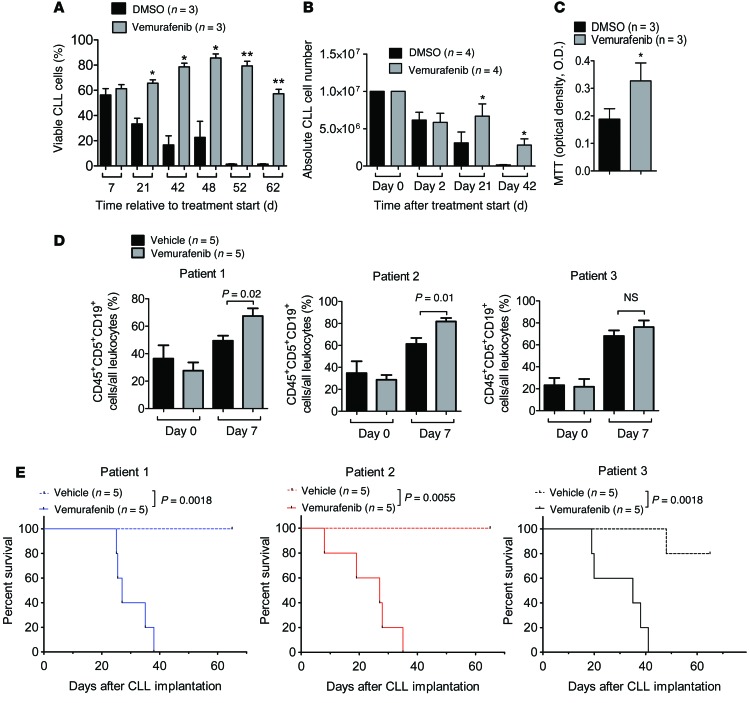
As vemurafenib treatment was paralleled by the expansion and subsequent decrease of the peripheral lymphocyte population, we suspected paradoxical ERK activation as a basis for exacerbation of preexisting CLL or MBL. Therefore, PBMCs were obtained from the patient 2 days after vemurafenib withdrawal, and highly enriched CLL cells (Supplemental Figure 1D) were cultured in the presence of the RAF inhibitors vemurafenib or dabrafenib and the MEK inhibitor trametinib, either singly or in combination. Trametinib was used because it has been successfully applied in clinical trials as a single agent or in combination with the BRAF inhibitor dabrafenib (19, 20). The rationale for this combination is based on the observation that most secondary drug-resistance phenomena observed in BRAF mutant melanoma, including those triggered by paradoxical ERK activation, are MEK dependent (12). Consistent with a growth-promoting effect of BRAF inhibitor, metabolic activity of CLL cells was increased in the presence of vemurafenib (Figure 2A). Levels of phosphorylated ERK (pERK) and total ERK (tERK) in enriched CD19+CD5+ CLL cells or CD14+ monocytes were measured by Western blot analysis. CLL cells exposed to dabrafenib or vemurafenib had an elevated pERK/tERK ratio as compared with vehicle (P < 0.01; Figure 2B). When CLL cells were isolated at later time points from the same patient (more than 5 days), the activation of ERK by BRAF inhibitor treatment was still detectable but less strong. These effects were only observed in the isolated CD19+CD5+ cells, but not in CD14+ myeloid cells (Figure 2C). Trametinib reduced ERK activation, despite BRAF inhibition, below the levels of activation in BRAF inhibitor–treated cells or cells treated with neither inhibitor (Figure 2D). Transfer of patient-derived CD19+CD5+ cells into immunodeficient Rag2–/–γC–/– mice treated with vehicle or vemurafenib led to engraftment of the CLL cells, which was followed by increased CLL numbers in the vemurafenib-treated group (Figure 2E) and reduced survival of mice in the vemurafenib compared with the vehicle group (Figure 2F). Because of the previously reported case of RAS-driven CMML under vemurafenib (10) and rare occurrences of RAS mutations in CLL (21), we investigated whether the CLL in our patient harbored an activating mutation upstream of RAF. However, 454 targeted resequencing of specimens derived from highly enriched CD5+CD19+ cells (>97% purity) from PBMCs revealed no mutations in NRAS or KRAS as well as in other genes recurrently mutated in CLL (Supplemental Table 2). Furthermore, whole-exome sequencing (WES) identified no alterations that could be linked to aberrant RAS activity or paradoxical RAF/MEK/ERK signaling (Supplemental Figure 2). We also identified 4 recurrent and characteristic copy number alterations by WES, including a small deletion at band 13q14 (Supplemental Figure 2). Therefore, CLL growth was likely driven by a signal other than mutation of a RAS pathway oncogene. As CLL, and in particular the subgroup with unmutated IgVH locus, is driven by chronic BCR signaling, we asked whether blocking a BCR proximal kinase would antagonize the paradoxical effects of BRAF inhibition. Given that BCR-induced activation of BRAF and RAF1 requires their binding to RAS, and as the tyrosine kinase SYK is essential for BCR-induced RAS/ERK activation in B lymphocytes (22, 23), we combined vemurafenib with the SYK inhibitor R406, which inhibits BCR- and stroma-induced signaling in CLL cells (24, 25). Indeed, R406 reversed BRAF inhibitor–provoked paradoxical ERK phosphorylation (Figure 3, A and B), which correlated with potent SYK inhibition (Figure 3C). Consistent with an effect on the BCR/RAS axis, we observed that the RAS-GTP/tRAS ratio in the CLL cells decreased upon SYK inhibition (Figure 3D), indicating reduced levels of active RAS. In vivo SYK inhibition with fostamatinib prevented vemurafenib-induced CLL proliferation and mortality (Figure 3, E and F). Since RTK upregulation represents an alternative mechanism driving paradoxical ERK activation (9) and to more precisely characterize the CLL cells of this individual patient, we analyzed the expression levels of different RTKs in highly enriched CLL cells. However, these analyses did not reveal any phosphorylated RTKs of the 49 investigated RTKs. (Supplemental Figure 3A). These findings argue against other mechanisms besides BCR signaling as the cause for the observed ERK activation upon BRAF inhibition.
Figure 2. Impact of BRAF and MEK inhibitor treatment on signaling and survival of CLL cells.
Highly purified (>97%) CD19+CD5+ cells or CD14+ myeloid cells obtained from the patient were cultured in the presence of the BRAF inhibitors (vemurafenib, dabrafenib), the MEK inhibitor trametinib, BRAF inhibitor combined with MEK inhibitor, or DMSO only at the indicated concentrations. (A) OD as an indicator for viability and metabolic activity of CLL cells under different vemurafenib concentrations in an MTT assay. The experiment was performed twice using in dependent samples from the patient with similar results. Levels of pERK and tERK in the CLL cells (B) or CD14+ cells (C) derived from PBMC were measured by Western blot. We used vemurafenib, dabrafenib, and trametinib (0.07 μM) as indicated. One of 3 independent experiments with similar results is shown. (D) Western blot analysis for pERK and tERK of the protein lysate of highly purified (>97%) patient-derived CD19+CD5+ cells at the indicated concentrations of dabrafenib (Dab; 6 μM) and trametinib (MEK-i; 0.03, 0.07, 0.14 μM). Quantification of the protein amount of the described groups shown as a bar diagram. The experiment was performed 3 times with similar results. (E) The amount of CLL cells in the peripheral blood of Rag2–/–γC–/– mice relative to start of treatment with vehicle or vemurafenib (24 mg/kg/d). (F) The survival of the Rag2–/–γC–/– mice treated as described in panel E is shown (P = 0.0004). Data from 3 independent experiments were pooled.
Figure 3. BRAF inhibition increases ERK phosphorylation and CLL proliferation in vivo, which can be reverted by SYK inhibition.
Highly purified (>97%) CD19+CD5+ cells (CLL) obtained from the patient were exposed to dabrafenib (6 μM) and R406 at increasing concentrations, as indicated, or DMSO as control. (A) A representative Western blot is shown. The experiment was performed twice with similar results. The pERK/tERK ratios (B) and the pSYK/tSYK ratios (C) are shown for the indicated conditions. (D) The patient’s CLL cells were exposed to dabrafenib, vemurafenib, R406, or DMSO as control at the indicated concentrations, and the resulting Western blot and the RAS-GTP/tRAS ratios are shown. One of 3 independent experiments with similar results is shown. (E) The amounts of CLL cells in the peripheral blood of Rag2–/–γC–/– mice on day 7 relative to treatment starting with vemurafenib (24 mg/kg/d) alone or vemurafenib and fostamatinib (60 mg/kg/d) are shown. On day 0, 2.5 × 107 patient-derived CD19+CD5+ cells were injected i.p. and i.v. (F) The survival of the Rag2–/–γC–/– mice treated as described in E is shown (P = 0.0001). Data from 2 experiments were pooled.
Paradoxical ERK activation and prolonged survival upon BRAF inhibition are found in primary CLL cells derived from multiple patients.
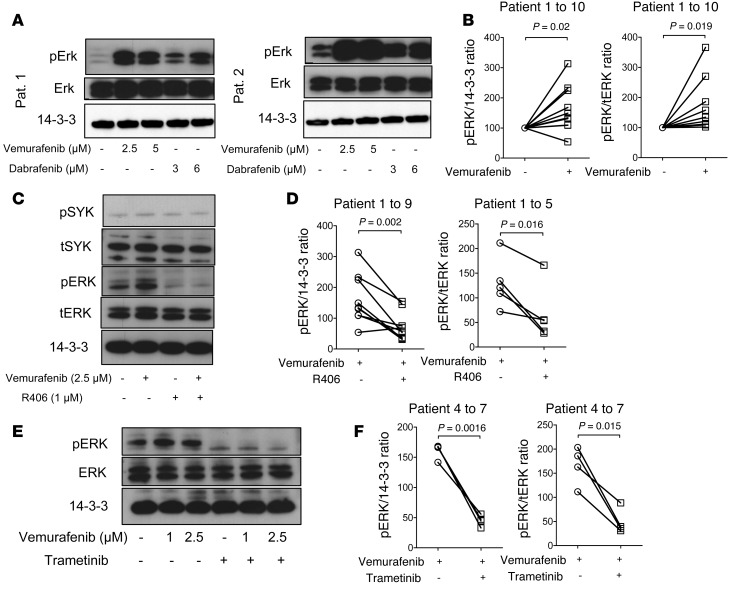
In order to understand whether the observed effects were only seen in primary CLL cells of this particular patient, we next turned to primary CLL cells from 10 other patients. The characteristics of the patients and the CLL cells are detailed in Supplemental Table 3. Levels of pERK and tERK in primary CLL cells were measured by Western blot analysis. Consistent with inhibition of the paradoxical ERK phosphorylation promoting effects of BCR activation by SYK inhibition, we observed higher pERK/tERK ratios in primary CLL cells when exposed to vemurafenib or dabrafenib compared with vehicle, as shown for 2 representative patients (Figure 4A), and for vemurafenib compared with vehicle in all analyzed patients (Figure 4B). The paradoxical ERK activation upon BRAF inhibition was abrogated by either SYK inhibition (Figure 4, C and D) or MEK inhibition (Figure 4, E and F). To analyze the effects of inhibiting more distal nodes of the BCR on RAS signaling and ERK activation, we blocked Bruton’s tyrosine kinase (BTK). In contrast with SYK inhibition, BTK inhibition did not reduce levels of pERK/tERK (Supplemental Figure 3B). As an additional readout of feedback activation of ERK signaling, we next studied pAKT levels in the presence of BRAF and SYK inhibition. We observed that vemurafenib caused increased pAKT/tAKT levels (Supplemental Figure 3C). Addition of SYK inhibition reduced the pAKT/tAKT ratio (Supplemental Figure 3C), which is consistent with the reduced pERK/ERK levels we observed under SYK inhibition. Consistent with the results in the CLL cells from the melanoma patient, we also found the RAS-GTP/tRAS ratio to be reduced by SYK inhibition when we used primary CLL cells from a second patient (Supplemental Figure 3D) as well as a third patient (data not shown).
Figure 4. Impact of BRAF and SYK/MEK inhibition on ERK activation in primary CLL cells.
(A–F) Primary CLL cells from multiple patients were highly purified (>97% CD19+CD5+) and exposed to the indicated inhibitors. (A) Representative Western blot analysis for pERK and tERK of the protein lysate at the indicated concentrations of vemurafenib and dabrafenib for patients 1 and 2. The experiment was performed 2 times with similar results. (B) Quantification of the pERK/14-3-3 ratios and pERK/tERK ratios of 10 different patients are displayed (Vem, 1 μM). (C) Addition of the SYK inhibitor R406 decreased the ERK phosphorylation as shown for the protein lysates for patient 3. The experiment was performed 2 times with similar results. (D) Quantification of the pERK/14-3-3 ratios and pERK/tERK ratios of 9 and 5 different patients, respectively, are displayed (Vem, 2.5 μM). (E) Western blot analysis for pERK and tERK of the protein lysate at the indicated concentrations of vemurafenib and trametinib (1 μM) for patient 4. The experiment was performed 3 times with similar results. (F) Quantification of the pERK/tERK ratios and pERK/14-3-3 ratios of all analyzed patients (n = 4) are displayed (Vem, 2.5 μM).
To understand whether this impact on signaling translated into functional differences, we next analyzed the viability of the primary CLL cells. We observed that cell viability and metabolic activity were significantly increased by the BRAF inhibitor vemurafenib (Figure 5, A–C). To clarify whether the observed effects of BRAF inhibition on viability, proliferation, and ERK activation of primary CLL cells in vitro would also translate into an effect in vivo, we next injected immunodeficient Rag2–/–γC–/– mice with primary CLL cells from 3 different patients and then treated the mice with vehicle or vemurafenib. The patients were selected because their CLL cells grew very strong in vitro when exposed to vemurafenib and not because of specific CLL-related risk factors.
Figure 5. Impact of BRAF inhibition on viability, proliferation, and in vivo expansion of primary CLL cells.
(A–C) Primary CLL cells from different patients were highly purified (>97% CD19+CD5+) and exposed to DMSO or vemurafenib. (A) The percentage of living cells relative to all cells (patients n = 3) or the absolute number of CLL cells (patients n = 4) (B) was determined for different time points. (C) Metabolic activity measured by MTT on day 52 of culture was higher in primary CLL cells (patients n = 3) when exposed to vemurafenib compared with DMSO. One of 3 independent experiments with similar results is shown. *P < 0.05; **P < 0.01. (D and E) On day 0, the amount of 2.5 × 107 patient-derived CD19+CD5+ cells was injected i.p. and i.v. (D) The amount of CLL cells in the peripheral blood of Rag2–/–γC–/– mice on days 0 and 7 relative to start of treatment. (E) The survival of the Rag2–/–γC–/– mice treated with DMSO or vemurafenib (24 mg/kg/d) is shown for the CLL cells derived from 3 different patients.
The amount of CLL cells in the peripheral blood of mice receiving primary CLL cells and treatment with vemurafenib was increased compared with that of mice treated with vehicle when CLL cells were derived from patients 1 and 2, and there was a comparable trend when CLL cells from patient 3 were used (Figure 5D). Consistently, survival of those mice receiving primary CLL cells from 1 of the 3 different patients and treatment with vemurafenib was significantly reduced compared with survival of mice treated with vehicle (Figure 5E).
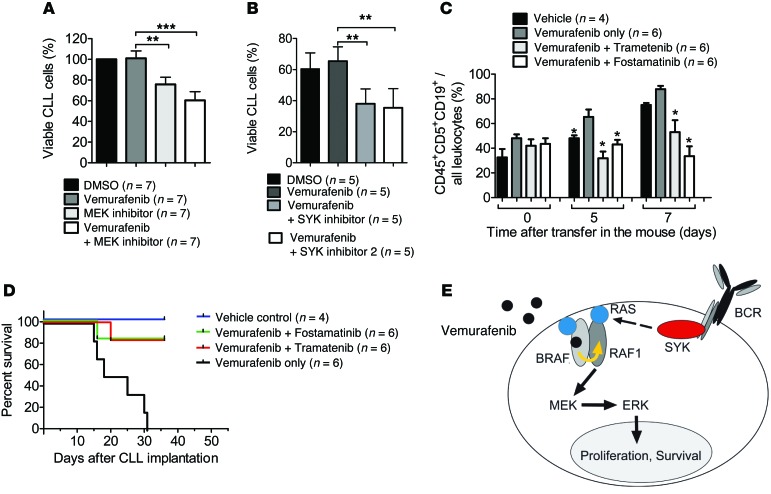
In order to determine whether the in vivo promoting effects of vemurafenib on primary CLL cells could be reversed, we additionally treated a group with a MEK (trametinib) or SYK inhibitor (fostamatinib). We observed that the viability of primary CLL cells from different patients decreased upon addition of MEK inhibitor (n = 7) or SYK inhibitor (n = 5) to vemurafenib (Figure 6, A and B). The amount of CLL cells in the peripheral blood of mice receiving primary CLL cells and treatment with vemurafenib and trametinib, or vemurafenib and fostamatinib, was decreased compared with that of mice treated with vemurafenib alone on days 5 and 7 after the start of treatment (Figure 6C). Consistently, survival of mice receiving primary CLL cells and treatment with vemurafenib and trametinib, or vemurafenib and fostamatinib, was improved compared with survival of mice treated with vemurafenib alone (Figure 6D).
Figure 6. Impact of SYK inhibition on BRAF inhibitor–induced primary CLL proliferation.
Primary CLL cells of different patients were exposed to DMSO or vemurafenib (1 μM), and the percentage of living cells relative to all cells (patients n = 7) when the MEK inhibitor (A) or the Syk inhibitor (B) was included, was determined for different time points. **P < 0.01; ***P < 0.001. (C) The amount of CLL cells in the peripheral blood of Rag2–/–γC–/– mice relative to start of treatment. * P < 0.05 when the group was compared with the vemurafenib-only group. (D) The survival of the Rag2–/–γC–/– mice treated with vemurafenib (24 mg/kg/d) alone or in combination with fostamatinib (60 mg/kg/d) or trametinib (1 mg/kg/every other day for 12 days) is shown (Vem vs. Vem/Fosta, P < 0.001; Vem vs. Vem/Tram, P < 0.001) (E) Proposed mechanism illustrating how BRAF inhibition could cooperate with SYK in paradoxical ERK activation. Vemurafenib binds to 1 protomer, e.g., BRAF, and stimulates a drug-free RAF molecule, e.g., RAF1, in an allosteric and RAS-dependent manner (yellow arrow). Active and RAF-binding competent RAS is supplied via SYK, which is hyperactivated due to the autonomous signaling capacity of the CLL-specific BCR.
Based on our findings, we propose the concept that BCR/SYK-activated RAS and drug-bound BRAF cooperatively induce paradoxical ERK activation in CLL cells (Figure 6E).
Discussion
We describe the effects of BRAF inhibition on primary CLL cells from multiple patients, based on a case of CLL developing during vemurafenib treatment in a patient with melanoma. In this patient and in the multiple investigated patients with CLL, we observed dependence of CLL cells on paradoxical, BRAF inhibitor–induced ERK signaling. The differential of vemurafenib-induced ERK activation (Figure 4B) was variable and could not be correlated with known risk factors of CLL, such as unmutated IgVH status or unfavorable cytogenetics (del 17p, del 11q). However, the extent of BCR signaling, which represents a major driver of CLL (17, 26), can be modulated by many mechanisms that are not genetically hard-wired, such as feedback loops and crosstalk with other signaling pathways (27). Consequently, one can expect a certain donor-to-donor variability of BCR signaling independent of known risk factors. We therefore hypothesize that different ground states, e.g., the degree of BCR signaling at the time point of BRAF inhibitor exposure, determine the strength of paradoxical ERK activation. However, given the relatively small number of patients analyzed in our studies (n = 10), further studies with more primary CLL samples and a correlation of BCR activity and BRAF inhibitor–induced ERK activation will be needed to identify the modulators of this event.
In vitro exposure of the CLL cells to the BRAF inhibitor caused a dose-dependent, reversible activation of ERK selectively in the CLL, but not in myeloid cells. We observed that CLL cells isolated more than 5 days after vemurafenib discontinuation in the melanoma patient still displayed activation of ERK by BRAF inhibitor treatment, but the effect was less pronounced. We hypothesize that the CLL cells that were best suited to react to vemurafenib with paradoxical ERK activation and clonal expansion lost their selective advantage after vemurafenib discontinuation and were subsequently overgrown by less vemurafenib-dependent subclones. Also, in vitro and in vivo expansion of CLL cells was specifically induced on treatment with vemurafenib. This effect was not restricted to the in vitro studies, but increased the in vivo expansion of CLL cells, while CLL-related death was also seen when primary CLL cells from multiple patients were implanted into immunodeficient mice. Our observation that the longevity of primary CLL cells can be markedly extended by BRAF inhibitors also represents a technical advance for the long-term cultivation of otherwise very short-lived CLL cells.
Importantly, our melanoma patient with a secondary vemurafenib-promoted CLL differs from the previously reported mutant RAS-driven CMML case (10) because no mutations in RAS genes were found. We recapitulated the effect of the RAF inhibitor on the CLL cells in the patient in vitro and a patient-derived xenograft mouse model. We found enhanced ERK signaling in the CLL cells exposed to BRAF inhibition, which was abrogated by combinatorial treatment with a MEK inhibitor and enhanced CLL progression upon vemurafenib treatment in vivo. Similar to what was seen in this melanoma patient, we also observed increased CLL progression due to vemurafenib treatment in patient-derived xenograft mouse models when using CLL cells from 3 different additional patients.
In light of these findings and because MEK inhibitors have recently been shown to prolong the survival of patients with BRAF V600E-mutated melanoma, alone (19) or in combination with dabrafenib (20, 28), we predict that if melanoma progression required systemic anti-melanoma treatment again, combined BRAF and MEK inhibition would prevent the progression of the CLL.
To understand the mechanism that underlies vemurafenib-induced paradoxical ERK activation in the primary CLL cells, we tested the role of the BCR, which was previously shown to provide autonomous signaling in CLL but not other B cell malignancies (29). When blocking BCR downstream signaling via inhibition of SYK, an essential BCR proximal kinase coupling the receptor to the RAS pathway, we observed partial abrogation of BRAF inhibitor–related ERK phosphorylation. This supports the concept that BCR/SYK-activated RAS and drug-bound BRAF cooperatively induce paradoxical ERK activation in CLL cells, in a similar manner to that described for RTKs in the drug-resistance scenarios (12). In addition, increased phosphorylation of RTKs that could have led to RAS/ERK activation was not found in the CLL cells from the melanoma patient. To better understand the role of the BCR/SYK axis, we investigated the effects of inhibiting a more distal node of the BCR by using the BTK inhibitor ibrutinib. We observed that BTK inhibition did not antagonize BRAF inhibitor–induced paradoxical ERK activation. However, besides the suspected link between BCR signaling and paradoxical ERK activation, a paracrine mechanism also remains possible.
Based on our observations, a SYK inhibitor such as fostamatinib, which is already successfully tested in clinical studies in patients with non-Hodgkin’s lymphoma (NHL) and CLL (30), could be a therapeutic alternative in our and other patients developing CLL progression during vemurafenib treatment. This may be important in the context of a recent report indicating that BRAF inhibitor–driven tumor proliferation in a KRAS-mutated colon carcinoma could not be overcome by MEK1/2 inhibition (31). The differences between the 2 BRAF inhibitors vemurafenib and dabrafenib are another clinically relevant aspect, as, for example, off-target effects of vemurafenib, such as cutaneous squamous cell carcinoma, which were less frequently observed with dabrafenib (32). Cutaneous squamous cell carcinoma rates were reported to be between 20% and 26% in trials with vemurafenib (1, 33), while the rates were 6% to 11% in dabrafenib trials (34–37).
Importantly, our study implicates the BCR signaling complex as a driver of paradoxical ERK activation. Given the increasing use of vemurafenib and dabrafenib in melanoma, and because malignant melanoma is overrepresented in the NHL population (~2.3–3.1 fold) (38, 39), the progression or development of CLL in such patients must be anticipated. Although BRAF-mutant melanoma and CLL can occur together, as seen in our patient, the typical patients with BRAF-mutant melanomas are different from the characteristic epidemiologic cohort, which includes CLL patients. Because in retrospect a monoclonal B cell population was detected in the melanoma patient before vemurafenib treatment, but no leukocytosis and lymphocytosis were observed, one would assume that vemurafenib promotes the expansion of cells with preexisting neoplastic lesions rather than inducing them.
Our study shows for what we believe is the first time that BRAF inhibition can promote progression of primary CLL cells that can be counteracted by SYK inhibitor treatment, which interferes with signals into the RAS/RAF/MEK/ERK pathway. Given the increasing clinical application of BRAF inhibitors, a therapeutic approach that can revert paradoxical ERK activation in CLL adds the SYK/BRAF inhibitor combination identified in this study to the therapeutic armamentarium against treatment-induced lymphoid neoplasms.
Methods
CLL samples.
Peripheral blood mononuclear cells (PBMCs) were separated by Ficoll gradient centrifugation, and CLL amount was determined by flow cytometric analysis with anti-CD19 and anti-CD5 (BD Biosciences). CD19-B cells were isolated by negative selection (B-cell Isolation Kit II; Miltenyi Biotec). CD14 cells were isolated by positive selection (Anti-CD14 Micro Beads, human; Miltenyi Biotec). Cells were either used fresh or cryopreserved in FCS/10% DMSO until use.
Mice.
Rag2–/–γC–/– mice were purchased from Charles River Laboratory and from the local stock of the animal facility at Freiburg University Medical Center (Freiburg, Germany). Mice were between 6 and 12 weeks of age. On day 0, patient-derived CD19+CD5+ cells were injected at the following doses per mouse: 2.5 × 107 cells (i.p.) and 2.5 × 107 cells (i.v.).
Cell culture and proliferation assay for the primary CLL cells.
The primary CLL cells from different patients (Supplemental Table 3) were cultured in RPMI 1640 (Sigma-Aldrich) supplemented with 10% FCS, 100 U/ml penicillin, and 0.1 mg/ml streptomycin at a concentration of 1 × 107 cells/ml in 24-well plates in the presence or absence of varying doses of vemurafenib (PLX4032, RG7204, catalog no. S1267; Selleckchem) or dabrafenib (GSK2118436, catalog no.S2807; Selleckchem) and/or R406 (catalog no.S1533; Selleckchem) for 24 to 72 hours. CLL cell proliferation was determined using a standard MTT assay as previously described (40).
Long-time culture of primary human CLL cells.
For long-time culture of primary human CLL cells, cells were cultured as described above in the presence of DMSO or vemurafenib (2.5–5 μM) in 6-well plates (total volume of 3 ml). Every third day, 1 ml of medium was replaced by new medium containing DMSO or vemurafenib at the indicated concentrations.
Immunoblotting.
Healthy B cells and freshly isolated or freshly thawed viable frozen CLL cells were lysed and subjected to Western blotting as described previously (24). Lysates were generated after 45 minutes of exposure to the respective drugs.
RTK array.
To analyze the expression levels of different RTKs, protein lysates from highly purified CLL cells from the melanoma patient were generated and analyzed in a Human Phospho-Receptor Tyrosine Kinase Array Kit as described in the technical data sheet (Proteome Profiler Array Human Phospho-Receptor Tyrosine Kinase Array Kit; ARY001B; R&D Systems).
Antibodies and reagents.
The following antibodies were used: phospho-p44/42 MAPK (pErk1/2) (Thr202/Tyr204) (D13.14.4E) XP rabbit mAb no. 4370; p44/42 MAPK (Erk1/2) antibody no. 9102; phospho- Syk (Tyr352)/ Zap-70 (Tyr319)/ (65E4) rabbit mAb no. 2717; phospho-Syk (Tyr525/526) (C87C1) rabbit mAb no. 2710; Syk (D1I5Q) rabbit mAb no. 12358; Ras (D2C1) rabbit mAb no. 8955; phospho-MEK1/2 (Ser217/221) (41G9) rabbit mAb no. 9154; MEK1 (61B12) mouse mAb no. 2352; phospho-AKT (Thr308) (D25E6) XP rabbit mAb no. 13038, AKT antibody no. 9272, β-actin (13E5) rabbit mAb no. 4970; and anti-rabbit IgG, HRP-linked antibody no. 7074. All were from Cell Signaling Technology. APC-conjugated mouse anti-human CD45 (HI30); FITC-conjugated mouse anti-human CD5 (UCHT2); and APC-conjugated mouse anti-human CD19 (HIB19) were from BD Biosciences. Mouse anti-human CD19-PE/TXRD (SJ25-C1) was from Southern Biotech. The following antibodies from BD Biosciences were used for the analysis of the patient cells: CD5 (L17F12), CD10 (HI10a), CD19 (4G7), CD20 (L27), CD22 (HIB22), CD23 (ML233), CD79b (SN8), and CD200 (MRC OX-104).
Pulldown assay for the detection of active Ras.
Patient CLL cells and the cell line MEC-1 were cultured in RPMI 1640 (Sigma-Aldrich) supplemented with 10% FCS, 100 U/ml penicillin, an 0.1 mg/ml streptomycin at a concentration of 1 × 107 cells/ml in 24-well plates in the presence or absence of varying doses of vemurafenib or dabrafenib and/or R406. After 45 minutes, the cells were lysed and Ras activity was determined as described in the Active Ras Detection Kit (no. 8821; Cell Signaling). Additionally, the cells were subjected to Western blotting to determine total Ras levels.
Isolation of analytes for sequencing.
DNA was extracted from highly purified (>97%) CD19+CD5+ cells obtained from the patient under vemurafenib treatment versus CD14+ germline control cells (>90% purity). DNA was isolated using a QIAGEN Allprep DNA/RNA/Protein Mini Kit according to the manufacturer’s protocol. After quality control of isolated DNA (gel electrophoresis and Agilent Bioanalyzer 2100), extracted nucleic acids were submitted to sequencing facilities.
DNA library preparation and sequencing.
Exome capturing was carried out with Agilent SureSelect Human All Exon version 5 with UTR in-solution capture reagents (vendor’s protocol v2.0.1). In brief, 1.5 μg of gDNA were fragmented to 150- to 200-bp (PE) insert-size with a Covaris S2 device. 250 ng of Illumina adapters containing libraries were hybridized with the exom baits at 65°C for 16 hours. Paired end sequencing (101 bp) was carried out with an Illumina HiSeq 2500 instrument in rapid mode.
Mapping and analysis.
Reads were mapped to the 1000 genomes phase-2 assembly of the human reference genome (NCBI build 37.1, downloaded from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/reference/phase2_reference_assembly_sequence/) using BWA (41) (version 0.6.2) with default parameters and maximum insert size set to 1000 bp. BAM files were sorted with SAMtools (42) (version 0.1.17), and duplicates were removed with Picard tools (http://broadinstitute.github.io/picard/), version 1.90). Average target coverage was 86% for the tumor and 70% for the control. In both, more than 80% of the targets had a coverage of at least ×30.
For detection of single nucleotide variant (SNV) and small insertions or deletions (indels), we applied our in-house analysis pipeline based on SAMtools mpileup and BCFtools with parameter adjustments to allow calling of somatic variants (43, 44). Initial SNV and indel candidates called on the tumor sample (by SAMtools mpileup -RE -q 20 -ug -m 2 and BCFtools -vcgN -p 2.0) were filtered for Illumina-specific error profiles and retained if the position was covered by at least 3 reads and the alternative allele frequency was 5% or more. On these positions, a pileup of the control sample was performed (by SAMtools mpileup -RAB -Q 0 -q 1) and parsed with a custom Perl script considering base quality.
We excluded variants that are located in regions of low mappability or overlapped with entries of the hiSeqDepthTop10Pct, Encode DAC Blacklisted Regions, or Duke Excluded Regions tracks from the UCSC genome browser (http://genome.ucsc.edu/). High-confidence somatic SNVs and indels were not allowed to overlap with any 2 of the following features at the same time: tandem repeats, simple repeats, low complexity, satellite repeats, or segmental duplications. In addition, they had to fulfill the following heuristic criteria: (a) at least 5 tumor reads at the position; (b) either more than 1 read supporting the variant per strand or at least 5 reads supporting the variant in total and total variant allele frequency greater than 0.1; (c) coverage at the position in the matching control sample at least 12 reads; (d) less than 1/30 of the control reads supporting the variant; (e) less than 500 reads at the corresponding position in the control; and (f) no nonreference, nonvariant bases at the corresponding position in the control. Variants were functionally annotated with RefSeq gene annotations using Annovar (45) (version November 2011) to select nonsilent coding events. Indels were manually checked in the Integrative Genome Viewer (46).
Copy number variants were identified by read depth plots and an in-house pipeline using the VarScan2 (47) copy number and CopyCaller modules. Regions were filtered for unmappable genomic stretches, merged by requiring at least 70 markers per called copy number event, and annotated with RefSeq genes using BEDTools (48). The sequencing data have been deposited in the European Genome-phenome Archive (EGA) (accession number EGAS00001000948).
Statistics.
GraphPad Prism version 5.03 was used for statistical analysis. For normally distributed data, a 2-tailed unpaired t test was utilized. The Mann-Whitney U test was used when the data did not conform to a normal distribution. Welch’s t test was applied if variances of the groups were not equal. Differences in animal survival (Kaplan-Meier survival curves) were analyzed by log-rank test. Data are presented as mean ± SEM. Differences were considered significant when the P value was less than 0.05.
Study approval.
We collected all human samples after approval by the Ethics Committee of Albert Ludwigs University (protocol number: 267/11) and after receiving written, informed consent. All animal protocols (G-11/36) were approved by the Federal Ministry for Nature, Environment, and Consumer Protection of the state of Baden-Württemberg, Germany.
Supplementary Material
Acknowledgments
We thank Hanno Glimm, Daniela Richter, Christoph von Kalle, Roland Eils, Peter Lichter, Stephan Wolf, Katja Beck, and Janna Kirchhof for infrastructure and program development within the DKFZ–Heidelberg Center for Personalized Oncology (DKFZ-HIPO) and the NCT–Precision Oncology Program (NCT-POP). We thank DKFZ-HIPO and NCT-POP (P021) for continued support. This study was supported by the Deutsche Forschungsgemeinschaft (DFG), Germany; a Heisenberg Professorship to R. Zeiser (DFG ZE 872/3-1); and in part by SFB 850 (B4, C6) and the Excellence Initiative of the German Federal and State Governments and EXC 294 BIOSS (to R. Zeiser and T. Brummer). T. Brummer is also supported by the DFG (Emmy-Noether-Program, BR3662/1-1; Heisenberg-Program, BR3662/2-1). T. Zenz is supported by a Mildred-Scheel Professorship of the “Dt. Krebshilfe,” and the study was supported by the Deutsches Konsortium für Translationale Krebsforschung (DKTK). R. Claus is supported by the German Cancer Aid (Max Eder stipend, DKH 110461).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2014;124(11):5074–5084. doi:10.1172/JCI76539.
References
- 1.Chapman PB, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heidorn SJ, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140(2):209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Röring M, et al. Distinct requirement for an intact dimer interface in wild-type, V600E and kinase-dead B-Raf signalling. EMBO J. 2012;31(11):2629–2647. doi: 10.1038/emboj.2012.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464(7287):427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hatzivassiliou G, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464(7287):431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 6.Robert C, Arnault JP, Mateus C. RAF inhibition and induction of cutaneous squamous cell carcinoma. Curr Opin Oncol. 2011;23(2):177–182. doi: 10.1097/CCO.0b013e3283436e8c. [DOI] [PubMed] [Google Scholar]
- 7.Zimmer L, et al. Atypical melanocytic proliferations and new primary melanomas in patients with advanced melanoma undergoing selective BRAF inhibition. J Clin Oncol. 2012;30(19):2375–2383. doi: 10.1200/JCO.2011.41.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su F, et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N Engl J Med. 2012;366(3):207–215. doi: 10.1056/NEJMoa1105358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nazarian R, Shi H, Wang Q, Kong X. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468(7326):973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Callahan MK, et al. Progression of RAS-mutant leukemia during RAF inhibitor treatment. N Engl J Med. 2012;367(24):2316–2321. doi: 10.1056/NEJMoa1208958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlino MS, et al. New RAS-mutant pancreatic adenocarcinoma with combined BRAF and MEK inhibition for metastatic melanoma. J Clin Oncol. doi: 10.1200/JCO.2013.51.5783. [published online ahead of print May 12, 2014]. doi: 10.1200/JCO.2013.51.5783. [DOI] [PubMed] [Google Scholar]
- 12.Röring M, Brummer T. Aberrant B-Raf signaling in human cancer — 10 years from bench to bedside. Crit Rev Oncog. 2012;17(1):97–121. doi: 10.1615/CritRevOncog.v17.i1.70. [DOI] [PubMed] [Google Scholar]
- 13.Lito P, Rosen N, Solit DB. Tumor adaptation and resistance to RAF inhibitors. Nat Med. 2013;19(11):1401–1409. doi: 10.1038/nm.3392. [DOI] [PubMed] [Google Scholar]
- 14.Balch CM, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27(36):6199–6206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ouillette P, et al. The prognostic significance of various 13q14 deletions in chronic lymphocytic leukemia. Clin Cancer Res. 2011;17(21):6778–6790. doi: 10.1158/1078-0432.CCR-11-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94(6):1848–1854. [PubMed] [Google Scholar]
- 17.Young RM, Staudt LM. Targeting pathological B cell receptor signalling in lymphoid malignancies. Nat Rev Drug Discov. 2013;12(3):229–243. doi: 10.1038/nrd3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanham S, Hamblin T, Oscier D, Ibbotson R, Stevenson F, Packham G. Differential signaling via surface IgM is associated with VH gene mutational status and CD38 expression in chronic lymphocytic leukemia. Blood. 2003;101(3):1087–1093. doi: 10.1182/blood-2002-06-1822. [DOI] [PubMed] [Google Scholar]
- 19.Flaherty KT, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med. 2012;367(2):107–114. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 20.Menzies AM, Long GV. Dabrafenib and trametinib, alone and in combination for BRAF-mutant metastatic melanoma. Clin Cancer Res. 2014;20(8):2035–2043. doi: 10.1158/1078-0432.CCR-13-2054. [DOI] [PubMed] [Google Scholar]
- 21.Jethwa A, et al. Targeted resequencing for analysis of clonal composition of recurrent gene mutations in chronic lymphocytic leukaemia. Br J Haematol. 2013;163(4):496–500. doi: 10.1111/bjh.12539. [DOI] [PubMed] [Google Scholar]
- 22.Brummer T, Shaw PE, Reth M, Misawa Y. Inducible gene deletion reveals different roles for B-Raf and Raf-1 in B-cell antigen receptor signalling. EMBO J. 2002;21(21):5611–5622. doi: 10.1093/emboj/cdf588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yasuda T, et al. Erk kinases link pre-B cell receptor signaling to transcriptional events required for early B cell expansion. Immunity. 2008;28(4):499–508. doi: 10.1016/j.immuni.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 24.Buchner M, et al. Spleen tyrosine kinase inhibition prevents chemokine- and integrin-mediated stromal protective effects in chronic lymphocytic leukemia. Blood. 2010;115(22):4497–4506. doi: 10.1182/blood-2009-07-233692. [DOI] [PubMed] [Google Scholar]
- 25.Quiroga MP, et al. B-cell antigen receptor signaling enhances chronic lymphocytic leukemia cell migration and survival: specific targeting with a novel spleen tyrosine kinase inhibitor, R406. Blood. 2009;114(5):1029–1037. doi: 10.1182/blood-2009-03-212837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buchner M, Müschen M. Targeting the B-cell receptor signaling pathway in B lymphoid malignancies. Curr Opin Hematol. 2014;21(4):341–349. doi: 10.1097/MOH.0000000000000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reth M, Brummer T. Feedback regulation of lymphocyte signalling. Nat Immunol. 2004;4(3):269–277. doi: 10.1038/nri1335. [DOI] [PubMed] [Google Scholar]
- 28.Flaherty KT, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367(18):1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dühren-von Minden M, et al. Chronic lymphocytic leukaemia is driven by antigen-independent cell-autonomous signalling. Nature. 2012;489(7415):309–312. doi: 10.1038/nature11309. [DOI] [PubMed] [Google Scholar]
- 30.Friedberg JW, et al. Inhibition of Syk with fostamatinib disodium has significant clinical activity in non-Hodgkin lymphoma and chronic lymphocytic leukemia. Blood. 2010;115(13):2578–2585. doi: 10.1182/blood-2009-08-236471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrews MC, et al. BRAF inhibitor-driven tumor proliferation in a KRAS-mutated colon carcinoma is not overcome by MEK1/2 inhibition. J Clin Oncol. 2013;31(35):e448–e451. doi: 10.1200/JCO.2013.50.4118. [DOI] [PubMed] [Google Scholar]
- 32.Vin H, et al. BRAF inhibitors suppress apoptosis through off-target inhibition of JNK signaling. eLife. 2013;2:e00969. doi: 10.7554/eLife.00969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flaherty KT, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363(9):809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ascierto PA, et al. Phase II trial (BREAK-2) of the BRAF inhibitor dabrafenib (GSK2118436) in patients with metastatic melanoma. J Clin Oncol. 2013;31(26):3205–3211. doi: 10.1200/JCO.2013.49.8691. [DOI] [PubMed] [Google Scholar]
- 35.Falchook GS, et al. Dabrafenib in patients with melanoma, untreated brain metastases, and other solid tumours: a phase 1 dose-escalation trial. Lancet. 2012;379(9829):1893–1901. doi: 10.1016/S0140-6736(12)60398-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hauschild A, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380(9839):358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 37.Long GV, et al. Dabrafenib in patients with Val600Glu or Val600Lys BRAF-mutant melanoma metastatic to the brain (BREAK-MB): a multicentre, open-label, phase 2 trial. Lancet Oncol. 2012;13(11):1087–1095. doi: 10.1016/S1470-2045(12)70431-X. [DOI] [PubMed] [Google Scholar]
- 38.Lorenzo Bermejo J, Pukkala E, Johannesen TB, Sundquist J, Hemminki K. Age-time risk patterns of solid cancers in 60 901 non-Hodgkin lymphoma survivors from Finland, Norway and Sweden. Br J Haematol. 2014;164(5):675–683. doi: 10.1111/bjh.12684. [DOI] [PubMed] [Google Scholar]
- 39.Brewer JD, Habermann TM, Shanafelt TD. Lymphoma-associated skin cancer: incidence, natural history, and clinical management. Int J Dermatol. 2014;53(3):267–274. doi: 10.1111/ijd.12208. [DOI] [PubMed] [Google Scholar]
- 40.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65(1):55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 41.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones DT, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488(7409):100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones DTW, et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45(8):927–932. doi: 10.1038/ng.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson JT, et al. Integrative genomics viewer. Nat Biotech. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koboldt DC, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;2(3):568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;15(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.