Abstract
Type 2 diabetes mellitus (T2DM) is growing in an epidemic manner across the world and India has the world’s largest number of diabetic subjects. The present study was carried out to investigate the association of glutathione-S-transferase (GSTM1, GSTT1) and fat mass and obesity associated (FTO) gene polymorphisms with T2DM patients and controls, and its role in increasing the susceptibility to T2DM. A total of 198 subjects (101 T2DM patients and 97 controls) participated in this study. GSTM1, GSTT1 and FTO gene polymorphisms in the patients and controls were evaluated by polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP). We observed significant association of GSTM1 positive (p = 0.046) and GSTM1 null (p = 0.046) genotypes with T2DM, while no significant association was found with the FTO gene polymorphism in our study. It seems that the GSTM1 gene polymorphism can be a predictive marker for early identification of a population at risk of T2DM. The potential role of GST and FTO gene polymorphisms as a marker of susceptibility to T2DM needs further studies in a larger number of patients.
Keywords: Glutathione-S-transferase (GSTM1, GSTT1); Fat mass and obesity associated (FTO) gene; Gene polymorphisms; Type 2 diabetes mellitus (T2DM)
INTRODUCTION
Diabetes has become a common global health problem that affects >170 million people worldwide. It is one of the leading causes of death and disability. It is estimated that by 2030, the number will rise to 366 million (www.who.int). According to Wild et al. [1] the ‘top’ three countries in terms of the number of individuals with type 2 diabetes mellitus (T2DM) are India (31.7 million in 2000; 79.4 million in 2030), China (20.8 million in 2000; 42.3 million in 2030); and the US (17.7 million in 2000; 30.3 million in 2030). Clearly, T2DM has become an epidemic in the 21st century with India leading the world with the largest number of diabetic subjects. Type 2 diabetes mellitus is a non autoimmune, complex, heterogeneous and polygenic metabolic disease in which the body fails to produce enough insulin, characterized by abnormal glucose homeostasis [2]. Its pathogenesis appears to involve complex interactions between genetic and environmental factors [2]. Identification of the genetic components of T2DM is the most important area of diabetes research because elucidation of the diabetes genes (alleles) will influence all efforts toward a mechanistic understanding of the disease, its complications, and its treatment, cure, and prevention [3].
Researchers have identified a number of genetic factors that may be responsible for selected or more general diabetic patients. It has also been reported that defects in antioxidant defense against oxidative stress play an important role in the etiology of diabetic complications [4,5]. Glutathione (GSH) is the major cellular antioxidant that protects against environmental toxicants as well as reactive oxygen species (ROS) mediated cell injury. Glutathione detoxifies ROS, reduces peroxides and detoxifies multiple compounds through glutathione-S-transferase (GST) conjunction [6]. Three of the GST genes, GSTM1, GSTT1 and GSTP1, have been found to have functional polymorphisms that are frequently present in the general population. There are several studies that reported significant association to T2DM for both null genotypes of GST [7,8] and others that verified no association between GSTT1 and GSTM1 polymorphisms and T2DM [9,10]. Some studies indicated that genetic variations of the GSTT1 enzyme are associated with the development of end-stage renal disease in diabetes mellitus patients [11]. Recently, part of a genome-wide association study found that several single nucleotide polymorphisms (SNPs) of the fat mass and obesity associated (FTO) gene were strongly associated with obesity and T2DM [12,13]. The FTO gene, which is located on chromosome 16q12.2, and has nine exons and emerged 450 million years ago [14]. The FTO gene is mainly expressed in the hypothalamus and encodes a 2-oxoglutarate-dependent nucleic acid demethylase. It may play an important role in the management of energy homeostasis [14,15], nucleic acid demethylation, and the regulation of body fat masses by lipolysis [16]. A recent meta-analysis [17] in East and South Asians, concluded that the FTO minor allele (or a proxy), the risk allele for obesity also increased the risk of T2DM. Similar results were reported in a Scandinavian population [18]. Currently, adequate data is not available dealing with GSTT1, GSTM1 and FTO gene polymorphisms and its susceptibility with T2DM patients among north Indian population; thus, we conceived the need for further studies on GSTT1, GSTM1 and FTO gene polymorphisms and its association with T2DM in north Indian population.
MATERIALS AND METHODS
Patient Selection
A total of 101 blood samples from T2DM patients and 97 healthy controls were collected at the Diabetic Clinic, Department of Medicine of Era’s Lucknow Medical College & Hospital, Lucknow, Uttar Pradesh, India. Written informed consent was obtained from all participants. Data collection was done for each patient on clinical variables including age, alcohol consumption, body mass index (BMI), height, weight, cigarette smoking, family history, etc., patients with overnight fasting plasma glucose of more than 126.0 mg/dL on two consecutive events were included in the T2DM category, while samples having a fasting blood glucose level below 110.0 mg/dL without a family history of diabetes were included in the study as controls. Patients with type 1 presentation, defined as diabetic ketoacidosis, acute presentation with heavy ketonuria (>3+), or uninterrupted requirement of insulin within 1 year of diagnosis, were excluded. Ethical Committee clearances were obtained from the respective departments, prior to the recruitment of subjects in this study.
Biochemical Estimations
The BMI was calculated according to Quetelet equation by using weight in kg, height in m2. Fasting plasma glucose and random blood sugar (glucose oxidase-peroxidase method). Serum creatinine concentration was assessed by a kinetic Jaffe’s method. Serum cholesterol (cholesterol oxidase-peroxidase), serum triglyceride (glycerol phosphate oxidase-peroxidaseamidopyrine method), high-density lipoprotein (HDL) cholesterol (immunoinhibition) were assessed by XL-300 Transasia Fully Auto Analyzer; Transasia, Mannheim, Germany. Low-density lipoprotein (LDL) cholesterol levels were calculated using the Friedewald formula [19]. Very low-density lipoprotein (VLDL) cholesterol was determined by an enzymatic method. Hb A1C was measured using a semi auto analyzer (Transasia).
DNA Extraction
Peripheral blood (5 mL) was collected from all the subjects in 0.5 M vacutainers with EDTA as anticoagulant. Genomic DNA was isolated from whole blood using the standard phenol-chloroform extraction method [20]. The DNA concentration was determined by spectrophotometer and stored at −20 °C.
ANALYSIS OF POLYMORPHISMS
GSTM1 and GSTT1 Gene Polymorphisms
GSTM1 and GSTT1 genetic polymorphisms were evaluated using a multiplex polymerase chain reaction (PCR) technique (MJ Mini Thermal Cycler; Bio-Rad Laboratories, Hercules, CA, USA). The PCR primers were synthesized according to Arand et al. [21]. Primers for the GSTM1 gene were: 5′-GAA CTC CCT GAA AAG CTAA AGC-3′ and 5′-GTT GGG CTC AAA TAT ACG GTG G-3′ and for GSTT1: 5′-TTC CTT ACT GGT CCT CAC ATC TC-3′ and 5′-TCA CCG GACAT GGC CAG CA-3′. The β-globin locus was used as an internal control to avoid false-negative readings. Primers for the β-globin locus were: 5′-CAA CTT CAT CCA CGT TCA CC-3′ and 5′-GAA GAG CCA AGG ACA GGT AC-3′. The PCR reaction was carried out in a total volume of 25 µL containing 10 pmol of each primer, 2.5 mmol/L of MgCl2, 0.2 mmol/L of each deoxynucleotide triphosphate, 1 unit of Taq polymerase (Bioline Ltd., London, UK) and 100 ng of genomic DNA. Amplification was performed by initial denaturation at 94 °C for 5 min., followed by 30 cycles at 94 °C for 1 min., 64 °C for 1 min. and 72 °C for 1 min., and a final extension of 72 °C for 7 min. The amplified products were identified by electrophoresis in a 1.5% agarose gel and stained with 0.5 mg/mL (concentration) ethidium bromide. The product lengths were 215, 480 and 268 bp for GSTM1, GSTT1 and β-globin locus, respectively.
FTO Gene Polymorphism
The FTO SNP (rs993 9609) was genotyped by PCR (MJ Mini Thermal Cycler; Bio-Rad Laboratories) and restriction fragment length polymorphism (RFLP), analyses. Genomic DNA (20 ng) was incubated in a 10 µL solution containing 1 × NH4 buffer, 2.5 mmol/L magnesium, 200 mmol/L each dNTP, 20 pmol forward (5′-AAC TGG CTC TTG AAT GAA ATA GGA TTC AGA-3′) and reverse (5′-AGA GTA ACA GAG ACT ATC CAA GTG CAG TAC-3′) oligonucleotide primers [22], and 0.5 U Taq DNA polymerase (Bioline Ltd.). The PCR mix was denaturated at 94 °C for 5 min., followed by 20 cycles of 45 seconds at 94 °C, 45 seconds at 61 °C (dropping 0.5 °C per cycle), and 45 seconds at 72 °C. After this, the PCR mix was incubated for 15 cycles at 45 seconds at 94 °C, 45 seconds at 51 °C, and 45 seconds at 72 °C, followed by a final extension at 72 °C for 10 min. This was then incubated at 37 °C for 16 hours with 2 U ScaI (New England Biolabs, Hitchin, Hertfordshire, UK). Upon running the final products on a 3.0% agarose gel, the T allele produced a 182 bp band and the A allele produced 154 and 28 bp bands.
Statistical Analyses
All the figures are presented as mean ± standard deviation (SD). The genotyping data were compared between patients and controls using the c2 test. Other variables were compared using the Student’s t-test for normally-distributed variables. All statistical tests were performed using the Statistical Package for the Social Sciences, version 12 software (SPSS Inc., Chicago, IL, USA).
RESULTS
Our study included 101 T2DM patients (65 males and 36 females) and 97 controls (53 males and 44 females). The mean age of the T2DM patients and controls were 41.32 ± 11.39 years and 40.13 ± 10.28 years, respectively. The age of onset in T2DM patients was 44.05 ± 11.20 years. Clinical and biochemical profiles of the patients and controls are shown in Table 1. The mean BMI, blood pressure, random blood sugar (RBS), fasting blood sugar (FBS), Hb A1C, total cholesterol, HDL cholesterol, VLDL cholesterol, triglyceride and serum creatinine levels in the diabetic patients were significantly higher than in the control subjects (p <0.001). We have found the significant correlation between the RBS-Hb A1C (p<0.001), RBS-serum cholesterol (p = 0.007), RBS-triglyceride (p = 0.049), serum creatinine-Hb A1C (p = 0.002), serum creatinine-triglyceride (p <0.001), serum creatinine-VLDL (p <0.001), serum cholesteroltriglyceride (p =0.001), serum cholesterol-HDL (p <0.001), HDL-VLDL (p = 0.019), VLDL-triglyceride (p <0.001). The frequency of GSTM1 and GSTT1 null genotypes was 38.61 and 7.92%, respectively, in T2DM patients, while it was 53.61 and 4.12%, respectively, in the controls. The frequency of positive GSTM1 and GSTT1 genotypes was 61.38 and 92.07%, respectively, in T2DM patients as compared to 46.39 and 95.87% in the controls. Odds ratio (OR) for GSTM1 null genotypes was 0.544 [95% confidence interval (CI) 0.31–0.96, p = 0.046, χ2 = 3.97, power = 0.923, Bonferroni corrected p = 0.092], for the GSTT1 null genotypes 2.0 OR (95% CI 0.58–6.87, p = 0.405, χ2 = 0.694, power = 0.869, Bonferroni corrected p = 0.810). The OR for the GSTM1 positive genotypes was 1.837 (95% CI 1.04–3.23, p = 0.046, χ2 = 3.97, power = 0.923, Bonferroni corrected p = 0.092), for the GSTT1 positive genotypes 0.5 OR (95% CI 0.15–1.72, p = 0.405, χ2 = 0.694, power = 0.869, Bonferroni corrected p = 0.810) and the GSTM1 and GSTT1 wild genotypes 1.569 (95% CI 0.89–2.75, p = 0.115, χ2 = 2.49, power = 0.918, Bonferroni corrected p = 0.345). The frequencies of the FTO genotypes AA, AT, TT in T2DM patients were 4.95, 71.28 and 23.76%, respectively, while in the control group these frequencies were 0.00, 75.25 and 24.74%, respectively. The frequency of the A and T alleles in T2DM patients was 40.59 and 59.40% as compared to 37.62 and 62.37% in the controls. The OR for AA was N/A(0) (95% CI NA, p = 0.077, χ2 = 3.12, power = 0.989, Bonferroni corrected p = 0.231), for AT 0.816 (95% CI 0.43–1.53, p = 0.528, χ2 = 0.40, power = 0.782, Bonferroni corrected p = 1.000), and for TT 0.948 (95% CI 0.50–1.82, p = 0.872, χ2 = 0.03, power = 0.509, Bonferroni corrected p = 0.509). The genotype, allele frequencies of the GSTM1, GSTT1 and FTO genes and statistical analysis of the patients and controls are also shown in Table 2.
Table 1.
Clinical and biochemical parameters of type 2 diabetes mellitus patients and controls.
| Parameters | T2DM Patients (n = 101) | Controls (n = 97) | p Values |
|---|---|---|---|
| Gender | 65 males; 36 females | 53 males; 44 females | 0.072 |
| Age (years) | 41.32 ± 11.39 | 40.1 ± 10.28 | 0.12 |
| BMI (kg/m2) | 26.01 ± 4.12 | 23.99 ± 2.36 | <0.001 |
| RBS (mg/dL) | 195.44 ± 91.66 | 118.33 ± 18.55 | <0.001 |
| FBS (mg/dL) | 190.81 ± 66.24 | 92.92 ± 14.03 | <0.001 |
| Systolic BP (mm Hg) | 136.23 ± 14.21 | 119.96 ± 5.06 | <0.001 |
| Diastolic BP (mm Hg) | 87.77 ± 6.77 | 78.22 ± 1.66 | <0.001 |
| Serum creatinine (mg/dL) | 1.03 ± 0.24 | 0.88 ± 0.12 | <0.001 |
| Hb A1C (%) | 6.43 ± 1.54 | 5.61 ± 0.38 | <0.001 |
| Serum cholesterol (mg/dL) | 194.92 ± 43.27 | 166.06 ± 20.52 | <0.001 |
| Triglyceride (mg/dL) | 237.77 ± 147.44 | 147.55 ± 40.33 | <0.001 |
| HDL (mg/dL) | 59.32 ± 12.22 | 57.33 ± 11.66 | <0.001 |
| LDL (mg/dL) | 99.22 ± 34.32 | 92.66 ± 18.88 | 0.133 |
| VLDL (mg/dL) | 47.55 ± 28.22 | 32.55 ± 10.22 | <0.001 |
For finding p values, the Student’s unpaired t-test was applied except in the case of gender comparison where the c2 test was applied. BMI: body mass index; RBS: random blood sugar; FBS: fasting blood sugar; BP: blood pressure; HDL: high-density lipoprotein; LDL: low-density lipoprotein; VLDL: very low-density lipoprotein.
Table 2.
Genotype and allele frequencies of the GSTT1, GSTM1 and FTO gene polymorphisms in type 2 diabetes mellitus patients and controls.
| n | Frequency (%) | n | Frequency (%) | OR | 95% CI | χ2 | p Value | Bonferroni Corrected p Value | Power | |
|---|---|---|---|---|---|---|---|---|---|---|
| GSTM1 positive | 45 | 46.39 | 62 | 61.38 | 1.837 | 1.04–3.23 | 3.970 | 0.046 | 0.092 | 0.923 |
| GSTM1 null | 52 | 53.61 | 39 | 38.61 | 0.544 | 0.31–0.96 | 3.970 | 0.046 | 0.092 | 0.923 |
| GSTT1 positive | 93 | 95.87 | 93 | 92.07 | 0.500 | 0.15–0.96 | 0.694 | 0.405 | 0.810 | 0.869 |
| GSTT1 null | 4 | 4.23 | 8 | 7.92 | 2.000 | 0.58–6.87 | 0.694 | 0.405 | 0.810 | 0.869 |
| GSTT1 positive; GSTM1 positive | 41 | 42.27 | 54 | 53.47 | 1.569 | 0.89–2.75 | 2.490 | 0.115 | 0.345 | 0.918 |
| GSTT1 positive; GSTM1 null | 52 | 53.61 | 39 | 38.61 | 0.544 | 0.31–0.96 | 4.480 | 0.034 | 0.102 | 0.898 |
| GSTT1 null; GSTM1 positive | 4 | 4.12 | 8 | 7.92 | 2.000 | 0.58–6.87 | 1.250 | 0.263 | 0.789 | 0.937 |
| AA | 0 | 0.00 | 5 | 4.95 | NA | NA | 3.120 | 0.077 | 0.231 | 0.989 |
| AT | 73 | 75.26 | 72 | 71.29 | 0.816 | 0.43–1.53 | 0.400 | 0.528 | 1.000 | 0.782 |
| TT | 24 | 24.74 | 24 | 23.76 | 0.948 | 0.50–1.82 | 0.030 | 0.872 | 1.000 | 0.509 |
| A | 73 | 37.63 | 82 | 40.59 | 1.133 | 0.76–1.70 | 0.360 | 0.546 | 1.000 | 0.825 |
| T | 121 | 62.27 | 120 | 59.41 | 0.833 | 0.59–1.32 | 0.360 | 0.546 | 1.000 | 0.825 |
NA: not available.
DISCUSSION
GSTM1 and GSTT1 Gene Polymorphisms
Type 2 diabetes mellitus is characterized as being a polygenic disorder and generally thought of as a syndrome, rather than a single specific entity. Oxidative stress plays a major role in the pathogenesis of T2DM [23]. Glutathione-S-transferases catalyze the conjugation of glutathione to a wide range of electrophiles and represent a protective mechanism against oxidative stress. The GST family of genes is critical in the protection of cells from ROS because they utilize as substrates a wide variety of products of oxidative stress [24]. The deletion polymorphisms of GSTT1 and GSTM1, which are associated with abolished enzyme activity [25], have been associated with T2DM when compared to control subjects [9,24]. The majority of case control studies of GSTM1 and GSTT1 null genotypes have reported risk associations with disease [26,27].The frequency of the GSTM1 null genotype in our study was 38.61%; it is little lower than reported in other populations (Egyptian 58.62%, Dubai 57.5%, North Indian 54.00%) [28–30]. The frequency of the GSTTI null genotype was 7.92% in T2DM patients, which is lower when compared to the Dubai (60.0%) and Egyptian (35.0%) populations [7,29]. We have also observed that the genotype frequency of the positive GSTT1 and GSTM1 genes was 92.07 and 61.38%, respectively, which is significantly higher in comparison with Dubai where the positive GSTT1 and GSTM1 genes was 40.0 and 42.5%, respectively [29].
The FTO Gene Polymorphism
The frequency of the FTO gene polymorphism is quite variable in different geographic and ethnic groups (Table 3). Previous studies have shown that T2DM risk associated with the A allele of the FTO variant (rs 9939609) was strongly mediated by BMI, and has been shown to potentially affect mass rather than height [31]. The FTO gene (rs 9939609) was shown to be strongly associated with T2DM risk (OR 1.27; p = 5 × 10−8) in a genome wide association (GWA) scan performed in the UK population [31]. In our study, we reported that the frequency of the AT genotype (Pro12Ala) was 71.29% in T2DM patients, which is significantly higher when compared with North Indians (47.6%), South Asian Indians (42.2%), South Africans (55.0%), Scots (48.8%) and Spanish (48.3%) populations [32–36]. The frequency of the FTO AA genotype described in other populations fluctuates between 14.0 to 18.0% in T2DM patients of North Indians (12.5%), South Asian Indians (14.2%), Scots (18.0%) and Spanish (17.8%) populations [32,33,35,36], but the frequency of the FTO AA genotype in our study group was 4.95%, which is considerably lower when compared to other reports. Many factors may account for the different results in similar studies. In our population, the frequency of the TT genotype was 23.76%, which is considerably lower than in South Asian Indians (43.6%) and North Indians (39.9%) [32,33], There may be differences even in the same population because of genetic, environmental factors and the number of samples analyzed.
Table 3.
GSTM1, GSTT1 and FTO gene polymorphisms in different ethnic groups.
| Alleles (%) | Significant | Genotypes (%) | Alleles (%) | Significant | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ethnicity | Ref. | GSTM1 Positive | GSTM1 Null | GSTT1 Positive | GSTT1 Null | Yes/No | TT | AT | AA | A | T | Yes/No |
| North Indian | this study | 61.38 | 38.61 | 92.07 | 7.92 | Yes | 23.76 | 71.29 | 4.95 | 40.59 | 59.41 | No |
| Egyptian | 7 | 38.00 | 62.00 | 65.00 | 35.00 | Yes | – | – | – | – | – | – |
| North Indian | 30 | 46.00 | 54.00 | 83.00 | 17.00 | Yes | – | – | – | – | – | – |
| Egyptian | 28 | 41.38 | 58.62 | 82.76 | 17.24 | No | – | – | – | – | – | – |
| Dubai | 29 | 42.50 | 57.50 | 40.00 | 60.00 | Yes | – | – | – | – | – | – |
| North Indian | 32 | – | – | – | – | – | 39.90 | 47.60 | 12.50 | 64.00 | 36.00 | Yes |
| South Asian Indian | 33 | – | – | – | – | – | 43.60 | 42.20 | 14.20 | 35.40 | 64.60 | Yes |
| South African | 34 | – | – | – | – | – | 15.00 | 55.00 | 30.00 | 57.00 | 43.00 | No |
| Spanish | 36 | – | – | – | – | – | 33.90 | 48.30 | 17.80 | 58.04 | 41.95 | No |
| Scots | 35 | – | – | – | – | – | 33.20 | 48.80 | 18.00 | 42.4 | 57.59 | Yes |
Our data suggest that the polymorphism in the GSTM1 gene might be a risk factor for T2DM, while no significant association was found with the FTO gene polymorphism. The potential role of GST and FTO gene polymorphisms as a marker of susceptibility for T2DM needs further studies in a larger number of patients.
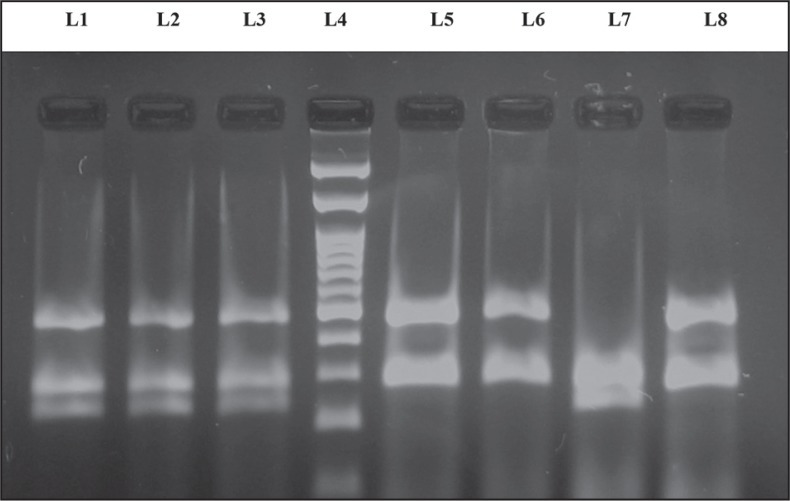
Figure 1.
The PCR products analyzed on 2.0% agarose gel. The presence or absence (null) of the GSTM1 and GSTT1 genes was detected by the presence or absence of a band at 480 bp (corresponding to GSTT1) and a band at 210 bp (corresponding to GSTM1). β-Globin is considered as an internal control (260 bp). Lane 4: 100 bp ladder; lanes 1, 2 and 3: GSTM1 and GSTT1 wild type genotype; lanes 5, 6 and 8: GSTM1 null genotype; lane 7: GSTT1 null genotype.
Figure 2.
Polyacrylamide gel picture showing digested PCR products for the FTO gene polymorphism. Lanes 2, 3, 5 and 7 show the AT genotype (182 and 154 bp); lane 6: shows the TT genotype (182 bp); lane 4 shows the 100 bp ladder; lane 1 shows the undigested PCR product of the FTO (182 bp) gene polymorphism.
Footnotes
Declaration of Interest. We are grateful for the support of the intramural grant from the Era’s Lucknow Medical College and Hospital, Lucknow, Uttar Pradesh, India. The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
REFERENCES
- 1.Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: Estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27(5):1047–1053. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 2.Gupta V, Khadgawat R, Saraswathy KN, Sachdeva MP, Kalla AK. Emergence of TCF7L2 as a most promising gene in predisposition of diabetes type II. Int J Hum Genet. 2008;8(1–2):199–215. [Google Scholar]
- 3.Das SK. Genetic epidemiology of adult onset T2Din Asian Indian population: Past, present and future. Int J Hum Genet. 2006;6(1):1–13. [Google Scholar]
- 4.Opara EC. Oxidative stress, micronutrients, diabetes mellitus and its complications. J R Soc Promot Health. 2002;122(1):28–34. doi: 10.1177/146642400212200112. [DOI] [PubMed] [Google Scholar]
- 5.Friedly LE, Philipson LH. Oxidative reactive species in cell injury: Mechanism in diabetes mellitus and therapeutic approaches. Ann N Y Acad Sci. 2005;1066:136–151. doi: 10.1196/annals.1363.019. [DOI] [PubMed] [Google Scholar]
- 6.Bekris LM, Shephard C, Peterson J, Hoehna B, Van Yserloo E, Rutledge F, et al. Glutathionestransferase M1 and T1 polymorphisms and associations with type 1 diabetes age-at-onset. Autoimmunity. 2005;38(8):567–575. doi: 10.1080/08916930500407238. [DOI] [PubMed] [Google Scholar]
- 7.Amer MA, Ghattas MH, Abo-ElMatty DM, Abou-El-Ela SH. Influence of glutathione S-transferase polymorphisms on type-2 diabetes mellitus risk. Genet Mol Res. 2011;10(4):3722–3730. doi: 10.4238/2011.October.31.14. [DOI] [PubMed] [Google Scholar]
- 8.Ramprasath T, Senthil Murugan P, Prabakaran AD, Gomathi P, Rathinavel A, Selvam GS. Potential risk modifications of GSTT1, GSTM1 and GSTP1 (glutathione-S transferase) variants and their association to CAD in patients with type-2 diabetes. Biochem Biophys Res Commun. 2011;407(1):49–53. doi: 10.1016/j.bbrc.2011.02.097. [DOI] [PubMed] [Google Scholar]
- 9.Hori M, Oniki K, Ueda, Goto S, Mihara S, Marubayashi T, et al. Combined glutathione S-transferase T1 and M1 positive genotypes afford protection against type 2 diabetes in Japanese. Pharmacogenomics. 2007;8(10):1307–1314. doi: 10.2217/14622416.8.10.1307. [DOI] [PubMed] [Google Scholar]
- 10.Datta SK, Kumar V, Pathak R, Tripathi AK, Ahmed RS, Kaira OP, et al. Association of glutathione S-transferase M1 and T1 gene polymorphism with oxidative stress in diabetic and nondiabetic chronic kidney disease. Ren Fail. 2010;32(10):1189–1195. doi: 10.3109/0886022X.2010.517348. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, Kao MT, Chang CC, Chen CM, Tsai JJ, Chang JG. Glutathione S-transferase T1 deletion is a risk factor for developing end-stage renal disease in diabetic patients. Int J Mol Med. 2004;14(5):855–859. [PubMed] [Google Scholar]
- 12.Hinney A. Genome wide association (GWA) study for early onset extreme obesity supports the role of fat mass and obesity associated gene (FTO) variants. PloS ONE. 2007;2(12):e1361. doi: 10.1371/journal.pone.0001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. Plos Genet. 2007;3(7):1200–1210. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fredriksson R, Hagglund M, Olszewski PK, Stephansson O, Jacobsson JA, Olszewska AM, et al. The obesity gene, FTO, is of ancient origin, up-regulated during food deprivation and expressed in neurons of feeding-related nuclei of the brain. Endocrinology. 2008;149(5):2062–2071. doi: 10.1210/en.2007-1457. [DOI] [PubMed] [Google Scholar]
- 15.Stratigopoulos G, Padilla SL, Leduc CA, Watson E, Hattersley AT, McCarthy MI, et al. Regulation of Fto/Ftm gene expression in mice and humans. Am J Physiol Regul Integr Comp Physiol. 2008;294(4):R1185–R1196. doi: 10.1152/ajpregu.00839.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wahlen K, Sjolin E, Hoffstedt J. The common rs9939609 gene variant of the fat mass- and obesity-associated gene FTO is related to fat cell lipolysis. J Lipid Res. 2008;49(3):607–661. doi: 10.1194/jlr.M700448-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Kilpeläinen TO, Liu C. Association of genetic variation in FTO with risk of obesity and type 2 diabetes with data from 96,551 East and South Asians. Diabetologia. 2012;55(4):981–995. doi: 10.1007/s00125-011-2370-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hertel JK, Johansson S, Sonestedt E, Jonsson A, Lie RT, Platou CG, et al. FTO, type 2 diabetes, and weight gain throughout adult life: A meta-analysis of 41,504 subjects from the Scandinavian HUNT, MDC, and MPP studies. Diabetes. 2011;60(5):1637–1644. doi: 10.2337/db10-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18(6):499–450. [PubMed] [Google Scholar]
- 20.Sambrook J, Frisch EF, Maniatis T, editors. Molecular Cloning: A Laboratory Manual. 2nd ed. New York, NY: Cold Spring Harbor Press; 1989. pp. 9.14–9.19. [Google Scholar]
- 21.Arand M, Mühlbauer R, Hengstler J, Jäger E, Fuchs J, Winkler L, et al. A multiplex polymerase chain reaction protocol for the simultaneous analysis of the glutathione S-transferase GSTM1 and GSTT1 polymorphisms. Anal Biochem. 1996;236(1):184–186. doi: 10.1006/abio.1996.0153. [DOI] [PubMed] [Google Scholar]
- 22.Hao K, Niu T, Sangokoya C, Li J, Xu X. SNP kit: An efficient approach to systematic evaluation of candidate single nucleotide polymorphisms in public databases. BioTechniques. 2002;33(4):822–848. doi: 10.2144/02334st06. [DOI] [PubMed] [Google Scholar]
- 23.Stephens JW, Khanolkar MP, Bain SC. The biological relevance and measurement of plasma markers of oxidative stress in diabetes and cardiovascular disease. Atherosclerosis. 2009;202(2):321–329. doi: 10.1016/j.atherosclerosis.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 24.Wang L, Zhang Q, Li Q. Genetic polymorphisms of GSTT1, GSTM1, and NQO1 genes and diabetes mellitus risk in Chinese population. Biochem Biophys Res Commun. 2006;341:310–313. doi: 10.1016/j.bbrc.2005.12.195. [DOI] [PubMed] [Google Scholar]
- 25.London SJ, Yuan JM, Chung FL, Gao YT. Isothiocyanates, glutathione S-transferase M1 and T1 polymorphisms, and lung-cancer risk: A prospective study of men in Shanghai, China. Lancet. 2000;356(9231):724–729. doi: 10.1016/S0140-6736(00)02631-3. [DOI] [PubMed] [Google Scholar]
- 26.Yuille M, Condie A, Hudson C, Kote-Jarai Z, Stone E, Eeles R, et al. Relationship between glutathione S-transferase M1, T1, and P1 polymorphisms and chronic lymphocytic leukemia. Blood. 2002;99(11):4216–4218. doi: 10.1182/blood.v99.11.4216. [DOI] [PubMed] [Google Scholar]
- 27.Chen K, Jiang QT, He HQ. Relationship between metabolic enzyme polymorphism and colorectal cancer. World J. Gastroenterol. 2005;11(3):331–335. doi: 10.3748/wjg.v11.i3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nowier SR, Kashmiry NK, Rasool HA, Morad H, Ismail S. Association of T2DMellitus and glutathione S transferase (GSTM1 and GSTT1) genetic polymorphism. Res J Med Med Sci. 2009;4(2):181–188. [Google Scholar]
- 29.Hossaini AM, Zamrroni IM, Kashem RA, Khan ZF. Polymorphism of glutathione S-transferases as genetic risk factors for the development of complications in T2D mellitus. J Crit Care. 2008;23(3):444–448. doi: 10.1016/j.jcrc.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 30.Bid HK, Konwar R, Saxena M, Chaudhari P, Agrawal CG, Banerjee M. Association of glutathione S-transferase (GSTM1, T1 and P1) gene polymorphisms with type 2 diabetes mellitus in north Indian population. J Postgrad Med. 2010;56(3):176–181. doi: 10.4103/0022-3859.68633. [DOI] [PubMed] [Google Scholar]
- 31.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanghera DK, Ortega L, Han S, Singh J, Ralhan SK, Wander GS, et al. Impact of nine common T2D risk polymorphisms in Asian Indian Sikhs: PPARG2 (Pro12 Ala), IGF2BP2, TCF7L2 and FTO variants confer a significant risk. BMC Med Genet. 2008;9:59. doi: 10.1186/1471-2350-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yajnik CS, Janipalli CS, Bhaskar S, Kulkarni SR, Freathy RM, Prakash S, et al. FTO gene variants are strongly associated with type 2 diabetes in South Asian Indians. Diabetologia. 2009;52(2):247–252. doi: 10.1007/s00125-008-1186-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pirie FJ, Motala AA, Pegoraro J, Paruk IM, Govender T, Rom L. Variants in PPARG, KCNJ11, TCF7L2, FTO, and HHEX genes in South African subjects of Zulu descent with type 2 diabetes. Afr J Diabetes Med. 2010;18(1):12–16. [Google Scholar]
- 35.Doney AS, Dannfald J, Kimber CH, Donnelly LA, Pearson E, Morris AD, et al. The FTO gene is associated with an atherogenic lipid profile and myocardial infarction in patients with type 2 diabetes: A Genetics of Diabetes Audit and Research Study in Tayside Scotland (Go-DARTS) study. Circ Cardiovasc Genet. 2009;2(3):255–259. doi: 10.1161/CIRCGENETICS.108.822320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ortega-Azorín C, Sorlí JV, Asensio EM, Coltell O, Martínez-González MÁ, Salas-Salvadó J, et al. Associations of the FTO rs9939609 and the MC4R rs17782313 polymorphisms with type 2 diabetes are modulated by diet, being higher when adherence to the Mediterranean diet pattern is low. Cardiovasc Diabetol. 2012;11:137. doi: 10.1186/1475-2840-11-137. [DOI] [PMC free article] [PubMed] [Google Scholar]