Phosphorylation and methylation of desmoplakin are required for proper junction assembly and adhesion strengthening, and inhibition of these modifications might contribute to skin and heart diseases.
Abstract
Intermediate filament (IF) attachment to intercellular junctions is required for skin and heart integrity, but how the strength and dynamics of this attachment are modulated during normal and pathological remodeling is poorly understood. We show that glycogen synthase kinase 3 (GSK3) and protein arginine methyltransferase 1 (PRMT-1) cooperate to orchestrate a series of posttranslational modifications on the IF-anchoring protein desmoplakin (DP) that play an essential role in coordinating cytoskeletal dynamics and cellular adhesion. Front-end electron transfer dissociation mass spectrometry analyses of DP revealed six novel serine phosphorylation sites dependent on GSK3 signaling and four novel arginine methylation sites including R2834, the mutation of which has been associated with arrhythmogenic cardiomyopathy (AC). Inhibition of GSK3 or PRMT-1 or overexpression of the AC-associated mutant R2834H enhanced DP–IF associations and delayed junction assembly. R2834H blocked the GSK3 phosphorylation cascade and reduced DP–GSK3 interactions in cultured keratinocytes and in the hearts of transgenic R2834H DP mice. Interference with this regulatory machinery may contribute to skin and heart diseases.
Introduction
Intercellular adhesive junctions structurally link neighboring cells to coordinate the establishment of cell polarity, cell migration, and the morphogenesis of developing embryos and tissues (Fuchs and Raghavan, 2002; Thomason et al., 2010). Essential for these functions is the ability of cell junctions to regulate the dynamics of the cortical cytoskeleton, a process that is tightly controlled by the spatiotemporal integration of mechanical and chemical signaling cues coming from adjacent cells or the environment (Jamora and Fuchs 2002; Simpson et al., 2011; Brieher and Yap, 2013).
Desmosomes are cell–cell adhesive junctions that confer structural integrity to tissues that undergo mechanical stress such as the skin and the heart (Kimura et al., 2007; Brooke et al., 2012). They perform this function by anchoring the keratin and desmin intermediate filament (IF) cytoskeleton to the plasma membrane–associated desmosomal plaque through an essential member of the plakin family of cytolinkers called desmoplakin (DP; Ruhrberg and Watt, 1997; Sonnenberg and Liem 2007; Kowalczyk and Green, 2013). DP is the sole essential desmosomal plakin (Gallicano et al., 1998). Its obligate nature is underscored by the early embryonic lethality of DP null mice and defects in embryonic heart, neuroepithelium, skin, and microvasculature in tetraploid rescued embryos (Gallicano et al., 2001). Genetic mutations in DP result in human disease ranging from lethal skin blistering disease to arrhythmogenic cardiomyopathy (AC), a cardiac disorder leading to sudden death (Jonkman et al., 2005; Lai-Cheong et al., 2007; Asimaki and Saffitz, 2014). Whether desmosomal disease is a result of the loss of mechanical functions or a product of altered signaling is still unknown (Garcia-Gras et al., 2006; Mahoney et al., 2010).
DP is composed of an N-terminal spectrin-repeat domain that links DP to desmosomal cadherins through associated armadillo proteins (Kowalczyk et al., 1997; Hatzfeld, 2007; Choi and Weis, 2011), a central coiled-coil domain (O’Keefe et al., 1989), and a C-terminal IF-binding domain with three plakin repeat domains (Kouklis et al., 1994; Bornslaeger et al., 1996; Choi et al., 2002). Loss of the C-terminal plakin repeat domains leads to IF detachment, compromising epithelial integrity leading to human cardiocutaneous disease (Norgett et al., 2000; Agullo-Pascual et al., 2014).
Association of DP with the IF cytoskeleton is dynamic and tightly regulated. Previous results have suggested that the DP C-tail, a 68-residue glycine-serine-arginine repeat–containing region at the very C terminus of DP, is important for this regulation (Stappenbeck et al., 1994; Godsel et al., 2005). 47% of the residues in this region are putative phosphosites. The C-tail also contains consensus sites for arginine methylation, a posttranslational modification (PTM) that has recently emerged as a critical regulatory feature of cytoplasmic protein–protein interactions (Bedford and Clarke, 2009; Cha et al., 2011; Xu et al., 2013). Multisite PTMs provide a mechanism for the rapid, reversible control of protein function (Deribe et al., 2010). The possibility that interplay between multiple PTMs in DP is important for cytoskeletal organization during development, tissue remodeling, and disease has never been addressed.
In this paper, we demonstrate that processive phosphorylation cascades coordinate with arginine methylation in the DP C-tail to mediate the dynamics of DP interactions with the IF cytoskeleton. We show further that DP PTMs are required for recruiting the enzymes that catalyze these modifications to the DP C-tail scaffold. Interfering with the DP PTM signaling machinery dramatically impairs junction assembly and adhesion strengthening and is a target for genetic mutations causing cardiocutaneous disease.
Results
Glycogen synthase kinase 3 (GSK3) signaling modulates DP–IF complexes
The last 68 residues of DP in the C-tail contain 29 Ser/Thr sites, of which residue S2849 and several other sites have been identified through unbiased screens for phosphoproteins (Beausoleil et al., 2004). To date, only S2849 has been functionally linked to regulation of DP–IF association, as mutation of this residue enhances DP’s association with IF and delays DP incorporation into forming junctions (Godsel et al., 2005). Among the putative phosphorylation sites are 15 consensus sites for GSK3 (Fig. 1 A). A diverse signaling mediator in Wnt signaling and wound-healing pathways, GSK3 is the primary kinase responsible for promoting processive phosphorylation cascades (Sun et al., 2009; Wu et al., 2011). To investigate if GSK3 regulates DP, we analyzed DP by immunofluorescence microscopy in the absence of GSK3 activity in SCC9 cell lines. We first applied a pharmacological approach to inhibit GSK3 where cells were treated with either LiCl or NaCl as a control (Cross et al., 1995). GSK3 inhibition led to a striking redistribution of DP into a filamentous pattern in the cytoplasm at the expense of DP at cell–cell interfaces, compared with control cells where DP remained at cell–cell borders (Fig. 1 B). To test if LiCl affects the closely related adherens junction, SCC9 cells were treated with 40 mM LiCl or NaCl and stained for the adherens junction components including p120-catenin, E-cadherin, and β-catenin. Junctional localization of these proteins was not detectably altered during GSK3 inhibition (Fig. S1, A and A′). Phospho-GSK3 and total GSK3 antibodies confirmed GSK3 inhibition in the presence of LiCl, indicated as an increase in the inactive phospho-form of GSK3. Total endogenous levels of both DP isoforms, DPI (332 kD) and DPII (260 kD), remain unchanged in treated cells (Fig. S1, B and B′).
Figure 1.
GSK3 signaling promotes desmosome assembly. (A) Schematic diagram of GSK3 consensus sites (bold) in DP. (B) NaCl- and LiCl-treated SCC9 cells stained with DP (NW6), assessed by immunofluorescence. Bars, 10 µm. (C) NaCl- and LiCl-treated SCC9 cells stained with DP (NW6) and keratin (K8), assessed by confocal immunofluorescence. (C′) Pearson’s correlation coefficients were calculated for DP and keratin. *, P < 0.001. Error bars indicate SEM. (D) LiCl-, Kenpaullone-, and siCtr- or siGSK3-treated SCC9s, assessed by immunofluorescence. (E and E′) NaCl- or LiCl-treated SCC9s transfected with control or caGSK3, assessed by immunofluorescence. DP fluorescence intensity was measured, normalized to background, and plotted. (F) caGSK3 was transfected into SCC9s and assessed by immunoblotting. (G) LiCl or NaCl SCC9s, fixed at 0, 1.0, and 2.5 h after being switched into high calcium media, were assessed by immunofluorescence. (G′) DP fluorescence intensity along cell borders was quantified (>200 cell borders) from three independent experiments. *, P < 0.001. Error bars indicate SEM. (H) Monolayers of SCC9 cells stably expressing WT DP-GFP (Godsel et al., 2005) were subjected to scratch wounding, treated with either 40 mM NaCl or LiCl, and imaged. Shown are stills of Videos 1 and 2 at 0 and 80 min. White arrows mark forming borders. (H′) Stills of LiCl treatment in WT DP-GFP cells from Video 3. Green circles denote DP particles appearing and being retained in the cytoplasm in a filamentous alignment. Time points indicate the minutes lapsed from initiation of cell–cell contact. Bars, 10 µm. (H″) Recruitment of DP-GFP to SCC9 cell–cell borders occurs in three temporally overlapping phases (Godsel et al., 2005) in control cells, whereas LiCl treatment decreases DP border intensity. Fluorescent intensities over time were calculated for representative borders from Videos 1 and 2 and correspond to borders shown in H. Results are representative of data obtained from >30 videos for each condition.
As DP is an IF-associated protein, to test whether the filamentous pattern represents DP alignment with IFs, we treated cells with two GSK3 inhibitors, LiCl and Kenpaullone, and costained with DP and keratin antibodies. Colocalization analyses indicated that GSK3 inhibition enhanced localization of DP along IF (Fig. 1, C and D). Pearson’s correlation coefficients calculated for DP and keratin demonstrated an ∼2.5-fold increase in DP colocalization with the keratin network after LiCl treatment (Fig. 1 C′). RNAi silencing approaches were performed to specifically knockdown GSK-3 α/β isoforms in SCC9 cells (Fig. S1 C). GSK3-specific RNAi treatment also led to enhanced localization of DP along IF (Fig. 1 D). Further, introducing a constitutively active GSK3 (caGSK3; S9A mutant) into cells by transient transfection was sufficient to partially rescue border formation in the presence of GSK3 inhibitors compared with control transfected cells (Fig. 1, E, E′, and F). To rule out possible global effects of GSK3 inhibition on the cytoskeleton, we addressed whether GSK3 plays a role in regulating actin or microtubule (MT) organization. To examine cortical actin, 40 mM LiCl-treated SCC9 cells were stained with phalloidin as a marker of F-actin. Quantification of the distances between actin bundles in contacting cells indicated that there were no significant changes in cortical actin bundle organization during GSK3 inhibition (Fig. S2, A and A′). As DP also regulates MT distribution (Sumigray et al., 2011; Patel el al., 2014) in keratinocytes, we compared the relative disposition and extent of colocalization between DP and IF versus DP and MT in LiCl- and NaCl-treated cells (Fig. S2, B and C). GSK3 inhibition did not significantly alter the extent to which DP localizes with MT compared with control (Fig. S2, B and C). Together, these data indicate that GSK3 plays a specific role in regulating DP’s association with the IF cytoskeleton.
Yeast two- and three-hybrid studies demonstrated that DP S2849G enhances DP–IF interactions and semiquantitative assays indicated by as much as ∼10-fold (Meng et al., 1997; Fontao et al., 2003). This led us to examine DP–IF in the absence of desmosomes. SCC9 cells maintained in low calcium media to disrupt cell junctions were treated with LiCl and stained for DP and keratin. Calculation of the Pearson’s correlation coefficient indicated that DP–IF colocalization in the cytoplasm increased approximately twofold in response to GSK3 inhibition, raising the possibility that GSK3 is required to promote the normal assembly of DP into desmosomes (Fig. S2, D and D′). To test this, we performed calcium switch assays in combination with GSK3 inhibition. For this experiment, desmosome assembly was initiated by switching cells from low calcium into normal calcium media, which resulted in accumulation of DP at new sites of cell–cell contact after 1 h in untreated cells; strong borders were formed after 2.5 h. GSK3 inhibition delayed DP redistribution to new sites of cell–cell contact (Fig. 1, G and G′). The kinetics of adherens junction formation were not altered by GSK3 inhibition (Fig. S1, D and D′).
To further investigate DP trafficking, we performed live cell imaging of confluent SCC9 cells expressing wild-type (WT) DP-GFP reforming junctions at the leading edge of scratch wounds after addition of 40 mM LiCl. After initial cell–cell contact, DP in control NaCl-treated cells exhibited DP trafficking kinetics comparable to a previously published study: DP first appeared at cell–cell interfaces within ∼10 min, followed by a plateau in fluorescence intensity of ∼20 min, and then another burst of intensity beginning at ∼40 min (Godsel et al., 2005; Fig. 1, H and H″; and Video 1). GSK3 inhibition delayed DP accumulation at forming borders, whereas the filamentous appearance of DP in the cytoplasm increased (Fig. 1 H, H′, and H″; and Videos 2 and 3). Together, these data establish a role for GSK3 signaling in mediating DP–IF interactions to promote desmosome assembly.
GSK3 associates with DP and modulates DP–IF complexes
Next, we addressed whether GSK3 physically associates with DP. HA-tagged GSK3 was transfected into HEK 293 cells, and RIPA lysates were used for HA coimmunoprecipitation assays. Precipitated products probed with HA and DP revealed that HA-tagged GSK3 is present in a complex with endogenous DP (Fig. 2 A). Colocalization analyses were then performed on SCC9 cells stained with GSK3 and DP antibodies. As reported previously, GSK3 staining is diffuse and cytoplasmic (Cross et al., 1995; Etienne-Manneville and Hall, 2003). To more directly address DP and GSK3 associations in situ we performed proximity ligation assay (PLA) analysis (DuoLink; Olink BioScience). This technique (Weibrecht et al., 2010) provides visualization of protein–protein proximities based on the detection of coupling between primary antibodies. Oligo-conjugated secondary antibodies form fluorescently labeled DNA adducts through rolling circle amplification when protein targets are within 40 to 100 nm. The number of fluorescent dots generated with DP and GSK3 antibodies was ∼20-fold greater than negative control pairings of anti-GSK3/rabbit IgG or anti-DP (NW6)/mouse IgG within the cell population (Fig. 2, B and B′).
Figure 2.
GSK3 is recruited to DP to modulate DP–IF complexes. (A) Endogenous DP associates with HA-GSK3 in HA immunoprecipitations in HEK 293 cells, assessed by immunoblotting. (B) SCC9 cells were fixed, and DP and GSK3 associations were analyzed by PLA analysis using anti-DP (NW6) and anti-GSK3 antibodies. Direct interactions (red) and nuclei (DAPI, blue). Bars, 10 µm. (B′) PLA fluorescent spots are counted and divided by the total number of nuclei in an image. Data were obtained from antibody pairs for >500 cells per experiment for three independent experiments. *, P < 0.001. Error bars indicate SEM. (C and C′) Phospho-DP in LiCl- or NaCl-treated SCC9s, assessed by immunoblotting. Full-length endogenous DP has two known isoforms, DPI (310 kD) and DPII (250 kD) that are generated by alternative splicing. Both of these contain S2849 and are recognized by the phospho-DP antibody. Densitometry quantification of three independent experiments. Error bars indicate SEM. (D) DP phosphorylation in siCtr or siGSK3-treated cells, assessed by immunoblotting. (E and E′) Phospho-DP in control or caGSK3 transfected cells, and lysates were assessed by immunoblotting. Densitometry quantification of three independent experiments. *, P < 0.001. Error bars indicate SEM. (F) S-tag affinity pull-down of HA-GSK3 and DP S-tag (DP CT) HEK 293 cells, assessed by immunoblotting. (G) NaCl-, LiCl-, or Ly-294,002 (GSK3 activator)–treated DP S-tag (DP CT) expressing cell lysates, assessed by immunoblotting. DP S-tag (DP CT) is present as a doublet where the lower band (32 kD) contains phosphorylation of S2849, whereas the higher band (red arrow, 36 kD) contains both phosphorylation of S2849 and downstream GSK3-phosphorylated sites (see cartoon in Fig. 3 A). (G′) Densitometry quantification of G from three independent experiments. *, P < 0.001. Error bars indicate SEM. (H) NaCl- or LiCl-treated DP S-tag (DP CT) cells were transfected with control or caGSK3, assessed by immunoblotting. Long exposure and short exposure of DP CT blot shown to visualize upper phospho-states of DP (36 kD). (H′) Densitometry quantification of phospho-DP divided by total DP from three independent experiments. Error bars indicate SEM.
We next tested if GSK3 alters DP phosphorylation using a DP-specific phospho-antibody, which recognizes phosphorylation at residue S2849 (Bouameur et al., 2013). Although total DP levels remained constant, gene silencing with siRNA targeting GSK3 or GSK3 inhibitors was sufficient to decrease phosphorylation of endogenous DP (Fig. 2, C, C′, and D). Further, caGSK3 expression enhanced DP phosphorylation (Fig. 2, E and E′).
To more closely examine phosphorylation of the C-tail we generated an S-tagged C-terminal truncation (32 kD) DP construct (DP S-tag or DP C terminus [DP CT]; Fig. S3 A). Consistent with full-length DP, DP S-tag associated with GSK3 in the S-tag pull-down product (Fig. 2 F). Next, we performed pharmacological and genetic manipulation of GSK3 in DP S-tag–transfected cells. Using the DP-specific phospho-antibody, we found that DP S-tag exists as a doublet at 32 and 36 kD (Fig. 2, G and G′). GSK3 inhibition with LiCl ablated the upper DP phospho-band (36 kD) compared with control cells (Fig. 2, G and G′). In contrast, treatment with a GSK3 activator, Ly-294,002, enhanced phosphorylation of DP (Fig. 2, G and G′). These data support the idea that the lower band (32 kD) is likely a DP isoform phosphorylated at S2849, previously shown to be important for plakin–IF interactions (Godsel et al., 2005; Bouameur et al., 2013), whereas the upper band (36 kD) is phosphorylated at multiple sites in response to GSK3 activation. Finally, we tested if the expression of a caGSK3 was sufficient to rescue the upper phospho-band after cells have been treated with a GSK3 inhibitor. LiCl treatment ablated the upper band (36 kD) compared with NaCl treatment. After expression of caGSK3 in LiCl-treated cells, DP S-tag phosphorylation is restored (Fig. 2, H and H′, red arrow). Together, these data introduce a novel role for GSK3 in the signaling machinery that modulates DP–IF interactions.
GSK3 promotes phosphorylation cascades within the DP C-tail
The glycine-serine-arginine repeat region in the DP C-tail eluded previous phosphorylation analyses by mass spectrometry (Fig. 3 A). To address these challenges, we applied tandem mass spectrometry (MS/MS) using front-end electron transfer dissociation (FETD) to map DP phosphorylation at the putative GSK3 sites (Syka et al., 2004; Earley et al., 2013). DP S-tag was expressed in HEK 293 cells and wash conditions for S-tag affinity purification were optimized as monitored by Coomassie blue staining (Fig. S3 B). In addition to S2849 (Beausoleil et al., 2004), FETD analyses identified six novel phosphorylation residues within the DP C-tail (Table S1), which fall within two GSK3 cascades (Fig. 3 A). FETD analyses also revealed that multiple phosphorylation modifications exist simultaneously, consistent with the processive nature of GSK3 phosphorylation (Table S1). To complement the mass spectrometry, we performed phos-tag gel analyses in which SDS-PAGE gels are run in the presence of a molecule that binds to phosphorylated proteins, thereby causing a mobility shift. This approach demonstrated that the DP S-tag protein appears as multiple bands representing different phospho-states (Fig. 3 B). Treatment with LiCl ablated upper phospho-states, consistent with the importance of GSK3 in generating these multiple phosphorylated species. Collectively, these data support the existence of processive GSK3-mediated phosphorylation cascades in the DP C-tail.
Figure 3.
GSK3 promotes phosphorylation cascades of the DP CT tail. (A) GSK3 cascade 1 (solid arrows) and 2 (dotted arrows) in WT, S2849G (red), and R2834H (*) DP. (B) NaCl- or LiCl-treated WT DP S-tag (DP CT) cell lysates were assessed by phos-tag immunoblot analyses (red arrow, 36 kD). (C and C′) Phosphorylation of DP S-tag (DP CT) point mutations, S2845A and T2853A, assessed by immunoblotting. Red arrows mark GSK3 phosphorylation of DP. Densitometry quantification of three independent experiments. *, P < 0.001. Error bars indicate SEM. (D and D′) NaCl-, LiCl-, DMSO-, or Ly-294,002–treated WT, S2845A, or T2853A DP S-tag (DP CT) lysates, assessed by immunoblotting. Densitometry quantification of three independent experiments. *, P < 0.001. Error bars indicate SEM. (E and E′) DMSO-, KT 5720-, Gö 6976-, or LiCl-treated DP S-tag (DP CT) lysates, assessed by immunoblotting (red arrow, 36 kD). Densitometry quantification of three independent experiments. Error bars indicate SEM. (F) DP S-tag (DP CT) expressing HEK 293 cells were treated with kinase inhibitors targeting CKI, CKII, MNKI, atypical PKC, and DYRK kinases, assessed by immunoblotting. (G) Control, 40 mM LiCl-, and PMA-treated SCC9s, assessed by immunofluorescence. Bars, 10 µm. (H and H′) GSK3 interactions with S2849G and WT DP S-tag (DP CT) in HEK 293 cells were examined after S-tag pull-down analyses where RIPA lysates (input) and pull-down products were analyzed by immunoblotting and quantified by densitometry. *, P < 0.03. Error bars indicate SEM.
As processive GSK3 cascades proceed in a C- to N-terminal direction (Fig. 3 A), we examined whether or not mutation of S2845, the first upstream serine after S2849 (DP-specific antibody site), inhibits the GSK3 phospho-states of DP (represented by the 36-kD band). S2845A and WT DP S-tag were transfected into HEK 293 cells and RIPA lysates were compared by immunoblotting. While total protein expression levels for the constructs were comparable, S2845A completely ablated the presence of the GSK3 phospho-state (36 kD) compared with WT DP S-tag (Fig. 3, C and C′, red arrow). In contrast, mutation of T2853, which lies just C-terminal to the putative cascade, had no effect on DP phosphorylation. Next, cells expressing S2845A and T2853A constructs were treated with GSK3 inhibitors and activators. Whereas the phospho-status of T2853A DP S-tag resembled that of WT DP, the 36-kD form of S2845A DP S-tag was absent even in the presence of GSK3 activation (Fig. 3, D and D′).
To further examine the kinase specificity of the 36-kD phospho-band we inhibited PKA and PKC, as these kinases have been implicated in desmosome assembly and stability (Bass-Zubek et al., 2008; Thomason et al., 2010; Kröger et al., 2013). Neither PKA nor PKC inhibition altered phosphorylation of WT DP S-tag (Fig. 3, E and E′). Further, DP S-tag cells were also treated with inhibitors of kinases with putative consensus motifs in the C-tail including atypical PKC, MNK1, CKI, CK II, CAMK, and DYRK. Compared with control-treated cells, none of these inhibitors alone were sufficient to decrease DP phosphorylation (Fig. 3 F). As PKC has been implicated in desmosome assembly and stability (Bass-Zubek et al., 2008; Thomason et al., 2010; Kröger et al., 2013), we next examined if PKC activation could override the requirement for GSK3. For this experiment, we treated cells with the PKC activator PMA in the presence or absence of GSK3 inhibition. Although DP localization at borders appeared similar or enhanced in PMA-treated versus control cells, LiCl treatment led to dramatic DP alignment with IF even in the presence of PMA (Fig. 3 G). These data indicate that GSK3 plays a specific and independent role in mediating DP dynamics that cannot be rescued by PKC.
To evaluate the dependence of GSK3 cascades on WT or S2849G, DP S-tag constructs were transfected into HEK 293 cells and RIPA lysates were examined by immunoblotting. In contrast to the doublet seen with WT DP S-tag, only a single band (32 kD) was observed for S2849G DP S-tag. Further, FETD mass spectrometry analyses of purified S2849G DP S-tag spectra confirm that S2849G ablated phosphorylation of downstream sites in the cascade (Table S1, Fig. S3 D, and Fig. 3 A). To examine if S2849 facilitates GSK3 binding to DP, we performed coimmunoprecipitation assays with DP and GSK3. Compared with WT DP S-tag, S2849G DP S-tag exhibited significantly weakened DP associations with GSK3 (Fig. 3, H and H′). Together, these data are consistent with a processive mechanism for GSK3 phosphorylation of DP where S2845 lies within the cascade after the phosphorylation of S2849.
Human disease mutation R2834H alters regulatory roles of the C-tail
A point mutation in DP (R2834H) identified in an AC patient occurs within the GSK3 cascade region of the C-tail (Yang et al., 2006). How this mutation contributes to AC has not been investigated. As altered GSK3 cascades enhanced DP–IF interactions, reduced DP incorporation into desmosomes, and delayed DP precursor trafficking to new sites of cell–cell contacts, we examined the behavior of R2834H DP compared with WT DP by immunofluorescence microscopy in SCC9 keratinocytes stably expressing these proteins. Cells were treated with siRNA specifically targeting the DP 3′UTR to knockdown the expression of the endogenous protein. Compared with WT DP-GFP, R2834H DP-GFP particles accumulated within the cytoplasm and exhibited enhanced localization along keratin IF and decreased junctional staining (Fig. 4, A and A′). Defects in adherens junctions (E-cadherin) or the actin cytoskeleton were not observed when comparing WT and R2834H DP stable lines (Fig. S4, A and A′).
Figure 4.
AC point mutant (R2834H) in C-tail alters DP border localization. (A) Colocalization of WT and R2834H DP-GFP SCC9 stable cell lines with keratin, assessed by immunofluorescence. Bars, 10 µm. (A′) Pearson’s correlation coefficients were calculated for DP and keratin where R2834H exhibits increased colocalization ∼2.5-fold compared with WT. *, P < 0.001. Error bars indicate SEM. (B) R2834H DP weakens intercellular adhesion compared with WT DP, assessed by dispase assay. Confluent cell monolayers were lifted with dispase enzyme and subjected to mechanical stress. (B′) Quantification of the number of total particles in each well between each condition was compared (t test; *, P < 0.001 and P < 0.005, respectively) from three independent experiments in which each condition was tested in triplicate. Error bars indicate SEM. (C) FRAP for WT (32% recovery after 11 min) and R2834H (29% recovery after 11 min) DP-GFP in stable lines. The bleach point is indicated as time 0. Images were taken at regular intervals (0.5–10 s) to monitor recovery. Representative series of FRAP regions depicted here. Bars, 1 µm. (C′) FRAP quantification. The means of the relative fluorescence intensities are plotted as functions of time for WT and R2834H DP-GFP from 0–11 min (n = 5 FRAP regions imaged of each condition from three independent experiments). (D and D′) WT and R2834H DP cells, fixed at 0, 0.5, and 2.5 h after being incubated in high calcium media, assessed by immunofluorescence. Bars, 10 µm. *, P < 0.001. Error bars indicate SEM. (E) Monolayers of SCC9 cells stably expressing WT, R2834H, or S2849G DP-GFP were subjected to scratch wounding and imaged. Shown are still frames from Videos 4–6. White arrows mark the forming borders. Red arrow in the R2834H DP image denotes filamentous alignment of DP, further highlighted in Video 5. Bars, 10 µm. (E′) Quantification of fluorescence intensities of DP-GFP expressing cell borders imaged under the same conditions. Fluorescent intensities over time were calculated for representative borders from Videos 4–6 and correspond to borders shown in E. Results are representative of data obtained from >30 videos for each condition. (F and F′) Phosphorylation (red arrow) of R2834H and WT DP S-tag (DP CT), assessed by immunoblotting. Densitometry quantification of three independent experiments. *, P < 0.001. Error bars indicate SEM. (G) WT, S2849G, and R2834H DP S-tag (DP CT) were compared by analyzing band shifts (red arrow denotes 36 kD), assessed by phos-tag gel immunoblotting.
To test if R2834H DP impacts intercellular adhesion, we performed an assay that tests the mechanical strength of confluent cell sheets, the “dispase” assay. For this, cells are grown to confluency and lifted as cell sheets with the dispase enzyme, which releases cell–substrate adhesions without affecting cell–cell adhesions (Hudson et al., 2004; Hobbs and Green, 2012). Cell monolayers are then subjected to mechanical stress and the numbers of fragments produced are quantified as a surrogate indicator of adhesive strength. R2834H cells exhibited significantly greater fragmentation compared with WT DP cells, suggesting that R2834H alters the integrity of intercellular junctions (Fig. 4, B and B′). S2849G DP-GFP cells were used as a positive control as a previous study demonstrated that S2849G DP leads to weakened intercellular adhesion in cell monolayers grown for 2 d (Hobbs and Green, 2012; Fig. 4, B and B′). These studies indicate that the presence of R2834H DP alters intercellular adhesion and weakens integrity of cell monolayers in a comparable manner to the phospho-deficient DP mutant cells.
To determine whether decreased junctional R2834H DP is caused by the instability of mature desmosomes at cell–cell interfaces, we performed FRAP and junction assembly analyses. Comparable regions of DP staining along the border were bleached, and then recovery was monitored over time for both WT and R2834H lines. DP fluorescence recovery along borders was similar for WT and R2834H DP cells, where after 11 min on average 32% of WT and 29% of R2834H was restored (Fig. 4, C and C′). These data are consistent with the possibility that the observed junctional defects are not caused by instability at the junction but instead are a result of impaired recruitment of R2834H DP into desmosomes. To address this idea, we performed calcium switch analyses and live cell imaging. Compared with WT, R2834H DP-GFP cells exhibited delayed border formation and altered DP trafficking kinetics in a calcium switch assay (Fig. 4 D). Live cell imaging of wounded R2834H DP-GFP cell monolayers was performed in parallel with WT and S2849G cells. These experiments revealed that, like DP S2849G (Godsel et al., 2005), R2834H exhibits aberrant particle dynamics and delayed kinetics of fluorescence intensity accumulation at sites of cell–cell contact, as compared with WT DP-GFP (Fig. 4 E′ and Videos 4–6).
DP phosphorylation cascades ablated by R2834H
The phenotypes of R2834H DP behavior, including the enhanced localization of DP along IF, a delayed incorporation of DP into forming desmosomes, and weakened intercellular adhesive strength represent phenotypes reminiscent of kinase-inhibited or phospho-deficient S2849G DP cells (Godsel et al., 2005). To test whether R2834H alters DP phosphorylation, we examined phosphorylation of R2834H DP S-tag. Whereas WT DP and R2834H DP S-tag were expressed at comparable levels, R2834H DP S-tag displayed a reduction in GSK3-dependent phosphorylation (36 kD; Fig. 4, F and F′). Additionally, FETD mass spectrometry analysis revealed that purified R2834H DP S-tag ablated phosphorylation of six serine residues within the GSK3 cascades (Fig. 3 A, Fig. S3 C, and Table S1). Further, the presence of R2834H decreased phosphorylation as assessed by phos-tag analysis compared with WT DP S-tag (Fig. 4 G).
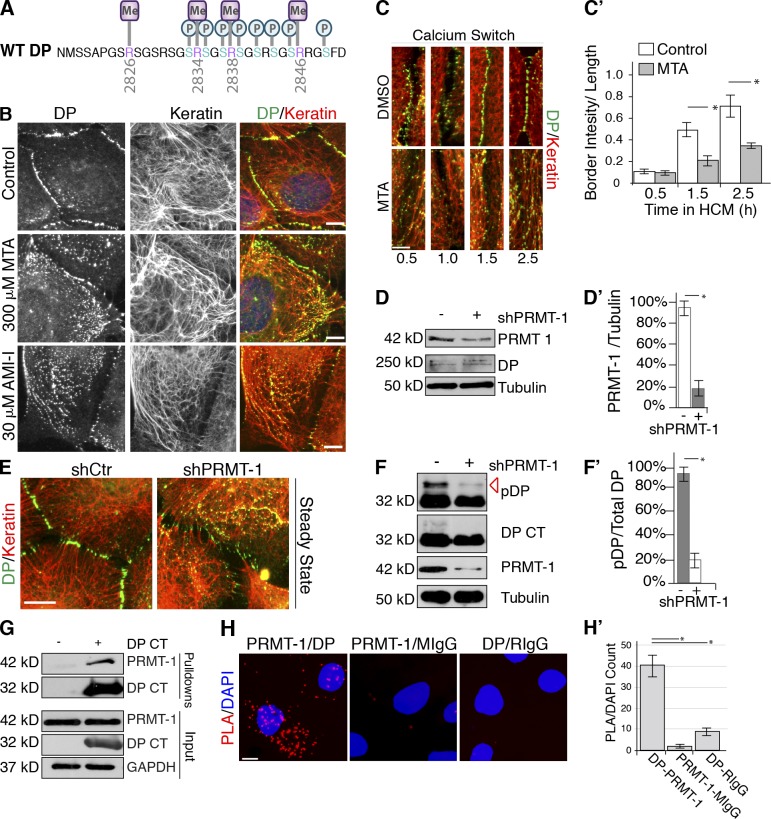
Arginine methylation occurs within the DP C-tail
The loss of DP phosphorylation in the presence of R2834H suggested a role for this arginine residue in tempering GSK3-dependent DP phosphorylation. R2834 and several other sites in the C-tail are consensus sites for arginine methylation, a PTM that has recently been implicated in a variety of cellular processes dependent on altering protein–protein interactions (Bedford and Clarke, 2009). This led us to investigate if the C-tail contains arginine methylation modifications. Using the purified WT DP S-tag protein, FETD mass spectrometry analyses revealed for the first time that R2834 is modified by asymmetric dimethylation, as are three other arginine residues within the DP C-tail (Fig. 5 A, Fig. S3 D, and Table S2). Further, peptides were identified that contain both methylated arginine residues and GSK3 phosphorylated residues, raising the possibility that these modifications coordinately regulate DP (Table S2 and Fig. 5 A).
Figure 5.
Arginine methylation contributes to the regulatory roles of the DP C-tail. (A) Cartoon of DP arginine methylation (purple) and phosphosites (blue) identified. (B) DMSO-, MTA-, or AMI-1–treated SCC9s, assessed by immunofluorescence. (C) DMSO- or MTA-treated SCC9s, fixed at 0, 0.5, and 2.5 h in high calcium media, were assessed by immunofluorescence. (C′) Mean fluorescence intensity of DP at cell borders was calculated. *, P < 0.001. Error bars SEM. (D and D′) shCtr and shPRMT-1–treated SCC9s. PRMT expression was assessed by immunoblotting. Densitometry quantification of three independent experiments. *, P < 0.001. Error bars indicate SEM. (E) shCtr and shPRMT-1–transfected SCC9s stained with DP (NW6) and keratin antibodies, assessed by immunofluorescence. (F and F′) Phosphorylation (red arrow) of WT DP S-tag (DP CT) expressing cells transfected with shCtr or shPRMT-1 was assessed by immunoblotting. Densitometry quantification of three independent experiments. *, P < 0.001. Error bars indicate SEM. (G) S-tag pull-down confirms that PRMT-1 associates with WT DP S-tag (DP CT), assessed by immunoblotting. (H) SCC9s incubated with primary antibody pairs DP (1G4)-PRMT-1, DP-RIgG, or PRMT-1-MIgG antibodies, assessed by PLA. Nuclei stained with DAPI (blue). Bars, 10 µm. (H′) Quantification of PLA signal divided by number of nuclei performed from three independent experiments. *, P < 0.03. Error bars indicate SEM.
Protein arginine methyltransferase 1 (PRMT-1) modulates DP–IF complexes
To investigate the importance of DP methylation, we treated cells with inhibitors of the enzymes that catalyze arginine methylation modifications, PRMTs (Cha et al., 2011; Xu et al., 2013). Global inhibition of arginine methylation in cells treated with methylthioadenosine (MTA) or arginine methyltransferase inhibitors I (AMI-1) enhanced DP association with IFs without inducing global changes in keratin organization (Fig. 5 B). Additionally, PRMT-inhibited cells exhibited delayed desmosome assembly in a manner reminiscent of kinase-inhibited cells (Fig. 5, C and C′).
PRMT enzymes catalyze the addition of mono- and dimethyl modifications to arginine residues. PRMTs fall into two classes, where Type I and Type II PRMTs catalyze asymmetric and symmetric dimethylation modifications, respectively. Of the eight PRMTs identified, PRMT-1 catalyzes the type I addition of asymmetric dimethylation arginine modifications to arginine/glycine-rich substrates (Zhang and Cheng, 2003). ETD analysis provided atomic-level resolution of PTMs within the DP C-tail where arginine dimethylation of R2834 occurs asymmetrically, with two methyl moieties on a single guanidinium nitrogen. This led us to investigate whether PRMT-1 contributes to arginine methylation of the DP C-tail. SCC9 cells were treated with shRNA targeting PRMT-1 (Fig. 5, D and D′). Compared with control cells, shPRMT-1–depleted cells exhibited enhanced alignment of DP along IF (Fig. 5 E). To investigate the role of PRMT-1 in mediating phosphorylation of DP, we performed shRNA depletion of PRMT-1 in HEK 293 cells expressing WT DP S-tag. Although total levels of DP remained constant, PRMT-1–depleted cells dramatically reduced the GSK3-band (36 kD) of WT DP S-tag (Fig. 5, F and F′). Further, PRMT-1 specifically associated with the DP CT in an S-tag pull-down assay from HEK 293 cells (Fig. 5 G). To examine endogenous DP and PRMT-1 associations in situ, we performed PLA analysis. Cells stained with PRMT-1 and DP (1G4) antibodies resulted in PLA fluorescent signals. The number of fluorescent dots generated with DP and PRMT-1 was ∼20-fold greater than the negative control pairings of anti-PRMT-1/mouse IgG or anti-DP/rabbit IgG within the cell population indicative of protein–protein associations (Fig. 5, H and H′). PRMT-1 antibodies with mouse IgG and 1G4 antibodies with rabbit IgG, used as negative controls, produced no visible signal. These data provide evidence that PRMT-1–mediated arginine methylation of DP coordinates with phosphorylation of the C-tail to modulate DP–IF interactions.
Loss of arginine methylation in R2834H alters GSK3 recruitment to DP
As inhibiting PRMT-1 interferes with GSK3-dependent DP phosphorylation, we next tested whether R2834H DP prevented binding of GSK3 to DP. Compared with WT, R2834H DP S-tag demonstrated reduced interactions with GSK3 by coimmunoprecipitation analyses, supporting the idea that arginine methylation facilitates GSK3 interactions with DP (Fig. 6, A and A′). In addition, PLA analyses of stable cell lines were performed after treatment with siRNA targeting the endogenous DP 3′UTR. Staining with GSK3 and GFP antibodies revealed a significant reduction in signal for both R2834H DP-GSK3 and S2849G DP-GSK3 compared with WT DP-GSK3 with R2834H DP exhibiting the most dramatic decrease in signal (Fig. 6, B and B′). Together, these data support a model whereby arginine methylation facilitates GSK3 interactions with DP to promote phosphorylation and where the presence of a human disease mutation at key arginine methylation site, R2834H, alters DP regulation.
Figure 6.
AC mutation alters DP phosphorylation by GSK3. (A) GSK3 interactions with R2834H and WT DP S-tag (DP CT) in HEK 293 cells were examined following S-tag pulldown analyses where RIPA lysates (input) and pull-down products were analyzed by immunoblotting. (A′) Densitometry quantification of GSK3 interactions with WT and R2834H DP S-tag (DP CT). *, P < 0.03. Error bars indicate SEM. (B) PLA of WT, R2834H, or S2849G DP-GFP cells incubated with primary antibody pairs of GFP-GSK3. Nuclei were stained with DAPI (blue). (B′) PLA fluorescence intensity divided by number of nuclei was performed for each cell line using primary antibody pairs of GFP-GSK3 and with controls GSK3-RIgG or GFP-MIgG. Data quantification was analyzed from three separate experiments (>500 nuclei per experiment). *, P < 0.05. Error bars indicate SEM. (C) Control or caGSK3-transfected and LiCl- or Ly-294,002 (GSK3 Act.)–treated DP-GFP cell lines, assessed by immunofluorescence. Bars, 10 µm. (D and D′) R2834H DP S-tag (DP CT) cells were treated with NaCl or LiCl or transfected with caGSK3, assessed by immunoblotting. Densitometry quantification of three independent experiments. *, P < 0.001. Error bars indicate SEM.
To examine if the manipulation of GSK3 activity can either enhance, or override, the observed increase in DP–IF interactions exhibited by DP R2834H or S2849G, we examined DP localization in stable cell lines treated with the GSK3 inhibitor LiCl, GSK3 activators, or caGSK3. LiCl enhanced the filamentous localization of DP in all cases and delayed border formation in a manner similar to endogenous DP (Fig. 6 C). E-Cadherin localization and the organization of actin bundles were not detectably affected after GSK3 inhibition (Fig. S4, B, B′, and B″). Increasing cellular levels of active GSK3 rescued R2834H DP-GFP localization at cell borders (Fig. 6 C). However, GSK3 activation failed to restore DP S2849G assembly into desmosomes, suggesting that phosphorylation of residue S2849 is required for the efficient initiation of GSK3-dependent processive phosphorylation and DP trafficking in cells.
We next examined whether GSK3 activation rescued R2834H DP-dependent defects by enhancing DP phosphorylation. For this, we examined phosphorylation of R2834H DP S-tag in cells transfected with caGSK3 by immunoblot analysis. As seen with WT, LiCl ablates the upper band (36 kD) compared with NaCl treatment (Fig. 6, D and D′). Further, caGSK3 was sufficient to rescue phosphorylation of R2834H DP to greater than control conditions (Fig. 6, D and D′, red arrows). These data suggest that increasing the stoichiometric ratio of active GSK3 enhances GSK3 phosphorylation of DP.
To examine the role of DP PTMs in vivo, we used a transgenic mouse model harboring full-length DP R2834H, which recapitulates features of the human disease AC. First, we investigated whether R2834H DP exhibits aberrant PTMs in vivo by examining phosphorylation of Flag-tagged DP in the mice. For this experiment, WT and R2834H DP transgenic hearts were lysed in urea sample buffer as performed previously and DP expression was assessed by immunoblot analysis (Yang et al., 2006). Total levels of DP expression were similar as shown by probing with anti-DP and anti-Flag antibodies. Phosphorylation of R2834H DP was dramatically decreased compared with WT DP using both the DP-specific phospho-antibody and a GSK3 substrate-specific phospho-antibody (R-X-X-pS; Fig. 7, A and A′). Tubulin was used as a loading control. These data support in vitro observations that R2834H DP decreases phosphorylation of DP compared with WT.
Figure 7.
GSK3 phosphorylation of DP altered in R2834H DP mice. (A) Transgenic mouse hearts were lysed in urea sample buffer and phosphorylation of WT and R2834H DP-Flag were analyzed by immunoblotting using DP phospho-specific and GSK3 substrate phospho-specific antibodies. Total levels of DP expression were assessed using anti-DP (NW6) and Flag. Tubulin was used as a loading control. (A′) Densitometry quantification indicates that although total levels of DP expression are similar, phosphorylation of R2834H DP was dramatically decreased compared with WT DP using both the DP-specific phospho-antibody and a GSK3 substrate-specific phospho-antibody. The data shown are from a single representative experiment out of three repeats (n = 3). Error bars represent SEM. *, P < 0.001. (B) R2834H DP-trangenic mice demonstrate altered cytoplasmic localization of DP and disruption of cell–cell contacts in cardiac tissues, assessed by confocal immunofluorescence. WT and R2834H DP cardiac sections were stained with anti-Flag and anti-desmin antibodies. (C) PLA analyses were performed with WT and R2834H DP cardiac sections incubated with primary antibody pairs DP (NW6)-GSK3, DP (NW6)-MIgG, or GSK3-RIgG. Colocalization allows for hybridization, ligation, and amplification of oligonucleotide adducts fused to secondary antibodies, ultimately producing a fluorescent spot (red) in situ. Blue DAPI staining marks nuclei. Bars, 10 µm. (C′) For the quantification of PLA, PLA spots were counted and divided by total number of nuclei in a frame. Quantification shows a statistically significant enhancement of signal for the DP-GSK3 antibody pairing in WT hearts compared with R2834H transgenic mouse hearts. *, P < 0.03; **, P < 0.001; from >500 cells counted from four independent experiments. Error bars indicate SEM.
Confocal microscopic cell biological analysis of cardiac tissues from WT DP and R2834H DP transgenic mice showed an enhanced localization of R2834H DP within the cytoplasm compared with WT DP hearts (Fig. 7 B). To investigate R2834H DP interactions with GSK3 in situ we applied PLA to the transgenic hearts. PLA analyses of frozen cardiac sections demonstrated a significantly lower PLA signal for the GSK3-R2834H DP pair compared with GSK3-WT DP pair, supporting the idea that the R2834H mutation impairs interactions with endogenous GSK3 in vivo (Fig. 7, C and C′). Together, these analyses demonstrate that DP acts as a scaffold for GSK3 in heart tissue and that this association is attenuated by the mutation of methylation site R2834 resulting in impaired DP dynamics and DP’s retention along desmin in the cardiac myocyte cytoplasm.
Discussion
In this paper, we have demonstrated a coordinated role for arginine methylation and processive phosphorylation in DP–IF cytoskeletal dynamics. The data support the idea that the proper distribution of PTMs depends on recruitment of GSK3 and PRMT-1 to the DP C-tail to regulate DP–IF complexes during the formation of intercellular junctions and establishment of cell–cell adhesion. This regulatory signaling machinery is disrupted in the presence of a point mutation in an arginine methylation site identified in AC, suggesting that this modification contributes to disease progression. We propose a model where preexisting arginine methylation aids GSK3 kinase recognition to initiate downstream PTMs that modulate DP–IF interactions during desmosome assembly.
GSK3 has recently emerged as a key regulator of cell polarity and cytoskeleton dynamics (Wu et al., 2011; Kumar et al., 2012). We now provide evidence that GSK3-mediated processive phosphorylation of the DP C-tail serves as a switch to regulate the dynamic association of DP and IF, thus facilitating DP’s trafficking and delivery to forming desmosomes. This mechanism may provide a way for ensuring that a sufficient pool of assembly-competent DP is available during tissue remodeling, for instance during wound healing, development, or pathogenic processes. Although we do not rule out that DP scaffolds GSK3 at cell–cell junctions for downstream GSK3 substrates, live cell imaging and calcium switch assays support the idea that a critical regulatory role of GSK3 is to modulate the dynamics of DP–IF interactions during desmosome assembly. That this function regulates the strength of DP’s association with keratin IFs is consistent with previous yeast two- and three-hybrid analyses demonstrating that inhibiting DP S2849 phosphorylation enhances DP–IF interactions up to 10-fold (Meng et al., 1997; Fontao et al., 2003). Once junctions are formed, the removal of phosphorylation by specific phosphatases at the cell membrane may facilitate the strengthening of DP–IF interactions in established junctions. Such an idea is consistent with our previous observation that when allowed to incorporate into junctions over a sufficiently extended time course, S2849G DP facilitates the formation of strong adhesions (Hobbs and Green, 2012). FRAP analysis of R2834H compared with WT DP is consistent with the idea that interference with processive phosphorylation through the alteration of DP methylation increases rather than decreases the stability of formed junctions.
Previous observations suggested that PKCα is recruited to DP by the armadillo protein PKP2, to facilitate DP’s incorporation into junctions (Bass-Zubek et al., 2008). Here we found that PKCα inhibition did not affect the GSK3 phosphorylation of DP. As IFs are functionally regulated by PKC, it is plausible that PKC regulates DP dynamics indirectly through the phosphorylation of IFs at DP–IF interfaces (Izawa and Inagaki, 2006; Pan et al. 2013; Snider and Omary, 2014). Inhibition of other kinases with putative consensus motifs within the C-tail did not alter DP phosphorylation, which is consistent with the idea that another kinase or combination of kinases phosphorylates S2849 (Bouameur et al., 2013).
Protein methylation has only recently begun to be appreciated as a regulator of cytoplasmic signal transduction in the initiation of signaling from cell surface receptors (BMP-induced Smad signaling) and Wnt signaling (by regulating Axin stability; Cha et al., 2011; Xu et al., 2013). We now present evidence that PRMT-1 plays a role in regulating intercellular junctions and IF cytoskeletal organization. Future studies on methylation in regulating intercellular junctions may reveal key aspects of the signaling machinery that promotes cellular adhesion. This could establish a novel paradigm for regulation of adhesion and cytoskeletal interactions and be more broadly applicable to plakin family members, such as ACF7/MACF (Leung et al., 1999; Karakesisoglou et al., 2000), which also harbor serine stretches phosphorylated by GSK3 (Wu et al., 2011).
Whether genetic mutations of DP promote the pathogenesis of AC through the loss of structural integrity of cardiac tissues or misregulated signaling is not well understood (Kiès et al., 2006; Asimaki and Saffitz, 2014). Analysis of individual disease mutations provides insights into the underlying molecular mechanism of AC pathogenesis. It was recently reported that N-terminal DP mutations interfere with DP binding to the MT-associated protein EB1, resulting in loss of MT cortical capture, impaired Cx43 trafficking, and gap junction dysfunction, which is known to occur in AC (Patel et al., 2014). In the current case, R2834H alters DP dynamics by enhancing DP associations with IF and interfering with DP’s assembly into desmosomes. This could exacerbate the response of the heart to mechanical stress on the tissue by obstructing desmosome-based adhesive strength. It is also plausible that loss of enzyme scaffolding functions of DP could contribute to disease. Here we demonstrate that DP acts as a scaffold for GSK3 in heart tissue and that this association is attenuated by a disease-causing mutation in methylation site R2834. In the absence of DP, a GSK3 substrate, it has been reported that plakoglobin is aberrantly transported into the nucleus, thus altering transcriptional pathways thought to be a contributing factor in AC pathogenesis (Garcia-Gras et al., 2006; Saffitz, 2006). Further, a recent study reports misregulated kinase signaling in AC and demonstrates that a kinase inhibitor of Wnt signaling, one of whose targets is GSK3, reverses AC phenotypes in zebrafish (Asimaki and Saffitz, 2014). Future studies of the roles of GSK3 and PRMT-1 signaling in AC could shed light on whether impairing DP scaffolding functions represents a global mechanism that is altered during the progression of AC.
In summary, we demonstrate that interplay between processive phosphorylation and arginine methylation dictates DP–IF cytoskeletal interactions during junction assembly to control cell–cell adhesion strengthening. These observations show that interference with GSK3 and PRMT-1 signal integration through mutations in a critical PTM site contributes to cardiocutaneous disease.
Materials and methods
Dispase assays
Stable cell monolayers were seeded in triplicate into 6-well plates. 24 h after reaching confluency, cultures were washed twice in DPBS and then incubated in 2 ml of dispase (2.4 U/ml; Roche) for 30 min (Hudson et al., 2004; Hobbs and Green, 2012). Released monolayers were subjected to rotational mechanical stress (e.g., 150 rpm for 5 min) to induce fragmentation. Fragments were counted using a dissecting microscope (MZ6; Leica). Statistical analysis was performed on the average of three separate experiments. Statistical significance (t test) was defined as P < 0.05.
PLA
Reagents used to conduct PLA were purchased from Sigma-Aldrich and used as described previously (Harmon et al., 2013). Cells were rinsed and fixed as described in Image acquisition and analysis for conventional fluorescence and live cell imaging. After incubation with primary antibody for 60 min at 37°C, samples were incubated with PLA secondary antibodies conjugated to DNA oligonucleotides for 60 min at 37°C. Samples were then subjected to a 30-min incubation at 37°C for ligation of nucleotides, followed by a 100-min incubation at 37°C for rolling circle polymerization, resulting in the production of red fluorescent dots if the antigens targeted by secondary antibodies were in close proximity (Fredriksson et al., 2002; Söderberg et al., 2006). DAPI is used to stain the nucleus. ImageJ software was used to quantify the number of PLA and DAPI signals per image field (endogenous DP–GSK3 interaction).
Generation of constructs
Full-length DP cDNA was used for the construction of DP S-tag into the mammalian pTriEx-Neo S-tag vector with a CMV promoter (EMD Millipore). Point mutations were generated using the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies). Cloning steps for full-length human DP were performed as described previously (Huen et al., 2002). The expression plasmid for HA-tagged CA-GSK3 was purchased from Addgene and was generated using a 1,300-bp insert of GSK3 with an S9A mutation that was cloned into the pcDNA3 vector backbone with a CMV promoter. For specific knockdown of endogenous DP, two oligonucleotides targeting the 3′ UTR of DP were purchased from IDT and were used as a mix (sense sequences: 5′-AAUUAAUACCAAACCAAA-3′ and 5′-GCAGUAGAGUGAUAGGA-3′). siRNA controls used a pool of siRNA with sequences: 5′-UAAGGCUAUGAAGAGAUAC-3′, 5′-AUGUAUUGGCCUGUAUUAG-3′, 5′-AUGAACGUGAAUUGCUCAA-3′, and 5′-UGGUUUACAUGUCGACUAA-3′. siRNA targeting GSK3 α/β isoforms were purchased from Cell Signaling Technology. Constructs of shRNA oligonucleotide targeting PRMT-1 were purchased from Sigma-Aldrich. SCC9 cells were transfected with siRNA oligonucleotides at a final concentration of 20 nM using DharmaFECT (GE Healthcare).
Antibodies and reagents
The following primary antibodies were used: NW6 Rabbit anti-DP was generated using anti-human DP CT fragment found in both human DPI and II (Angst et al., 1990), anti–phospho-S2849 DP antibody was generated using rabbits raised against a DP peptide phosphorylated at S2849 (Bouameur et al., 2013), mouse anti-S-Tag (EMD Millipore), M2 mouse anti-FLAG (Abcam), rabbit anti-FLAG (Abcam), DM1A mouse anti–α-tubulin (Abcam), HECD-1 mouse anti–E-cadherin (Abcam), rabbit anti-PRMT-1 (Abcam), mouse anti-desmin (Abcam), rabbit anti-GAPDH (Sigma-Aldrich), rabbit anti-HA (Sigma-Aldrich), KSB17.2 mouse anti-K8/K18 (Sigma-Aldrich), C2206 rabbit anti–β-catenin (Sigma-Aldrich), mouse anti-tubulin (University of Iowa Developmental Studies Hybridoma Bank), mouse anti-GSK3 and rabbit anti-pGSK3 (Cell Signaling Technology), JL-8 mouse anti-EGFP (Takara Bio Inc.), and 110B7E rabbit anti-phospho Substrate (RXXS*/T*; Cell Signaling Technology). The following secondary antibodies were used: Alexa Fluor 568 goat anti–mouse IgG at 1:300 (Life Technologies) and HRP-conjugated goat anti–mouse and goat anti–rabbit IgG at 1:5,000 (KLP Labs). Dispase Enzyme was purchased from Roche.
Cell lines, culture conditions, and transfections
HEK 293 cells were grown in modified Eagle’s medium and DMEM, respectively, supplemented with 10% FBS (Life Technologies). Human-derived oral squamous cell carcinoma SCC9 cell lines (a gift from J. Rheinwald, Harvard Medical School, Boston, MA) were maintained in DMEM/F12, 10% FBS, and 1% penicillin/streptomycin. Stable cell lines were maintained in the same culture, with the addition of 400 µg/ml G418. All cell cultures were maintained at 37°C in a humidified atmosphere of 5% CO2. R2834H DP-GFP SCC9 stable line was generated by transfecting cells with R2834H DP-GFP DNA using ExGen 500 transfection reagent (Fermentas Life Sciences) according to the manufacturer’s protocol, selected with 400 µg G418, ring cloned, and screened by direct fluorescence and immunoblotting. For siRNA or shRNA transfection assays, HEK 293 or SCC9 cells were plated at 3.5 × 105 cell per well and transfected with Dharmafect (Thermo Fisher Scientific). 24 h after transfection, cells were lysed and analyzed.
Affinity purification of DP S-tag
For each experiment HEK 293 cells were plated onto five 15-cm plates, grown to 80% confluency, and transiently transfected with DP S-tag DNA using Lipofectamine 2000 (Life Technologies). After 24 h, cells were lysed on ice for 15 min in 500 µl of lysis buffer (50 mM Tris-HCl, pH 7.9, 1% IGEPAL CA-630, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 20 mM β-glycerophosphate, 1× protease inhibitor mixture V [EMD Millipore], 1 mM PMSF, and 1 mM dithiothreitol). The lysate was centrifuged at 13,000 rpm for 15 min, and the supernatant was incubated with S-protein agarose for 3 h at 4°C. The agarose beads were washed twice by centrifugation with MS lysis buffer containing 500 mM NaCl and no protease inhibitors, twice with 50 mM Tris, pH 7.9, and 500 mM NaCl, and once with Tris buffer and 150 mM NaCl. Samples were resolved using SDS-AGE electrophoresis 10% gels (Bio-Rad Laboratories) and transferred to nitrocellulose membrane. Membranes were incubated with primary antibodies, washed, and incubated with secondary antibodies. Immunoblots were visualized by enhanced chemiluminescence.
Immunoprecipitations and preparation of cell lysates
For immunoprecipitation, DP S-tag expressing HEK 293 cells were harvested 24 or 48 h after transfection and lysed in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 0.1% NP-40, 10% glycerol, and protease inhibitor cocktail). Lysates were subjected to immunoprecipitation with anti-HA antibody or affinity purification with S-tag agarose. Immune complexes were washed three times with lysis buffer and subjected to immunoblotting with anti-HA, anti–S-tag, or NW6 antibodies. For immunoprecipitation of endogenous DP protein, HEK 293 cells were grown to 80% confluence in100-mm tissue culture dishes, washed with PBS, and lysed in lysis buffer. The lysates were precleared with rabbit IgG (Jackson ImmunoResearch Laboratories, Inc.) or mouse monoclonal anti-GFP antibody (Sigma-Aldrich) and Sepharose, followed by immunoprecipitation with anti-HA (Cell Signaling Technology). Immune complexes were precipitated with protein A/G sepharose and separated by SDS-PAGE followed by immunoblotting. Whole cell lysates in RIPA sample buffer were resolved by 7.5% SDS-PAGE and immunoblotted as previously described (Angst et al., 1990). Immunoreactive proteins were visualized using enhanced chemiluminescence.
Image acquisition and analysis for conventional fluorescence and live cell imaging
SCC9 cells were seeded onto collagen-coated glass coverslips for 24 h. Coverslips were washed in PBS, fixed in anhydrous methanol for 2 min at −20°C, air dried, and mounted on slides for observation of direct fluorescence. Frozen sections of transgenic hearts were fixed in 4% paraformaldehyde and permeabilized in 0.05% Triton X-100. Primary antibodies were detected using Alexa Fluor 488 goat anti–rabbit and 568 goat anti–mouse IgG (Life Technologies) and mounted in polyvinyl alcohol (Sigma-Aldrich). Fixed cells were visualized with a microscope (model DMR; Leica) fitted with a 40× (PlanFluotar; NA 1.0) objective and a confocal microscope (LSM 510; Carl Zeiss) fitted with 63× or 100× objective lenses (Plan Apochromat, NA 1.32 and NA 1.4; Carl Zeiss). DMR images were captured using a charge-coupled device camera (C4742-95; Orca 100; Hamamatsu Photonics) and analyzed using imaging software (Metamorph 7.7; Molecular Devices). LSM 510 images were processed using LSM 510 and ImageJ software. For live cell imaging, SCC9 cells stably expressing GFP-tagged DP (Godsel et al., 2005) were plated on Lab-Tek chambered coverglass slides (Thermo Fisher Scientific). Cells were switched to imaging media (Hanks’ balanced salt solution, 20 mM Hepes, 1% FBS, 2 mM l-glutamine, 4.5 g/liter glucose, and 1× amino acids) and imaged on a microscope (DMI6000; Leica) housed within a 37°C environmental control system chamber using 100-W mercury halide fiber optic illumination. Green channel was captured at 2-min intervals with a 63× objective lens (HCX Plan Apochromat, NA 1.4) using a MAG Dual-Cam adaptor fitted with two ORCA-ER AG cameras (Hamamatsu Photonics). Images were captured using Simple PCI version 6.0 (Hamamatsu Photonics) and processed using Photoshop (Adobe) and ImageJ software. Differential interference contrast was visualized to ensure that cells used for imaging were in contact with neighboring cells at the end of a movie. Deconvolution was performed using a blind deconvolution synthetic algorithm, and z-stacks from each time point were assembled into multi-page projections using ImageJ.
FRAP
FRAP was performed on live cells housed within a 37°C chamber using a spinning disk confocal microscope (XDI Revolution equipped with a Yokogawa CSU-X1; Andor Technology). Images were acquired with a Nikon Plan Apo 100×1.49NA TIRF objective and an Andor iXon3 EMCCD camera using MetaMorph 7.7 (Molecular Devices). Three prebleach images were acquired. Photobleaching of 75% was performed for three iterations at the appropriate wavelength. Image acquisitions of fluorescence recovery were performed 5 s after initial FRAP for 100 s. Imaging intervals were then increased to every 60 s to assure complete recovery after photobleaching. The intensity within the bleached region and a nonbleached region (as a control) were first measured using Fiji/ImageJ and then plotted to obtain the t1/2 values.
Calcium switch and drug treatments
SCC9 cells expressing GFP-tagged DP proteins were incubated in low calcium medium (DMEM with 0.05 mM CaCl2) for 16 h, switched to normal growth media containing ∼1.8 mM Ca2+ to induce cell junction assembly for time periods ranging up to 3 h, and processed for immunofluorescence analysis. For GSK3 inhibition experiments, SCC9 cells were treated with 40 mM LiCl (Sigma-Aldrich) or NaCl and 40 µM Kenpaullone or DMSO for 3 h at 37°C. For PKA and PKC inhibition experiments, HEK 293 cells were treated for 3 h with either 5 µM KT 5720 (Sigma-Aldrich) or 20 µM Gö 6976 (Sigma-Aldrich), respectively, or DMSO. PI3K inhibition was performed by treating HEK 293 or SCC9 cells with 20 µM Ly-294,002 (Sigma-Aldrich) for 3 h. Additional kinase inhibitors include W-7 (Santa Cruz Biotechnology, Inc.), INDY (R&D Systems), ZIP (R&D Systems), (R)-CR8 (R&D Systems), and TBB (R&D Systems). Phos-tag gels were performed using phos-tag acrylamide (Wako Pure Chemical Industries) AAL-107. Global PRMT inhibition was performed by treating SCC9 or HEK 293 cells with 30 µM AMI-1 (Sigma-Aldrich), 300 µM MTA (Sigma-Aldrich), or DMSO for 3 h. PKCα activation was performed as described previously, where PKC activation was stimulated by adding 15 nM PMA for 30 min (Bass-Zubek et al., 2008).
Phos-tag gel analysis
Phosphate-affinity SDS-PAGE was performed using Phos-tag (Wako Pure Chemical Industries). Procedure was performed following manufacturer protocol (http://www.phos-tag.com). For this procedure, 50 µM of phos-tag chemical is added during the preparation of a 15% wt/vol polyacrylamide gel. Subsequent SDS-PAGE electrophoresis and immunoblotting were performed as described in Affinity purification of DP S-tag. The acrylamide pendant binds to phosphorylated proteins and thus separate phosphorylation states can be visualized by gel or Western blot as up-shifted migration bands.
Fluorescence intensity of cell–cell borders and densitometric analysis of immunoblots
Fluorescence pixel intensity at cell borders over time was determined by multiplying the mean pixel intensity by the area of the defined border divided by the border length. Background intensity was subtracted from border intensity. P-values were calculated using two-tailed, two-sample unequal variance Student’s t tests. P-values <0.05 were considered statistically significant. Error bars represent SEM. All calculations were performed using ImageJ software. Densitometric analyses were performed of scanned immunoblots using ImageJ software. Pearson’s correlation coefficients were calculated using ImageJ software to assess the degree of colocalization between DP and IF.
Generation of transgenic mice
The aforementioned transgenic mice were generated as follows. The full-length WT and mutant DSP cDNA sequences were cloned into a vector containing the mouse α-myosin heavy chain promoter (provided by J. Robbins, University of Cincinnati, Cincinnati, OH). The linearized cDNAs were injected into fertilized oocytes (two-cell blastocyst stage) derived from C57BL6J mice and then the oocytes were transferred into the oviducts of pseudopregnant FVB mice. This was performed by the Transgenic and Homologous Recombination Core Facility of Baylor College of Medicine. The genotype and copy number of offspring were determined by PCR and Southern blot analysis of genomic DNA. Embryonic day 12.5–17.5 embryos were isolated from pseudo-pregnant mothers after microinjection and genotypes were determined by analysis of DNA extracts from the yolk sac (Yang et al., 2006).
Analysis of protein expression in transgenic hearts
Mouse hearts were snap frozen in liquid nitrogen and homogenized in lysis buffer containing 1% Triton X-100 (Yang et al., 2006). The lysates were clarified by centrifugation at 20,000 g for 30 min and the Triton X-100–insoluble fraction was solubilized using the lysis buffer containing 9 M urea. DP mutations and ARVD/C containing 9 M urea. Protein concentration was determined by BCA protein assay kit (Thermo Fisher Scientific).
In-solution protein cleavage and sample preparation for LC-MS/MS analyses
Affinity-purified DP S-tag was retained on agarose beads and washed three times with 100 µl of water. DP S-tag was first cleaved C-terminal to Met using cyanogen bromide (Sigma-Aldrich). The DP S-tag beads were suspended in 50 µl of 70% TFA (Sigma-Aldrich) in water and one small crystal (<1 mg) of cyanogen bromide was added to the solution. The reaction proceeded 20 h at 4°C. After cleavage, the supernatant containing peptides was collected and dried down in a centrivap concentrator (Labconco). The sample was then resuspended in 30 µl of 100 mM ammonium bicarbonate (Sigma-Aldrich) and AspN (Roche) was added at an enzyme/substrate ratio of 1:20. The digestion was performed for 6 h at room temperature and then quenched by adding acetic acid (Sigma-Aldrich) to adjust the pH to 3.
HPLC capillary column assembly
An analytical column was constructed using 360 (outside diameter) × 50-µm (inside diameter) fused silica capillary (Polymicro Technologies, LLC) with a 2-mm Kasil 1624 (PQ Corporation) frit. The analytical column was packed with 7 cm of Reprosil-Pur 120 C18 resin (3-µm diameter; Dr. Maisch GmbH) and equipped with a laser-pulled (P-2000 microcapillary laser puller; Sutter Instrument) electrospray emitter tip. A precolumn was constructed using 360 (outside diameter) × 75-µm (inside diameter) fused silica capillary (Polymicro Technologies, LLC) with a 2-mm Kasil 1624 frit and packed with 5 cm of Reprosil-Pur 120 C18 resin (5-µm diameter). Both columns were conditioned with several picomoles of angiotensin (Sigma-Aldrich) and histone H2A (New England Biolabs, Inc.) tryptic digests three times to optimize chromatographic performance. The two columns were connected with a microcapillary union (New Objective). The chromatographic performance was examined by analyzing 1 pmol of histone H2A tryptic digests before use to assure separation efficiency.
HPLC-MS/MS analyses
A 5% fraction of the DP S-tag digest (800 fmol) was pressure loaded onto the precolumn on a pressure bomb at a flow rate of 1 µl/min. The sample was desalted by rinsing the precolumn with 0.1 M acetic acid on an HPLC (1100 series; Agilent Technologies) for 10 min at a flow rate of 1 µl/min. The precolumn was then connected to the analytical column using a microcapillary union. Peptides were gradient eluted at a flow rate of 60 nl/min from the HPLC column and electrosprayed into the mass spectrometer (LTQ-Orbitrap Velos modified with FETD; Thermo Fisher Scientific). The LC gradient was as follows: 0–60% of solvent B in 60 min, 60–100% solvent B in 10 min, 100% solvent B hold for 5 min, 100% solvent B in 5 min, and 100% solvent A for 10 min (solvent A: 0.1 M acetic acid in water; solvent B: 70% acetonitrile [Mallinckrodt Pharmaceuticals] and 0.1 M acetic acid in water).
Data-dependent MS/MS analyses included a high resolution (60,000 at m/z 400) MS1 scan in the Orbitrap, and the top five most abundant ions were selected for MS2 analysis in the LTQ. Parameters for data dependence MS/MS data acquisition included monoisotopic precursor selection enabled, charge state rejection enabled for z = +1 and unassigned species, and dynamic exclusion enabled: repeat count of 3, repeat duration of 30 s, exclusion list duration of 20 s, exclusion list size of 500, and precursor isolation window of 3 m/z. ETD reaction time was 30 ms. FTMS AGC target was 500,000; ITMSn AGC target was 60,000; and ETD reagent target was 300,000.
Targeted ETD MS analysis included high resolution MS1 (60,000 at m/z 400) and MS2 (30,000 at m/z 400) scan in the Orbitrap. Precursor isolation window for MS2 was 3 m/z. ETD reaction time was 30 ms. Fragment ions resulting from 10 rounds of ETD were accumulated in the C-trap before detection in the Orbitrap. FTMS AGC target was 100,000, FTMSn AGC target was 50,000, and ETD reagent AGC target was 300,000.
Mass spectrometric data analyses
MS/MS data from data-dependent experiments were searched against a database containing the sequence of DP S-tag or its mutations using the Open Mass Spectrometry Search Algorithm software. MS/MS data were searched using c and z• fragment ions from ETD spectra. Database searches used no enzyme specificity and an E-value cutoff of 1. Variable modifications included in the database search were oxidation of Met and phosphorylation of Ser, Thr, and Tyr. A precursor mass tolerance of ±0.01 D was used for MS1 data and a fragment ion mass tolerance of ±0.35 D was used for MS2 data. The removal of reduced charge species within ETD data before searches was selected. All peptide sequences were manually validated by inspection of the accurate precursor mass and the corresponding ETD spectra. Some peptide sequences were also determined by de novo interpretation of the ETD spectra.
MS/MS data from targeted ETD experiments were manually validated by inspection of the accurate precursor mass and the corresponding ETD spectra.
Online supplemental material
Online supplemental material includes additional immunofluorescence analyses and additional details about PTM residues identified by mass spectrometry analyses. Fig. S1 demonstrates that the assembly kinetics and localization of adherens junctions are not dramatically altered after GSK3 inhibition. Fig. S2 includes immunofluorescence analysis of actin and microtubule cytoskeletal organization during GSK3 inhibition. Fig. S3 includes silver stain analysis of DP S-tag protein used for mass spectrometry analysis and FETD mass spectra identifying methylated residues within the DP C-tail. Fig. S4 demonstrates that R2834H DP-GFP stable line does not affect adherens junctions or the actin cytoskeleton. Table S1 identifies the phosphorylated residues identified in DP S-tag constructs. Table S2 identifies residue numbers for DP methylation and phosphosites. Videos 1–3 depict DP-GFP during scratch-wound assays in NaCl- and GSK3-treated SCC9 cells. Videos 4–6 depict stable cell lines expressing WT, S2849G, and R2834H DP-GFP during scratch-wound assays. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201406020/DC1.
Supplementary Material
Acknowledgments
Thanks go to the members of the Green laboratory and Cara Gottardi, Douglas Freymann, and Mitchel Denning for insightful discussions during development of this manuscript. Traditional sequencing services were performed at the Northwestern University Genomics Core Facility. Keratinocytes were obtained from the Tissue Engineering Core of the Northwestern University Skin Disease Research Center (P30AR057216).
L.V. Albrecht and K.J. Green designed the research. L.V. Albrecht performed the research excluding the mass spectrometry. L. Zhang, J. Shabanowitz, and D.F. Hunt performed and analyzed data from mass spectrometry. E. Purevjav and J.A. Towbin contributed tissues from transgenic mice. L.V. Albreacht and K.J. Green analyzed the data and wrote the paper.
This work was supported by National Institutes of Health (R01 AR041836 and AR43380 [K.J. Green] and GM037537 [D.F. Hunt]) and a Leducq Transatlantic Network grant (principal investigators J. Jalife and S. Hatem), with partial support from CA122151 (K.J. Green). Additional support was provided by the JL Mayberry endowment to K.J. Green. L.V. Albrecht is supported by T32 GM008061, awarded by the National Institutes of Health, and in part by the Malkin Scholars Program from the Robert H. Lurie Comprehensive Cancer Center of Northwestern University. FRAP imaging was performed at the Northwestern University Cell Imaging Facility supported by National Cancer Institute (NCI) grant CCSG P30 CA060553 (Robert H. Lurie Comprehensive Cancer Center).
Any opinions, findings, and conclusions or recommendations expressed in this material are of the authors and do not necessarily reflect the views of the National Institutes of Health/National Institute of Arthritis and Musculosckeletal and Skin Diseases/NCI.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AC
- arrhythmogenic cardiomyopathy
- AMI-1
- arginine methyltransferase inhibitors I
- caGSK3
- constitutively active GSK3
- DP
- desmoplakin
- DP CT
- DP C terminus
- FETD
- front-end electron transfer dissociation
- GSK3
- glycogen synthase kinase 3
- IF
- intermediate filament
- MS/MS
- tandem mass spectrometry
- MT
- microtubule
- MTA
- methylthioadenosine
- PLA
- proximity ligation assay
- PRMT-1
- protein arginine methyltransferase 1
- PTM
- posttranslational modification
- WT
- wild type
References
- Agullo-Pascual E., Cerrone M., and Delmar M.. 2014. Arrhythmogenic cardiomyopathy and Brugada syndrome: diseases of the connexome. FEBS Lett. 588:1322–1330 10.1016/j.febslet.2014.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angst B.D., Nilles L.A., and Green K.J.. 1990. Desmoplakin II expression is not restricted to stratified epithelia. J. Cell Sci. 97:247–257. [DOI] [PubMed] [Google Scholar]
- Asimaki A., and Saffitz J.E.. 2014. Remodeling of cell-cell junctions in arrhythmogenic cardiomyopathy. Cell Commun. Adhes. 21:13–23 10.3109/15419061.2013.876016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass-Zubek A.E., Hobbs R.P., Amargo E.V., Garcia N.J., Hsieh S.N., Chen X., Wahl J.K. III, Denning M.F., and Green K.J.. 2008. Plakophilin 2: a critical scaffold for PKCα that regulates intercellular junction assembly. J. Cell Biol. 181:605–613 10.1083/jcb.200712133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil S.A., Jedrychowski M., Schwartz D., Elias J.E., Villén J., Li J., Cohn M.A., Cantley L.C., and Gygi S.P.. 2004. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. USA. 101:12130–12135 10.1073/pnas.0404720101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford M.T.M., and Clarke S.G.S.. 2009. Protein arginine methylation in mammals: who, what, and why. Mol. Cell. 33:1–13 10.1016/j.molcel.2008.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornslaeger E.A., Corcoran C.M., Stappenbeck T.S., and Green K.J.. 1996. Breaking the connection: displacement of the desmosomal plaque protein desmoplakin from cell–cell interfaces disrupts anchorage of intermediate filament bundles and alters intercellular junction assembly. J. Cell Biol. 134:985–1001 10.1083/jcb.134.4.985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouameur J.-E., Schneider Y., Begré N., Hobbs R.P., Lingasamy P., Fontao L., Green K.J., Favre B., and Borradori L.. 2013. Phosphorylation of serine 4,642 in the C-terminus of plectin by MNK2 and PKA modulates its interaction with intermediate filaments. J. Cell Sci. 126:4195–4207 10.1242/jcs.127779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieher W.M., and Yap A.S.. 2013. Cadherin junctions and their cytoskeleton(s). Curr. Opin. Cell Biol. 25:39–46 10.1016/j.ceb.2012.10.010 [DOI] [PubMed] [Google Scholar]
- Brooke M.A., Nitoiu D., and Kelsell D.P.. 2012. Cell–cell connectivity: desmosomes and disease. J. Pathol. 226:158–171 10.1002/path.3027 [DOI] [PubMed] [Google Scholar]
- Cha B., Kim W., Kim Y.K., Hwang B.N., Park S.Y., Yoon J.W., Park W.S., Cho J.W., Bedford M.T., and Jho E.-H.. 2011. Methylation by protein arginine methyltransferase 1 increases stability of Axin, a negative regulator of Wnt signaling. Oncogene. 30:2379–2389 10.1038/onc.2010.610 [DOI] [PubMed] [Google Scholar]
- Choi H.-J., and Weis W.I.. 2011. Crystal structure of a rigid four-spectrin-repeat fragment of the human desmoplakin plakin domain. J. Mol. Biol. 409:800–812 10.1016/j.jmb.2011.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.-J., Park-Snyder S., Pascoe L.T., Green K.J., and Weis W.I.. 2002. Structures of two intermediate filament-binding fragments of desmoplakin reveal a unique repeat motif structure. Nat. Struct. Biol. 9:612–620. [DOI] [PubMed] [Google Scholar]
- Cross D.A., Alessi D.R., Cohen P., Andjelkovich M., and Hemmings B.A.. 1995. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 378:785–789 10.1038/378785a0 [DOI] [PubMed] [Google Scholar]
- Deribe Y.L., Pawson T., and Dikic I.. 2010. Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 17:666–672 10.1038/nsmb.1842 [DOI] [PubMed] [Google Scholar]
- Earley L., Anderson L.C., Bai D.L., Mullen C., Syka J.E.P., English A.M., Dunyach J.-J., Stafford G.C. Jr, Shabanowitz J., Hunt D.F., and Compton P.D.. 2013. Front-end electron transfer dissociation: a new ionization source. Anal. Chem. 85:8385–8390 10.1021/ac401783f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S., and Hall A.. 2003. Cdc42 regulates GSK-3β and adenomatous polyposis coli to control cell polarity. Nature. 421:753–756 10.1038/nature01423 [DOI] [PubMed] [Google Scholar]
- Fontao L., Favre B., Riou S., Geerts D., Jaunin F., Saurat J.-H., Green K.J., Sonnenberg A., and Borradori L.. 2003. Interaction of the bullous pemphigoid antigen 1 (BP230) and desmoplakin with intermediate filaments is mediated by distinct sequences within their COOH terminus. Mol. Biol. Cell. 14:1978–1992 10.1091/mbc.E02-08-0548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson S., Gullberg M., Jarvius J., Olsson C., Pietras K., Gústafsdóttir S.M., Ostman A., and Landegren U.. 2002. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 20:473–477 10.1038/nbt0502-473 [DOI] [PubMed] [Google Scholar]
- Fuchs E., and Raghavan S.. 2002. Getting under the skin of epidermal morphogenesis. Nat. Rev. Genet. 3:199–209 10.1038/nrg758 [DOI] [PubMed] [Google Scholar]
- Gallicano G.I., Kouklis P., Bauer C., Yin M., Vasioukhin V., Degenstein L., and Fuchs E.. 1998. Desmoplakin is required early in development for assembly of desmosomes and cytoskeletal linkage. J. Cell Biol. 143:2009–2022 10.1083/jcb.143.7.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallicano G.I., Bauer C., and Fuchs E.. 2001. Rescuing desmoplakin function in extra-embryonic ectoderm reveals the importance of this protein in embryonic heart, neuroepithelium, skin and vasculature. Development. 128:929–941. [DOI] [PubMed] [Google Scholar]
- Garcia-Gras E., Lombardi R., Giocondo M.J., Willerson J.T., Schneider M.D., Khoury D.S., and Marian A.J.. 2006. Suppression of canonical Wnt/β-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. J. Clin. Invest. 116:2012–2021 10.1172/JCI27751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godsel L.M., Hsieh S.N., Amargo E.V., Bass A.E., Pascoe-McGillicuddy L.T., Huen A.C., Thorne M.E., Gaudry C.A., Park J.K., Myung K., et al. 2005. Desmoplakin assembly dynamics in four dimensions: multiple phases differentially regulated by intermediate filaments and actin. J. Cell Biol. 171:1045–1059 10.1083/jcb.200510038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon R.M., Simpson C.L., Johnson J.L., Koetsier J.L., Dubash A.D., Najor N.A., Sarig O., Sprecher E., and Green K.J.. 2013. Desmoglein-1/Erbin interaction suppresses ERK activation to support epidermal differentiation. J. Clin. Invest. 123:1556–1570 10.1172/JCI65220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzfeld M.2007. Plakophilins: Multifunctional proteins or just regulators of desmosomal adhesion? Biochim. Biophys. Acta. 1773:69–77 10.1016/j.bbamcr.2006.04.009 [DOI] [PubMed] [Google Scholar]
- Hobbs R.P., and Green K.J.. 2012. Desmoplakin regulates desmosome hyperadhesion. J. Invest. Dermatol. 132:482–485 10.1038/jid.2011.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson T.Y., Fontao L., Godsel L.M., Choi H.-J., Huen A.C., Borradori L., Weis W.I., and Green K.J.. 2004. In vitro methods for investigating desmoplakin–intermediate filament interactions and their role in adhesive strength. Methods Cell Biol. 78:757–786 10.1016/S0091-679X(04)78026-7 [DOI] [PubMed] [Google Scholar]
- Huen A.C., Park J.K., Godsel L.M., Chen X., Bannon L.J., Amargo E.V., Hudson T.Y., Mongiu A.K., Leigh I.M., Kelsell D.P., et al. 2002. Intermediate filament–membrane attachments function synergistically with actin-dependent contacts to regulate intercellular adhesive strength. J. Cell Biol. 159:1005–1017 10.1083/jcb.200206098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa I., and Inagaki M.. 2006. Regulatory mechanisms and functions of intermediate filaments: a study using site- and phosphorylation state-specific antibodies. Cancer Sci. 97:167–174 10.1111/j.1349-7006.2006.00161.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamora C., and Fuchs E.. 2002. Intercellular adhesion, signalling and the cytoskeleton. Nat. Cell Biol. 4:E101–E108 10.1038/ncb0402-e101 [DOI] [PubMed] [Google Scholar]
- Jonkman M.F., Pasmooij A.M.G., Pasmans S.G.M.A., van den Berg M.P., Ter Horst H.J., Timmer A., and Pas H.H.. 2005. Loss of desmoplakin tail causes lethal acantholytic epidermolysis bullosa. Am. J. Hum. Genet. 77:653–660 10.1086/496901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakesisoglou I., Yang Y., and Fuchs E.. 2000. An epidermal plakin that integrates actin and microtubule networks at cellular junctions. J. Cell Biol. 149:195–208 10.1083/jcb.149.1.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiès P., Bootsma M., Bax J., Schalij M.J., and van der Wall E.E.. 2006. Arrhythmogenic right ventricular dysplasia/cardiomyopathy: screening, diagnosis, and treatment. Heart Rhythm. 3:225–234 10.1016/j.hrthm.2005.10.018 [DOI] [PubMed] [Google Scholar]
- Kimura T.E., Merritt A.J., and Garrod D.R.. 2007. Calcium-independent desmosomes of keratinocytes are hyper-adhesive. J. Invest. Dermatol. 127:775–781 10.1038/sj.jid.5700643 [DOI] [PubMed] [Google Scholar]
- Kouklis P.D., Hutton E., and Fuchs E.. 1994. Making a connection: direct binding between keratin intermediate filaments and desmosomal proteins. J. Cell Biol. 127:1049–1060 10.1083/jcb.127.4.1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk A.P., and Green K.J.. 2013. Structure, function, and regulation of desmosomes. Prog. Mol. Biol. Transl. Sci. 116:95–118 10.1016/B978-0-12-394311-8.00005-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk A.P., Bornslaeger E.A., Borgwardt J.E., Palka H.L., Dhaliwal A.S., Corcoran C.M., Denning M.F., and Green K.J.. 1997. The amino-terminal domain of desmoplakin binds to plakoglobin and clusters desmosomal cadherin–plakoglobin complexes. J. Cell Biol. 139:773–784 10.1083/jcb.139.3.773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger C., Loschke F., Schwarz N., Windoffer R., Leube R.E., and Magin T.M.. 2013. Keratins control intercellular adhesion involving PKC-α–mediated desmoplakin phosphorylation. J. Cell Biol. 201:681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Chimenti M.S., Pemble H., Schönichen A., Thompson O., Jacobson M.P., and Wittmann T.. 2012. Multisite phosphorylation disrupts arginine-glutamate salt bridge networks required for binding of cytoplasmic linker-associated protein 2 (CLASP2) to end-binding protein 1 (EB1). J. Biol. Chem. 287:17050–17064 10.1074/jbc.M111.316661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai-Cheong J.E., Arita K., and McGrath J.A.. 2007. Genetic diseases of junctions. J. Invest. Dermatol. 127:2713–2725 10.1038/sj.jid.5700727 [DOI] [PubMed] [Google Scholar]
- Leung C.L., Sun D., Zheng M., Knowles D.R., and Liem R.K.. 1999. Microtubule actin cross-linking factor (MACF): a hybrid of dystonin and dystrophin that can interact with the actin and microtubule cytoskeletons. J. Cell Biol. 147:1275–1286 10.1083/jcb.147.6.1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney M.G., Sadowski S., Brennan D., Pikander P., Saukko P., Wahl J., Aho H., Heikinheimo K., Bruckner-Tuderman L., Fertala A., et al. 2010. Compound heterozygous desmoplakin mutations result in a phenotype with a combination of myocardial, skin, hair, and enamel abnormalities. J. Invest. Dermatol. 130:968–978 10.1038/jid.2009.357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J.J., Bornslaeger E.A., Green K.J., Steinert P.M., and Ip W.. 1997. Two-hybrid analysis reveals fundamental differences in direct interactions between desmoplakin and cell type-specific intermediate filaments. J. Biol. Chem. 272:21495–21503 10.1074/jbc.272.34.21495 [DOI] [PubMed] [Google Scholar]
- Norgett E.E., Hatsell S.J., Carvajal-Huerta L., Cabezas J.C., Common J., Purkis P.E., Whittock N., Leigh I.M., Stevens H.P., and Kelsell D.P.. 2000. Recessive mutation in desmoplakin disrupts desmoplakin–intermediate filament interactions and causes dilated cardiomyopathy, woolly hair and keratoderma. Hum. Mol. Genet. 9:2761–2766 10.1093/hmg/9.18.2761 [DOI] [PubMed] [Google Scholar]
- O’Keefe E.J., Erickson H.P., and Bennett V.. 1989. Desmoplakin I and desmoplakin II. Purification and characterization. J. Biol. Chem. 264:8310–8318. [PubMed] [Google Scholar]
- Pan X., Hobbs R.P., and Coulombe P.A.. 2013. The expanding significance of keratin intermediate filaments in normal and diseased epithelia. Curr. Opin. Cell Biol. 25:47–56 10.1016/j.ceb.2012.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D.M., Dubash A.D., Kreitzer G., and Green K.J.. 2014. Disease mutations in desmoplakin inhibit Cx43 membrane targeting mediated by desmoplakin–EB1 interactions. J. Cell Biol. 206:779–797 10.1083/jcb.201312110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhrberg C., and Watt F.M.. 1997. The plakin family: versatile organizers of cytoskeletal architecture. Curr. Opin. Genet. Dev. 7:392–397 10.1016/S0959-437X(97)80154-2 [DOI] [PubMed] [Google Scholar]
- Saffitz J.E.2006. Adhesion molecules: why they are important to the electrophysiologist. J. Cardiovasc. Electrophysiol. 17:225–229 10.1111/j.1540-8167.2006.00365.x [DOI] [PubMed] [Google Scholar]
- Simpson C.L., Patel D.M., and Green K.J.. 2011. Deconstructing the skin: cytoarchitectural determinants of epidermal morphogenesis. Nat. Rev. Mol. Cell Biol. 12:565–580 10.1038/nrm3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider N.T., and Omary M.B.. 2014. Post-translational modifications of intermediate filament proteins: mechanisms and functions. Nat. Rev. Mol. Cell Biol. 15:163–177 10.1038/nrm3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.G., and Landegren U.. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 3:995–1000 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- Sonnenberg A., and Liem R.K.H.. 2007. Plakins in development and disease. Exp. Cell Res. 313:2189–2203 10.1016/j.yexcr.2007.03.039 [DOI] [PubMed] [Google Scholar]
- Stappenbeck T.S., Lamb J.A., Corcoran C.M., and Green K.J.. 1994. Phosphorylation of the desmoplakin COOH terminus negatively regulates its interaction with keratin intermediate filament networks. J. Biol. Chem. 269:29351–29354. [PubMed] [Google Scholar]
- Sumigray K.D., Chen H., and Lechler T.. 2011. Lis1 is essential for cortical microtubule organization and desmosome stability in the epidermis. J. Cell Biol. 194:631–642 10.1083/jcb.201104009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T., Rodriguez M., and Kim L.. 2009. Glycogen synthase kinase 3 in the world of cell migration. Dev. Growth Differ. 51:735–742 10.1111/j.1440-169X.2009.01141.x [DOI] [PubMed] [Google Scholar]
- Syka J.E.P., Coon J.J., Schroeder M.J., Shabanowitz J., and Hunt D.F.. 2004. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. USA. 101:9528–9533 10.1073/pnas.0402700101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason H.A., Scothern A., McHarg S., and Garrod D.R.. 2010. Desmosomes: adhesive strength and signalling in health and disease. Biochem. J. 429:419–433. [DOI] [PubMed] [Google Scholar]
- Weibrecht I., Leuchowius K.-J., Clausson C.-M., Conze T., Jarvius M., Howell W.M., Kamali-Moghaddam M., and Söderberg O.. 2010. Proximity ligation assays: a recent addition to the proteomics toolbox. Expert Rev. Proteomics. 7:401–409 10.1586/epr.10.10 [DOI] [PubMed] [Google Scholar]
- Wu X., Shen Q.-T., Oristian D.S., Lu C.P., Zheng Q., Wang H.-W., and Fuchs E.. 2011. Skin stem cells orchestrate directional migration by regulating microtubule–ACF7 connections through GSK3β. Cell. 144:341–352 10.1016/j.cell.2010.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Wang A.H., Oses-Prieto J., Makhijani K., Katsuno Y., Pei M., Yan L., Zheng Y.G., Burlingame A., Brückner K., and Derynck R.. 2013. Arginine methylation initiates BMP-induced Smad signaling. Mol. Cell. 51:5–19 10.1016/j.molcel.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Bowles N.E., Scherer S.E., Taylor M.D., Kearney D.L., Ge S., Nadvoretskiy V.V., DeFreitas G., Carabello B., Brandon L.I., et al. 2006. Desmosomal dysfunction due to mutations in desmoplakin causes arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circ. Res. 99:646–655 10.1161/01.RES.0000241482.19382.c6 [DOI] [PubMed] [Google Scholar]
- Zhang X., and Cheng X.. 2003. Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure. 11:509–520 10.1016/S0969-2126(03)00071-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.