Recurrent supra-ribosomal building blocks separated by ribosome-free mRNA regions form polysomes and reflect cellular translational activity.
Abstract
Translation is increasingly recognized as a central control layer of gene expression in eukaryotic cells. The overall organization of mRNA and ribosomes within polysomes, as well as the possible role of this organization in translation are poorly understood. Here we show that polysomes are primarily formed by three distinct classes of ribosome assemblies. We observe that these assemblies can be connected by naked RNA regions of the transcript. We show that the relative proportions of the three classes of ribosome assemblies reflect, and probably dictate, the level of translational activity. These results reveal the existence of recurrent supra-ribosomal building blocks forming polysomes and suggest the presence of unexplored translational controls embedded in the polysome structure.
Introduction
The ribosome is the basic machinery for shaping cell phenotypes through protein synthesis. Small and large ribonucleoprotein subunits assemble on mRNAs to convert the linear information encoded in genes into three-dimensionally structured proteins. The interaction of mRNA and isolated ribosomes was shown for the first time 50 years ago by sedimentation analysis and transmission electron micrographs of polysomes from rabbit reticulocytes, demonstrating that mRNA is indeed the template for protein synthesis (Warner et al., 1962). After this, the organization of ribosomes into higher-order structures was recognized in several different organisms (Palade, 1955; Warner et al., 1962; Wettstein et al., 1963). Warner et al. (1962) called these actively translating ribosomal assemblies “polyribosomes” or “polysomes,” and they are currently understood to exist in an equilibrium among the three stages of translation (initiation, elongation, and termination), as well as ribosome recycling.
Ever since its discovery, the ribosome has been extensively characterized (Schluenzen et al., 2000; Harms et al., 2001; Schmeing and Ramakrishnan, 2009; Melnikov et al., 2012), providing fundamental molecular insights into the dynamics of the three phases of translation and the role of the accessory protein factors involved (Ramakrishnan, 2002; Schmeing and Ramakrishnan, 2009). Even though some work has proposed hypothetical models of mRNA organization inside polysomes (Kopeina et al., 2008; Brandt et al., 2009; Afonina et al., 2013), our understanding of their topology with respect to mRNA has not significantly progressed since initial descriptions of the circular (Palade, 1955; Warner et al., 1962; Wettstein et al., 1963; Yazaki et al., 2000; Madin et al., 2004), spiral (Palade, 1955), rosette (Palade, 1955; Warner et al., 1962; Wettstein et al., 1963; Madin et al., 2004), staggered line (Daneholt et al., 1977), and caterpillar-like double-rowed (Kopeina et al., 2008; Afonina et al., 2013) shapes observed in micrographs. In recent years, cryo-EM has been successfully used to reconstruct the 3D shape of prokaryotic and human polysomes in cell-free lysates (Brandt et al., 2009) and in intact cells (Brandt et al., 2010), respectively. These reconstructions revealed a nonrandom set of spatial ‘‘pseudohelical’’ and ‘‘pseudoplanar’’ configurations of neighboring ribosome arrangements within polysomes. Moreover, tomographic reconstruction of mammalian polysomes obtained from rough ER microsomes out of dog pancreas suggested that ER-embedded polysomes display a flexible spatial organization within the membrane milieu (Pfeffer et al., 2012). During the revision of this paper, the pseudoatomic modeling of very large eukaryotic polysomes was published, providing details about inter-ribosome contacts (Myasnikov et al., 2014). Despite these significant advances in describing the 3D arrangement of ribosomes within polysomes, to date no study has yet analyzed the overall polysome organization in systematic detail. In particular, the previously reported structures of polysomes could not resolve naked mRNA filaments connecting ribosomes on the same transcript.
Here we took advantage of atomic force microscopy (AFM), which has the unique capacity to resolve both single RNA molecules (Hansma et al., 2004) and ribosomes (Yoshida et al., 1997; Mikamo et al., 2005; Mikamo-Satoh et al., 2009). Highlight results from AFM imaging include the visualization of single- and double-stranded nucleic acids (Hansma et al., 2004; Condon, 2006) and of telomere and nucleosome formation (Hansma et al., 2004). In other work, Tetrahymena polysomes were imaged by AFM after fixation, and a comparison with EM images was performed (Yoshida et al., 1997), whereas polysomes isolated from Saccharomyces cerevisiae were observed in solution, including an image showing an exposed RNA strand (Mikamo-Satoh et al., 2009). Therefore, although AFM cannot distinguish the relative orientations of ribosome subunits within polysomes as cryo-EM does (Brandt et al., 2009, 2010), the deep gap in information about the relative organization of ribosome and mRNA can be filled using this complementary technique.
In this study we asked the following: (a) whether polysome shapes are nonstochastic, i.e., whether recurrent ribosomal organizations indicate that specific assemblies are embedded within polysomes; (b) whether the transcripts are completely and homogeneously covered by ribosomes; and (c) whether nonstochastic clusters reflect the translational state of the cell. We combined EM with AFM and high-resolution light microscopy using a stimulated emission depletion (STED) approach (Hell and Wichmann, 1994; Vicidomini et al., 2014) to examine functioning cellular polysomes. The data from these imaging techniques were analyzed by unsupervised classification, statistical analysis, and 3D reconstruction and complemented by cellular and biochemical functional assays. We showed that three distinct clusters of ribosomes recur within polysomes, forming assemblies within them separated by ribosome-free mRNA filaments. We additionally demonstrated that the relative proportions of the three different ribosomal arrangements change in relation to the metabolic state of the cells in which they reside.
Results
Ribosomes are tightly arranged in complex shapes
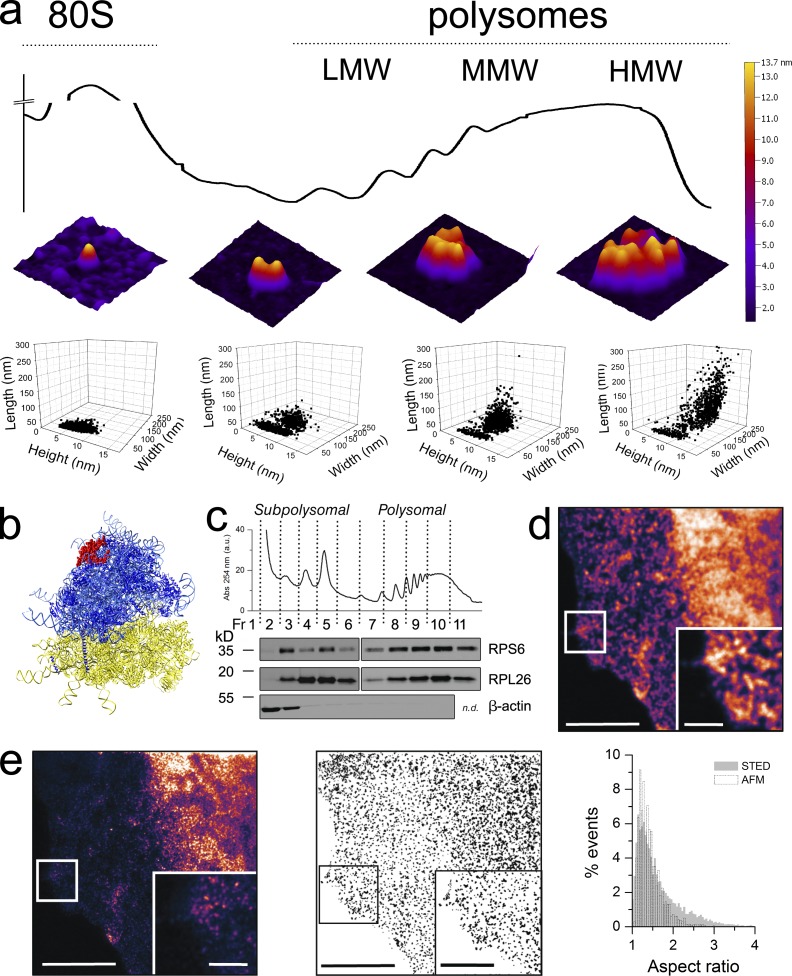
Sucrose density gradient centrifugation followed by fractionation is a standard approach for the gentle purification of polysomes, trapping the ribosomes on mRNAs with the antibiotic cycloheximide. We extracted polysomes from a human cell line (MCF-7) by applying a cytoplasmic lysate to a 15–50% sucrose gradient. The collected fractions were monitored by UV absorption, producing a typical profile (Fig. 1 a). Fractions corresponding to isolated ribosomes (monosomes; 80S) and to low-, medium-, and high-molecular-weight (LMW, MMW, and HMW, respectively) polysomes were individually observed by AFM after deposition on mica (Fig. 1 a). The distributions of the ribosomal size values were obtained by grain analysis (see Materials and methods) to determine a height of 9.1 ± 1.2 nm and a width of 34.1 ± 3.9 nm in air (Fig. 1 a and Table S1). In agreement with an estimated diameter of around 23–25 nm (Warner et al., 1962; Yazaki et al., 2000), the isolated ribosome height measured in solution was 18.5 ± 2.5 nm (Table S1) and 25 nm in polysomes (Fig. S1 b). The observed discrepancy with air AFM height and width, also observed by other groups (Yoshida et al., 1997; Mikamo et al., 2005; Mikamo-Satoh et al., 2009), was expected because of air drying and broadening of the microscope tip. To further ascertain that the observed assemblies were native ribosome groups, sucrose fractions obtained by treating lysates with EDTA or puromycin were adsorbed on mica and observed by AFM (Fig. S1 a). EDTA induced the disassembly of ribosomal subunits, and RNase brought about an increase in free 80S particles, whereas puromycin made for the complete absence of assemblies. The deposition on mica of the fraction corresponding to HMW polysomes pretreated in each of these ways confirmed that the images in Fig. 1 a portray ribosomes and polysomes on the mica surface. We then derived the polysome lateral sizes and compared the obtained values with those for polysomes visualized by transmission EM in tissue slices and cells (Steward and Levy, 1982; Ostroff et al., 2002), as well as in purified samples (Warner et al., 1962; Daneholt et al., 1977; Yoshida et al., 1997; Yazaki et al., 2000; Kopeina et al., 2008; Brandt et al., 2009), finding a good agreement (Table S1).
Figure 1.
Native human polysomes observed by AFM preserve their in vivo organization and reveal tight ribosome interactions. (a) Typical sedimentation profile of polysomes from MCF-7 cell lysates obtained in a concave 15–50% sucrose gradient. The absorbance peaks corresponding to 40S and ribonucleoparticles are not shown for clarity. Fractions containing 80S, LMW, MMW, and HMW polysomes observed by AFM in air are shown as 3D projections (middle row). In the bottom row, the grain analysis of AFM structures is reported, showing the height, width, and length scatterplot for the 80S, LMW, MMW, and HMW polysomes. The 80S fraction data look to be grouped in a single cluster, which is also present in the other three fractions, alongside a second cluster that displays an increase of width and length along the gradient. The total number of objects considered are 1,319 (80S), 1,319 (LMW), 925 (MMW), and 1,187 (HMW) obtained from one to three independent polysomal profiles. (b) Side view of the crystal structure of the human ribosome (Anger et al., 2013) showing the position of RPL26 (or RPL24 according with the new nomenclature proposed by Jenner et al., 2012; in red) at the top of the 60S subunit. (c) RPS6 and RPL26, core proteins of the small and large ribosomal subunit, respectively, are present along sucrose fractions as revealed by Western blotting analysis. Actin is used as a control for nonribosomal fractions and does not cosediment with ribosomes, ribosomal subunits, and polysomes. (d) Magnification of MCF7 cytoplasm immunolabeled with ATTO-488 against RPL26 and imaged by confocal microscopy. (e, left) The same image as in d imaged with g-STED and magnification of cytoplasmic granules. The subdiffraction x–y resolution at 488 nm is <45 nm. The image shown is from a single representative experiment out of three repeats. For the further analysis, we used nine images. (middle) Example of MCF7 image processing for particle analysis and extraction of shape parameters after binarizing and thresholding (see Materials and methods for details). (right) AFM and STED shapes of polysomes are compared plotting the normalized distributions of the aspect ratio (ratio of the major axis to the minor axis) of each particle (3,219 and 8,757 objects, respectively). Bars: (d and e) 5 µm; (d, inset) 1 µm; (e, inset) 2 µm.
By observing the overall organization of ribosomes in polysomes, we distinguished tight ribosome–ribosome interactions and complex clusters of ribosomes with variable shapes, ribosomal arrangements, and 3D stacking, similar to those observed in the earliest micrographs (Warner et al., 1962; Slayter et al., 1963) and the cryo-electron tomography (ET) reconstructions (Brandt et al., 2010; Myasnikov et al., 2014) of human polysomes and ribosome protection assays (Wolin and Walter, 1988).
To further assess whether these multiple shapes reflect the native in vivo organization of polysomes, we used STED nanoscopy. This technique has the advantage of a better resolution (<45 nm) than conventional confocal microscopy and of being fast, with easy sample preparation and data acquisition. We used the ribosomal protein RPL26 (or L24 according to the new nomenclature proposed by Jenner et al. [2012]) as a convenient ribosome tag for immunofluorescence analysis because it is located on the surface of the 60S subunit (Anger et al., 2013), far away from the site of mRNA interaction (Fig. 1, b and c). The achieved resolution (<45 nm) was adequate to identify polysomes but not sharp enough to visualize single ribosomes. In fact, in the polysome the ribosome–ribosome center-to-center distance is 20 nm in AFM but can reach 30–35 nm in EM, depending on the imaging approach (Warner et al., 1962). With STED nanoscopy, we identified clearly defined elongated spots in the cytoplasmic milieu and performed single particle analysis to measure the overall polysome geometric parameters (Fig. 1, d and e). These were then directly compared with the purified polysomes observed using AFM. The comparison demonstrated a clear superimposition in the size distributions of the observed objects (Fig. 1 e). Finally, using the STED point spread function, we started from AFM images of purified polysomes to obtain a simulation of these images in STED and performed a comparison between the obtained aspect ratios (Fig. 2). Dimensions, shapes, and fluorescence intensity in STED magnifications of ribosomal assemblies were very similar to the simulations based on AFM images, confirming that polysomes within cells appear not to differ from those observed by AFM.
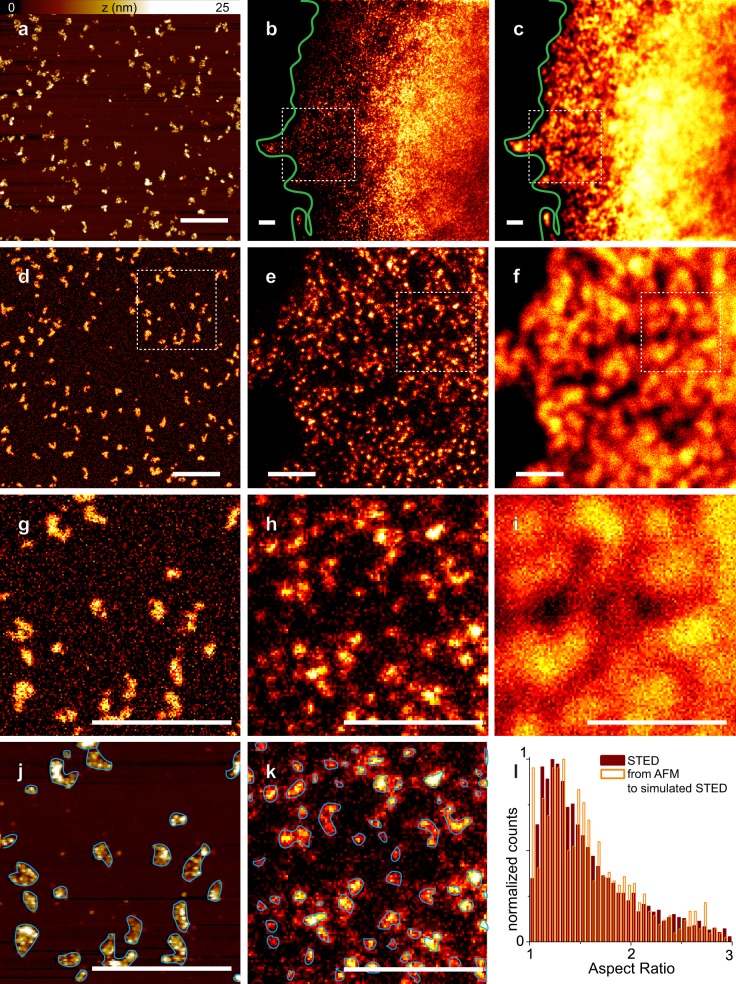
Figure 2.
STED simulations of AFM ribosome clusters display aspect ratios overlapping with objects observed by STED in cells. (a) Example of a typical AFM image. (b and c) STED and confocal images of MCF-7 cells, respectively, where RPL26 has been immunolabeled with ATTO-488. (d) Applying a threshold to the raw AFM image, we generated a binary image that we convolved with the simulated STED point spread function (PSF) after noise addition. The amount of noise was calculated measuring the mean standard deviation of counts in the background of the STED images. (e and f) Zoom of the squared regions of interest in b and c, respectively. The zoom has been chosen to match the size of the AFM image. (g–i) Zoom of the squared regions of interest in d–f, respectively. Fluorescence spots in STED (e and h) are comparable with simulated images of HMW polysomes obtained by convolution of AFM scans with STED PSF (d and g). (j and k) Shown are the result of the object localization procedure (see Materials and methods) applied to the g and h images, respectively. The blue lines show the perimeter of the objects identified. (l) Distribution of the aspect ratio of the objects that arise from the analysis of 10 STED images and 10 simulated STED images from AFM originals. The two graphs are normalized by the whole number of counted objects, respectively, and show a good matching. All images were acquired at 1024 × 1024 pixels with 14-nm pixel size. Bars, 1 µm.
Polysomes can be classified in three main different ribosome clusters
By extensive use of AFM, we then acquired thousands of polysome images (LMW, MMW, and HMW) for analysis by unsupervised object classification. This allowed us to address our first question, whether polysomes display preferential and recurrent shapes. The aim was to unearth possible common conformational features, detected as rotational symmetries, embedded in AFM images. To perform this analysis, we used the standardized single-particle image analysis package Xmipp (Scheres et al., 2008).
To efficiently uncover differences in the rotational symmetry for classification, the original AFM scans were examined for isolated polysomes (Fig. 3 a). Then, the images were aligned translationally and rotationally, and their rotational power spectra were computed (Crowther and Amos, 1971). We then used the Xmipp KerDenSOM algorithm to group the images in classes (or clusters) of objects sharing common rotational symmetries (Pascual-Montano et al., 2001). This method allows the objective identification of groups of images (classes or clusters) sharing similar properties without, importantly, the need for any a priori knowledge on the number of groups or their properties. We found that the separate analysis of the LMW, MMW, and HMW polysomes distinguished in each case three main classes of features, characterized by distinctive rotational spectra, i.e., different levels of rotational symmetry (Fig. 3 b for HMW and Fig. S2 for LMW and MMW).
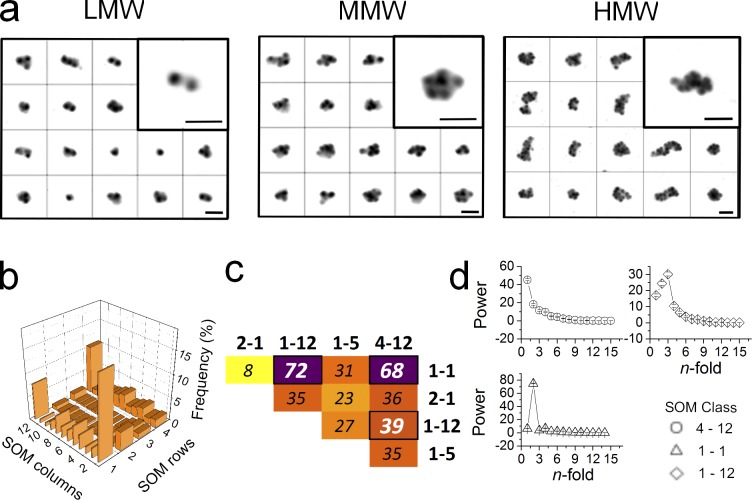
Figure 3.
Polysome clustering by RSC and MMD reveals three recurrent classes. (a) Galleries of LMW, MMW, and HMW polysomes before classification. For each sample under consideration, manual object selection was used to generate galleries of images. We aligned rotationally and translationally 544 objects for the LMW sample, 567 objects for the MMW sample, and 700 objects for the HMW sample. Bars, 100 nm. (b) To identify the most populated classes of polysomes according to their rotational symmetry, the frequency of polysome classes in the SOM (4 rows, 12 columns) was obtained. Three classes appeared to be the most frequent among 674 HMW polysomes (for LMW and HMW refer to Fig. S2). (c) Heat map displaying the top scoring inter-class differences based on the MMD test. The largest differences (thicker stripes) are found between classes 1-1 and 1-12 and between 1-1 and 4-12 (corresponding to the most frequent classes in b). Classes 1-12 and 4-12 show a slightly lower difference. (d) Image classification in b sorted out images into clusters characterized by similar rotational symmetry. Each set of images corresponding to the three most populated classes (i.e., 1-1, 1-12, and 4-12, where the first number refers to the row and the second to the column position in the SOM) was used to obtain the characteristic mean rotational power spectra (mean ± SEM is displayed).
We then studied the statistical significance of the differences between the emerging clusters of ribosomes in polysomes to identify the most diverse classes and statistically assess their differences. We applied the distribution-free two-sample statistical test maximum mean discrepancy (MMD). This test allows evaluation of the diversity between two populations, again without making any assumption on their characteristics. In our case, the higher the value of discrepancy, the higher the difference between the shapes of the polysome classes. The highest diversities obtained for the classification of HMW polysomes are displayed as heat maps (Fig. 3 c and Fig. S2 a for LMW and MMW). The MMD test confirmed that the three emerging groups, corresponding to the three most populated corners of the self-organizing maps (SOMs), were indeed maximally diverse. Less pronounced but still statistically significant differences were also observed for the classes that appeared to be characterized by intermediate rotational symmetries. These classes were less populated, showing a combination of the features characterizing the three main and most abundant classes.
To understand the underlying structural features of the three maximally diverse classes, we explored the corresponding rotational folds of symmetry, showing that each class had a distinctive type (Fig. 3 d for HMW and Fig. S2 c for LMW and MMW). In particular we found a class with onefold symmetry (round shape), a class with twofold symmetry (rectangular shape), and a class with threefold symmetry (triangular shape). None of the intermediate and less populated classes showed additional n-fold symmetries, meaning that the n-fold symmetries n = 1, 2, and 3 are the only ones to emerge from the data. Therefore, these polysomes with intermediate rotational symmetries are not a fourth rotational class, but rather likely transition forms during polysomal organization. Consequently, we decided not to follow up these intermediate classes so as to avoid an unavoidable and arbitrarily subjective intervention in the class selection. The presence of three main fold of symmetries demonstrates that the three classes, associated with the three most frequent and most diverse ribosomal organizations, correspond to different polysomal shapes, or, in other words, to different supraorganizations of ribosomes in the polysome. The fact that the averaged rotational power spectrum of the three main classes revealed the presence of well-resolved peaks corresponding to one-, two-, and threefold symmetry prompted us to call these three classes of polysomes rounded, rectangular, and triangular (Fig. 4 a and Fig. S2 c for galleries), respectively. The same classes were also identified independently in the smaller polysomes (LMW and MMW; Fig. S2), indicating that the propensity to organize these shapes is independent of the number of ribosomes per polysome. Indeed, these shapes may correspond to previously observed organizations of ribosomes in polysomes. In fact, the round shapes most probably resemble the so-called “circular” polysomes (Palade, 1955; Warner et al., 1962; Wettstein et al., 1963; Yazaki et al., 2000; Madin et al., 2004; Afonina et al., 2013), the well-known “rosette” (Palade, 1955; Warner et al., 1962; Wettstein et al., 1963; Madin et al., 2004), and “helical” polysomes (Brandt et al., 2010; Myasnikov et al., 2014); the rectangular shapes are very similar to the “staggered line” (Daneholt et al., 1977), “caterpillar-like double-row” (Kopeina et al., 2008; Afonina et al., 2013), and “pseudohelical” and “pseudoplanar” (Brandt et al., 2009) polysomes; the triangular shapes can be compatible with “spirals” (Palade, 1955; Brandt et al., 2010), “variables” (Brandt et al., 2009), and “loose-helical” polysomes (Brandt et al., 2010). The difference with most of the previous work, performed using in vitro translation systems and specific constructs, is that here we observed polysome populations derived from whole cell lysates. Using then an unbiased approach on thousands of objects, we describe the emergence of common conformations in complex populations.
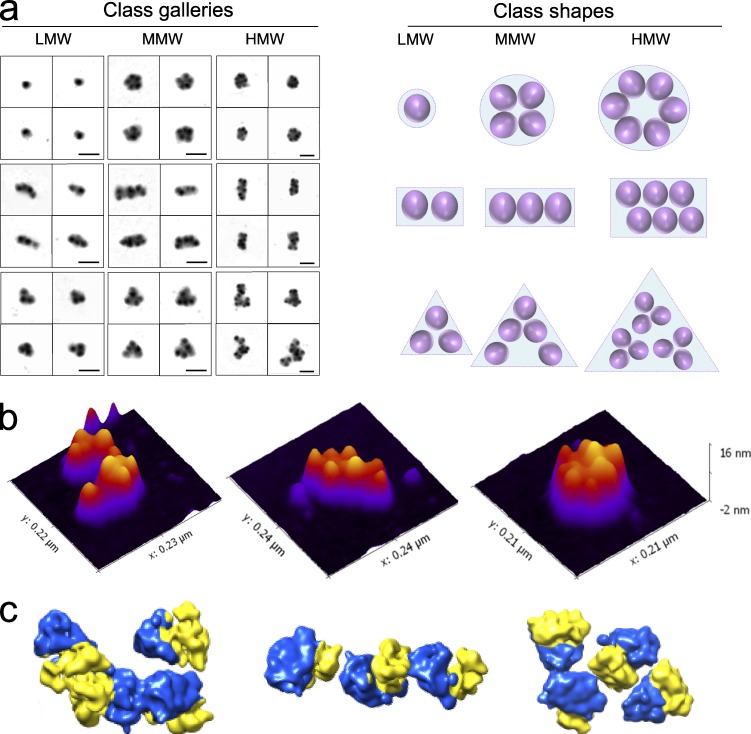
Figure 4.
Three recurrent classes of polysomes display specific conformations. (a, left) Galleries of LMW, MMW, and HMW polysomes showing examples belonging to the three main classes obtained after KerDenSOM classification. (right) Schematic representations of possible ribosome arrangements typical of each class (from top to bottom: rounded, rectangular, and triangular). Bars, 100 nm. (b) Representative images of polysomes for each class are shown by 3D AFM projections (from left to right: triangular, rectangular, and rounded). (c) Representative images of polysomes for each class are shown by cryo-EM, where arrangements of ribosomes were determined by placing a consensus map of all the ribosomes imaged at the positions and in the orientations determined (40S in yellow, 60S in blue; from left to right: triangular, rectangular, and rounded).
To exclude the possibility that the observed polysomal structures were the result of preferential ribosome drop-off, we covalently stabilized the polysome complexes through the GraFix procedure (Kastner et al., 2008), in which a cross-linker is present at low concentration in the sucrose gradient. After analyzing the GraFix-treated HMW fractions, we again obtained three main classes identical to those observed for the uncross-linked samples (Fig. S3, a–e). This excludes the possibility that the observed shapes were artifacts produced by drop-off during sample preparation. Moreover, we ruled out the presence of adverse effects after the drying of polysomes already adherent to our surfaces by comparing the appearance of hydrated samples with that of dried samples. As shown by Fig. S1 b, the dimension of single ribosomes is affected by the drying process (the main consequence being a height decrease of the visualized structures by a factor of 2), but the same polysomal shapes are found in both the hydrated and dried state after classification (Fig. S2 c). Importantly, the same frequencies of the three classes were also observed in both liquid (10% round, 16% rectangular, and 9% triangular) and air-dried polysomes (Fig. 3 b).
To ascertain that the three classes were indeed present in surface-free conditions, as well as when imaged on mica, we observed polysomes by cryo-EM in vitreous ice. After removal of the sucrose, the ribosomes appeared less tightly interacting. Nevertheless, we were able to observe polysomes as localized assemblies, indicating that the ribosome–ribosome interactions were not an effect of the action of osmotic protectant or air drying procedure. This result is in line with the previously observed organization of neighboring ribosomes in the polysome, where they adopt preferential relative orientations that suggest conserved supramolecular organization (Brandt et al., 2009, 2010). We then identified individual ribosomes within micrographs of polysomes and assigned their orientations by projection matching. This yielded the reconstruction of the mammalian 80S ribosome shown in Fig. S4 (a and b). The 80S ribosome was then used to assign the positions and orientations of the individual ribosomes in the polysomes. The overall organization of polysomes obtained with this approach (Fig. 4 c) was very similar to that observed in the AFM images (Fig. 4 b), resembling rectangular, rounded, and triangular shapes. Therefore, these three types of assemblies appear to be intrinsic ultrastructural arrangements of native polysomes.
Various ribosome–ribosome orientations are found in HMW polysomes
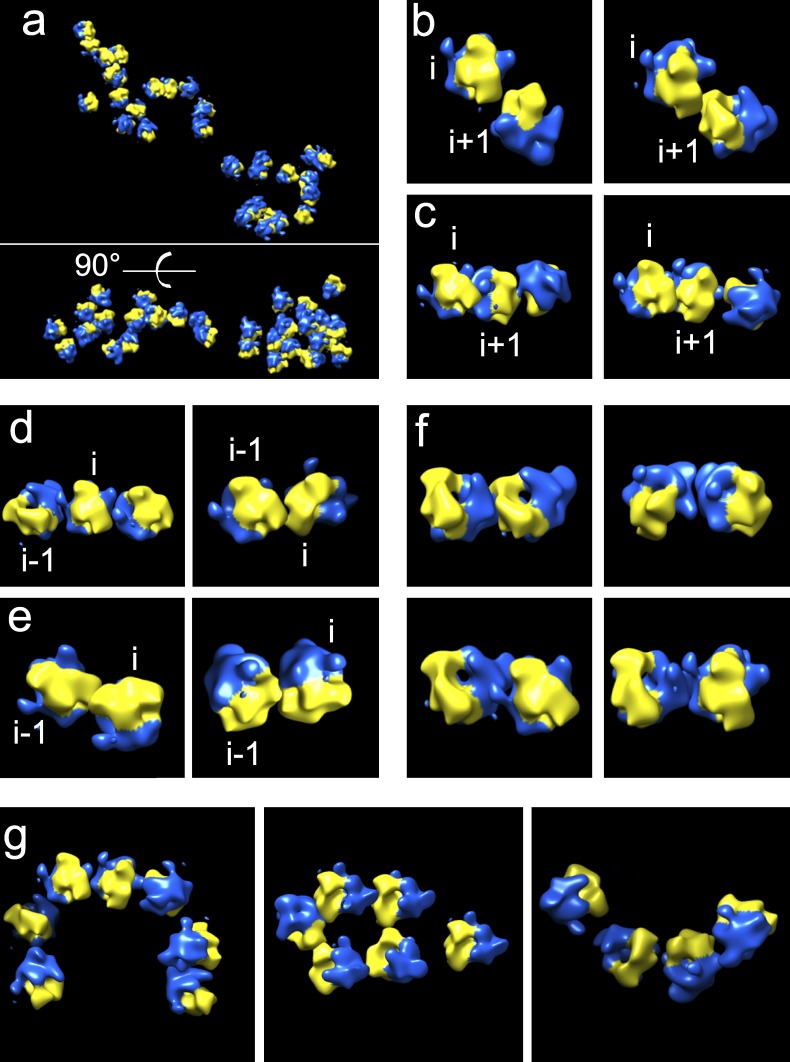
We wondered whether the three polysomal shapes arise from specific ribosome orientations. To this end, electron tomograms of HMW polysomes were collected. We compared the previously described top-top (t-t) and top-bottom (t-b) ribosome arrangements (Brandt et al., 2009, 2010) with our data and asked whether these were responsible for the observed polysome shapes (Fig. 5 a). We identified t-t but not t-b arrangements (Fig. 5, b–e), which were indeed found in bacterial polysomes (Brandt et al., 2009) but not in mammalian cells (Brandt et al., 2010). From this point of view, our results completely agree with the analysis of orientations of mammalian ribosomes observed in situ in cells by Brandt et al. (2010). In many cases (and especially the defined i/i+1 and i/i−1 pairs), the mRNA exit and entrance ribosomes in the + or − direction, respectively, are adjacent, facilitating the threading of the message out of one ribosome and into another.
Figure 5.
Different ribosome–ribosome orientations in HMW polysomes. (a) Example of area of cryo-tomograms that has been fitted with a low-resolution 80S reconstruction (yellow, small subunit; blue, large subunit). (b) Close-up of fitted ribosomes with relative orientations between the ith and i+1 ribosome similar to that described for the t-t class previously (equivalent to cluster 118 in Brandt et al., 2010). (c) Close-up of fitted ribosomes with relative arrangements of the ith and i+1 ribosomes similar to the alternative (cluster 30 in Brandt et al., 2010) arrangement seen previously. Note in addition that these are all mixed assemblies, the ith and i+1 ribosomes are followed by an i+2 ribosome in various relative arrangements: in three cases a basis inversion compared with the other two in the cluster (left and bottom right) and in the other at 90° to them (top right). (d) Close-up of fitted ribosomes with relative orientations between the ith and i−1 ribosome similar to that described for the t-t orientation (see cluster 42 in Brandt et al., 2010). (e) Fitted ribosomes with i to i−1 arrangements apparently similar to an alternative arrangement seen in a previous study (cluster 125 in Brandt et al., 2010). (f) Pairwise arrangements of ribosomes within polysomes that do not appear to correlate to one of the previously described major clusters. (g) Extended, mixed close assemblies of ribosomes identified within the polysomal tomograms.
Linear arrangements of ribosomes, classified as rectangular shapes by rotational classification of AFM images, are consistent with t-t assemblies mixed with other orientations. The round shape, comprising rings of ribosomes, requires mixed arrangements, whereas in the triangular class the t-t arrangement is found for two of the member monosomes, and then a different relative arrangement is responsible for the third monosome. We would like to stress here that the i to i±1 t-t arrangement previously described is one subset of the arrangements observed by Brandt et al. (2010). Interestingly and in addition to the principal relative orientations described in detail before (Brandt et al., 2010), some novel arrangements are repeatedly seen (Fig. 5 f). These novel orientations give rise to mixed configurations of ribosomes in polysomes (Fig. 5 g). Indeed, among the arrangements we were able to see examples of ring-like assemblies, as also seen by AFM, and of helical half-turns, as predicted by Brandt et al. (2010).
Larger polysomes can be formed by combinations of the three types of ribosome assemblies connected by free RNA segments
Complementarily to cryo-ET, AFM can be used to simultaneously observe free RNA and ribosomes. Therefore, we addressed the question of the unknown relative organization of mRNA and ribosomes in polysomes, taking advantage of the good height resolution of AFM and of its unique capacity to clearly detect nucleic acids lying on the mica surface. When the Z-scale was decreased to upper values <2 nm, 25% of polysomes in the HMW fraction showed thin ribbon features that were either protruding outwards or connecting ribosome clusters (Fig. 6, a and b; and Video 1). In agreement with AFM and EM measurements of free RNA (Mikamo-Satoh et al., 2009), the height of the ribbons was found to be ∼0.6–1 nm by cross-section analysis (Fig. 6 b), demonstrating that these structures are bona fide single RNA filaments. The same type of free filaments connecting ribosome clusters were also found on GraFix samples, excluding the possibility that the RNA was naked as the result of artifactual drop-off of ribosomes (Fig. S3 f).
Figure 6.
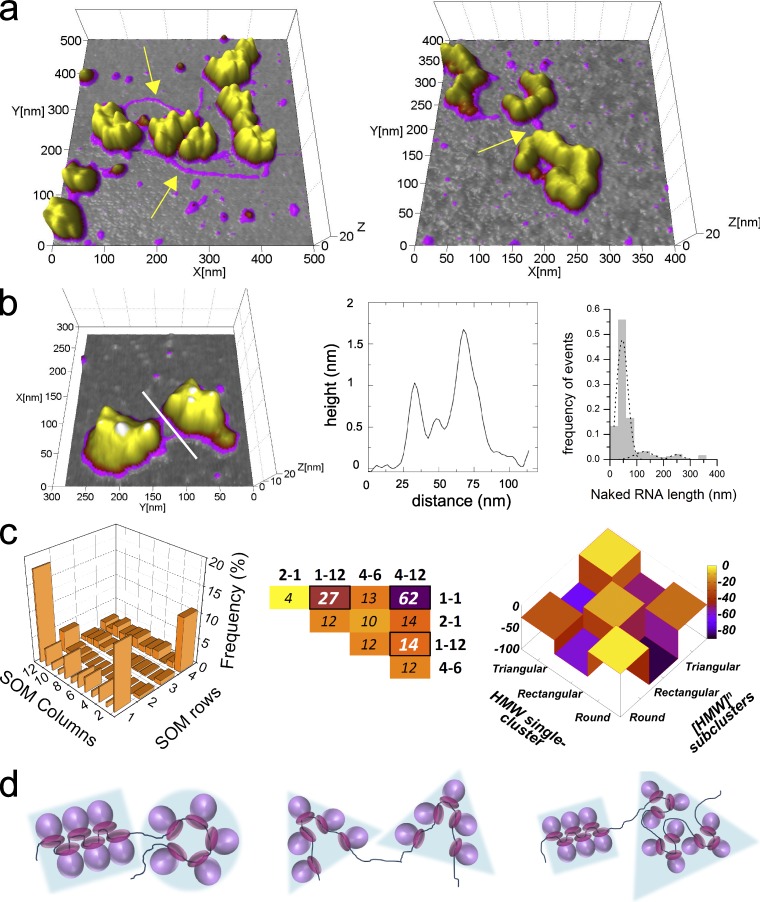
Large polysomes are formed by ribosome clusters connected by naked RNA. (a) AFM 3D projections of large HMW polysomes images. To appreciate both ribosome and RNA height levels, the image is shown using different Z-ranges for the color scale (0–0.5 nm gray [background], 0.5–2 nm magenta [RNA], and 2–10 nm yellow [ribosomes]). Thin strands, with height values <2 nm (see yellow arrows), connect clusters of ribosomes ([HMW]n, 25% of the HMW fraction). (b) 3D projection of HMW polysome formed by two clusters of ribosomes linked by free RNA that appears as a magenta ribbon. Cross-section profiles (solid white line) of RNA connecting adjacent ribosome clusters are shown with corresponding height profile compatible with free RNA. In the right panel, the distribution of naked RNA lengths for 259 [HMW]n polysomes is shown. (c) Ribosome clusters were considered as single objects, isolated, and classified to unveil the underlying rotational symmetry. (left) The distribution of the clusters forming [HMW]n polysomes after rotational spectra computation and KerDenSOM classification is shown. Three classes appeared to be the most represented and the MMD was performed. (middle) The top diversities resulting from the MMD test are displayed as a heat map. (right) To highlight any significant similarity in the ribosome organization between these classes ([HMW]n subclusters) with those found in HMW polysomes (HMW single cluster polysomes) described in Figs. 3 and 4, we compared the corresponding power spectra and ran the MMD over these two populations. Because in this case we are looking for similarities rather than dissimilarities, we plotted the discrepancy values as negative, so the higher the value the higher the similarity. (d) Proposed models of [HMW]n polysomes (n = 2) formed by assemblies of rounded, rectangular, and triangular clusters of ribosomes (cliques) bridged by free RNA.
We then measured the length of naked RNA regions, whose distribution is displayed in Fig. 6 c. The first and more abundant population has a mean value of 45.6 ± 18 nm, corresponding to ∼140 nt of nonprotected RNA. The other two populations are less frequent, with mean values of 128 ± 34 nm and 247.5 ± 21 nm (corresponding to 488 nt and 750 nt of naked RNA, respectively). The frequencies of these populations are likely to be underestimated because it is possible that long naked RNA regions may break during the purification procedure as the result of mechanical stress. Nevertheless, these data may explain or shed light on some incongruences that arise under the hypothesis of homogeneous coverage of the specific transcripts. For example, wheat germ polysomes described by Madin et al. (2004) appear as circular beads on a string with tightly packed ribosomes loaded on a transcript of 1,650 nt. From the images, a maximum of 25 ribosomes per polysome can be counted. Given a ribosome footprinting of around ∼30 nt (Steitz, 1969; Ingolia et al., 2009), a polysome with 25 ribosomes and without free RNA in the coding region should match a transcript of around 750–1,000 nt, which is far below the 1,650 nt coding sequence showed by Madin et al. (2004). The RNA that is missing (around 650–900 nt) has a compatible length with the third population observed in Fig. 6 d. In contrast, considering the evaluation of ribosome occupancy and inter-ribosomal RNA length of 110 nt proposed by Brandt et al. (2009), a polysome with 25 ribosomes should match a transcript 3,750 nt long, which is two times the length of the coding sequence used in Madin et al. (2004). Also, in the hypothesis of beads on a string model, where ribosomes are equally distant and the inter-ribosomal distance ranges between 7 and 15 nm (Rich et al., 1963), the length of the transcript and the number of ribosomes per polysome from several micrographs is longer than the ribosome coverage can explain. A possibility is therefore that some images of polysomes lack naked RNA, giving us an incomplete picture of the native polysome in its complexity. The presence of long naked RNA in the polysome may be instrumental to release possible torsion forces and/or ribosomal crowding, which would flood the translation machinery. In agreement with this observation, a study described irregular distances between ribosomes (Friedberg et al., 1975) and the presence of sparsely stacked ribosome clusters on mRNA after nuclease digestion, suggesting implicitly the presence of free RNA (Wolin and Walter, 1988). Interestingly, Warner et al. (1962) showed EM micrographs of polysomes absorbed on mica where naked RNA of 100–200 nm connects noncontinuous ribosome clusters, similarly to what we observed and is reported in Fig. 6 (a and b). There, the authors hypothesized that long RNA strands emerge as a result of ribosomal dropping out, which is quite unlikely in our case given the use of cycloheximide and the GraFix stabilization (Fig. S3).
The extent of naked RNA in polysomes suggests a model for sparse and isolated assemblies of ribosomes forming composite polysomes (we call these HMW complexes, formed by n subassemblies, [HMW]n complexes). It has to be stressed here that polysomes composed of subassemblies (or subclusters) have not been considered in the previous rotational classification. Therefore, we wondered whether these assemblies showed the very same fold of symmetries found for the single-cluster polysomes (HMW) analyzed before. We used the same computational analysis based on rotational classification and MMD test. First, we separated each subassembly and considered it as a unique object. Then we applied our classification approach to identify classes of objects with a similar rotational symmetry. We obtained again three main classes (Fig. 6 c) displaying the same rotational properties we already identified: rounded, rectangular, and triangular. These clusters gave rise to [HMW]n polysomes in whatever combination with n = 2 or 3. Second, by using MMD, we compared the similarity between the subassemblies forming the [HMW]n polysomes and the single-cluster HMW polysomes in terms of shape similarity. The comparison confirmed that there is a substantial similarity between the rounded, rectangular, and triangular shape classes found in single-cluster HMW polysomes and the corresponding classes found in the subassemblies of [HMW]n complexes (Fig. 6 c, right).
Collectively, these results clearly support the idea that the three classes of ribosome assemblies we identified are the most likely building blocks of larger, composite polysomes (Fig. 6, c and d). We call these suprastructural assemblies of ribosomes forming polysomes “cliques” because of the tendency of their composing ribosomes to establish ordered connections that most likely arise from specific ribosome–ribosome interactions. This result has an obvious consequence with respect to previously published cryo-EM structures: clusters linked by naked RNA regions may belong to the same polysome but could not be identified as separate polysomes if the RNA strand cannot be detected.
The relative frequency distribution of the three polysome classes depends on the translational state of the cell
The observation that polysomes consist of single or combined clusters defined by specific shapes has led us to propose the existence of three repeating structural units of translation or ribosome cliques. This in turn raises the question of whether ribosome cliques also act as functional units within polysomes, similarly to what nucleosomes represent for DNA topology and availability in the context of transcription. To explore this hypothesis and address our third question, i.e., whether nonstochastic clusters reflect the translational state of the cell, we considered the mTOR pathway because it is known to potently regulate translation initiation and integrate translational activity with cell environmental cues (Kang et al., 2013; Kim et al., 2013). We inhibited translation with rapamycin, which affects mTORC1 complex activity (Dowling et al., 2010; Kang et al., 2013; Kim et al., 2013) on its downstream substrates S6K and 4E-BP1/eIF4E (Fig. 7 a, top left). To interrogate the possible role of ribosome clusters in the translational state of the cell, we first identified a convenient rapamycin time–dose condition that would induce a block of translation without an impairment in the number of ribosomes engaged in polysomes.
Figure 7.
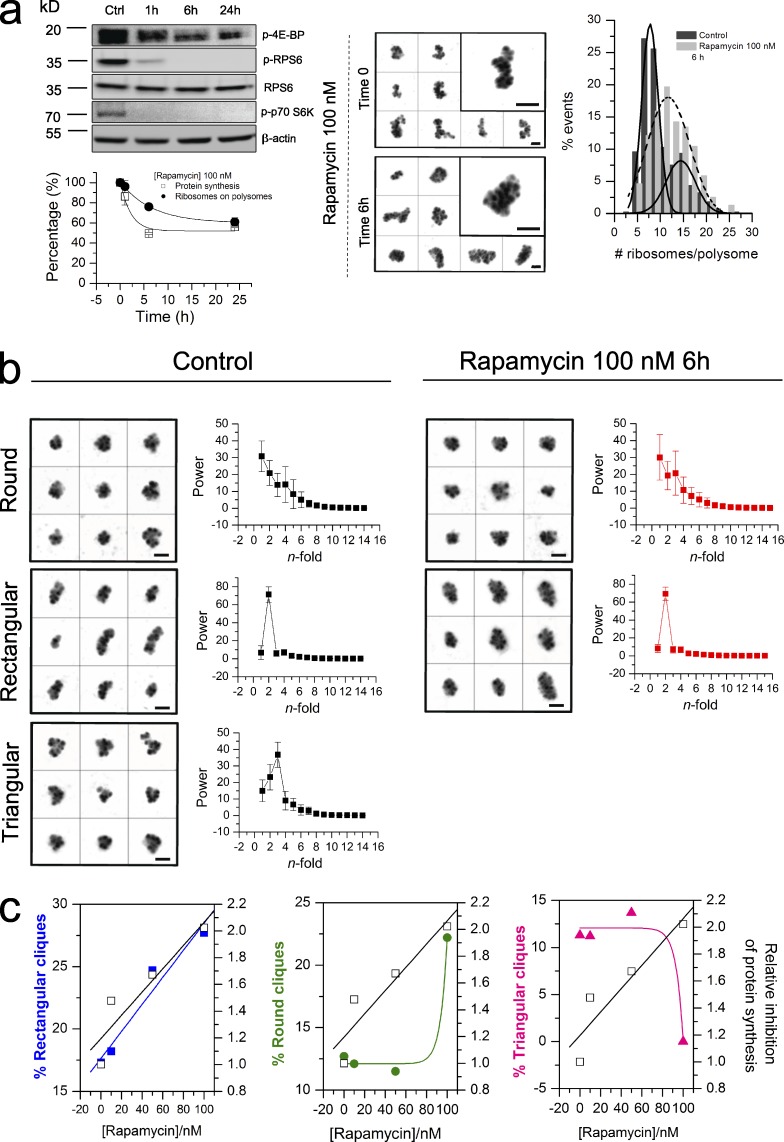
Frequency changes of the three cliques after translation inhibition by rapamycin. (a, top left) Phosphorylation responses by immunoblotting after 1-, 6-, and 24-h treatment with 100 nM rapamycin of known mTORC1 downstream targets (4E-BP, RPS6, and p70-RPS6K) in MCF7 cells. (bottom left) The decrease of protein synthesis was measured by means of AHA incorporation using Click-iT AHA Alexa Fluor 488. The methionine analogue AHA in methionine-free DMEM was added to cells for 30 min. Cells were fixed and incubated with fluorescent alkyne to label the AHA incorporated into nascent proteins. The relative AHA incorporation was assessed by using Operetta HCS. Experiments were run in biological triplicate (mean ± SD is displayed), and 700–1,000 cells were considered for each sample. The kinetics of protein synthesis inhibition (open squares) and the corresponding decrease in polysome content (closed circles) are shown. (middle) Galleries of unclassified HMW polysomes before and after treatment of MCF7 with 100 nM rapamycin. (right) HMW polysomes purified from control cells and cells treated with 100 nM rapamycin for 6 h were adsorbed on the mica and the number of ribosomes in each polysome was counted. The corresponding distributions of the numbers of ribosomes per transcript are shown. (b) Comparison between galleries of classes of HMW polysomes before and after 100 nM rapamycin treatment for 6 h. After grouping the 2D rotational spectra images, the differences in rotational symmetry for distinguishing classes are shown (mean ± SD is displayed). The classification has been performed on 502 and 197 objects purified from MCF7 polysomes before and after rapamycin treatment. (a and b) Bars, 100 nm. (c) Frequencies of rectangular (blue), rounded (green), and triangular cliques (magenta) observed at different rapamycin concentrations are compared with the corresponding protein inhibition (open squares). Protein inhibition is relative to the control (no rapamycin) and was determined by AHA incorporation.
Therefore, we estimated the level of translation inhibition by means of homoazidoalanine (AHA) incorporation and compared it with the corresponding overall polysome content during various time and dose exposures to rapamycin (Fig. 7 a, bottom left). After 6 h of 50 and 100 nM rapamycin treatment, an exponential decrease of the AHA incorporation was observed. At this time point, translation initiation reached the steady-state, whereas the amount of ribosomes engaged in translation, as observed by AFM, was only slightly decreased (∼20%; Fig. 7 a, bottom left). The different time constants (τprotein synthesis = 2.1 h and τpolysome = 6.7 h, for the higher rapamycin dose) suggested that an uncoupling between the changes in the engagement of ribosomes, i.e., in the organization of the protein synthesis machinery, and the protein synthesis functionality was occurring. In other words, at this time point translation was significantly reduced but the translational machinery was still engaged with the template mRNAs. This condition should allow us to observe any fine change occurring in ribosome organization upon translation inhibition. We therefore again used AFM to study the organization of the polysomes purified from rapamycin-treated cells.
We counted the number of ribosomes per polysome and the distribution of polysome classes in control and treated cells. The immediate impression gained was that rapamycin-treated HMW polysomes appeared larger, with more loosely packed conformations (Fig. 7 a, middle). The number of ribosomes engaged in HMW polysomes in standard cell growth conditions displayed a bimodal distribution (Fig. 7 a, right), with peaks at ∼7 and 15 ribosomes per polysome. The results suggest a previously undescribed feature of polysomes: the presence of two distinct peaks in the distribution of ribosomes in polysomes. This finding may also represent an indirect clue in favor of a modular organization of polysomes, with a preferential number of ribosomes needed to form stable assemblies. Remarkably, after adding rapamycin, the bimodality is lost in favor of a monomodal distribution. In this distribution, the mean number of ribosomes per polysome is around 10, owing possibly to diffused stalling of ribosomes on the transcript or to other mechanisms yet to be understood, as proposed by Dahlberg et al. (1973).
We wondered whether the relative distribution of ribosome cliques was affected by rapamycin and whether this might uncover an association with the translational state of the cell. We therefore used the KerDenSOM classification on images of control and rapamycin-treated polysomes (Fig. 7 b) and found that the stronger the translation inhibition induced by rapamycin, the higher the frequency of the rectangular class (Fig. 7 c, left). This result suggests that this shape may reflect a state of inefficient or even inhibited translation. In contrast, the rounded and triangular classes were almost insensitive to low doses of rapamycin. However, at higher rapamycin concentration (100 nM), triangular cliques completely disappeared in favor of rounded cliques, which became abundant (Fig. 7 c, middle and right). This suggests that triangular cliques could correspond to translationally highly active polysomes, whereas rectangular cliques are translationally silent. This latter result might be in agreement with what recently observed by Myasnikov et al. (2014) for “heavy” polysomes obtained from translation of a single transcript with a continuous cell-free wheat germ translation system.
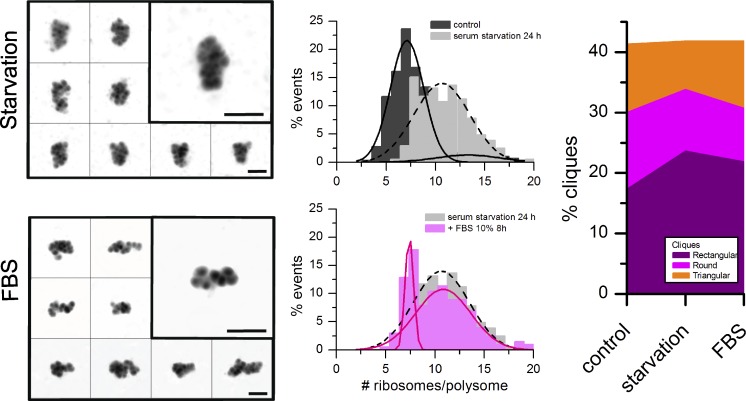
To ensure that the observed clique switches were a general mechanism of translational control rather than a specific effect caused by rapamycin-induced decrease in translational activity, we tested whether the induction of another well-known translation-repressing stimulus, serum withdrawal, could provoke a similar effect and, if so, whether this effect was reversible. After 24 h of serum deprivation, HMW polysomes showed the same loosened conformations and the same increase in the mean number of ribosomes per polysome (Fig. 8, left), as previously observed after rapamycin treatment. Notably, the bimodality of the distribution of the number of ribosomes per polysome was again lost during starvation but restored after serum readdition (Fig. 8, middle). In accordance, starvation led to an increase in rectangular and rounded cliques, whereas serum readdition induced an almost complete recovery of the initial clique frequencies (Fig. 8, right; and Fig. S5), demonstrating that polysome shapes depend on the functionality of cellular translation.
Figure 8.
Frequency changes of the three cliques are reversible. (left) Cells were starved for 24 h, and then 10% FBS was added. After 12 h of serum addition, polysomes obtained from serum-starved cells and after FBS readdition were adsorbed on the mica and imaged by AFM. (The data shown are from a single representative experiment out of three repeats. For the experiment shown, n = 484 and 496, for starvation and FBS readdition, respectively.) Galleries of unclassified HMW polysomes are shown. Bars, 100 nm. (top middle) Comparison between the distribution of the number of ribosomes per polysome in the control (before serum withdrawal) and in starved cells. (bottom middle) Comparison between the distributions of the number of ribosomes per polysome obtained before and after FBS readdition to the starved cells. (right) RSC was run, and the frequency of each clique was calculated in the three conditions.
Discussion
Translation is increasingly recognized as a primary determinant of gene expression control in mammalian cells (Vogel et al., 2010; Schwanhäusser et al., 2011; Tebaldi et al., 2012). Current evidence identifies translation factors (Sonenberg and Hinnebusch, 2009), RNA-binding proteins (Kong and Lasko, 2012), noncoding RNAs (Fabian et al., 2010; Yoon et al., 2012; Memczak et al., 2013), ribosomes themselves (Kondrashov et al., 2011), and the process of mRNA circularization (Craig et al., 1998) as molecular players in such control. Although eukaryotic translation is a hub where several controls of gene expression converge to shape cell phenotypes, no clear data have been available about the topological organization of coding and noncoding regions of mRNAs in polysomes. As reported in the Introduction, polysomes have to date been imaged by EM (Palade, 1955; Warner et al., 1962; Wettstein et al., 1963; Yazaki et al., 2000; Madin et al., 2004) and cryo-EM (Brandt et al., 2009, 2010; Pfeffer et al., 2012; Afonina et al., 2014; Myasnikov et al., 2014) as ribosome assemblies, but their detailed organization has not been statistically characterized nor related to their functional roles.
Prompted by these motivations, we searched for evidence of recurrent ribosome assemblies in polysomes and then looked for their role in translational modulation. We used three different nano-resolution imaging approaches (AFM, STED nanoscopy, and cryo-EM) and found that native human polysomes display heterogeneous but compact conformations. We extensively used AFM because it can be considered a complementary technique with respect to EM and cryo-EM. Compared with these microscopies, AFM has the advantage that no fixation or dying is required, and the sample can be imaged in a liquid environment at room temperature. Unfortunately, AFM cannot visualize ribosome details and needs a flat surface where the sample has to be absorbed. In fact, it is well known that the immobilization of biomolecules on a suitable surface is a prerequisite for their successful visualization by AFM. This fact has both pros and cons. Among the pros is that the use of flat surfaces allows the visualization of thin RNA filaments. For this precise reason in this work, we imaged polysomes on mica pretreated with Ni2+ ions, which are recognized to effectively bind nucleic acids (Hansma and Laney, 1996). The cons include that when a complex biomolecule binds to a surface, the possibility of a distortion of its shape after the onset of interactions with the surface should be considered. Actually, when polyelectrolytes, like nucleic acids, are bound to surfaces, a severe distortion can arise if long-range adhesion forces are present (Sushko et al., 2006). If this is not the case, as in the conditions used in this work, the biomolecule still maintains a 3D conformation after its adhesion to the surface (Pastré et al., 2003). Some relaxation may indeed occur, but a preservation of its general shape is expected. This outcome was also confirmed by a comparison of the polysomal shapes obtained in this work (Fig. S1 b) with those obtained by cryo-EM tomography (Brandt et al., 2009, 2010; Afonina et al., 2014). As a consequence, the shapes of the HMW polysomes in our images appeared different from the particle assemblies described in previous AFM studies (Mikamo et al., 2005; Mikamo-Satoh et al., 2009) but very similar to those seen in cryo-ET (Brandt et al., 2009, 2010; Myasnikov et al., 2014). The complete length of the HMW polysomes observed was similar to that observed in EM, both in tissue slices and cells (Steward and Levy, 1982; Ostroff et al., 2002; Brandt et al., 2010) and in purified samples (Warner et al., 1962; Daneholt et al., 1977; Yazaki et al., 2000; Mikamo et al., 2005; Kopeina et al., 2008; Myasnikov et al., 2014). We found, based on cryo-EM images, that the ribosome orientations in polysomes most probably minimize the probability of contacts between nascent amino acidic chains on ribosomes, as previously suggested (Brandt et al., 2009). This organization confers a tightly packed structure to polysomes in agreement with that suggested in a ribosome protection assay on preprolactin mRNA (Wolin and Walter, 1988). We also demonstrated that the assemblies of ribosomes in polysomes can be divided into three preferential classes, corresponding to distinct shapes, which emerge as building blocks of polysomes. Interestingly, we found by cryo-ET novel orientations between ribosomes in addition to the previously described t-t arrangement (Brandt et al., 2009, 2010). Moreover, ribosome-free mRNA filaments connect these assemblies in a combinatorial way.
In the context of translational control, it has been demonstrated that ribosomes, through ribosomal proteins (Kondrashov et al., 2011; Xue and Barna, 2012), play an additional role to that exerted by translation initiation and elongation (Tuller et al., 2010; Shah et al., 2013; Chu et al., 2014). This may suggest the hypothesis that “specialized ribosomes” with unique protein compositions, or bearing specific posttranscriptional and/or posttranslational modifications so that they have specialized activity, may be a general phenomenon (Topisirovic and Sonenberg, 2011; Xue and Barna, 2012). We therefore reasoned that the possible existence of such ribosomal heterogeneity should give rise to different kinds of ribosome–ribosome interactions, which in turn could influence the global polysome organization. The best-known pathway controlling the activation of key components of translation in response to extracellular stimuli is the AKT–mTORC1 axis (Ma and Blenis, 2009). Well-studied downstream targets of the mTORC1 complex include the eukaryotic initiation factor 4E-BP1 and the 40S ribosomal protein S6, RPS6. Upon phosphorylation, 4E-BP1 dissociates from the cap-binding protein eukaryotic initiation factor 4E (eIF4E), allowing translation initiation to proceed (Sonenberg and Hinnebusch, 2009). mTORC1 also activates S6K1, which is responsible for the phosphorylation of the co-helicase initiator factor 4B (eIF4B) and again, interestingly, of RPS6 (Ma and Blenis, 2009; Sonenberg and Hinnebusch, 2009; Silvera et al., 2010). However, to date, the role played by the phosphorylation state of the ribosomal protein RPS6 has still to be clarified. Clues suggest that structural elements of protein synthesis may be directly involved in translational control of gene expression (Topisirovic and Sonenberg, 2011). 10 years ago the role of ribosomal subunits as additional regulatory elements that could be able to mediate specific interactions between mRNAs and other components of the translation machinery led to the so-called “filter hypothesis” (Mauro and Edelman, 2002). More recently, the dynamic regulation of individual ribosomal proteins in mouse embryos has been demonstrated (Kondrashov et al., 2011). Moreover, in line with the filter hypothesis, the murine ribosomal protein RPL38 has been found to specifically impact translation with the appearance in knockout mice of characteristic phenotypes (Kondrashov et al., 2011). Even if evidence of the direct involvement of ribosome constituents in translational control is emerging, to date, the role in the process of polysomal organization, if any, is unknown. We therefore used two common stimuli able to strongly modulate translation, rapamycin and serum starvation, to understand whether they were able to modify the frequency of the three types of polysomal assemblies we observed. We found that the higher the protein synthesis inhibition, the higher the frequency of the rectangular clique, suggesting that this class reflects a silent translational state triggered by rapamycin in a dose-dependent fashion. The existence of this low-translation class agrees with the fact that ribosomes are known to stall even during physiological translation, owing to secondary mRNA structures, rare codons (Chu et al., 2014), or substrate paucity (Buchan and Stansfield, 2007), or during protein synthesis regulation, posttranslational or transcriptional modifications of ribosomal proteins, or metabolite deprivation (Ortiz et al., 2010). In contrast, the increase in the triangular cliques suggests this class is more prone to characterize translationally efficient polysomes.
The results we present on the morphology and modulation of polysomes do not reveal the molecular mechanisms responsible for these changes, and their elucidation remains a future challenge. As far as we can see, there are no ways to selectively purify polysomes or polysomal subassemblies with specific ribosomal organization, i.e., to isolate specific clique classes. Nevertheless, given our results, we speculate that such molecular mechanisms exist and that they could be induced by posttranscriptional and posttranslational modifications in response to cell signaling events. These modifications may either directly impact the ribosome structure or affect other targets that indirectly promote the remodeling of ribosome–ribosome interactions. Alternatively, the polysomal organization could be driven by mRNA code characteristics (Tuller et al., 2010; Novoa and Ribas de Pouplana, 2012) that impact translational elongation and initiation rates (Shah et al., 2013), by ribosome pausing (Wolin and Walter, 1988), or by the recently proposed “start codon clearance” control (Chu et al., 2014). More studies are therefore required to completely uncover the involvement of specific posttranslational or posttranscriptional modifications as molecular determinants of clique interchange. A particularly intriguing hypothesis is that the RPS6 phosphorylation by mTORC1 could be directly or indirectly involved in the underlying mechanism.
In conclusion, the novelty of our results stands in discovering that mammalian polysomes adopt recurrent nonstochastic shapes, where ribosomes do not homogeneously cover mRNAs, rather leaving naked RNA regions. These nonstochastic shapes reflect the cell translational state of the cell. We therefore propose a novel model of polysomes as spaced assemblies of three distinct ribosome cliques, which can be reversibly modulated in structure as a function of well-known stimuli that affect protein synthesis. Notably, our model opens up the idea that conformational switches in polysomes can be a target of regulation mechanisms. This perspective could trigger a new area of investigation, into whether ribosome cliques can affect proteome fluctuations in ways that could be as important for shaping the cell phenotype as the intensively studied changes in chromatin topology.
Materials and methods
Chemicals
All solution used were prepared in RNase-free water containing 10 µg/ml cycloheximide to prevent ribosome subunit disassembly. All reagents, unless otherwise cited, were molecular biological grade and purchased from Sigma-Aldrich.
Cell culture and polysome purification
MCF-7 breast carcinoma cells were seeded at a density of 2.5 × 104 cells/cm2 and maintained for 3 d in the growth medium (DMEM supplemented with 10% FBS, 2 mM glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin at 37°C, 5% CO2). Once the 80% confluence was reached, cells were incubated for 3–4 min with 10 µg/ml cycloheximide at 37°C to interfere with the translocation step during protein synthesis and to block translational elongation trapping the ribosomes on the mRNA. Cells were washed with PBS + 10 µg/ml cycloheximide and scraped directly on the plate with 300 µl lysis buffer (10 mM NaCl, 10 mM MgCl2, 10 mM Tris-HCl, pH 7.5, 1% Triton X-100, 1% sodium deoxycholate, 0.2 U/µl RNase inhibitor [Fermentas], 10 µg/ml cycloheximide, and 1 mM dithiothreitol) and transferred to an Eppendorf tube. After a few minutes of incubation on ice with occasional vortexing, nuclei and cellular debris were removed by centrifugation for 5 min at 12,000 g at 4°C. The supernatant was directly transferred onto a 15–50% linear sucrose gradient containing 30 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 10 mM MgCl2 and centrifuged in a Sorvall ultracentrifuge on a swinging rotor for 100 min at 180,000 g at 4°C. The fraction corresponding to the 80S peak and those corresponding to the polysomes were collected, monitoring the absorbance at 254 nm. Each fraction was aliquoted, flash frozen in liquid N2, and stored at −80°C for further deposition on mica and AFM imaging. Two negative controls for polysome formation were considered: (1) the cells were supplemented with 100 mg/ml puromycin 10 min before harvesting the lysate and (2) the lysate was supplemented with 100 mM EDTA before the sucrose gradient separation of polysomes.
AFM imaging
For AFM imaging, a few microliters of MCF-7 breast carcinoma 80S or polysome fractions was adsorbed for 3 min on freshly cleaved mica pretreated with Ni2+ for 3 min. The sample was then covered with 10 mM Hepes, pH 7.4, 100 mM NaCl, 10 mM MgCl2, 20 µg/ml cycloheximide, and 3% (wt/vol) sucrose and imaged in liquid. For dried samples, after 1 h of incubation at 20°C, the sample was extensively and gently washed with DEPC-water containing 10 µg/ml cycloheximide and dried at 20°C for at least 1 h.
Imaging was performed at 30°C using a Cypher AFM (Asylum Research) in AC mode, using Asylum routines for the IGOR software environment (WaveMetrics). BL-AC40TS tips (Olympus) were used when imaging in liquid, and OMCL-AC240TS tips (Olympus) were used for dried samples. Nominal spring constants were 0.1 N/m and 2 N/m, respectively. The scanning parameters were as follows: typical excitation frequency 25–30 KHz in liquid, 70 KHz in air, and scanning rate 1–2 Hz. AFM images were levelled line by line and rendered using the SPIP Metrology (Image Metrology) and Gwyddion software packages. Grain analysis was performed with Gwyddion. Image classification was performed with Xmipp. Video 1 was rendered with Blender version 2.66, using a custom shader to obtain the same color scale used in Fig. 6.
Immunofluorescence, STED nanoscopy, and particle analysis
In brief, cells were plated on glass coverslips 24 h before fixation. Cells were pretreated with cycloheximide for 3–4 min, fixed with formaldehyde, permeabilized, blocked, and incubated according to the dilution suggested by the manufacturer’s instructions with 0.01 µg/ml rabbit polyclonal antibody against the N terminus of RPL26 (Abcam) for 1 h at room temperature. Cells were washed extensively and incubated with the secondary antibody goat anti–rabbit ATTO-488 (0.005–1 µg/ml; Sigma-Aldrich) for 45 min. Nuclei were stained while mounting the coverslip with DAPI-Prolong antifade (Invitrogen). Confocal and STED images were acquired at 23°C with a TCS SP5 STED gated (Leica) operated with Leica’s microscope imaging software. All of the images have 14-nm pixel size and 37-µs pixel dwell time. The ATTO-488 fluorescence was excited at 488 nm by means of a supercontinuum-pulsed laser system, and the fluorescence depletion was performed by a 592-nm cw-laser beam. The maximal focal power of the STED beam was 120 mW. Both beams were focused into the 1.4 NA objective lens (HCX PL APO 100× 1.40 NA Oil STED Orange; Leica). Fluorescence was collected by the same lens, filtered with a 592 notch filter, and imaged in the spectral range 500–550 nm by hybrid detector with a time gating of 1.5 ns.
We performed the analysis of polysome clusters in human cells over 26 images of 16 different cells. Image analysis was performed using the Fiji software. We applied the Bersen auto local threshold plugin to the images to isolate the fluorescent particles. The parameter was chosen to set as positive the pixels that have a gray level higher than the half of the local maximum. The plugin generates a binary image that we analyzed with the “analyze particles” function. For each closed region of positive pixels, the plugin allows the measurement of the shape descriptors and the ellipse fit of the particles. Finally, we plotted the distribution of the major (we called length) and minor (we called width) axis of the ellipse, the area, the aspect ratio, and the solidity of the analyzed particles.
Cryo-EM
Preparations of polysomes were plunge-frozen on 300-mesh lacey carbon grids in liquid ethane. EM of polysomes for single-particle analysis was performed using an FEI F30 cryo-microscope, capturing images at 300 keV at 36,000 magnification on SO-163 film. The films were scanned using an SCAI scanner (pixel size 7 µm, 1.8 Å at the specimen; Carl Zeiss). Ribosomes within polysomes were subsequently identified using Boxer (EMAN suite; Ludtke et al., 1999) and excised for analysis in Spider (Frank et al., 1996). A 3D reconstruction was determined by projection matching using as a model a rabbit reticulocyte ribosome reconstruction filtered to 80-Å resolution. This 3D map was then used to interpret electron tomograms of the same sample and for visualization of the relative orientations of ribosomes within polysomal assemblies. Sections through the map are shown in Fig. S4 a.
Electron tomograms of polysomes (dosed with 10-nm gold markers to act as fiducials in alignment) were collected using a Polara cryo-microscope (FEI) operating at 300 keV and fitted with a charge-coupled device camera for data capture (Ultrascan 4000SP; Gatan). The microscope was fitted with an energy filter (GIF 2002; Gatan) operating in zero-energy-loss mode with a slit width of 20 eV, and images were taken at 95,000 magnification with a pixel size of 4 Å. Single-axis tilt series were collected over an angular range of approximately −65° to 65° at 2° increments using the program SerialEM under low-dose conditions (Mastronarde, 2005). The tomograms were reconstructed using IMOD software (Kremer et al., 1996) by weighted back projection (Sandberg et al., 2003). Tomogram maps were visualized using UCSF Chimera software (Pettersen et al., 2004; Guichard et al., 2010), with density inversion and applying a low-pass filter of 40 Å, and a low-resolution 3D 80S reconstruction was automatically fitted into regions of continuous electron density within the Chimera software.
2D rotational spectra classification (RSC)
To classify the AFM images with 2D RSC by the Xmipp software (Scheres et al., 2008), single objects (ribosomes or polysomes) were manually picked and aligned rotationally and translationally from 2-µm × 2-µm images acquired at 1,024 pixels × 1,024 pixels corresponding to an x–y pixel dimension of 1.9-nm resolution. The obtained images were analyzed by Kernel Probability Density Estimator SOM (KerDenSOM) clustering applied to individual rotational spectra to identify structurally homogeneous image sets (Pascual-Montano et al., 2001). This method allows identification of different classes according to their rotational symmetry. The most diverse groups of rotational spectra tend to be pushed toward the corners of the KerDenSOM output map. The images belonging to each corner for each sample considered (LMW, MMW, and HMW polysomes) were used to produce a gallery of polysomes with defined rotational characteristics.
Protein synthesis assay
10,000 cells were grown for 24 h in 96-well plates and then (a) treated with Rapamycin (10, 50, or 100 nm; for 1, 6, or 24 h), (b) serum starved (0.5% FBS for 24 h), or (c) serum starved (0.5% FBS for 24 h) and then stimulated with 10% FBS for 8 h. Protein synthesis was measured using Click-iT AHA Alexa Fluor 488 Protein Synthesis HCS Assay (Life Technologies) according to the manufacturer’s protocol.
Statistics
The diversity between the groups identified by the KerDenSOM output maps was further evaluated by MMD, a powerful nonparametric two-sample test determining whether two samples are drawn from different distributions (Gretton et al., 2012). The significance level α was set to 0.01, with Bonferroni correction for multiple testing. For each sample, the 10 pairwise comparisons with the highest statistic (all statistically significant) were graphically arranged as heat maps.
Online supplemental material
Fig. S1 shows controls of AFM imaging: ribosome assemblies disappear after EDTA and puromycin treatments and display the expected height when measured in liquid. Fig. S2 shows the classification of LMW, MMW, and HMW polysomes in air and HMW polysomes in liquid by KerDenSOM maps. Fig. S3 shows the AFM characterization and RSC of the HMW polysome fraction after GraFix. Fig. S4 shows the cryo-EM and cryo-ET ribosome and a gallery of polysomes. Fig. S5 shows RSC of polysomes after serum starvation and 10% FBS readdition (relative to Fig. 8). Table S1 lists the distribution of size values for 80S in liquid and air and LMW, MMW, and HMW polysomes obtained using grain analysis of AFM images (relative to Fig. 1). Video 1 shows the gallery of a 3D collection of composite polysomes with naked RNA as 3D objects where polysomes show subclusters of ribosomes. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201406040/DC1.
Supplementary Material
Acknowledgments
We thank W. Filipowicz and R. Mendez for fruitful discussions. We also wish to thank Valentina Adami and the High-Throughput Screening Core Facility of the Centre for Integrative Biology (CIBIO), University of Trento, for the experiments of AHA incorporation and Matteo Gaglio and Natalia Arseni for technical assistance.
R.J. Gilbert was a Royal Society University Research Fellow, and the Oxford Division of Structural Biology is part of the Wellcome Trust Centre for Human Genetics, Wellcome Trust Core Award grant number 090532/Z/09/Z. We acknowledge the University of Trento CIBIO start-up grant for funding.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AFM
- atomic force microscopy
- AHA
- homoazidoalanine
- ET
- electron tomography
- HMW
- high molecular weight
- LMW
- low molecular weight
- MMD
- maximum mean discrepancy
- MMW
- medium molecular weight
- RSC
- rotational spectra classification
- SOM
- self-organizing map
- STED
- stimulated emission depletion
References
- Afonina ZhA., Myasnikov A.G., Khabibullina N.F., Belorusova A.Y., Menetret J.-F., Vasiliev V.D., Klaholz B.P., Shirokov V.A., and Spirin A.S.. 2013. Topology of mRNA chain in isolated eukaryotic double-row polyribosomes. Biochemistry Mosc. 78:445–454 10.1134/S0006297913050027 [DOI] [PubMed] [Google Scholar]
- Afonina Z.A., Myasnikov A.G., Shirokov V.A., Klaholz B.P., and Spirin A.S.. 2014. Formation of circular polyribosomes on eukaryotic mRNA without cap-structure and poly(A)-tail: a cryo electron tomography study. Nucleic Acids Res. 42:9461–9469 10.1093/nar/gku599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anger A.M., Armache J.-P., Berninghausen O., Habeck M., Subklewe M., Wilson D.N., and Beckmann R.. 2013. Structures of the human and Drosophila 80S ribosome. Nature. 497:80–85 10.1038/nature12104 [DOI] [PubMed] [Google Scholar]
- Brandt F., Etchells S.A., Ortiz J.O., Elcock A.H., Hartl F.U., and Baumeister W.. 2009. The native 3D organization of bacterial polysomes. Cell. 136:261–271 10.1016/j.cell.2008.11.016 [DOI] [PubMed] [Google Scholar]
- Brandt F., Carlson L.-A., Hartl F.U., Baumeister W., and Grünewald K.. 2010. The three-dimensional organization of polyribosomes in intact human cells. Mol. Cell. 39:560–569 10.1016/j.molcel.2010.08.003 [DOI] [PubMed] [Google Scholar]
- Buchan J.R., and Stansfield I.. 2007. Halting a cellular production line: responses to ribosomal pausing during translation. Biol. Cell. 99:475–487 10.1042/BC20070037 [DOI] [PubMed] [Google Scholar]
- Chu D., Kazana E., Bellanger N., Singh T., Tuite M.F., and von der Haar T.. 2014. Translation elongation can control translation initiation on eukaryotic mRNAs. EMBO J. 33:21–34 10.1002/embj.201385651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon A.2006. Designed DNA molecules: principles and applications of molecular nanotechnology. Nat. Rev. Genet. 7:565–575 10.1038/nrg1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig A.W., Haghighat A., Yu A.T., and Sonenberg N.. 1998. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 392:520–523 10.1038/33198 [DOI] [PubMed] [Google Scholar]
- Crowther R.A., and Amos L.A.. 1971. Harmonic analysis of electron microscope images with rotational symmetry. J. Mol. Biol. 60:123–130 10.1016/0022-2836(71)90452-9 [DOI] [PubMed] [Google Scholar]
- Dahlberg A.E., Lund E., and Kjeldgaard N.O.. 1973. Some effects of antibiotics on bacterial polyribosomes as studied by gel electrophoresis. J. Mol. Biol. 78:627–636 10.1016/0022-2836(73)90284-2 [DOI] [PubMed] [Google Scholar]
- Daneholt B., Anderson K., and Fagerlind M.. 1977. Large-sized polysomes in Chironomus tentans salivary glands and their relation to Balbiani ring 75S RNA. J. Cell Biol. 73:149–160 10.1083/jcb.73.1.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling R.J.O., Topisirovic I., Fonseca B.D., and Sonenberg N.. 2010. Dissecting the role of mTOR: lessons from mTOR inhibitors. Biochim. Biophys. Acta. 1804:433–439 10.1016/j.bbapap.2009.12.001 [DOI] [PubMed] [Google Scholar]
- Fabian M.R., Sonenberg N., and Filipowicz W.. 2010. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79:351–379 10.1146/annurev-biochem-060308-103103 [DOI] [PubMed] [Google Scholar]
- Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., and Leith A.. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190–199 10.1006/jsbi.1996.0030 [DOI] [PubMed] [Google Scholar]
- Friedberg I., Herzberg M., Zarivatch S., and Ron E.Z.. 1975. Polysomes in Escherichia coli during amino acid starvation: structural change observed by electron microscopy. FEBS Lett. 56:108–110 10.1016/0014-5793(75)80121-9 [DOI] [PubMed] [Google Scholar]
- Gretton A., Borgwardt K.M., Rasch M.J., Schölkopf B., and Smola A.. 2012. A kernel two-sample test. J. Mach. Learn. Res. 13:723–773. [Google Scholar]
- Guichard P., Chrétien D., Marco S., and Tassin A.-M.. 2010. Procentriole assembly revealed by cryo-electron tomography. EMBO J. 29:1565–1572 10.1038/emboj.2010.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansma H.G., and Laney D.E.. 1996. DNA binding to mica correlates with cationic radius: assay by atomic force microscopy. Biophys. J. 70:1933–1939 10.1016/S0006-3495(96)79757-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansma H.G., Kasuya K., and Oroudjev E.. 2004. Atomic force microscopy imaging and pulling of nucleic acids. Curr. Opin. Struct. Biol. 14:380–385 10.1016/j.sbi.2004.05.005 [DOI] [PubMed] [Google Scholar]
- Harms J., Schluenzen F., Zarivach R., Bashan A., Gat S., Agmon I., Bartels H., Franceschi F., and Yonath A.. 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 107:679–688 10.1016/S0092-8674(01)00546-3 [DOI] [PubMed] [Google Scholar]
- Hell S.W., and Wichmann J.. 1994. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 19:780–782 10.1364/OL.19.000780 [DOI] [PubMed] [Google Scholar]
- Ingolia N.T., Ghaemmaghami S., Newman J.R.S., and Weissman J.S.. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 324:218–223 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner L., Melnikov S., Garreau de Loubresse N., Ben-Shem A., Iskakova M., Urzhumtsev A., Meskauskas A., Dinman J., Yusupova G., and Yusupov M.. 2012. Crystal structure of the 80S yeast ribosome. Curr. Opin. Struct. Biol. 22:759–767 10.1016/j.sbi.2012.07.013 [DOI] [PubMed] [Google Scholar]
- Kang S.A., Pacold M.E., Cervantes C.L., Lim D., Lou H.J., Ottina K., Gray N.S., Turk B.E., Yaffe M.B., and Sabatini D.M.. 2013. mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science. 341:1236566 10.1126/science.1236566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner B., Fischer N., Golas M.M., Sander B., Dube P., Boehringer D., Hartmuth K., Deckert J., Hauer F., Wolf E., et al. . 2008. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods. 5:53–55 10.1038/nmeth1139 [DOI] [PubMed] [Google Scholar]
- Kim S.G., Buel G.R., and Blenis J.. 2013. Nutrient regulation of the mTOR complex 1 signaling pathway. Mol. Cells. 35:463–473 10.1007/s10059-013-0138-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov N., Pusic A., Stumpf C.R., Shimizu K., Hsieh A.C., Xue S., Ishijima J., Shiroishi T., and Barna M.. 2011. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 145:383–397 10.1016/j.cell.2011.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J., and Lasko P.. 2012. Translational control in cellular and developmental processes. Nat. Rev. Genet. 13:383–394 10.1038/nrg3184 [DOI] [PubMed] [Google Scholar]
- Kopeina G.S., Afonina Z.A., Gromova K.V., Shirokov V.A., Vasiliev V.D., and Spirin A.S.. 2008. Step-wise formation of eukaryotic double-row polyribosomes and circular translation of polysomal mRNA. Nucleic Acids Res. 36:2476–2488 10.1093/nar/gkm1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., and McIntosh J.R.. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Ludtke S.J., Baldwin P.R., and Chiu W.. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82–97 10.1006/jsbi.1999.4174 [DOI] [PubMed] [Google Scholar]
- Ma X.M., and Blenis J.. 2009. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 10:307–318 10.1038/nrm2672 [DOI] [PubMed] [Google Scholar]
- Madin K., Sawasaki T., Kamura N., Takai K., Ogasawara T., Yazaki K., Takei T., Miura K., and Endo Y.. 2004. Formation of circular polyribosomes in wheat germ cell-free protein synthesis system. FEBS Lett. 562:155–159 10.1016/S0014-5793(04)00221-2 [DOI] [PubMed] [Google Scholar]
- Mastronarde D.N.2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- Mauro V.P., and Edelman G.M.. 2002. The ribosome filter hypothesis. Proc. Natl. Acad. Sci. USA. 99:12031–12036 10.1073/pnas.192442499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov S., Ben-Shem A., Garreau de Loubresse N., Jenner L., Yusupova G., and Yusupov M.. 2012. One core, two shells: bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 19:560–567 10.1038/nsmb.2313 [DOI] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., et al. . 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 495:333–338 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- Mikamo E., Tanaka C., Kanno T., Akiyama H., Jung G., Tanaka H., and Kawai T.. 2005. Native polysomes of Saccharomyces cerevisiae in liquid solution observed by atomic force microscopy. J. Struct. Biol. 151:106–110 10.1016/j.jsb.2005.05.002 [DOI] [PubMed] [Google Scholar]
- Mikamo-Satoh E., Takagi A., Tanaka H., Matsumoto T., Nishihara T., and Kawai T.. 2009. Profiling of gene-dependent translational progress in cell-free protein synthesis by real-space imaging. Anal. Biochem. 394:275–280 10.1016/j.ab.2009.07.033 [DOI] [PubMed] [Google Scholar]
- Myasnikov A.G., Afonina Z.A., Ménétret J.-F., Shirokov V.A., Spirin A.S., and Klaholz B.P.. 2014. The molecular structure of the left-handed supra-molecular helix of eukaryotic polyribosomes. Nat. Commun. 5:5294 10.1038/ncomms6294 [DOI] [PubMed] [Google Scholar]
- Novoa E.M., and Ribas de Pouplana L.. 2012. Speeding with control: codon usage, tRNAs, and ribosomes. Trends Genet. 28:574–581 10.1016/j.tig.2012.07.006 [DOI] [PubMed] [Google Scholar]
- Ortiz J.O., Brandt F., Matias V.R.F., Sennels L., Rappsilber J., Scheres S.H.W., Eibauer M., Hartl F.U., and Baumeister W.. 2010. Structure of hibernating ribosomes studied by cryoelectron tomography in vitro and in situ. J. Cell Biol. 190:613–621 10.1083/jcb.201005007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostroff L.E., Fiala J.C., Allwardt B., and Harris K.M.. 2002. Polyribosomes redistribute from dendritic shafts into spines with enlarged synapses during LTP in developing rat hippocampal slices. Neuron. 35:535–545 10.1016/S0896-6273(02)00785-7 [DOI] [PubMed] [Google Scholar]
- Palade G.E.1955. A small particulate component of the cytoplasm. J. Biophys. Biochem. Cytol. 1:59–68 10.1083/jcb.1.1.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-Montano A., Donate L.E., Valle M., Bárcena M., Pascual-Marqui R.D., and Carazo J.M.. 2001. A novel neural network technique for analysis and classification of EM single-particle images. J. Struct. Biol. 133:233–245 10.1006/jsbi.2001.4369 [DOI] [PubMed] [Google Scholar]
- Pastré D., Piétrement O., Fusil S., Landousy F., Jeusset J., David M.-O., Hamon L., Le Cam E., and Zozime A.. 2003. Adsorption of DNA to mica mediated by divalent counterions: a theoretical and experimental study. Biophys. J. 85:2507–2518 10.1016/S0006-3495(03)74673-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., and Ferrin T.E.. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Pfeffer S., Brandt F., Hrabe T., Lang S., Eibauer M., Zimmermann R., and Förster F.. 2012. Structure and 3D arrangement of endoplasmic reticulum membrane-associated ribosomes. Structure. 20:1508–1518 10.1016/j.str.2012.06.010 [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V.2002. Ribosome structure and the mechanism of translation. Cell. 108:557–572 10.1016/S0092-8674(02)00619-0 [DOI] [PubMed] [Google Scholar]
- Rich A., Penman S., Becker Y., Darnell J., and Hall C.. 1963. Polyribosomes: Size in normal and Polio-infected HeLa cells. Science. 142:1658–1663 10.1126/science.142.3600.1658 [DOI] [PubMed] [Google Scholar]
- Sandberg K., Mastronarde D.N., and Beylkin G.. 2003. A fast reconstruction algorithm for electron microscope tomography. J. Struct. Biol. 144:61–72 10.1016/j.jsb.2003.09.013 [DOI] [PubMed] [Google Scholar]
- Scheres S.H.W., Núñez-Ramírez R., Sorzano C.O.S., Carazo J.M., and Marabini R.. 2008. Image processing for electron microscopy single-particle analysis using XMIPP. Nat. Protoc. 3:977–990 10.1038/nprot.2008.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluenzen F., Tocilj A., Zarivach R., Harms J., Gluehmann M., Janell D., Bashan A., Bartels H., Agmon I., Franceschi F., and Yonath A.. 2000. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 102:615–623 10.1016/S0092-8674(00)00084-2 [DOI] [PubMed] [Google Scholar]
- Schmeing T.M., and Ramakrishnan V.. 2009. What recent ribosome structures have revealed about the mechanism of translation. Nature. 461:1234–1242 10.1038/nature08403 [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., and Selbach M.. 2011. Global quantification of mammalian gene expression control. Nature. 473:337–342 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- Shah P., Ding Y., Niemczyk M., Kudla G., and Plotkin J.B.. 2013. Rate-limiting steps in yeast protein translation. Cell. 153:1589–1601 10.1016/j.cell.2013.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvera D., Formenti S.C., and Schneider R.J.. 2010. Translational control in cancer. Nat. Rev. Cancer. 10:254–266 10.1038/nrc2824 [DOI] [PubMed] [Google Scholar]
- Slayter H.S., Warner J.R., Rich A., and Hall C.E.. 1963. The visualization of polyribosomal structure. J. Mol. Biol. 7:652–657 10.1016/S0022-2836(63)80112-6 [DOI] [PubMed] [Google Scholar]
- Sonenberg N., and Hinnebusch A.G.. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 136:731–745 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz J.A.1969. Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature. 224:957–964 10.1038/224957a0 [DOI] [PubMed] [Google Scholar]
- Steward O., and Levy W.B.. 1982. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J. Neurosci. 2:284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sushko M.L., Shluger A.L., and Rivetti C.. 2006. Simple model for DNA adsorption onto a mica surface in 1:1 and 2:1 electrolyte solutions. Langmuir. 22:7678–7688 10.1021/la060356+ [DOI] [PubMed] [Google Scholar]
- Tebaldi T., Re A., Viero G., Pegoretti I., Passerini A., Blanzieri E., and Quattrone A.. 2012. Widespread uncoupling between transcriptome and translatome variations after a stimulus in mammalian cells. BMC Genomics. 13:220 10.1186/1471-2164-13-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topisirovic I., and Sonenberg N.. 2011. Translational control by the eukaryotic ribosome. Cell. 145:333–334 10.1016/j.cell.2011.04.006 [DOI] [PubMed] [Google Scholar]
- Tuller T., Carmi A., Vestsigian K., Navon S., Dorfan Y., Zaborske J., Pan T., Dahan O., Furman I., and Pilpel Y.. 2010. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 141:344–354 10.1016/j.cell.2010.03.031 [DOI] [PubMed] [Google Scholar]
- Vicidomini G., Hernández I.C., d’Amora M., Zanacchi F.C., Bianchini P., and Diaspro A.. 2014. Gated CW-STED microscopy: a versatile tool for biological nanometer scale investigation. Methods. 66:124–130 10.1016/j.ymeth.2013.06.029 [DOI] [PubMed] [Google Scholar]
- Vogel C., Abreu R.S., Ko D., Le S.Y., Shapiro B.A., Burns S.C., Sandhu D., Boutz D.R., Marcotte E.M., and Penalva L.O.. 2010. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol. Syst. Biol. 6:400 10.1038/msb.2010.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J.R., Rich A., and Hall C.E.. 1962. Electron microscope studies of ribosomal clusters synthesizing hemoglobin. Science. 138:1399–1403 10.1126/science.138.3548.1399 [DOI] [PubMed] [Google Scholar]
- Wettstein F.O., Staehelin T., and Noll H.. 1963. Ribosomal aggregate engaged in protein synthesis: characterization of the ergosome. Nature. 197:430–435 10.1038/197430a0 [DOI] [PubMed] [Google Scholar]
- Wolin S.L., and Walter P.. 1988. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 7:3559–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S., and Barna M.. 2012. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 13:355–369 10.1038/nrm3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki K., Yoshida T., Wakiyama M., and Miura K.. 2000. Polysomes of eukaryotic cells observed by electron microscopy. J. Electron Microsc. (Tokyo). 49:663–668 10.1093/oxfordjournals.jmicro.a023856 [DOI] [PubMed] [Google Scholar]
- Yoon J.-H., Abdelmohsen K., Srikantan S., Yang X., Martindale J.L., De S., Huarte M., Zhan M., Becker K.G., and Gorospe M.. 2012. LincRNA-p21 suppresses target mRNA translation. Mol. Cell. 47:648–655 10.1016/j.molcel.2012.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Wakiyama M., Yazaki K., and Miura K.. 1997. Transmission electron and atomic force microscopic observation of polysomes on carbon-coated grids prepared by surface spreading. J. Electron Microsc. (Tokyo). 46:503–506 10.1093/oxfordjournals.jmicro.a023550 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.