Abstract
We developed a TaqMan-based real-time quadriplex polymerase chain reaction (PCR) to simultaneously detect Treponema pallidum subspecies pallidum, T. pallidum subsp. pertenue, and T. pallidum subsp. endemicum, the causative agents of venereal syphilis, yaws, and bejel, respectively. The PCR assay was applied to samples from skin ulcerations of clinically presumptive yaws cases among children on Tanna Island, Vanuatu. Another real-time triplex PCR was used to screen for the point mutations in the 23S rRNA genes that have previously been associated with azithromycin resistance in T. pallidum subsp. pallidum strains. Seropositivity by the classical syphilis serological tests was 35.5% among children with skin ulcerations clinically suspected with yaws, whereas the presence of T. pallidum subsp. pertenue DNA was only found in lesions from 15.5% of children. No evidence of T. pallidum subsp. pertenue infection, by either PCR or serology was found in ∼59% of cases indicating alternative causes of yaws-like lesions in this endemic area.
Introduction
Syphilis and the endemic treponematoses are caused by closely related, motile, spiral bacteria of the genus Treponema. Treponema pallidum subsp. pallidum (TP-pallidum) is the etiologic agent of venereal syphilis, whereas Treponema pallidum subsp. pertenue (TP-pertenue) and T. pallidum subsp. endemicum (TP-endemicum) are the causative agents of the non-venereal or endemic treponematoses yaws and bejel, respectively. Unlike syphilis, yaws is spread by direct skin-to-skin contact predominantly affecting children < 15 years of age living in poor socioeconomic conditions in certain rural, wet, tropical areas of West and Central Africa, South East Asia, and the South Pacific islands.1 Bejel is also largely transmitted between children, often as a result of sharing drinking vessels, but in dry, more temperate areas of the world.2 Despite differences in the mode of transmission, geographic distribution, propensity to invade the central nervous system, or infect the fetus, the three subspecies share striking similarities at both the genomic and antigenic level.2–5 These treponemes cannot be serially cultivated in vitro, are indistinguishable by darkfield microscopy, and cannot be differentiated by classical syphilis serological methods. However, subspecies-specific genetic signatures permit molecular differentiation using methods that involve polymerase chain reaction (PCR), restriction fragment length polymorphism (RFLP), and DNA sequencing.3,6–9 These existing molecular methods are inherently technically complex resulting in inevitable delays in reporting results. We therefore developed a TaqMan-based real-time quadriplex PCR assay to rapidly and simultaneously differentiate between TP-pallidum, TP-pertenue, and TP-endemicum in a single test that can be used for screening and investigation of individual cases.
Yaws remains endemic in parts of Vanuatu in the South Pacific with more than 1,500 cases reported each year between 2008 and 2010.10 The World Health Organization (WHO) has recently launched a worldwide yaws eradication program based on community mass drug administration (MDA) using the oral macrolide antibiotic, azithromycin, followed by clinical case management for individuals subsequently developing skin lesions.10 To measure the burden of disease before initiation of MDA using azithromycin on Tanna Island, Vanuatu, serological testing and a TaqMan-based real-time multiplex PCR test on swabs taken from lesions were used to investigate the disease among a series of children presenting with skin ulcerations clinically suspected with yaws.
Materials and methods
Study participants and specimen collection.
In May 2013, a study was undertaken to assess the prevalence of yaws in Tanna Island, Vanuatu before initiation of MDA. A total of 244 children < 15 years of age who presented with clinically suspected yaws-like skin ulcerations at randomly selected primary schools, villages, and health centers were examined by clinicians. Of these, 155 children with active skin ulcerations defined by the presence of a significant break in the skin filled with necrotic tissue and/or pus were enrolled in the sub-study for molecular detection and characterization of TP-pertenue. The remaining 89 children with small or healing skin ulcerations and/or papillomatous lesions were excluded as a result of difficulties in collection of ulcer exudates. Informed written consent was obtained from children's parents or guardians. Ethical approval for the study was granted by the ethics committees of the Ministry of Health in Vanuatu, the WHO, and the U.S. Centers for Disease Control and Prevention (CDC).
Venous blood samples were collected for both non-treponemal and treponemal serological testing. Samples for PCR were collected from skin lesions using two dacron-tipped swabs, one was suspended in AssayAssure transport medium (Thermo Fisher Scientific, Waltham, MA), whereas another was rolled onto an FTA micro Elute card (Thermo Fisher Scientific). Genomic DNA was extracted from a 350-μL AssayAssure sample using iPrep PureLink gDNA blood kit and the iPrep purification instrument (Life Technologies, Grand Island, NY). The DNA was extracted from three 3-mm discs punched from the FTA cards. Briefly, the discs were washed in sterile water and the DNA eluted by incubating in 10 mM Tris buffer (pH 8.0) at 95°C for 30 min.
Serological methods.
Serum samples were tested quantitatively for non-treponemal antibodies using the Wampole Impact Syphilis Rapid Plasma Reagin (RPR) test (Alere North America, Inc., Orlando, FL), and for Treponema-specific antibodies by a T. pallidum passive particle agglutination assay (Serodia TP-PA, Fujirebio Diagnostics Inc., Malvern, PA) according to the manufacturer's instructions.
Real-time quadriplex PCR testing and screening for azithromycin resistance markers.
The molecular differentiation of TP-pertenue from TP-pallidum and TP-endemicum was achieved using a real-time quadriplex PCR containing primers and TaqMan probes targeting the tp0858 gene, which encodes a hypothetical protein and two areas of the tprI (tp0620) gene. Amplification of the human ribonuclease P gene (RNase P) was included as an internal control to monitor PCR inhibition and confirm sample adequacy. The quadriplex PCR was performed with a 10-μL sample of DNA in a 25-μL reaction volume containing 12.5-μL of PerfeCTa Multiplex qPCR SuperMix (Quanta Biosciences, Gaithersburg, MD), and the appropriate volume of each primer and probe. The PCR targets, amplicon sizes, primer and probe sequences, and their final concentrations and the fluorescent dye used for each specific probe are shown in Table 1. The assay was performed on a Rotor-Gene Q real-time PCR instrument (Qiagen Inc., Valencia, CA) using the following thermocycling conditions: 95°C for 4 min, followed by 50 cycles of 95°C for 20 s, and 65°C for 1 min. Purified genomic DNA from TP-pertenue, TP-pallidum, or TP-endemicum; or custom synthesized DNA fragments encompassing unique genetic signature areas within tp0858 or tprI (the size of each insert is ∼250 to 400 bp) cloned into pIDTSmart plasmids (Integrated DNA Technologies, Coralville, IA) were used as positive controls. Both positive and no-template controls (NTC) were included to standardize the threshold cutoff in each run. The analytic sensitivity of the real-time quadriplex PCR assay was determined using 10-fold serial dilutions of purified TP-pertenue genomic DNA (CDC-1 strain). The analytical specificity of the PCR assay was verified using DNA purified from 10 laboratory strains of TP-pallidum8,9 (Nichols, SS 14, Mexico A, JV1, DAL-1, Madras, 1 strain from Maryland, 3 strains from Minnesota) and 11 clinical strains from South Africa,11 13 laboratory strains of TP-pertenue8,9 (CDC-1, CDC-2, CDC-2575, Samoa D, Samoa F, Ghana 051, Gauthier, and 6 strains from Indonesia), a clinical strain from Democratic Republic of the Congo,6 and 2 strains of TP-endemicum (Bosnia A and Iraq B). In addition, a previously used panel of nonpathogenic treponemes (T. denticola, T. refringens, and T. phagedenis) and other microorganisms was included to determine the specificity of the assay.12
Table 1.
Primers and probes for molecular detection of the T. pallidum subspecies and human DNA control
| Organism/Control | Primer or probe | Sequence (5′ to 3′) | Final concentration (nmol/L) | Gene target | Product (bp) |
|---|---|---|---|---|---|
| TP-endemicum | FP* | TTATTAACCCCGCGTACACC | 200 | tp0620 | 173 |
| Probe | CalRed610†-ATGCCCATGTAGGGAACATCGGAG-BHQ2‡ | 200 | |||
| RP§ | GCCAAAGTAACGCTCAGACC | 200 | |||
| TP-pallidum | FP | CACTCCTGTGGGGAGTAGGA | 300 | tp0620 | 136 |
| Probe | FAM†-CGATTACCTGCATCGGCAGGGTC-BHQ1 | 300 | |||
| RP | GAGCTCCCCGTTGCCA | 300 | |||
| TP-pertenue | FP | AGTCCTGCTGCAACGGTAGTAC | 200 | tp0858 | 93 |
| Probe | Quasar705†-TGCTGCACGAAGAAGTGCGAAGG-BHQ3 | 200 | |||
| RP | CGTACACCGAACCTTTGTCTT | 200 | |||
| Internal control | FP | CCA AGT GTG AGG GCT GAA AAG | 80 | Human RNase P | 80 |
| Probe | Quasar670†- CC CCA GTC TCT GTC AGC ACT CCC TTC-BHQ3 | 80 | |||
| RP | TGT TGT GGC TGA TGA ACT ATA AAA GG | 80 |
Forward primer.
Fluorescent dye.
BHQ, black hole quencher.
Reverse primer.
The stability of the tp0858 PCR target was also tested with the real-time quadriplex PCR using DNA from the TP-pertenue strain CDC-1 obtained from multiple rabbit passages performed sequentially seven times in 1986 and three times in 1987.
Samples that tested positive for TP-pertenue DNA by the real-time quadriplex PCR were further tested with a TaqMan real-time triplex PCR assay to detect the A2058G and A2059G point mutations in the 23S rRNA gene that are associated with azithromycin resistance in TP-pallidum, as previously described.12
DNA sequencing.
Confirmation of the T. pallidum subsp. was done as previously described using PCR and sequencing of a 280-bp intergenic spacer (IGR19) between the fliG gene (tp0026) and a putative hemolysin gene (tp0027) and a 493-bp specific region within the tprI (tp0620) gene, which is present in all three subspecies.6 The DNA sequencing was performed using the BigDye Terminator v3.1 cycle sequencing kit and an ABI 3130-XL sequencer. Lasergene Software (DNASTAR, Inc., Madison, MI) was used to assemble and align DNA sequences.
Statistical analysis.
Data were captured in a pre-coded database using Microsoft Excel. Statistical analysis was done using the SPSS version 13.0 (SPSS Inc., Chicago, IL). Conditional maximum likelihood estimate of odds ratio (OR) and 95% confidence intervals (CIs) were calculated using OpenEpi version 3.0, and the Mid-P exact test was used to measure the associations between serological reactivity and PCR positivity.
Results
Syphilis serology.
Both RPR and TPPA testing were performed on 155 sera. Of these, 55 (35.5%) were reactive in both RPR and TPPA tests, 9 (5.8%) were reactive by the TPPA alone and 91 (58.7%) were non-reactive by both the RPR and TPPA (Table 2). No biological false positive (RPR+/TPPA−) reactions were detected. The distribution of PCR-positive and -negative samples by RPR titer (R1-R128) in the total sample of children tested is shown in Table 3. A total of 22 (40%) RPR-reactive sera were found to have low titers (< R4) of non-treponemal antibody.
Table 2.
Distribution of TP-pertenue PCR positivity by RPR and TPPA (N = 155)
| Serology results | TP-pertenue-PCR+ (%) |
|---|---|
| RPR+/TPPA+ | 23/55 (41.8) |
| RPR+/TPPA− | 0/0 |
| RPR−/TPPA+ | 0/9 |
| RPR−/TPPA− | 1/91 (1.1) |
RPR = rapid plasma reagin; TPPA = Treponema pallidum passive particle agglutination assay.
Table 3.
Distribution of TP-pertenue PCR results by RPR titer (N = 155)
| RPR titer | TP-pertenue PCR+ve | TP-pertenue PCR−ve | ||
|---|---|---|---|---|
| No. of sera | Cumulative (%) | No. of sera | Cumulative (%) | |
| Non-reactive | 1 | 1 (4.2) | 99 | 99 (75.6) |
| R1 (undiluted) | 3 | 4 (16.7) | 14 | 113 (86.3) |
| R2 (1:2) | 1 | 5 (20.8) | 4 | 117 (89.3) |
| R4 (1:4) | 3 | 8 (33.3) | 1 | 118 (90.1) |
| R8 (1:8) | 3 | 11 (45.8) | 2 | 120 (91.6) |
| R16 (1:16) | 1 | 12 (50.0) | 5 | 125 (95.4) |
| R32 (1:32) | 2 | 14 (58.3) | 0 | 125 (95.4) |
| R64 (1:64) | 6 | 20 (83.3) | 4 | 129 (98.4) |
| R128 (1:128) | 4 | 24 (100) | 2 | 131 (100) |
PCR = polymerase chain reaction; RPR = rapid plasma reagin.
Performance characteristics of the real-time quadriplex PCR.
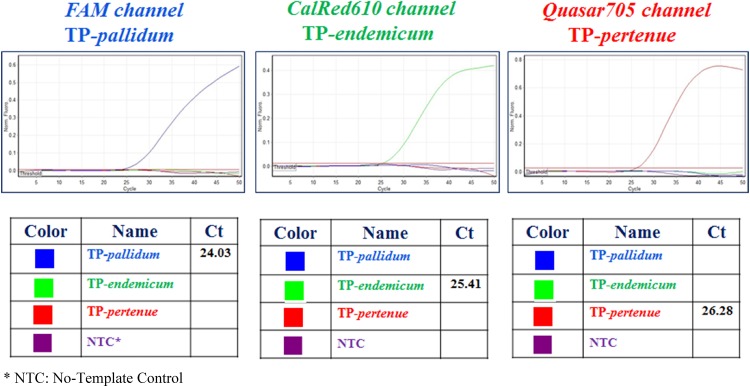
The analytical sensitivity of the real-time quadriplex assay was determined to be 10–100 genomic copies per reaction using serial dilutions of TP-pertenue genomic DNA with an amplification efficiency of 97.6% and a correlation coefficient of 0.982 (data not shown). Representative amplification curves and interpretation of results are shown in Figure 1. Briefly, the fluorescent dye (FAM)-labeled TaqMan probe hybridized specifically to the TP-pallidum DNA template, whereas positive fluorescence signals in the CalRead610 channel was observed only with the TP-endemicum DNA. Similarly, samples containing TP-pertenue DNA were detected only in the Quasar705 channel.
Figure 1.
Representative real-time quadriplex PCR amplification curves and results for simultaneous detection of TP-pallidum, TP-endemicum, and TP-pertenue.
Detection of TP-pertenue DNA by real-time quadriplex-PCR.
The DNA from the 155 paired AssayAssure samples and FTA cards were tested by the real-time quadriplex PCR. Overall, TP-pertenue DNA was detected in skin lesions from 24 of 155 (15.5%) of children. Twenty-three of 24 (95.8%) TP-pertenue-PCR positive AssayAssure samples were also positive using the corresponding FTA card samples. Neither TP-pallidum nor TP-endemicum DNA was detected in any of the skin lesions. The laboratory strains of TP-pertenue, together with positive controls for TP-pallidum and TP-endemicum were all correctly identified by the assay. The RNase P gene target used as an internal control was amplified from all AssayAssure and FTA card samples tested (data not shown). The DNA from nonpathogenic treponemes, or other pathogenic and commensal genital tract microorganisms, and normal skin flora tested negative by the quadriplex PCR, indicating that the assay was highly specific for the respective treponemal DNA sequences (data not shown). The DNA obtained from CDC-1 strain from 10 rabbit passages all tested positive specifically for TP-pertenue (data not shown) suggesting that the tp0858 DNA sequence used in our assay is stable, although the data are limited to only one strain evaluated and with a < 2-year time frame.
Relationship between TP-pertenue-PCR results and serology.
The TP-pertenue DNA was detected in skin lesions from 24 of 155 (15.5%) of children and among them 23 (95.8%) had both reactive RPR and TPPA (Table 2). There was only one PCR-confirmed yaws case with dually non-reactive RPR/TPPA. The proportion of PCR-confirmed yaws cases that were RPR/TPPA dually reactive was significantly higher than those that were dually non-reactive (23 of 55 [41.8%] versus 1 of 91 [1.1%], OR 62.9, 95% CI 11–1359, P < 0.001). The distribution of RPR titer by TP-pertenue-PCR results is shown in Table 3. Children with dually reactive RPR and TPPA who had RPR titers ≥ R4 were more likely to be PCR-confirmed yaws cases than those with titers < R4 (19 of 33 [57.6%] versus 4 of 22 [18.2%], OR 5.9, 95% CI 1.7–24.3, P < 0.01). The rate of high titer RPR seropositivity (≥ R4) was found to be significantly greater in those cases whose lesions were TP-pertenue-PCR-positive than those that were TP-pertenue-PCR-negative (19 of 24 [79.2%] versus 14 of 131 [10.7%], OR 30.5, 95% CI 10.2–104, P < 0.0001).
Among children with dual reactive RPR/TPPA and PCR-positive AssayAssure samples, only one corresponding FTA card sample failed to be detected by PCR.
Detection of azithromycin resistance markers by real-time triplex-PCR.
None of the TP-pertenue-PCR positive samples tested positive for either the A2058G or A2059G point mutation in the 23S rRNA gene, which have previously been associated with macrolide (and thus azithromycin) resistance in TP-pallidum (data not shown).
Confirmation of subspecies-specific DNA by sequencing.
The differentiation of the three T. pallidum subsp. using the real-time quadriplex PCR was confirmed by verifying both corresponding IGR19 and tprI gene sequences using laboratory strains from a specificity panel that included 21 strains of TP-pallidum, 14 strains of TP-pertenue, and 2 strains of TP-endemicum (data not shown). The DNA sequence analysis within IGR19 ruled out the possibility of a TP-pallidum subspecies in the clinical specimens, whereas unique genetic signature sequences of tprI ruled out TP-endemicum as previously described.6
Discussion
Previously reported molecular methods to identify T. pallidum to the subspecies level involved the use of nested PCR combined with sequencing and/or RFLP of multiple treponemal targets.2–5 Although these methods are useful for developing and evaluating new assays, they are costly, laborious, and time-consuming. Consequently, these methods are not suitable for rapid screening applications. In this report, we describe a real-time quadriplex PCR for the simultaneous detection of the etiologic agents of yaws, syphilis, and bejel in a single assay. As part of the baseline yaws survey in Tanna Island, Vanuatu, where the burden of disease is known to be high, serological testing together with two real-time multiplex PCR assays were used to assess the seroprevalence of yaws, to verify the presence of TP-pertenue DNA in lesions, and to detect mutations associated with macrolide resistance in those lesions found to harbor yaws treponemes among children presenting with skin ulcerations clinically suspected with the disease.
The real-time quadriplex PCR described in this study confirmed a diagnosis of yaws by detecting TP-pertenue-specific DNA in skin lesions from ∼15% of children presenting with skin lesions suspected of yaws. Because the test had a high analytical sensitivity and also proved to be specific, it is not surprising that all but one had reactive RPR tests that could be confirmed by a more specific TPPA. Indeed, the detection of TP-pertenue-specific DNA in skin lesions was significantly associated with rates of detection of both non-treponemal and treponemal antibody compared with those who were TP-pertenue DNA negative. Furthermore, among those that were seroreactive by both RPR and TPPA, the RPR titers were significantly higher in those that were PCR positive. This combination of PCR and serological results suggests that those children with PCR-positive lesions have more active disease and are likely to be more infectious than those children who were PCR negative.
In this assessment, which was done before initiation of MDA using azithromycin, TP-pertenue-specific DNA was found in lesions of ∼42% of children who had reactive RPR and TPPA serology. The finding of a significant proportion of low titer RPR seropositivity (< R4) among PCR-negative cases may indicate previously treated cases of yaws that remain serofast for both non-treponemal and treponemal antibody. This situation has been described previously in successfully treated cases of syphilis in high prevalence settings.13 This could further complicate the interpretation of disease status when using serological tests alone. The absence of TP-pertenue strains carrying the point mutations associated with macrolide resistance suggests that azithromycin can be used successfully for MDA in Vanuatu.
This study also indicates that the diagnosis of yaws based on clinical presentation alone appears to have a lower accuracy than anticipated. In this yaws-endemic region, 59% of children presenting with suspected yaws lesions were found to be both PCR negative and non-reactive by both TPPA and RPR serological tests, indicating no current or previous exposure to the disease and strongly suggests the existence of other possible causes for skin ulcers in this marginalized population. The addition of serological tests that detect both treponemal and non-treponemal antibodies greatly improves the odds of identifying suspected yaws cases; however, the etiology of many skin lesions remains uncertain and co-infections cannot be excluded. What is the significance of those ulcers that are PCR negative, but seropositive, either dually, or by the TPPA alone? The possible explanations for these findings include previous use of antibiotics, healing ulcers with numbers of TP-pertenue in lesions below the PCR detection limit, or ulcers that were caused by another etiological agent or by trauma in a subject who had previously had yaws. The unexpected finding of a high percentage of symptomatic children without an identifiable etiology and those not having non-treponemal or treponemal antibodies might have implications for the WHO yaws eradication strategy—especially if they are found to be refractory to the antibiotic therapy provided in the MDA. There have been a few cases reports on Haemophilus ducreyi-associated skin lesions in the south Pacific islands14–16 and more recently a study in Papua New Guinea, a yaws-endemic country, where 60% of lesions in children were found to be H. ducreyi-positive by PCR.17 Further molecular studies to explore the full range of etiologies of skin ulcers in Vanuatu and other yaws-endemic regions are needed to improve our current understanding of yaws epidemiology and other tropical ulcer diseases and to better inform disease control efforts.
The FTA Elute card is a paper-based specimen collection method designed to lyse cellular materials and to preserve DNA and RNA within the matrix, thus rendering samples non-infectious.18–20 Because yaws-endemic communities are most likely located in remote settings, the FTA Elute card can provide a simple way to collect, store, and transport skin lesion specimens at ambient temperature in the absence of a cold chain for molecular testing. Although the sample size was small in our evaluation, the PCR results obtained using FTA Elute cards was in good agreement with liquid AssayAssure transport media. Although there was a lapse of more than 6 months between specimen collection in Vanuatu and PCR testing of the FTA cards at the CDC laboratory, previous studies using DNA from blood and buccal samples applied to FTA cards showed that the DNA remained stable at ambient temperatures for as many as 20 years.21
In conclusion, the real-time quadriplex PCR described here, is a fast, highly sensitive, and specific assay for the molecular differentiation of T. pallidum subspecies and should be particularly useful in areas where both syphilis and yaws or bejel are endemic and in determining the extent of treponemal infections worldwide to support WHO's endemic treponematoses eradication efforts. The use of a combination of classical serological testing and cutting-edge molecular techniques such as the quadriplex PCR will help determine the current geographic distribution of yaws, increase the ability to identify the most potentially infectious cases, and detect treatment failures or reinfections following mass treatment with azithromycin. When used in combination with a previously published molecular assay to screen for point mutations associated with azithromycin resistance and/or treatment failures,8 the quadriplex PCR assay could be used to monitor the possible emergence of macrolide-resistant TP-pertenue strains when azithromycin usage increases as a result of implementation of the WHO's yaws eradication strategy.
ACKNOWLEDGMENTS
Treponema pallidum subsp. pertenue strains from Indonesia were kindly provided by Gerda Noordhoek.
Disclaimer: The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention. The authors declare no conflicts of interests.
Footnotes
Authors' addresses: Cheng-Yen Chen, Kai-Hua Chi, Damien Danavall, Allan Pillay, Eli Nachamkin, and Tun Ye, Centers for Disease Control and Prevention, Division of STD Prevention, Atlanta, GA, E-mails: cyc1@cdc.gov, krc2@cdc.gov, bpu8@cdc.gov, ajp7@cdc.gov, ecn1@cdc.gov, and gwr5@cdc.gov. Ronald C. Ballard, CDC, Center for Global Health, Atlanta, GA, E-mail: ztp7@cdc.gov. Fasihah Taleo, Ministry of Health, Health Department, NTD Program, Port Vila, Vanuatu, E-mail: ftaleo@vanuatu.gov.vu. Jacob L. Kool, World Health Organization, Office for the South Pacific, Port Vila, Vanuatu, E-mail: KoolJ@wpro.who.int. Lasse S. Vestergaard, WHO, Western Pacific Regional Office, Manila, Philippines, E-mail: vestergaardl@wpro.who.int. Kingsley Asiedu, World Health Organization, Neglected Tropical Diseases, Geneva, Switzerland, E-mail: asieduk@who.int. David Fegan, WHO, Consultant, Brisbane, Australia, E-mail: djfegan@gmail.com.
References
- 1.Rinaldi A. Yaws eradication: facing old problems, raising new hopes. PLoS Negl Trop Dis. 2012;6:e21837. doi: 10.1371/journal.pntd.0001837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antal GM, Lukehart SA, Meheus AZ. The endemic treponematoses. Microbes Infect. 2002;4:83–94. doi: 10.1016/s1286-4579(01)01513-1. [DOI] [PubMed] [Google Scholar]
- 3.Čejková D, Zobaníková M, Chen L, Pospíšilová P, Strouhal M, Qin X, Mikalová L, Norris SJ, Muzny DM, Gibbs RA, Fulton LL, Sodergren E, Weinstock GM, Šmajs D. Whole genome sequences of three Treponema pallidum ssp. pertenue strains: yaws and syphilis treponemes differ in less than 0.2% of the genome sequence. PLoS Negl Trop Dis. 2012;6:e1471. doi: 10.1371/journal.pntd.0001471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker-Zander SA, Lukehart SA. Molecular basis of immunological cross-reactivity between Treponema pallidum and Treponema pertenue. Infect Immun. 1983;42:634–638. doi: 10.1128/iai.42.2.634-638.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker-Zander SA, Lukehart SA. Antigenic cross-reactivity between Treponema pallidum and other pathogenic members of the family Spirochaetaceae. Infect Immun. 1984;46:116–121. doi: 10.1128/iai.46.1.116-121.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pillay A, Chen CY, Reynolds MG, Mombouli JV, Castro AC, Louvouezo D, Steiner B, Ballard RC. Laboratory-confirmed case of yaws in a 10-year-old boy from the Republic of the Congo. J Clin Microbiol. 2011;49:4013–4015. doi: 10.1128/JCM.01121-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centurion-Lara A, Castro C, Castillo R, Shaffer JM, Van Voorhis WC, Lukehart SA. The flanking region sequences of the 15-kDa lipoprotein gene differentiate pathogenic treponemes. J Infect Dis. 1998;177:1036–1040. doi: 10.1086/515247. [DOI] [PubMed] [Google Scholar]
- 8.Centurion-Lara A, Molini BJ, Godornes C, Sun E, Hevner K, Van Voorhis WC, Lukehart SA. Molecular differentiation of Treponema pallidum subspecies. J Clin Microbiol. 2006;44:3377–3380. doi: 10.1128/JCM.00784-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harper KN, Liu H, Ocampo PS, Steiner BM, Martin A, Levert K, Wang D, Sutton M, Armelagos GJ. The sequence of the acidic repeat protein (arp) gene differentiates venereal from nonvenereal Treponema pallidum subspecies, and the gene has evolved under strong positive selection in the subspecies that causes syphilis. FEMS Immunol Med Microbiol. 2008;53:322–332. doi: 10.1111/j.1574-695X.2008.00427.x. [DOI] [PubMed] [Google Scholar]
- 10.The World Health Organization Eradication of yaws - the Morges strategy. Wkly Epidemiol Rec. 2012;87:189–194. [PubMed] [Google Scholar]
- 11.Pillay A, Liu H, Ebrahim S, Chen CY, Lai W, Fehler G, Ballard RC, Steiner B, Sturm AW, Morse SA. Molecular typing of Treponema pallidum in South Africa: cross-sectional studies. J Clin Microbiol. 2002;40:256–258. doi: 10.1128/JCM.40.1.256-258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen CY, Chi KH, Pillay A, Nachamkin E, Su JR, Ballard RC. Detection of the A2058G and A2059G 23S rRNA gene point mutations associated with azithromycin resistance in Treponema pallidum by use of a TaqMan real-time multiplex PCR assay. J Clin Microbiol. 2013;51:908–913. doi: 10.1128/JCM.02770-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seña AC, Wolff M, Behets F, Van Damme K, Martin DH, Leone P, McNeil L, Hook EW. Response to therapy following retreatment of serofast early syphilis patients with benzathine penicillin. Clin Infect Dis. 2013;56:420–422. doi: 10.1093/cid/cis918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peel TN, Bhatti D, De Boer JC, Stratov I, Spelman DW. Chronic cutaneous ulcers secondary to Haemophilus ducreyi infection. Med J Aust. 2010;192:348–350. doi: 10.5694/j.1326-5377.2010.tb03537.x. [DOI] [PubMed] [Google Scholar]
- 15.McBride WJ, Hannah RC, Le Cornec GM, Bletchly C. Cutaneous chancroid in a visitor from Vanuatu. Australas J Dermatol. 2008;49:98–99. doi: 10.1111/j.1440-0960.2008.00439.x. [DOI] [PubMed] [Google Scholar]
- 16.Ussher JE, Wilson E, Campanella S, Taylor SL, Roberts SA. Haemophilus ducreyi causing chronic skin ulceration in children visiting Samoa. Clin Infect Dis. 2007;44:e85–e87. doi: 10.1086/515404. [DOI] [PubMed] [Google Scholar]
- 17.Mitjà O, Lukehart SA, Pokowas G, Moses P, Kapa A, Godornes C, Robson J, Cherian S, Houinei W, Kazadi W, Siba P, de Lazzari E, Bassat Q. Haemophilus ducreyi as a cause of skin ulcers in children from a yaws-endemic area of Papua New Guinea: a prospective cohort study. Lancet Global Health. 2014;2:e235–e241. doi: 10.1016/S2214-109X(14)70019-1. [DOI] [PubMed] [Google Scholar]
- 18.Cortes AL, Montiel ER, Gimeno IM. Validation of Marek's disease diagnosis and monitoring of Marek's disease vaccines from samples collected in FTA cards. Avian Dis. 2009;54:510–516. doi: 10.1637/8871-041009-Reg.1. [DOI] [PubMed] [Google Scholar]
- 19.Abdelwhaba EM, Lüschowa D, Harderb TC, Hafez HM. The use of FTA filter papers for diagnosis of avian influenza virus. J Virol Methods. 2011;174:120–122. doi: 10.1016/j.jviromet.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 20.Healthcare GE, Life Sciences FTA Elute Sample Collection Cards. 2014. http://www.gelifesciences.com/webapp/wcs/stores/servlet/catalog/en/GELifeSciences-us/products/AlternativeProductStructure_18503/ Available at. Accessed October 23, 2014.
- 21.Healthcare GE, Life Sciences Long-term stability of DNA in buccal and blood samples on FTA. 2014. http://www.gelifesciences.com/gehcls_images/GELS/Related%20Content/Files/1326706518989/litdoc29005583_20120725211651.pdf Available at. Accessed October 23, 2014.