Abstract
In this study, three molecular assays (real-time multiplex polymerase chain reaction [PCR], merozoite surface antigen gene [MSP]-multiplex PCR, and the PlasmoNex Multiplex PCR Kit) have been developed for diagnosis of Plasmodium species. In total, 52 microscopy-positive and 20 malaria-negative samples were used in this study. We found that real-time multiplex PCR was the most sensitive for detecting P. falciparum and P. knowlesi. The MSP-multiplex PCR assay and the PlasmoNex Multiplex PCR Kit were equally sensitive for diagnosing P. knowlesi infection, whereas the PlasmoNex Multiplex PCR Kit and real-time multiplex PCR showed similar sensitivity for detecting P. vivax. The three molecular assays displayed 100% specificity for detecting malaria samples. We observed no significant differences between MSP-multiplex PCR and the PlasmoNex multiplex PCR kit (McNemar's test: P = 0.1489). However, significant differences were observed comparing real-time multiplex PCR with the PlasmoNex Multiplex PCR Kit (McNemar's test: P = 0.0044) or real-time multiplex PCR with MSP-multiplex PCR (McNemar's test: P = 0.0012).
Introduction
Malaria is a devastating disease that affects people in many tropical and subtropical regions. The World Health Organization has estimated that 627,000 people died of malaria in 2012.1 Five Plasmodium species are known to cause malaria in humans: P. falciparum, P. vivax, P. malariae, P. ovale, and P. knowlesi. Among these species, P. falciparum causes the greatest morbidity and mortality in humans.2 Also, the simian malaria infecting humans, P. knowlesi, is known to produce rapid hyperparasitemia because of its ability to replicate every 24 hours, which can result in life-threatening complications and death.3
In endemic regions, individuals have been found to display simultaneous infections involving more than one human malaria species.4 These mixed infections, which are often unrecognized and underestimated, produce non-specific clinical manifestations of malaria that contribute to presumptive diagnosis and treatment of patients.5 For this reason, laboratory tools that are capable of accurately diagnosing all five human malaria species are essential for estimating clinical prognosis and monitoring therapeutic responses, particularly within geographical areas that harbor drug-resistant parasites.
Existing methods of malaria diagnosis include microscopy, rapid diagnostic tests (RDTs), and molecular tools.6 However, for more than a century, microscopy has been the gold standard for malaria diagnosis in endemic areas, because it is relatively inexpensive and allows for quantification of parasitemia. Nevertheless, this method requires highly experienced personnel to accurately diagnose and quantify malaria parasites. Indeed, two- to threefold discrepancies in parasite quantification can be observed between individuals. In fact, even experienced microscopists have been found to misdiagnose cases involving low parasitemia or mixed infections.7–9 Moreover, morphological similarities between P. knowlesi and P. malariae often result in misdiagnosis.10 Although RDTs are generally specific for detecting P. falciparum and P. vivax infections, they are non-specific for other malaria species.11 Additionally, RDTs are known to produce false-positive results because of residual antigen, which can persist for weeks after treatment and parasite clearance.12
Molecular methods, such as nested polymerase chain reaction (PCR) and real-time PCR, allow accurate species identification and are valuable for distinguishing species in cases of mixed malaria infection.13 In this regard, Snounou and others14 developed a nested PCR method, which targets the 18S ribosomal DNA (rDNA) gene and allows for the discrimination between distinct malaria species based on differentially sized PCR products. Although this method is widely used, it is expensive and laborious, because it requires separate PCR reactions for each human malaria species. Therefore, efforts have been made to develop single-reaction assays for the rapid and specific identification of all five human malaria species. In this regard, protocols for seminested multiplex15 and single-round multiplex PCR16 were recently established based on targeting a region of the 18S gene. Notably, in a recent report published by Mixson-Hayden and others,12 the seminested multiplex assay was found to display similar sensitivity to the nested assay for the detection of P. falciparum and P. vivax but more sensitivity for identifying P. malariae (0.04 parasites [p]/μL). Its detection threshold for P. ovale was found to be 4 p/μL. However, the detection limits for the multiplex single-round protocol were found to be 4 p/μL for P. ovale and 40 p/μL for P. malariae and P. vivax. In addition, a high-throughput multiplex 5′ nuclease quantitative PCR (qPCR) assay was recently developed by Reller and others17 for clinical diagnosis of human malaria species. This method was reported to be highly sensitive for all five Plasmodium species, showing a detection limit of 1–6 p/μL blood. Furthermore, the PlasmoNex Multiplex PCR Kit, which was commercially introduced in 2012, claims to detect all five human malaria species and requires a DNA concentration of at least 35 ng/μL. Nevertheless, preliminary results obtained in our laboratory have indicated that this kit shows low sensitivity for detecting P. knowlesi infection. In this study, we have evaluated the sensitivity and specificity of three molecular methods (the PlasmoNex Multiplex PCR Kit, in-house merozoite surface antigen gene (MSP) -multiplex PCR, and real-time multiplex PCR) using microscopy and nested PCR as gold standards.
Materials and Methods
Clinical samples.
Fifty-two malaria-positive blood samples were collected from the University Malaya Medical Center (UMMC) in Malaysia. These samples were evaluated using microscopy and nested PCR assay. In addition, 20 blood samples were collected from healthy donors. Ethical approval for this study was obtained from the Medical Ethics Committee of the UMMC (reference number 817.18).
Microscopic examination.
Thick and thin blood films were prepared and examined by skilled personnel who had extensive experience in identifying malaria parasites. Parasitemia was assessed per 1,000 erythrocytes in thin films and per 200 white blood cells (WBCs) in thick films. Films were considered as negative if no parasites were observed after counting 500 leukocytes.
Rapid diagnostic kit.
The BinaxNOW Malaria Kit (Inverness Medical International, United Kingdom) was used to analyze microscopy-confirmed cases. The manufacturer's protocol was, however, modified to use a drop of ethylenediaminetettraacetic acid (EDTA)-treated whole blood. Test results were analyzed after 15 minutes.
DNA extraction from whole blood.
For nested PCR and multiplex PCR, DNA was extracted from the blood samples of malaria-infected and non-infected patients using the DNeasy Blood and Tissue Kit based on the manufacturer's protocol (Qiagen, Hilden, Germany). Briefly, proteinase K (20 μL) was added to EDTA-treated whole blood (100 μL). Buffer AL (200 μL) and ethanol (200 μL) were subsequently added. The sample was transferred to a DNeasy spin column and centrifuged. The flow-through was discarded, and buffer AW1 (500 μL) was added. The column was centrifuged again, and the flow-through was discarded. This step was repeated using buffer AW2. The column was placed into a fresh tube and the DNA was eluted using buffer AE (50 μL). Finally, the purified DNA was stored at −20°C until further use.
Nested PCR assay.
Species identification for the malaria-infected samples was achieved through nested PCR, which targets the Plasmodium small subunit ribosomal RNA (ssrRNA) gene. The primers used for the assay have been previously described.10,18 The reaction mixture for the first PCR step included 4 μL DNA template, 250 nmol/L each primer (Primer for 18S Plasmodium sp.) (rPLU1 and rPLU5 in PCR buffer [50 mmol/L KCl, 10 mmol/L Tris-HCl]), 200 mmol/L each deoxyribonucleotide triphosphate (dNTP), 1.25 U Taq polymerase (iDNA, Singapore), and water (added to a final volume of 25 μL). The primary amplification conditions were as follows: 94°C for 4 minutes, 35 cycles of denaturation at 94°C for 30 seconds, annealing at 55°C for 1 minute, and elongation at 72°C for 1 minute, and final extension at 72°C for 4 minutes.
For each 20 μL secondary amplification, 4 μL first amplification product was used as the DNA template. The conditions and concentrations used in the secondary amplifications were identical to those used in the primary reactions, except for the annealing temperature (58°C) and the amount of Taq polymerase (0.5 U).
Real-time multiplex PCR.
The real-time multiplex PCR was performed using the C1000 Thermal Cycler with a CFX96 Real-Time PCR System (Bio-Rad, Hercules, CA). Reaction mixtures were prepared with 2.5 μL genomic DNA (gDNA), 200 nM each primer and probe, and 10 μL 2× iQ Multiplex Powermix (Bio-Rad) in a final volume of 20 μL. A standard two-step reaction was performed; an initial denaturation step at 95°C for 3 minutes was followed by 40 cycles of denaturation at 95°C for 10 seconds and annealing/extension at 55°C for 30 seconds. Real-time assay for human β-actin (ACTB) was performed separately.
PlasmoNex Multiplex PCR Kit.
For analysis using the PlasmoNex Multiplex PCR Kit (Reszon Diagnostics, Selangor, Malaysia), 1 μL diluted Taq polymerase (supplied with the kit) was added to each PCR tube, which contained PlasmoNex PCR mix. DNA was extracted from the blood sample (1.5 μL) and added to the PCR mix. PCR amplification was initiated at 95°C for 5 minutes followed by 35 cycles of denaturation at 95°C for 30 seconds, annealing at 58.5°C for 30 seconds, and elongation at 72°C for 40 seconds. Final extension was performed at 72°C for 5 minutes, after which the PCR products were analyzed by electrophoresis using 3% (wt/vol) agarose gels.
Primer design.
Based on GenBank sequence alignments, multiplex PCR primers were designed to specifically amplify a distinct region of the MSP for each of the human Plasmodium species (Table 1).
Table 1.
MSP-multiplex PCR primers
| Primers | Sequence (5′ to 3′) | Amplicon (bp) |
|---|---|---|
| P. knowlesi forward | GCACCCACAGAACCAGTAA | 1,069 |
| P. knowlesi reverse | TCCGTTGCATTCTCTGTG | |
| P. ovale forward | ACTCCAGCGACAGCACA | 850 |
| P. ovale reverse | GGAACATTACCTTGTGTAGCG | |
| P. vivax forward | GAAGACAAATTGCCCAACC | 677 |
| P. vivax reverse | CTTTGCCACTATATCTGCCG | |
| P. falciparum forward | AGGTGCAAGTGCTCAAAG | 508 |
| P. falciparum reverse | CGTCTAATTCATTTGCACG | |
| P. malariae forward | GGGTCAGATGACGAAGAT | 291 |
| P. malariae reverse | AGAGGCAGATGCTTCACTAC |
MSP-Multiplex PCR.
Initially, a monoplex PCR was carried out for each primer set to determine specificity, which was followed by rigorous optimization to develop our multiplex PCR detection assay. Briefly, multiplex PCR was performed in a 25 μL reaction mixture that contained 4 μL template DNA, 1× buffer, 0.4 μM each P. knowlesi, P. ovale, P. vivax, and P. falciparum primers, 0.8 μM P. malariae primers, 200 μM dNTP mix, 1.5 mM MgCl2, and 1 U Taq polymerase (Promega, Madison, WI). PCR amplification was initiated at 95°C for 6 minutes, which was followed by 30 cycles of denaturation at 95°C for 30 seconds, annealing at 53°C for 30 seconds, and elongation at 72°C for 2 minutes. Final extension was performed at 72°C for 5 minutes. The amplified PCR products were viewed on 1% (wt/vol) agarose gels, which were stained with SYBR Safe DNA Gel Stain (Invitrogen) and visualized with the Gel Doc system (Bio-Rad).
Analytical sensitivity of MSP-multiplex PCR.
The DNeasy Blood and Tissue Kit (Qiagen) was used to extract DNA from infected blood samples (100 μL), which were obtained from patients with 0.04% parasitemia (2 × 103 p/μL) for each species. The DNA was eluted in 50 μL sterile Tris-EDTA (TE) buffer. Therefore, 1 μL sample contained DNA equivalent to 4,000 parasites. The stock DNA was aliquoted and stored at −20°C, from which six 10-fold serial dilutions were prepared to achieve a final concentration equivalent to 0.004 p/μL. Assay detection limits were established for samples corresponding to individual Plasmodium species as well as mixed samples (i.e., DNA cocktails of multiple species), which were done in triplicate Ultimately, 1 μL each sample was analyzed by real-time PCR, MSP-multiplex PCR, and the PlasmoNex Multiplex PCR Kit.
Analytical specificity of MSP-multiplex PCR.
Specificity of all three assays was tested using gDNAs of all five species of human malaria.
Clinical sensitivity and specificity.
The clinical sensitivity and specificity of three assays was calculated using 72 whole-blood samples with microscopy and nested PCR as the reference method. Sensitivity was calculated as (number of true positives)/(number of true positives + number of false negatives), and specificity was calculated as (number of true negatives)/(number of true negatives + number of false positives).
Statistical analysis.
Results from the comparisons between MSP-multiplex PCR and the PlasmoNex Multiplex PCR Kit, between the PlasmoNex Multiplex PCR Kit and real-time PCR, and between MSP-multiplex PCR and real-time PCR assays are shown in a 2 × 2 table. Agreement of results between the two methods was assessed using Cohen's κ-test19 for concordance and McNemar's test for discordance.
Results
Our analysis revealed that the real-time multiplex PCR was more sensitive (81%) for detecting P. vivax, P. falciparum, and P. knowlesi infections compared with the PlasmoNex Multiplex PCR Kit (62%) and MSP-multiplex PCR (50%). Indeed, of 52 malaria samples that tested positive by both microscopy and nested PCR, 42 samples were correctly identified by real-time multiplex PCR. In contrast, the PlasmoNex Multiplex PCR Kit and MSP-multiplex PCR were able to accurately detect 32 and 25 samples, respectively.
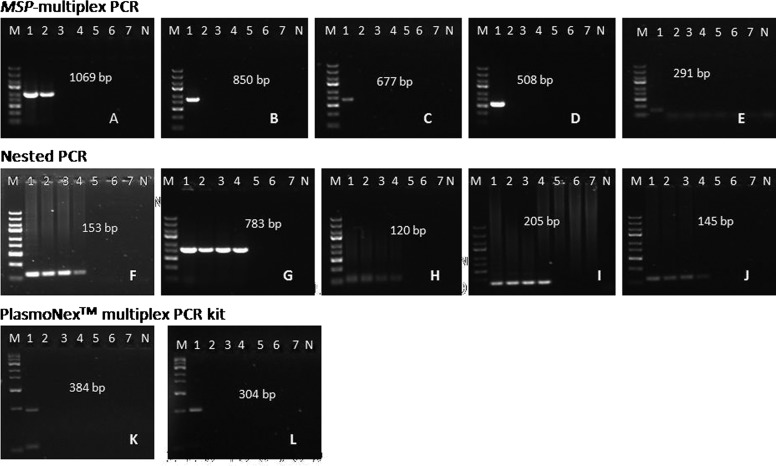
Although the detection limit of the MSP-multiplex PCR assay was more sensitive than that of the PlasmoNex Multiplex PCR Kit for detecting P. knowlesi and other Plasmodium species (Figure 1A), it was only capable of detecting down to 400 and 4,000 p/μL, respectively. In contrast, the nested PCR assay displayed a detection threshold of 4 p/μL for all Plasmodium species, constituting an approximately 1,000-fold difference in sensitivity (Figures 1F–J). However, the real-time multiplex PCR assay could be used to detect 1–6 p/μL. Because we relied on microscopic parasite counts for determining our initial species concentrations (in parasites per microliter), it is possible that these values did not directly correspond to known DNA quantities because of the presence of schizonts. Thus, this potential discrepancy might have affected our ability to replicate previously reported sensitivity results. Notably, all three assays showed 100% specificity, because we observed no amplification of other gDNA.
Figure 1.
Detection limits for the three multiplex assays based on 10-fold serial dilutions of five Plasmodium species from 4,000 p/μL stocks. Lane 1 = 4,000 p/μL; lane 2 = 400 p/μL; lane 3 = 40 p/μL; lane 4 = 4 p/μL; lane 5 = 0.4 p/μL; lane 6 = 0.04 p/μL; lane 7 = 0.004 p/μL. One microliter of each dilution was amplified using the various methods. Representative results are shown for (A–E) MSP-multiplex PCR assay, (F–J) nested PCR method, and (K and L) the PlasmoNex Multiplex PCR Kit. The nested PCR method amplified up to 4 p/μL for each Plasmodium species. The MSP-multiplex PCR assay amplified up to 400 p/μL for P. knowlesi and 4,000 p/μL for P. ovale, P. falciparum, P. vivax, and P. malariae. The PlasmoNex Multiplex PCR Kit amplified up to 4,000 p/μL for both P. knowlesi and P. ovale, whereas no amplification was observed for other Plasmodium species. The positive process control (109 bp) was used to validate the results. (A, F, and K) P. knowlesi. (B, G, and L) P. ovale. (C and H) P. vivax. (D and I) P. falciparum. (E and J) P. malariae. M = GeneRuler Express DNA Ladder (Thermo Scientific, Waltham, MA). N = negative control (distilled water).
Furthermore, we found that each of the multiplex assays was incapable of simultaneously amplifying all five Plasmodium species from mixed samples (4,000 p/μL each species). Therefore, these molecular tools were more effective when analyzing monoinfected samples, suggesting that primer competition might contribute to their diminished performance in heterogeneous samples. Therefore, in cases of suspected mixed infection, our in-house MSP-multiplex method could be performed using individual species-specific primers to enhance specificity. In terms of clinical sensitivity and specificity, real-time PCR detected all of the P. knowlesi-positive samples, whereas the PlasmoNex Multiplex PCR Kit and MSP-multiplex PCR only identified 9 of 15 specimens each. In addition, the MSP-multiplex PCR assay performed slightly better than real-time multiplex PCR and the PlasmoNex Multiplex PCR Kit with regard to P. ovale, with a detection limit of 5,000 p/μL. However, among 18 P. vivax-positive samples, MSP-multiplex PCR only detected 9 samples, whereas real-time PCR and the PlasmoNex Multiplex PCR Kit were each capable of identifying 14 samples. Furthermore, both the PlasmoNex Multiplex PCR Kit and MSP-multiplex PCR assay performed poorly in the detection of P. falciparum, identifying 9 and 7 of 17 total samples, respectively. In contrast, real-time multiplex PCR distinguished 13 of 17 P. falciparum samples. Nevertheless, all three methods were found to be 100% specific for detecting malaria infection (Table 2). Results from our comparison between the PlasmoNex Multiplex PCR Kit and MSP-multiplex PCR assay are presented in Table 3. Cohen's κ-value of 0.656 indicated good agreement between the two methods.20 Also, McNemar's test confirmed that there was no significant difference between the two methods (P = 0.1489). However, when the PlasmoNex Multiplex PCR Kit was compared with the real-time multiplex PCR assay (Table 4), a significant difference was observed (McNemar's test value: P = 0.0044), despite the fact that Cohen's κ-value (0.727) indicated good agreement. Comparing real-time PCR with MSP-multiplex PCR (Table 5), Cohen's κ-value revealed moderate agreement (Cohen's κ-value: 0.568), and the difference was statistically significant (P value = 0.0012).
Table 2.
Sensitivity and specificity of the PlasmoNex Multiplex PCR Kit, MSP-multiplex PCR, and real-time multiplex PCR
| Methods | P. falciparum | P. vivax | P. knowlesi | P. malariae | P. ovale | Healthy donor | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|---|---|
| Microscopy | ||||||||
| Positive | 17 | 18 | 15 | 1 | 1 | 0 | ||
| Negative | 0 | 0 | 0 | 0 | 0 | 20 | ||
| Nested PCR | ||||||||
| Positive | 17 | 18 | 15 | 1 | 1 | 0 | ||
| Negative | 0 | 0 | 0 | 0 | 0 | 20 | ||
| PlasmoNex Multiplex PCR Kit | ||||||||
| Positive | 9 | 14 | 9 | 0 | 0 | 0 | 62 | 100 |
| Negative | 8 | 4 | 6 | 1 | 1 | 20 | ||
| MSP-multiplex PCR | ||||||||
| Positive | 7 | 9 | 9 | 0 | 1 | 0 | 50 | 100 |
| Negative | 10 | 9 | 6 | 1 | 0 | 20 | ||
| Real-time PCR | ||||||||
| Positive | 13 | 14 | 15 | 0 | 0 | 0 | 81 | 100 |
| Negative | 4 | 4 | 0 | 1 | 1 | 20 | ||
Table 3.
Comparison between the PlasmoNex Multiplex PCR Kit and MSP-multiplex PCR
| MSP-multiplex PCR | PlasmoNex Multiplex PCR Kit | ||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 23 | 3 | 26 |
| Negative | 9 | 37 | 46 |
| Total | 32 | 40 | 72 |
Number of microscopy-positive samples = 52; Cohen's κ = 0.656; McNemar's test: P = 0.1489 (difference between the two methods is not statistically significant).
Table 4.
Comparison between PlasmoNex Multiplex PCR Kit and real-time multiplex PCR
| Real-time multiplex PCR | PlasmoNex Multiplex PCR Kit | ||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 32 | 10 | 42 |
| Negative | 0 | 30 | 30 |
| Total | 32 | 40 | 72 |
Number of microscopy-positive samples = 52; Cohen's κ = 0.727; McNemar's test: P = 0.0044 (difference between the two methods is considered to be very statistically significant).
Table 5.
Comparison between MSP-multiplex PCR and real-time multiplex PCR
| Real-time multiplex PCR | MSP-multiplex PCR | ||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 25 | 15 | 40 |
| Negative | 1 | 31 | 32 |
| Total | 26 | 46 | 72 |
Number of microscopy-positive samples = 52; Cohen's κ = 0.568; McNemar's test: P = 0.0012 (difference between the two methods is considered to be very statistically significant).
In addition, the cost of each assay, including the price of reagents and plastic consumables, was calculated based on a single sample. In this regard, the real-time multiplex PCR was found to be the most expensive ($14.80 US per sample) followed by the PlasmoNex Multiplex PCR Kit ($9.00 US per sample), nested PCR ($1.00 US per sample ), and MSP-multiplex assay ($0.50 US per sample). Finally, we considered the time required for each of the assays based on PCR cycles: real-time multiplex PCR (60 minutes), the PlasmoNex Multiplex PCR Kit (70 minutes), nested PCR (6 hours), and MSP-multiplex PCR (101 minutes).
Discussion
Although nested PCR represents the most consistent and accurate method for detecting low levels of malaria parasites, it is tedious and time-consuming. Therefore, the generation of rapid, highly sensitive/specific, cheap, and widely accessible detection assays is essential for effectively combating malaria transmission.21 In this regard, comparative studies evaluating available diagnostic assays are required to determine the most efficacious methods. Therefore, in this study, we have assessed three distinct molecular methods for Plasmodium detection, including an in-house multiplex PCR assay.
The development of novel assays for malaria detection is fundamental for several reasons. First, humans can harbor malaria parasites without displaying clinical manifestations, a phenomenon that is mainly attributed to partial immunity. Indeed, these subclinical or asymptomatic infections are prevalent in malaria-endemic regions, and diagnosing these cases can be challenging because of the lack of symptoms and low parasitemia.22 As a result, these subclinical cases are often missed by microscopy and/or RDTs, which are capable of detecting parasitemia of up to 50–100 p/μL. Furthermore, RDTs are only capable of specifically detecting P. falciparum and P. vivax and can detect other species in a panspecific manner. Although these malaria infections are subpatent, they remain transmissible, complicating attempts to eradicate malaria in endemic regions. In addition, malaria-infected patients can present with unusual clinical features in cases of immunity and/or antimalarial drug resistance.10 Nevertheless, there remains a lack of awareness concerning these atypical manifestations, which are often diagnosed late or not at all, ultimately resulting in severe complications or death. Thus, optimization of current techniques along with the development new technologies should ultimately yield rapid and sensitive detection assays that can facilitate diagnosis of subclinical cases in the field, thereby reducing malaria transmission and morbidity.
Second, it is known that mixed infections can produce non-specific clinical manifestations of malaria, which can contribute to presumptive diagnosis and treatment. Although these mixed infections are underreported because of limitations in detection methods,4,5,23,24 PCR-based techniques have verified that low-level mixed infections are actually quite common.24 Thus, assays that can specifically detect individual Plasmodium species within mixed samples should lead to improved patient management after diagnosis. Moreover, improved detection of mixed infections might enhance epidemiological studies regarding malaria transmission and prognosis.
In this study, we have compared the sensitivity of three multiplex PCR methods for malaria detection using conventional nested PCR and microscopy as gold standards. Our findings suggest that improved sensitivity might be achieved through optimization of primers and reaction conditions. Moreover, our investigation has indicated that care should be taken when considering the use of these molecular assays in clinical diagnostic settings because of the likelihood of false-negative results. Nevertheless, use of these assays may be acceptable in geographical regions with few circulating Plasmodium species (i.e., lower chance of mixed infection). However, nested PCR currently represents the most appropriate technique for detecting malaria in endemic areas containing a variety of Plasmodium species.
Notably, although alternative molecular methods using loop-mediated isothermal amplification (LAMP) have been reported,25–27 this technology has not been sufficiently evaluated in field conditions. Also, to date, multiplex LAMP assays remain to be developed. Thus, advancement of this technique could yield novel clinical diagnostic tools for field-based detection of malaria.
In conclusion, the development of sensitive, simple, and accessible molecular diagnostic tools, such as multiplex assays, is critical for combatting malaria. Our comparative study has shown that MSP-multiplex PCR assay and the PlasmoNex Multiplex PCR Kit display poor sensitivity compared with real-time multiplex PCR. Therefore, these two assays should not be used in diagnostic settings requiring more than 80% sensitivity. Future studies involving larger sample sizes may be needed to verify these findings. Nevertheless, our results suggest that additional optimization of real-time PCR sensitivity could enhance on-time and point-of-care diagnosis of malaria patients in endemic regions.
Footnotes
Financial support: The study was funded by High-Impact Research Grant UM-MOHE UM.C/625/1/HIR/MOHE/MED/16 and UM.C/625/1/HIR/MOHE/CHAN/14/3 from the Ministry of Higher Education Malaysia and University Malaya Research Grant RG233/10HTM.
Authors' addresses: Yee Ling Lau, Meng Yee Lai, Claudia N. Anthony, Phooi Yee Chang, Vanitha Palaeya, Mun Yik Fong, and Rohela Mahmud, Tropical Infectious Disease Research and Education Center (TIDREC), Department of Parasitology, Faculty of Medicine, University of Malaya, Kuala Lumpur, Malaysia, E-mails: lauyeeling@um.edu.my, mengylai11@yahoo.com, claudia.anthony88@gmail.com, phooiyee@gmail.com, caprirox@yahoo.com, fongny@um.edu.my, and rohela@ummc.edu.my.
References
- 1.World Health Organization . The World Malaria Report 2012. Geneva: World Health Organization; 2012. [Google Scholar]
- 2.Tahar R, Boudin C, Thiery I, Bourgouin C. Immune response of Anopheles gambiae to the early sporogonic stages of the human malaria parasite Plasmodium falciparum. EMBO J. 2002;21:6673–6680. doi: 10.1093/emboj/cdf664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barber BE, William T, Grigg MJ, Piera K, Yeo TW, Anstey NM. Evaluation of the sensitivity of a pLDH-based and an aldolase-based rapid diagnostic test for diagnosis of uncomplicated and severe malaria caused by PCR-confirmed Plasmodium knowlesi, Plasmodium falciparum and Plasmodium vivax. J Clin Microbiol. 2013;51:1118–1123. doi: 10.1128/JCM.03285-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zimmerman PA, Mehlotra RK, Kasehagen LJ, Kazura JW. Why do we need to know more about mixed Plasmodium species infections in humans? Trends Parasitol. 2008;20:440–447. doi: 10.1016/j.pt.2004.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayxay M, Pukrittayakamee S, Newton PN, White NJ. Mixed-species malaria infections in humans. Trends Parasitol. 2004;20:233–240. doi: 10.1016/j.pt.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Bronzan RN, McMorrow ML, Kachur SP. Diagnosis of malaria: challenges for clinicians in endemic and non-endemic regions. Mol Diagn Ther. 2008;12:299–306. doi: 10.1007/BF03256295. [DOI] [PubMed] [Google Scholar]
- 7.O'Meara WP, Barcus M, Wongsrichanalai C. Reader technique as a source of variability in determining malaria parasite density by microscopy. Malar J. 2006;5:118. doi: 10.1186/1475-2875-5-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanscheid T. Current strategies to avoid misdiagnosis of malaria. Clin Microbiol Infect. 2003;9:497–504. doi: 10.1046/j.1469-0691.2003.00640.x. [DOI] [PubMed] [Google Scholar]
- 9.Payne D. Use and limitations of light microscopy for diagnosing malaria at the primary health care level. Bull World Health Organ. 1988;66:621–626. [PMC free article] [PubMed] [Google Scholar]
- 10.Singh B, Kim Sung L, Matusop A, Radhakrishnan A, Shamsull SS, Cox-Singh J, Thomas A, Conway DJ. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363:1017–1024. doi: 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- 11.Shokoples SE, Ndao M, Kowalewska-Grochowska K, Yanow SK. Multiplexed real-time PCR assay for discrimination of Plasmodium species with improved sensitivity for mixed infections. J Clin Microbiol. 2009;47:975–980. doi: 10.1128/JCM.01858-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mixson-Hayden T, Lucchi NW, Udhayakumar V. Evaluation of three PCR-based diagnostic assays for detecting mixed Plasmodium infection. BMC Res Notes. 2010;3:88. doi: 10.1186/1756-0500-3-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mens P, Spieker N, Omar S, Heijnen M, Schalling H, Kager PA. Is molecular biology the best alternative for diagnosis of malaria to microscopy? A comparison between microscopy, antigen detection and molecular tests in rural Kenya and urban Tanzania. Trop Med Int Health. 2007;12:238–244. doi: 10.1111/j.1365-3156.2006.01779.x. [DOI] [PubMed] [Google Scholar]
- 14.Snounou GS, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, do Rosario VE, Thaithong S, Brown KN. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-b. [DOI] [PubMed] [Google Scholar]
- 15.Rubio JM, Benito A, Berzosa PJ, Roche J, Puente S, Subirats M, Lopez-Velez R, Garcia L, Alavar J. Usefulness of seminested multiplex PCR in surveillance of imported malaria in Spain. J Clin Microbiol. 1999;37:3260–3264. doi: 10.1128/jcm.37.10.3260-3264.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Padley D, Moody AH, Chiodini PL, Saldanha J. Use of a rapid, single round, multiplex PCR to detect malarial parasites and identify the species present. Ann Trop Med Parasitol. 2003;97:131–137. doi: 10.1179/000349803125002977. [DOI] [PubMed] [Google Scholar]
- 17.Reller ME, Chen WH, Dalton J, Lichay MA, Dumler JS. Multiplex 5′nuclease quantitative real-time PCR for clinical diagnosis of malaria and species level identification and epidemiologic evaluation of malaria-causing parasites, including Plasmodium knowlesi. J Clin Microbiol. 2013;51:2931–2938. doi: 10.1128/JCM.00958-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, Rahman HA. A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg. 1999;60:687–692. doi: 10.4269/ajtmh.1999.60.687. [DOI] [PubMed] [Google Scholar]
- 19.Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas. 1960;20:37–46. [Google Scholar]
- 20.Altman DG. Practical Statistics for Medical Research. London, United Kingdom: Chapman and Hall; 1991. [Google Scholar]
- 21.Vafa M, Troye-Blomberg M, Anchang J, Garcia A, Migot-Nabias F. Multiplicity of Plasmodium falciparum infection in asymptomatic children in Senegal: relation to transmission, age and erythrocyte variants. Malar J. 2008;7:17. doi: 10.1186/1475-2875-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bottius E, Guanzirolli A, Trape JF, Rogier C, Konate L, Druilhe P. Malaria: even more chronic in nature than previously thought; evidence for subpatent parasitaemia detectable by the polymerase chain reaction. Trans R Soc Trop Med Hyg. 1996;90:15–19. doi: 10.1016/s0035-9203(96)90463-0. [DOI] [PubMed] [Google Scholar]
- 23.Mueller I, Widmer S, Michel D, Maraga S, McNamara DT, Kiniboro B, Sie A, Smith TA, Zimmerman PA. High sensitivity dection of Plasmodium species reveals positive correlations between infections of different species, shifts in age distribution and reduced local variation in Papua New Guinea. Malar J. 2009;8:41. doi: 10.1186/1475-2875-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mueller I, Zimmerman PA, Reeder JC. Plasmodium malariae and Plasmodium ovale– the “bashful” malaria parasites. Trends Parasitol. 2007;23:278–283. doi: 10.1016/j.pt.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han ET, Watanabe R, Sattabongkot J, Khuntirat B, Sirichaisinthop J, Iriko H, Jin L, Takeo S, Tsuboi T. Detection of four Plasmodium species by genus and species- specific loop-mediated isothermal amplification for clinical diagnosis. J Clin Microbiol. 2007;45:2521–2528. doi: 10.1128/JCM.02117-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poon LL, Wong BW, Ma EH, Chan KH, Chow LM, Abeyewickreme W, Tangpukdee N, Yuen KY, Guan Y, Looareesuwan S, Pieris JS. Sensitive and inexpensive molecular test for falciparum malaria: detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clin Chem. 2006;52:303–306. doi: 10.1373/clinchem.2005.057901. [DOI] [PubMed] [Google Scholar]
- 27.Aonuma H, Suzuki M, Iseki H, Perera N, Nelson B, Igarashi I, Yagi T, Kanuka H, Fukumoto S. Rapid identification of Plasmodium-carrying mosquitoes using loop-mediated isothermal amplification. Biochem Biophys Res Commun. 2008;376:671–676. doi: 10.1016/j.bbrc.2008.09.061. [DOI] [PubMed] [Google Scholar]