Abstract
Parasite antigen diversity poses an obstacle to developing an effective malaria vaccine. A protein microarray containing Plasmodium falciparum apical membrane antigen 1 (AMA1, n = 57) and merozoite surface protein 1 19-kD (MSP119, n = 10) variants prevalent at a malaria vaccine testing site in Bandiagara, Mali, was used to assess changes in seroreactivity caused by seasonal and lifetime exposure to malaria. Malian adults had significantly higher magnitude and breadth of seroreactivity to variants of both antigens than did Malian children. Seroreactivity increased over the course of the malaria season in children and adults, but the difference was more dramatic in children. These results help to validate diversity-covering protein microarrays as a promising tool for measuring the breadth of antibody responses to highly variant proteins, and demonstrate the potential of this new tool to help guide the development of malaria vaccines with strain-transcending efficacy.
Plasmodium falciparum uses antigenic variation to evade host immunity, including vaccine-induced immunity.1,2 The protective immunity against P. falciparum malaria disease that is acquired after years of exposure is believed to reflect, in part, the gradual acquisition of allele-specific immune responses against a repertoire of parasite antigens, and, importantly, against diverse variants of these antigens.3 Apical membrane antigen 1 (AMA1) and merozoite surface protein 1 (MSP1) are immunogenic parasite surface antigens considered vital for erythrocyte attachment and invasion. Antibodies against these proteins are associated with protection against malaria, and several subunit AMA1 and MSP1 vaccines have been developed and tested.1,4–6 Both AMA1 and MSP119, the C-terminus portion of the MSP1 protein used in subunit vaccines, are highly variable, with sequence diversity encoded by single nucleotide polymorphisms.7,8 The highest prevalence of any one of the 440 AMA1 haplotypes observed from sequencing the ama1 gene among more than 1,400 field infections diagnosed during prospective incidence studies and vaccine trials was less than 4%.1,8,9
Although less extreme, MSP119 also has substantial diversity, with 17 haplotypes detected among more than 1,300 infections.7 Because vaccines that include dozens or hundreds of antigen variants are not feasible, malaria vaccine development would be aided by the identification of a manageable number of serodominant, cross-protective haplotypes among AMA1 and MSP119 variants. Developing such strain-transcending vaccines is hindered by the limited availability of diverse parasite antigens and the low throughput of standard assays such as enzyme-linked immunosorbent assay for measuring antibody responses.
Protein microarrays offer the possibility of overcoming these obstacles to enable high throughput evaluations of seroreactivity to large numbers of antigen variants. Plasmodium falciparum proteins expressed in a cell-free Escherichia coli translation system and spotted onto microarrays are recognized by serum antibodies of persons exposed to malaria.10–12 To date, this platform has been used to measure seroreactivity to large numbers of proteins derived from the genome of the P. falciparum reference strain 3D7.10,12–14 To examine antibody responses to diverse variants of highly polymorphic P. falciparum antigens, we amplified, expressed and printed dozens of field-derived variants of AMA1 and MSP119 on a protein microarray.
Antigen genes were amplified by polymerase chain reaction from DNA extracted from dried blood spots collected in a cohort study and an AMA1 vaccine trial in Mali, Africa. The resulting amplicons were analyzed by using the BigDye® Terminator v3.1 Cycle Direct Sequencing Kit (Applied Biosciences, Foster City, CA).4,8,15 Sequence contigs were compiled by using Sequencher software (Gene Codes, Ann Arbor, MI).9,16 Sixty of the most prevalent AMA1 ectodomain haplotypes and 10 MSP119 haplotypes were selected for inclusion on the prototype protein microarray. Microarray construction has been described.17
Serum samples were obtained from 18 adults 18–55 years of age enrolled in the control arm of a phase 1 trial of an AMA1-based vaccine conducted during 2004–2005 in Bandiagara, Mali.18 Similarly, serum samples were randomly obtained from 24 children 1–6 years of age enrolled in the control arm of a phase 1 trial of an AMA1-based malaria vaccine during 2006–2007 at the same site.6,19 Bandiagara, Mali has high seasonal malaria transmission coinciding with the June–November rainy season, with entomologic inoculation rates of 50–150 infected bites/person/season.8 For each participant, paired serum samples from two time points corresponding to the pre-malaria (May–June) and post-malaria (December–January) seasons were probed in random order to eliminate batch and slide effects.
Two microliters of serum from individual participant samples was diluted 1:200 with 10% E. coli lysate in blocking buffer and hybridized onto separate protein arrays overnight. Arrays were stained, washed, and scanned by using a Perkin-Elmer (Waltham, MA) ScanArray Express HT microarray scanner. Probing protocols have been described.10,12,17 Fluorescence was quantified by using the ScanArray Express Suite (Perkin-Elmer). Median fluorescent intensities (MFIs) were calculated by using an adaptive capture feature to account for varying spot size, and the per-spot local background was subtracted.
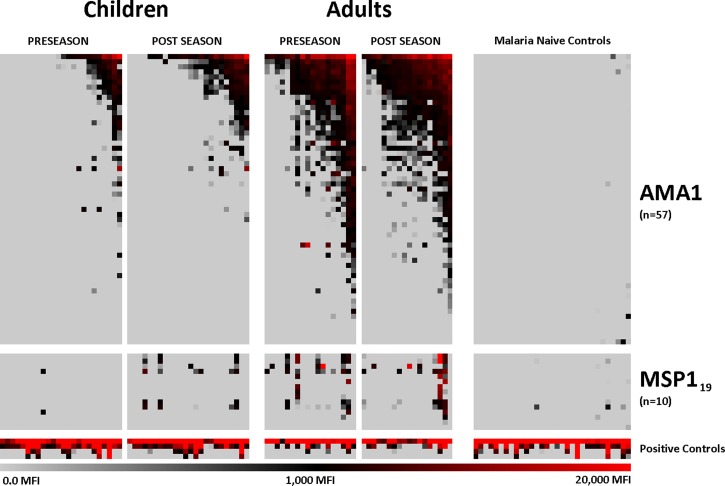
Raw MFIs were asinh-transformed to convert the MFI values to a Gaussian (normal) distribution, and biological variance contributed by seroreactivity to the E. coli components included in the translation protocol was subtracted by taking the average of eight empty vector, translated, negative controls (i.e., the no DNA controls). Residual non-biological variance between slides, batches, and individual arrays was corrected by using robust linear modeling (RLM) with respect to the negative and positive controls.20 Asinh-transformed, RLM-scaled data were reverted to fluorescent intensities by sinh reverse-transformation and plotted on a heat map to show global trends (Figure 1).
Figure 1.
Heat map of seroreactivity to Plasmodium falciparum apical membrane antigen 1 (AMA1) and merozoite surface protein 1–19 (MSP119) variants. Each column represents serum from an individual Malian child or adult, or a malaria-naive North American adult, and each row represents an antigen variant. Gray, black, and red indicate low, moderate and high seroreactivity, respectively, to probed antigens. Four serial dilutions of IgG peptide were printed and probed as positive probing controls and used as a standardization parameter for robust linear model scaling as a measurement of total IgG. MFI = median fluorescent intensity.
Parametric statistical hypotheses were tested on asinh-transformed, RLM-scaled data by using BioconductR (http://www.bioconductor.org/index.html, Excel (Microsoft, Redmond, WA), and SAS/STAT software (SAS Institute, Cary, NC). The magnitude of seroreactivity was measured by mean MFI using matched pair Student's t tests (Table 1). Breadth of seroreactivity was calculated by counting the number of variants recognized by each serum sample at each study time point that were 2 SD above the average of 31 North American malaria-naive controls for each antigen variant, and compared using Poisson generalized linear regression analysis. One putatively malaria-naive control was excluded from the analysis as an extreme outlier. Four different protocols were compared with maximize signal to noise ratios. Six AMA1 variants that did not show any seroreactivity above background in any cohort were excluded from statistical analyses.
Table 1.
Seroreactivity to recombinant Plasmodium falciparum apical membrane antigen 1 and merozoite surface protein 1 C-terminal 19-kD variants by protein microarray*
| Antigen | Cohort | Mean MFI (95% CI) | Serorecognition (%, 95% CI) | ||
|---|---|---|---|---|---|
| Pre-season | Post-season | Pre-season | Post-season | ||
| AMA1 | Children | 296 (226–365) | 434 (348–522) | 13.1 (5.8–20.4) | 18.6 (11.1– 26.1) |
| Adults | 1,506 (1,324–1,687) | 1,428 (1,271–1,585) | 49.3 (37.0–61.6) | 53.2 (42.7–63.7) | |
| Controls | 8.78 (5.22–12.4) | – | – | – | |
| MSP119 | Children | 20.9 (4.40–37.5) | 191 (110–273) | 2.5 (–0.9 to 5.9) | 9.4 (3.5–15.4) |
| Adults | 627 (345–909) | 940 (474–1,406) | 21.1 (11.4–30.9) | 18.9 (6.3–31.4) | |
| Controls | 21.4 (16.8–26.05) | – | – | – | |
Serum samples from Malian children and adult were considered positive for a given antigen variant if the mean MFI was 2 SD above the average of 30 malaria-naive North American controls. Serorecognition is given as the average percentage of AMA1 and MSP119 variants recognized by each cohort at each time point.
MFI = median fluorescence intensity; CI = confidence interval; AMA1 = apical membrane antigen 1; MSP119 = merozoite surface protein 1 C-terminal 19 kD.
Seroreactivity profiles to AMA1 and MSP119 variants were compared for Malian adults, Malian children, and malaria-naive North American adults. At the beginning of the transmission season, Malian adult serum samples had significantly higher magnitude and breadth of seroreactivity to AMA1 and MSP119 variants than Malian pediatric serum samples and North American malaria-naive control serum samples (P < 0.001, for all comparisons). Post-season serum samples had a higher magnitude and breadth of AMA1 reactivity than pre-season serum samples from Malian children (P < 0.01 for both comparisons). The magnitude and breadth of antibody seroreactivity to MSP119 variants was significantly higher in Malian children at the post-season time point compared with pre-season (P < 0.001). No seasonal difference was observed in seroreactivity to MSP119 variants for adults in magnitude or breadth of antibody response. Assessments of significance were calculated using matched Student's t test for differences in magnitude data and Poisson regression analysis for the comparison of antibody breadth.
Multiplicity of infection is a concern when working with field samples from an area where 64–87% of malaria infections in adults are multiple clone infections with respect to the ama1 haplotype.21 Among 600 malaria infections, 21% were polyclonal with respect to ama1, making haplotype determination impossible.15 We used single-colony isolation to sequence clonal AMA1 and MSP119 plasmid-containing E. coli and evaluated a kit to translate proteins with disulfide bonds (RTSS) (5 Prime, Hilden, Germany). Single-colony-extracted proteins with disulfide bonds had significantly higher signal-to-noise ratios than any other preparation protocol, despite the increased background seroreactivity of negative controls.
Despite using antigen variants and serum samples from the same site, six variants on the array were unrecognized above background at any time point. This lack of seroreactivity could represent allele specificity, or alternatively incorrect folding of reactive epitopes in these particular variants during translation. We also detected eight AMA1 variants that were recognized by most serum samples from adults.
A previous study in southern Mali found no differences in seroreactivity to AMA1 and MSP1 in children who had none versus one or more clinical malaria episodes, but this analysis used a protein microarray with the P. falciparum reference proteome 3D7.12 The lack of association could be because the parasites causing illness had AMA1 and MSP1 variants distinct from those in the reference genome. In contrast, our diversity-covering protein array makes it possible for the first time to study the strain-specificity of antibody responses in a high throughput fashion.
In the 3D7 protein microarray study, serum from persons exposed to malaria had higher reactivity against a full-length MSP1 protein than against AMA1, whereas the seroreactivity to MSP119 in this study was significantly less than to AMA1.12 This finding may be explained by the relative sizes of the protein fragments. In this study, we translated the entire AMA1 ectodomain, but used only the 19-kD post-cleavage fragment of MSP1 that serves as a vaccine antigen. We will compare seroreactivity to different-sized fragments of MSP1 and AMA1 in subsequent studies.
Protein microarrays have limitations. They cannot measure antibody function, and seroreactivity to any one of the large number of antigenic variants on an array does not provide the level of confidence obtained from results of an enzyme-linked immunosorbent assay that measures antibody responses to a protein that has been shown to be correctly folded. Nevertheless, this prototype array represents an enabling technology that will make it possible to evaluate antibody responses to large numbers of antigenic variants among large numbers of individual serum samples to assess immunologic responses in relation to important phenotypes, such as clinical immunity or vaccine-induced protection at a population level. This study was not powered to correlate antibody reactivity to clinical outcomes. The next iteration of the array will include many more variants of AMA1 and other antigens, and will be used to explore the role of allele-specific antibody responses in naturally acquired and vaccine-induced immune protection. Next-generation P. falciparum protein arrays will be used to study allele-specific naturally acquired and vaccine-induced immunity and to inform the design of broadly efficacious strain-transcending malaria vaccines.
Footnotes
Authors' addresses: Jason A. Bailey, Mark A. Travassos, Amed Ouattara, Matthew B. Laurens, Shannon L. Takala-Harrison, Kirsten E. Lyke, and Christopher V. Plowe, Center for Vaccine Development, University of Maryland School of Medicine, Baltimore, MD, E-mails: jbail006@umaryland.edu, aouattara@medicine.umaryland.edu, mlaurens@medicine.umaryland.edu, stakala@medicine.umaryland.edu, klyke@medicine.umaryland.edu, and cplowe@medicine.umaryland.edu. Jozelyn Pablo, Algis Jasinskas, Rie Nakajima-Sasaki, and Philip L. Felgner, Division of Infectious Diseases, University of California, Irvine, CA, E-mails: jozelynp@gmail.com, ajasinsk@uci.edu, and rie3@uci.edu. Amadou Niangaly, Drissa Coulibaly, and Mahamadou A. Thera, Faculty of Medicine, Malaria Research and Training Center Bamako, University of Bamako, Bamako, Mali, and Malaria Research and Training Center, Mali University of Science, Techniques, and Technology of Bamako, Bamako, Mali, E-mails: niangaly@icermali.org, coulibalyd@icermali.org, and mthera@icermali.org. Jeff Skinner, Laboratory of Immunogenetics, National Institute of Allergy and Infectious Diseases, National Institutes of Health Rockville, MD, E-mail: skinnerj@niaid.nih.gov. Bourema Kouriba and Ogobara K. Doumbo, Malaria Research and Training Center, University of Science, Techniques, and Technology of Bamako, Bamako, Mali, E-mails: kouriba@icermali.org and okd@icermali.org.
References
- 1.Ouattara A, Mu J, Takala-Harrison S, Saye R, Sagara I, Dicko A, Niangaly A, Duan J, Ellis RD, Miller LH, Su XZ, Plowe CV, Doumbo OK. Lack of allele-specific efficacy of a bivalent AMA1 malaria vaccine. Malar J. 2010;9:175. doi: 10.1186/1475-2875-9-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takala SL, Plowe CV. Genetic diversity and malaria vaccine design, testing and efficacy: preventing and overcoming 'vaccine resistant malaria'. Parasite Immunol. 2009;31:560–573. doi: 10.1111/j.1365-3024.2009.01138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hviid L. Naturally acquired immunity to Plasmodium falciparum malaria in Africa. Acta Trop. 2005;95:270–275. doi: 10.1016/j.actatropica.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Thera MA, Doumbo OK, Coulibaly D, Laurens MB, Ouattara A, Kone AK, Guindo AB, Traore K, Traore I, Kouriba B, Diallo DA, Diarra I, Daou M, Dolo A, Tolo Y, Sissoko MS, Niangaly A, Sissoko M, Takala-Harrison S, Lyke KE, Wu Y, Blackwelder WC, Godeaux O, Vekemans J, Dubois MC, Ballou WR, Cohen J, Thompson D, Dube T, Soisson L, Diggs CL, House B, Lanar DE, Dutta S, Heppner DG, Jr, Plowe CV. A field trial to assess a blood-stage malaria vaccine. N Engl J Med. 2011;365:1004–1013. doi: 10.1056/NEJMoa1008115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sagara I, Dicko A, Ellis RD, Fay MP, Diawara SI, Assadou MH, Sissoko MS, Kone M, Diallo AI, Saye R, Guindo MA, Kante O, Niambele MB, Miura K, Mullen GE, Pierce M, Martin LB, Dolo A, Diallo DA, Doumbo OK, Miller LH, Saul A. A randomized controlled phase 2 trial of the blood stage AMA1-C1/Alhydrogel malaria vaccine in children in Mali. Vaccine. 2009;27:3090–3098. doi: 10.1016/j.vaccine.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thera MA, Doumbo OK, Coulibaly D, Diallo DA, Kone AK, Guindo AB, Traore K, Dicko A, Sagara I, Sissoko MS, Baby M, Sissoko M, Diarra I, Niangaly A, Dolo A, Daou M, Diawara SI, Heppner DG, Stewart VA, Angov E, Bergmann-Leitner ES, Lanar DE, Dutta S, Soisson L, Diggs CL, Leach A, Owusu A, Dubois MC, Cohen J, Nixon JN, Gregson A, Takala SL, Lyke KE, Plowe CV. Safety and immunogenicity of an AMA-1 malaria vaccine in Malian adults: results of a phase 1 randomized control trial. PLoS One. 2008;3:e1465. doi: 10.1371/journal.pone.0001465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takala SL, Coulibaly D, Thera MA, Dicko A, Smith DL, Guindo AB, Kone AK, Traore K, Ouattara A, Djimde AA, Sehdev PS, Lyke KE, Diallo DA, Doumbo OK, Plowe CV. Dynamics of polymorphism in a malaria vaccine antigen at a vaccine-testing site in Mali. PLoS One. 2007;4:e93. doi: 10.1371/journal.pmed.0040093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takala SL, Coulibaly D, Thera MA, Batchelor AH, Cummings MP, Escalante AA, Ouattara A, Traore K, Niangaly A, Djimde AA, Doumbo OK, Plowe CV. Extreme polymorphism in a vaccine antigen and risk of clinical malaria: implications for vaccine development. Sci Transl Med. 2009;2:2ra5. doi: 10.1126/scitranslmed.3000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ouattara A, Takala-Harrison S, Thera MA, Coulibaly D, Niangaly A, Saye R, Tolo Y, Dutta S, Heppner DG, Soisson L, Diggs CL, Vekemans J, Cohen J, Blackwelder WC, Dube T, Laurens MB, Doumbo OK, Plowe CV. Molecular basis of allele-specific efficacy of a blood-stage malaria vaccine: vaccine development implications. J Infect Dis. 2013;207:511–519. doi: 10.1093/infdis/jis709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doolan DL, Mu Y, Unal B, Sundaresh S, Hirst S, Valdez C, Randall A, Molina D, Liang X, Freilich DA, Oloo JA, Blair PL, Aguiar JC, Baldi P, Davies DH, Felgner PL. Profiling humoral immune responses to P. falciparum infection with protein microarrays. Proteomics. 2008;8:4680–4694. doi: 10.1002/pmic.200800194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barry AE, Trieu A, Fowkes FJ, Pablo J, Kalantari-Dehaghi M, Jasinskas A, Tan X, Kayala MA, Tavul L, Siba PM, Day KP, Baldi P, Felgner PL, Doolan DL. The stability and complexity of antibody responses to the major surface antigen of Plasmodium falciparum are associated with age in a malaria endemic area. Mol Cell Proteomics. 2011;10:M111. doi: 10.1074/mcp.M111.008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crompton PD, Kayala MA, Traore B, Kayentao K, Ongoiba A, Weiss GE, Molina DM, Burk CR, Waisberg M, Jasinskas A, Tan X, Doumbo S, Doumtabe D, Kone Y, Narum DL, Liang X, Doumbo OK, Miller LH, Doolan DL, Baldi P, Felgner PL, Pierce SK. A prospective analysis of the Ab response to Plasmodium falciparum before and after a malaria season by protein microarray. Proc Natl Acad Sci USA. 2010;107:6958–6963. doi: 10.1073/pnas.1001323107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trieu A, Kayala MA, Burk C, Molina DM, Freilich DA, Richie TL, Baldi P, Felgner PL, Doolan DL. Sterile protective immunity to malaria is associated with a panel of novel P. falciparum antigens. Mol Cell Proteomics. 2011;10:M111. doi: 10.1074/mcp.M111.007948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baum E, Badu K, Molina DM, Liang X, Felgner PL, Yan G. Protein microarray analysis of antibody responses to Plasmodium falciparum in western Kenyan highland sites with different transmission levels. PLoS One. 2013;8:e82246. doi: 10.1371/journal.pone.0082246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ouattara A, Takala-Harrison S, Thera MA, Coulibaly D, Niangaly A, Saye R, Tolo Y, Dutta S, Heppner DG, Soisson L, Diggs CL, Vekemans J, Cohen J, Blackwelder WC, Dube T, Laurens MB, Doumbo OK, Plowe CV. Molecular basis of allele-specific efficacy of a blood-stage malaria vaccine: vaccine development implications. J Infect Dis. 2013;207:511–519. doi: 10.1093/infdis/jis709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takala SL, Smith DL, Thera MA, Coulibaly D, Doumbo OK, Plowe CV. Short report: rare Plasmodium falciparum merozoite surface protein 1 19-kda (msp-1(19)) haplotypes identified in Mali using high-throughput genotyping methods. Am J Trop Med Hyg. 2007;76:855–859. [PMC free article] [PubMed] [Google Scholar]
- 17.Davies DH, Liang X, Hernandez JE, Randall A, Hirst S, Mu Y, Romero KM, Nguyen TT, Kalantari-Dehaghi M, Crotty S, Baldi P, Villarreal LP, Felgner PL. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci USA. 2005;102:547–552. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thera MA, Doumbo OK, Coulibaly D, Diallo DA, Sagara I, Dicko A, Diemert DJ, Heppner DG, Jr., Stewart VA, Angov E, Soisson L, Leach A, Tucker K, Lyke KE, Plowe CV. Safety and allele-specific immunogenicity in Malian adults: results of a phase I randomized trial. PLoS Clin Trials. 2006;1:e34. doi: 10.1371/journal.pctr.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thera MA, Doumbo OK, Coulibaly D, Laurens MB, Kone AK, Guindo AB, Traore K, Sissoko M, Diallo DA, Diarra I, Kouriba B, Daou M, Dolo A, Baby M, Sissoko MS, Sagara I, Niangaly A, Traore I, Olotu A, Godeaux O, Leach A, Dubois MC, Ballou WR, Cohen J, Thompson D, Dube T, Soisson L, Diggs CL, Takala SL, Lyke KE, House B, Lanar DE, Dutta S, Heppner DG, Plowe CV. Safety and immunogenicity of an AMA-1 malaria vaccine in Malian adults: results of a phase 1 randomized control trial. PLoS One. 2010;5:e9041. doi: 10.1371/journal.pone.0001465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sboner A, Karpikov A, Chen G, Smith M, Mattoon D, Freeman-Cook L, Schweitzer B, Gerstein MB. Robust-linear-model normalization to reduce technical variability in functional protein microarrays. J Proteome Res. 2009;8:5451–5464. doi: 10.1021/pr900412k. [DOI] [PubMed] [Google Scholar]
- 21.Vafa M, Troye-Blomberg M, Anchang J, Garcia A, Migot-Nabias F. Multiplicity of Plasmodium falciparum infection in asymptomatic children in Senegal: relation to transmission, age and erythrocyte variants. Malar J. 2008;7:17. doi: 10.1186/1475-2875-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]