Key Points
Filamin A interacts directly with the third intracellular loop and the C-terminal tail of CXCR4.
Disruption of FLNA binding to the ICL3 attenuates signaling and restores CXCL12-mediated endocytosis of WHIM-like CXCR4 receptor mutants.
Abstract
Warts, hypogammaglobulinemia, infections, and myelokathexis (WHIM) syndrome is a rare congenital immunodeficiency often caused by mutations in the last 10 to 19 C-terminal amino acids of CXCR4. These mutations impair CXCR4 internalization and increase responsiveness to CXCL12. The CXCR4 C-terminal domain (C-tail) also has a binding site for the actin-binding protein filamin A (FLNA); it is not known whether FLNA binds to WHIM CXCR4 mutants or whether this interaction is implicated in the hyperfunction of these receptors. Here we show that, in addition to interacting with the CXCR4 C-tail, FLNA interacted with a region in the receptor third intracellular loop (ICL3) spanning amino acids 238 to 246. This interaction involved specific FLNA repeats and was sensitive to Rho kinase inhibition. Deletion of the 238-246 motif accelerated CXCL12-induced wild-type (WT) receptor endocytosis but enabled CXCL12-mediated endocytosis and normalized signaling by the WHIM-associated receptor CXCR4R334X. CXCL12 stimulation triggered CXCR4R334X internalization in FLNA-deficient M2 cells but not in the FLNA-expressing M2 subclone A7; this suggests a role for FLNA in stabilization of WHIM-like CXCR4 at the cell surface. FLNA increased β-arrestin2 binding to CXCR4R334X in vivo, which provides a molecular basis for FLNA-mediated hyperactivation of WHIM receptor signaling. We propose that FLNA interaction with ICL3 is central for endocytosis and signaling of WT and WHIM-like CXCR4 receptors.
Introduction
The warts, hypogammaglobulinemia, infections, and myelokathexis (WHIM) syndrome is a rare congenital immunodeficiency characterized by severe neutropenia, B-cell lymphopenia, delayed antibody class switching to immunoglobulin G, hypogammaglobulinemia, recurrent bacterial infections, and warts that develop after early exposure to human papillomavirus. In most cases, WHIM syndrome is associated with the dominant inheritance of variants of the chemokine receptor CXCR4 with mutations in the last 10 to 19 C-terminal amino acids.1-4 These mutations lead to a hyperfunctional receptor with impaired internalization and increased responsiveness to CXCL12.2,5 Dysfunctional CXCR4-mediated signaling is a common manifestation in WHIM patients, even in cases in which the disorder is not genetically associated with CXCR4 mutations.6 The causal role of CXCR4 in WHIM syndrome was confirmed in several cell and animal models,7,8 including a knockin mouse bearing the common WHIM syndrome–causing mutation CXCR4R334X in heterozygotes.9 Treatment with a CXCR4 antagonist transiently reverses most associated immunologic anomalies in WHIM patients.10,11
Evidence pinpoints the C-terminal region (C-tail) as an important determinant of CXCR4 endocytosis, desensitization, and recycling.12,13 CXCL12 binding to the receptor triggers phosphorylation of serine (Ser) and threonine (Thr) residues at the C-tail by G-protein–coupled receptor kinases and other Ser/Thr kinases.14,15 Initial studies indicated that Ser phosphorylation at positions 324, 325, 338, and 339 is critical for CXCR4 endocytosis.16 Nonetheless, recent work suggests hierarchical Ser/Thr phosphorylation at the C-tail, by which the 3-amino-acid Ser346-348 motif regulates Ser324/325 and Ser338/339 phosphorylation17; this finding might explain why WHIM-associated mutations involving the last 10 amino acids impair receptor endocytosis.
One effect of Ser/Thr phosphorylation at the C-tail is recruitment of the adaptor protein β-arrestin (β-arr) 1/2, which links CXCR4 to a clathrin lattice. β-arr binding uncouples heterotrimeric G proteins from the receptor, leading to its desensitization, and engages adaptor protein 2 (AP-2) and clathrin, two important elements of the endocytic machinery.18 β-arr also acts as a scaffold for activation of other signaling pathways downstream of CXCR4, such as those of the p38 mitogen-activated protein kinase (MAPK) and the p44/p42 extracellular-regulated kinases (ERK1/2).19 Although C-tail phosphorylation is a major signal for β-arr–CXCR4 interaction, β-arr also bind the third intracellular loop (ICL3) of the receptor,20 which is linked to prolonged CXCR4 signaling in WHIM receptor mutants.21
We previously reported a CXCR4 C-tail interaction with filamin A (FLNA).22 FLNA is a dimeric protein, each subunit of which has an N-terminal spectrin-related actin-binding domain and 24 immunoglobulin-like β-sheet tandem repeats separated into 2 rods by 2 hinge regions.23 Repeat 24 mediates protein dimerization and endows the dimer with a structure that assists orthogonal branching of actin filaments. FLNA activity nonetheless extends beyond its F-actin crosslinking ability because it acts as a scaffold for the binding of more than 90 partners,23 most of which interact with the second rod (repeats 16 to 23). The chemokine receptors CXCR4 and CCR2b are among the FLNA binding partners. FLNA interaction with CXCR4 triggers and stabilizes receptor clustering at the cell surface during HIV-1 infection22; its binding to CCR2b is necessary for efficient receptor endocytosis and intracellular trafficking of endocytic vesicles.24,25 Indeed, FLNA has dual effects on the endocytosis of other receptors; it stabilizes rectifying K+ channels,26 calcitonin,27 or cystic fibrosis transmembrane conductance regulator28 receptors at the plasma membrane but sustains dopamine D3 receptor endocytosis.29 Whether FLNA interacts with WHIM mutants and the influence of this putative interaction on β-arr binding and receptor endocytosis are not known but are potentially important in the dysfunction of WHIM-associated CXCR4 mutants.
Here we show that, in addition to its interaction with the C-tail, FLNA bound the 238LKTTVILIL246 motif in the CXCR4 ICL3 domain. Deletion of this motif accelerated CXCL12-mediated endocytosis of wild-type (WT) CXCR4 and permitted CXCL12-induced internalization of the WHIM-like CXCR4R334X mutant receptor. Loss of the FLNA-ICL3 interaction also attenuated ligand-induced CXCR4R334X receptor signaling, restoring normal CXCL12-induced migration and ERK1/2 activation kinetics. Whereas FLNA and β-arr2 competed for ICL3 binding in vitro, FLNA cooperated with β-arr2 binding to CXCR4R334X in cells. Our results identify the FLNA-ICL3 interaction as a regulatory element in CXCR4 internalization and signaling, with potential implications for WHIM syndrome.
Methods
Human embryonic kidney (HEK) –293 and HEK-293T cells were obtained from the American Type Culture Collection. The M2 (FLNA-deficient) and the isogenic A7 (FLNA-expressing) melanoma cell lines were maintained in minimal essential medium (Invitrogen) supplemented with 5% fetal calf serum; A7 cells also received 0.5 mg/mL G418 (Sigma-Aldrich). X4WT and X4WHIM complementary DNA cloned in the m5p retroviral system were kindly provided by M. Kallikourdis and A. Viola.30 ICL3 mutations were generated with the Quickchange Site-Directed Mutagenesis Kit (Stratagene) by using specific primers (see supplemental Methods available at the Blood Web site for more detail). These X4 constructs as well as the pnglv-gag-pol and envVSVG plasmids were transfected in HEK-293T cells by using jetPEI (Polyplus Transfection) to generate retroviral particles used to transduce the cell lines, as described.31,32 X4KG2A and X4S324X mutants were generated by site-directed mutagenesis using specific primers. The X4S324X mutant was further engineered to generate ICL3 mutations as above.
For pull-down assays, recombinant FLNA constructs were expressed and purified as described,33 and biotinylated ICL3 peptides were synthesized at the Centro Nacional de Biotecnología Core Facility. Glutathione S-transferase–β-arr2 was obtained by restriction-free cloning34 using pGEX 4T1 (GE Healthcare) and rat β-arr2–enhanced green fluorescent protein as templates and Phusion DNA polymerase (Thermo Scientific). Immunoblotting was performed by using anti-FLNA monoclonal antibody1-735 or horseradish peroxidase–conjugated anti-His monoclonal antibody (Sigma-Aldrich). Surface plasmon resonance (SPR) experiments were performed by using Biacore 3000 (GE Healthcare), and sensograms were analyzed with BIAevaluation Software 4.1. Data sets were processed by a double-referencing method. For immunoprecipitation, we used specific antibodies and HEK-293 or green fluorescent protein (GFP) –β-arr2-HEK-293 cells that expressed the indicated receptors (supplemental Methods).
CXCL12-induced receptor endocytosis was analyzed by fluorescence-activated cell sorter (FACS) (Cytomics FC500; Beckman Coulter). Receptor endocytosis was calculated as the quotient of the geometric mean fluorescence intensity of treated cells and the mean fluorescence intensity of unstimulated cells expressed as a percentage; 100% corresponds to receptor expression on the surface of unstimulated cells.
HEK-293T cells were transiently transfected with X4WHIM and X4WHIM-ICL3ΔCt plasmids and were analyzed by fluorescence microscopy (Olympus Fluoview FV-1000 microscope fitted with a 60× NA1.4 objective) using anti-early endosome antigen 1 (EEA1) (BD Biosciences) or anti-lysosomal-associated membrane protein-1 (LAMP-1) antibodies (Abcam) for costaining. Images were processed with ImageJ software (National Institutes of Health). ERK activation, actin polymerization, and chemotaxis assays were performed as described.36,37
Results
CXCR4 C-tail is not the only FLNA binding motif
Studies have shown that the CXCR4 C-tail binds to the atypical FLNA repeat 10.22,38 Sequence analysis predicted a small motif at the CXCR4 C-tail, with charge and secondary structure similar to those identified in CD4 as important for binding to FLNA repeat 10 (supplemental Figure 1A). This motif coincides in CXCR4 and CD4 with a phorbol ester-activated endocytosis signal and bears an AP-2 clathrin adaptor-reactive sequence.15 In CD4, residues E416 and K417, located in a helix turn (1Q68 structure), are critical for FLNA binding. We thus generated a CXCR4 mutant in which the homologous residues K331 and G332 were replaced with alanine (X4KG2A); a mutant lacking the entire motif (X4S324X) was also engineered.
Cell surface expression of X4KG2A and X4S324X mutants was comparable to that of the X4WT receptor (not shown), although CXCL12-induced internalization was reduced (supplemental Figure 1B). As predicted, the X4S324X deletion mutant was completely resistant to CXCL12-induced internalization because it lost most S/T residues involved in receptor endocytosis.16 The X4KG2A receptor internalization rate was reduced, and it was endocytosed only after lengthy incubation with CXCL12. This severe impairment in X4KG2A internalization might be the result of mutation-dependent conformational changes in adjacent regions, particularly the α-helix, which bears the di-leucine (I328L329) motif implicated in CXCR4 endocytosis.15,16
FLNA is important for transduction of some CXCR4-mediated signaling pathways, including CXCL12-induced F-actin polymerization.22 Analysis of this process indicated that CXCL12-induced F-actin formation was clearly enhanced by both of our C-tail mutants compared with X4WT (supplemental Figure 1C), which paralleled the impaired endocytosis of mutant CXCR4 receptors with their enhanced signaling ability.
We used immunoprecipitation assays to test whether the X4 mutations affected interaction with FLNA. The X4WT receptor as well as the X4KG2A and X4S324X mutants coprecipitated with endogenous FLNA (supplemental Figure 1D), although X4S324X lacks the putative FLNA-binding motif. FLNA interaction with the X4S324X mutant appeared to be enhanced compared with the X4WT receptor. The results suggest that FLNA-CXCR4 interaction involves receptor domains in addition to the C-tail.
ICL3 bears a consensus FLNA-binding motif
All FLNA-binding sites thus far characterized at the atomic level consist of a β-strand formed by alternating hydrophobic residues pointing toward the CD groove of an FLNA β-sheet repeat.23,39 By using the FLNA-binding motif, we searched for a potential FLNA-binding site on CXCR4 and found a sequence in the C-terminal part of the ICL3 (Figure 1A).
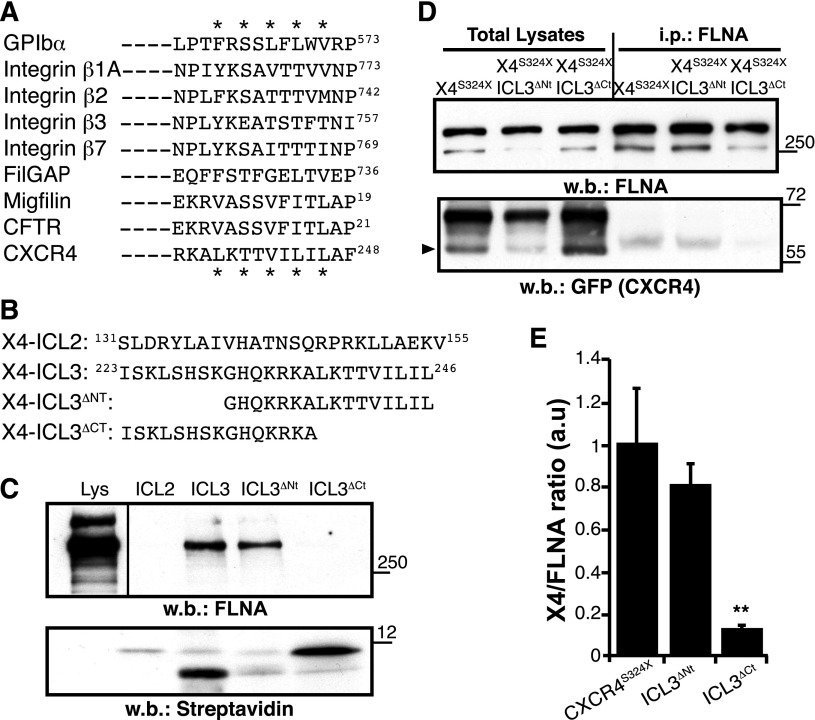
Figure 1.
The CXCR4 ICL3 domain bears an FLNA-binding motif. (A) Alignment of distinct FLNA partners, showing the consensus binding sequence; (*) indicates relevant amino acids. (B) Scheme of the CXCR4 peptides designed for pull-down assays and their position in the CXCR4 sequence. (C) Pull-down assay. Biotinylated peptides were incubated with HEK-293 cell lysates (Lys) and pull-down was performed. Immunoblot of total lysates (left) and peptide-bound proteins using anti-FLNA antibody (top right) and streptavidin (bottom right). A representative experiment is shown (n = 5). (D) Total lysates of HEK-293 cells expressing indicated GFP-fused receptors were immunoprecipitated (i.p.) with anti-FLNA antibody. Total lysates (left) and FLNA-bound proteins (right) were detected with anti-FLNA (top) and anti-GFP antibodies (bottom); arrowhead indicates the CXCR4 band. (E) Immunoblots as in (D) were quantified by densitometry, and the CXCR4:FLNA ratio was determined. Values were normalized to those for the CXCR4S324X mutant, which has an unmodified ICL3 domain. Data are mean ± standard error of the mean (SEM; n = 3). **P < .01, 2-tailed Student t test. w.b., western blot.
To test whether the predicted CXCR4 motif binds to FLNA, we performed pull-down experiments using synthetic peptides with (ICL3) or without (ICL3ΔCt) the LKTTVILIL motif (Figure 1B) as bait; an ICL3 peptide lacking the N-terminal residues (ICL3ΔNt) and another resembling the second intracellular loop (ICL2) were used as controls. Peptides were biotinylated at the N terminus to allow directed immobilization on streptavidin beads. ICL3 and ICL3ΔNt peptides readily precipitated FLNA from HEK-293 cell lysates, whereas ICL3ΔCt did not (Figure 1C).
We analyzed the role of the LKTTVILIL motif in FLNA interaction in living cells by deleting these residues in the ICL3 region of the X4S324X receptor, which lacks the putative FLNA repeat 10 interaction motif at the C-tail. Whereas the X4S324X receptor retained FLNA binding, deletion of the ICL3 C-terminal amino acids (residues 238 to 246; X4S324X-ICL3ΔCt) disrupted interaction with the actin-binding protein (Figure 1D-E). Deletion of the ICL3 N-terminal residues (amino acids 223 to 230; X4S324X-ICL3ΔNt) had little or no effect on FLNA coprecipitation (Figure 1D-E), coinciding with the pull-down assay data. The results suggest that the ICL3 C-terminal region constitutes an unreported CXCR4 motif for FLNA binding in vitro and in vivo.
CXCR4 ICL3 interacts with specific FLNA repeats
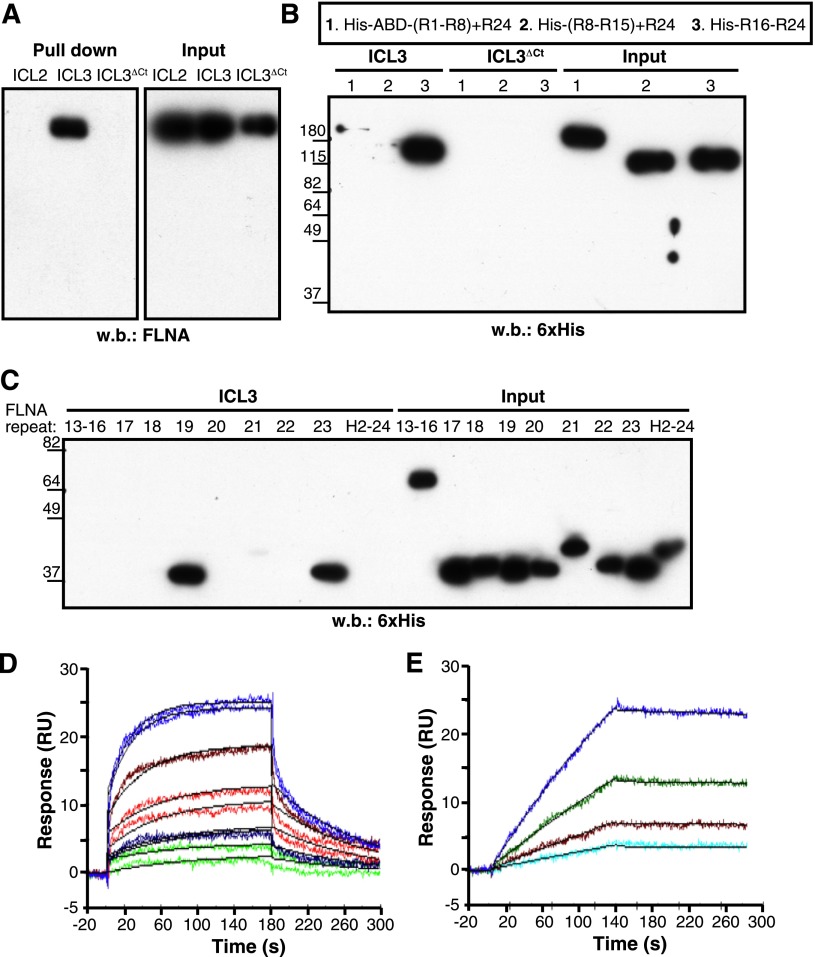
Direct interaction between the ICL3 C-terminal region and FLNA was confirmed by the pull-down of ICL3, but not of ICL3ΔCt peptides, with recombinant full-length FLNA protein (Figure 2A). Pull-down experiments with His-tagged FLNA fragments covering the entire molecule narrowed the CXCR4-ICL3 interaction domain of FLNA to C-terminal repeats 16 to 24 (Figure 2B). Further analyses identified repeats 19 and 23 as major ICL3 binding domains, whereas repeat 21 showed a weak interaction with this CXCR4 loop (Figure 2C).
Figure 2.
FLNA repeats 19 and 23 bind to the CXCR4 ICL3 domain. (A) Pull-down assay of the indicated CXCR4 peptides with recombinant full-length FLNA protein. (B-C) ICL3 peptide pull-down assay with (B) purified His-tagged FLNA fragments or (C) individual FLNA repeats. Bound fragments or repeats were detected by immunoblot with peroxidase-labeled anti-His antibody. (D) Representative sensorgrams for FLNA repeat 19 binding kinetics to immobilized ICL3 peptide, monitored as relative units (RU). The concentrations of FLNA repeat 19 tested were 4 (violet), 2 (brown), 1 (red), 0.5 (blue), and 0.25 μM (green). Binding of FLNA repeat to the reference sensorchip (immobilized ICL3ΔCt) was subtracted for each experimental condition. Two sensorgrams are shown for each repeat 19 concentration. Langmuir fitting is shown in black. (E) Sensorgram for FLNA(16-23) binding kinetics to immobilized ICL3 peptide as in (D). The FLNA (16-23) concentrations tested were 155 (blue), 77.5 (green), 38.7 (brown), and 19.3 nM (cyan). The black line represents the fitting to the 1:1 model. H2-24, hinge region 2 + repeat 24; R1-R8, repeats 1-8.
We analyzed the ICL3 binding kinetics of repeat 19 by SPR. Repeat 19-ICL3 interaction fitted well to the 1:1 Langmuir model, yielding association and dissociation rate constants of ka = 6.66 × 103 ± 0.052 M–1s−1 and kd = 1.01 × 10−2 ± 5.6 × 10−5 s−1 (n = 2), corresponding to an equilibrium dissociation constant of KD = 1.51 μM (Figure 2D). SPR analysis showed that the FLNA(16-23) fragment also bound to ICL3 at 1:1 stoichiometry. Compared with repeat 19, FLNA(16-23)-binding affinity was stronger (KD = 7.8 nM), mainly because of a much slower dissociation rate (ka = 2.5 × 104 ± 0.022 M–1s−1 and kd = 1.95 × 10−4 ± 1.3 × 10−5 s−1; Figure 2E), probably because of the high valency of FLNA(16-23) (bearing repeats 19, 21, and 23, which bind ICL3).
FLNA-ICL3 interaction is sensitive to p160ROCK inhibition
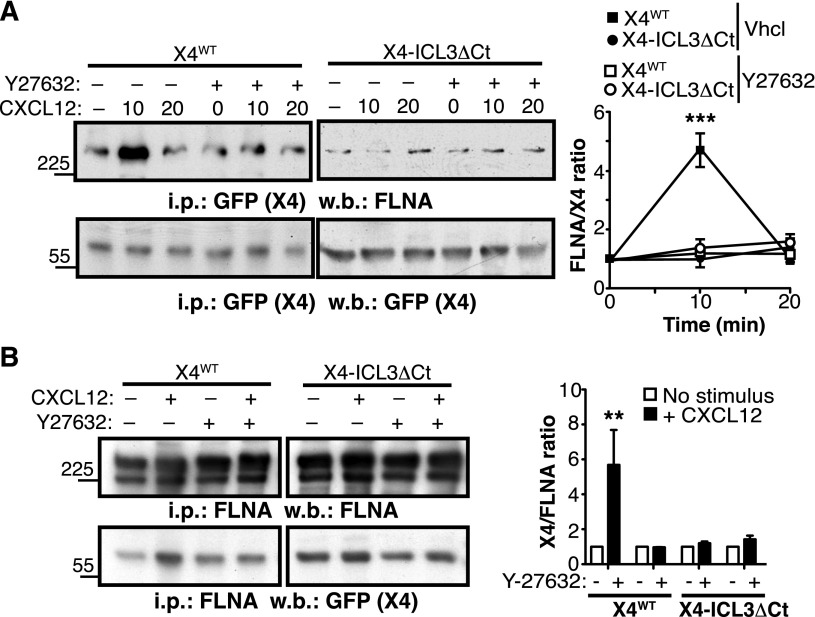
The p160 Rho kinase (ROCK1) participates in CXCR4 signaling,40 and FLNA has been implicated in CXCR4-mediated activation of the RhoA-ROCK pathway.22 Because FLNA interaction with some partners is dependent on actomyosin contraction, which confers mechanical force and exposes cryptic FLNA binding sites for its ligands,41,42 we tested whether the ROCK pathway, a well-characterized upstream activator of myosin II, affected CXCR4-FLNA binding. In cells expressing the X4WT receptor, CXCL12 stimulation induced a transient increase in CXCR4-bound FLNA. This was prevented by cell pretreatment with the ROCK inhibitor Y27632 (Figure 3A), suggesting that F-actin stabilization and subsequent actomyosin contraction affected the FLNA-CXCR4 interaction. In contrast, CXCL12 stimulation did not enhance FLNA interaction with an X4 receptor mutant that lacked the ICL3 C-terminal region (X4-ICL3ΔCt) (Figure 3A). Results were similar when FLNA was immunoprecipitated; we found that CXCL12-induced Y27632-sensitive FLNA association with X4WT was not observed in X4-ICL3ΔCt–expressing cells (Figure 3B). These findings suggest that ligand-induced conformational changes promote FLNA-CXCR4 interaction in an ROCK- and ICL3-dependent manner.
Figure 3.
CXCL12 enhances CXCR4-FLNA interaction in a ROCK- and ICL3-dependent manner. (A) X4WT- or X4-ICL3ΔCt–expressing HEK-293 cells were pretreated with the ROCK inhibitor Y27632 or vehicle (Vhcl; dimethylsulfoxide), then left untreated or stimulated for 10 or 20 minutes with CXCL12. Cell lysates were immunoprecipitated with anti-GFP (CXCR4 tag) antibody and immunoblotted with anti-FLNA (top) and anti-GFP (bottom) antibodies. The relative ratio is shown for CXCR4:FLNA (FLNA), calculated by densitometric analysis (right); values were normalized to the ratio in unstimulated cells. (B) HEK-293 cell were pretreated as in (B) and stimulated with CXCL12 for 5 minutes or unstimulated. Cell lysates were immunoprecipitated with anti-FLNA, and resolved proteins were blotted with anti-FLNA and anti-GFP antibodies. The relative CXCR4:FLNA ratio was calculated as above. (A-B) Data are mean ± SEM (n = 3). **P < .01; ***P < .001; 2-tailed Student t test.
FLNA-ICL3 interaction regulates CXCL12-induced CXCR4 endocytosis
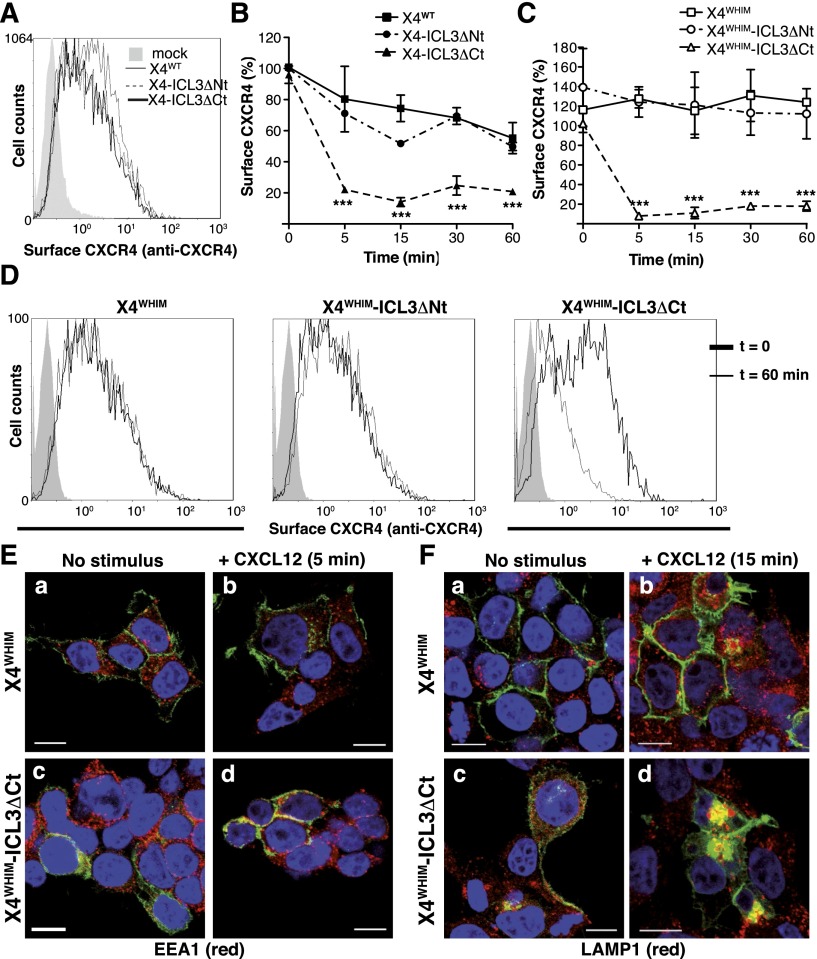
Because FLNA regulates CCR2b internalization,24,43 we tested whether the FLNA-ICL3 interaction affects CXCR4 endocytosis. In unstimulated cells, receptor expression at the cell surface was comparable (Figure 4A). CXCL12 stimulation triggered endocytosis of all receptors, but X4-ICL3ΔCt internalization was more rapid and persistent than that of X4WT or X4-ICL3ΔNt (Figure 4B).
Figure 4.
ICL3-FLNA interaction regulates CXCL12-induced internalization of WHIM-like CXCR4 receptors. (A) FACS analysis of cell surface expression of X4WT, X4-ICL3ΔNt, and X4-ICL3ΔCt in unstimulated HEK-293 cells; a representative experiment is shown (n = 4). (B) Kinetics of X4WT, X4-ICL3ΔNt, and X4-ICL3ΔCt internalization in HEK-293 cells after CXCL12 stimulation. Cell surface expression was determined with an anti-CXCR4 antibody in the GFP+ cell population and calculated as indicated (see “Methods”). (C) HEK-293 cells were transfected with X4WHIM, X4WHIM-ICL3ΔNt, and X4WHIM-ICL3ΔCt, and CXCL12-induced internalization kinetics were analyzed as above. (D) Representative histograms of cell surface levels of X4WHIM, X4WHIM-ICL3ΔNt, and X4WHIM-ICL3ΔCt before and after 60 minutes of CXCL12 stimulation. (B-C) n = 4. ***P < .001, 2-way analysis of variance (ANOVA) with Bonferroni posttest. (E) HEK-293T cells were transiently transfected with GFP-tagged X4WHIM and X4WHIM-ICL3ΔCt (green), and colocalization with the EEA1 (red) was analyzed by immunofluorescence (a,c) before and (b,d) after 5 minutes of CXCL12 stimulation. (F) Colocalization of the aforementioned receptors (green) with LAMP1 (red) in (a,c) resting and (b,d) CXCL12 cells stimulated for 15 minutes. (E-F) Cells shown are representative of at least 10 fields; bar = 10 μm.
Given that lack of FLNA-ICL3 binding appeared to accelerate receptor endocytosis, we tested whether this interaction also affected internalization of a WHIM-like CXCR4 receptor. By using CXCR4R334X, a common WHIM syndrome–causing mutation that renders an endocytosis-resistant receptor,2 we deleted the ICL3 N- or C-terminal region (X4WHIM-ICL3ΔNt and X4WHIM-ICL3ΔCt); these constructs were expressed in HEK-293 cells, and CXCL12-induced endocytosis was analyzed by FACS. X4WHIM was not internalized after CXCL12 stimulation, which was also the case for X4WHIM-ICL3ΔNt; in contrast, X4WHIM-ICL3ΔCt was endocytosed shortly after CXCL12 stimulation (Figure 4C). Histograms for each construct before and after 60 minutes of stimulation are shown from a representative experiment in Figure 4D.
We analyzed CXCL12-induced receptor endocytosis by immunofluorescence in the presence of specific markers of the endocytic pathway. In accordance with its lack of internalization in FACS analyses, X4WHIM receptor showed membrane-like staining; it did not colocalize with EEA1 (Figure 4Ea,b) or LAMP1 (Figure 4Fa,b) at 5 or 15 minutes after stimulation, respectively. In contrast, X4WHIM-ICL3ΔCt colocalized with EEA1 after 5 minutes of stimulation (Figure 4Ec,d) and with LAMP1 at longer times (Figure 4Fc,d), which coincides with the endocytic trafficking reported for CXCR4.43 FLNA-ICL3 interaction appears to act as a major regulator of CXCL12-induced CXCR4 internalization, even for WHIM-like mutant receptors.
FLNA deficiency enhances CXCL12-induced endocytosis of WHIM-like CXCR4
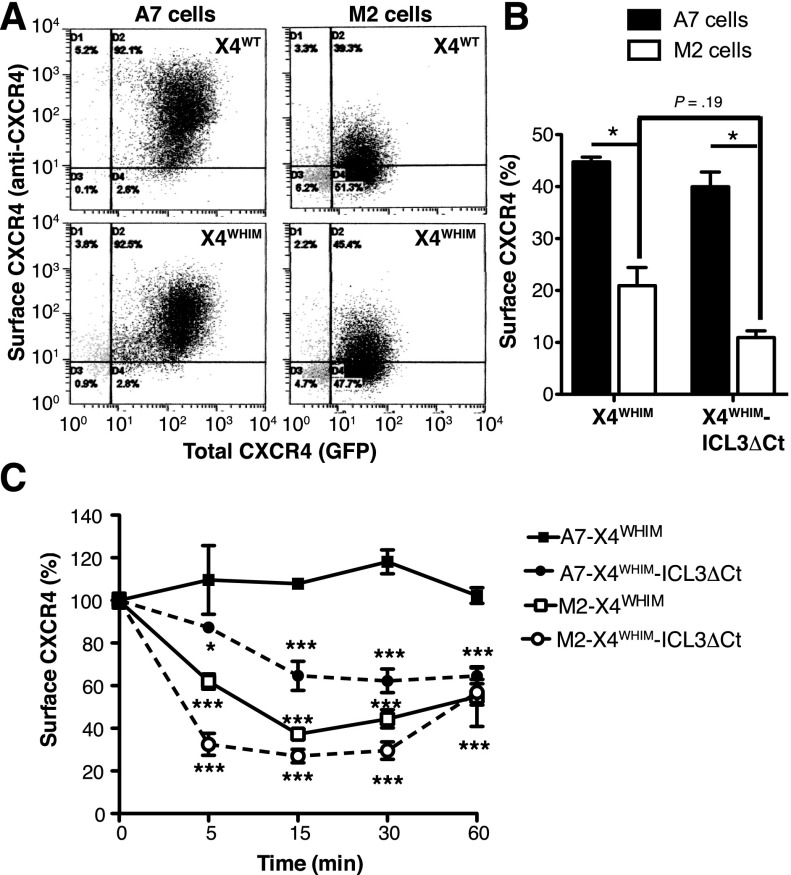
To further study the role of FLNA in the endocytosis of WHIM-like receptors, we compared CXCR4 internalization in A7 (FLNA-expressing) and M2 (FLNA-deficient) cells. FLNA deficiency impaired basal cell surface levels of X4WT and X4WHIM receptors. Whereas stably transfected A7 and M2 cells showed comparable total levels of these receptors (as indicated by GFP tag fluorescence), FLNA deficiency reduced cell surface localization (as shown by anti-CXCR4 antibody staining) (Figure 5A). We observed similar FLNA-dependent modulation of basal cell surface expression for X4WHIM and X4WHIM-ICL3ΔCt transiently transfected in A7 and M2 cells (Figure 5B). In both cell types, cell surface levels of X4WHIM-ICL3ΔCt tended to be lower than those of X4WHIM, although values were not significant.
Figure 5.
FLNA deficiency enhances tonic and CXCL12-induced internalization of WHIM-like CXCR4 receptors. (A) Dot plots of unstimulated A7 and M2 cells stably expressing GFP-tagged X4WT and X4WHIM stained with anti-CXCR4 antibody. (B) Basal cell surface levels of X4WHIM and X4WHIM-ICL3ΔCt in A7 (solid bars) and M2 cells (open bars) determined by anti-CXCR4 staining. The ratio is shown (×100) between the percentage of anti-CXCR4–stained and GFP+ cells for each construct (n = 3). *P < .05, 2-tailed Student t test. (C) Kinetics of CXCL12-induced X4WHIM and X4WHIM-ICL3ΔCt receptor internalization in A7 (solid symbols) and M2 cells (open symbols). In all cases, data are mean ± SEM of a representative experiment (n = 3). *P < .05; ***P < .001, 2-way ANOVA with Bonferroni posttest.
In addition to the role of FLNA in stabilizing CXCR4 at the plasma membrane in basal conditions, experiments with M2 and A7 cells confirmed FLNA participation in CXCL12-induced receptor endocytosis. The X4WHIM mutant, which was insensitive to CXCL12-induced internalization in A7 cells, was endocytosed efficiently in M2 cells (Figure 5C). Moreover, as in HEK-293 cells, CXCL12 induced X4WHIM-ICL3ΔCt internalization in FLNA-expressing and -deficient cells, although receptor endocytosis was more robust in cells that did not express FLNA (Figure 5C). These results confirm the importance of FLNA interaction with the ICL3 domain for CXCL12-induced internalization of WHIM-like CXCR4 mutants.
FLNA-ICL3 interaction is responsible for amplified X4WHIM signaling
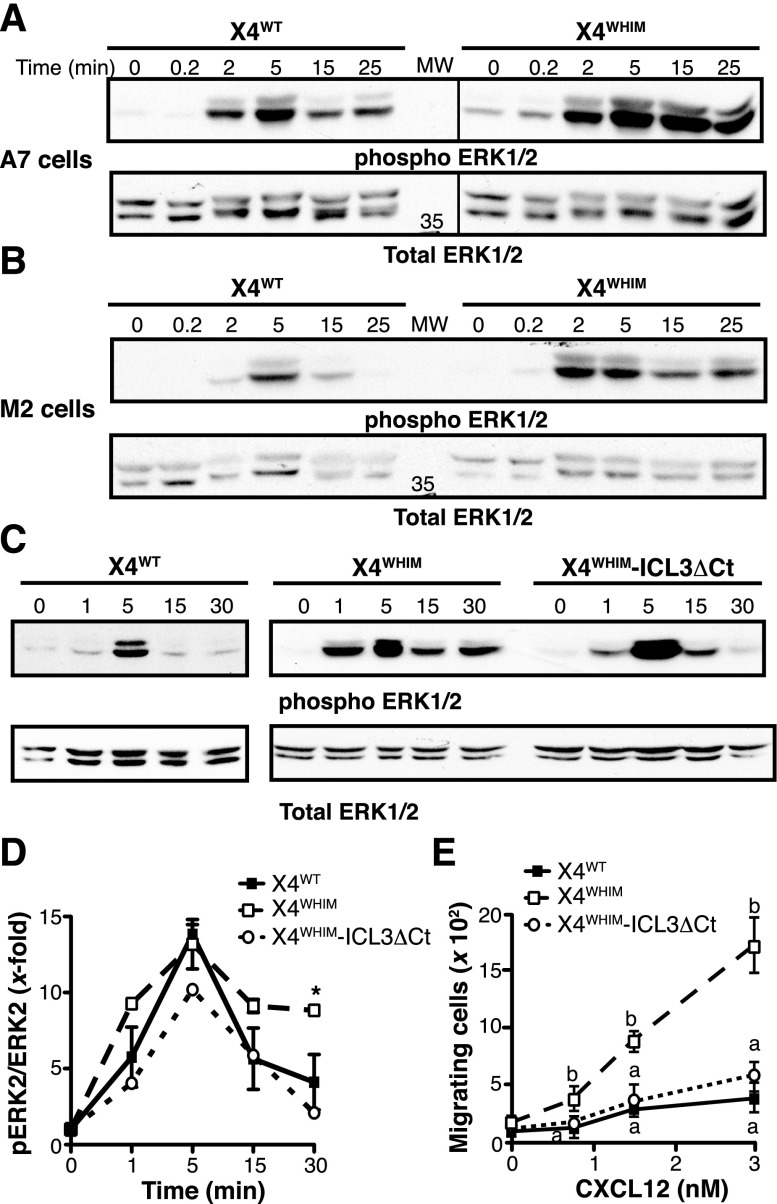
WHIM-associated CXCR4 receptors show increased responsiveness to CXCL12. To test whether FLNA also affects WHIM-like receptor hyperfunction, we analyzed CXCL12-induced activation of ERK1/2 in X4WT- and X4WHIM-expressing M2 and A7 cells. In A7 cells (FLNA+), CXCL12-induced ERK1/2 phosphorylation appeared earlier and was more robust and persistent in cells expressing X4WHIM than the X4WT receptor (Figure 6A), which is consistent with X4WHIM hyperfunction in these cells. CXCL12-induced ERK1/2 activation in M2 cells (FLNA–) was weaker and less persistent than in A7 cells, independently of the receptor expressed (Figure 6B). Compared with M2-X4WT cells, M2-X4WHIM cells also showed enhanced ERK activation after CXCL12 stimulation. Nonetheless, phospho-ERK levels in M2-X4WHIM cells declined at 15 minutes after stimulation (Figure 6A) in contrast to the sustained activation observed in A7-X4WHIM cells. The results thus suggest that FLNA is involved in the duration of ERK activation downstream of WHIM-like receptors.
Figure 6.
ICL3-FLNA interaction regulates CXCR4-induced ERK1/2 activation. (A-B) X4WT- and X4WHIM-expressing (A) A7 and (B) M2 cells were stimulated with CXCL12 for the times indicated. Phosphorylated (top) and total (bottom) ERK1/2 levels were determined by immunoblot with specific antibodies (n = 3). (C) Phosphorylated (top) and total (bottom) ERK1/2 levels after CXCL12 stimulation were determined as above in HEK-293 cells transfected with X4WHIM and X4WHIM-ICL3ΔCt. A representative experiment is shown (n = 3). (D) Kinetics of CXCL12-induced ERK1/2 activation in HEK-293 cells as in (B), determined by densitometric analysis. Values were normalized by using the phosphoERK:ERK ratio in unstimulated cells for each receptor. Data shown as mean ± SEM; *P < .05, 2-way ANOVA with Bonferroni posttest. (E) HEK-293 chemotaxis was assayed in transwell devices by using the indicated doses of CXCL12 as a chemoattractant. Data are mean ± SEM in a representative experiment. Curves with distinct letters differ significantly from one another (n = 3; Student-Newman-Keuls test, P < .05).
Because FLNA binds many partners that regulate ERK activation directly,23 we sought a cell system to specifically analyze how FLNA-ICL3 interaction might regulate hyperactivation of WHIM receptors. We overexpressed X4WT-, X4WHIM-, or X4WHIM-ICL3ΔCt receptors in HEK-293 cells and analyzed CXCL12-induced ERK activation kinetics. As in A7 cells, CXCL12 stimulation of HEK-293-X4WHIM cells induced more robust, sustained ERK1/2 activation than in X4WT-expressing cells; X4WHIM-ICL3ΔCt expression resulted in transient ERK1/2 activation, which resembled that of X4WT (Figure 6C). Densitometric quantification of immunoblots indicated a receptor-independent ERK activation peak at the same time and of approximately the same degree; however, we observed a decrease in phospho-ERK levels only in cells expressing X4WHIM-ICL3ΔCt receptors (Figure 6D). Likewise, we did not find the amplification of the CXCL12-induced chemotaxis that is characteristic of WHIM receptors in cells that overexpressed the X4WHIM-ICL3ΔCt receptor (Figure 6E). These data indicate that FLNA-ICL3 interaction is necessary for the WHIM mutant receptor gain-of-function phenotype.
FLNA regulates β-arr2 interaction with ICL3
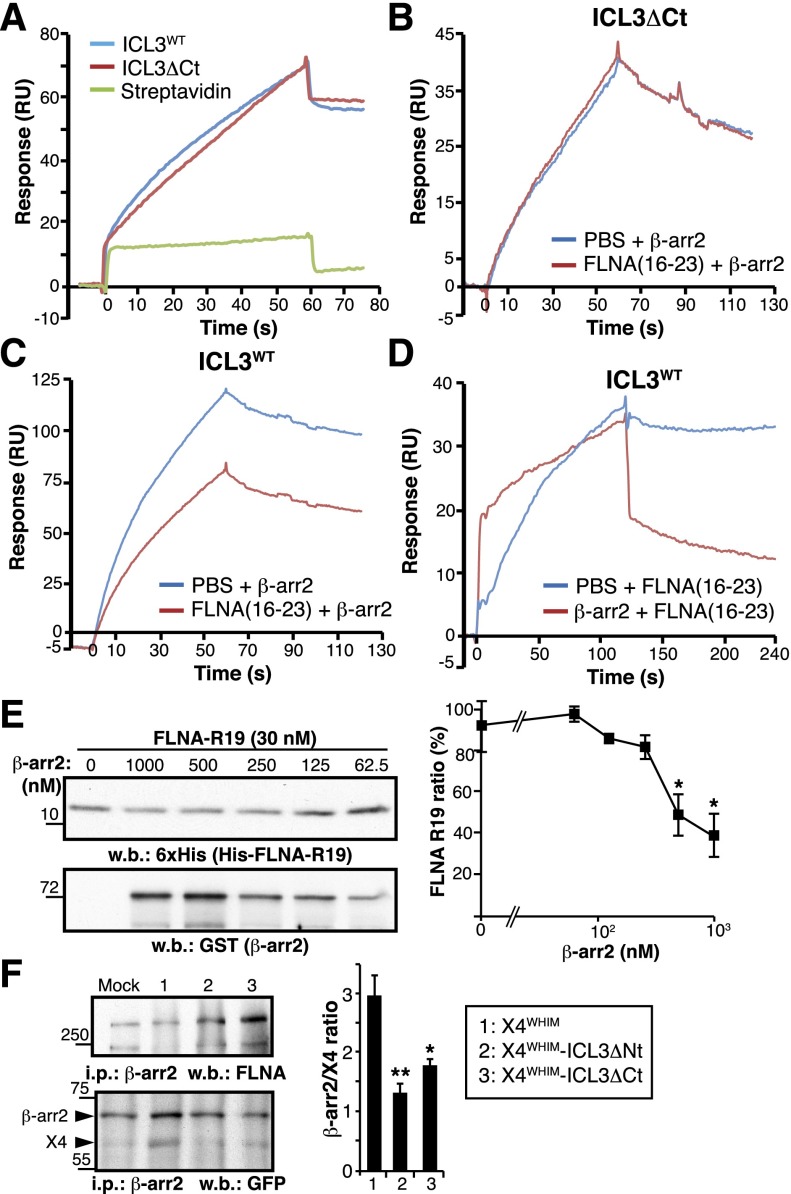
In addition to the FLNA-binding site described here, ICL3 bears a β-arr2 interaction motif.20,21 By using SPR, we found that recombinant β-arr2 interacted with biotinylated ICL3 and ICL3ΔCt peptides immobilized on streptavidin, but not with streptavidin alone (Figure 7A), confirming the presence of a β-arr2–binding site in the ICL3 N-terminal region. Because of the proximity of FLNA and β-arr2–binding motifs, we tested whether both molecules can bind simultaneously to ICL3. The FLNA(16-23) fragment was injected onto immobilized ICL3 or ICL3ΔCt (FLNA-negative control) prior to β-arr2 injection. As anticipated, FLNA(16-23) did not affect β-arr2 interaction with ICL3ΔCt (Figure 7B), but it partially inhibited β-arr2 binding to ICL3 (Figure 7C); we were unable to saturate ICL3 at the FLNA(16-23) concentration used (400 nM), which might explain this partial inhibition.
Figure 7.
FLNA-ICL3 interaction enhances β-arr2 association with WHIM receptor in cells. (A) BiaCore sensorgrams for β-arr2 binding to immobilized ICL3, ICL3ΔCt peptides, and streptavidin (reference), monitored as RU. The β-arr2 concentration was 20 nM in all cases. (B-C) SPR analysis of FLNA interference with β-arr2 binding to ICL3. The FLNA(16-23) fragment (400 nM) and β-arr2 (20 nM) were injected sequentially onto flow cells with immobilized (B) ICL3ΔCt or (C) ICL3WT peptides. Curves depict β-arr2 binding (in RU) to each peptide after subtraction of the RU signal for FLNA(16-23) fragment binding. (D) Analysis of simultaneous β-arr2 and FLNA binding to ICL3. β-arr2 (200 nM; red curve) or buffer (blue curve) were injected onto ICL3WT, followed by injection of the FLNA(16-23) fragment (250 nM). The sensorgrams show the association and dissociation phases of the FLNA(16-23) fragment in each condition, after subtraction of β-arr2 and buffer RU values. (E) Analysis of FLNA interference with β-arr2 binding to ICL3 by pull-down assay with purified His-tagged FLNA repeat 19 fragment and glutathione S-transferase (GST) –β-arr2 at indicated concentrations. Representative immunoblots are shown with anti-6xHis and anti-GST antibodies and densitometric quantification. (F) Analysis of FLNA and β-arr2 binding to X4WHIM receptor variants in living cells. GFP-tagged X4WHIM, X4WHIM-ΔNt, and X4WHIM-ΔCt receptors were transfected in HEK-293 cells stably expressing GFP-β-arr2. Equal amounts of cell lysates were immunoprecipitated with anti-β-arr2 antibody, and the precipitated proteins were analyzed with anti-FLNA and anti-GFP antibodies. Bands corresponding to β-arr2 and CXCR4 were quantified by densitometry (right), and the ratio was calculated by using the values for mock cells as reference. (E-F) Data are mean ± SEM (n = 3). **P < .02; *P < .05, 1-tailed Student t test compared with X4WHIM cells.
β-arr2 injection onto immobilized ICL3 also interfered with FLNA(16-23) binding to the peptide (Figure 7D). In this case, FLNA(16-23) showed more rapid association rates and slower dissociation rates to the β-arr2-ICL3 complex than to ICL3 alone (Figure 7D), suggesting that the FLNA fragment interacts with β-arr2 rather than with ICL3. Nevertheless, β-arr2 dissociation from ICL3 precluded reliable determination of FLNA(16-23) binding kinetics to the β-arr2-ICL3 complex.
SPR experiments suggested relative steric hindrance in FLNA and β-arr2 binding to ICL3. To better understand this competition, we used pull-down assays to analyze FLNA repeat 19 binding to ICL3 in the presence of distinct concentrations of recombinant β-arr2; FLNA repeat 19 does not bind to β-arr2.44 β-arr2 showed dose-dependent partial inhibition of FLNA repeat 19 binding to ICL3 (Figure 7E). SPR and pull-down assays thus suggest some competition between β-arr2 and FLNA for ICL3 binding in vitro.
We next analyzed whether β-arr2 and FLNA also compete in vivo. We expressed X4WHIM, X4WHIM-ICL3ΔNt, and X4WHIM-ICL3ΔCt receptors in HEK-293 cells stably expressing β-arr2-GFP; β-arr2 was specifically precipitated. A significantly larger amount of β-arr2 coprecipitated with X4WHIM compared with X4WHIM-ICL3ΔNt or X4WHIM-ICL3ΔCt receptors (Figure 7F); there was a tendency toward more β-arr2 binding to X4WHIM-ICL3ΔCt than to X4WHIM-ICL3ΔNt, which is consistent with loss of the β-arr2 motif in the latter.21 β-arr2 coprecipitation with FLNA was observed in all conditions, indicating that this interaction was direct and did not require prior β-arr2 binding to CXCR4. These results suggest that FLNA does not compete with but rather facilitates β-arr2 binding to ICL3 in vivo.
Discussion
Here we report identification of an FLNA-binding motif in the CXCR4 ICL3 as an important regulatory element in CXCL12-induced internalization not only of the WT receptor but also of the endocytosis-resistant CXCR4R334X mutant, the most common WHIM-associated receptor. We base this conclusion on the observations that (1) recombinant FLNA repeats 19 and/or 23 bound in vitro with high affinity to a synthetic peptide representing ICL3, (2) truncation of the ICL3 FLNA-binding motif severely inhibited FLNA association with CXCR4 in cells, (3) deletion of the ICL3 FLNA-binding motif accelerated CXCL12-induced endocytosis of WT CXCR4 and enabled internalization of the CXCR4R334X mutant in FLNA-expressing cells, and (4) FLNA deficiency enhanced tonic and CXCL12-induced endocytosis of WT and mutant CXCR4. Removal of the ICL3 FLNA-binding motif also led to normalization of CXCL12-induced ERK signaling and chemotaxis of WHIM-expressing cells. FLNA binding to ICL3 might thus be central to CXCR4 internalization and signaling in physiological and pathological contexts.
We previously showed that FLNA repeat 10 interacts with the CXCR4 C-tail,22 and our results suggest that the ICL3 is also a binding motif for FLNA repeats 19 and 23. This ICL3 FLNA-binding motif contains hydrophobic amino acid that can fit the hydrophobic cleft formed by CD strands of FLNA repeats (Figure 1A).23,39 The ICL3 binding motif includes 6 residues predicted to lie within the receptor transmembrane domain.45,46 To the best of our knowledge, there is no precedent in undisrupted cells for interaction between a cytoplasmic protein and one partially embedded in the membrane. Further structural analysis is necessary to determine precisely how FLNA gains access to this region.
Although these two binding sites are independent, they probably work in concert for CXCR4 interaction after CXCL12 stimulation. We hypothesize that the C-tail masks ICL3 in the resting state, but receptor activation removes this steric hindrance.47 CXCL12 increased FLNA binding to complete CXCR4, but not to a truncated ICL3 mutant still able to bind FLNA through its C-tail domain (Figure 3A). The increase in FLNA binding to CXCR4 was ROCK-dependent, and FLNA is involved in CXCL12-induced RhoA activation and myosin light chain phosphorylation.22 We thus propose a model of sequential interaction between FLNA and CXCR4, in which constitutive FLNA repeat 10-CXCR4–C-tail binding initiates a CXCL12-induced signaling cascade that culminates in myosin contraction and enables binding of FLNA repeats 19 to 23 to ICL3. This model also explains the enhanced FLNA coprecipitation with C-tail–truncated CXCR4 compared with WT in resting conditions (supplemental Figure 1D), because C-tail truncation would expose ICL3 for FLNA binding. Each individual CXCR4 domain nonetheless appears to be dispensable for FLNA binding, because FLNA coprecipitated with CXCR4 mutants that lack the C-tail but have an intact ICL3 (Figure 1D), as well as with ICL3-truncated mutants that maintain the C-tail (Figure 3A-B).
How myosin contraction enhances FLNA-CXCR4 interaction is not entirely clear. F-actin rearrangements could induce conformational changes in both CXCR4 and FLNA. CXCL12 modifies CXCR4 conformation, leading to receptor dimerization and activation48,49; both processes require actin cytoskeleton dynamics.50 Since the ICL3 FLNA-binding motif is predicted to lie partially within the lipid bilayer, CXCL12-induced actomyosin contraction might induce conformational changes that expose the ICL3 motif, permitting its interaction with FLNA repeats 19 to 23.
Myosin contraction might also promote FLNA-CXCR4 interaction by directly affecting the structure of FLNA repeats involved in ICL3 binding. Repeats 19 and 21 are in an autoinhibitory conformation because of the occupancy of their CD faces (used to bind to partners) with strand A of their preceding repeats.23,39 This autoinhibition must be removed for binding to partners. For example, myosin contraction enables FLNA repeat 21 to interact with the cytoplasmic tail of β-integrins by inducing local FLNA deformation.41 We cannot rule out the possibility that a similar mechanism initiates ICL3-FLNA interaction after CXCL12 stimulation, at least for the WT receptor.
Our study identifies a role for FLNA in stabilizing CXCR4 at the cell surface. Abrogation of the FLNA-ICL3 interaction accelerated CXCR4 internalization (Figure 4B) and, more importantly, allowed CXCL12-induced endocytosis of a WHIM-like receptor (Figure 4C). The endocytosed X4WHIM-ICL3ΔCt mutant colocalized with early endosome and lysosomal markers in a time-dependent manner. The role of FLNA in stabilizing CXCR4 at the cell surface is probably multifaceted. Part of the effect might be explained by FLNA-mediated regulation of the β-arr2–ICL3 interaction; indeed, β-arr2 binding to ICL3 stabilizes cell surface WHIM receptors.21 Reduced expression of CXCR4 WT and WHIM mutants at the unstimulated M2 cell membrane nonetheless suggests that FLNA has additional functions in receptor stabilization at the cell surface. One of these functions, supported by our previous data, could be stabilization of the cortical actin cytoskeleton. The actin cytoskeleton has a dual effect on clathrin-mediated endocytosis; whereas F-actin formation is necessary for endocytosis,51 inhibition of actin turnover also prevents receptor internalization.52 Precise control of F-actin remodeling appears to be central to endocytosis, and FLNA seems to be an important element in reinforcing the submembrane cortical cytoskeleton.53 FLNA participates in this process by enabling cofilin phosphorylation and inactivation,22 which stabilizes F-actin and promotes HIV-1–induced CD4 and CXCR4 clustering at the cell surface. FLNA could thus act as a mechanotransducer that regulates endocytosis of WHIM-like receptors by affecting rigidity of the cortical cytoskeleton.
Our data show that FLNA not only links CXCR4 to the actin cytoskeleton, but it also shapes receptor signaling through its interaction with ICL3; indeed, abrogation of FLNA-ICL3 interaction attenuated ERK signaling and chemotaxis. These effects on CXCR4 signaling might rely on FLNA-mediated regulation of the β-arr2–ICL3 interaction. Previous studies showed that β-arr2 binds separately to the CXCR4 C-tail and to the ICL3 N-terminal SHSK motif.20,21 β-arr2 binding to these two sites has distinct functional consequences; whereas β-arr2–C-tail interaction mediates receptor desensitization and endocytosis, its binding to ICL3 is important for β-arr2–mediated signaling.20 β-arr2 interaction with the WHIM-like CXCR4S338X mutant is preserved and the receptor is not internalized; instead, it enhances β-arr2–mediated ERK activation and chemotaxis,21 two pathways critically dependent on β-arr2.19,20
Our in vitro data using SPR and pull-down assays suggest that β-arr2 and FLNA compete for binding to ICL3. The in vivo data nonetheless indicate that β-arr2 binding to X4WHIM depends on FLNA-ICL3 binding because more β-arr2 coprecipitated with X4WHIM than with X4WHIM-ICL3ΔCt. Although β-arr2 levels are similar in FLNA-deficient M2 cells and the FLNA-expressing A7 subclone, CXCL12-induced ERK activation is lower in M2 than in A7 cells (Figure 7A), indicating the functional importance of FLNA–β-arr2 crosstalk. FLNA might present β-arr2 to ICL3, thus stabilizing the interaction among the three partners. β-arr2 did not coprecipitate with X4WHIM-ICL3ΔNt, although this receptor conserves the FLNA-binding domain and FLNA–β-arr2 interaction, which supports a model in which FLNA and β-arr2 cooperate to bind ICL3 at high affinity. This docking is probably more critical in C-tail–truncated receptors, in which β-arr2–C-tail interaction is lost. Our model would explain the parallel enhancement of FLNA (this study) and β-arr2 binding21 to C-tail–truncated CXCR4 mutants compared with WT receptors.
CXCR4 and FLNA have been linked independently to cancer metastasis. Our results suggest that FLNA might contribute to metastasis by direct regulation of CXCR4. FLNA-ICL3 interaction would reduce CXCR4 endocytosis and endow tumor cells with more sustained adhesive and migratory capacities, thus aiding metastasis. Whereas the CXCR4 contribution to the metastatic process is not debated, FLNA appears to have a dual role.54 FLNA reduction could conceivably render tumor cells less invasive, more sensitive to mechanical stress, and less adhesive, which would affect metastasis initiation and colonization of distant organs. Some studies have results that coincide with this view; for instance, FLNA depletion reduces in vitro migration and invasion of melanoma cells55 and reduces the metastatic potential of breast cancer and hepatocarcinoma cells in mouse models.56,57 Reduced FLNA expression is also associated with decreased lymph node metastasis, neural invasion, and enhanced distant metastasis-free survival in breast cancer patients.57,58 In contrast, FLNA absence correlates with reduced lymph node metastasis and poor prognosis in nasopharyngeal cancer.59 The role of FLNA in metastasis thus appears to be dependent on tumor type; whether CXCR4 expression is associated with this tumor specificity remains to be determined.
CXCR4 hyperactivation is proposed as a major underlying pathogenic mechanism in the WHIM syndrome.60 A recent phase 1 trial showed the clinical effectiveness of the CXCR4 antagonist plerixafor (AMD3100) in partially restoring the immune functions of 3 patients.61 On the basis of our results, it is tempting to hypothesize blockade of the FLNA-ICL3 interaction as a new therapeutic strategy for modulating surface expression and normalizing signaling of CXCR4 mutants in the WHIM syndrome.
Acknowledgments
The authors thank A. Viola, M. Kallikourdis, and F. Bachelerie for reagents and helpful discussions, M.C. Moreno and S. Escudero for assistance with fluorescence-activated cell sorting and analysis, M. Martín-Monzón and the Protein Tool service for help with surface plasmon resonance assays, L. Carmona for technical help, and C. Mark for editorial assistance.
This work was supported by the Spanish Ministry of Science and Innovation (SAF2011-24553), the Comunidad de Madrid (IMMUNOTHERCAN, S2010/BMD-2326), the European Union ERAnet program (e-rare PI07/1314), and the National Institutes of Health, National Heart, Lung, and Blood Institute (grant HL19749).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: T.P.S., F.N., and S.M. conceived the research; C.G.-M., T.P.S., F.N., and S.M. designed the experiments; C.G.-M., R.M.P., S.J.-B., F.N., and S.M. performed the experiments and analyzed the data; T.F. performed immunofluorescence experiments; F.N. and S.M. wrote the manuscript; and all authors read, added comments to, and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Santos Mañes, Centro Nacional de Biotecnologia/Consejo Superior de Investigaciones Científicas, Campus Universidad Autonoma de Madrid, Cantoblanco, 28049 Madrid, Spain; e-mail: smanes@cnb.csic.es.
References
- 1.Bachelerie F. CXCL12/CXCR4-axis dysfunctions: Markers of the rare immunodeficiency disorder WHIM syndrome. Dis Markers. 2010;29(3-4):189–198. doi: 10.3233/DMA-2010-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balabanian K, Lagane B, Pablos JL, et al. WHIM syndromes with different genetic anomalies are accounted for by impaired CXCR4 desensitization to CXCL12. Blood. 2005;105(6):2449–2457. doi: 10.1182/blood-2004-06-2289. [DOI] [PubMed] [Google Scholar]
- 3.Hernandez PA, Gorlin RJ, Lukens JN, et al. Mutations in the chemokine receptor gene CXCR4 are associated with WHIM syndrome, a combined immunodeficiency disease. Nat Genet. 2003;34(1):70–74. doi: 10.1038/ng1149. [DOI] [PubMed] [Google Scholar]
- 4.Liu Q, Chen H, Ojode T, et al. WHIM syndrome caused by a single amino acid substitution in the carboxy-tail of chemokine receptor CXCR4. Blood. 2012;120(1):181–189. doi: 10.1182/blood-2011-12-395608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gulino AV, Moratto D, Sozzani S, et al. Altered leukocyte response to CXCL12 in patients with warts hypogammaglobulinemia, infections, myelokathexis (WHIM) syndrome. Blood. 2004;104(2):444–452. doi: 10.1182/blood-2003-10-3532. [DOI] [PubMed] [Google Scholar]
- 6.Balabanian K, Levoye A, Klemm L, et al. Leukocyte analysis from WHIM syndrome patients reveals a pivotal role for GRK3 in CXCR4 signaling. J Clin Invest. 2008;118(3):1074–1084. doi: 10.1172/JCI33187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawai T, Choi U, Cardwell L, et al. WHIM syndrome myelokathexis reproduced in the NOD/SCID mouse xenotransplant model engrafted with healthy human stem cells transduced with C-terminus-truncated CXCR4. Blood. 2007;109(1):78–84. doi: 10.1182/blood-2006-05-025296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walters KB, Green JM, Surfus JC, Yoo SK, Huttenlocher A. Live imaging of neutrophil motility in a zebrafish model of WHIM syndrome. Blood. 2010;116(15):2803–2811. doi: 10.1182/blood-2010-03-276972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balabanian K, Brotin E, Biajoux V, et al. Proper desensitization of CXCR4 is required for lymphocyte development and peripheral compartmentalization in mice. Blood. 2012;119(24):5722–5730. doi: 10.1182/blood-2012-01-403378. [DOI] [PubMed] [Google Scholar]
- 10.Dale DC, Bolyard AA, Kelley ML, et al. The CXCR4 antagonist plerixafor is a potential therapy for myelokathexis, WHIM syndrome. Blood. 2011;118(18):4963–4966. doi: 10.1182/blood-2011-06-360586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDermott DH, Liu Q, Ulrick J, et al. The CXCR4 antagonist plerixafor corrects panleukopenia in patients with WHIM syndrome. Blood. 2011;118(18):4957–4962. doi: 10.1182/blood-2011-07-368084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Busillo JM, Benovic JL. Regulation of CXCR4 signaling. Biochim Biophys Acta. 2007;1768(4):952–963. doi: 10.1016/j.bbamem.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roland J, Murphy BJ, Ahr B, et al. Role of the intracellular domains of CXCR4 in SDF-1-mediated signaling. Blood. 2003;101(2):399–406. doi: 10.1182/blood-2002-03-0978. [DOI] [PubMed] [Google Scholar]
- 14.Haribabu B, Richardson RM, Fisher I, et al. Regulation of human chemokine receptors CXCR4. Role of phosphorylation in desensitization and internalization. J Biol Chem. 1997;272(45):28726–28731. doi: 10.1074/jbc.272.45.28726. [DOI] [PubMed] [Google Scholar]
- 15.Signoret N, Rosenkilde MM, Klasse PJ, et al. Differential regulation of CXCR4 and CCR5 endocytosis. J Cell Sci. 1998;111(Pt 18):2819–2830. doi: 10.1242/jcs.111.18.2819. [DOI] [PubMed] [Google Scholar]
- 16.Orsini MJ, Parent JL, Mundell SJ, Marchese A, Benovic JL. Trafficking of the HIV coreceptor CXCR4. Role of arrestins and identification of residues in the c-terminal tail that mediate receptor internalization. J Biol Chem. 1999;274(43):31076–31086. doi: 10.1074/jbc.274.43.31076. [DOI] [PubMed] [Google Scholar]
- 17.Mueller W, Schütz D, Nagel F, Schulz S, Stumm R. Hierarchical organization of multi-site phosphorylation at the CXCR4 C terminus. PLoS ONE. 2013;8(5):e64975. doi: 10.1371/journal.pone.0064975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchese A, Trejo J. Ubiquitin-dependent regulation of G protein-coupled receptor trafficking and signaling. Cell Signal. 2013;25(3):707–716. doi: 10.1016/j.cellsig.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Y, Cheng Z, Ma L, Pei G. Beta-arrestin2 is critically involved in CXCR4-mediated chemotaxis, and this is mediated by its enhancement of p38 MAPK activation. J Biol Chem. 2002;277(51):49212–49219. doi: 10.1074/jbc.M207294200. [DOI] [PubMed] [Google Scholar]
- 20.Cheng ZJ, Zhao J, Sun Y, et al. beta-arrestin differentially regulates the chemokine receptor CXCR4-mediated signaling and receptor internalization, and this implicates multiple interaction sites between beta-arrestin and CXCR4. J Biol Chem. 2000;275(4):2479–2485. doi: 10.1074/jbc.275.4.2479. [DOI] [PubMed] [Google Scholar]
- 21.Lagane B, Chow KY, Balabanian K, et al. CXCR4 dimerization and beta-arrestin-mediated signaling account for the enhanced chemotaxis to CXCL12 in WHIM syndrome. Blood. 2008;112(1):34–44. doi: 10.1182/blood-2007-07-102103. [DOI] [PubMed] [Google Scholar]
- 22.Jiménez-Baranda S, Gómez-Moutón C, Rojas A, et al. Filamin-A regulates actin-dependent clustering of HIV receptors. Nat Cell Biol. 2007;9(7):838–846. doi: 10.1038/ncb1610. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura F, Stossel TP, Hartwig JH. The filamins: organizers of cell structure and function. Cell Adhes Migr. 2011;5(2):160–169. doi: 10.4161/cam.5.2.14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minsaas L, Planagumà J, Madziva M, et al. Filamin a binds to CCR2B and regulates its internalization. PLoS ONE. 2010;5(8):e12212. doi: 10.1371/journal.pone.0012212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Planagumà J, Minsaas L, Pons M, Myhren L, Garrido G, Aragay AM. Filamin A-hinge region 1-EGFP: a novel tool for tracking the cellular functions of filamin A in real-time. PLoS ONE. 2012;7(8):e40864. doi: 10.1371/journal.pone.0040864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sampson LJ, Leyland ML, Dart C. Direct interaction between the actin-binding protein filamin-A and the inwardly rectifying potassium channel, Kir2.1. J Biol Chem. 2003;278(43):41988–41997. doi: 10.1074/jbc.M307479200. [DOI] [PubMed] [Google Scholar]
- 27.Seck T, Baron R, Horne WC. Binding of filamin to the C-terminal tail of the calcitonin receptor controls recycling. J Biol Chem. 2003;278(12):10408–10416. doi: 10.1074/jbc.M209655200. [DOI] [PubMed] [Google Scholar]
- 28.Thelin WR, Chen Y, Gentzsch M, et al. Direct interaction with filamins modulates the stability and plasma membrane expression of CFTR. J Clin Invest. 2007;117(2):364–374. doi: 10.1172/JCI30376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim KM, Gainetdinov RR, Laporte SA, Caron MG, Barak LS. G protein-coupled receptor kinase regulates dopamine D3 receptor signaling by modulating the stability of a receptor-filamin-beta-arrestin complex. A case of autoreceptor regulation. J Biol Chem. 2005;280(13):12774–12780. doi: 10.1074/jbc.M408901200. [DOI] [PubMed] [Google Scholar]
- 30.Kallikourdis M, Trovato AE, Anselmi F, et al. The CXCR4 mutations in WHIM syndrome impair the stability of the T-cell immunologic synapse. Blood. 2013;122(5):666–673. doi: 10.1182/blood-2012-10-461830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mira E, Lacalle RA, González MA, et al. A role for chemokine receptor transactivation in growth factor signaling. EMBO Rep. 2001;2(2):151–156. doi: 10.1093/embo-reports/kve027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.García Lopez MA, Aguado Martínez A, Lamaze C, Martínez-A C, Fischer T. Inhibition of dynamin prevents CCL2-mediated endocytosis of CCR2 and activation of ERK1/2. Cell Signal. 2009;21(12):1748–1757. doi: 10.1016/j.cellsig.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 33.Playford MP, Nurminen E, Pentikäinen OT, et al. Cystic fibrosis transmembrane conductance regulator interacts with multiple immunoglobulin domains of filamin A. J Biol Chem. 2010;285(22):17156–17165. doi: 10.1074/jbc.M109.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van den Ent F, Löwe J. RF cloning: a restriction-free method for inserting target genes into plasmids. J Biochem Biophys Methods. 2006;67(1):67–74. doi: 10.1016/j.jbbm.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 35.Nakamura F, Osborn TM, Hartemink CA, Hartwig JH, Stossel TP. Structural basis of filamin A functions. J Cell Biol. 2007;179(5):1011–1025. doi: 10.1083/jcb.200707073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lacalle RA, Gómez-Moutón C, Barber DF, et al. PTEN regulates motility but not directionality during leukocyte chemotaxis. J Cell Sci. 2004;117(Pt 25):6207–6215. doi: 10.1242/jcs.01545. [DOI] [PubMed] [Google Scholar]
- 37.Tardáguila M, Mira E, García-Cabezas MA, et al. CX3CL1 promotes breast cancer via transactivation of the EGF pathway. Cancer Res. 2013;73(14):4461–4473. doi: 10.1158/0008-5472.CAN-12-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Page RC, Clark JG, Misra S. Structure of filamin A immunoglobulin-like repeat 10 from Homo sapiens. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67(Pt 8):871–876. doi: 10.1107/S1744309111024249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Razinia Z, Mäkelä T, Ylänne J, Calderwood DA. Filamins in mechanosensing and signaling. Annu Rev Biophys. 2012;41:227–246. doi: 10.1146/annurev-biophys-050511-102252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao M, Discipio RG, Wimmer AG, Schraufstatter IU. Regulation of CXCR4-mediated nuclear translocation of extracellular signal-related kinases 1 and 2. Mol Pharmacol. 2006;69(1):66–75. doi: 10.1124/mol.105.016923. [DOI] [PubMed] [Google Scholar]
- 41.Ehrlicher AJ, Nakamura F, Hartwig JH, Weitz DA, Stossel TP. Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature. 2011;478(7368):260–263. doi: 10.1038/nature10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakamura F, Song M, Hartwig JH, Stossel TP. Documentation and localization of force-mediated filamin A domain perturbations in moving cells. Nat Commun. 2014;5:4656. doi: 10.1038/ncomms5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marchese A. Endocytic trafficking of chemokine receptors. Curr Opin Cell Biol. 2014;27:72–77. doi: 10.1016/j.ceb.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scott MG, Pierotti V, Storez H, et al. Cooperative regulation of extracellular signal-regulated kinase activation and cell shape change by filamin A and beta-arrestins. Mol Cell Biol. 2006;26(9):3432–3445. doi: 10.1128/MCB.26.9.3432-3445.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu B, Chien EY, Mol CD, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330(6007):1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Etzkorn M, Raschle T, Hagn F, et al. Cell-free expressed bacteriorhodopsin in different soluble membrane mimetics: biophysical properties and NMR accessibility. Structure. 2013;21(3):394–401. doi: 10.1016/j.str.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ueda Y, Neel NF, Schutyser E, Raman D, Richmond A. Deletion of the COOH-terminal domain of CXC chemokine receptor 4 leads to the down-regulation of cell-to-cell contact, enhanced motility and proliferation in breast carcinoma cells. Cancer Res. 2006;66(11):5665–5675. doi: 10.1158/0008-5472.CAN-05-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mellado M, Rodríguez-Frade JM, Mañes S, Martínez-A C. Chemokine signaling and functional responses: the role of receptor dimerization and TK pathway activation. Annu Rev Immunol. 2001;19:397–421. doi: 10.1146/annurev.immunol.19.1.397. [DOI] [PubMed] [Google Scholar]
- 49.Percherancier Y, Berchiche YA, Slight I, et al. Bioluminescence resonance energy transfer reveals ligand-induced conformational changes in CXCR4 homo- and heterodimers. J Biol Chem. 2005;280(11):9895–9903. doi: 10.1074/jbc.M411151200. [DOI] [PubMed] [Google Scholar]
- 50.Zoughlami Y, Voermans C, Brussen K, et al. Regulation of CXCR4 conformation by the small GTPase Rac1: implications for HIV infection. Blood. 2012;119(9):2024–2032. doi: 10.1182/blood-2011-06-364828. [DOI] [PubMed] [Google Scholar]
- 51.Kaur S, Fielding AB, Gassner G, Carter NJ, Royle SJ. An unmet actin requirement explains the mitotic inhibition of clathrin-mediated endocytosis. eLife. 2014;3:e00829. doi: 10.7554/eLife.00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Volovyk ZM, Wolf MJ, Prasad SV, Rockman HA. Agonist-stimulated beta-adrenergic receptor internalization requires dynamic cytoskeletal actin turnover. J Biol Chem. 2006;281(14):9773–9780. doi: 10.1074/jbc.M511435200. [DOI] [PubMed] [Google Scholar]
- 53.Glogauer M, Arora P, Chou D, Janmey PA, Downey GP, McCulloch CA. The role of actin-binding protein 280 in integrin-dependent mechanoprotection. J Biol Chem. 1998;273(3):1689–1698. doi: 10.1074/jbc.273.3.1689. [DOI] [PubMed] [Google Scholar]
- 54.Savoy RM, Ghosh PM. The dual role of filamin A in cancer: can’t live with (too much of) it, can’t live without it. Endocr Relat Cancer. 2013;20(6):R341–R356. doi: 10.1530/ERC-13-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gawecka JE, Griffiths GS, Ek-Rylander B, Ramos JW, Matter ML. R-Ras regulates migration through an interaction with filamin A in melanoma cells. PLoS ONE. 2010;5(6):e11269. doi: 10.1371/journal.pone.0011269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ai J, Huang H, Lv X, et al. FLNA and PGK1 are two potential markers for progression in hepatocellular carcinoma. Cell Physiol Biochem. 2011;27(3-4):207–216. doi: 10.1159/000327946. [DOI] [PubMed] [Google Scholar]
- 57.Jiang X, Yue J, Lu H, et al. Inhibition of filamin-A reduces cancer metastatic potential. Int J Biol Sci. 2013;9(1):67–77. doi: 10.7150/ijbs.5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tian HM, Liu XH, Han W, Zhao LL, Yuan B, Yuan CJ. Differential expression of filamin A and its clinical significance in breast cancer. Oncol Lett. 2013;6(3):681–686. doi: 10.3892/ol.2013.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun GG, Lu YF, Cheng YJ, Hu WN. Absent expression of FLNA is correlated with poor prognosis of nasopharyngeal cancer. Tumour Biol. 2014;35(4):2967–2974. doi: 10.1007/s13277-013-1381-z. [DOI] [PubMed] [Google Scholar]
- 60.Diaz GA. Released on a WHIM. Blood. 2011;118(18):4764–4765. doi: 10.1182/blood-2011-08-375162. [DOI] [PubMed] [Google Scholar]
- 61.McDermott DH, Liu Q, Velez D, et al. A phase 1 clinical trial of long-term, low-dose treatment of WHIM syndrome with the CXCR4 antagonist plerixafor. Blood. 2014;123(15):2308–2316. doi: 10.1182/blood-2013-09-527226. [DOI] [PMC free article] [PubMed] [Google Scholar]