Abstract
Recently, there has been a dramatic increase in the detection and characterization of insect-specific viruses in field-collected mosquitoes. Evidence suggests that these viruses are ubiquitous in nature and that many are maintained by vertical transmission in mosquito populations. Some studies suggest that the presence of insect-specific viruses may inhibit replication of a super-infecting arbovirus, thus altering vector competence of the mosquito host. Accordingly, we screened our laboratory mosquito colonies for insect-specific viruses. Pools of colony mosquitoes were homogenized and inoculated into cultures of Aedes albopictus (C6/36) cells. The infected cells were examined by electron microscopy and deep sequencing was performed on RNA extracts. Electron micrograph images indicated the presence of three different viruses in three of our laboratory mosquito colonies. Potential implications of these findings for vector competence studies are discussed.
Introduction
Arthropod-borne viral (arboviral) diseases are a major public health burden worldwide. Several mosquito-borne arboviruses, such as dengue, West Nile, and Chikungunya viruses have recently increased in importance as human pathogens because of geographic expansion and adaptation to new vector species.1 Recent studies have shown that the microbiota of mosquitoes can affect their ability to become infected with and to vector certain pathogens.2–5 There is a growing realization that mosquito populations are naturally infected with a wide range of bacteria,6,7 fungi,8 and viruses9–15; and this recognition has raised interest in the potential effects of these microbes on mosquito ecology and on pathogen transmission.
During the past decade, there has been a dramatic increase in the detection and description of novel insect-specific viruses found in field-collected mosquitoes. The term “insect-specific” refers to viruses that replicate in mosquitoes and mosquito cells but not in vertebrates or vertebrate cells, in contrast to classical arboviruses that can replicate in both. It is now apparent that these viruses are widespread in nature and that many are probably maintained by vertical transmission.16–19 Some studies also suggest that superinfection exclusion (cells infected with one virus are refractory to infection by a second related virus) may occur between some insect-specific viruses and pathogenic arboviruses,20,21 altering the vector competence of the mosquito host. In vivo and in vitro studies investigating interactions between two insect-specific flaviviruses (Culex flavivirus [CxFV]20,22 and Palm Creek virus [PCV]21) with West Nile virus (WNV), have shown modulation of WNV replication and dissemination in dually infected mosquitoes, suggesting that some insect-specific viruses may alter the mosquito's ability to serve as a competent vector for certain pathogenic arboviruses. These findings prompted us to screen our 16 laboratory mosquito colonies for infection with insect-specific viruses.
Materials and Methods
Testing mosquito colonies for viral infection.
Our mosquito insectary is maintained at 27°C and a relative humidity of 80% under a 16-h light:8-h dark photoperiod, as previously described.23 Each of our laboratory colonies (Table 1) were screened for viral infection by homogenizing two pools of 50 male and 50 female mosquitoes and attempting virus isolation in the C6/36 cell line. Briefly, mosquito pools were homogenized in 3 mL of minimum essential medium (MEM) supplemented with penicillin (100 U/mL) and streptomycin (100 μg/mL) and 20% fetal bovine serum (FBS), using a TissueLyser (Qiagen, Hidden, Germany). After centrifugation at 10,000 rpm for 10 min, 200 μL of each supernatant was inoculated into separate 25 cm2 flask cultures of C6/36 cells. After 2 hrs of absorption at 28°C, 10 mL of maintenance medium was added. Cultures were held in an incubator at 28°C for 7 days and examined daily for cytopathic effect (CPE). All cultures were also subsequently screened by electron microscopy (EM) for the presence of virus-like particles. If CPE was observed or virus-like structures were seen by EM, total RNA was then extracted from the C6/36 cell culture supernate and submitted for deep sequencing using an Illumina platform.
Table 1.
Laboratory mosquito colonies tested for persistent insect virus infections
| Species | Strain | Origin | Evidence of viral infection | Virus isolated |
|---|---|---|---|---|
| Aedes aegypti | RexD/Higgs white eye | Puerto Rico | None | |
| Aedes aegypti | Iquitos | Iquitos, Peru | None | |
| Aedes aegypti | Galveston | Galveston | Viral particles by EM, Sequence | Flavivirus: Cell fusing agent virus |
| Aedes aegypti | Thailand | Bangkok, Thailand | None | |
| Aedes albopictus | Galveston | Galveston, TX | Viral particles by EM | Putative reovirus |
| Aedes albopictus | La Reunion | La Reunion | None | |
| Aedes albopictus | Thailand | Bangkok, Thailand | Viral particles by EM, Sequence | Flavivirus: Aedes flavivirus |
| Aedes albopictus | Venezuela | Venezuela | None | |
| Aedes triseriatus | Galveston | Galveston, TX | None | |
| Aedes sollicitans | Galveston | Galveston, TX | None | |
| Aedes taeniorhynchus | Florida | Florida | None | |
| Anophles gambiae | G3 | Gambia | None | |
| Culex tarsalis | n/a | Kern County, CA | None | |
| Culex quinquefasciatus | Sebring | Sebring, FL | None | |
| Culex taeniopus | Mexico | Mexico | None | |
| Culiseta melanura | n/a | Connecticut | None |
EM = electron microscopy.
Transmission electron microscopy.
Transmission electron microscopy was used to initially confirm the presence of virus in all cultures of C6/36 cells. For ultrastructural analyses, infected cells were fixed for at least 1 hr in a mixture of 2.5% formaldehyde prepared from paraformaldehyde powder, and 0.1% glutaraldehyde in 0.05 M cacodylate buffer pH 7.3 to which 0.03% picric acid and 0.03% CaCl2 were added. The monolayers were washed in 0.1 M cacodylate buffer, cells were scraped off and processed further as a pellet. The pellets were post-fixed in 1% OsO4 in 0.1 M cacodylate buffer pH 7.3 for 1 hr, washed with distilled water, and en bloc stained with 2% aqueous uranyl acetate for 20 min at 60°C. The pellets were dehydrated in ethanol, processed through propylene oxide, and embedded in Poly/Bed 812 (Polysciences, Warrington, PA). Ultrathin sections were cut on Leica EM UC7 μL tramicrotome (Leica Microsystems, Buffalo Grove, IL), stained with lead citrate, and examined in a Philips 201 transmission electron microscope (Philips Electron Optics, Eindhoven, The Netherlands) at 60 kV.
Next generation sequencing.
The C6/36 cells grown to 90% confluence in 25 cm2 plastic culture flasks were inoculated with culture supernatants from mosquito homogenates exhibiting CPE in C6/36 cells. Virus harvest and isolation of vRNA for next generation genome sequencing were done as described previously.14
The de novo assembly program ABySS24 was used to assemble the reads into contigs, using several different sets of reads, and k values from 20 to 40. A nearly full-length contig for the Ae. aegypti-Galveston sample was obtained from 250,000 paired-end reads with a k value of 23, whereas 500,000 paired-end reads at a k value of 39 were used for the Ae. albopictus-Thailand sample. Reads were mapped back to the contigs using bowtie2,25 and visualized with the Integrated Genomics Viewer26 to verify that the assembled contig was correct. About 6.3% and 28% of the read-pairs in the sample mapped to the viral contigs, from about 5.8 and 4.3 million total reads for the Ae. aegypti-Galveston and Ae. albopictus-Thailand samples, respectively.
Phylogenetic analysis.
Selected flavivirus sequences, representing a 1900 nt segment of the polymerase gene, were downloaded from GenBank. These sequences, together with the corresponding sequences of the viruses detected in our laboratory mosquito colonies, were combined and aligned manually using the Se-Al application based on amino acid sequence alignments. A Neighbor-Joining tree was built based on this alignment, using the PAUP* v4.0b package (Sinauer Associates, Sunderland, MA) as a guide tree. Overly sampled sequences from a single location and year were deleted to accelerate the analysis without sacrificing overall genetic diversity. This led to a final data set of 83 samples. A maximum likelihood tree was then inferred using PAUP* based on the best-fit substitution model estimated from Modeltest version 3.06.27 The optimal maximum likelihood tree was estimated using the appropriate model and a heuristic search with tree-bisection-reconstruction branch swapping and 1,000 replicates, estimating variable parameters from the data, where necessary. Bootstrap replicates were calculated for each data set under the same models mentioned previously. Additionally, Bayesian analysis was undertaken using MrBayes v3.1,28,29 and data sets were run for 10 million generations until they reached congruence. The models used were HKY+G and HKY+I+G. Trees obtained by either analysis exhibited similar topologies.
Characterization of virus-infected mosquito colonies.
To investigate the level of virus infection within the infected laboratory mosquito colonies, individual adult mosquitoes (∼5–7 days old) were homogenized in 0.5 mL of diluent (phosphate buffered saline supplemented with 10% FBS) using a TissueLyser. The RNA extractions were performed using a QIAamp viral RNA Mini kit (Qiagen, Inc., Valencia, CA) according to the manufacturer's instructions and tested using a standard reverse transcription-polymerase chain reaction (RT-PCR) assay and flavivirus-specific primers. Samples producing bands of the expected size, compared with positive controls generated from virus stocks, were sequenced for confirmation. A subset of individual female mosquitoes (∼5–7 days old) was further examined by assaying infection rates of the legs and saliva. Briefly, mosquitoes were cold anesthetized by exposure to ice and one hind leg was removed and homogenized in 0.5 mL of diluent. Wings and remaining legs were removed and discarded. Saliva was then collected from the insects by inserting the proboscis (with mosquito attached) into a capillary tube23 containing 2.5% FBS and 25% sucrose solution. Mosquitoes were allowed to salivate for at least 15 min and then capillary tubes (containing saliva) and bodies were placed into separate microcentrifuge tubes containing 0.5 mL of diluent. To process these samples for viral detection, one steel BB was added to each tube containing mosquito bodies or legs. Samples were homogenized at 24 cycles/second for 4 min using a TissueLyser and then clarified by centrifuging at 3,000 × g for 3 min. Saliva samples were centrifuged at 3,000 × g for 3 min to expel fluid from capillary tubes. All samples were then tested using a standard RT-PCR assay.
Results
Virus isolation in C6/36 cells.
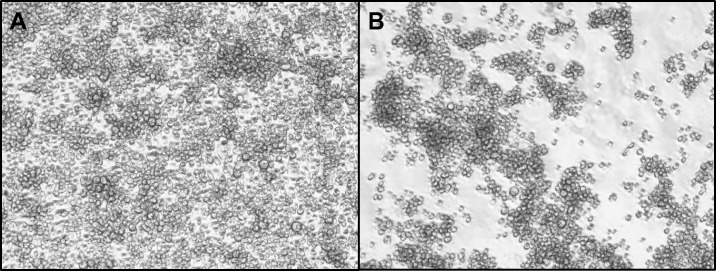
Cytopathic effects were observed in cultures of C6/36 cells inoculated with mosquito homogenates from three laboratory colonies. Cultures from an Ae. albopictus-Thailand colony showed marked growth inhibition and cell aggregation compared with controls (Figure 1). Similar CPE was also observed in C6/36 cells inoculated with homogenates from Ae. aegypti and Ae. albopictus laboratory colonies originally established from mosquitoes collected in Galveston, Texas.
Figure 1.
Phase contrast micrographs of (A) control C6/36 cells and (B) Aedes flavivirus (AeFV)-infected cells at 72 h post-infection.
Transmission electron microscopy.
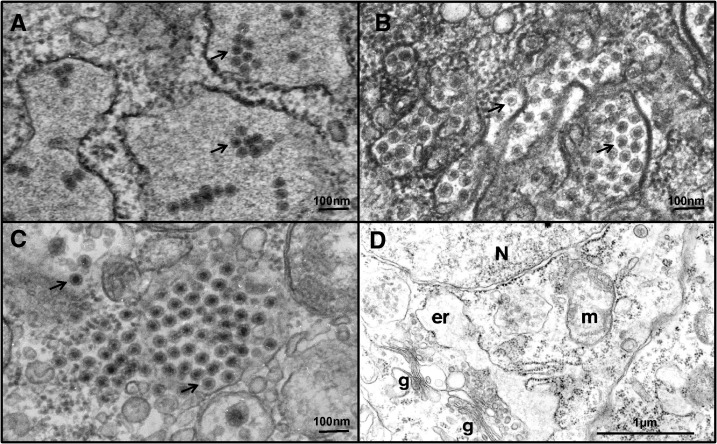
Electron micrographs of ultrathin sections of C6/36 cells, from cultures showing CPE, showed evidence of virus-like structures within the cells (Table 1). Samples from two colonies, (Ae. aegypti-Galveston and Ae. albopictus-Thailand) revealed the presence of spherical virions of ∼40 nm in diameter with typical flavivirus morphology (Figure 2A and B). These virions were mainly localized inside expanded cisternae of granular endoplasmic reticulum. Virus-like particles were also observed in C6/36 cells inoculated with pools of an Ae. albopictus-Galveston mosquito colony. They were ∼70 nm in diameter, located in clusters in the cytosol, and showed characteristic reovirus morphology (Figure 2C). Control C6/36 cells were devoid of any viruses (Figure 2D).
Figure 2.
Transmission electron microscopy photographs showing evidence of viral infection in C6/36 cell cultures inoculated with homogenates of laboratory colony mosquitoes. (A) Aedes aegypti-Galveston and (B) Aedes albopictus-Thailand with arrows indicating flavivirus virions, and (C) Aedes albopictus-Galveston with arrows indicating reovirus-like structures, and (D) ultrastructure of uninfected C6/36 cells in culture. N = nucleus, er = swollen cistern of granular endoplasmic reticulum, g = Golgi apparatus, m mitochondrion.
Next generation sequencing.
Illumina sequencing confirmed the presence of RNA from a single flavivirus in the Ae. albopictus-Thailand colony. Blast results showed 91% and 98% nucleotide and amino acid identities, respectively, when compared with the Aedes flavivirus (AeFV), Narita-21 strain, an insect-specific flavivirus first isolated in Japan in 2003.30 Viral sequences were also detected in the Ae. aegypti-Galveston colony, which confirmed the presence of cell fusing agent virus (CFAV), a previously described insect-specific flavivirus, first isolated from an Ae. aegypti cell line in 1975.31 The CFAV has subsequently been found in Ae. aegypti populations in many parts of the world. Sequencing of cultures prepared from the Ae. albopictus-Galveston colony confirmed the presence of a plant reovirus, likely related to rice dwarf virus. (GenBank accession nos. are Aedes flavivirus-Bangkok strain: KJ741266 and CFAV-Galveston strain: KJ741267.)
Phylogenetic analysis.
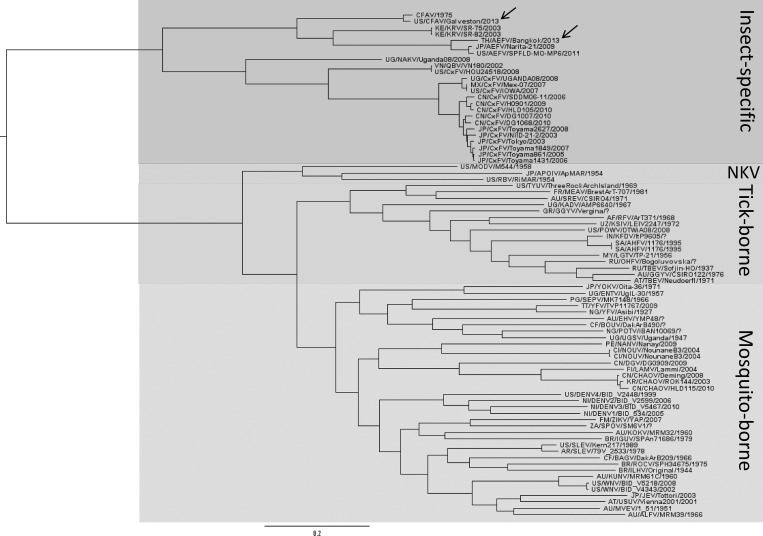
The phylogenetic relationships between the two insect-specific flaviviruses detected in our laboratory colonies and other selected flaviviruses were assessed by performing Bayesian analysis based on a 1900 nt region of the polymerase gene. The resulting tree topology (Figure 3) showed that the AeFV strain isolated from the Ae. albopictus-Thailand colony groups with other Aedes-associated insect-specific flaviviruses, forming a clade with AeFV isolates from Japan30 and Missouri.19 The CFAV isolate from the Ae. aegypti-Galveston colony also groups with the Aedes-associated insect-specific flaviviruses and forms a clade with the prototype CFAV isolate, originally described in 1975.31
Figure 3.
Phylogenetic tree inferred from Bayesian analysis of a 1900 nucleotide section of the polymerase gene of two insect-specific flaviviruses (indicated by arrows), cell fusing agent virus (CFAV) and Aedes flavivirus (AeFV), isolated from our laboratory mosquito colonies. Both viruses group with other Aedes-associated insect-specific flaviviruses. Bootstrap P values are 100% at all major nodes. GenBank accession numbers used for analysis are listed in a supplemental file.
Examination of persistently infected colonies.
Individual mosquitoes from the two flavivirus-infected laboratory colonies were tested by standard RT-PCR to estimate infection rates. In the Ae. albopictus-Thailand colony, AeFV viral RNA was detected in 85% of females (N = 11/13) and 43% of males (N = 3/7) sampled, suggesting vertical transmission as the probable mechanism for viral maintenance within the colony. Viral RNA was also detected in the legs (60%, N = 3/5) and saliva (60%, N = 3/5) from individual females of the Thailand colony. Examination of the Ae. aegypti-Galveston colony indicated that it is also persistently infected with CFAV. Viral RNA was detected in 100% of females (N = 10/10) and 90% of males (N = 9/10) from the colony, suggesting that CFAV is also maintained in the colony by vertical transmission. Viral RNA was detected in 100% of the legs from CFAV-infected females (N = 10/10). Interestingly, we were unable to detect CFAV RNA in the saliva (N = 0/10), as was seen with the AeFV-infected colony mosquitoes.
Discussion
Three of 16 laboratory mosquito colonies tested showed evidence of viral infection. Sequence results confirmed the presence of CFAV in the Galveston Ae. aegypti colony. Cell fusing agent virus was the first insect-specific virus described and was originally isolated in 1975 from an Ae. aegypti cell line that showed massive syncytia formation when co-cultivated with Ae. albopictus cells.31 The complete nucleotide sequence for CFAV was determined ∼15 years later and was found to be distantly related to other flaviviruses, with sequence identities between CFAV and other flaviviruses highest for NS5 and NS3 genes, with 45% and 34% identities, respectively. More recently, isolates of CFAV have been detected in field-caught Aedes and Culex spp. mosquitoes from Puerto Rico,32 Thailand,33 Mexico, and the United States (R. B. Tesh, unpublished data). The CFAV strain detected in our Galveston Ae. aegypti colony shares 97% nucleotide sequence identity with the prototype 1975 strain; but unlike the prototype, our strain did not show syncytia formation when inoculated onto C6/36 cells. A second insect-specific flavivirus, AeFV, was detected in our Ae. albopictus colony established from eggs originally collected in Bangkok, Thailand. This is the first report of AeFV isolated from mosquitoes collected in Thailand. Aedes flavivirus was first isolated from Ae. albopictus and Ae. flavopictus mosquitoes collected in Japan.30 It has subsequently been isolated from Ae. albopictus mosquitoes collected in Italy34 and Missouri.19 Phylogenetic analysis, based on a portion of the polymerase gene, grouped the AeFV Bangkok strain with isolates from Japan and Missouri, sharing 91% sequence identity at the nucleotide level with these strains. A third virus was detected by EM in C6/36 cell cultures prepared from homogenates of the Ae. albopictus Galveston colony; it appears to be a reovirus based on virion size and morphology. Preliminary sequence analysis indicates that the latter virus consists of 12 segments, typical of reoviruses, and analysis of segment 1 indicates distant relationship (22% nucleotide identity) with rice dwarf virus. Studies are ongoing to identify and characterize this virus.
Preliminary studies with the Ae. albopictus-Thailand and Ae. aegypti-Galveston colonies suggest that the progenitor mosquitoes were infected with the respective insect-specific flaviviruses at the time the colonies were established. Mosquitoes from these two colonies have been tested by us multiple times over the past several years and the respective viral infections have persisted. Interestingly, these two naturally infected colonies have been maintained in the same insectary space for over 8 years with other non-infected mosquito colonies (including Ae. aegypti and Ae. albopictus colonies from other geographic regions); and thus far, no cross-contamination has been detected in these other colonies, providing supporting evidence for vertical transmission. The infection rates for CFAV in the Ae. aegypti-Galveston colony were higher (90–100%) compared with the infection rates observed with AeFV in the Ae. albopictus-Thailand (43–85%). It is unclear at this time why infection rates were different between the two naturally infected colonies. One potential reason to consider is the variable sensitivity of the virus-specific primer sets used for detection of the two viruses. Viral RNA was detected in both males and females from the two colonies. Infection rates observed in the Ae. albopictus-Thailand colony were variable between males (43%) and females (85%), which does not support vertical transmission as the primary mechanism of transmission, as we would expect both sexes to have similar infection rates. However, this unexpected finding could be a result of testing only a small portion of the cage population (13 females and 7 males). Vertical transmission has been reported previously with other insect-specific flaviviruses, based on the detection of viral RNA in adults reared from field-caught immature stages16,33 and in field-collected male mosquitoes.17,19,32,35
When laboratory colonies are established from wild-caught mosquitoes or stocks are obtained from other laboratory colonies, especially eggs, it is generally assumed that the emerging adults are “pathogen-free.” However, as the results of this study show, that may not be a valid assumption, because the mosquitoes may be infected with insect-specific viruses, which may not be detrimental to the mosquitoes, but may interfere with other types of viruses being tested in the colonies. Observations of infected legs and saliva from mosquitoes in our colonies suggest that some of these insect-specific viruses potentially have similar mosquito tissue tropisms as medically important arboviruses. Studies of the tissue tropisms of CxFV,35 another insect-specific flavivirus, in naturally infected Culex pipiens mosquitoes revealed a pervasive infection, with viral RNA detected in all tissues tested (salivary glands, ovaries, testes, head, fat body, and midgut).18 Culex flavivirus RNA was also detected in all life stages (eggs, larvae, pupae, and adults from a naturally infected laboratory colony), with viral titers estimated to be as high as 8.95 log10 RNA copies per individual.20 Interestingly, one study22 reported that saliva collected from Culex quinquefasciatus mosquitoes co-inoculated with CxFV (Izabal strain) and WNV contained both viruses, but no CxFV was detected in the saliva from singly infected mosquitoes. This observation suggests there could be a potential “piggybacking” mechanism of CxFV with WNV.22 Interestingly, we detected AeFV in the saliva of Ae. albopictus-Thailand colony mosquitoes, but did not detect CFAV in the saliva collected from Ae. aegypti-Galveston mosquitoes. This observation suggests either a salivary gland infection barrier exists for CFAV in this Ae. aegypti colony or the number of virus particles present were below our level of detection. Further studies are needed to more clearly define the insect-specific viral load in naturally infected mosquitoes and the potential effects that such infection may have on life-history traits of the vector and on pathogen transmission.
There is accumulating evidence that the insect microbiota can influence pathogen transmission by activating the vector immune response or by directly inhibiting pathogen development.36–40 With the growing realization of the diversity of the mosquito microbiome, it is important to understand the nature of these microorganisms and the role they may play in vector ecology and arbovirus transmission. Few studies have been done to evaluate potential interactions between insect-specific viruses with arboviruses in mosquitoes. A field study conducted in Chicago found a positive ecological association between the infection rates with WNV and CxFV, in Culex pipiens mosquito pools.41 In contrast, Crockett and others42 found no evidence to support an association between WNV and CxFV prevalence rates in Culex quinquefasciatus populations in the southeastern United States. In vitro and in vivo studies looking at the potential interaction of WNV with insect-specific flaviviruses in mosquito cells and in mosquitoes have also produced conflicting results. Studies looking at sequential infections in C6/36 (Ae. albopictus) cells, first with CxFV and followed by WNV 48 h later, resulted in significantly reduced WNV titers in co-infected cells, compared with controls.20 A similar study looking at WNV replication kinetics in cells co-infected with CxFV showed reduced WNV titers, but these differences were not significant.22 More recent studies in Australia with a newly described insect-specific flavivirus, PCV, showed suppression of WNV (Kunjin strain) and Murray Valley encephalitis virus replication in cells that were persistently infected with PCV.21 Kenney and others43 reported that Nahuirim virus, a newly characterized flavivirus from Brazil, reduces replication of WNV, Japanese encephalitis, and St. Louis encephalitis viruses in dually infected mosquito cell cultures. All of these in vitro experiments were conducted using C6/36 cells, which, unlike live mosquitoes, do not have a functional antiviral RNAi response,44 thus the biological relevance of these in vitro results is uncertain.
To our knowledge, there are few published studies that have looked at the effects of insect-specific virus infection on vector competence for arboviruses using live mosquitoes. Bolling and others20 compared the vector competence for WNV, using a Culex pipiens colony from Colorado (naturally infected with CxFV) and compared it to a C. pipiens colony from Iowa that was CxFV-free, and showed significantly reduced dissemination rates of WNV at 7 days post infection (dpi) in the co-infected mosquitoes. A similar study by Kent and others,22 also investigating vector competence for WNV, compared a Florida strain of C. quinquefasciatus mosquitoes experimentally infected with CxFV to uninfected mosquitoes. No significant effects on WNV replication were observed; however when a Honduran C. quinquefasciatus strain was inoculated simultaneously with CxFV and WNV, enhanced transmission of WNV was found at 14 dpi. The results from these experiments have been inconclusive and conflicting, suggesting that interactions between insect-specific viruses and pathogenic arboviruses may be species- and strain-specific, depending on the mosquito used. Such observed differences have frequently been explained as “genetic differences” among mosquito populations, but they could also be the result of differences in the mosquitoes' microbiome and to the effect of other unrecognized bacteria or viruses in the insect.
With recent advances in molecular tools for viral detection, there has been a dramatic increase in the isolation and characterization of insect-specific viruses in mosquitoes. This group of viruses is very diverse, representing many different virus families and genera (Togaviridae, Flaviviridae, Bunyaviridae, Rhabdoviridae, Reoviridae, Mesoniviridae, Negevirus) and other still unclassified RNA viruses.9–12,14,45,46 Data are lacking on the basic ecology of most insect-specific viruses in nature and their potential effects on mosquito life-history traits and the arboviruses they transmit. When conducting vector competence experiments, investigators should consider screening laboratory mosquito colonies for persistent insect-specific viral infections, and endosymbiotic bacteria, to be aware of potential variables that could alter resulting data sets. Studies are ongoing to characterize insect-specific viruses in our mosquito laboratory colonies and to further evaluate their potential effects on vector competence.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jing Huang, David Romero, and Krystal Walker for technical assistance.
Footnotes
Financial support: This study was supported by NIH T-32 training grant A1007536 and by NIH contract HHSN27220100040I/HHSN27200004/DO4.
Authors' addresses: Bethany G. Bolling, Nikos Vasilakis, Hilda Guzman, Steven G. Widen, Thomas G. Wood, Vsevolod L. Popov, Saravanan Thangamani, and Robert B. Tesh, Department of Pathology, University of Texas Medical Branch, Galveston, TX, E-mails: bethanybolling@gmail.com, nivasila@utmb.edu, hguzman@utmb.edu, sgwiden@utmb.edu, tgwood@utmb.edu, vpopov@utmb.edu, sathanga@utmb.edu, and rtesh@utmb.edu. Present address: Bethany G. Bolling, Texas Department of State Health Services, Laboratory Services Section, Austin, TX, E-mail: bethany.bolling@dshs.state.tx.us.
References
- 1.Weaver SC, Reisen WK. Present and future arboviral threats. Antiviral Res. 2010;85:328–345. doi: 10.1016/j.antiviral.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cirimotich CM, Dong Y, Clayton AM, Sandiford SL, Souza-Neto JA, Mulenga M, Dimopoulos G. Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science. 2011;332:855–858. doi: 10.1126/science.1201618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glaser RL, Meola MA. The native Wolbachia endosymbionts of Drosophila melanogaster and Culex quinquefasciatus increase host resistance to West Nile virus infection. PLoS ONE. 2010;5:e11977. doi: 10.1371/journal.pone.0011977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hedges LM, Brownlie JC, O'Neill SL, Johnson KN. Wolbachia and virus protection in insects. Science. 2008;322:702. doi: 10.1126/science.1162418. [DOI] [PubMed] [Google Scholar]
- 5.Ramirez JL, Souza-Neto J, Torres Cosme R, Rovira J, Ortiz A, Pascale JM, Dimopoulos G. Reciprocal tripartite interactions between the Aedes aegypti midgut microbiota, innate immune system and dengue virus influences vector competence. PLoS Negl Trop Dis. 2012;6:e1561. doi: 10.1371/journal.pntd.0001561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chouaia B, Rossi P, Montagna M, Ricci I, Crotti E, Damiani C, Epis S, Faye I, Sagnon N, Alma A, Favia G, Daffonchio D, Bandi C. Molecular evidence for multiple infections as revealed by typing of Asaia bacterial symbionts of four mosquito species. Appl Environ Microbiol. 2010;76:7444–7450. doi: 10.1128/AEM.01747-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terenius O, Lindh JM, Eriksson-Gonzales K, Bussière L, Laugen AT, Bergquist H, Titanji K, Faye I. Midgut bacterial dynamics in Aedes aegypti. FEMS Microbiol Ecol. 2012;80:556–565. doi: 10.1111/j.1574-6941.2012.01317.x. [DOI] [PubMed] [Google Scholar]
- 8.Bishop-Lilly KA, Turell MJ, Willner KM, Butani A, Nolan NM, Lentz SM, Akmal A, Mateczun A, Brahmbhatt TN, Sozhamannan S, Whitehouse CA, Read TD. Arbovirus detection in insect vectors by rapid, high-throughput pyrosequencing. PLoS Negl Trop Dis. 2010;4:e878. doi: 10.1371/journal.pntd.0000878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Attoui H, Mohd Jaafar F, Belhouchet M, Biagini P, Cantaloube J-F, de Micco P, de Lamballerie X. Expansion of family Reoviridae to include nine-segmented dsRNA viruses: isolation and characterization of a new virus designated Aedes pseudoscutellaris reovirus assigned to a proposed genus (Dinovernavirus) Virology. 2005;343:212–223. doi: 10.1016/j.virol.2005.08.028. [DOI] [PubMed] [Google Scholar]
- 10.Cook S, Moureau G, Kitchen A, Gould EA, de Lamballerie X, Holmes EC, Harbach RE. Molecular evolution of the insect-specific flaviviruses. J Gen Virol. 2012;93:223–234. doi: 10.1099/vir.0.036525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nasar F, Palacios G, Gorchakov RV, Guzman H, Da Rosa AP, Savji N, Popov VL, Sherman MB, Lipkin WI, Tesh RB, Weaver SC. Eilat virus, a unique alphavirus with host range restricted to insects by RNA replication. Proc Natl Acad Sci USA. 2012;109:14622–14627. doi: 10.1073/pnas.1204787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quan P-L, Junglen S, Tashmukhamedova A, Conlan S, Hutchison SK, Kurth A, Ellerbrok H, Egholm M, Briese T, Leendertz FH, Lipkin WI. Moussa virus: a new member of the Rhabdoviridae family isolated from Culex decens mosquitoes in Côte d'Ivoire. Virus Res. 2010;147:17–24. doi: 10.1016/j.virusres.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tyler S, Bolling BG, Blair CD, Brault AC, Pabbaraju K, Armijos MV, Clark DC, Calisher CH, Drebot MA. Distribution and phylogenetic comparisons of a novel mosquito flavivirus sequence present in Culex tarsalis mosquitoes from western Canada with viruses isolated in California and Colorado. Am J Trop Med Hyg. 2011;85:162–168. doi: 10.4269/ajtmh.2011.10-0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasilakis N, Forrester NL, Palacios G, Nasar F, Savji N, Rossi SL, Guzman H, Wood TG, Popov V, Gorchakov R, González AV, Haddow AD, Watts DM, da Rosa AP, Weaver SC, Lipkin WI, Tesh RB. Negevirus: a proposed new taxon of insect-specific viruses with wide geographic distribution. J Virol. 2013;87:2475–2488. doi: 10.1128/JVI.00776-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marklewitz M, Zirkel F, Rwego IB, Heidemann H, Trippner P, Kurth A, Kallies R, Briese T, Lipkin WI, Drosten C, Gillespie TR, Junglen S. Discovery of a unique novel clade of mosquito-associated bunyaviruses. J Virol. 2013;87:12850–12865. doi: 10.1128/JVI.01862-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sang RC, Gichogo A, Gachoya J, Dunster MD, Ofula V, Hunt AR, Crabtree MB, Miller BR, Dunster LM. Isolation of a new flavivirus related to cell fusing agent virus (CFAV) from field-collected flood-water Aedes mosquitoes sampled from a dambo in central Kenya. Arch Virol. 2003;148:1085–1093. doi: 10.1007/s00705-003-0018-8. [DOI] [PubMed] [Google Scholar]
- 17.Bolling BG, Eisen L, Moore CG, Blair CD. Insect-specific flaviviruses from Culex mosquitoes in Colorado, with evidence of vertical transmission. Am J Trop Med Hyg. 2011;85:169–177. doi: 10.4269/ajtmh.2011.10-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saiyasombat R, Bolling BG, Brault AC, Bartholomay LC, Blitvich BJ. Evidence of efficient transovarial transmission of Culex flavivirus by Culex pipiens (Diptera: Culicidae) J Med Entomol. 2011;48:1031–1038. doi: 10.1603/me11043. [DOI] [PubMed] [Google Scholar]
- 19.Haddow AD, Guzman H, Popov VL, Wood TG, Widen SG, Haddow AD, Tesh RB, Weaver SC. First isolation of Aedes flavivirus in the Western Hemisphere and evidence of vertical transmission in the mosquito Aedes (Stegomyia) albopictus (Diptera: Culicidae) Virology. 2013;440:134–139. doi: 10.1016/j.virol.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 20.Bolling BG, Olea-Popelka FJ, Eisen L, Moore CG, Blair CD. Transmission dynamics of an insect-specific flavivirus in a naturally infected Culex pipiens laboratory colony and effects of co-infection on vector competence for West Nile virus. Virology. 2012;427:90–97. doi: 10.1016/j.virol.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hobson-Peters J, Yam AW, Lu JW, Setoh YX, May FJ, Kurucz N, Walsh S, Prow NA, Davis SS, Weir R, Melville L, Hunt N, Webb RI, Blitvich BJ, Whelan P, Hall RA. A new insect-specific flavivirus from northern Australia suppresses replication of West Nile virus and Murray Valley encephalitis virus in co-infected mosquito cells. PLoS ONE. 2013;8:e56534. doi: 10.1371/journal.pone.0056534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kent RJ, Crabtree MB, Miller BR. Transmission of West Nile virus by Culex quinquefasciatus Say infected with Culex Flavivirus Izabal. PLoS Negl Trop Dis. 2010;4:e671. doi: 10.1371/journal.pntd.0000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higgs S. Care, maintenance, and experimental infection of mosquitoes. In: Marquardt WC, editor. Biology of Disease Vectors. Second edition. Burlington, MA: Elsevier; 2005. pp. 733–739. [Google Scholar]
- 24.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 28.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 29.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 30.Hoshino K, Isawa H, Tsuda Y, Sawabe K, Kobayashi M. Isolation and characterization of a new insect flavivirus from Aedes albopictus and Aedes flavopictus mosquitoes in Japan. Virology. 2009;391:119–129. doi: 10.1016/j.virol.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 31.Stollar V, Thomas L. An agent in the Aedes aegypti cell line (Peleg) which causes fusion of Aedes albopictus cells. Virology. 1975;64:367–377. doi: 10.1016/0042-6822(75)90113-0. [DOI] [PubMed] [Google Scholar]
- 32.Cook S, Bennett SN, Holmes EC, De Chesse R, Moureau G, de Lamballerie X. Isolation of a new strain of the flavivirus cell fusing agent virus in a natural mosquito population from Puerto Rico. J Gen Virol. 2006;87:735–748. doi: 10.1099/vir.0.81475-0. [DOI] [PubMed] [Google Scholar]
- 33.Yamanaka A, Thongrungkiat S, Ramasoota P, Konishi E. Genetic and evolutionary analysis of cell-fusing agent virus based on Thai strains isolated in 2008 and 2012. Infect Genet Evol. 2013;19:188–194. doi: 10.1016/j.meegid.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 34.Roiz D, Vázquez A, Rosso F, Arnoldi D, Girardi M, Cuevas L, Perez-Pastrana E, Sánchez-Seco MP, Tenorio A, Rizzoli A. Detection of a new insect flavivirus and isolation of Aedes flavivirus in northern Italy. Parasit Vectors. 2012;5:223. doi: 10.1186/1756-3305-5-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoshino K, Isawa H, Tsuda Y, Yano K, Sasaki T, Yuda M, Takasaki T, Kobayashi M, Sawabe K. Genetic characterization of a new insect flavivirus isolated from Culex pipiens mosquito in Japan. Virology. 2007;359:405–414. doi: 10.1016/j.virol.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 36.Cirimotich CM, Ramirez JL, Dimopoulos G. Native microbiota shape insect vector competence for human pathogens. Cell Host Microbe. 2011;10:307–310. doi: 10.1016/j.chom.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiss B, Aksoy S. Microbiome influences on insect host vector competence. Trends Parasitol. 2011;27:514–522. doi: 10.1016/j.pt.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye YH, Woolfit M, Rancès E, O'Neill SL, McGraw EA. Wolbachia-associated bacterial protection in the mosquito Aedes aegypti. PLoS Negl Trop Dis. 2013;7:e2362. doi: 10.1371/journal.pntd.0002362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kambris Z, Cook PE, Phuc HK, Sinkins SP. Immune activation by life-shortening Wolbachia and reduced filarial competence in mosquitoes. Science. 2009;326:134–136. doi: 10.1126/science.1177531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bian G, Joshi D, Dong Y, Lu P, Zhou G, Pan X, Xu Y, Dimopoulos G, Xi Z. Wolbachia invades Anopheles stephensi populations and induces refractoriness to Plasmodium infection. Science. 2013;340:748–751. doi: 10.1126/science.1236192. [DOI] [PubMed] [Google Scholar]
- 41.Newman CM, Cerutti F, Anderson TK, Hamer GL, Walker ED, Kitron UD, Ruiz MO, Brawn JD, Goldberg TL. Culex flavivirus and West Nile virus mosquito coinfection and positive ecological association in Chicago, United States. Vector Borne Zoonotic Dis. 2011;11:1099–1105. doi: 10.1089/vbz.2010.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crockett RK, Burkhalter K, Mead D, Kelly R, Brown J, Varnado W, Roy A, Horiuchi K, Biggerstaff BJ, Miller B, Nasci R. Culex flavivirus and West Nile virus in Culex quinquefasciatus populations in the southeastern United States. J Med Entomol. 2012;49:165–174. doi: 10.1603/me11080. [DOI] [PubMed] [Google Scholar]
- 43.Kenney J, Solberg OD, Langevin SA, Brault AC. Characterization of a novel insect-specific flavivirus from Brazil: potential for inhibition of infection of arthropod cells with medically important flaviviruses. J Gen Virol. 2014 doi: 10.1099/vir.0.068031-0. [Epub ahead of print 2014 Aug 21], doi:10.1099/vir.0.068031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brackney DE, Scott JC, Sagawa F, Woodward JE, Miller NA, Schilkey FD, Mudge J, Wilusz J, Olson KE, Blair CD, Ebel GD. C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response. PLoS Negl Trop Dis. 2010;4:e856. doi: 10.1371/journal.pntd.0000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kolodziejek J, Pachler K, Bin H, Mendelson E, Shulman L, Orshan L, Nowotny N. Barkedji virus, a novel mosquito-borne flavivirus identified in Culex perexiguus mosquitoes, Israel, 2011. J Gen Virol. 2013;94:2449–2457. doi: 10.1099/vir.0.056200-0. [DOI] [PubMed] [Google Scholar]
- 46.Zirkel F, Roth H, Kurth A, Drosten C, Ziebuhr J, Junglen S. Identification and characterization of genetically divergent members of the newly established family Mesoniviridae. J Virol. 2013;87:6346–6358. doi: 10.1128/JVI.00416-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.