Abstract
Cryptosporidium is an important diarrhea-associated pathogen, however the correlation between parasite burden and diarrhea severity remains unclear. We studied this relationship in 10 experimentally infected calves using immunofluorescence microscopy and real-time polymerase chain reaction (qPCR) (N = 124 fecal samples). The qPCR data were corrected for extraction/amplification efficiency and gene copy number to generate parasite counts. The qPCR and microscopic oocyst quantities exhibited significant correlation (R2 = 0.33, P < 0.05), however qPCR had increased sensitivity. Upon comparison with diarrhea severity scores (from 0 to 3), a PCR-based count of ≥ 2.6 × 105 parasites or an immunofluorescence microscopy count of ≥ 4.5 × 104 oocysts were discriminatory predictors of moderate-to-severe diarrhea (versus no-to-mild diarrhea), with accuracies and predictive values of 72–82%. In summary, a quantitative approach for Cryptosporidium can refine predictive power for diarrhea and appears useful for distinguishing clinical cryptosporidiosis versus subclinical infection.
Introduction
Cryptosporidium represents a major cause of infectious diarrhea worldwide affecting all age groups. Impact is most severe in children < 2 years of age where diarrheal incidence is highest,1,2 and Cryptosporidium was recently found to be a predictor of mortality in toddlers, with an observed hazard ratio of 2.3.3
Although it has been previously shown in experimental systems, both in humans and cattle, that there is a dose-dependent relationship between oocyst ingestion and diarrhea,4,5 the correlation between oocyst shedding/detection in feces and diarrheal severity remains unclear. Some data suggest a quantitative load of Cryptosporidium may correlate with disease severity.6 However, this information largely derives from acquired immunodeficiency syndrome (AIDS)-related cryptosporidiosis and has not been consistently seen in childhood diarrhea.7
Because diarrheal symptoms are non-specific and many pathogens may be responsible for a diarrheal episode, diagnosis requires laboratory testing. For Cryptosporidium this is generally achieved through microscopy involving acid-fast staining, fluorescent antibodies, or through enzyme immunoassays.8 Use of nucleic acid-based technologies is of increasing importance and several groups have reported successful real-time polymerase chain reaction (qPCR) systems for the detection of Cryptosporidium targeting genes such as 18S rRNA and COWP.9,10 In this study, we compared immunofluorescence microscopy and quantitative PCR to quantitatively detect Cryptosporidium in serially collected feces samples from 10 calves experimentally infected with Cryptosporidium parvum. We also compared our quantitative data to diarrhea severity scores obtained on each day.
Materials and Methods
Cryptosporidium parvum oocysts.
Oocysts used in the oral challenge were purified as previously described.11 Briefly, feces were collected from naturally infected 6- to 14-day-old calves from a commercial dairy operation. Fecal matter was processed by continuous-flow differential density flotation and stored at 4°C until needed. Oocyst DNA was genotyped as C. parvum by PCR.12 Before inoculation, oocysts were treated with 0.6% sodium hypochlorite, washed with phosphate buffered saline (PBS), quantified using a hemocytometer, and viability determined using a dye permeability assay as previously described.11,13,14 Oocysts used for dosing were at least 87% viable.
Calf enrollment and management.
Ten calves were purchased from a local dairy farm and enrolled in the study as they were born. Each birth was attended by study personnel, and were performed as clean catch as previously described.15 Briefly, calves were delivered onto single-use plastic sheets to prevent manure contamination and were transported individually to a Biosafety Level 2 facility. Calves experimentally infected in this study were distinct from those from which oocysts were obtained. Calves received an oral challenge of 1 × 105 C. parvum oocysts within the first 24 hours of life. Each dose was administered in a 5 mL suspension of oocysts by the rigid portion of an oroesophageal feeding tube, followed by 120 mL of water to ensure all of the oocyst suspension was delivered to the calf. A fecal sample was collected from each calf every 24 h after oral challenge, for a period of 21 days post challenge. Health status and fecal consistency was recorded daily in accordance with previous methods.16 Fecal consistency data were recorded on a scale of 0–3, with score increasing severity (0 = normal feces, 3 = severe diarrhea). All calves were housed in a BL-2 facility in individual concrete box stalls and calves were cared for in compliance with the Cornell University Institutional Animal Care and Use Committee (IACUC).
Immunofluorescent microscopy.
Immunofluorescence microscopy was performed using the MeriFluor Crypto/Giardia immunofluorescence antibody detection reagent from Meridian Diagnostics (Cincinnati, OH)17 using a modified procedure as previously described.5,18 Briefly, a 0.1 g portion of feces was mixed into 10 mL of PBS and then 100 μL of suspension was mixed with 5 μL MeriFluor and incubated in the dark at room temperature for at least 30 min and stored at 4°C until examination. During microscopy, 10.5 μL of incubated sample was examined under the 20× objective on a fluorescent microscope (460–490 wavelength, Olympus model BX41 [Olympus America Inc., Center Valley, PA]) to count the number of labeled oocysts. The obtained count was multiplied by 104 to yield the number of oocysts per gram of feces.
DNA extraction, qPCR, and quantification.
The DNA was extracted from calf fecal samples using the Qiagen QIAamp Mini Stool Kit (Qiagen, Valencia, CA) using a previously described protocol.9 Briefly, for each sample 200 mg of feces was lysed using buffer ASL before beating with 212–300 μM glass beads (Sigma, St. Louis, MO) for a total of 3 minutes. Lysed samples were then heated at 95°C for 5 minutes before proceeding according to manufacturer's instructions. All samples were spiked with 106 copies of Phocine herpesvirus (PhHV, used as an extraction/amplification control; a gift from Martin Schutten, Erasmus MC, Department of Virology, Rotterdam, The Netherlands) during the extraction process.
All qPCR was carried out in the Bio-Rad CFX-96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA). The primer-probe set used to detect Cryptosporidium is a TaqMan adaptation of those found in Stroup and others, 200619 with the probe labeled with HEX and BHQ2 at its 5′ and 3′ ends, respectively. The PhHV primer-probe set was as previously described,9 but with a probe labeled with Quasar 670 and BHQ2 at its 5′ and 3′ ends, respectively. All primers were obtained from Integrated DNA Technologies (IDT, Coralville, IA). The Cryptosporidium probe was obtained from Eurofins MWG Operon (Huntsville, AL) and the PhHV probe was obtained from BioSearch Technologies (Petaluma, CA). Each 25 μL reaction contained 12.5 μL 2× Bio-Rad iQ Multiplex Mastermix, 0.4 μM each of Cryptosporidium F- and R-primers, 0.2 μM Cryptosporidium probe, 0.8 μM each of PhHV F- and R-primers, 0.2 μM PhHV probe, and 3 μL of extracted DNA. Each qPCR run included a dilution series of known amounts of Cryptosporidium genomic DNA and PhHV (acting as both positive controls and standard curves) and a negative control (3 μL nuclease-free water used as sample).
All qPCR data were collected and analyzed using CFX Manager Software, ver. 3.0 (Bio-Rad), with fluorescence thresholds set manually. An analytical cutoff of 35 cycles was applied to all Cryptosporidium data; any sample whose amplification curve crossed the threshold at or above 35.0 cycles was considered negative. Subsequent analyses were carried out in Microsoft Excel 2010 (Microsoft Corp., Redmond, WA) and in IBM SPSS Statistics, version 21 (IBM, Armonk, NY). The Cryptosporidium Cq value was converted to a genome count by comparison to the standard curve, standardized to the amount of detected PhHV, and reported per gram of feces. Genome counts were converted to oocyst counts by dividing by 4 (because there are 4 nuclei per oocyst) to give a PCR-based parasite count per gram feces.
Results
Comparison of Cryptosporidium detection by microscopy and qPCR.
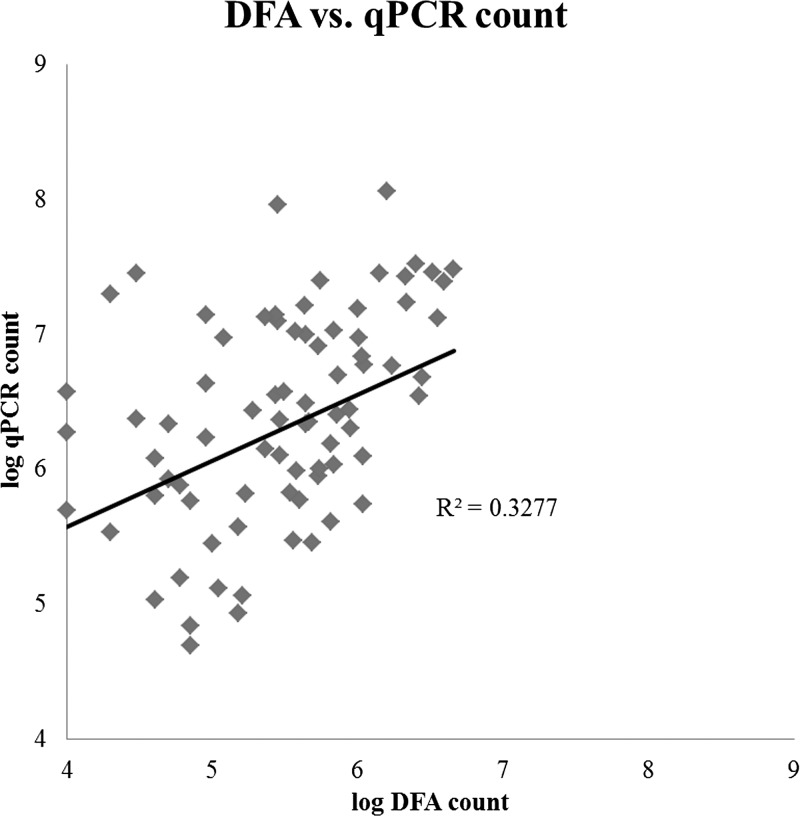
This study evaluated and compared direct fluorescent antibody microscopy (DFA) and a laboratory-developed real-time quantitative PCR assay for detection of Cryptosporidium. Overall, there was reasonable agreement between the two methods (Figure 1 , R2 = 0.33, P < 0.01). As shown in Table 1 , qPCR detected Cryptosporidium in more samples than did DFA. There was an expected quantitative relationship whereby DFA detected all higher burden specimens, whereas PCR additionally detected lower burden specimens, as revealed by qPCR Ct or qPCR-based parasite count. Overall the parasite count enumerated in each sample by PCR was higher than the oocyst count enumerated by DFA (Figure 1). For instance, on PCR/microscopy positive specimens, qPCR estimated a mean of 2.4 × 106 parasites per gram of feces, whereas DFA estimated 3.7 × 105 oocysts.
Figure 1.
Comparison of oocyst quantification by DFA and real-time polymerase chain reaction (qPCR). Oocyst counts obtained through immunofluorescence microscopy (DFA) and qPCR are shown for 78 individual samples (the R2 is similar whether applied to these 78 or all 124 fecal samples).
Table 1.
Comparison of Cryptosporidium detection between microscopy and qPCR
| DFA (+) | DFA (−) | |
|---|---|---|
| qPCR (+) | 78 | 29 |
| qPCR Ct (median) | 28 (IQR = 26.1–30.7) | 32 (IQR = 28.4–32.5) |
| qPCR parasite count (median) | 2.4 × 106 (IQR = 6.9 × 105–1.1 × 107) | 1.7 × 105 (IQR = 1.1 × 105–2.4 × 106) |
| DFA oocyst count (median) | 3.7 × 105 (IQR = 9.3 × 104–8.4 × 105) | n/a |
| qPCR (−) | 2 | 15 |
| qPCR Ct (median) | n/a | n/a |
| qPCR parasite count (median) | n/a | n/a |
| DFA oocyst count (range) | 1 × 104 – 4 × 104 | n/a |
DFA = direct fluorescent antibody; qPCR = real-time polymerase chain reaction; IQR = interquartile range.
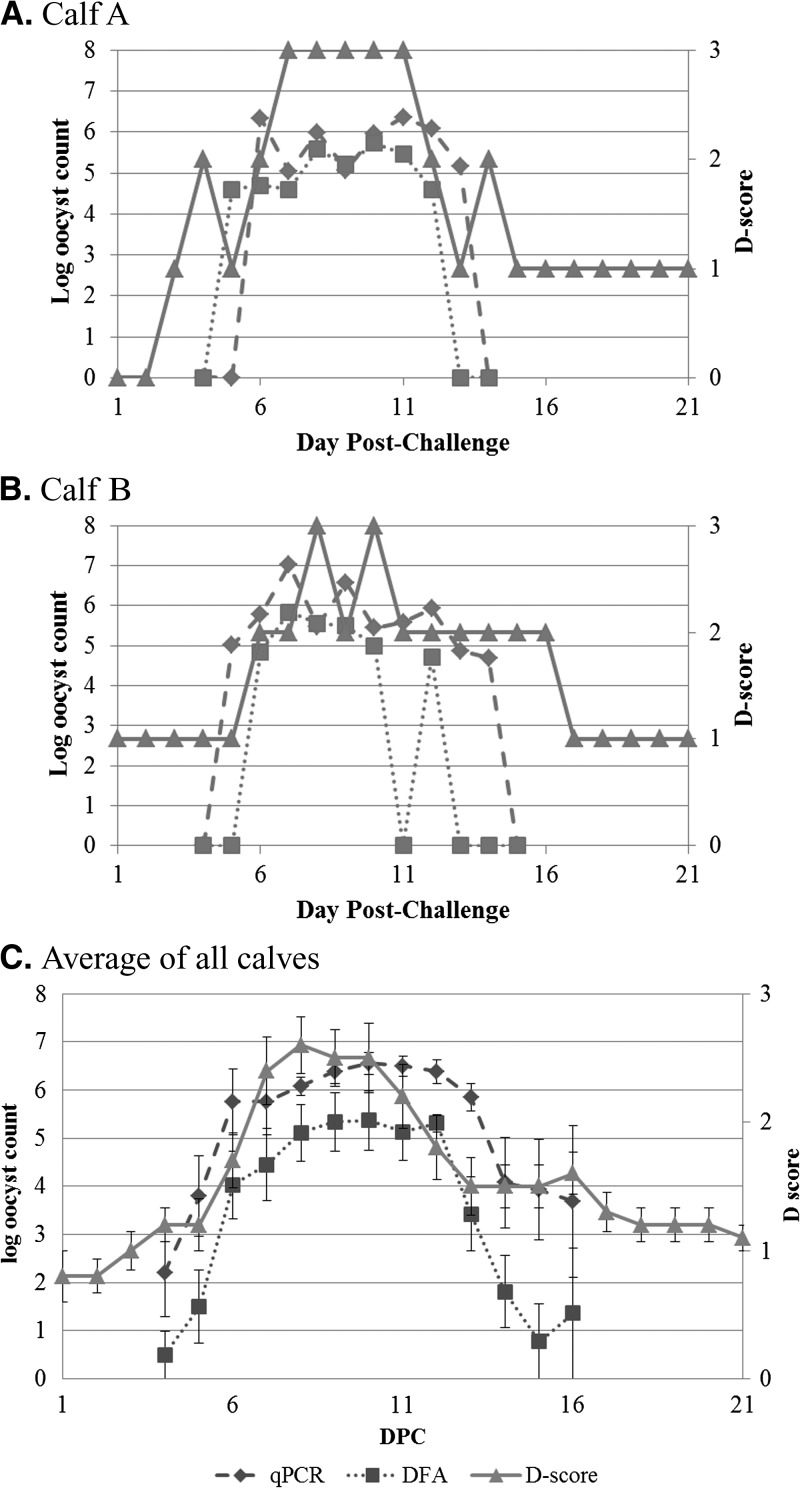
We then examined clinical scores. Fecal consistency was recorded daily using a standard 0–3 scoring system. Kinetics of diarrhea score and quantitative Cryptosporidium measurements were evaluated in each animal and in the aggregate (Figure 2
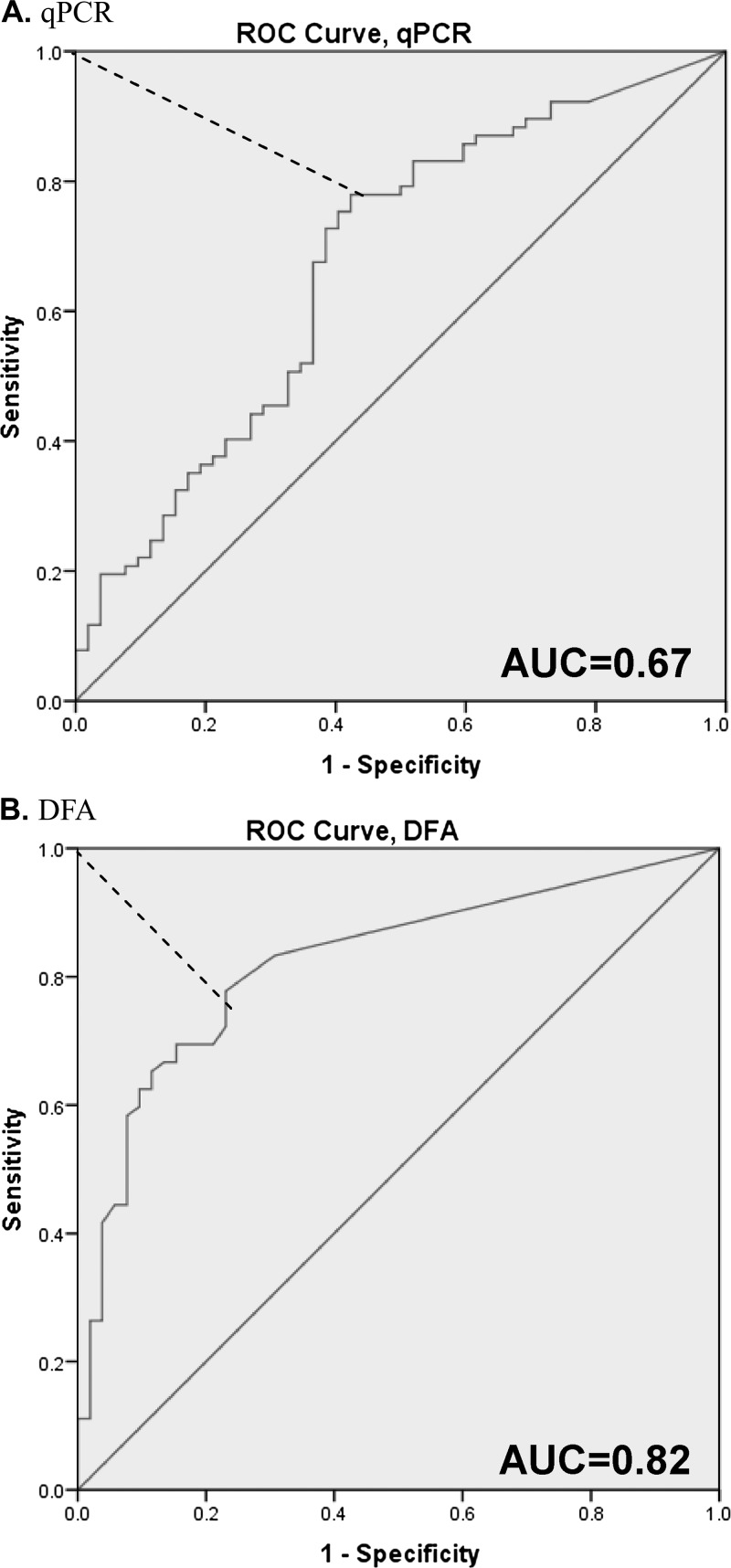
Figure 3.
(A) real-time polymerase chain reaction (qPCR). (B) Direct fluorescent antibody (DFA) receiver operating characteristic (ROC) analysis of qPCR and DFA versus diarrhea score. These ROC plots show sensitivity and (1-specificity) across oocyst counts, using the diarrhea score as a gold standard for positivity. AUC, area under the curve. The red dotted line depicts the Youden index of maximum (sensitivity + specificity-1) value.
). Overall the parasite curves tracked well with an increasing and decreasing diarrhea score. Specifically, the parasite score rose ∼1 day before the rise in diarrhea score, and fell ∼2 days after the decrease in diarrhea score. We analyzed parasite quantities as predictors of diarrhea (i.e., diarrhea scores of two and above). Without cutoffs, microscopy and qPCR detection yielded sensitivities for diarrhea (scores of 2–3 versus 0–1) of 84.7% and 93.1% and specificities of 63.5% and 23.1%, respectively. Using receiver operating characteristic (ROC) analysis and the Youden index (Figure 3 ), we identified that a count of 4.5 × 104 oocysts by microscopy and 2.6 × 105 parasites by qPCR could optimize the specificities to 76.9% and 53.9% for microscopy and qPCR, respectively, with little cost to sensitivity, corresponding to positive and negative predictive values of 82% and 71% for microscopy and 71% and 68% for qPCR, respectively (Table 2 ).
Figure 2.
(A) Calf A. (B) Calf B. (C) Average of all calves Oocyst count versus diarrhea score. Oocyst counts from both immunofluorescence microscopy (DFA, squares, dotted line) and real-time polymerase chain reaction (qPCR) (diamonds, dashed line) along with the diarrhea score (D-score, triangles, solid line) were plotted against the day post-challenge. Fecal data were available for all 10 calves at all time points except Days 15 and 16 where only 5–9 samples were available. Data shown as mean + standard error.
Figure 3.
(A) real-time polymerase chain reaction (qPCR). (B) Direct fluorescent antibody (DFA) receiver operating characteristic (ROC) analysis of qPCR and DFA versus diarrhea score. These ROC plots show sensitivity and (1-specificity) across oocyst counts, using the diarrhea score as a gold standard for positivity. AUC, area under the curve. The red dotted line depicts the Youden index of maximum (sensitivity + specificity-1) value.
Table 2.
Diagnostic characteristics of PCR and DFA, post-ROC analysis
| D-score 2,3 | D-score 1 | Sensitivity/specificity, % | PPV/NPV, % | |
|---|---|---|---|---|
| qPCR ≥ 2.6 × 105 | 59 | 24 | 82/54 | 71/68 |
| qPCR < 2.6 × 105 | 13 | 28 | ||
| DFA ≥ 4.5 × 104 | 56 | 12 | 78/77 | 82/71 |
| DFA < 4.5 × 104 | 16 | 40 | ||
PCR = polymerase chain reaction; DFA = direct fluorescent antibody; ROC = receiver operating characteristic; PPV = positive predictive value; NPV = negative predictive value.
Discussion
Cryptosporidium is now recognized as a major contributor to diarrheal disease worldwide. The results from a recent global study found Cryptosporidium to be among the top four most attributable pathogens to severe diarrhea, along with rotavirus, enterotoxigenic Escherichia coli and Shigella.3 This study also found a high rate of mixed enteropathogen infection, with two or more pathogens detected in 45% of cases. This raises important questions as to whether one pathogen is contributing entirely, partially, or not at all to a given diarrheal episode. Hence, this study was undertaken to determine whether symptoms could be predicted on the basis of Cryptosporidium quantity, not just detection.
First, we found that Cryptosporidium could be accurately quantified by either DFA microscopy or qPCR, with greater sensitivity and higher parasite estimations with the latter. This was expected, because increased sensitivity through PCR has been noted frequently for Cryptosporidium20,21 and other enteric pathogens.22 Both methods, however, showed consistent trends between Cryptosporidium quantity, kinetics, and associated diarrhea. In this study, we found that a quantitative cutoff of 4.5 × 104 oocysts by microscopy or 2.6 × 105 parasites by qPCR (or simply a Cryptosporidium Cq of 31.4, data not shown) would predict diarrhea with optimized accuracy. Of course, we would not expect perfect sensitivity and specificity, because the curves clearly showed increased numbers of oocysts both before and after the symptomatic period, and the advantage to the experimental calf system is that timing was controlled and no other confounding pathogens were likely present. In fact, these calves tested negative for coronavirus, rotavirus, and Salmonella (data not shown). Either DFA or qPCR appear useful for this purpose. Our preference is qPCR because of the ability to test for other pathogens as well.
That said, whether these cutoffs obtained in calves are applicable in the setting of childhood diarrhea is unknown. We propose that such studies would be useful, with close daily measurements of symptoms and parasite. If a similarly tight correlation between parasite load and symptoms is seen, we would advocate such a quantitative approach as a clinical diagnostic, particularly in resource-limited settings where asymptomatic carriage is common.23 Additionally, Cryptosporidium has limited treatment options and no approved agents exist for the first year of life where incidence is highest.3 New therapeutic options are needed, and we believe use of such a quantitative approach will be useful to screen candidates, either in the calf model or in children, because perhaps an agent that reduces burden by even a few logs (e.g., from 106 to 104) could have substantial efficacy.
Footnotes
Authors' addresses: Darwin J. Operario and Eric R. Houpt, Division of Infectious Diseases and International Health, Department of Medicine, University of Virginia, Charlottesville, VA, E-mails: do2s@virginia.edu and erh6k@virginia.edu. Lauren S. Bristol, Janice Liotta, and Daryl V. Nydam, Department of Population Medicine and Diagnostic Science, College of Veterinary Medicine, Cornell University, Ithaca, NY, E-mails: lls92@cornell.edu, jll55@cornell.edu, and dvn2@cornell.edu.
References
- 1.Chalmers RM, Davies AP. Minireview: clinical cryptosporidiosis. Exp Parasitol. 2010;124:138–146. doi: 10.1016/j.exppara.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Bern C, Martines J, de Zoysa I, Glass RI. The magnitude of the global problem of diarrheal disease: a ten-year update. Bull World Health Organ. 1992;70:705–714. [PMC free article] [PubMed] [Google Scholar]
- 3.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acacio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. Burden and aetiology of diarrheal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 4.Chappell CL, Okhuysen PC, Langer-Curry R, Widmer G, Akiyoshi DE, Tanriverdi S, Tzipori S. Cryptosporidium hominis: experimental challenge of healthy adults. Am J Trop Med Hyg. 2006;75:851–857. [PubMed] [Google Scholar]
- 5.Zambriski JA, Nydam DV, Wilcox ZJ, Bowman DD, Mohammed HO, Liotta JL. Cryptosporidium parvum: determination of ID(5)(0) and the dose-response relationship in experimentally challenged dairy calves. Vet Parasitol. 2013;197:104–112. doi: 10.1016/j.vetpar.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodgame RW, Kimball K, Ou CN, White AC, Jr, Genta RM, Lifschitz CH, Chappell CL. Intestinal function and injury in acquired immunodeficiency syndrome-related cryptosporidiosis. Gastroenterology. 1995;108:1075–1082. doi: 10.1016/0016-5085(95)90205-8. [DOI] [PubMed] [Google Scholar]
- 7.Haque R, Mondal D, Karim A, Molla IH, Rahim A, Faruque AS, Ahmad N, Kirkpatrick BD, Houpt E, Snider C, Petri WA., Jr Prospective case-control study of the association between common enteric protozoal parasites and diarrhea in Bangladesh. Clin Infect Dis. 2009;48:1191–1197. doi: 10.1086/597580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chalmers RM, Katzer F. Looking for Cryptosporidium: the application of advances in detection and diagnosis. Trends Parasitol. 2013;29:237–251. doi: 10.1016/j.pt.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Gratz J, Amour C, Kibiki G, Becker S, Janaki L, Verweij JJ, Taniuchi M, Sobuz SU, Haque R, Haverstick DM, Houpt ER. A laboratory-developed TaqMan Array Card for simultaneous detection of 19 enteropathogens. J Clin Microbiol. 2013;51:472–480. doi: 10.1128/JCM.02658-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taniuchi M, Verweij JJ, Noor Z, Sobuz SU, Lieshout L, Petri WA, Jr, Haque R, Houpt ER. High throughput multiplex PCR and probe-based detection with Luminex beads for seven intestinal parasites. Am J Trop Med Hyg. 2011;84:332–337. doi: 10.4269/ajtmh.2011.10-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenkins MB, Anguish LJ, Bowman DD, Walker MJ, Ghiorse WC. Assessment of a dye permeability assay for determination of inactivation rates of Cryptosporidium parvum oocysts. Appl Environ Microbiol. 1997;63:3844–3850. doi: 10.1128/aem.63.10.3844-3850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang J, Alderisio KA, Xiao L. Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl Environ Microbiol. 2005;71:4446–4454. doi: 10.1128/AEM.71.8.4446-4454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anguish LJ, Ghiorse WC. Computer-assisted laser scanning and video microscopy for analysis of Cryptosporidium parvum oocysts in soil, sediment, and feces. Appl Environ Microbiol. 1997;63:724–733. doi: 10.1128/aem.63.2.724-733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell AT, Robertson LJ, Smith HV. Viability of Cryptosporidium parvum oocysts: correlation of in vitro excystation with inclusion or exclusion of fluorogenic vital dyes. Appl Environ Microbiol. 1992;58:3488–3493. doi: 10.1128/aem.58.11.3488-3493.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ollivett TL, Nydam DV, Linden TC, Bowman DD, Van Amburgh ME. Effect of nutritional plane on health and performance in dairy calves after experimental infection with Cryptosporidium parvum. J Am Vet Med Assoc. 2012;241:1514–1520. doi: 10.2460/javma.241.11.1514. [DOI] [PubMed] [Google Scholar]
- 16.Bellosa ML, Nydam DV, Liotta JL, Zambriski JA, Linden TC, Bowman DD. A comparison of fecal percent dry matter and number of Cryptosporidium parvum oocysts shed to observational fecal consistency scoring in dairy calves. J Parasitol. 2011;97:349–351. doi: 10.1645/GE-2475.1. [DOI] [PubMed] [Google Scholar]
- 17.Xiao L, Herd RP. Quantitation of Giardia cysts and Cryptosporidium oocysts in fecal samples by direct immunofluorescence assay. J Clin Microbiol. 1993;31:2944–2946. doi: 10.1128/jcm.31.11.2944-2946.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burton AJ, Nydam DV, Jones G, Zambriski JA, Linden TC, Cox G, Davis R, Brown A, Bowman DD. Antibody responses following administration of a Cryptosporidium parvum rCP15/60 vaccine to pregnant cattle. Vet Parasitol. 2011;175:178–181. doi: 10.1016/j.vetpar.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Stroup SE, Roy S, McHele J, Maro V, Ntabaguzi S, Siddique A, Kang G, Guerrant RL, Kirkpatrick BD, Fayer R, Herbein J, Ward H, Haque R, Houpt ER. Real-time PCR detection and speciation of Cryptosporidium infection using Scorpion probes. J Med Microbiol. 2006;55:1217–1222. doi: 10.1099/jmm.0.46678-0. [DOI] [PubMed] [Google Scholar]
- 20.De Waele V, Berzano M, Berkvens D, Speybroeck N, Lowery C, Mulcahy GM, Murphy TM. Age-stratified Bayesian analysis to estimate sensitivity and specificity of four diagnostic tests for detection of Cryptosporidium oocysts in neonatal calves. J Clin Microbiol. 2011;49:76–84. doi: 10.1128/JCM.01424-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geurden T, Berkvens D, Geldhof P, Vercruysse J, Claerebout E. A Bayesian approach for the evaluation of six diagnostic assays and the estimation of Cryptosporidium prevalence in dairy calves. Vet Res. 2006;37:671–682. doi: 10.1051/vetres:2006029. [DOI] [PubMed] [Google Scholar]
- 22.Amar CF, East CL, Gray J, Iturriza-Gomara M, Maclure EA, McLauchlin J. Detection by PCR of eight groups of enteric pathogens in 4,627 fecal samples: re-examination of the English case-control Infectious Intestinal Disease Study (1993–1996) Eur J Clin Microbiol Infect Dis. 2007;26:311–323. doi: 10.1007/s10096-007-0290-8. [DOI] [PubMed] [Google Scholar]
- 23.Taniuchi M, Sobuz SU, Begum S, Platts-Mills JA, Liu J, Yang Z, Wang XQ, Petri WA, Jr, Haque R, Houpt ER. Etiology of diarrhea in Bangladeshi infants in the first year of life analyzed using molecular methods. J Infect Dis. 2013;208:1794–1802. doi: 10.1093/infdis/jit507. [DOI] [PMC free article] [PubMed] [Google Scholar]