Abstract
Epidemic cholera was reported in Haiti in 2010, with no information available on the occurrence or geographic distribution of toxigenic Vibrio cholerae in Haitian waters. In a series of field visits conducted in Haiti between 2011 and 2013, water and plankton samples were collected at 19 sites. Vibrio cholerae was detected using culture, polymerase chain reaction, and direct viable count methods (DFA-DVC). Cholera toxin genes were detected by polymerase chain reaction in broth enrichments of samples collected in all visits except March 2012. Toxigenic V. cholerae was isolated from river water in 2011 and 2013. Whole genome sequencing revealed that these isolates were a match to the outbreak strain. The DFA-DVC tests were positive for V. cholerae O1 in plankton samples collected from multiple sites. Results of this survey show that toxigenic V. cholerae could be recovered from surface waters in Haiti more than 2 years after the onset of the epidemic.
Introduction
Toxigenic Vibrio cholerae serogroup O1 and O139 are etiologic agents associated with epidemic cholera. Water contaminated with toxigenic V. cholerae is considered the primary source of the causative agent of cholera, which can spread rapidly in poverty-stricken areas with poor sanitation and limited access to safe drinking water.1 Haiti is the poorest country in the Western Hemisphere, and it is estimated that in 2011 only 64% of the population had access to improved drinking water sources and 26% had access to improved sanitation.2 However, epidemic cholera had not been diagnosed in Haiti until the onset of the outbreak in October 2010.3 During the first 2 years of the outbreak, more than 600,000 cases of cholera were reported, with more than 7,000 deaths.4 Because of the absence of reported cholera outbreaks in Haiti before 2010, there is little information on the potential presence of toxigenic V. cholerae in surface waters in Haiti beyond detections reported during the acute phase of the 2010 outbreak and an environmental surveillance study performed in the Ouest Department in Haiti.5,6
Vibrio cholerae is present in the environment both as free-living cells and attached to chitinous exoskeletons of zooplankton as a commensal of copepods.7,8 Detection of toxigenic V. cholerae can be difficult because of an abundance of non-toxigenic V. cholerae in natural water systems, and the inability to isolate viable but non-culturable bacteria using traditional culture methods.9 When V. cholerae cannot be detected by traditional culture methods, viable cells can be detected in environmental samples using fluorescent antibody and direct viable count methods (DFA-DVC).10,11 Because DFA-DVC allows for the detection of plankton-associated V. cholerae, data obtained from DFA-DVC can be used to evaluate whether V. cholerae may be present in an environmental niche. Additionally, gene sequences specific to toxigenic V. cholerae can be used to determine the presence of viable V. cholerae cells in enrichment cultures.5,12
To assess the presence of toxigenic V. cholerae in Haitian water resources, water samples from 19 fresh and marine water sites were collected during four field visits between October 2011 and January 2013. Multiple water sample collection and detection methods were used to detect and isolate V. cholerae in Haiti. The DFA-DVC was used to obtain a direct count of viable free-living and plankton-associated V. cholerae O1 and O139 cells. Traditional culture methods and polymerase chain reaction (PCR) were used to detect culturable cells of V. cholerae using ompW, toxR, ctxA, tcpA, and serogroup O1 and O139 gene targets. Molecular characterization was performed on recovered V. cholerae isolates for comparison to the epidemic strain. The goal of this surveillance study was to obtain data on the environmental presence of epidemic V. cholerae that could be used in conjunction with epidemiological data to guide decisions about cholera prevention efforts in Haiti.
Methods
Sample collection.
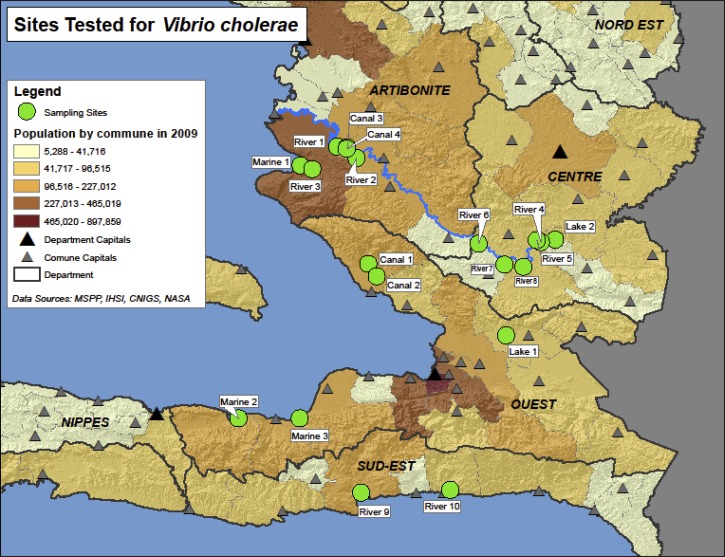
Field visits were conducted in October 2011, March and August 2012, and January 2013. During each field visit, 16 freshwater and 3 marine water samples were collected from 19 sites in the Artibonite, Center, Ouest, and Sud-Est Departments (Figure 1). Six of the sites were sampled at the beginning of the cholera outbreak in October and November 2010.5 The remaining sites were chosen to represent various water types and geographic locations, with several tributaries of the Artibonite River included. At each site, 100 L of water was concentrated using dead-end hollow-fiber ultrafiltration (DEUF).13,14 A 1-L grab sample was also collected to determine whether this simple, small-volume collection method could be as effective as a more labor-intensive large-volume method such as DEUF. Plankton samples (PLK) containing zooplankton and phytoplankton were collected by filtering 100 L through a 20-μm mesh plankton net fitted with a polyvinyl chloride (PVC) cod end bucket (Aquatic Research Instruments, Lehmi, ID) to obtain a final volume of < 1 L. A plankton net-filtered water (PFW) fraction containing free-living bacteria and plankton < 20 μm was obtained by collecting 1 L of the flow-through from the plankton net. Turbidity, specific conductance, salinity, temperature, and pH were measured in the field. Turbidity was measured with a Hach 2100P portable turbidimeter (Hach, Loveland, CO). Specific conductance, salinity, and temperature were measured with a YSI 556MPS multi-parameter instrument (YSI Inc., Yellow Springs, OH). Water pH was measured with a Hanna HI9812 portable meter (Hanna Instruments, Woonsocket, RI). Fecal indicator bacteria (Escherichia coli for freshwater and enterococci for marine water) were quantified from a 100-mL grab sample using the USEPA-approved IDEXX Quanti-Tray/2000 method. Escherichia coli and enterococci analyses were conducted within 6–34 hours of sample collection. Water samples for V. cholerae testing were held in a cooler at ambient temperature for 24 hours to improve detection by culture.15 Some samples were held for 48 hours before processing as a result of logistical constraints.
Figure 1.
Name and locations of sites sampled during the study.
Sample processing and culture.
At the Haiti National Public Health Laboratory (LNSP), V. cholerae were recovered from ultrafilters by backflushing with 400 mL of a surfactant solution to generate a final backflush volume of ∼500 mL. This backflush was added to an equal volume of 2× alkaline peptone water (APW). The PLK samples were passed through a 20-μm mesh filter to obtain a final volume of 10–50 mL, depending on water quality.16 The samples were vortexed and the plankton were homogenized with a tissue grinder. Two milliliters of the homogenized plankton sample were added to 18 mL of 1× APW, whereas 100 μL was spread plated directly onto thiosulfate citrate bile salts sucrose agar (TCBS) and CHROMagar Vibrio (CV) and incubated overnight at 37°C. Because of the logistics of plating on multiple agar media, only TCBS agar culture was performed for the August and January sampling events. The PFW samples (1-L) were filtered through 0.2-μm polycarbonate membrane filters. The filters were placed in 10–20 mL of 0.01 M PBS (1× strength) and vortexed to release the cell material from the surface of the filter. Two milliliters of the resulting solution were added to 18 mL of 1× APW. Nine hundred milliliters from the 1-L grab sample were added to 100 mL of 10× APW. Alkaline peptone water cultures were incubated overnight at 37°C. To extract DNA from APW cultures, duplicate 0.8-mL aliquots were removed from the broth surface and added to an equal volume of lysis buffer17 and stored at 4°C. In addition, a 10-μL inoculating loop was used to remove the pellicle at the surface and the enrichment culture was streaked for isolation onto TCBS and CV. Agar plates were incubated overnight at 37°C. Five or more presumptive V. cholerae colonies from each agar plate were placed into tryptic soy agar (TSA) transport tubes and incubated overnight at 37°C. For DFA-DVC testing, 10 μL each of 2.5% yeast extract and 0.2% nalidixic acid were added to 1-mL aliquots of homogenized PLK and PFW samples.11 After overnight incubation in the dark at 37°C, samples were fixed with 112 μL of formaldehyde (37% by weight), and stored at 4°C. Alkaline peptone water enrichments in lysis buffer, culture isolates in TSA transport vials, and DFA samples were shipped to the Centers for Disease Control and Prevention (CDC) in Atlanta, GA for subsequent analysis. One replicate set of the APW enrichments in lysis buffer and the DFA-DVC samples were shipped to the University of Maryland (UMD).
Vibrio cholerae detection and characterization.
Nucleic acid was extracted from replicate APW enrichment broth samples at CDC and UMD. At UMD, samples were tested by two conventional multiplex PCR assays for toxR, ctxA, and genes specific to the O-antigen biosynthesis region of serogroup O1 (wbeT gene) and O139 (wbfR gene).18,19 For the toxR multiplex assay, oligonucleotide concentrations were 600 nM for the universal forward primer and 400 nM for each reverse primer. For the ctxA/O1/O139 multiplex assay, oligonucleotide concentrations were 170 nM for each ctxA primer, 500 nM for each O1 primer, and 270 nM for each O139 primer. Each multiplex PCR assay was performed using PromegaGoTaq Green Master Mix with 1 μL template DNA in 25-μL reactions with the following thermal cycling conditions: one cycle of denaturation at 94°C for 10 min, followed by 36 cycles of denaturation at 94°C for 30 sec, 55°C annealing for 30 sec, extension at 72°C for 60 sec, with a final extension at 72°C for 10 min.
At CDC, APW enrichments were analyzed by multiplex real-time PCR (qPCR) for ctxA and ompW genes. The primer and TaqMan probe sequences for these assays are shown in Supplemental Table S1. For both qPCR assays, oligonucleotide concentrations were 625 nM for each primer and 125 nM for each probe. Each qPCR assay was performed using ABI Environmental Master Mix 2.0 and 2 μL template DNA in 20 μL reactions with the following thermal cycling conditions: one cycle of denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 sec, and then annealing, extension, and fluorescence acquisition at 60°C for 1 min.
Presumptive V. cholerae culture isolates from TSA transport vials were streaked for isolation on the corresponding agar they were picked from in the field and incubated overnight at 37°C. Isolated colonies were picked and streaked on Luria Bertani (LB) agar and incubated overnight at 37°C. Part of an isolated colony was picked with an inoculating loop and added to 200 μL of TE buffer and heated at 95°C for 10 minutes for multiplex qPCR for ompW and ctxA genes using Qiagen QuantiTect Probe PCR Master Mix. Isolates positive for ompW or ctxA were further analyzed by multiplex qPCR for tcpAEl Tor and tcpAClassical. The primer and TaqMan probe sequences for these assays are shown in Supplemental Table S1. Oligonucleotide concentrations and qPCR cycling conditions were the same as for the ompW and ctxA multiplex assay.
A subset of isolates positive for ctxA or tcpA were further characterized.20 Tests included agglutination in serogroup O1 antiserum, sequence determination of tcpA and/or ctxAB genes, and PCR amplification of rstR and the seventh pandemic marker smp. Serogroup O1-positive isolates were tested for agglutination in Inaba and Ogawa serotype antisera. Gene sequences were processed using LaserGene software (DNASTAR, Madison, WI) and compared with prototypical V. cholerae O1 ctxAB and tcpA sequences using MEGA5.21 Toxigenic isolates were also subjected to whole genome sequencing on a MiSeq using the Nextera XT library prep kit following manufacturer's recommendations (Illumina Inc., San Diego, CA). Whole genome assemblies were taken either from National Center for Biotechnology Information (NCBI) or assembled in-house using the Computational Genomics Pipeline (CG-Pipeline) v. 0.3.22 Genomes without gene predictions were analyzed with the CG-Pipeline module. Genomes were compared with a closed early-outbreak genome 2010EL-1786 (accessions CP003069 and CP003070) using Mauve.23 Homologous genes were identified using USEARCH at 95% identity24 and were aligned with MUSCLE,25 and these individual multiple sequence alignments were concatenated into whole genome multiple sequence alignment. Sites where any genome had a gap or N, or where the site was invariant, were removed. The maximum likelihood tree was constructed with RAxML using rapid bootstrap analysis.26
The DFA-DVC was performed to obtain a direct viable count of V. cholerae O1 and O139 from PLK and PFW samples.11 At UMD, formalin-fixed samples were analyzed by microscopy using DFA kits for V. cholerae O1 and O139 (New Horizons Diagnostics, Baltimore, MD).
Statistical analyses.
To evaluate the association between measured water quality parameters and ctxA gene detection rates in APW enrichments and V. cholerae O1 cells by DFA-DVC, a marginal logistic model using the generalized estimating equation (GEE) approach27,28 was applied. The model accounted for correlations stemming from repeated measures from the same collection site. The water quality analyses were conducted for freshwater only, because of substantial differences in some measured water quality parameters for freshwater and marine water. Separate analyses for marine water could not be performed because of the small sample size.
The performance of TCBS and CV for isolation of V. cholerae was evaluated by comparing the number of V. cholerae detections out of the total number of presumptive isolates tested. The performances of the large volume DEUF method and small volume grab sample method were evaluated by comparing the number of V. cholerae detections out of the total number of presumptive isolates tested and the number of ctxA detections from APW enrichments for each collection method. These comparisons were made using the GEE approach. For the TCBS versus CV comparison and DEUF versus grab culture isolation comparison, the GEE model accounted for correlations stemming from repeated measurements for the same collection site by sample type combination. For the DEUF versus grab ctxA comparison, the model accounted for collection site by test combination. Independent variables included: sample type (DEUF, PLK, PFW, grab), tests (TCBS or CV), month of collection, and collection site.
The exchangeable working correlation structure was assumed in all GEE models. The GEE analyses were done using a two-sided hypothesis test, and analyses were performed using SAS version 9.3 (SAS Institute, Inc., Cary, NC). A P value < 0.05 was considered statistically significant.
Results
Water quality.
Measured water quality parameters for each of the water sample types are presented in Table 1. Median physical water quality parameters are presented with minimum and maximum values, and the geometric means of bacterial parameters are presented with the SD. Water temperature, pH, salinity, and specific conductance were found to be consistent at each of the sampling sites. However, turbidity and bacterial indicators were more variable. Escherichia coli and enterococci data were consistent with substantial fecal contamination at most sites. Of the freshwater sites, 100% of canal samples, 25% of lake samples, and 74% of river samples fell into the high-risk category for E. coli levels (> 101 MPN/100 mL), according to WHO guidelines for drinking water.29
Table 1.
Median physical water quality parameters (Min, Max) and geometric mean (SD) of Escherichia coli and enterococci for each water type
| Parameter | Canal N = 16* | Lake N = 8† | Marine N = 12‡ | River N = 39§ |
|---|---|---|---|---|
| Temperature (°C) | 26.6 (21.5, 29.4) | 28.8 (26.3, 32.3) | 30.1 (27.1, 32.4) | 27.7 (24.1, 34.2) |
| pH | 8.2 (7.8, 8.3) | 8.2 (7.9, 8.5) | 8.0 (7.1, 8.7) | 8.1 (7.6, 8.6) |
| Turbidity (NTU) | 16 (0.89, 120) | 30 (1.8, 360) | 3.5 (0.92, 31) | 9.0 (1.3, 270) |
| Salinity (mg/L) | 0.15 (0.12, 0.18) | 0.41 (0.14, 1.6) | 36 (23, 39) | 0.16 (0.08, 0.26) |
| Specific conductance (μS/cm) | 308 (245, 382) | 821 (298, 3,140) | 54,200 (36,800, 58,900) | 342 (161, 568) |
| E. coli (MPN/100 mL) | 600 (4.2) | 22 (6.3) | NA¶ | 500 (6.0) |
| Enterococci (MPN/100 mL) | NA | NA | 180 (18) | NA |
N = 12 for pH.
N = 6 for pH.
N = 9 for pH.
N = 29 for pH.
Not analyzed.
Vibrio cholerae PCR detection.
When pooled, culture data for all sample collection methods showed that V. cholerae genes could be detected in APW enrichments at every site during each field visit (Table 2). The ompW gene was detected at every site in every month, whereas toxR was detected at 18 sites in October 2011, 17 sites in March 2012, 16 sites in August 2012, and 7 sites January 2013. Toxigenic V. cholerae, indicated by detection of the ctxA gene, was found at 11 sites in October, 5 sites in August, and 6 sites in January. One of the ctxA detections in January 2013 was for a PLK sample collected from River 8, which was the only incidence of ctxA-positive PLK sample (data not shown). The ctxA gene was not detected in samples collected in March 2012. Detections of the ctxA gene in APW enrichments were associated with higher concentrations of E. coli and lower pH and dissolved oxygen levels (Table 3). The V. cholerae wbeT gene (O1) was detected at only one site in October, although the wbfR gene (O139) was not detected in any of the samples collected during the study.
Table 2.
Detection of ompW, toxR, and ctxA genes in APW enrichments
| Site | October 2011 | March 2012 | August 2012 | January 2013 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ompW | toxR | ctxA | ompW | toxR | ctxA | ompW | toxR | ctxA | ompW | toxR | ctxA | |
| Canal 1 | + | + | + | + | + | − | + | + | − | + | − | − |
| Canal 2 | + | + | + | + | + | − | + | + | − | + | − | + |
| Marine 1 | + | + | − | + | + | − | + | − | − | NA* | NA | NA |
| River 1 | + | + | + | + | + | − | + | + | + | + | − | + |
| River 2 | + | + | + | + | + | − | + | + | − | + | − | − |
| Canal 3† | + | + | + | + | + | − | + | + | − | + | − | + |
| Canal 4 | + | + | + | + | + | − | + | + | + | + | − | − |
| River 3 | + | + | + | + | + | − | + | + | − | + | − | + |
| Lake 1 | + | + | − | + | + | − | + | + | − | + | + | − |
| Lake 2 | + | + | − | + | + | − | + | + | − | + | + | − |
| River 4 | + | + | − | + | + | − | + | + | − | + | + | − |
| River 5 | + | + | − | + | + | − | + | + | − | + | − | + |
| River 6 | + | + | + | + | + | − | + | + | + | + | + | − |
| River 7 | + | + | + | + | + | − | + | + | + | + | + | − |
| River 8 | + | + | + | + | + | − | + | + | − | + | + | + |
| River 9 | + | + | − | + | + | − | + | + | + | + | + | − |
| River 10 | + | + | + | + | + | − | + | + | − | + | − | − |
| Marine 2 | + | + | − | + | − | − | + | − | − | + | − | − |
| Marine 3 | + | − | − | + | − | − | + | − | − | + | − | − |
NA = not analyzed.
Detection of V. cholerae O1 gene in October 2011.
Table 3.
Logistic regression analysis for association between water quality parameters and detection rates of the ctxA gene in APW enrichments and viable V. cholerae O1 cells by DFA-DVC for plankton (PLK) and plankton net-filtered (PFW) water samples
| Water quality parameter | Units | APW ctxA | DFA-DVC PLK | DFA-DVC PFW | |||
|---|---|---|---|---|---|---|---|
| Odds ratio (95% CI) | P value | Odds ratio (95% CI) | P value | Odds ratio (95% CI) | P value | ||
| E. coli | Log CFU/100 mL | 1.54 (1.24–1.90)* | < 0.0001 | 1.08 (0.76–1.53) | 0.67 | 0.99 (0.81–1.22) | 0.95 |
| Salinity | 0.1 mg/L | 0.89 (0.70–1.13) | 0.34 | 0.26 (0.06–1.23) | 0.09 | 1.11 (1.06–1.13) | < 0.0001 |
| Temperature | 1°C | 1.08 (0.86–1.36) | 0.52 | 1.04 (0.85–1.28) | 0.69 | 1.25 (1.03–1.50) | 0.02 |
| Specific conductance | 100 μS/cm | 0.94 (0.83–1.07) | 0.38 | 0.52 (0.25–1.08) | 0.08 | 1.06 (1.03–1.08) | < 0.0001 |
| pH | 0.1 log[H+] | 0.75 (0.61–0.93) | 0.01 | 1.07 (0.85–1.33) | 0.56 | 1.00 (0.74–1.36) | 0.99 |
| Turbidity | 50 NTU | 1.64 (0.86–3.13) | 0.13 | 1.75 (1.27–2.41) | 0.15 | 1.75 (1.27–2.41) | 0.001 |
Bold type indicates a significant association.
Detection rates of V. cholerae genes were compared for DEUF and grab sample collection methods. The ompW gene was detected in 66 DEUF and 51 grab samples throughout the study. There were 18 ctxA detections associated with the DEUF method and 8 ctxA detections associated with grab samples (data not shown). This difference in ctxA detection for DEUF and grab samples was statistically significant, with ctxA detections more likely from DEUF-collected samples (odds ratio [OR] 2.60, 95% confidence interval [CI] 1.10–6.17).
Vibrio cholerae culture detection.
A total of 2,353 presumptive colonies of V. cholerae were examined by PCR for ompW, ctxA, and tcpA genes (Table 4). Although 936 isolates (40%) were V. cholerae, only three isolates from two sites were positive for ctxA. One toxigenic isolate was recovered from the River 1 sampling site in the Artibonite Department in October 2011 and two others from the River 3 site in the Artibonite Department in January 2013. In October, March, and August, the tcpA gene was detected in 5, 19, and 10 non-toxigenic V. cholerae isolates, respectively, from the River 3 site. Only tcpAEl Tor isolates were detected in October and August, and only tcpAClassical isolates were detected in March.
Table 4.
Characterization of presumptive V. cholerae culture isolates
| Month | Isolates tested | ompW-positive (%) | ctxA-positive (%) | tcpA-positive (%)* |
|---|---|---|---|---|
| October | 584 | 186 (32) | 1 (0.17) | 5 (0.86) |
| March | 911 | 370 (41) | 0 | 19 (2.1) |
| August | 468 | 220 (47) | 0 | 10 (2.1) |
| January | 390 | 160 (41) | 2 (0.51) | 2 (0.51) |
| Total | 2,353 | 936 (40) | 3 (0.13) | 36 (1.5) |
tcpAEl Tor detected in October, August, January; tcpAClassical detected in March.
Isolate characterization.
Results of genetic characterization of selected ctxA and tcpA-positive V. cholerae isolates from different sample sites are shown in Table 5. The toxigenic isolates matched the profile for the Haiti epidemic strain as characterized in a previous study.20 The isolates were positive by PCR for El Tor rstR and smp, and the ctxAB and tcpA sequences matched 100% to the epidemic strain. The whole genome sequence of two water sample isolates (GenBank accession no. JNEW01000000) clustered with Haiti clinical isolates (Supplemental Figure S1).30 The tcpA gene was sequenced for a total of six isolates that were tcpA-positive but ctxA-negative by qPCR (Table 5). The tcpA sequences did not match prototypical El Tor and classical tcpA sequences (Supplemental Figure S2). The allele detected by qPCR as El Tor differed from the prototypical allele at 14 nucleotide sites (2.1%), whereas the allele detected as classical differed from the prototypical allele at two nucleotide sites (0.3%).
Table 5.
Characterization of V. cholerae isolates
| Isolate ID | Location | Month | Serogroup | Serotype | ctxA | tcpA | smp | rstR |
|---|---|---|---|---|---|---|---|---|
| 2012EL-1094 | River 3 | Oct. | Non-O1/O139 | NA | − | El Tor* | − | Classical |
| 2012EL-1098 | River 1 | Oct. | O1 | Ogawa | + | El Tor | + | El Tor |
| 2012EL-1759 | River 3 | March | Non-O1/O139 | NA | − | classical† | − | Calcutta |
| 2012EL-1762 | River 3 | March | Non-O1/O139 | NA | − | classical† | − | Calcutta |
| 2012EL-1765 | River 3 | March | Non-O1/O139 | NA | − | classical† | − | Calcutta |
| 2012EL-1768 | River 3 | March | Non-O1/O139 | NA | − | classical† | − | Calcutta |
| 2012EL-1774 | River 3 | March | Non-O1/O139 | NA | − | classical† | − | Calcutta |
| 2013EL-1241 | River 3 | January | O1 | Ogawa | + | El Tor | + | El Tor |
Additional genetic characterization determined this allele differed from the prototypical El Tor allele by 2.1%.
Additional genetic characterization determined this allele differed from the prototypical classical allele by 0.3%.
DFA-DVC.
Viable cells of V. cholerae O1 were detected by DFA-DVC at 15 sites in October 2011, 7 sites in August 2012, and 3 sites in January 2013 (Table 6). Vibrio cholerae O1 was not detected in March 2012. Vibrio cholerae O139 was not detected in any of the samples collected in the study. The DFA-DVC detections of V. cholerae O1 from PFW samples were associated with higher salinity, temperature, specific conductance, and turbidity (Table 3). However, no significant correlations were found between water quality parameters and DFA-DVC detection of V. cholerae O1 from PLK samples. Statistical analyses using the DFA-DVC concentration data presented in Table 5 were attempted using alternative modeling approaches, but none resulted in a good fit because of data characteristics (e.g., highly skewed, zero-inflated distribution, potential outliers).
Table 6.
DFA-DVC counts (cells/liter) of Vibrio cholerae O1 from plankton (PLK) and plankton net-filtered water (PFW) fractions
| Site | October 2011 | March 2012 | August 2012 | January 2013 | ||||
|---|---|---|---|---|---|---|---|---|
| PLK | PFW | PLK | PFW | PLK | PFW | PLK | PFW | |
| Canal 1 | < 20 | < 2,000 | < 40 | < 2,000 | 72 | < 2,000 | < 26 | < 2,000 |
| Canal 2 | 80 | 32,000 | < 40 | < 2,000 | < 40 | < 2,000 | < 20 | < 2,000 |
| Marine 1 | < 20 | < 2,000 | < 70 | < 2,000 | < 20 | < 2,000 | ND* | ND |
| River 1 | 3,080 | 112,000 | < 40 | < 2,000 | < 34 | < 2,000 | 40 | 528,000 |
| River 2 | 40 | 312,000 | < 40 | < 2,000 | 132 | < 2,000 | < 24 | < 2,000 |
| Canal 3 | < 20 | 8,000 | < 40 | < 2,000 | < 40 | < 2,000 | < 25 | < 2,000 |
| Canal 4 | 180 | 210,000 | < 40 | < 2,000 | < 44 | < 2,000 | < 20 | < 2,000 |
| River 3 | < 20 | < 2,000 | < 40 | < 2,000 | < 34 | 8,000 | < 20 | < 2,000 |
| Lake 1 | < 20 | 4,000 | < 40 | < 4,000 | < 180 | 8,000 | < 100 | < 2,000 |
| Lake 2 | 100 | 10,000 | < 40 | < 4,000 | < 40 | < 2,000 | < 20 | < 2,000 |
| River 4 | 140 | 42,000 | < 40 | < 4,000 | 40 | 8,000 | 72 | < 2,000 |
| River 5 | < 20 | 4,000 | < 40 | < 2,000 | < 40 | 2,000 | < 25 | < 2,000 |
| River 6 | 260 | 134,000 | < 40 | < 2,000 | < 40 | 4,000 | < 20 | < 2,000 |
| River 7 | < 20 | < 2,000 | < 40 | < 4,000 | < 40 | < 2,000 | < 20 | < 2,000 |
| River 8 | 80 | 170,000 | < 40 | < 4,000 | < 40 | < 2,000 | < 20 | < 2,000 |
| River 9 | 200 | 2,000 | < 80 | < 2,000 | < 40 | < 2,000 | < 20 | 4,000 |
| River 10 | 40 | < 2,000 | < 40 | < 2,000 | < 40 | < 2,000 | < 25 | < 2,000 |
| Marine 2 | 280 | < 2,000 | < 40 | < 2,000 | < 40 | < 2,000 | < 28 | < 2,000 |
| Marine 3 | 260 | 6,000 | < 40 | < 2,000 | < 40 | < 2,000 | < 24 | < 2,000 |
ND = not done.
Method comparisons.
Results from TCBS and CV agar plating in October and March provided a comparison of the effectiveness of each agar for recovery of V. cholerae (Table 7). A higher percentage of isolates were recovered from TSA transport vials originally from TCBS (75–94%), compared with CV (58–77%). Additionally, a higher percentage of isolates from TCBS were confirmed as V. cholerae (34–48%) compared with CV (28–31%). The GEE analysis comparing TCBS versus CV, adjusting for sample type, found a statistically significant difference in detection rates between these two methods (OR 1.91, 95% CI 1.31–2.77). A comparison of V. cholerae detection rates for the DEUF and grab sample collection methods using TCBS agar (data not shown) indicated no significant difference between the two methods for recovering V. cholerae (OR 1.37, 95% CI 0.58–3.25).
Table 7.
Comparison of TCBS and CHROMagar Vibrio for recovery of V. cholerae (ompW) isolates
| Month | CHROMagar Vibrio | TCBS | ||||
|---|---|---|---|---|---|---|
| Picked | Recovered (%) | Positive (%) | Picked | Recovered (%) | Positive (%) | |
| October | 389 | 226 (58) | 63 (28) | 480 | 359 (75) | 123 (34) |
| March | 528 | 405 (77) | 125 (31) | 541 | 506 (94) | 245 (48) |
| Total | 917 | 630 (69) | 188 (30) | 1021 | 865 (85) | 368 (43) |
Discussion
Results of this study show that toxigenic V. cholerae O1 can be detected in Haitian freshwater systems, with detections as recently as January 2013. Although toxigenic V. cholerae was isolated from only two sites during the course of this surveillance study, ctxA genes were found in APW enrichments 22 times at 12 sites, and V. cholerae O1 was detected by DFA-DVC 25 times at 17 sites. Detection of ctxA genes in APW broths is an indication that culturable toxigenic V. cholerae cells were present in the enriched water samples. Therefore, detection of viable toxigenic V. cholerae and V. cholerae O1 was enhanced by APW qPCR and DFA-DVC compared with culture isolation alone. These findings are not unexpected, as other investigators have found that isolation of V. cholerae O1 from the environment is difficult, even when the bacteria can be detected by other methods.32,33
Detection of plankton-associated V. cholerae O1 by DFA-DVC throughout the study period is evidence of the presence of the bacteria in the environment because V. cholerae is a commensal of copepods and cladocerans. The association of V. cholerae with zooplankton can protect the bacteria from environmental stresses and low nutrient levels and enhance their distribution in the environment.7,8,34
The toxigenic V. cholerae detection data from this study are supported by the research results of Alam and others6 who reported isolating the toxigenic V. cholerae O1 El Tor biotype from three water samples collected between April 2012 and March 2013 in the Ouest Department near the towns of Gressier and Leogane. However, other researchers, using only culture methods, did not isolate or detect toxigenic V. cholerae from water samples collected in July 2012 from 36 aquatic locations in the Ouest and Artibonite Departments.35
Non-toxigenic V. cholerae non-O1/O139 isolates that were tcpA-positive were repeatedly recovered from the River 3 site in the Artibonite Department during the surveillance project. The tcpA gene encodes the major subunit of the toxin co-regulated pilus (TCP), the receptor for lysogenic bacteriophage (CTXΦ) carrying cholera toxin genes.1,36 In other countries, virulence genes, including tcpA, have been found in V. cholerae non-O1/O139 from the environment, notably in cholera-endemic areas.37,38 Although initial PCR results suggested these strains carried El Tor and classical tcpA alleles, sequence determination of the tcpA gene and phylogenetic analysis showed that the alleles were related to (but different from) the prototypical alleles (Supplemental Figure S2). The ctxA-negative isolates were also positive by PCR for different alleles of rstR, suggesting they could each possess a different variant of pre-CTX prophage (the CTX prophage devoid of the ctxAB genes).39 These results show that at least two non-toxigenic lineages of tcpA-positive V. cholerae exist in Haitian waters. These strains could conceivably acquire the ctxAB genes and become toxigenic, but experimental evidence to support this premise is lacking. Nonetheless, the results highlight the need to characterize environmental V. cholerae so that the source of non-toxigenic strains recovered from human cases, or new toxigenic strains should they appear, can be better understood.
Logistical regression indicated a negative association for ctxA detections with pH, which appears to contradict the established theories about conditions that support V. cholerae survival in the environment. However, the range of pH values for freshwater samples was small (7.6–8.6) and was within the range of favorable pH values for V. cholerae. Positive associations between ctxA detections and E. coli concentrations, and DFA-DVC detections and turbidity, may be indicative of the introduction of V. cholerae into surface waters through fecal contamination. The positive associations between DFA-DVC detections and salinity, temperature, and specific conductance are in concordance with the established paradigm of V. cholerae as autochthonous to the aquatic environment.34
During this study, TCBS agar resulted in a significantly higher percentage of V. cholerae positive isolates compared with CV agar. However, these findings warrant further investigation; there are several brands of TCBS on the market and there are differences in performance between brands.40,41 There was no significant difference in the ability of a large volume sample collection method, such as DEUF, to recover V. cholerae from water samples compared with a 1-L grab sample. The DEUF method resulted in a greater number of ctxA detections from APW enrichments than the grab sample method, indicating that this large volume sample collection method can be effective for increasing detection rates of toxigenic V. cholerae in water samples. However, in three instances the grab sample was ctxA positive, whereas the DEUF sample was negative (data not shown). Taken together, these data indicate that the use of large and small volume sample collection methods may improve detection and isolation of V. cholerae in water samples.16
There were several limitations associated with this study, because only four visits to Haiti could be conducted. As a result of logistical constraints and testing capacity at LNSP, presumptive V. cholerae isolates were sent to CDC in Atlanta for confirmation. Only a portion of these isolates could be recovered from the transport media after shipment (Table 7). Additionally, the distance of some collection sites resulted in some samples being held for 48 hours instead of the desired 24 hours before processing. A 24-hour holding time has been shown to be beneficial for V. cholerae culture.15
Using multiple sample collection and analytical methods, toxigenic V. cholerae O1 was detected in Haitian water sources more than 2 years after the start of the cholera epidemic. The association of V. cholerae ctxA gene detections with higher E. coli concentrations and turbidity indicate that V. cholerae contamination may have been associated with inadequate sanitation and fecal runoff into water bodies. However, detections of plankton-associated V. cholerae O1 throughout the study and the detection of the ctxA gene in a PLK sample in January 2013 show that V. cholerae O1 can be found in a known environmental niche (plankton) in Haitian freshwater. The data from this study suggest that ongoing environmental surveillance of V. cholerae could provide valuable data for characterizing the potential for cholera transmission by untreated water consumption. Additional study of the ecology of V. cholerae in Haiti would provide useful data for water-related exposure risk management and informed decision making regarding potential public health measures such as vaccination campaigns and distribution of safe drinking water supplies and informational materials. Continued environmental surveillance is important to understand the ongoing ecological dynamics of V. cholerae and the contribution of waterborne transmission to cholera in Haiti.
Supplementary Material
ACKNOWLEDGMENTS
We thank the following colleagues for their contributions to this study: Emmanuel Rossignol (LNSP); Carrie Whitworth, Michele B. Parsons, Nancy Garrett, Cheryl Bopp, Mike Frace (CDC); Marsha McCary, Amber Dismer, Wilson Dolne, and Nora Purcell (CDC Haiti).
Disclaimer: The use of trade names and names of commercial sources is for identification only and does not imply endorsement by the Centers for Disease Control and Prevention or the U.S. Department of Health and Human Services. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Financial support: Centers for Disease Control and Prevention, partial support at the University of Maryland came from NIH grant no. 2RO1A1039129-11A2 and NSF grant no. 0813066.
Authors' addresses: Amy M. Kahler, Bonnie J. Mull, Cheryl L. Tarr, Maryann Turnsek, Lee S. Katz, Michael S. Humphrys, Gordana Derado, and Vincent R. Hill, Division of Foodborne, Waterborne, and Environmental Diseases, Centers for Disease Control and Prevention, Atlanta, GA, E-mails: akahler@cdc.gov, bonniejmull@gmail.com, ctarr@cdc.gov, HUD4@cdc.gov, GZU2@cdc.gov, msh06f@gmail.com, uwx8@cdc.gov, and vhill@cdc.gov. Bradd J. Haley, Arlene Chen, Rita R. Colwell, and Anwar Huq, Maryland Pathogen Research Institute and Center for Bioinformatics and Computational Biology, University of Maryland, College Park, MD, E-mails: bradd.haley@gmail.com, achen87@umd.edu, rcolwell@umiacs.umd.edu, and huqanwar@gmail.com. Nicole Freeman and Jacques Boncy, National Public Health Laboratory, Haitian Ministry of Public Health and Population, Port-au-Prince, Haiti, E-mails: nicolemfreeman@gmail.com and jboncy2001@yahoo.fr.
References
- 1.Faruque SM, Albert MJ, Mekalanos JJ. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO/United Nations Children's Fund JMPfWSaS Estimates on the Use of Water Sources and Sanitation Facilities. 2013. http://www.wssinfo.org/documents-links/documents/?tx_displaycontroller[type]=country_files Available at. Accessed July 15, 2013.
- 3.Jenson D, Szabo V, Duke FH. Cholera in Haiti and other Caribbean regions, 19th Century. Emerg Infect Dis. 2011;17:2130–2135. doi: 10.3201/eid1711.110958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barzilay EJ, Schaad N, Magloire R, Mung KS, Boncy J, Dahourou GA, Mintz ED, Steenland MW, Vertefeuille JF, Tappero JW. Cholera surveillance during the Haiti epidemic–the first 2 years. N Engl J Med. 2013;368:599–609. doi: 10.1056/NEJMoa1204927. [DOI] [PubMed] [Google Scholar]
- 5.Hill VR, Cohen N, Kahler AM, Jones JL, Bopp CA, Marano N, Tarr CL, Garrett NM, Boncy J, Henry A, Gomez GA, Wellman M, Curtis M, Freeman MM, Turnsek M, Benner RA, Dahourou G, Espey D, DePaola A, Tappero JW, Handzel T, Tauxe RV. Toxigenic Vibrio cholerae O1 in water and seafood, Haiti. Emerg Infect Dis. 2011;17:2147–2150. doi: 10.3201/eid1711.110748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alam MT, Weppelmann TA, Weber CD, Johnson JA, Rashid MH, Birch CS, Brumback BA, de Rochars V, Morris JG, Ali A. Monitoring water sources for environmental reservoirs of toxigenic Vibrio cholerae O1, Haiti. Emerg Infect Dis. 2014;20:356–363. doi: 10.3201/eid2003.131293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huq A, Small EB, West PA, Huq MI, Rahman R, Colwell RR. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl Environ Microbiol. 1983;45:275–283. doi: 10.1128/aem.45.1.275-283.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamplin ML, Gauzens AL, Huq A, Sack DA, Colwell RR. Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl Environ Microbiol. 1990;56:1977–1980. doi: 10.1128/aem.56.6.1977-1980.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colwell RR, Brayton PR, Grimes DJ, Roszak DB, Huq SA, Palmer LM. Viable but non-culturable Vibrio cholerae and related pathogens in the environment–implications for release of genetically engineered microorganisms. Biotechnology (N Y) 1985;3:817–820. [Google Scholar]
- 10.Brayton PR, Tamplin ML, Huq A, Colwell RR. Enumeration of Vibrio cholerae O1 in Bangladesh waters by fluorescent antibody direct viable count. Appl Environ Microbiol. 1987;53:2862–2865. doi: 10.1128/aem.53.12.2862-2865.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chowdhury MA, Xu B, Montilla R, Hasan JA, Huq A, Colwell RR. A simplified immunofluorescence technique for detection of viable cells of Vibrio cholerae O1 and O139. J Microbiol Methods. 1995;24:165–170. [Google Scholar]
- 12.Blackstone GM, Nordstrom JL, Bowen MD, Meyer RF, Imbro P, DePaola A. Use of a real time PCR assay for detection of the ctxA gene of Vibrio cholerae in an environmental survey of Mobile Bay. J Microbiol Methods. 2007;68:254–259. doi: 10.1016/j.mimet.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Smith CM, Hill VR. Dead-end hollow-fiber ultrafiltration for recovery of diverse microbes from water. Appl Environ Microbiol. 2009;75:5284–5289. doi: 10.1128/AEM.00456-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mull B, Hill VR. Recovery of diverse microbes in high turbidity surface water samples using dead-end ultrafiltration. J Microbiol Methods. 2012;91:429–433. doi: 10.1016/j.mimet.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alam M, Sadique A, Nur AH, Bhuiyan NA, Nair GB, Siddique AK, Sack DA, Ahsan S, Huq A, Sack RB, Colwell RR. Effect of transport at ambient temperature on detection and isolation of Vibrio cholerae from environmental samples. Appl Environ Microbiol. 2006;72:2185–2190. doi: 10.1128/AEM.72.3.2185-2190.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huq A, Haley BJ, Taviani E, Chen A, Hasan NA, Colwell RR. Detection, isolation, and identification of Vibrio cholerae from the environment. Curr Prot Microbiol. 2012;26:6A.5.1–6A.5.51. doi: 10.1002/9780471729259.mc06a05s26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill VR, Kahler AM, Jothikumar N, Johnson TB, Hahn D, Cromeans TL. Multistate evaluation of an ultrafiltration-based procedure for simultaneous recovery of enteric microbes in 100-liter tap water samples. Appl Environ Microbiol. 2007;73:4218–4225. doi: 10.1128/AEM.02713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bauer A, Rorvik LM. A novel multiplex PCR for the identification of Vibrio parahaemolyticus, Vibrio cholerae and Vibrio vulnificus. Lett Appl Microbiol. 2007;45:371–375. doi: 10.1111/j.1472-765X.2007.02195.x. [DOI] [PubMed] [Google Scholar]
- 19.Hoshino K, Yamasaki S, Mukhopadhyay AK, Chakraborty S, Basu A, Bhattacharya SK, Nair GB, Shimada T, Takeda Y. Development and evaluation of a multiplex PCR assay for rapid detection of toxigenic Vibrio cholerae O1 and O139. FEMS Immunol Med Microbiol. 1998;20:201–207. doi: 10.1111/j.1574-695X.1998.tb01128.x. [DOI] [PubMed] [Google Scholar]
- 20.Talkington D, Bopp C, Tarr C, Parsons MB, Dahourou G, Freeman M, Joyce K, Turnsek M, Garrett N, Humphrys M, Gomez G, Stroika S, Boncy J, Ochieng B, Oundo J, Klena J, Smith A, Keddy K, Gerner-Smidt P. Characterization of toxigenic Vibrio cholerae from Haiti, 2010–2011. Emerg Infect Dis. 2011;17:2122–2129. doi: 10.3201/eid1711.110805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kislyuk AO, Katz LS, Agrawal S, Hagen MS, Conley AB, Jayaraman P, Nelakuditi V, Humphrey JC, Sammons SA, Govil D, Mair RD, Tatti KM, Tondella ML, Harcourt BH, Mayer LW, Jordan IK. A computational genomics pipeline for prokaryotic sequencing projects. Bioinformatics. 2010;26:1819–1826. doi: 10.1093/bioinformatics/btq284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 25.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 27.Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- 28.Zeger SL, Liang KY. The analysis of discrete and continuous longitudinal data. Biometrics. 1986;42:121–130. [PubMed] [Google Scholar]
- 29.WHO . Guidelines for Drinking-Water Quality. Geneva: World Health Organization Library Cataloguing-in-Publication Data; 2011. [Google Scholar]
- 30.Katz LS, Petkau A, Beaulaurier J, Tyler S, Antonova ES, Turnsek MA, Guo Y, Wang S, Paxinos EE, Orata F, Gladney LM, Stroika S, Folster JP, Rowe L, Freeman MM, Knox N, Frace M, Boncy J, Graham M, Hammer BK, Boucher Y, Bashir A, Hanage WP, Van Domselaar G, Tarr CL. Evolutionary dynamics of Vibrio cholerae O1 following a single-source introduction to Haiti. MBio. 2013;4:e00398. doi: 10.1128/mBio.00398-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reimer AR, Van Domselaar G, Stroika S, Walker M, Kent H, Tarr C, Talkington D, Rowe L, Olsen-Rasmussen M, Frace M, Sammons S, Dahourou GA, Boncy J, Smith AM, Mabon P, Petkau A, Graham M, Gilmour MW, Gerner-Smidt P, Task VC. Comparative genomics of Vibrio cholerae from Haiti, Asia, and Africa. Emerg Infect Dis. 2011;17:2113–2121. doi: 10.3201/eid1711.110794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huq A, Colwell RR, Rahman R, Ali A, Chowdhury MA, Parveen S, Sack DA, Russekcohen E. Detection of Vibrio cholerae O1 in the aquatic environment by fluorescent monoclonal antibody and culture methods. Appl Environ Microbiol. 1990;56:2370–2373. doi: 10.1128/aem.56.8.2370-2373.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gil AI, Louis VR, Rivera IN, Lipp E, Huq A, Lanata CF, Taylor DN, Russek-Cohen E, Choopun N, Sack RB, Colwell RR. Occurrence and distribution of Vibrio cholerae in the coastal environment of Peru. Environ Microbiol. 2004;6:699–706. doi: 10.1111/j.1462-2920.2004.00601.x. [DOI] [PubMed] [Google Scholar]
- 34.Colwell RR, Huq A. Environmental reservoir of Vibrio cholerae–the causative agent of cholera. In: Wilson ME, Levins R, Spielman A, editors. Disease in Evolution: Global Changes and Emergence of Infectious Diseases. New York: New York Academy Sciences; 1994. pp. 44–54. [DOI] [PubMed] [Google Scholar]
- 35.Baron S, Lesne J, Moore S, Rossignol E, Rebaudet S, Gazin P, Barrais R, Magloire R, Boncy J, Piarroux R. No evidence of significant levels of toxigenic V. cholerae O1 in the Haitian aquatic environment during the 2012 rainy season. PLoS Curr. 2013;13:5. doi: 10.1371/currents.outbreaks.7735b392bdcb749baf5812d2096d331e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 37.Chakraborty S, Mukhopadhyay AK, Bhadra RK, Ghosh AN, Mitra R, Shimada T, Yamasaki S, Faruque SM, Takeda Y, Colwell RR, Nair GB. Virulence genes in environmental strains of Vibrio cholerae. Appl Environ Microbiol. 2000;66:4022–4028. doi: 10.1128/aem.66.9.4022-4028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faruque SM, Chowdhury N, Kamruzzaman M, Dziejman M, Rahman MH, Sack DA, Nair GB, Mekalanos JJ. Genetic diversity and virulence potential of environmental Vibrio cholerae population in a cholera-endemic area. Proc Natl Acad Sci USA. 2004;101:2123–2128. doi: 10.1073/pnas.0308485100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maiti D, Das B, Saha A, Nandy RK, Nair GB, Bhadra RK. Genetic organization of pre-CTX and CTX prophages in the genome of an environmental Vibrio cholerae non-O1, non-O139 strain. Microbiology-Sgm. 2006;152:3633–3641. doi: 10.1099/mic.0.2006/000117-0. [DOI] [PubMed] [Google Scholar]
- 40.Morris GK, Merson MH, Huq I, Kibrya AK, Black R. Comparison of four plating media for isolating Vibrio cholerae. J Clin Microbiol. 1979;9:79–83. doi: 10.1128/jcm.9.1.79-83.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor JA, Barrow GI. A non-pathogenic vibrio for the routine quality control of TCBS cholera medium. J Clin Pathol. 1981;34:208–212. doi: 10.1136/jcp.34.2.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.