New roles for the N-terminal histone tail and folded core of CENP-A are revealed by monitoring early steps in centromere establishment.
Abstract
The centromere—defined by the presence of nucleosomes containing the histone H3 variant, CENP-A—is the chromosomal locus required for the accurate segregation of chromosomes during cell division. Although the sequence determinants of human CENP-A required to maintain a centromere were reported, those that are required for early steps in establishing a new centromere are unknown. In this paper, we used gain-of-function histone H3 chimeras containing various regions unique to CENP-A to investigate early events in centromere establishment. We targeted histone H3 chimeras to chromosomally integrated Lac operator sequences by fusing each of the chimeras to the Lac repressor. Using this approach, we found surprising contributions from a small portion of the N-terminal tail and the CENP-A targeting domain in the initial recruitment of two essential constitutive centromere proteins, CENP-C and CENP-T. Our results indicate that the regions of CENP-A required for early events in centromere establishment differ from those that are required for maintaining centromere identity.
Introduction
Genetic inheritance at cell division relies upon a region of the chromosome called the centromere. In many eukaryotes, centromeres—including new ones (i.e., neocentromeres) that must be established if the original centromere is lost—are not defined by a particular DNA sequence but rather by the presence of nucleosomes containing a histone H3 variant, CENP-A (Black and Cleveland, 2011). CENP-A is targeted to centromeres once per cell cycle via a self-propagation mechanism wherein the existing pool of CENP-A nucleosomes is at the top of a hierarchy of protein–protein interactions that culminate in the local assembly of newly expressed CENP-A (Westhorpe and Straight, 2015). CENP-A nucleosomes are also at the top of the hierarchy for recruitment of proteins required for kinetochore function during chromosome segregation. To dissect the roles of CENP-A in centromere maintenance, gain-of-function histone H3 chimeras containing various regions unique to CENP-A have been particularly helpful (Black et al., 2004, 2007; Carroll et al., 2009, 2010; Foltz et al., 2009; Fachinetti et al., 2013). Gene replacement with these chimeras in budding yeast (Black et al., 2007), fission yeast (Fachinetti et al., 2013), and human cells (Fachinetti et al., 2013) has shown that the CENP-A targeting domain (CATD; all organisms tested; Black et al., 2007; Fachinetti et al., 2013), the N terminus (budding and fission yeasts; Black et al., 2007; Fachinetti et al., 2013), the C terminus (budding yeast; Black et al., 2007), or either terminus (human cells; Fachinetti et al., 2013) are essential for viability. Such gene replacement experiments yield information about the requirements for regions of CENP-A at existing centromeres where a group of 16 proteins, termed the constitutive centromere-associated network (CCAN; Cheeseman, 2014), is constitutively engaged in a network of interactions on or near CENP-A nucleosomes. Whether or not the required sequence determinants of CENP-A are the same during centromere establishment as once a centromere is fully formed remains unknown.
Results and discussion
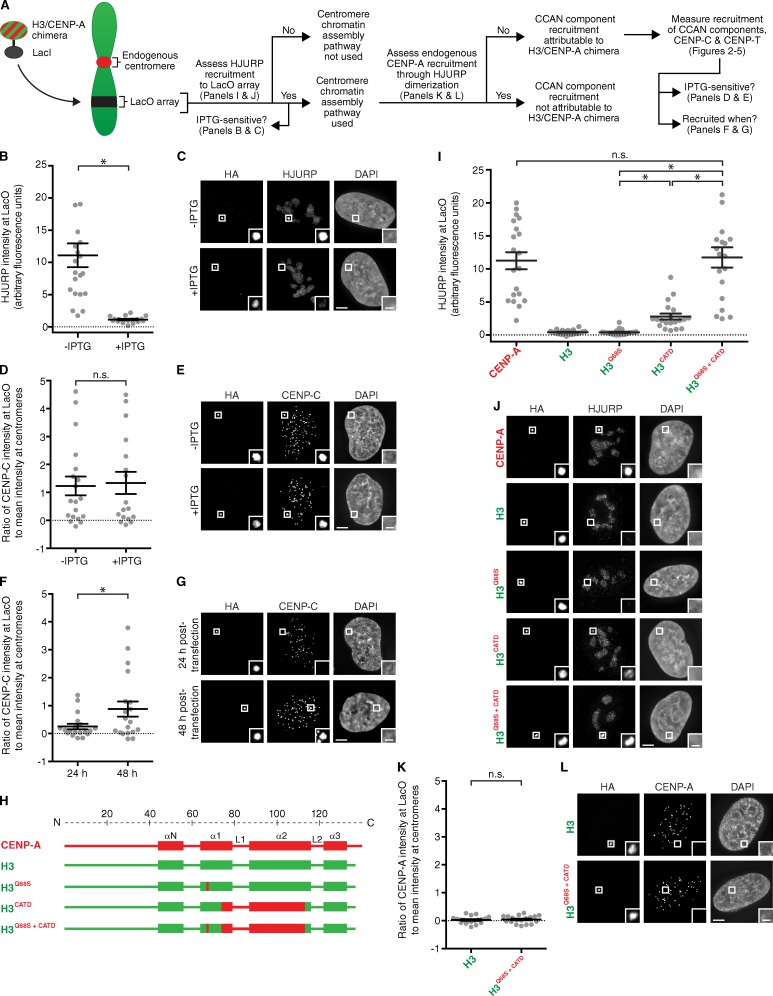
Recruitment of HJURP to the LacO array by LacI-tagged CENP-A is IPTG sensitive, but recruitment of CENP-C is IPTG insensitive
One way to generate a new centromere is to artificially assemble CENP-A nucleosomes at a Lac operator (LacO)–containing array. LacO-directed centromeric chromatin assembly can be accomplished by expression of fusion proteins combining the CENP-A chaperone, HJURP (Barnhart et al., 2011), or CENP-A itself (Mendiburo et al., 2011) with Lac repressor (LacI). Direct LacI–CENP-A fusions provide an opportunity to investigate the determinants of CENP-A for early steps in centromere establishment, as we do here in human cells (Fig. 1 A). We first sought to determine whether LacI-tagged CENP-A is incorporated into chromatin at a LacO-containing array in U2OS cells. Direct targeting of LacI-tagged CENP-A could, in principle, generate two pools of CENP-A at the LacO array: one bound to LacO through its LacI tag and one that is assembled into nucleosomes. The former pool is predicted to be sensitive to the LacI allosteric effector molecule IPTG and available for binding to the CENP-A chaperone, HJURP, whose binding to a CENP-A–histone H4 dimer is incompatible with nucleosome formation (Hu et al., 2011). The latter pool is predicted to be IPTG insensitive and also available for binding to CENP-C, whose interface with CENP-A nucleosomes includes contacts to other nucleosome components (Kato et al., 2013). We found that both HJURP and CENP-C are recruited by LacI-tagged CENP-A to the LacO array (Fig. 1, B–E). HJURP recruitment at the LacO array overrode cell cycle regulation of its targeting to centromeres: it occurs in the majority of asynchronous cells, at times in the cell cycle outside of the brief period when HJURP transiently visits centromeres for chromatin assembly in G1 (Dunleavy et al., 2009; Foltz et al., 2009). Indeed, we found HJURP accumulation is almost entirely IPTG sensitive, whereas CENP-C is retained in an IPTG-insensitive manner (Fig. 1, B–E). More than half of the pool of HA-LacI–CENP-A protein is retained after this IPTG treatment, indicating that a substantial fraction of the targeting protein has assembled into chromatin at or near the LacO array (Fig. 1, C and E; and Fig. S1, A and B).
Figure 1.
Recruitment of HJURP to the LacO array by LacI-tagged CENP-A is IPTG sensitive, but recruitment of CENP-C is IPTG insensitive. (A) Experimental scheme for this study. (B and D) Quantitation of HJURP intensity (B) and CENP-C intensity (D) at the LacO array normalized to the mean intensity at endogenous centromeres for HA-LacI–tagged CENP-A ±IPTG treatment. (C and E) Representative images of HJURP (C) or CENP-C (E) recruitment to the LacO array by HA-LacI–tagged CENP-A ±IPTG treatment. (F) Quantitation of CENP-C intensity at the LacO array 24 and 48 h after transfection of HA-LacI–tagged CENP-A. (G) Representative images of CENP-C recruitment to the LacO array 24 and 48 h after transfection of HA-LacI–tagged CENP-A. (H) Diagram of chimeras used in I–L. (I) Quantitation of HJURP intensity at the LacO array for the indicated chimeras. (J) Representative images of HJURP recruitment to the LacO array by the indicated chimeras fused to HA-LacI. (K) Quantitation of CENP-A intensity at the LacO array for the indicated chimeras. Error bars show SEM. (L) Representative images of CENP-A recruitment to the LacO array by the indicated chimeras. Bars: (main images) 5 µm; (insets) 1 µm. Insets show magnification of the boxed regions. For all graphs, an asterisk denotes significant differences (*, P < 0.05), with others marked as not significant (n.s.). See Materials and methods for details of statistical analysis.
To establish the appropriate time frame to measure centromere assembly steps at the LacO array, we determined the earliest time point at which LacI-tagged CENP-A recruits detectable levels of CENP-C. At 24 h after transfection of LacI-CENP-A, CENP-C is rarely detectable at the LacO array, but many cells have recruited CENP-C by 48 h (Figs. 1, F and G; and S1 C). Because we observe, albeit rarely, cells that have missegregated the LacI–CENP-A–tethered LacO-containing chromosome (Fig. S1 D) even at 48 h, it would be problematic to wait an additional day for more cells to recruit detectable levels of CENP-C. Therefore, the time point of 48 h was chosen for all H3/CENP-A chimera experiments.
The CATD is sufficient to recruit HJURP, but not endogenous CENP-A, to the LacO array
We predicted that the LacI-tagged H3 chimera containing the CATD (Fig. 1 H) would bind to HJURP and ferry it to the LacO array for centromere chromatin assembly. With these constructs (Fig. 1, H–J) the LacO–I interaction is dominant—there is major enrichment at the LacO array (Fig. 1 J)—but proper localization to endogenous centromeres, albeit at low levels, is apparent for all chimeras containing the CATD (Fig. S1 E). Even chimeras that lack the CATD are incorporated into chromatin at the LacO array, remaining largely insensitive to IPTG treatment in which HA-LacI alone is IPTG sensitive (Fig. S1, F and G).
Interestingly, the CATD alone does not recruit HJURP to the same high level as does CENP-A, itself (Fig. 1, I and J). HJURP has another close contact with a CENP-A–specific residue outside of the CATD, S68 (Hu et al., 2011), that is neither necessary nor sufficient for recognition by HJURP, as seen in binding studies with recombinant proteins and in cell-based experiments (Bassett et al., 2012). We found that H3Q68S does not recruit HJURP to the LacO array; however, the combination of this substitution with the CATD recruits HJURP to the same high levels as observed with wild-type CENP-A (Fig. 1, I and J). This observation could be explained because HJURP residues 63–80 form an extended β-sheet that makes a strong interface with the α1 helix of CENP-A, where HJURPW66 and CENP-AS68 closely contact (Hu et al., 2011). This contact surface transmits stability through most of the helices of CENP-A and its histone partner, H4, rigidifying both CENP-A and H4 in the complex (Bassett et al., 2012). Indeed, we observed that the CENP-AS68Q substitution reduces stability within the HJURP–CENP-A–H4 trimer, even in regions of helices of H4 that are distinct from those that comprise the HJURP interface with CENP-A–H4 (Fig. S1, H–K). We note a recent proposal that phosphorylation of CENP-AS68 in early mitosis causes a transient release of HJURP from CENP-A (Yu et al., 2015) before subsequent G1 assembly by HJURP. The importance of this proposed transient release step remains unclear, however, since another effort reported that endogenous HJURP remains bound to CENP-A in early mitosis (Foltz et al., 2009), and because the H3CATD chimera lacking S68 is sufficient for proper G1 centromere chromatin assembly (Bodor et al., 2013), even 9 d after removal of the endogenous CENP-A gene (Fachinetti et al., 2013).
Because high levels of HJURP are recruited to the LacO array by the CATD-containing constructs (Fig. 1, I and J), and because HJURP is known to dimerize (Zasadzińska et al., 2013; i.e., one monomer of an HJURP dimer could conceivably bind to the chimera containing the CATD with another monomer bound to endogenous CENP-A), we considered the possibility that endogenous CENP-A molecules may be corecruited to the LacO array. If this occurred at the time of our measurements of centromere component recruitment to the array, a potential contribution of endogenous CENP-A would complicate the analysis (Fig. 1 A). Using a CENP-A mAb raised against its N terminus (aa 3–19), we found that the CATD-containing chimeras do not recruit detectable endogenous CENP-A to the LacO array (Fig. 1, K and L; and Fig. S2, A–C). This includes H3Q68S+CATD (Fig. 1, K and L), which recruits the same high levels of HJURP to the array as does wild-type CENP-A (Fig. 1, I and J). Thus, HJURP recruitment via H3/CENP-A chimeras does not lead to corecruitment of endogenous CENP-A.
In addition to the C-terminal tail of CENP-A, the CATD has a role in CENP-C recruitment during centromere establishment distinct from recruiting CENP-N
Current models for CENP-A nucleosome assembly suggest that CENP-C can recruit HJURP to centromeres (Moree et al., 2011; Hori et al., 2013) via the Mis18 complex (Fujita et al., 2007; Maddox et al., 2007). To test whether this cross-bridge also functions in reverse (i.e., CENP-A→HJURP→Mis18→CENP-C), we measured the extent to which the H3 chimeras containing the CATD recruit CENP-C to the array (Figs. 2 and S2, A, D, and E). We found that the CATD-containing chimeras (H3Q68S+CATD and H3CATD) do not recruit detectable levels of CENP-C (Figs. 2 and S2, A, D, and E), indicating that even high levels of HJURP at the LacO array do not suffice to recruit endogenous CENP-C. This is further in agreement with our finding that CENP-C levels remain unaffected by IPTG treatment, which effectively removed HJURP (Fig. 1, B–E).
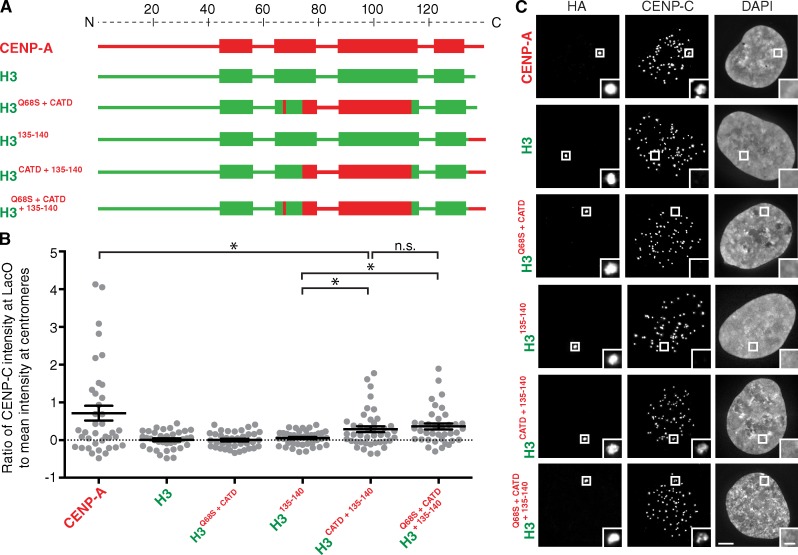
Figure 2.
Recruitment of detectable levels of CENP-C to the LacO array requires the CATD and C terminus of CENP-A. (A) Diagram of the chimeras assessed for recruitment of CENP-C. (B) Quantitation of CENP-C intensity at the LacO array. Error bars show SEM. *, P < 0.05. (C) Representative images of CENP-C recruitment by the indicated chimeras fused to HA-LacI. Insets show magnification of the boxed regions. Bars: (main images) 5 µm; (insets) 1 µm.
In nucleosome reconstitution experiments with recombinant CENP-C (Carroll et al., 2010; Kato et al., 2013) or with CENP-C from Xenopus laevis egg extracts (Guse et al., 2011), an H3 chimera containing only the C-terminal six amino acids of CENP-A (aa 135–140) is sufficient to recruit CENP-C. Surprisingly, we found that the H3135–140 chimera is insufficient to recruit CENP-C to the LacO array (Fig. 2). In contrast, constructs containing both aa 135–140 of CENP-A and the CATD recruit substantial amounts of CENP-C (Fig. 2). Importantly, we noted that even with both of these domains, the H3 chimeras tested in this experiment do not recruit CENP-C to the array to the same high levels achieved by CENP-A itself (Fig. 2). Furthermore, H3Q68S+CATD does not provide additional CENP-C recruitment beyond what is achieved by H3CATD (Figs. 2 and S2, D and E); thus, regions of CENP-A outside the CATD and C terminus must be involved.
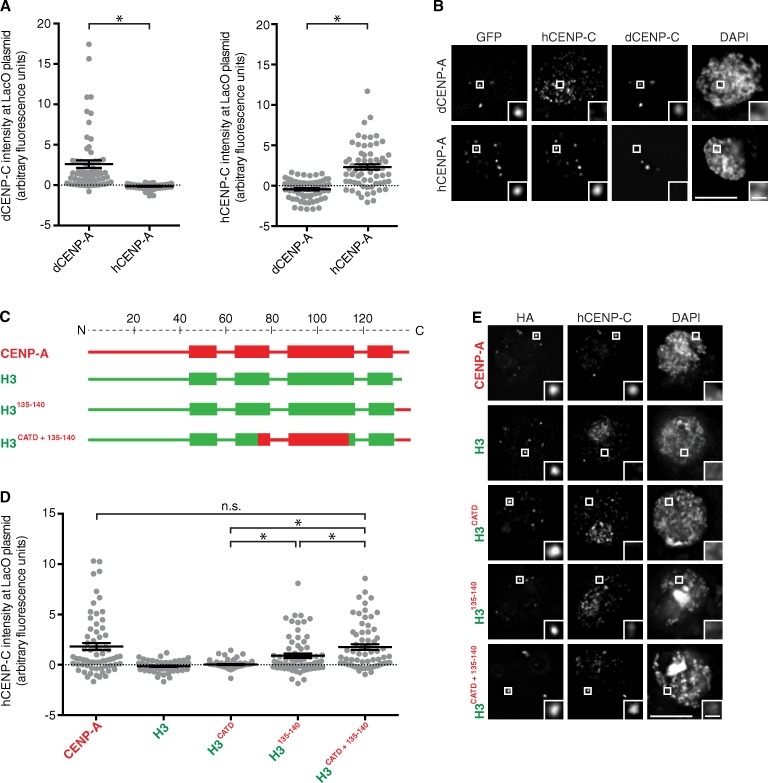
Given the contribution of the CATD in CENP-C recruitment, we considered a role for CENP-N, as it is known to bind to the CENP-A nucleosome directly and recognize it via the CATD in metazoans (Carroll et al., 2009). CENP-N is also known to form a heterodimer with CENP-L that binds to CENP-CMif2 in budding yeast (Hinshaw and Harrison, 2013). We found that the CATD is sufficient to recruit CENP-N but not CENP-C to the LacO array (Fig. S2, A and F), providing evidence that the CATD has independent roles in CENP-C versus CENP-N recruitment during centromere establishment. To test this notion, we interrogated the recruitment of CENP-C using a heterologous system we developed in Drosophila melanogaster S2 cells (Fig. 3). There is no CENP-N orthologue nor any other CCAN component except for CENP-C reported in Drosophila (Drinnenberg et al., 2014). Therefore, any human CENP-C recruitment driven by the CATD in this system can be considered independent of CENP-N. This system uses a LacO plasmid similar to that used in previous centromere establishment experiments using LacI-fused Drosophila CENP-A (dCENP-A; Mendiburo et al., 2011). We found that LacI-fused human CENP-A (hCENP-A) recruits human CENP-C but not Drosophila CENP-C (Fig. 3, A and B). Using this approach, we reproduced the previously reported interaction between the chimera containing the C terminus of CENP-A and CENP-C (Guse et al., 2011), albeit at low levels. Interestingly, when combining the C terminus and the CATD of CENP-A, we achieved a significantly higher level of CENP-C recruitment similar to the levels observed with wild-type CENP-A (Fig. 3, C–E). Basal levels of CENP-C recruitment by H3135–140 may only be detectable in systems devoid of the CCAN group of proteins, in which CENP-C doesn’t compete with any other protein for binding CENP-A. These data suggest that the CATD functions in CENP-C recruitment independently of CENP-N.
Figure 3.
The CATD directly contributes to CENP-C recruitment to a LacO array-containing plasmid in a heterologous Drosophila system. (A) Quantitation of dCENP-C or hCENP-C intensity with either GFP-LacI–dCENP-A or GFP-LacI–hCENP-A targeted to the LacO array plasmids. (B) Representative images of hCENP-C or dCENP-C recruitment to the LacO array plasmids by dCENP-A or hCENP-A fused to GFP-LacI. (C) Diagram of the human chimeras assessed for recruitment of human CENP-C to a LacO array-containing plasmid in Drosophila S2 cells. (D) Quantitation of hCENP-C intensity in S2 cells with indicated HA-LacI–tagged chimeras targeted to the LacO array plasmids. Error bars show SEM. (E) Representative images of hCENP-C recruitment to the LacO array plasmids by the indicated HA-LacI–tagged chimeras. *, P < 0.05. Insets show magnification of the boxed regions. Bars: (main images) 5 µm; (insets) 0.5 µm.
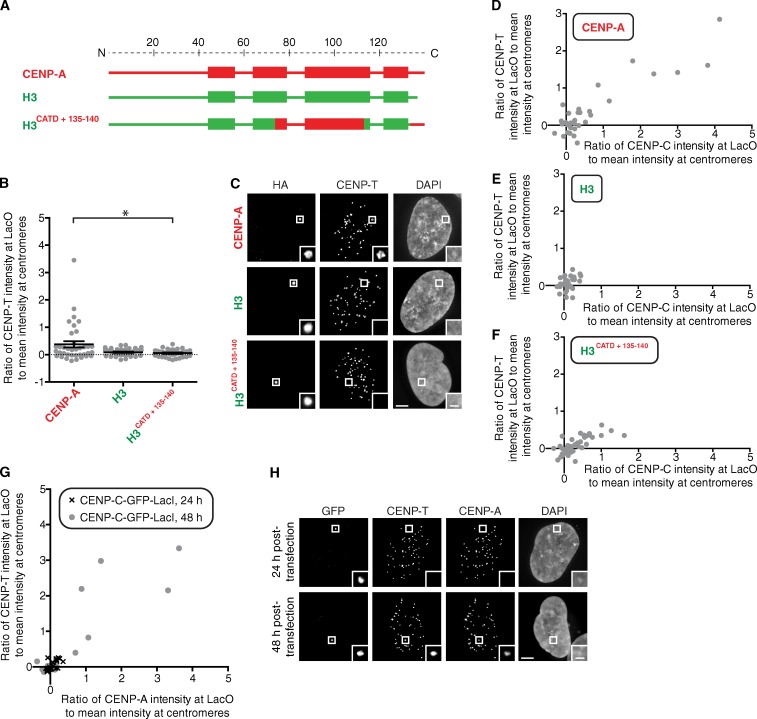
CENP-T is not recruited by an H3/CENP-A chimera that recruits detectable levels of CENP-C and CENP-N
CENP-T and its binding partners (CENP-W, CENP-S, and CENP-X) are histone fold proteins that heterotetramerize and bind DNA (Nishino et al., 2012). They have been proposed to represent a separate branch from CENP-C within the CCAN for building a functional centromere (Gascoigne et al., 2011; Nishino et al., 2012). On the other hand, depletion of CENP-C also reduces CENP-T/W levels at centromeres, suggesting a single branch with CENP-T/W/S/X downstream of CENP-A nucleosomes and CENP-C (Basilico et al., 2014). We first found that CENP-A targeting to the LacO array directs the recruitment of CENP-T but that the H3CATD+135–140 chimera does not (Fig. 4, A–C) despite its ability to recruit CENP-C (Fig. 2). Although we found that higher levels of CENP-C correspond to instances of CENP-T detection on the LacO array with LacI–CENP-A (Fig. 4, D–F; and Fig. S2 G), H3CATD+135–140 only ever recruits very low levels of CENP-T (lower than that found at an endogenous centromere, in all cases; Fig. 4, C and F). We also found that targeting CENP-C, by itself, is not sufficient to recruit CENP-T (Fig. 4, G and H). 1 d after transfection, there is no detectable CENP-T recruited by LacI–CENP-C, and no recruitment of endogenous CENP-A through the Mis18–HJURP chromatin assembly pathway is detectable (Fig. 4, G and H). 2 d after transfection, however, both CENP-T and CENP-A are recruited by LacI–CENP-C to the LacO array, with CENP-T only detectable in cells with at least as much CENP-A as is present on endogenous centromeres (Fig. 4, G and H). Together, these data suggest that CENP-T is only recruited during centromere establishment when both CENP-A nucleosomes and CENP-C are present.
Figure 4.
CENP-C and regions of CENP-A outside of its CATD and C terminus are required to recruit CENP-T to the LacO array. (A) Diagram of the H3/CENP-A chimeric histones assessed for recruitment of CENP-T. (B) Quantitation of CENP-T intensity at the LacO array for the indicated chimeras. Error bars show SEM. *, P < 0.05. (C) Representative images of CENP-T recruitment to the LacO array by the indicated chimeras fused to HA-LacI. (D–F) Plots of CENP-C versus CENP-T intensity at the LacO array for each of the indicated chimeras targeted to the LacO array. (G) Plot of CENP-A versus CENP-T intensity at the LacO array 24 and 48 h after transfection of CENP-C–GFP-LacI. (H) Representative images of CENP-A and CENP-T intensity at the LacO array 24 and 48 h after transfection of CENP-C–GFP-LacI. Insets show magnification of the boxed regions. Bars: (main images) 5 µm; (insets) 1 µm.
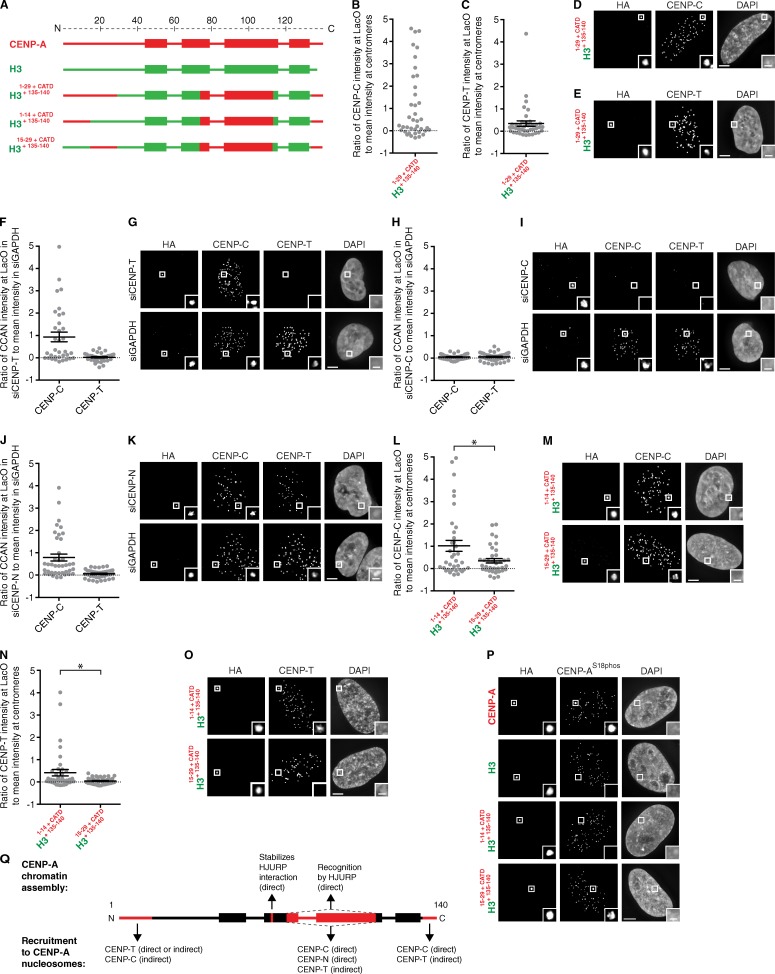
A portion of the N-terminal tail of CENP-A, along with the C-terminal tail and the CATD, is required for CENP-T recruitment to the LacO array
Because the CATD and the C-terminal tail of CENP-A are insufficient for CENP-T recruitment (Fig. 4, B, C, and F), we considered that the N-terminal tail of CENP-A might play a role. H31–29+CATD is sufficient to confer viability to cells lacking endogenous CENP-A (Fachinetti et al., 2013). The proposed role of the N-terminal tail is to recruit high levels of CENP-B protein (Fachinetti et al., 2013) to typical human centromeres that contain repetitive α-satellite housing CENP-B binding sites (the 17-mer CENP-B boxes; Masumoto et al., 1989). The LacO array used in our experiments lacks CENP-B boxes of any sort, and, indeed, there is no CENP-B recruited even with the targeting of LacI-CENP-A (Fig. S3 A). We tested a series of H3 chimeras containing the N-terminal tail of CENP-A for recruitment of CENP-C and CENP-T (Fig. 5, A–E; and Fig. S3, B–D). We first observed that the inclusion of aa 1–29 with the CATD and the C-terminal tail of CENP-A leads to high levels of CENP-C recruitment (Fig. 5, B and D). There is no additional recruitment of human CENP-C contributed by the N-terminal tail of human CENP-A (aa 1–29) in the Drosophila cell system (Fig. S3, E and F), suggesting that the additional recruitment we see during centromere establishment steps in human cells (Fig. 5, B and D) is not via direct binding of CENP-C. We also observed that H31–29+CATD+135–140 is capable of recruiting CENP-T to similarly high levels as does wild-type CENP-A in human cells (Fig. 4, B–D; and Fig. 5, C and E; the mean CENP-T recruitment at LacO relative to endogenous centromeres is 0.38 ± 0.12 for wild-type CENP-A [Fig. 4 B], 0.058 ± 0.02 for H3CATD+135–140 [Fig. 3 B], and 0.34 ± 0.12 for H31–29+CATD+135–140 [Fig. 5 C]). All three regions—the N-terminal tail, the CATD, and the C-terminal tail—are required for CENP-T recruitment, as no substantial CENP-T recruitment occurs with versions of H3 chimeras containing any of these three regions individually or in pairs (Fig. S3, B–D). Thus, our findings reveal a new function for the N-terminal tail of CENP-A in recruiting CENP-T.
Figure 5.
A portion of the N-terminal tail of CENP-A recruits CENP-C and CENP-T to the LacO array. (A) Diagram of the chimeric histones targeted to the LacO array in U2OS-LacO cells and assessed for recruitment of CENP-C and CENP-T. (B and C) Quantitation of CENP-C (B) and CENP-T (C) intensity at the LacO array normalized to the mean intensity at endogenous centromeres. (D and E) Representative images of CENP-C (D) and CENP-T (E) recruitment to the LacO by H31–29+CATD+135–140 fused to LacI and an HA tag. (F, H, and J) Quantitation of CENP-C and CENP-T intensity at the LacO array for HA-LacI–fused H31–29+CATD+135–140 in cells treated with CENP-T (F), CENP-C (H), and CENP-N (J) siRNA normalized to the mean intensity at the LacO array in cells treated with GAPDH siRNA. (G, I, and K) Representative images of CENP-C and CENP-T recruitment to the LacO array by HA-LacI–fused H31–29+CATD+135–140 in cells treated with CENP-T (G), CENP-C (I), and CENP-N (K) siRNA. (L and N) Quantitation of CENP-C (L) and CENP-T (N) intensity at the LacO array. Error bars show SEM. *, P < 0.05. (M and O) Representative images of CENP-C (M) and CENP-T (O) recruitment to the LacO array by the indicated chimeras. (P) Representative images of CENP-AS18phos detection at the LacO array for each of the indicated chimeras. Insets show magnification of the boxed regions. Bars: (main images) 5 µm; (insets) 1 µm. (Q) Summary diagram.
We considered the possibility that CENP-T, itself, contributes to the increase in CENP-C recruitment to the LacO array provided by the N terminus of CENP-A (Fig. 5, B and D). CENP-T depletion, however, does not affect CENP-C recruitment to the LacO array (Fig. 5, F and G). CENP-C depletion, on the other hand, abrogates CENP-T recruitment to the LacO array (Fig. 5, H and I). More surprisingly, CENP-N depletion, which only has a minor effect on CENP-T levels at centromeres, potently disrupts CENP-T recruitment to the LacO array but has no effect on CENP-C recruitment (Fig. 5, J and K). Together, with the findings that the CATD is sufficient to recruit CENP-N but not CENP-C to the LacO array (Figs. 2 and S2 F) and that the CATD and the C terminus directly interact with CENP-C in Drosophila cells (Fig. 3), our experiments provide evidence that the CATD functions independently in recruiting CENP-C and CENP-N to centromeres. We further subdivided the N terminus and found that the activity in recruiting CENP-C and CENP-T is located within aa 1–14 (Fig. 5, L–O). To rule out the recruitment of endogenous CENP-A by this chimera (H31–14+CATD+135–140), we used an anti–CENP-AS18phos pAb (S18 is constitutively phosphorylated on >90% of CENP-A molecules; Bailey et al., 2013) and found that CENP-A was undetectable (Fig. 5 P). Thus, all of the CENP-T recruitment activity of H31–14+CATD+135–140 we measure is attributable to itself without any contribution from endogenous CENP-A. Together, our results strongly suggest that CENP-T requires CENP-C, CENP-N, and the N-terminal tail of CENP-A for initial recruitment during centromere establishment.
Conclusions
Our observations of the early steps of centromere establishment have revealed that three elements within CENP-A—the N- and C-terminal tails and the CATD within the structured histone fold domain—work together to recruit key centromere components (Fig. 5 Q), supporting a model for initial recruitment of CCAN components that reflects the interconnected protein network that exists at the centromere. For the contribution of the N-terminal tail, the simplest prediction from our surprising findings for its CENP-B–independent role in centromere establishment is that this region of CENP-A serves as an additional docking site for the CENP-T–W–S–X complex, itself. Alternatively, it could recruit another CCAN component that stabilizes CENP-C binding, thereby directly or indirectly promoting CENP-T recruitment. Together with a very recent study in fission yeast of a requirement for the N-terminal tail of CENP-A to recruit CENP-T (Folco et al., 2015), our findings here emphasize the need for future biochemical reconstitution—starting with CENP-A nucleosomes and purified CCAN components—to differentiate between these distinct scenarios.
After a centromere is fully formed, the requirements for both termini of CENP-A change from what we observe during centromere establishment. H3 chimeras lacking the CENP-A N-terminal tail then recruit wild-type levels of CENP-C and CENP-T, and in stark contrast to what we find early on in centromere establishment, the C-terminal tail of CENP-A becomes dispensable for recruiting wild-type levels of CENP-T (Fachinetti et al., 2013). Collectively, these conclusions about centromere establishment versus maintenance lead to a model in which it is difficult and rare to seed a new centromere. Then, once formed, multiple intermolecular contacts between several centromere protein subunits have to be broken to lose centromere identity. We think that this model for centromere establishment and maintenance is a satisfactory reflection of biology, in which a 50-fold difference in CENP-A density is what separates the rest of the genome from a functional centromere (Bodor et al., 2014) and in which either spontaneous formation of a new centromere on the chromosome arm or loss of a functional centromere is a rare event in healthy cells. The rarity of these events is advantageous because either would be catastrophic to the fate of the chromosome.
Materials and methods
Cell culture
U2OS cells containing 200 copies of an array of 256 tandem repeats of the 17-bp LacO sequence on chromosome 1 (a gift from S. Janicki, The Wistar Institute, Philadelphia, PA; Janicki et al., 2004) were cultured in DMEM supplemented with 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin. U2OS-LacO cells were maintained at 37°C in a humidified incubator with 5% CO2. Schneider S2 cells were grown at 25°C in Schneider’s Drosophila medium (Serva) supplemented with 10% FBS (Sigma-Aldrich) and antibiotics (300 µg/ml penicillin, 300 µg/ml streptomycin, and 750 µg/ml amphotericin B).
Plasmids
All mammalian expression vectors used in this study were constructed in a pcDNA3.1 vector containing a cytomegalovirus promoter and a HA tag and a LacI tag fused to the N terminus of the CENP-A, H3, or H3/CENP-A chimeric histone. To construct pcDNA3.1-HA-LacI-CENP-A, the LacI gene was amplified by PCR from pN2-GFP-LacI, a derivative of pAFS135 (a gift from A. Straight, Stanford University, Stanford, CA; Straight et al., 1996) and inserted into pcDNA3.1-HA-CENP-A (a gift from D. Cleveland, University of California, San Diego, San Diego, CA) at an EcoRI site present between the HA tag and CENP-A. To construct pcDNA3.1-HA-LacI-H3, pcDNA3.1-H3CATD, and pcDNA3.1-H31–29+CATD, the H3, H3CATD, and H31–29+CATD chimeras were amplified by PCR from pYFP-H3 (Black et al., 2004), pYFP-H3CATD (Black et al., 2004), and pYFP-H31−29+CATD (Fachinetti et al., 2013), respectively, and inserted into pcDNA3.1-HA at the EcoRI–ApaI sites. Then, the LacI gene was amplified by PCR from pN2-GFP-LacI and inserted into the plasmids at the EcoRI site. To construct pcDNA3.1-HA-LacI-H3135–140, pcDNA3.1-H3CATD+135–140, and pcDNA3.1-H31–29+CATD+135–140, PCR site-directed mutagenesis using QuikChange (Agilent Technologies) was performed on pcDNA3.1-HA-LacI-H3, pcDNA3.1-H3CATD, and pcDNA3.1-H31–-29+CATD, respectively, to mutate the C terminus of H3 to CENP-A residues. Similarly, to construct pcDNA3.1-HA-LacI-H3Q68S, pcDNA3.1-H3Q68S+CATD, and pcDNA3.1-H31–29+CATD+135–140, PCR site-directed mutagenesis was performed on pcDNA3.1-HA-LacI-H3 and pcDNA3.1–H3CATD, respectively, to mutate the glutamine in H3 to a serine. To construct pcDNA3.1-HA-LacI-H31–29, the C-terminal region of H3 was digested out of pcDNA3.1-HA-LacI-H3 at the AfeI–SacII sites and inserted into pcDNA3.1-HA-LacI-H31–29+CATD at the same restriction sites. To construct pcDNA3.1-HA-LacI-H31–-14+CATD+135–140 and pcDNA3.1-HA-LacI-H315–29+CATD+135–140, pcDNA3.1-HA-LacI-H31–29+CATD+135–140 was amplified by PCR with phosphorylated primers containing the N-terminal chimeric sequences, and the resulting PCR fragments were purified and ligated. All Drosophila expression vectors used in this study were constructed in a pMT/V5-Hygro vector, except for the LacO array-containing plasmid, which was cloned in a pAFS52 vector that uses a pUC19 plasmid backbone, as originally described (Straight et al., 1996; Mendiburo et al., 2011). pMT_GFP-LacI-hCENP-A was constructed by PCR amplification of LacI–CENP-A from pCDNA3.1-HA-LacI-CENP-A and insertion into the pMT_GFP-LacI vector (Mendiburo et al., 2011) at the BstEII–AgeI sites. pMT-dCENP-A-GFP-LacI was constructed as originally described (Mendiburo et al., 2011). pMT_HA-LacI-CENP-A was constructed by replacing GFP-LacI with HA-LacI using KpnI–BstEII sites. All other pMT_HA-LacI-H3/CENP-A chimera vectors were constructed by inserting NheI–SacII fragments from the respective pcDNA3.1 vector into the SpeI–SacII-digested pMT/V5-Hygro vector. The pMT–hCENP-C construct was cloned by PCR amplification of hCENP-C from HEK293 cDNA and insertion into SpeI–NotI sites in the pMT/V5-Hygro vector. All bacterial expression vectors used in this study for the histones were constructed in a pET12 vector containing a T7 bacterial promoter. Construction of the bacterial expression vectors were described previously (Black et al., 2004; Bassett et al., 2012), except for pET-CENP-AS68Q–H4, which was generated by performing PCR site-directed mutagenesis on pET-CENP-A–H4 (Black et al., 2004) to mutate the serine in CENP-A to a glutamine. All plasmids used in this study were verified by DNA sequencing.
Transfections and indirect immunofluorescence
U2OS cells containing an array of LacO sequences were transfected with plasmids encoding the indicated fusion proteins using FuGENE6 (Roche) and processed for indirect immunofluorescence at the indicated time points. For experiments involving CENP-A, CENP-C, and CENP-T immunofluorescence, cells were fixed in 4% formaldehyde in PBS for 10 min, extracted with 0.5% Triton X-100 in PBS for 5 min, and then washed three times in 0.1% Tween in PBS for 5 min each before blocking and antibody incubation. For experiments involving HJURP or CENP-AS18phos immunofluorescence, cells were preextracted with 0.5% Triton X-100 in PBS for 3 min, fixed with 4% formaldehyde in PBS for 10 min, and then washed with 0.1% Tween in PBS for 5 min. All cells were blocked in 2% FBS, 2% BSA, and 0.1% Tween in PBS and then incubated with primary and secondary antibodies. An anti-HA mouse monoclonal antibody (HA.11; 16B12; Covance) was used at 1 µg/ml in most experiments, and an anti-HA goat polyclonal antibody (ab9134; Abcam) was used at 1 µg/ml in experiments in which mouse antibodies were used to codetect an endogenous protein. An anti-HJURP rabbit polyclonal antibody (Foltz et al., 2009), anti–CENP-A mouse monoclonal antibody (ADI-KAM-CC006; Enzo Life Sciences), and anti–CENP-B rabbit polyclonal antibody (H-65; Santa Cruz Biotechnology, Inc.) were each used at 1 µg/ml. Affinity-purified anti–CENP-C rabbit polyclonal antibody (Bassett et al., 2010) was used at 0.75 µg/ml in most experiments, and anti–CENP-C mouse monoclonal ascites (Bodor et al., 2013) was used at a 1:10,000 dilution in experiments in which CENP-T was codetected. An anti–CENP-T rabbit polyclonal antibody (a gift from I. Cheeseman, Massachusetts Institute of Technology, Cambridge, MA; Gascoigne et al., 2011) was used at 1 µg/ml. An anti-GFP rabbit polyclonal antibody was used at 0.1 µg/ml to enhance detection of TetR-GFP. An anti–CENP-AS18phos rabbit polyclonal antibody (61483; Active Motif) was used at a 1:2,500 dilution. Goat secondary antibodies conjugated to FITC or Cy3, bovine secondary antibodies conjugated to FITC, and donkey secondary antibodies conjugated to Cy3 or Cy5 (Jackson ImmunoResearch Laboratories, Inc.) were used at a 1:200 dilution. Cells were stained with DAPI and mounted with Vectashield mounting medium (Vector Laboratories) onto glass slides before imaging. For U20S experiments involving IPTG treatment, 48 h after transfection, cells were treated or untreated with 15 mM IPTG (Sigma-Aldrich) for 1 h and processed for indirect immunofluorescence, as described earlier in this section. For siRNA experiments, cells were treated with 20 µM CENP-T siRNAs (Silencer Select; catalog no. 4392420, siRNA ID s36976; target sequence 5′-AGAAGUGCCUAGAUAAAUA-3′; Life Technologies), CENP-C siRNAs (ON-TARGETplus SMARTpool; catalog no. L-003251; target sequences 5′-GCGAAUAGAUUAUCAAGGA-3′, 5′-CCAUAAACCUCACCCAGUA-3′, 5′-UUUGAUAGGUGAAACAGUA-3′, and 5′-CGAAGUUGAUAGAGGAUGA-3′; GE Healthcare), CENP-N siRNAs (siGENOME SMARTpool; catalog no. M-015872-02-005; target sequences 5′-CUACCUACGUGGUGUACUA-3′, 5′-AUACACCGCUUCUGGGUCA-3′, 5′-GAUCCAUCUGUGUGAGGAA-3′, and 5′-GUAAAUUUCCGACAGAGAA-3′; GE Healthcare), or GAPDH siRNAs (ON-TARGETplus SMARTpool; catalog no. D-001830; target sequence 5′-GUCAACGGAUUUGGUCGUA-3′; GE Healthcare). CENP-T, CENP-C, and GAPDH siRNAs were a gift of D. Foltz (University of Virginia, Charlottesville, VA). 8–24 h after siRNA treatment, cells were transfected with pcDNA3.1-HA-LacI-H31–29+CATD+135–140 and processed for indirect immunofluorescence 48 h later, as described earlier in this section. Schneider S2 cells were cotransfected with pLacO-LexA, hCENP-C, and the relevant LacI-tagged H3, CENP-A, or H3/CENP-A chimera construct using XtremeGENE DNA transfection reagent (Roche). Cells were harvested 48 h after transfection and fixed for 10 min in 3.7% formaldehyde in PBS and 0.1% Triton X-100. Primary antibodies were incubated overnight at 4°C at the following dilutions or concentrations: rat anti-HA (1:20; clone 3F10; Roche), rabbit affinity-purified anti–hCENP-C (0.75 µg/ml; Bassett et al., 2010), mouse anti–hCENP-C (1:100; ab50974; Abcam), rabbit anti–dCENP-C (1:100; a gift from C. Lehner, University of Zurich, Zurich, Switzerland; Heeger et al., 2005). Goat secondary antibodies conjugated to Alexa Fluor 555 or Alexa Fluor 647 (Invitrogen) were incubated for 1 h at room temperature at a dilution of 1:100. DNA was counterstained with DAPI at 5 µg/ml.
Image acquisition and quantitation
U2OS-LacO cells were imaged on an inverted fluorescence microscope (DMI6000 B; Leica) equipped with a charge-coupled device camera (ORCA AG; Hamamatsu Photonics) and a 100×, 1.4 NA oil immersion objective. HA-LacI fusion protein expression varies from cell to cell for each transfection, as expected; cells with moderate expression were chosen, and very high-expressing cells were specifically excluded. Furthermore, to ensure that none of the differences in centromere establishment behavior of the H3/CENP-A chimeras reported in this study are caused by wide variations in copy number of HA-LacI fusion proteins at the LacO array, all HA-LacI proteins in this study expressed to high enough levels to have many cells in the moderate expression range (1,000–3,000 maximal pixel intensity in a scale of 0–4,095 for a 50-ms exposure in the channel used for anti-HA detection). At least 20 individual cells were imaged for each individual experiment, with experiments repeated multiple times in all cases. Data are shown from one or more experiments that are representative of two or more independent experimental repeats. For instance, the data in Fig. 4 G is from a single experiment representative of two independently performed experiments. Images were acquired at room temperature in 0.2-µm z sections and deconvolved using LAS software (Leica). Images were maximum projected into single two-dimensional images using ImageJ (Version 1.46r; National Institutes of Health). Deconvolved and two-dimensionally projected images were used for all fluorescence intensity measurements. The fluorescence intensity at the LacO array was measured by placing a 20 × 20–pixel box around the LacO array (as indicated by the HA signal) and measuring the total pixel intensity within the box in the channel of the relevant centromere or kinetochore protein. The mean fluorescence intensity at centromeres was measured using CRaQ ver1.12, a modified version of the ImageJ macro, CRaQ (Bodor et al., 2012), in which a 7 × 7–pixel box was placed around the centroid position of each centromere, and the total pixel intensity within the box was measured and averaged over the total number of centromeres in each cell. Mean background intensity was measured at nuclear sites that were not coincident with the LacO array or centromeres and was subtracted from the LacO array fluorescence intensity and mean centromeric fluorescence intensity. In the case of HJURP, which does not localize to centromeres during most of the cell cycle, the intensity at the LacO array was measured by summing the total pixel intensity in a 20 × 20–pixel box and subtracting the mean background intensity in the nucleoplasm. For experiments with S2 cells, the samples were imaged on a microscope (DeltaVision RT Elite) and deconvolved using softWoRx Explorer Suite (Applied Precision). Images were acquired as 50 z stacks of 0.2-µm increments using a 100× oil immersion objective and a monochrome camera (CoolSNAP HQ; Photometrics). Quantification of hCENP-C and dCENP-C fluorescence intensities was performed using CRaQ ver1.12. The mean ± SEM for all data points is plotted in each graph. Tests of significance for HJURP, CENP-C, and CENP-T recruitment were performed using an unpaired t test in Prism (GraphPad Software), and only results with significant differences (P < 0.05) are reported as different from one another (denoted with asterisks in the figures).
Recombinant protein preparation, hydrogen/deuterium exchange (HX) reactions, protein fragmentation, mass spectrometry, and data analysis
All proteins used in the HX experiments were expressed, purified, and reconstituted as previously described (Bassett et al., 2012). In brief, Maltose-binding protein–tagged HJURP proteins were expressed in a BL21 Rosetta(DE3) bacteria strain. Cells were lysed by sonicating in a buffer containing 25 mM Tris-HCl, pH 7, 300 mM NaCl, and 5 mM β-mercaptoethanol. Lysates were incubated with amylose resin (New England Biolabs, Inc.) at 4°C for 2 h and eluted with 15 mM maltose. The human histone proteins were expressed and purified from bacteria as previously described (Black et al., 2004; Sekulic et al., 2010). The Maltose-binding protein–HJURP and human histones were mixed in an equimolar ratio (20 µM each in 2 ml volume). After incubation on ice for 30 min, the protein mixtures were injected onto a HiLoad 16/60 Superdex200 column (GE Healthcare) with running buffer identical to sample buffer. For the HX reactions, 30 µl of protein mixtures at 1 mg/ml of the indicated protein complex sample was mixed with 90 µl D2O and incubated at 4°C. Final buffer conditions in HX reactions are 0.25 mg/ml protein mix, 10 mM sodium phosphate, pH 7, 1.25 mM β-mercaptoethanol, and 500 mM NaCl. At the indicated time point, 20 µl of each HX reaction was withdrawn and added to 30 µl of ice-cold quench buffer (1.66 M guanidinium-HCl, 0.8% formic acid, and 10% glycerol). Samples were immediately frozen in liquid N2 and stored at −80°C before analysis. After HX, the proteins were fragmented by injecting the samples onto an immobilized pepsin column at an initial flow rate of 50 µl/min for 3 min followed by 150 µl/min for another 3 min. Pepsin (Sigma-Aldrich) was coupled to Poros 20 AL support (Applied Biosystems) and packed into column housings of 2 mm × 2 cm dimensions (IDEX). Peptides were collected onto a C18 HPLC trap column (2.5 × 0.5 mm; LC Packings) and eluted through an analytical C18 HPLC column (0.3 × 75 mm; Agilent Technologies) by a linear 12–55% buffer B gradient at 6 µl/min (buffer A: 0.1% formic acid; buffer B: 0.1% formic acid and 99.9% acetonitrile). The effluent was electrosprayed into the mass spectrometer (LTQ Orbitrap XL; Thermo Fisher Scientific). Nondeuterated samples were used to identify the likely sequence of the parent peptides in SEQUEST (Bioworks v3.3.1) software program (Thermo Fisher Scientific). The data were analyzed using ExMS program (Kan et al., 2011), and the broad deprotection from HX reported for CENP-AS68Q–H4 is representative of results obtained in two independently performed experiments.
Online supplemental material
Fig. S1 shows that a significant subset of LacI-tagged chimeras remain at the LacO array upon IPTG treatment while LacI alone is removed, low levels of CENP-C are recruited to the LacO array by LacI–CENP-A at 24 h, missegregation of the LacO-containing chromosome begins to occur at 48 h, LacI-fused H3/CENP-A chimeras containing the CATD localize to endogenous centromeres, and Ser68 of CENP-A transmits stability to distant helices within histone H4. Fig. S2 shows that LacI-H3CATD does not recruit detectable levels of CENP-A or CENP-C to the LacO array, CATD-containing constructs recruit CENP-N to the LacO array, and LacI-H3CATD+135–140 recruits CENP-C but not CENP-T to the LacO array. Fig. S3 shows that CENP-B is not recruited to the LacO array by LacI–CENP-A, LacI-tagged chimeric histones containing combinations of the CATD and N- and C-terminal tails of CENP-A do not recruit CENP-T to the LacO array, LacI-H31–29+CATD+135–140 does not enhance CENP-C recruitment to the LacO-containing plasmid in Drosophila S2 cells, and CENP-T, CENP-C, and CENP-N are depleted after siRNA treatment. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201412011/DC1.
Supplementary Material
Acknowledgments
We thank D. Foltz (Virginia) and S. Janicki (Wistar) for advice and reagents. We also thank A. Straight (Stanford), D. Cleveland (University of California, San Diego), C. Lehner (Zurich), and I. Cheeseman (Massachusetts Institute of Technology) for the kind gift of reagents. We thank M. Lampson (University of Pennsylvania [UPenn]) for helpful discussions and comments on the manuscript and A. Straight for sharing unpublished results. We thank S. Kemmer (Max Planck Institute, Freiburg) and T. van Emden (University of Edinburgh) for technical assistance.
This work was supported by National Institutes of Health grants GM082989 (B.E. Black), GM007229 (UPenn Cell and Molecular Biology Training Grant; G.A. Logsdon), GM08275 (UPenn Structural Biology Training Grant; E.A. Bassett, L.Y. Guo, and T. Panchenko), CA186430 (L.Y. Guo), GM108360 (J.M. Dawicki-McKenna), European Research Council Starting-Consolidator Grant 311674–BioSynCEN (E.J. Barrey and P. Heun), and an American Heart Association predoctoral fellowship (E.A. Bassett). The Wellcome Trust Centre for Cell Biology is supported by core funding from the Wellcome Trust (092076).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CATD
- CENP-A targeting domain
- CCAN
- constitutive centromere-associated network
- HX
- hydrogen/deuterium exchange
- LacI
- Lac repressor, LacO, Lac operator
References
- Bailey A.O., Panchenko T., Sathyan K.M., Petkowski J.J., Pai P.-J., Bai D.L., Russell D.H., Macara I.G., Shabanowitz J., Hunt D.F., et al. 2013. Posttranslational modification of CENP-A influences the conformation of centromeric chromatin. Proc. Natl. Acad. Sci. USA. 110:11827–11832 10.1073/pnas.1300325110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart M.C., Kuich P.H.J.L., Stellfox M.E., Ward J.A., Bassett E.A., Black B.E., and Foltz D.R.. 2011. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol. 194:229–243 10.1083/jcb.201012017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basilico F., Maffini S., Weir J.R., Prumbaum D., Rojas A.M., Zimniak T., De Antoni A., Jeganathan S., Voss B., van Gerwen S., et al. 2014. The pseudo GTPase CENP-M drives human kinetochore assembly. eLife. 3:e02978 10.7554/eLife.02978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett E.A., Wood S., Salimian K.J., Ajith S., Foltz D.R., and Black B.E.. 2010. Epigenetic centromere specification directs aurora B accumulation but is insufficient to efficiently correct mitotic errors. J. Cell Biol. 190:177–185 10.1083/jcb.201001035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett E.A., DeNizio J., Barnhart-Dailey M.C., Panchenko T., Sekulic N., Rogers D.J., Foltz D.R., and Black B.E.. 2012. HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly. Dev. Cell. 22:749–762 10.1016/j.devcel.2012.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B.E., and Cleveland D.W.. 2011. Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell. 144:471–479 10.1016/j.cell.2011.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B.E., Foltz D.R., Chakravarthy S., Luger K., Woods V.L. Jr, and Cleveland D.W.. 2004. Structural determinants for generating centromeric chromatin. Nature. 430:578–582 10.1038/nature02766 [DOI] [PubMed] [Google Scholar]
- Black B.E., Jansen L.E.T., Maddox P.S., Foltz D.R., Desai A.B., Shah J.V., and Cleveland D.W.. 2007. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell. 25:309–322 10.1016/j.molcel.2006.12.018 [DOI] [PubMed] [Google Scholar]
- Bodor D.L., Rodríguez M.G., Moreno N., and Jansen L.E.T.. 2012. Analysis of protein turnover by quantitative SNAP-based pulse-chase imaging. Curr. Protoc. Cell Biol. Chapter 8:Unit 8.8. [DOI] [PubMed] [Google Scholar]
- Bodor D.L., Valente L.P., Mata J.F., Black B.E., and Jansen L.E.T.. 2013. Assembly in G1 phase and long-term stability are unique intrinsic features of CENP-A nucleosomes. Mol. Biol. Cell. 24:923–932 10.1091/mbc.E13-01-0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor D.L., Mata J.F., Sergeev M., David A.F., Salimian K.J., Panchenko T., Cleveland D.W., Black B.E., Shah J.V., and Jansen L.E.. 2014. The quantitative architecture of centromeric chromatin. eLife. 3:e02137 10.7554/eLife.02137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.W., Silva M.C.C., Godek K.M., Jansen L.E.T., and Straight A.F.. 2009. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 11:896–902 10.1038/ncb1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.W., Milks K.J., and Straight A.F.. 2010. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189:1143–1155 10.1083/jcb.201001013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M.2014. The kinetochore. Cold Spring Harb. Perspect. Biol. 6:a015826 10.1101/cshperspect.a015826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg I.A., deYoung D., Henikoff S., and Malik H.S.. 2014. Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects. eLife. 3 10.7554/eLife.03676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy E.M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., Daigo Y., Nakatani Y., and Almouzni-Pettinotti G.. 2009. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 137:485–497 10.1016/j.cell.2009.02.040 [DOI] [PubMed] [Google Scholar]
- Fachinetti D., Folco H.D., Nechemia-Arbely Y., Valente L.P., Nguyen K., Wong A.J., Zhu Q., Holland A.J., Desai A., Jansen L.E.T., and Cleveland D.W.. 2013. A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol. 15:1056–1066 10.1038/ncb2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco H.D., Campbell C.S., May K.M., Espinoza C.A., Oegema K., Hardwick K.G., Grewal S.I., and Desai A.. 2015. The CENP-A N-tail confers epigenetic stability to centromeres via the CENP-T branch of the CCAN in fission yeast. Curr. Biol. 25:348–356 10.1016/j.cub.2014.11.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E.T., Bailey A.O., Yates J.R. III, Bassett E.A., Wood S., Black B.E., and Cleveland D.W.. 2009. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell. 137:472–484 10.1016/j.cell.2009.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Hayashi T., Kiyomitsu T., Toyoda Y., Kokubu A., Obuse C., and Yanagida M.. 2007. Priming of centromere for CENP-A recruitment by human hMis18α, hMis18β, and M18BP1. Dev. Cell. 12:17–30 10.1016/j.devcel.2006.11.002 [DOI] [PubMed] [Google Scholar]
- Gascoigne K.E., Takeuchi K., Suzuki A., Hori T., Fukagawa T., and Cheeseman I.M.. 2011. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 145:410–422 10.1016/j.cell.2011.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse A., Carroll C.W., Moree B., Fuller C.J., and Straight A.F.. 2011. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 477:354–358 10.1038/nature10379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeger S., Leismann O., Schittenhelm R., Schraidt O., Heidmann S., and Lehner C.F.. 2005. Genetic interactions of separase regulatory subunits reveal the diverged Drosophila Cenp-C homolog. Genes Dev. 19:2041–2053 10.1101/gad.347805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw S.M., and Harrison S.C.. 2013. An Iml3-Chl4 heterodimer links the core centromere to factors required for accurate chromosome segregation. Cell Reports. 5:29–36 10.1016/j.celrep.2013.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Shang W.-H., Takeuchi K., and Fukagawa T.. 2013. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J. Cell Biol. 200:45–60 10.1083/jcb.201210106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Liu Y., Wang M., Fang J., Huang H., Yang N., Li Y., Wang J., Yao X., Shi Y., et al. 2011. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 25:901–906 10.1101/gad.2045111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki S.M., Tsukamoto T., Salghetti S.E., Tansey W.P., Sachidanandam R., Prasanth K.V., Ried T., Shav-Tal Y., Bertrand E., Singer R.H., and Spector D.L.. 2004. From silencing to gene expression: real-time analysis in single cells. Cell. 116:683–698 10.1016/S0092-8674(04)00171-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Z.-Y., Mayne L., Chetty P.S., and Englander S.W.. 2011. ExMS: data analysis for HX-MS experiments. J. Am. Soc. Mass Spectrom. 22:1906–1915 10.1007/s13361-011-0236-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Jiang J., Zhou B.-R., Rozendaal M., Feng H., Ghirlando R., Xiao T.S., Straight A.F., and Bai Y.. 2013. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science. 340:1110–1113 10.1126/science.1235532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P.S., Hyndman F., Monen J., Oegema K., and Desai A.. 2007. Functional genomics identifies a Myb domain–containing protein family required for assembly of CENP-A chromatin. J. Cell Biol. 176:757–763 10.1083/jcb.200701065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H., Masukata H., Muro Y., Nozaki N., and Okazaki T.. 1989. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J. Cell Biol. 109:1963–1973 10.1083/jcb.109.5.1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiburo M.J., Padeken J., Fülöp S., Schepers A., and Heun P.. 2011. Drosophila CENH3 is sufficient for centromere formation. Science. 334:686–690 10.1126/science.1206880 [DOI] [PubMed] [Google Scholar]
- Moree B., Meyer C.B., Fuller C.J., and Straight A.F.. 2011. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J. Cell Biol. 194:855–871 10.1083/jcb.201106079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T., Takeuchi K., Gascoigne K.E., Suzuki A., Hori T., Oyama T., Morikawa K., Cheeseman I.M., and Fukagawa T.. 2012. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell. 148:487–501 10.1016/j.cell.2011.11.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulic N., Bassett E.A., Rogers D.J., and Black B.E.. 2010. The structure of (CENP-A-H4)2 reveals physical features that mark centromeres. Nature. 467:347–351 10.1038/nature09323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A.F., Belmont A.S., Robinett C.C., and Murray A.W.. 1996. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6:1599–1608 10.1016/S0960-9822(02)70783-5 [DOI] [PubMed] [Google Scholar]
- Westhorpe F.G., and Straight A.F.. 2015. The centromere: epigenetic control of chromosome segregation during mitosis. Cold Spring Harb. Perspect. Biol. 7:a015818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Zhou X., Wang W., Deng W., Fang J., Hu H., Wang Z., Li S., Cui L., Shen J., et al. 2015. Dynamic phosphorylation of CENP-A at Ser68 orchestrates its cell-cycle-dependent deposition at centromeres. Dev. Cell. 32:68–81 10.1016/j.devcel.2014.11.030 [DOI] [PubMed] [Google Scholar]
- Zasadzińska E., Barnhart-Dailey M.C., Kuich P.H.J.L., and Foltz D.R.. 2013. Dimerization of the CENP-A assembly factor HJURP is required for centromeric nucleosome deposition. EMBO J. 32:2113–2124 10.1038/emboj.2013.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.