Following prolonged genotoxic stress, DNA2 and WRN functionally interact to degrade reversed replication forks and promote replication restart, thereby preventing aberrant processing of unresolved replication intermediates
Abstract
Accurate processing of stalled or damaged DNA replication forks is paramount to genomic integrity and recent work points to replication fork reversal and restart as a central mechanism to ensuring high-fidelity DNA replication. Here, we identify a novel DNA2- and WRN-dependent mechanism of reversed replication fork processing and restart after prolonged genotoxic stress. The human DNA2 nuclease and WRN ATPase activities functionally interact to degrade reversed replication forks with a 5′-to-3′ polarity and promote replication restart, thus preventing aberrant processing of unresolved replication intermediates. Unexpectedly, EXO1, MRE11, and CtIP are not involved in the same mechanism of reversed fork processing, whereas human RECQ1 limits DNA2 activity by preventing extensive nascent strand degradation. RAD51 depletion antagonizes this mechanism, presumably by preventing reversed fork formation. These studies define a new mechanism for maintaining genome integrity tightly controlled by specific nucleolytic activities and central homologous recombination factors.
Introduction
The accurate replication of our genome is an essential requirement for the high-fidelity transmission of genetic information to daughter cells. DNA replication forks are constantly challenged and arrested by DNA lesions, induced by endogenous and exogenous agents, and by a diverse range of intrinsic replication fork obstacles, such as transcribing RNA polymerases, unusual DNA structures or tightly bound protein–DNA complexes (Carr and Lambert, 2013). An emerging model of how stalled or damaged forks are processed is that replication forks can reverse to aid repair of the damage (Atkinson and McGlynn, 2009; Ray Chaudhuri et al., 2012; Berti et al., 2013). This model implies significant remodeling of replication fork structures into four-way junctions and the molecular determinants required for reversed fork processing and restart are just beginning to be elucidated. The first evidence that supports the physiological relevance of this DNA transaction during replication stress in human cells arose from studies with DNA topoisomerase I (TOP1) inhibitors (Ray Chaudhuri et al., 2012). Additional studies established that the human RECQ1 helicase promotes the restart of replication forks that have reversed upon TOP1 inhibition by virtue of its ATPase and branch migration activities (Berti et al., 2013). These observations were recently extended to show that the RECQ1 mechanism of reversed fork restart is a more general response to a wide variety of replication challenges (Zellweger et al., 2015). Nonetheless, new lines of evidence point to alternative mechanisms and factors that might mediate either formation or processing of reversed replication forks (Bétous et al., 2012; Gari et al., 2008). These putative mechanisms likely include nucleases that are capable of processing stalled replication intermediates upon genotoxic stress (Cotta-Ramusino et al., 2005; Schlacher et al., 2011; Hu et al., 2012; Ying et al., 2012).
Here, we investigate the contribution of the human DNA2 nuclease/helicase in reversed fork processing. DNA2 is a highly conserved nuclease/helicase initially identified in Saccharomyces cerevisiae screening for mutants deficient in DNA replication (Kuo et al., 1983; Budd and Campbell, 1995). Yeast Dna2 plays an essential role in Okazaki fragment maturation during lagging strand DNA replication (Budd and Campbell, 1997; Bae et al., 2001; Ayyagari et al., 2003). However, increasing evidence suggests that DNA2 has important—albeit yet undefined—roles in DNA replication stress response and DNA repair, which go beyond its postulated role in Okazaki fragment processing (Duxin et al., 2012; Karanja et al., 2012; Peng et al., 2012). The notion that DNA2 is important for DNA replication is strengthened by the observation that DNA2 forms a complex with various replication core components, including the replisome protein And-1 (Wawrousek et al., 2010; Duxin et al., 2012). Moreover, human DNA2 seems to play a partially redundant role with human exonuclease I (EXO1) in replication-coupled repair (Karanja et al., 2012), whereas a recent study in S. pombe suggested that the nuclease activity of DNA2 is required to prevent stalled forks from reversing upon HU treatment (Hu et al., 2012).
DNA2 also has an independent function in dsDNA break repair. Two distinct pathways act redundantly to mediate processive DSB resection downstream from the MRE11-RAD50-NBS1 (MRN) and CtIP factors in eukaryotic cells: one requires DNA2 and the other EXO1 (Gravel et al., 2008; Mimitou and Symington, 2008; Zhu et al., 2008; Nicolette et al., 2010). Specifically, DNA2 and EXO1 resect the 5′ ends of double-strand DNA breaks (DSBs) to generate 3′ single-stranded overhangs, which are essential to initiate homologous recombination. In yeast, DNA2-dependent dsDNA-end resection reaction requires the Sgs1 helicase to unwind the DNA from the break (Zhu et al., 2008; Cejka et al., 2010; Niu et al., 2010). This mechanism appears to be largely conserved in mammalian cells where DNA2 cooperates with the human BLM helicase to resect dsDNA ends in vitro (Nimonkar et al., 2011). However, mammalian cells possess five human RecQ homologues (RECQ1, RECQ4, RECQ5, BLM, and WRN) and WRN can also assist DNA2-dependent end resection, suggesting that BLM might not be the sole RecQ homologue required for this process (Liao et al., 2008; Sturzenegger et al., 2014). The ability of DNA2 and EXO1 to process dsDNA ends might also be relevant in the context of DNA replication to prevent the accumulation of replication-associated DSBs by promoting homologous recombination (HR) repair (Peng et al., 2012). Alternatively, these nucleases might be involved in the recovery of replication fork blockage by processing specific stalled replication fork structures.
This work uncovers a new DNA2- and WRN-dependent mechanism that mammalian cells use to process replication forks that have reversed as a result of replication inhibition. Importantly, it also shows that this mechanism is tightly regulated by human RECQ1 and the HR factor RAD51. Our observations shed light on a novel pathway for the suppression of chromosomal instability in mammalian cells and provide important new insight into the mechanisms of replication stress response associated with chemotherapeutic drug damage.
Results
DNA2 is required for stalled fork processing and restart
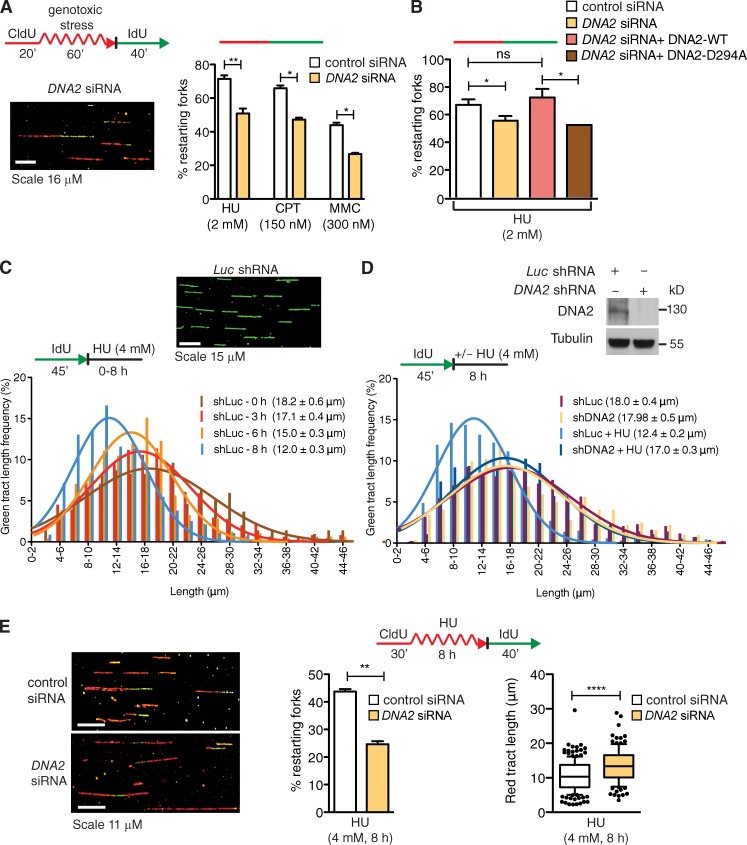
To begin elucidating the role of human DNA2 during replication stress, we monitored replication perturbation by genome-wide single-molecule DNA fiber replication assays. We pulse-labeled human osteosarcoma (U-2 OS) cells with the thymidine analogue CldU for 20 min, followed by a 60-min exposure to a selected genotoxic agent during the CldU labeling period, and by labeling with the second thymidine analogue, IdU, for an additional 40 min after removal of the genotoxic drug. We found that DNA2 plays an important role in restarting replication forks after treatment with the ribonucleotide reductase inhibitor hydroxyurea (HU), the topoisomerase I inhibitor camptothecin (CPT), and the interstrand cross-linking agent mitomycin C (MMC) (Fig. 1 A). In addition, DNA2 depletion increased the percentage of origin firing, but not of fork termination events (Fig. S1 A). Genetic knockdown–rescue experiments confirmed that complementation in DNA2-depleted U-2 OS cells with siRNA-resistant WT DNA2 abrogated the effect of DNA2 depletion on replication fork restart upon HU treatment. Moreover, expression of the nuclease-deficient DNA2 mutant D294A in DNA2-depleted cells revealed that the nuclease activity of DNA2 was essential for its role in replication fork restart (Fig. 1 B and Fig. S1 B).
Figure 1.
DNA2 is required for replication fork restart and stalled fork processing upon genotoxic stress. (A) Schematic of DNA fiber tract analysis. U-2 OS cells were transfected with control siRNA or DNA2 siRNA before CldU or IdU labeling. Red tracts, CldU; curved red tracts, CldU with genotoxic agents (HU or CPT or MMC); green tracts, IdU. (bottom) Representative DNA fiber image. (right) quantification of red-green contiguous tracts (restarting forks). Mean shown, n = 3. Error bars, standard error. ns, not significant; *, P < 0.05; **, P < 0.01 (paired t test). (B) Quantification of restarting forks in DNA2-depleted cells expressing DNA-WT or DNA2-D294A. ns, not significant; *, P < 0.05 (paired Student’s t test). (C, top) Representative DNA fiber image. (bottom) Representative IdU tract length distributions in Luc-depleted cells during different exposure time to HU (out of 3 repeats; n ≥ 300 tracts scored for each dataset). Mean tract lengths are indicated in parentheses. (D) Top, DNA2 expression after shRNA knockdown. Bottom, representative IdU tracts in DNA2-depleted U-2 OS cells in the presence or absence of HU (out of 2 repeats; n ≥ 700 scored for each dataset). (E, left) Representative DNA fiber images. (middle) Quantification of red-green contiguous tracts (restarting forks) after 8 h of HU. Mean shown, n = 3. Error bars, standard error. **, P < 0.01 (paired Student’s t test). (right) Statistical analysis of CldU tracts detected within contiguous red-green tracts. Whiskers the 10th and 90th percentiles. ****, P < 0.0001 (Mann-Whitney test).
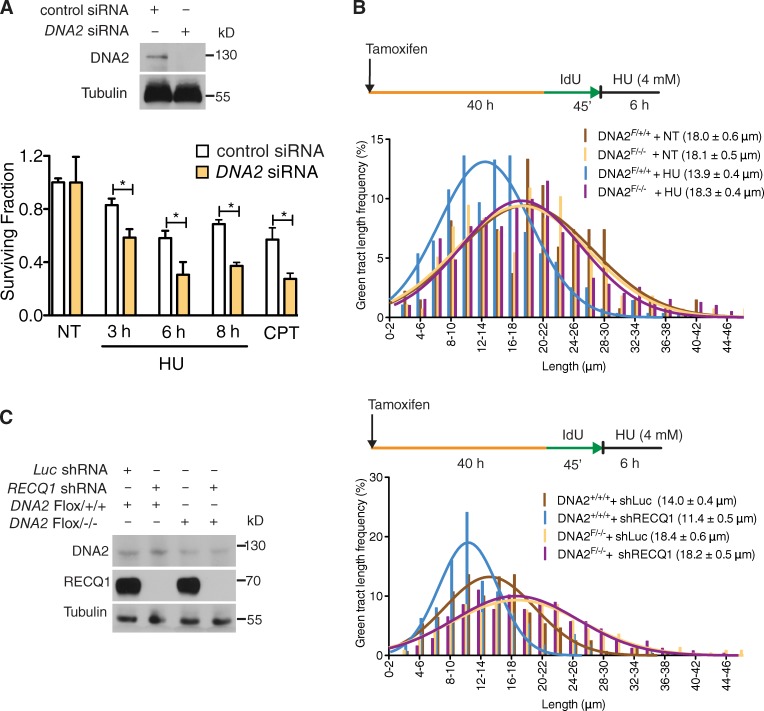
We next measured whether DNA2 uses its nuclease activity to process stalled replication intermediates by monitoring the integrity of the newly synthesized DNA after HU treatment. To this purpose, we changed the DNA labeling scheme. We first pulsed U-2 OS cells with IdU for 45 min, and then varied the exposure time to HU from 0 to 8 h. The mean length of the IdU tracts progressively decreased during HU treatment from 18.2 µm (0 h) to 12.0 µm (8 h; Fig. 1 C). However, shRNA-mediated DNA2 depletion largely prevented IdU tract shortening, confirming that DNA2 is responsible for the observed nascent strand degradation (Fig. 1 D). Double-labeling experiments confirmed that the observed nascent tract shortening is indeed caused by the DNA2-dependent processing of ongoing replication forks and that this degradation is important to mediate efficient replication fork restart upon prolonged HU treatment (Fig. 1 E). Clonogenic analysis of U-2 OS cells treated with the same HU concentration used for the DNA fiber experiments showed a significantly reduced cell survival upon DNA2 depletion, indicating that the DNA2-dependent processing of stalled replication intermediates is critical for recovery from replication fork blockage (Fig. 2 A). The results obtained with the shRNA DNA2-depleted U-2 OS cells were validated using a new conditional knockout human colorectal carcinoma cell line (HCT116) where addition of tamoxifen to the culture medium led to DNA2-null cells. Analysis of the mean tract lengths confirmed that DNA2 knockout in HCT116 cells abrogates the prominent degradation observed upon HU treatment (Fig. 2 B). Collectively, these results indicate that human DNA2 degrades nascent strands at stalled replication forks to facilitate fork restart and promote viability after genotoxic stress induction.
Figure 2.
DNA2 processes stalled replication forks. (A, top) DNA2 expression after siRNA knockdown. (bottom) Colony-forming assays in control and DNA2-depleted U-2 OS cells treated with 4 mM HU for the indicated time. (B) Representative IdU tracts in DNA2 conditional knockout HCT116 cells (out of two repeats). Tamoxifen was added to generate conditional knockout cells (see Materials and methods). (C, left) Expression of DNA2 and RECQ1 in tamoxifen-treated HCT116 cells. Right, representative IdU tracts in DNA2 conditional knockout HCT116 cells depleted for Luc or RECQ1 (out of three repeats). n ≥ 300 tracts scored for each dataset shown in B and C.
RECQ1 regulates the fork processing activity of DNA2
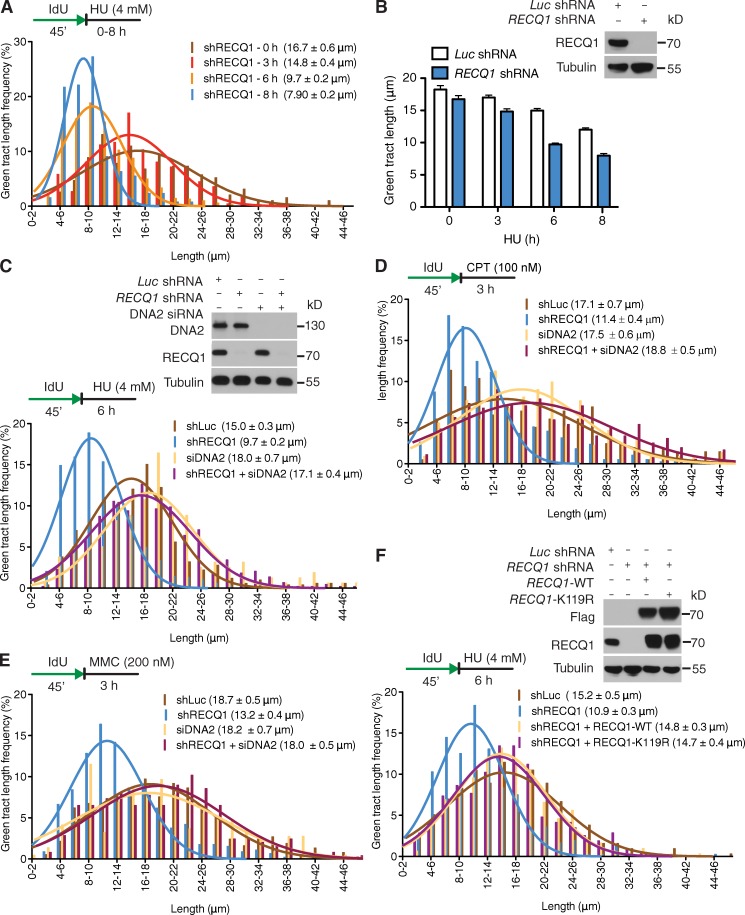
On the basis of the recent discovery that RECQ1 is required to restart replication forks that have reversed upon genotoxic stress induction (Berti et al., 2013), we investigated whether RECQ1 regulates the fork processing activity of DNA2. Nascent IdU tracts were substantially shorter in RECQ1-depleted cells compared with control when replication forks were stalled with HU (after 8 h of HU treatment, the mean tract lengths were 7.9 and 12.0 µm, respectively; P < 0.0001; Fig. 3, A and B). In agreement with results from luciferase-depleted cells, DNA2 was also responsible for the nascent strand degradation phenotype observed in RECQ1-deficient U-2 OS cells (Fig. 3 C). Analogous results were obtained using the conditional DNA2 knockout HCT116 cell line (Fig. 2 C). In addition, we confirmed that the DNA2-dependent nascent strand degradation observed in the absence of RECQ1 is not limited to a specific replication inhibitor by replacing HU with CPT or MMC (Fig. 3, D and E).
Figure 3.
RECQ1 regulates the DNA2-dependent degradation of stalled forks. (A) Representative IdU tracts in RECQ1-depleted U-2 OS cells during different exposure time to HU (out of 2 repeats; n ≥ 350 tracts scored for each dataset). (B) Bar graph represents the mean values of each time point from Figs. 1 C and 2 A. (top) RECQ1 expression after shRNA knockdown. (C, D, and E) Representative IdU tracts in RECQ1-, DNA2-, or RECQ1/DNA2-codepleted U-2 OS cells in the presence of HU (C), CPT (D), and MMC (E; out of 2 repeats; n ≥ 300 tracts scored for each dataset).(top) RECQ1 and DNA2 expression after shRNA or siRNA knockdown. (F) Representative IdU tracts in RECQ1-depleted U-2 OS cells complemented with shRNA-resistant WT RECQ1 (WT) or ATPase-deficient (K119R) RECQ1 (out of 2 repeats; n ≥ 325 tracts scored for each dataset). (top) Expression of Flag-tagged RECQ1-WT and RECQ1-K119R in RECQ1-depleted cells.
Genetic knockdown–rescue experiments confirmed that complementation in RECQ1-depleted U-2 OS cells with shRNA-resistant WT RECQ1 abrogates the effect of RECQ1 depletion on replication fork processing upon HU treatment (Fig. 3 F). Interestingly, expression of the ATPase-deficient RECQ1 mutant K119R in RECQ1-depleted cells also abrogated the effect of RECQ1 depletion indicating that the ATPase activity of RECQ1 was not required for its role in protecting stalled forks from DNA2-dependent degradation (Fig. 3 F). These results point to an additional role of RECQ1 in protecting replication forks from extensive DNA2-dependent degradation, which is independent of RECQ1 ATPase activity.
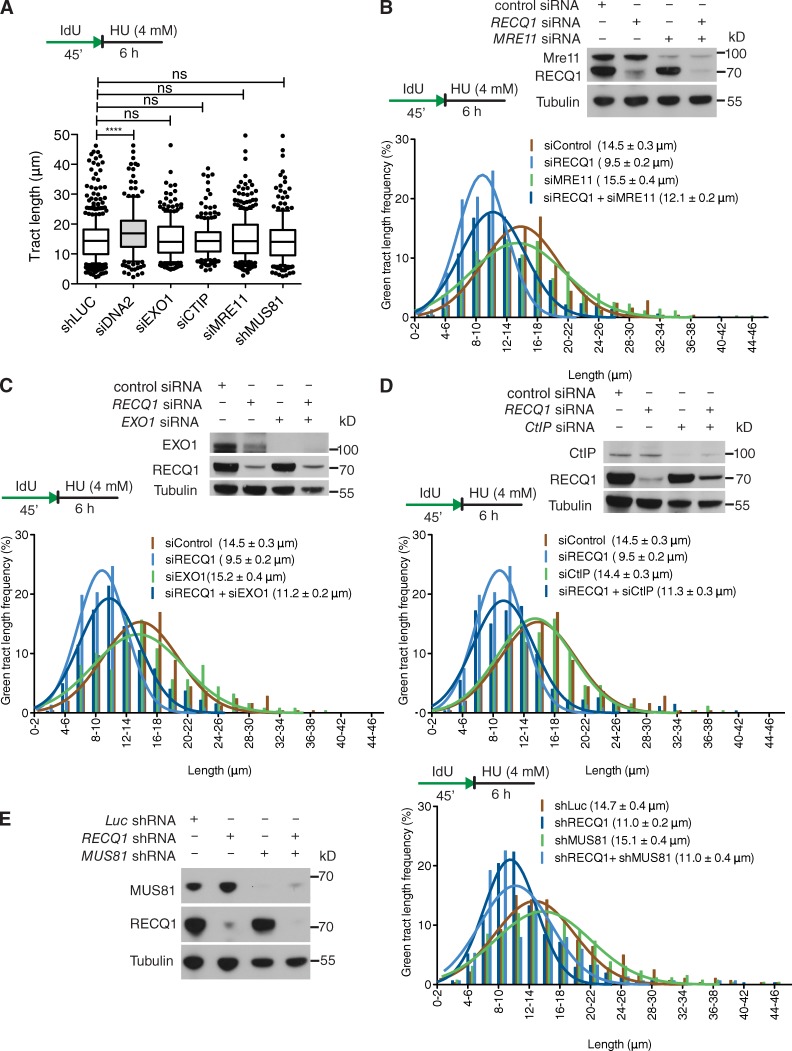
DNA2 function in stalled fork processing is distinct from EXO1, Mre11, and CtIP
Next, we tested whether other nucleases share a function similar to DNA2 in stalled fork processing. To address this point, we depleted Mre11, EXO1, and CtIP in U-2 OS cells with siRNA-mediated technologies. We found that none of these nucleases share the same phenotype of DNA2 in RECQ1-proficient cells (Fig. 4 A). Furthermore, depletion of these nucleases had only a marginal effect on the rescue of the prominent nascent strand degradation phenotype observed in the absence of RECQ1, indicating that DNA2 has a unique function in reversed fork processing that is not shared by these human nucleases (Fig. 4, B–D). MUS81 is another structure-specific nuclease that plays a critical role in replication fork rescue by converting stalled replication forks into DNA DSBs that can be processed by Homology Directed Repair (HDR) (Hanada et al., 2007; Franchitto et al., 2008). This raised the possibility that the DNA2-dependent degradation originated from the processing of MUS81-dependent DSBs. However, MUS81 depletion did not prevent nascent strand degradation, indicating that DNA2 is not processing stalled replication intermediates that are cleaved by MUS81 (Fig. 4 E).
Figure 4.
EXO1, MRE11, CtIP, and MUS81 depletion does not affect stalled fork processing. (A) Statistical analysis of IdU tracts from U-2 OS cells depleted for the indicated proteins in the presence of 4 mM HU. (B) Representative IdU tracts in control, RECQ1-, MRE11-, or RECQ1/MRE11-codepleted U-2 OS cells (out of 2 repeats). (top) Expression of RECQ1 and MRE11 after siRNA knockdown. (C) Representative IdU tracts in control, RECQ1-, EXO1-, or RECQ1/EXO1-codepleted U-2 OS cells (out of 2 repeats). (top) Expression of RECQ1 and EXO1 after siRNA knockdown. (D) Representative IdU tracts in control, RECQ1-, CtIP-, or RECQ1/CtIP-codepleted U-2 OS cells (out of 2 repeats). (top) Expression of RECQ1 and CtIP after siRNA knockdown. (E) Representative IdU tracts in Luc-, RECQ1-, MUS81-, or RECQ1/MUS81-codepleted U-2 OS cells in the presence of HU (out of 2 repeats). (left) Expression of RECQ1 and MUS81 after shRNA knockdown. n ≥ 300 tracts scored for each dataset shown in A–E.
DNA2 and WRN act together to process stalled replication forks
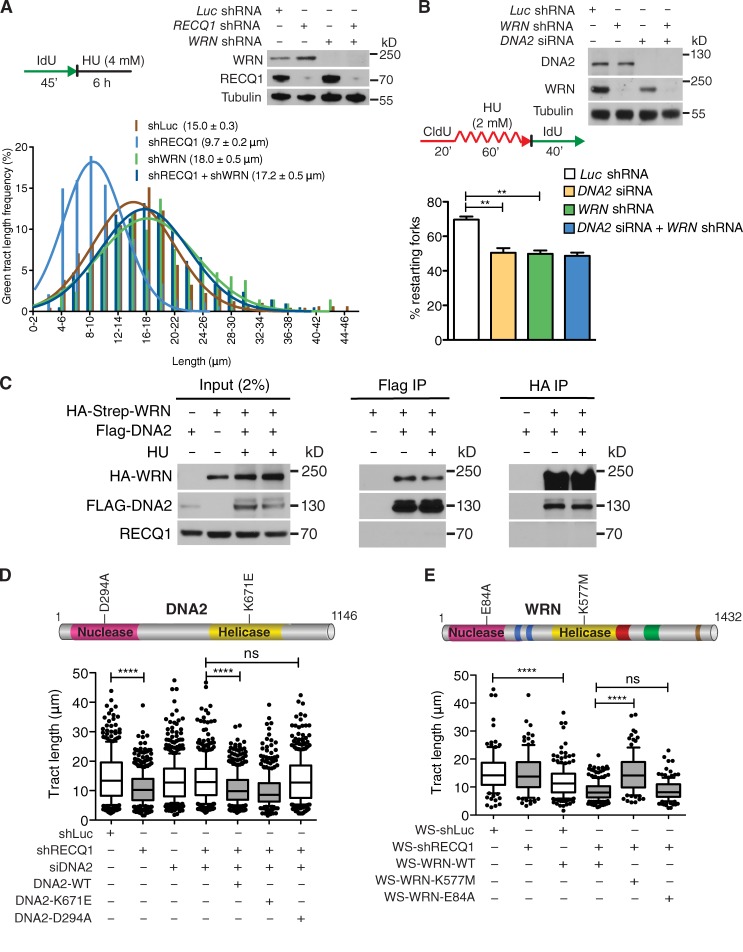
DNA2-dependent dsDNA-end resection needs the support of a RecQ helicase to unwind the DNA from the break (Cejka et al., 2010; Niu et al., 2010; Nimonkar et al., 2011). To determine the identity of the helicase that acts in conjunction with DNA2 in stalled fork processing, we measured the extent of nascent strand degradation in BLM-, WRN-, and RECQ4-depleted U-2 OS cells. Our DNA fiber analysis showed that WRN depletion mimicked the effect of DNA2-depletion, completely abrogating the prominent nascent strand degradation phenotype observed in RECQ1-depleted U-2 OS cells (Fig. 5 A). The same results were confirmed using WRN and DNA2 codepleted cells, suggesting that DNA2 and WRN are epistatic in nucleolytic processing of stalled forks (Fig. S1 C). The partial nascent strand degradation observed in RECQ1-proficient U-2 OS cells was also abrogated by WRN depletion (Fig. S1 D). Conversely, BLM depletion had only a marginal effect on the nascent strand degradation phenotype observed in RECQ1-depleted cells, whereas RECQ4 depletion had no effect (Fig. S2, A and B). Thus, the WRN helicase plays a prominent role in assisting DNA2-dependent degradation of stalled replication forks.
Figure 5.
DNA2 and WRN are epistatic in stalled fork processing and replication restart. (A) Representative IdU tracts in RECQ1-, WRN-, or RECQ1/WRN-codepleted U-2 OS cells (out of 2 repeats; n ≥ 300 tracts scored for each dataset). (top) RECQ1 and WRN expression after shRNA knockdown. (B) Quantification of restarting forks in DNA2-, WRN-, or DNA2/WRN-codepleted cells. Mean shown, n = 3. Error bars, standard error. *, P < 0.05; **, P < 0.01 (paired t test). (top) Expression of WRN and DNA2 after shRNA knockdown. (C) Co-IP experiments in HEK293T cells transfected with empty vectors, Flag-DNA2, or Strep-HA-WRN. Cells were treated with 4 mM HU (3 h) where indicated. Whole-cell extracts were analyzed before (input) and after IP. (D) Statistical analysis of IdU tracts from RECQ1/DNA2-codepleted U-2 OS cells complemented with WT, ATPase-deficient (K671E), or nuclease-deficient (D294A) DNA2, when indicated. (E) Statistical analysis of IdU tracts from RECQ1-depleted WS cells complemented with WT, ATPase-deficient (K577M), or nuclease-deficient (E84A) WRN. Whiskers in D and E indicate the 10th and 90th percentiles. ns, not significant; ****, P < 0.0001 (Mann-Whitney test). n ≥ 300 tracts scored for each dataset shown in D and E.
We next compared the percentage of restarting replication forks in DNA2-depleted, WRN-depleted, and DNA2/WRN-codepleted cells. WRN depletion leads to a decrease in restarting forks (69 to 50%; P = 0.0068). These results are almost identical to those obtained with the DNA2-depleted or DNA2/WRN-codepleted cells, implying that WRN and DNA2 are epistatic also in the restart process (Fig. 5 B). The notion that DNA2 and WRN functionally interact to process stalled replication intermediates is further supported by our observation that the two proteins form a complex both in the presence and absence of replication stress (Fig. 5 C). Of note, RECQ1 is not part of the WRN:DNA2 complex. Collectively, these results suggest that DNA2 cooperates with WRN to promote nascent strand processing and fork restart after HU treatment.
The nuclease activity of DNA2 and the ATPase activity of WRN are essential to process stalled replication forks
DNA2 is characterized by an N-terminal nuclease domain and by a C-terminal helicase domain, but the function of its helicase activity is still debated (Masuda-Sasa et al., 2006). To assess the roles of these two activities in stalled fork processing, we performed genetic knockdown-rescue experiments where we depleted DNA2 and then attempted to rescue fork processing by expressing a Flag-tagged siRNA resistant WT DNA2 control, nuclease-deficient DNA2-D294A, or ATPase-deficient DNA2-K671E. All the experiments were performed in RECQ1-depleted cells, where the effect of DNA2 is more apparent. DNA fiber analysis showed that complementation with nuclease-deficient DNA2 prevents fork processing, whereas complementation with WT or ATPase-deficient DNA2 leads to the same fork processing phenotype observed in DNA2-proficient cells (Fig. 5 D and Fig. S2 C). Therefore, the nuclease, but not the ATPase activity of DNA2, is necessary for fork processing.
Next, we used a Werner Syndrome (WS) fibroblast cell line (AG11395) expressing missense mutant forms of WRN, which inactivate either the exonuclease (WRN-E84A) or the ATPase (K577M) activity of WRN (Pirzio et al., 2008). The ATPase, but not the nuclease activity of WRN, was important for fork processing (Fig. 5 E and Fig. S2 D). These findings were validated by genetic knockdown-rescue experiments where we complemented WRN-depleted U-2 OS cells either with an shRNA resistant WT WRN control or the ATPase-deficient WRN-K577M mutant and found that complementation with the ATPase-deficient mutant prevented fork processing (Fig. S2, E and F). Collectively, these results show that human DNA2 needs the support of the ATPase activity of WRN to promote degradation of the nascent DNA strands.
DNA2 processes reversed replication forks
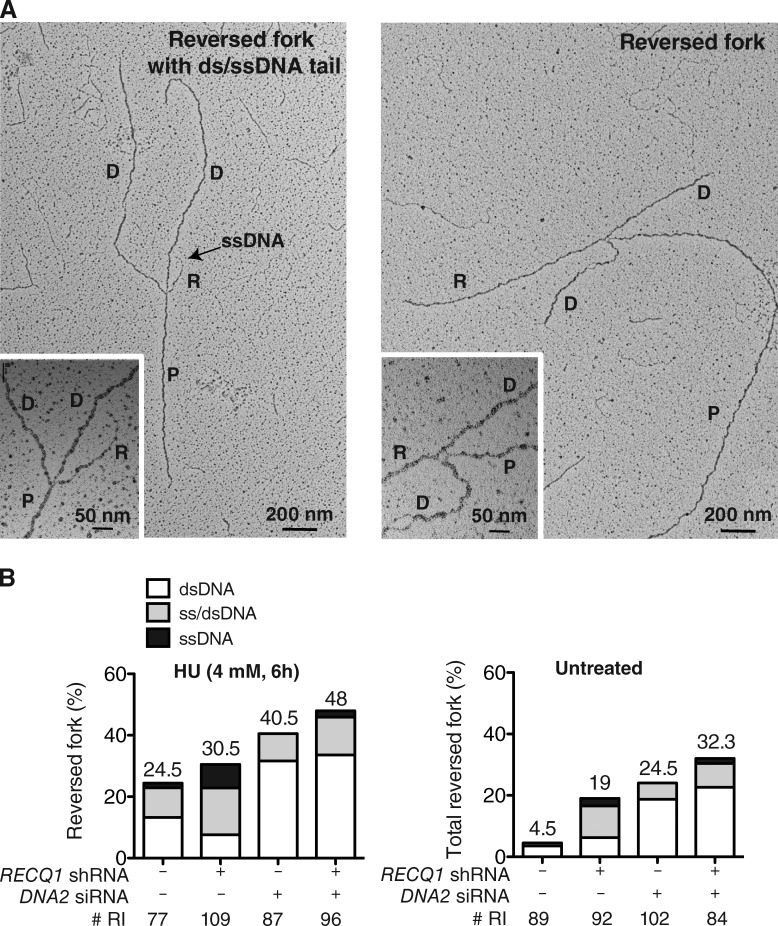
To gain insight into the actual replication structures processed by DNA2, we inspected the fine architecture of the replication intermediates using a combination of in vivo psoralen cross-linking and EM (Neelsen et al., 2014). Our analysis showed a substantial fraction of reversed replication forks (∼24% of molecules analyzed) in control U-2 OS cells treated with 4 mM HU. RECQ1-depletion, and to an even greater extent DNA2-depletion, resulted in a higher frequency of fork reversal events (∼30 and 40%, respectively) compared with HU-treated cells. Co-depletion of RECQ1 and DNA2 further increased the frequency of reversed forks (∼50%), suggesting that RECQ1 and DNA2 are involved into two distinct mechanisms of reversed fork processing. Interestingly, RECQ1 and/or DNA2 depletion also led to a significant amount of fork reversal events in unperturbed U-2 OS cells (Fig. 6, A and B). WRN-depletion phenocopied DNA2-depletion in terms of reversed fork accumulation, both the presence and in the absence of HU. Moreover, DNA2/WRN-codepletion did not cause a further increase in reversed fork frequency, thus supporting our conclusion that DNA2 and WRN work together in reversed fork processing (Fig. S3 A).
Figure 6.
DNA2 resects reversed replication forks. (A) Electron micrograph of a partially single-stranded (left) and entirely double-stranded (right) reversed fork observed on genomic DNA upon HU-treatment. The black arrow points to the ssDNA region on the reversed arm. Inset, magnified four-way junction at the reversed replication fork. D, Daughter strand; P, Parental strand; R, Reversed arm. (B) Frequency of fork reversal and ssDNA composition of the reversed arms in RECQ1- or DNA2-depleted U-2 OS cells treated with HU (left) or in unperturbed conditions (right). The percentage values are indicated on the top of the bar. “# RI” indicates the number of analyzed replication intermediates. Data in B are reproduced with very similar results in at least one independent experiment.
Next, we evaluated the single-strand composition of the regressed arms. To measure ssDNA, we carefully inspected the frequency and length of ssDNA regions on the regressed arms by detecting local difference in filament thickness. DNA2 depletion led to a higher frequency of reversed forks with a dsDNA arm—and a corresponding decrease of partially or entirely single-stranded reversed forks—in both RECQ1-proficient and deficient cells (Fig. 6). Thus, DNA2-mediated resection is directed to completely or partially digest one strand of the reversed arm leading to reversed forks that are either entirely single stranded or have a protruding ssDNA tail. However, prolonged stalling by HU was associated with accumulation of postreplicative ssDNA gaps on replicated duplexes, which was maximal in RECQ1-depleted cells and suppressed by DNA2 depletion (Fig. S3, B and C). Consequently, ssDNA gaps may reflect additional activity of the same nucleolytic apparatus along the postreplicated duplexes or restart of partially resected reversed forks.
As an alternative readout for DNA2-dependent resection, we examined the phosphorylation status of RPA and the checkpoint kinase Chk1 (Zeman and Cimprich, 2014). DNA2 depletion caused a reduction in RPA and Chk1 phosphorylation in both RECQ1-proficient and RECQ1-deficient U-2 OS cells, suggesting that the DNA2-dependent resection of nascent strands might also contribute to checkpoint activation (Fig. S3 D).
RAD51 promotes DNA2-dependent degradation of reversed replication forks
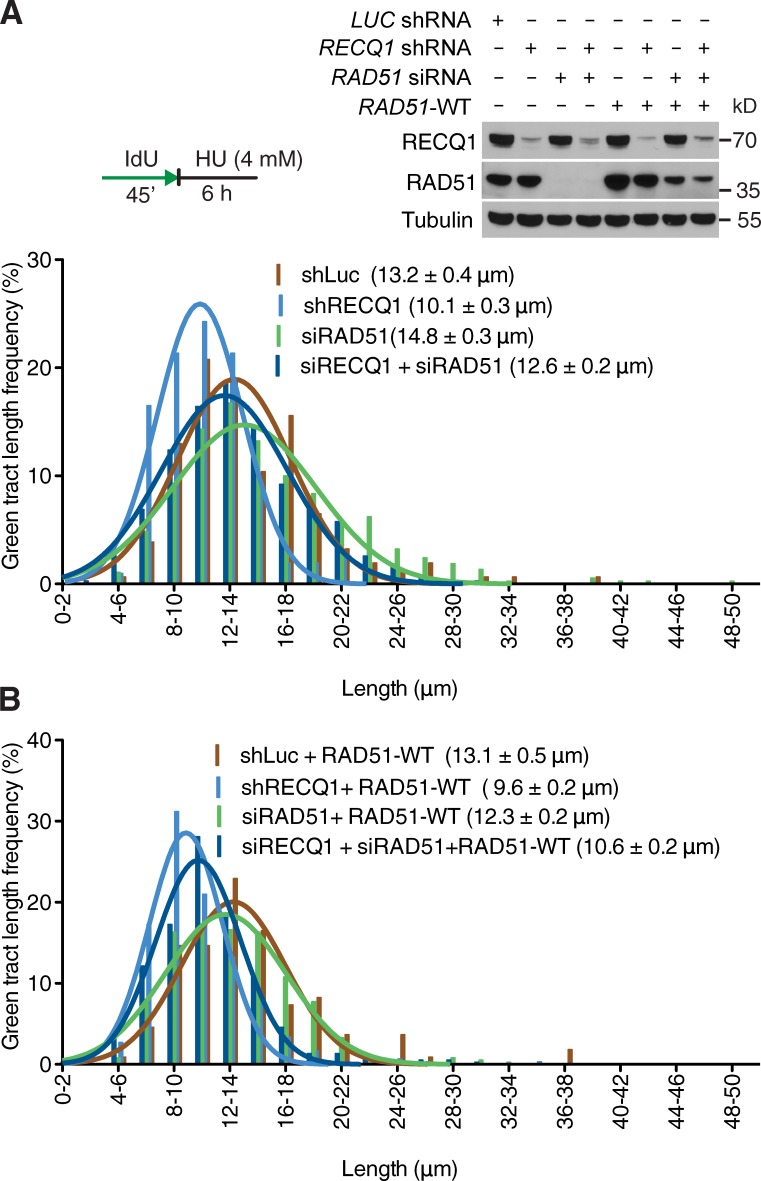
The central recombinase factor RAD51 is directly implicated in reversed fork formation upon genotoxic stress (Zellweger et al., 2015). Thus, we investigated whether RAD51 depletion may affect the reversed fork processing activity of DNA2. We found that RAD51 knockdown largely prevents DNA2 nucleolytic processing both in RECQ1 proficient and RECQ1-deficient cells (Fig. 7 A). Genetic knockdown–rescue experiments confirmed that expression of exogenous RAD51 in RAD51-depleted U-2 OS cells restored the fork processing phenotype (Fig. 7 B). These results indicate that DNA2-dependent nucleolytic processing is specifically targeted to reversed fork structures because it is not detected in a genetic background that prevents reversed fork formation—i.e., RAD51 knockdown.
Figure 7.
RAD51 promotes DNA2-depedent degradation of reversed replication forks. (A) Representative IdU tracts in RECQ1-, RAD51-, or RECQ1/RAD51-codepleted U-2 OS cells (out of 2 repeats). Above, RECQ1 and RAD51 expression after siRNA knockdown RAD51-WT are U-2 OS cells stably expressing siRNA resistant exogenous RAD51. (B) Representative IdU tracts in U-2 OS cells expressing exogenous RAD51 (out of 2 repeats). n ≥ 300 tracts scored for each dataset shown in A and B.
DNA2 preferentially degrades reversed fork structures with a 5′-to-3′ polarity
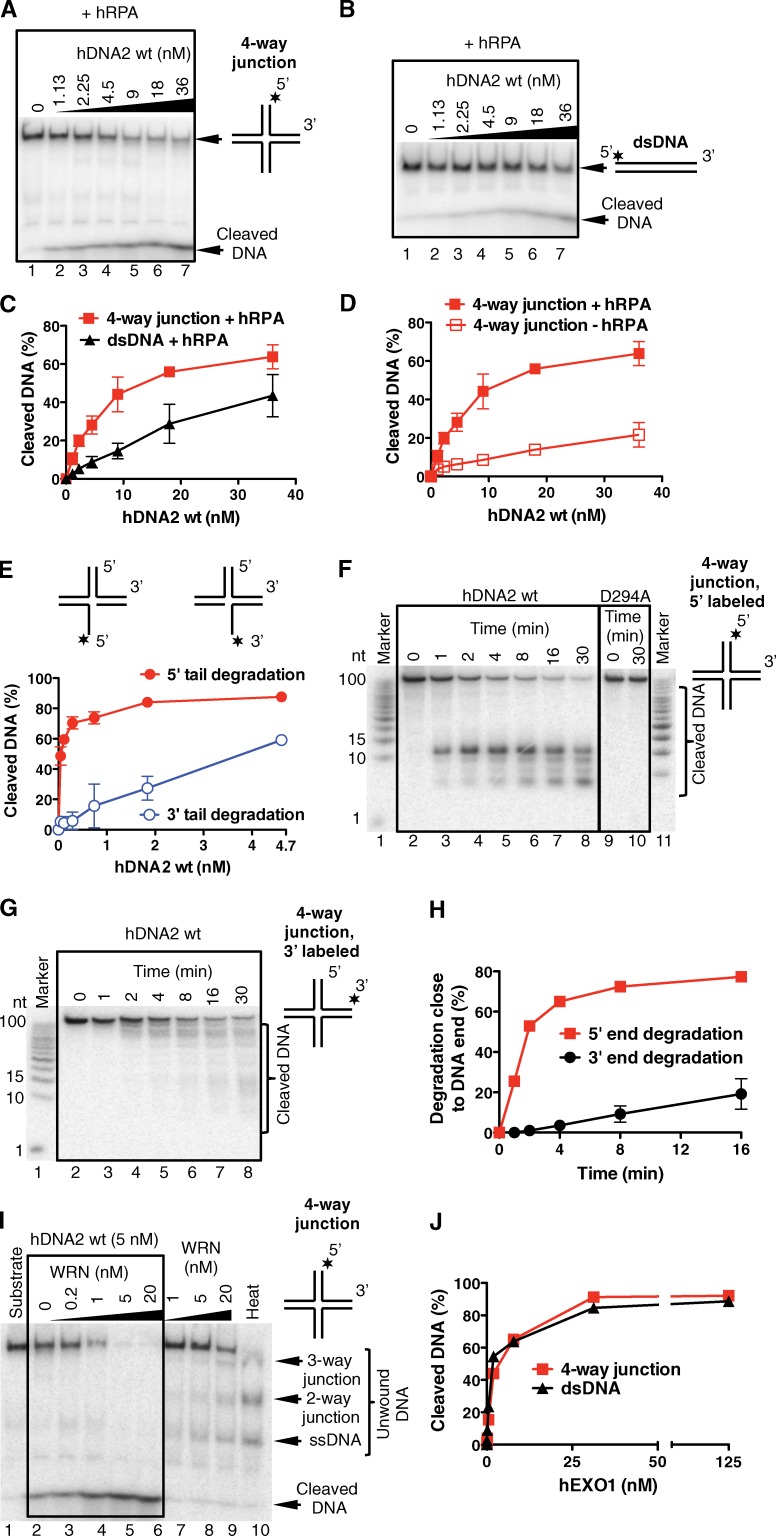
The notion that DNA2 end resection has a preferential polarity in vivo is consistent with biochemical studies showing that even though DNA2 has the intrinsic capacity to degrade both 5′- and 3′-terminated ssDNA, RPA enforces a primarily 5′-to-3′ end-resection bias (Cejka et al., 2010; Niu et al., 2010; Nimonkar et al., 2011). Thus, we set up new biochemical assays to test whether human DNA2 prefers four-way junction substrates—i.e., reversed replication forks—versus linear DNA duplexes and whether it degrades these substrates with a 5′-to-3′ polarity in the presence of RPA (Fig. 8, A and B). The sequences of the four arms of the four-way junction substrates are mutually heterologous to prevent four-way junction branch migration. DNA2-degraded four-way junction substrates more efficiently than linear dsDNA duplexes, with 20 nM DNA2 required to degrade ∼60% of the four-way junction substrates versus only ∼30% of the linear duplex (Fig. 8 C). Importantly, supplementing the reaction with RPA greatly stimulated the degradation activity of human DNA2 (Fig. 8 D and Fig. S4 A). Additional experiments using either 5′-end or 3′-end 32P-labeled four-way junctions confirmed that human DNA2 had a strong 5′-to-3′ bias in end resection in the presence of RPA (Fig. 8, E–H; and Fig. S4, B and C). Catalytically dead DNA2 D294A had no capacity to degrade DNA, showing that the nuclease activity is inherent to WT DNA2 (Fig. 8 F). The same results were recapitulated using purified yeast DNA2 (Fig. S5, A–F). Interestingly, addition of the ATPase-deficient RECQ1 mutant (RECQ1-K119R) to the reaction mix significantly inhibited the four-way junction degradation activity of human DNA2 (Fig. S4, D and E). These results suggest that the binding of RECQ1 to stalled replication forks limits the fork processing activity of DNA2, as inferred by our cellular studies. However, we cannot rule out the possibility that the inhibitory effect observed in the biochemical assays is simply associated with competition for substrate recognition between the two proteins. In agreement with our in vivo data, we show that WRN promoted the degradative capacity of DNA2 on nicked, gapped, or four-way junction substrates (Fig. 8 I and Fig. S4, F and G); similar behavior was observed when yeast Dna2 was coupled with the Sgs1 helicase (Fig. S5, G and H). DNA was degraded by WRN and DNA2 in a remarkably synergistic manner: 5 nM concentration of either WRN or DNA2 alone led only to a minor DNA unwinding/degradation (Fig. 8 I, lanes 2 and 8). When combined, both enzymes completely degraded the four-way junction DNA (Fig. 8 I, lane 5). In contrast, no such synergy was observed when human DNA2 was combined with the noncognate yeast meiotic Mer3 helicase (Fig. S4 H), suggesting that the species-specific interaction between DNA2 and WRN results in a vigorous DNA degradation. Similarly, WT RECQ1 did not promote DNA degradation by DNA2 (Fig. S4 I).
Figure 8.
Human DNA2 preferentially degrades branched DNA in a 5′-3′ direction in reactions stimulated by WRN. (A) Degradation of a four-way junction by human DNA2 (hDNA2) in the presence of hRPA (native 6% polyacrylamide gel) (B) Experiment as in A, but with dsDNA. (C) Quantitation of data from A and B. Averages shown ± SEM; n = 2. (D) DNA degradation is stimulated by hRPA. The data points from +hRPA condition are the same as in C. Averages shown ± SEM; n = 2. (E) Quantitation of degradation of a 3′ or 5′ ssDNA-tailed three-way junction by hDNA2. The reactions were performed in 3 mM magnesium acetate and 22.3 nM hRPA. Averages shown ± SEM; n = 2. (F) Kinetics of degradation of a four-way junction by hDNA2 (9 nM) in the presence of hRPA (denaturing 20% polyacrylamide gel). The substrate was labeled at the 5′ end (*). D294A, nuclease-dead variant of hDNA2. (G) Experiment as in F, but using a four-way junction labeled at the 3′ end. (H) Quantitation of DNA cleavage near (less than 15 nt) a 5′ or 3′ DNA end from experiments of F and G. Averages shown ± SEM; n = 2. (I) WRN and hDNA2 degrade four-way junction DNA in a synergistic manner. Reactions with indicated hDNA2 and/or WRN concentrations and 65 nM hRPA were analyzed on a 6% native polyacrylamide gel. Heat, partially heated DNA substrate indicating the positions of DNA unwinding intermediates. (J) Quantitation of four-way junction and dsDNA degradation by human EXO1 (hEXO1). Averages shown ± SEM; n = 2.
On the basis of our results that DNA2 does not share the same function of EXO1 in reversed fork processing, we decided to compare the end-resection activities of human DNA2 and human EXO1 using the four-way junction substrates. EXO1—unlike DNA2—degraded both four-way junction substrates and linear duplexes with equal efficiency (Fig. 8 J and Fig. S4, J and K). The use of yeast variants of Dna2 and Exo1 yielded analogous results (Fig. S5, I–K). Collectively, these studies further implicate DNA2, and its nuclease activity, in reversed replication fork degradation—that is specifically stimulated by WRN—and point to an important difference in substrate preference between DNA2 and EXO1. Moreover, the polarity of reversed fork degradation by DNA2 measured in the presence of RPA displays the same bias anticipated from the EM analysis of the replication intermediates.
Discussion
The present work uncovers a new mechanism for reversed fork processing and restart that requires the coordinated activities of the human DNA2 nuclease and WRN helicase (Fig. 9). The DNA2-dependent end resection leads to partially single-stranded reversed forks and is required for efficient replication fork restart under conditions of persistent replication blockage. WRN interacts with DNA2 and its ATPase activity is needed for DNA2-dependent degradation, presumably to transiently open the dsDNA arm of the reversed replication forks.
Figure 9.
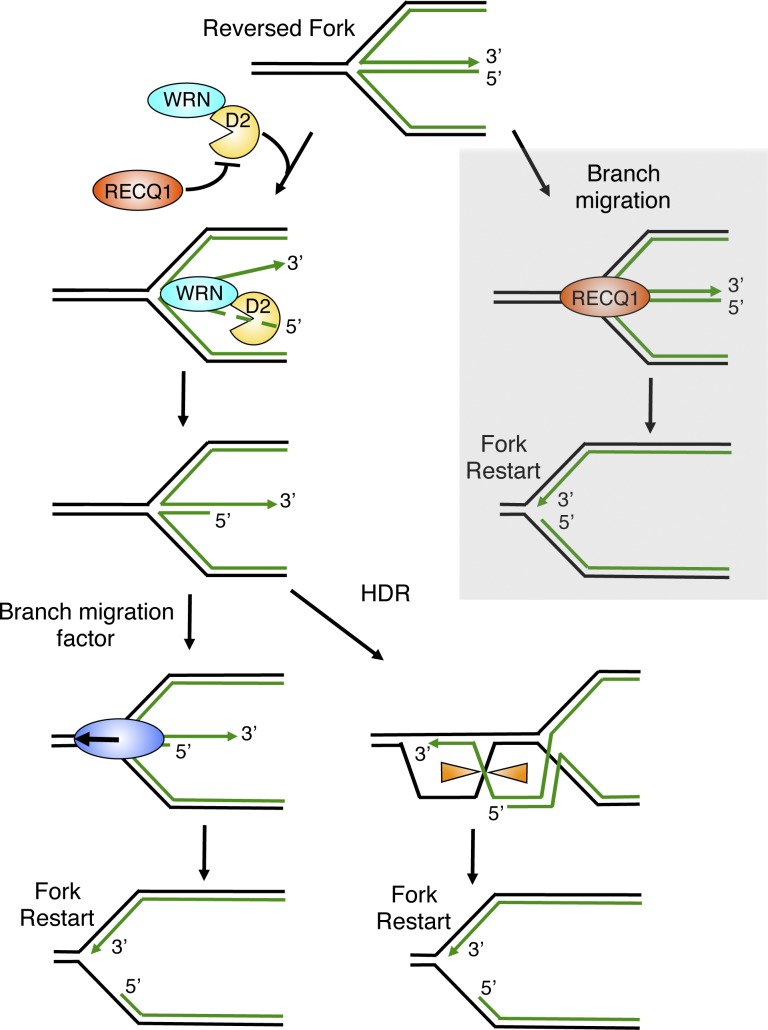
Schematic model for the combined roles of DNA2 and WRN in reversed fork processing. DNA2 and WRN functionally interact to process reversed forks. DNA2 degrades reversed forks with a 5′-to-3′ polarity. WRN ATPase activity assists DNA2 degradation possibly by promoting the opening of the reversed arm of the fork. RECQ1 limits DNA2 activity by an ATPase-independent function. Branch migration factors specifically recognize the partially resected reversed forks to promote fork restart. Alternatively, the newly formed 3′ overhang of the reversed fork invades the duplex ahead of the fork, resulting in Holliday junction structures that can be resolved by specific resolvases or dissolvases to promote fork restart. Gray box, RECQ1 can independently restart reversed forks by virtue of its ATPase and branch migration activity.
To date, we have identified two mechanisms of reversed replication fork resolution, one dependent on RECQ1 ATPase and branch migration activity (Berti et al., 2013) and the other on DNA2 nuclease and WRN ATPase activity. Moreover, the DNA2/WRN mechanism is tightly regulated by an ATPase-independent function of RECQ1 that might limit DNA2 activity by binding to reversed forks. Of note, our EM experiments show that reversed replication forks accumulate in RECQ1- and DNA2-depleted cells also in unperturbed conditions suggesting that fork reversal is remarkably frequent when DNA replication faces intrinsic replication fork obstacles, and that RECQ1 and DNA2 have a conserved role in restarting reversed forks in unperturbed S-phase.
DNA2 function during DNA replication is vital for maintenance of genome stability (this study; Duxin et al., 2012; Karanja et al., 2012). These findings indicate that the controlled DNA2-dependent degradation of reversed replication forks is a physiologically relevant mechanism to provide resistance to prolonged genotoxic treatments. This mechanism is distinct from the pathological MRE11-dependent degradation of stalled replication intermediates detected in the absence of crucial Fanconi Anemia (FA)/HR factors (Schlacher et al., 2011, 2012; Hashimoto et al., 2012; Ying et al., 2012).
We find that depletion of the central recombinase factor RAD51 prevents nascent strand degradation. This finding, coupled with the recent observation that RAD51 is directly implicated in reversed fork formation (Zellweger et al., 2015), reinforce our conclusion that the DNA2-dependent pathway starts from the reversed arm of stalled replication forks and acts downstream of the RAD51-mediated replication fork reversal. Given that RAD51 is required for reversed fork formation (Zellweger et al., 2015), we speculate that the MRE11-dependent pathway is only uncovered in the absence of fork reversal—i.e., via a perturbation in RAD51 function—and likely attacks unprotected and nonreversed forks upon prolonged stalling. A crucial challenge for future studies will be to investigate why we do not observe a contribution of the MRE11 pathway in nascent strand degradation upon RAD51 depletion. It is tempting to speculate that RAD51 depletion might interfere with MRE11-dependent fork processing, in addition to preventing fork reversal. Conversely, perturbation of RAD51 function—e.g., via BRCA2 depletion (Schlacher et al., 2011)—might be sufficient to prevent fork reversal—hence DNA2-dependent degradation—but still allow residual RAD51 loading to promote MRE11-dependent degradation.
Our DNA fiber analysis suggests that DNA2 degrades stalled replication intermediates beyond the maximum length of the reversed arms measured by EM (up to several kilobases). A possible interpretation of these results is that after the initial DNA2/WRN-mediated regressed arm degradation is complete, other nucleolytic activities or DNA2 itself may codegrade both sides of the replication fork, thus leading to extensive degradation events detectable by DNA fibers. In this scenario, our EM images likely represent snapshots of the “slow steps” of this reaction—i.e., the DNA2/WRN-mediated degradation of the regressed arms—resulting in the drastic increase in reversed fork frequency observed in the absence of DNA2. Once the regressed arm has been resolved, the nucleolytic degradation might quickly proceed to degrade nascent strands behind the junction—as suggested by the DNA2-dependent increase in ssDNA gaps behind the observed forks—finally leading to reannealing of the parental strands and backtracking of the fork (Fig. S3 E). A new reversal event may occur when this extensive degradation leads to asymmetric ssDNA accumulation at the fork (Zellweger et al., 2015), resetting the backtracked fork to the slow step of the process. However, fork backtracking is only one possible model to explain the extensive degradation detected by DNA fibers and further work would be required to uncover additional nucleolytic activities that might be involved in this process.
Biochemical studies suggested that Schizosaccharomyces pombe Dna2 cleaves the leading and lagging reversed strands of a model replication fork with similar efficiency in the absence of replication protein A (Hu et al., 2012). However, it is likely that only the 5′-to-3′ directionality is important in vivo, because RPA is known to stimulate the 5′-to-3′ and inhibit the 3′-to-5′ nuclease activity of yeast DNA2 (Cejka et al., 2010; Niu et al., 2010). In agreement with this conclusion, our biochemical data show that DNA2-dependent end resection proceeds with a 5′ to 3′ polarity in the presence of RPA. Moreover, our EM experiments clearly show that DNA2 depletion affects the frequency of reversed forks that are either entirely or partially single-stranded supporting the notion that DNA2-dependent degradation of reversed forks occurs with a preferential polarity in vivo.
The resection activity of human DNA2 was postulated to activate the ATR/Chk1 checkpoint under conditions of replication stress (Karanja et al., 2012). Indeed, we find that DNA2 depletion prevents ATR checkpoint activation after HU treatment. Moreover, the increased origin firing observed upon DNA2 depletion is consistent with observations that the deregulation of checkpoint activity leads to a large increase in the number of newly initiated origins (Couch et al., 2013). However, the extent of ATR activation does not necessarily reflect the amount of ssDNA detected at replication forks, whether at the junction, at ssDNA gaps, or at regressed arms (Zellweger et al., 2015). In light of these findings, we rather suggest that DNA2-dependent ATR activation may reflect DNA2 recruitment to the stalled forks per se, or subtle changes of fork architecture that are associated with its recruitment but possibly escape our EM analysis. This interpretation is supported by the recent discovery that yeast Dna2 has a direct role in Mec1 activation (the ortholog of human ATR), independent from its nuclease or helicase activity (Kumar and Burgers, 2013). Of note, the increased origin firing frequency observed upon DNA2 depletion is not associated to a parallel increase in the frequency of termination events (Fig. S1 A) possibly because the defects in replication fork restart associated with DNA2 depletion limit the number of termination events even under conditions of increased origin firing.
WRN plays an important—albeit mechanistically ill-defined—role in the recovery from replication blockage, and mutations in the WRN gene are linked to the cancer predisposition disorder Werner Syndrome (Sidorova et al., 2008; Murfuni et al., 2012). Our studies infer that the high genomic instability of WRN-deficient cells may result from aberrant processing of reversed replication intermediates. In particular, given the consolidated role of WRN at difficult-to-replicate regions—e.g., telomeres and fragile sites (Crabbe et al., 2004; Murfuni et al., 2012)—we speculate that WRN, in conjunction with DNA2, is required to process reversed forks arising spontaneously at these genomic loci. Biochemical studies pointed to a putative role of WRN in fork reversal and/or restart by showing that WRN efficiently promotes both the formation and restoration of oligonucleotide-based reversed fork substrates (Machwe et al., 2011). We show that WRN ATPase activity is needed for the DNA2-dependent degradation of reversed replication forks. Our interpretation for the role of WRN ATPase activity is that it facilitates DNA2-dependent degradation of the reversed forks by transiently opening the dsDNA arm of the reversed fork. This mechanism is reminiscent to the DNA2-dependent mechanism of DSB resection where the yeast Sgs1 helicase is required to transiently open the DNA duplex to generate a 5′ ssDNA tail that is in turn degraded by DNA2 (Zhu et al., 2008; Cejka et al., 2010; Niu et al., 2010). We suggest that WRN is the functional homologue of Sgs1 in mammalian cells, at least in the context of DNA2-dependent reversed replication fork processing. However, BLM was also shown to interact and cooperate with DNA2 to resect dsDNA ends in vitro opening the possibility that other human RecQ helicases might substitute for WRN, depending on the nature of the DNA lesion being processed or the particular cellular context (Nimonkar et al., 2011; Sturzenegger et al., 2014). This mechanism seems to be well-conserved throughout evolution because it is highly reminiscent of the stalled fork processing pathway described in E. coli where the RecJ nuclease cooperates with bacterial RecQ to process blocked replication intermediates (Courcelle et al., 2003). In addition, the prokaryotic RecBCD helicase-nuclease plays an important role in resecting replication forks after reversal (Seigneur et al., 1998) and DNA2 is of the same family of nucleases as RecB. Whether the DNA2/WRN-mediated resection activity can degrade additional stalled replication intermediates other than reversed forks is worth future investigation.
EXO1, MRE11, and CtIP play central roles in DNA repair and are also implicated in the recovery from replication fork blockage (Cotta-Ramusino et al., 2005; Schlacher et al., 2011; Yeo et al., 2014). None of these nucleases, however, participates in the DNA2-dependent processing of reversed replication forks pointing to a specific role of DNA2 that, unlike its function in DSB resection, is not shared by other nucleases. A possible interpretation of these results is that the reversed forks are characterized by a particular structure of the terminal end that does not require the trimming activity of other nucleases to promote DNA2-dependent resection. However, some of these nucleases might still be able to access stalled forks under specific genetic backgrounds. For example, MRE11 degrades stalled replication intermediates only in a BRCA2-deficent background, as already discussed (Schlacher et al., 2011). Moreover, the cleavage of unresolved replicative intermediates by the structure-specific MUS81 endonuclease is a late response to replicative stress, which becomes activated only when other attempts to overcome stalled replication have been exhausted (Hanada et al., 2007; Franchitto et al., 2008). Thus, MUS81 might still resolve reversed replication forks as a back-up system to unlink sister chromatids and facilitate mitotic segregation in the absence of DNA2 or WRN.
Collectively, these studies highlight a new important mechanism for the recovery from replication blockage. This mechanism relies on the DNA2-dependent processing of reversed forks—leading to ssDNA stretches on the regressed arms—which appear to promote efficient fork restart. A possible explanation for the need of partially single-stranded DNA structures to promote fork restart is that they represent a key intermediate to activate an HDR-like mechanism of reversed fork restart, as recently proposed in S. pombe (Carr and Lambert, 2013). In particular, the newly formed 3′ overhang of the reversed fork might invade the duplex ahead of the fork resulting in Holliday junction structures that can be resolved by specific resolvases or dissolved by the combined action of the BLM helicase (Sgs1 in yeast) and the type I topoisomerase TOP3 (Fig. 9). Alternatively, resumption of DNA replication might be obtained by reverse branch migration, where the partially resected reversed fork structures might be specifically recognized by a motor protein—e.g., SMARCAL1 (Béous et al., 2013) or a human RecQ helicase—to promote the branch migration-assisted reestablishment of a functional replication fork.
Materials and methods
Cell lines, culture conditions, and reagents
U-2 OS, HEK 293, and Werner Syndrome fibroblast (AG11395) cells were grown in DMEM supplemented with 10% FBS at 37°C in 5% CO2. HCT116 cells were grown in McCoy’s 5A medium supplemented with 10% FBS. CldU, IdU, BrdU, hydroxyurea, mitomycin C, camptothecin, tamoxifen, puromycin, and hygromycin were obtained from Sigma-Aldrich.
DNA2 conditional knockout HCT116 cells
To examine the response of cells to the complete absence of DNA2, we used a DNA2 conditional knockout cell line where exon 2 of the DNA2 gene is deleted (Karanja et al., 2014). The colorectal carcinoma HCT116 cell line carries 3 copies of DNA2 due to a duplication on chromosome 10. Two chromosomal copies were disrupted using rAAV-mediated gene targeting technology and exon 2 of the third allele was replaced with a conditional exon where the exon was flanked by loxP sites (DNA2flox/−/−). To create a conditional cell line these cells were stably transduced with a tamoxifen (4-OHT)-inducible Cre recombinase. Thus, the cell line is viable and can be propagated. The addition of tamoxifen to the culture media leads to excision of the endogenous DNA2 and the generation of a true DNA2-null cell. Complete loss of DNA2 occurs after 72 h of tamoxifen treatment. However, the DNA fiber experiments were performed after 40 h of tamoxifen treatment to have enough S-phase cells for DNA labeling.
Antibodies
Anti-DNA2 rabbit polyclonal (ab96488; 1:1,000), anti-MUS81 mouse monoclonal (ab14387; 1:1,000), and anti-CldU/BrdU rat monoclonal (ab6326; 1:6) antibodies (all from Abcam); anti-CtIP rabbit polyclonal (A300-488A; 1:1,000), anti-EXO1 rabbit polyclonal (A302-639A; 1:1,000), anti-pRPA32 (S4/S8) rabbit polyclonal (A300-245A; 1:1,000), and anti-pRPA32 (S33) rabbit polyclonal (300-246A; 1:2,000; all from Bethyl); anti-WRN rabbit polyclonal (NB100-471; 1:1,000); and anti-MRE11 rabbit polyclonal (NB100-142; 1:2,000; Novus); anti-RAD51 (H-92) rabbit polyclonal (sc-8349; 1:1,000) and anti-RECQ1 rabbit polyclonal (sc-25547; 1:2,000) from Santa Cruz; anti-rat Alexa (594-A11007; 1:1,000); and anti–mouse Alexa Flour (488-A11001; 1:1,000; Invitrogen); anti-rabbit (31460; 1:10,000; Thermo Fisher Scientific); anti-tubulin mouse monoclonal (T5168; 1:5,000; Sigma-Aldrich); anti-IdU/BrdU mouse monoclonal (347580; 1:6) from BD; anti-Chk1 mouse monoclonal (sc-8408; 1:1,000; Santa Cruz Biotechnology, Inc.); anti-p-Chk1 (S345) rabbit monoclonal (2348; 1:1,000; Cell Signaling Technology); anti-RPA32 mouse monoclonal (NA19L; 1:1,000) from EMD Millipore; anti-RECQ1 rabbit polyclonal, raised against residues 634–649 of human RECQ1, is custom made (Mendoza-Maldonado et al., 2011); anti-BLM rabbit polyclonal, raised against residues 1–449 of human BLM (Wu and Hickson, 2003), was a gift from I. Hickson (University of Copenhagen, Copenhagen, Denmark); and anti-RECQ4 rabbit polyclonal, raised against residues 60–111 of human RECQ4 (Yin et al., 2004), was a gift from W. Wang (National Institute on Aging, Baltimore, MD).
Recombinant proteins
Yeast Dna2 was expressed in yeast WDH668 strain from pGAL:DNA2 vector (Budd et al., 2000) and purified as previously described (Levikova et al., 2013). In brief, the cells were lysed and Dna2 was purified by affinity chromatography on Ni-NTA agarose (QIAGEN) and anti-Flag M2 affinity gel (Sigma-Aldrich). Yeast RPA was expressed in yeast BJ5464 strain containing three plasmids coding for Rfa1, Rfa2, and Rfa3 and purified as previously described (Kantake et al., 2003). In brief, the cells were lysed and yeast RPA was purified by affinity on ssDNA cellulose column (USB corporation) and by ion exchange chromatography using HiTrap Q column (GE Healthcare). Human DNA2 was expressed in Sf9 cells from a pFastBac:hDNA2 vector (polyhedrin promoter) provided by J. Campbell (Masuda-Sasa et al., 2006). The soluble extracts were obtained by salt extraction as previously described for Sgs1 (Cejka and Kowalczykowski, 2010). The subsequent purification of hDNA2 was performed as previously described for yeast Dna2 (Levikova et al., 2013) by affinity chromatography using Ni-NTA agarose (QIAGEN) and Anti-Flag M2 affinity gel (Sigma-Aldrich). Human RPA was expressed from p11d-tRPA vector (Henricksen et al., 1994) in BL21 E. coli cells and purified as described (Henricksen et al., 1994). In brief, hRPA was first bound to HiTrap Blue column (GE healthcare) and then to HiTrap Q column. The sequence coding for yeast Mer3 helicase was amplified from yeast genomic DNA (SK1 strain) using primers Mer3FO (5′-GCGCGCGGGCCCATGAAAACAAAGTTTGATCGCCTCGGTACAGGAAAAAGAAGTAGACCCTCTCCAAATAATATTGACTTTAACGACCAG-3′) and Mer3RE (5′-CGCGCGCTCGAGTTCAAACTCTATATCGGAAC-3′). The PCR product was digested with ApaI and XhoI restriction endonucleases (both from New England Biolabs) and cloned into corresponding sites in pFB-MBP-Sgs1-his after the polyhedrin promoter, creating pFB-MBP-Mer3-his vector. Mer3 was then expressed in Sf9 cells and purified using affinity chromatography as previously described for Sgs1 (Cejka and Kowalczykowski, 2010). In brief, MBP-tagged Mer3 was first bound to amylose resin (New England Biolabs), eluted and digested with PreScission protease to cleave the MBP tag. Mer3 was further purified by affinity on Ni-NTA agarose (QIAGEN) exploiting the 10x His-tag at its C-terminus. Sequence information is available on request.
Genetic knock-down-rescue experiments
RECQ1, DNA2, and RAD51 genetic knockdown-rescue experiments were performed using the procedure described (Berti et al., 2013; Yata et al., 2012). In brief, RECQ1 is depleted using the pLKO.1-puro-shRECQ1 (5′-GAGCTTATGTTACCAGTTA-3′) construct and rescue experiments are performed using the shRNA resistant pIRES-RECQ1-WT or K119R (ATPase dead) constructs as described (Berti et al., 2013). DNA2 is depleted using an siRNA targeting the 3′UTR of DNA2 (5′-CAGUAUCUCCUCUAGCUAG-3′). At least one isoform of DNA2 is not targeted by this sequence. DNA2 rescue experiments are performed using the pBabe-hygro-3xFLAG-DNA2 WT, D294A (Nuclease dead), or K671E (helicase dead) constructs. RAD51 is depleted using siRNAs targeting the 3′UTR (5′-GACUGCCAGGAUAAAGCUU-3′ and 5′-GUGCUGCAGCCUAAUGAGA-3′) in U-2 OS stable cell lines expressing WT RAD51 as described (Yata et al., 2012). WRN depletions were achieved using pRS-puro-shWRN (5′-AGGCAGGTGTAGGAATTGAAGGAGATCAG-3′; sequence ID: TI333414) and exogenous expression is done with the shRNA resistant Flag-pCMVTag2B-WRN WT or K577M (helicase dead) constructs. Constructs for WRN depletion and overexpression of WT WRN and ATPase-deficient WRN (WRN-K577M) were kind gifts from Dr. Pietro Pichierri (Insituto Superiore di Sanità, Rome, Italy). All transfections were done with Lipofectamine 2000 (Life technologies Catalog no: 11668027). An shRNA targeting luciferase (5′-ACGCTGAGTACTTCGAAATGT-3′) was used for control shRNA experiments. The silencer select negative control (Life technologies, Catalog no. 4390843) or an siRNA targeting luciferase (5′-CGUACGCGGAAUACUUCGA-3′) were used for control siRNA experiments, as indicated. Lentiviral mediated shRNA depletions were achieved using the following sequences cloned into the pLKO.1 lentiviral shRNA expression vector: BLM (5′-CGAAGGAAGTTGTATGCACTA-3′), WRN (5′-GCTGGCAATTACCAGAACAAT-3′), and MUS81 (5′-CACGCGCTTCGTATTTCAGAA-3′). The procedure for lentiviral generation and transduction has been described (Berti et al., 2013). Transduced U-2 OS cells were selected with 6 µg/ml puromycin. siRNA-mediated depletions were achieved using the following siRNAs from Invitrogen: DNA2 (5′-AUAGCCAGUAGUAUUCGAU-3′), CtIP (5′-CGAAUCUUAGAUGCACAAA-3′), EXO1 (Invitrogen-HSS113557), and RAD51 (Invitrogen-1299001). In brief, siRNAs were transfected using Lipofectamine RNAiMAX (Life Technologies) following the manufacturer’s protocol. MRE11 (5′-GAAAGGCUCUAUCGAAUGU-3′) and RECQ4 (SMART pool) siRNAs were from Dharmacon and were transfected as previously described (Thangavel et al., 2010).
Microfluidic-assisted DNA fiber stretching
For DNA replication fork restart analysis, asynchronous cells were pulse-labeled with 50 µM CldU for 20–30 min. 2 mM HU, 300 nM MMC, or 150 nM CPT was added to the CldU containing media and incubated for the indicated times. Cells were washed three times with medium and released with 50 µM IdU for 40 min. For nascent strand degradation analysis, asynchronous cells were pulse-labeled with 50 µM IdU for 45 min, washed three times with medium, incubated with 4 mM HU, 100 nM CPT, 200 nM MMC, or medium for times indicated. The pulse-labeled cells were trypsin collected and lysed in agarose plugs to prevent any mechanical breakage of replication tracts. Microfluidic platform for stretching the high-molecular weight DNA, coverslips, immunostaining and image acquisition of replication tracts were performed as described (Sidorova et al., 2009; Berti et al., 2013). In brief, polydimethylsiloxane (PDMS) stamps with microchannels were Oxygen plasma treated and reversibly sealed to the silanized coverslips. High-molecular weight DNA was loaded and stretched by capillary force into the microchannels. PDMS stamps were peeled-off and coverslips were left drying overnight. For immunostaining, DNA-stretched coverslips were denatured (2.5N HCL for 45 min), neutralized (0.1 M sodium borate and 3 washes with PBS), blocked (5% BSA and 0.5% Tween 20 in PBS for 30 min), incubated with primary antibodies (Anti-IdU/BrdU or both anti-IdU/BrdU and anti-CldU/BrdU for 30 min), washed (1% BSA and 0.1% Tween 20 in PBS, 3 times 5 min each) and incubated with secondary antibodies (anti–mouse Alexa Fluor 488-conjugated, or both anti–mouse Alexa Fluor 488–conjugated and anti–rat Alexa Fluor 594–conjugated for 1 h). Washed slides were mounted in prolong gold anti-fade reagent (Life Technologies) and images were sequentially acquired (for double-label) with LAS AF software using TCS SP5 confocal microscope (Leica). A 63×/1.4 oil immersion objective was used. Images were captured at room temperature. n ≥ 300 fiber tracts scored for each dataset. The DNA tract lengths were measured using ImageJ and the pixel length values were converted into micrometers using the scale bars created by the microscope. Statistical analysis was done using GraphPad Prism.
Clonogenic survival assay
Colony-forming assays were performed as previously described (Franken et al., 2006). In brief, 1,000 cells were plated per well and treated on the next day with 4 mM HU for 3, 6, and 8 h or 100 nM CPT for 6 h. Colonies were fixed, stained, and quantified 10 d after release from genotoxic stress. The plating efficiency and survival fraction were calculated as previously described (Franken et al., 2006). In brief, colonies were counted using an inverted stereomicroscope and the plating efficiency was calculated using the following formula: Plating Efficiency (PE) = (no. of colonies formed/no. of cells seeded) × 100%. From the plating efficiency, the surviving fraction (SF) was calculated as: SF = (no. of colonies formed after treatment/no. of cells seeded) × PE. The experiments were performed in triplicate and the statistical analysis was performed using GraphPad Prism.
Western blotting
Cells were washed with PBS and lysed either in standard RIPA buffer (PBS, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 10 µg/ml aprotinin, 10 µg/ml PMSF, 1 mM Na3VO4, and 1 mM NaF) or MCL buffer (50 mM Tris, pH 8.0, 5 mM EDTA, 0.5% NP-40, 100 mM NaCl, 2 mM DTT, and freshly added protease and phosphatase inhibitors from Roche (1 tablet/10 ml of buffer). Cell lysates were resolved by SDS-PAGE and transferred to PVDF membrane (GE Healthcare). Incubation with antibodies was performed overnight at 4°C. Proteins were visualized using ECL (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Co-immunoprecipitation experiments
HEK293T cells were transfected with empty vectors, FLAG-DNA2, and Strep-HA-WRN by calcium phosphate. 48 h after transfection, cells were treated with 4 mM HU for 3 h, lysed in benzonase lysis buffer (50 mM Tris-HCl, pH 7.5, 75 mM KCl, 2 mM MgCl2, 20 mM NaF, 10 mM β-glycerophosphate, 0.2 mM Na3VO4, and 0.2% Triton X-100) supplemented with protease inhibitors (EDTA-free tablet; Sigma-Aldrich) by passing 10 times through a 26-G syringe needle and incubated 1 h at 4°C with 2 U/µl Benzonase (Sigma-Aldrich) to digest genomic DNA. KCl and EDTA concentrations were adjusted to 120 and 3 mM, respectively, and lysates were centrifuged at 14,000 rpm for 30 min. Immunoprecipitations of clarified lysates were performed with FLAG M2 or HA affinity agarose resin (Sigma-Aldrich) overnight at 4°C. Beads were washed 5 times with wash buffer (50 mM Tris-HCl, pH 7.5, 150 mM KCl, 3 mM EDTA, 2 mM MgCl2, 20 mM NaF, 10 mM β-glycerophosphate, 0.2 mM Na3VO4, and 0.2% Triton X-100) and bound proteins were eluted by boiling in SDS-PAGE sample buffer.
EM analysis of genomic DNA in mammalian cells
EM analysis of replication intermediates has been described in detail (Ray Chaudhuri et al., 2012; Neelsen et al., 2014), including a description of the important parameters to consider specifically for the identification and the scoring of reversed forks (Neelsen et al., 2014). In brief, 5–10 × 106 U-2 OS cells were harvested and genomic DNA was cross-linked by two rounds of incubation in 10 µg/ml 4,5′,8-trimethylpsoralen (Sigma-Aldrich) and 3 min of irradiation with 366 nm UV light on a precooled metal block. Cells were lysed and genomic DNA was isolated from the nuclei by proteinase K (Roche) digestion and phenol-chloroform extraction. DNA was purified by isopropanol precipitation, digested with PvuII HF in the proper buffer for 3–5 h at 37°C, and replication intermediates were enriched on a benzoylated naphthoylated DEAE–cellulose (Sigma-Aldrich) column. EM samples were prepared by spreading the DNA on carbon-coated grids in the presence of benzyl-dimethyl-alkylammonium chloride and visualized by platinum rotary shadowing. Images were acquired on a transmission electron microscope (JOEL 1200 EX) with side-mounted camera (AMTXR41 supported by AMT software v601) and analyzed with ImageJ (National Institutes of Health).
Preparation of oligonucleotide-based DNA substrates
DNA oligonucleotides were purchased from Microsynth and 32P-labeled either at the 5′ terminus with [γ-32P] ATP and T4 polynucleotide kinase (New England Biolabs), or at the 3′ end with [α-32P] cordycepin-5′-triphosphate and terminal transferase (New England Biolabs) according to manufacturer’s instructions. Unincorporated nucleotides were removed using MicroSpin G25 columns (GE Healthcare). The substrates were prepared by heating the respective oligonucleotides at 95°C and gradually cooling to room temperature. The following oligonucleotides were used for the preparation of the four-way junction (X12-3 TOP L, HJ 1, HJ 2, and HJ 3), three-way junction with 3′ tail (X12-3 TOP L, HJ 1, HJ 2Sb, and HJ 3), three-way junction with 5′ ssDNA tail (X12-3 TOP L, HJ 1S, HJ 2, and HJ 3), nicked four-way junction (X12-3 TOP L, HJ 1, HJ 2Sa, HJ 2Sb, and HJ 3), replication fork (X12-3 TOP L, HJ 1S, HJ 2Sb, and HJ 3), and dsDNA (X12-3 TOP L and Bottom LC), respectively. The sequences of the oligonucleotides were: X12-3 TOP L (93 nt), 5′-GACGTCATAGACGATTACATTGCTAGGACATGCTGTCTAGAGACTATCGCGACTTACGTTCCATCGCTAGGTTATTTTTTTTTTTTTTTTTTT-3′ X12-3 HJ 1 (93 nt), 5′-AAAAAAAAAAAAAAAAAAATAACCTAGCGATGGAACGTAAGTCGCGATGGGCTTAACTAGGATGCTACTGGCCCCGAATCAACCGTACTTGGG-3′ X12-3 HJ 1S (48 nt), 5′-AAAAAAAAAAAAAAAAAAATAACCTAGCGATGGAACGTAAGTCGCGAT-3′ X12-3 HJ 2 (93 nt), 5′-CCCAAGTACGGTTGATTCGGGGCCAGTAGCATCCTAGTTAAGCCCATTACGATTCGTTACCCATTCACTGTCAGAAGGCACCAGATAGATCTC-3′ X12-3 HJ 2Sa (45 nt), 5′-CCCAAGTACGGTTGATTCGGGGCCAGTAGCATCCTAGTTAAGCCC-3′ X12-3 HJ 2Sb (48 nt), 5′-ATTACGATTCGTTACCCATTCACTGTCAGAAGGCACCAGATAGATCTC-3′ X12-3 HJ 3 (93 nt), 5′-GAGATCTATCTGGTGCCTTCTGACAGTGAATGGGTAACGAATCGTAATAGTCTCTAGACAGCATGTCCTAGCAATGTAATCGTCTATGACGTC-3′ X12-3 BOTTOM LC, 5′-AAAAAAAAAAAAAAAAAAATAACCTAGCGATGGAACGTAAGTCGCGATAGTCTCTAGACAGCATGTCCTAGCAATGTAATCGTCTATGACGTC-3′.
Nuclease assays
The experiments were performed in a 15-µl volume in 25 mM Tris-acetate (pH 7.5), 2 mM magnesium acetate, 1 mM ATP, 1 mM dithiothreitol, 0.1 mg/ml BSA (New England Biolabs), 1 mM phosphoenolpyruvate, 80 U/ml pyruvate kinase, 1 nM DNA substrate (molecules), and recombinant proteins, as indicated. The reactions were assembled on ice and incubated for 30 min at 30°C for yeast proteins and at 37°C for human proteins. Unless indicated otherwise, RPA was present in the reactions at saturating concentrations corresponding to a threefold excess over DNA, assuming all DNA was single-stranded and a DNA-binding site size of 25 nt for hRPA and of 20 nt for yRPA. The reactions were terminated by adding 5 µl Stop buffer (150 mM EDTA, 2% SDS, 30% glycerol, and 0.01% bromophenol blue), incubated for 30 min at room temperature and separated on polyacrylamide gels in TBE buffer under native conditions. Alternatively, for denaturing conditions, the reaction were terminated by adding 15 µl Formamide stop buffer (95% (vol/vol) formamide, 20 mM EDTA, 0.01% bromophenol blue), denatured by heating at 95°C for 5 min and separated on 20% denaturing polyacrylamide gels in TBE buffer. Gels were fixed, dried, exposed to a storage phosphor screen, and analyzed on Typhoon phosphor imager (GE Healthcare).
Online supplemental material
Fig. S1 shows quantification of stalled forks, new origins, and termination events in DNA2-depleted cells upon genotoxic stress induction, as well as the statistical analysis of IdU tracts from RECQ1-, DNA2-, WRN-, RECQ1/DNA2-, RECQ1/WRN-, WRN/DNA2-, and RECQ1/WRN/DNA2-depleted U-2 OS cells. Fig. S2 shows the IdU tract length distribution in BLM- and RECQ4-depleted cells, respectively, as well as statistical analysis of IdU tracts from RECQ1/WRN-codepleted cells complemented with WT WRN or with ATPase-deficient WRN. Fig. S3 shows additional EM analysis, as well as the Western blot analysis of ATR-checkpoint activation in RECQ1- and/or DNA2-depleted U-2 OS cells. Fig. S4 shows additional biochemical analysis of substrate specificity of human DNA2 and human EXO1. TFig. S5 shows biochemical assays of substrate specificity of yeast Dna2 and yeast Exo1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201406100/DC1.
Supplementary Material
Acknowledgments
We are grateful to Pietro Pichierri (Istituto Superiore di Sanità, Rome) for providing the WS cells and WRN constructs, Damian Dalcher for his help with the EM analysis, Stephanie Felscher (University of Zurich) for kindly providing human EXO1 protein, Marc Wold (University of Iowa) for human RPA expression construct, Lepakshi Ranjha (University of Zurich) for Mer3 protein, Fumiko Esashi (University of Oxford) for the Rad51 siRNAs and the U-2 OS cells stably expressing exogenous RAD51, and Judith Campbell (California Institute of Technology) for human and yeast DNA2/Dna2 expression constructs. We thank the Research Microscopy Core Facility of Saint Louis University for technical support.
This work was supported by National Institutes of Health grant R01GM108648 to A. Vindigni, by startup funding from the Doisy Department of Biochemistry and Molecular Biology and from the Saint Louis University Cancer Center to A. Vindigni, by grants from the President’s Research Fund of Saint Louis University and by the GLIOMA-Interreg (Slovenian-Italian Cooperation 2007-2013) project to A. Vindigni, by the Swiss National Science Foundation grants 31003A_146924 to M. Levikova and PP00P3 133636 to P. Cejka, by National Institutes of Health grant GM0088351 and CA15446 to E.A. Hendrickson, and by a research contract from Horizon Discovery, Ltd to E.A. Hendrickson. M. Berti was supported by an EMBO short-term fellowship to perform EM experiments in M. Levikova laboratory.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CPT
- camptothecin
- DSB
- double-strand DNA break
- EXO1
- human exonuclease I
- HDR
- Homology directed repair
- HR
- homologous recombination
- HU
- hydroxyurea
- MMC
- mitomycin C
- MRN
- MRE11-RAD50-NBS1
- TOP1
- DNA topoisomerase I
References
- Atkinson J., and McGlynn P.. 2009. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 37:3475–3492 10.1093/nar/gkp244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyagari R., Gomes X.V., Gordenin D.A., and Burgers P.M.. 2003. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 AND DNA2. J. Biol. Chem. 278:1618–1625 10.1074/jbc.M209801200 [DOI] [PubMed] [Google Scholar]
- Bae S.H., Bae K.H., Kim J.A., and Seo Y.S.. 2001. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature. 412:456–461 10.1038/35086609 [DOI] [PubMed] [Google Scholar]
- Berti M., Ray Chaudhuri A., Thangavel S., Gomathinayagam S., Kenig S., Vujanovic M., Odreman F., Glatter T., Graziano S., Mendoza-Maldonado R., et al. 2013. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat. Struct. Mol. Biol. 20:347–354 10.1038/nsmb.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bétous R., Mason A.C., Rambo R.P., Bansbach C.E., Badu-Nkansah A., Sirbu B.M., Eichman B.F., and Cortez D.. 2012. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 26:151–162 10.1101/gad.178459.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bétous R., Couch F.B., Mason A.C., Eichman B.F., Manosas M., and Cortez D.. 2013. Substrate-selective repair and restart of replication forks by DNA translocases. Cell Reports. 3:1958–1969 10.1016/j.celrep.2013.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd M.E., and Campbell J.L.. 1995. A yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc. Natl. Acad. Sci. USA. 92:7642–7646 10.1073/pnas.92.17.7642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd M.E., and Campbell J.L.. 1997. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol. Cell. Biol. 17:2136–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd M.E., Choe W., and Campbell J.L.. 2000. The nuclease activity of the yeast DNA2 protein, which is related to the RecB-like nucleases, is essential in vivo. J. Biol. Chem. 275:16518–16529 10.1074/jbc.M909511199 [DOI] [PubMed] [Google Scholar]
- Carr A.M., and Lambert S.. 2013. Replication stress-induced genome instability: the dark side of replication maintenance by homologous recombination. J. Mol. Biol. 425:4733–4744 10.1016/j.jmb.2013.04.023 [DOI] [PubMed] [Google Scholar]
- Cejka P., and Kowalczykowski S.C.. 2010. The full-length Saccharomyces cerevisiae Sgs1 protein is a vigorous DNA helicase that preferentially unwinds holliday junctions. J. Biol. Chem. 285:8290–8301 10.1074/jbc.M109.083196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P., Cannavo E., Polaczek P., Masuda-Sasa T., Pokharel S., Campbell J.L., and Kowalczykowski S.C.. 2010. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 467:112–116 10.1038/nature09355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotta-Ramusino C., Fachinetti D., Lucca C., Doksani Y., Lopes M., Sogo J., and Foiani M.. 2005. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol. Cell. 17:153–159 10.1016/j.molcel.2004.11.032 [DOI] [PubMed] [Google Scholar]
- Couch F.B., Bansbach C.E., Driscoll R., Luzwick J.W., Glick G.G., Bétous R., Carroll C.M., Jung S.Y., Qin J., Cimprich K.A., and Cortez D.. 2013. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 27:1610–1623 10.1101/gad.214080.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J., Donaldson J.R., Chow K.H., and Courcelle C.T.. 2003. DNA damage-induced replication fork regression and processing in Escherichia coli. Science. 299:1064–1067 10.1126/science.1081328 [DOI] [PubMed] [Google Scholar]
- Crabbe L., Verdun R.E., Haggblom C.I., and Karlseder J.. 2004. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 306:1951–1953 10.1126/science.1103619 [DOI] [PubMed] [Google Scholar]
- Duxin J.P., Moore H.R., Sidorova J., Karanja K., Honaker Y., Dao B., Piwnica-Worms H., Campbell J.L., Monnat R.J. Jr, and Stewart S.A.. 2012. Okazaki fragment processing-independent role for human Dna2 enzyme during DNA replication. J. Biol. Chem. 287:21980–21991 10.1074/jbc.M112.359018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchitto A., Pirzio L.M., Prosperi E., Sapora O., Bignami M., and Pichierri P.. 2008. Replication fork stalling in WRN-deficient cells is overcome by prompt activation of a MUS81-dependent pathway. J. Cell Biol. 183:241–252 10.1083/jcb.200803173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken N.A., Rodermond H.M., Stap J., Haveman J., and van Bree C.. 2006. Clonogenic assay of cells in vitro. Nat. Protoc. 1:2315–2319 10.1038/nprot.2006.339 [DOI] [PubMed] [Google Scholar]
- Gari K., Décaillet C., Delannoy M., Wu L., and Constantinou A.. 2008. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc. Natl. Acad. Sci. USA. 105:16107–16112 10.1073/pnas.0804777105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S., Chapman J.R., Magill C., and Jackson S.P.. 2008. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 22:2767–2772 10.1101/gad.503108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K., Budzowska M., Davies S.L., van Drunen E., Onizawa H., Beverloo H.B., Maas A., Essers J., Hickson I.D., and Kanaar R.. 2007. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 14:1096–1104 10.1038/nsmb1313 [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., Puddu F., and Costanzo V.. 2012. RAD51- and MRE11-dependent reassembly of uncoupled CMG helicase complex at collapsed replication forks. Nat. Struct. Mol. Biol. 19:17–24 10.1038/nsmb.2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henricksen L.A., Umbricht C.B., and Wold M.S.. 1994. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 269:11121–11132. [PubMed] [Google Scholar]
- Hu J., Sun L., Shen F., Chen Y., Hua Y., Liu Y., Zhang M., Hu Y., Wang Q., Xu W., et al. 2012. The intra-S phase checkpoint targets Dna2 to prevent stalled replication forks from reversing. Cell. 149:1221–1232 10.1016/j.cell.2012.04.030 [DOI] [PubMed] [Google Scholar]
- Kantake N., Sugiyama T., Kolodner R.D., and Kowalczykowski S.C.. 2003. The recombination-deficient mutant RPA (rfa1-t11) is displaced slowly from single-stranded DNA by Rad51 protein. J. Biol. Chem. 278:23410–23417 10.1074/jbc.M302995200 [DOI] [PubMed] [Google Scholar]
- Karanja K.K., Cox S.W., Duxin J.P., Stewart S.A., and Campbell J.L.. 2012. DNA2 and EXO1 in replication-coupled, homology-directed repair and in the interplay between HDR and the FA/BRCA network. Cell Cycle. 11:3983–3996 10.4161/cc.22215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanja K.K., Lee E.H., Hendrickson E.A., and Campbell J.L.. 2014. Preventing over-resection by DNA2 helicase/nuclease suppresses repair defects in Fanconi anemia cells. Cell Cycle. 13:1540–1550 10.4161/cc.28476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., and Burgers P.M.. 2013. Lagging strand maturation factor Dna2 is a component of the replication checkpoint initiation machinery. Genes Dev. 27:313–321 10.1101/gad.204750.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C., Nuang H., and Campbell J.L.. 1983. Isolation of yeast DNA replication mutants in permeabilized cells. Proc. Natl. Acad. Sci. USA. 80:6465–6469 10.1073/pnas.80.21.6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levikova M., Klaue D., Seidel R., and Cejka P.. 2013. Nuclease activity of Saccharomyces cerevisiae Dna2 inhibits its potent DNA helicase activity. Proc. Natl. Acad. Sci. USA. 110:E1992–E2001 10.1073/pnas.1300390110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S., Toczylowski T., and Yan H.. 2008. Identification of the Xenopus DNA2 protein as a major nuclease for the 5′->3′ strand-specific processing of DNA ends. Nucleic Acids Res. 36:6091–6100 10.1093/nar/gkn616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machwe A., Karale R., Xu X., Liu Y., and Orren D.K.. 2011. The Werner and Bloom syndrome proteins help resolve replication blockage by converting (regressed) holliday junctions to functional replication forks. Biochemistry. 50:6774–6788 10.1021/bi2001054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda-Sasa T., Imamura O., and Campbell J.L.. 2006. Biochemical analysis of human Dna2. Nucleic Acids Res. 34:1865–1875 10.1093/nar/gkl070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Maldonado R., Faoro V., Bajpai S., Berti M., Odreman F., Vindigni M., Ius T., Ghasemian A., Bonin S., Skrap M., et al. 2011. The human RECQ1 helicase is highly expressed in glioblastoma and plays an important role in tumor cell proliferation. Mol. Cancer. 10:83 10.1186/1476-4598-10-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E.P., and Symington L.S.. 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 455:770–774 10.1038/nature07312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfuni I., De Santis A., Federico M., Bignami M., Pichierri P., and Franchitto A.. 2012. Perturbed replication induced genome wide or at common fragile sites is differently managed in the absence of WRN. Carcinogenesis. 33:1655–1663 10.1093/carcin/bgs206 [DOI] [PubMed] [Google Scholar]
- Neelsen K.J., Chaudhuri A.R., Follonier C., Herrador R., and Lopes M.. 2014. Visualization and interpretation of eukaryotic DNA replication intermediates in vivo by electron microscopy. Methods Mol. Biol. 1094:177–208 10.1007/978-1-62703-706-8_15 [DOI] [PubMed] [Google Scholar]
- Nicolette M.L., Lee K., Guo Z., Rani M., Chow J.M., Lee S.E., and Paull T.T.. 2010. Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat. Struct. Mol. Biol. 17:1478–1485 10.1038/nsmb.1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar A.V., Genschel J., Kinoshita E., Polaczek P., Campbell J.L., Wyman C., Modrich P., and Kowalczykowski S.C.. 2011. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 25:350–362 10.1101/gad.2003811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H., Chung W.H., Zhu Z., Kwon Y., Zhao W., Chi P., Prakash R., Seong C., Liu D., Lu L., et al. 2010. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 467:108–111 10.1038/nature09318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G., Dai H., Zhang W., Hsieh H.J., Pan M.R., Park Y.Y., Tsai R.Y., Bedrosian I., Lee J.S., Ira G., and Lin S.Y.. 2012. Human nuclease/helicase DNA2 alleviates replication stress by promoting DNA end resection. Cancer Res. 72:2802–2813 10.1158/0008-5472.CAN-11-3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirzio L.M., Pichierri P., Bignami M., and Franchitto A.. 2008. Werner syndrome helicase activity is essential in maintaining fragile site stability. J. Cell Biol. 180:305–314 10.1083/jcb.200705126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A., Hashimoto Y., Herrador R., Neelsen K.J., Fachinetti D., Bermejo R., Cocito A., Costanzo V., and Lopes M.. 2012. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat. Struct. Mol. Biol. 19:417–423 10.1038/nsmb.2258 [DOI] [PubMed] [Google Scholar]
- Schlacher K., Christ N., Siaud N., Egashira A., Wu H., and Jasin M.. 2011. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 145:529–542 10.1016/j.cell.2011.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K., Wu H., and Jasin M.. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 22:106–116 10.1016/j.ccr.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigneur M., Bidnenko V., Ehrlich S.D., and Michel B.. 1998. RuvAB acts at arrested replication forks. Cell. 95:419–430 10.1016/S0092-8674(00)81772-9 [DOI] [PubMed] [Google Scholar]
- Sidorova J.M., Li N., Folch A., and Monnat R.J. Jr. 2008. The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle. 7:796–807 10.4161/cc.7.6.5566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova J.M., Li N., Schwartz D.C., Folch A., and Monnat R.J. Jr. 2009. Microfluidic-assisted analysis of replicating DNA molecules. Nat. Protoc. 4:849–861 10.1038/nprot.2009.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturzenegger A., Burdova K., Kanagaraj R., Levikova M., Pinto C., Cejka P., and Janscak P.. 2014. DNA2 cooperates with the WRN and BLM RecQ helicases to mediate long-range DNA end resection in human cells. J. Biol. Chem. 289:27314–27326 10.1074/jbc.M114.578823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangavel S., Mendoza-Maldonado R., Tissino E., Sidorova J.M., Yin J., Wang W., Monnat R.J. Jr, Falaschi A., and Vindigni A.. 2010. Human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol. Cell. Biol. 30:1382–1396 10.1128/MCB.01290-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrousek K.E., Fortini B.K., Polaczek P., Chen L., Liu Q., Dunphy W.G., and Campbell J.L.. 2010. Xenopus DNA2 is a helicase/nuclease that is found in complexes with replication proteins And-1/Ctf4 and Mcm10 and DSB response proteins Nbs1 and ATM. Cell Cycle. 9:1156–1166 10.4161/cc.9.6.11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., and Hickson I.D.. 2003. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 426:870–874 10.1038/nature02253 [DOI] [PubMed] [Google Scholar]
- Yata K., Lloyd J., Maslen S., Bleuyard J.Y., Skehel M., Smerdon S.J., and Esashi F.. 2012. Plk1 and CK2 act in concert to regulate Rad51 during DNA double strand break repair. Mol. Cell. 45:371–383 10.1016/j.molcel.2011.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo J.E., Lee E.H., Hendrickson E.A., and Sobeck A.. 2014. CtIP mediates replication fork recovery in a FANCD2-regulated manner. Hum. Mol. Genet. 23:3695–3705 10.1093/hmg/ddu078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Kwon Y.T., Varshavsky A., and Wang W.. 2004. RECQL4, mutated in the Rothmund-Thomson and RAPADILINO syndromes, interacts with ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Hum. Mol. Genet. 13:2421–2430 10.1093/hmg/ddh269 [DOI] [PubMed] [Google Scholar]
- Ying S., Hamdy F.C., and Helleday T.. 2012. Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res. 72:2814–2821 10.1158/0008-5472.CAN-11-3417 [DOI] [PubMed] [Google Scholar]
- Zellweger R., Dalcher D., Mutreja K., Herrador R., Berti M., Vindigni A., and Lopes M.. 2015. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol. 208:563–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman M.K., and Cimprich K.A.. 2014. Causes and consequences of replication stress. Nat. Cell Biol. 16:2–9 10.1038/ncb2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Chung W.H., Shim E.Y., Lee S.E., and Ira G.. 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 134:981–994 10.1016/j.cell.2008.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.