DENND2B, in a complex with the Rab13 effector MICAL-L2, activates Rab13 at the cell periphery, promoting the dynamic remodeling of the cell’s leading edge during tumor cell migration both in vitro and in vivo.
Abstract
The small guanosine triphosphatase Rab13 functions in exocytic vesicle trafficking in epithelial cells. Alterations in Rab13 activity have been observed in human cancers, yet the mechanism of Rab13 activation and its role in cancer progression remain unclear. In this paper, we identify the DENN domain protein DENND2B as the guanine nucleotide exchange factor for Rab13 and develop a novel Förster resonance energy transfer–based Rab biosensor to reveal activation of Rab13 by DENND2B at the leading edge of migrating cells. DENND2B interacts with the Rab13 effector MICAL-L2 at the cell periphery, and this interaction is required for the dynamic remodeling of the cell’s leading edge. Disruption of Rab13-mediated trafficking dramatically limits the invasive behavior of epithelial cells in vitro and the growth and migration of highly invasive cancer cells in vivo. Thus, blocking Rab13 activation by DENND2B may provide a novel target to limit the spread of epithelial cancers.
Introduction
The formation of carcinomas (epithelial cell cancers) involves loss of cell–cell contact, dynamic changes in cell morphology, increased proliferation, and enhanced cell migration and invasion (Goldenring, 2013). Membrane trafficking underlies all of these events. Rab GTPases are master regulators of membrane trafficking controlling the formation of vesicles, vesicle transport via the actin and microtubule cytoskeletons, and vesicle tethering and fusion (Stenmark, 2009). As such, Rabs are key regulators of physiological processes that drive cancer cell biology (Caswell and Norman, 2006, 2008; Mellman and Yarden, 2013).
Enhanced cell migration and the ability to invade through a matrix are perhaps the most obvious manifestations of carcinogenesis. Rab-mediated membrane trafficking controls both normal cell migration, such as that seen in development, and the migratory capacity of invasive carcinomas in at least two distinct ways (Goldenring, 2013). First, Rabs regulate the delivery of protein cargo required for a balance between cell adhesion and cell migration. For example, Rab35 mediates recycling of cadherins from endosomes, and loss of Rab35 function leads to loss of cell–cell contact and enhanced cell migration (Allaire et al., 2013). In addition, cell migration and invasion require a dynamic plasma membrane and the extension of cell protrusions at the migratory leading edge. These processes involve actin assembly, creating protrusive forces to push the membrane forward, coupled with movement of membrane into the growing protrusion to relieve membrane tension (Ridley, 2011; Goldenring, 2013). In fact, invasive cells take on a distinct paradigm of polarity with directed membrane delivery to the leading edge (Goldenring, 2013). One example is seen with Rab8, which controls an endosomal-recycling pathway that delivers membrane tubules to the growing leading edge, and disruption of Rab8 function blocks the formation of membrane protrusions (Hattula et al., 2006). However, whether specific Rab GTPases link new membrane insertion to actin remodeling at the leading edge during carcinoma cell invasion and metastasis is unclear.
Rabs are activated by guanine-nucleotide exchange factors (GEFs) that catalyze the exchange of GDP for GTP. The DENN (differentially expressed in normal and neoplastic cells) domain is an evolutionarily ancient and structurally conserved protein module (Levivier et al., 2001; Harsay and Schekman, 2007; Marat et al., 2011; Wu et al., 2011; Nookala et al., 2012) that functions as a GEF for multiple Rabs (Allaire et al., 2010; Marat and McPherson, 2010; Yoshimura et al., 2010). Currently, there are 26 identified DENN domain proteins (Marat et al., 2011; Zhang et al., 2012; Levine et al., 2013), the majority of which are poorly characterized. One such protein, DENND2B, is a member of the DENND2A–D subfamily that contains a C-terminal DENN domain but few other distinguishing features (Marat et al., 2011). DENND2B was originally identified by screening a cDNA expression library for gene products that suppress the tumorigenicity of HeLa cells in nude mice and was named ST5 (suppression of tumorigenicity 5; Oshimura et al., 1990; Lichy et al., 1992, 1996). More recently, DENND2B was shown to function as a GEF for Rab9 (Yoshimura et al., 2010). However, the mechanism by which DENND2B regulates tumorigenicity and the potential role of its GEF activity in the control of membrane trafficking in this process remain unknown.
In exploring the role of DENND2B in membrane trafficking, we have now discovered, through the use of multiple complementary approaches, that Rab13 is a specific substrate for DENND2B. Rab13 functions in exocytic membrane trafficking from the TGN to the cell surface via recycling endosomes in polarized epithelial cells (Nokes et al., 2008). Interestingly, Rab13 has also been indirectly implicated in cancer. For example, Rab13 levels are altered in both glioblastoma and carcinoma (Mo et al., 2013; Li et al., 2014), and Rab13 mRNA is found in membrane protrusions of breast cancer cells (Jakobsen et al., 2013).
Here, we demonstrate that DENND2B localizes to the cell surface in association with the actin regulatory protein MICAL-L2 (molecule interacting with CasL-like 2), a Rab13 effector that induces membrane ruffles when bound to active Rab13 (Baldassarre et al., 2009; Sakane et al., 2013). We develop and optimize a novel Förster resonance energy transfer (FRET)–based Rab13 biosensor that reveals selective activation of Rab13 on membrane ruffles at the dynamic leading edge of migrating cells, and we demonstrate that Rab13 activation by DENND2B at the cell surface is required for cell migration and invasion. Finally, we find that disruption of the Rab13–DENND2B trafficking pathway dramatically reduces the migration of highly aggressive breast cancer cells in vivo. These findings provide evidence for a DENND2B-centered module that appears critical for the metastasis of cancer cells.
Results
DENND2B is a GEF for Rab13
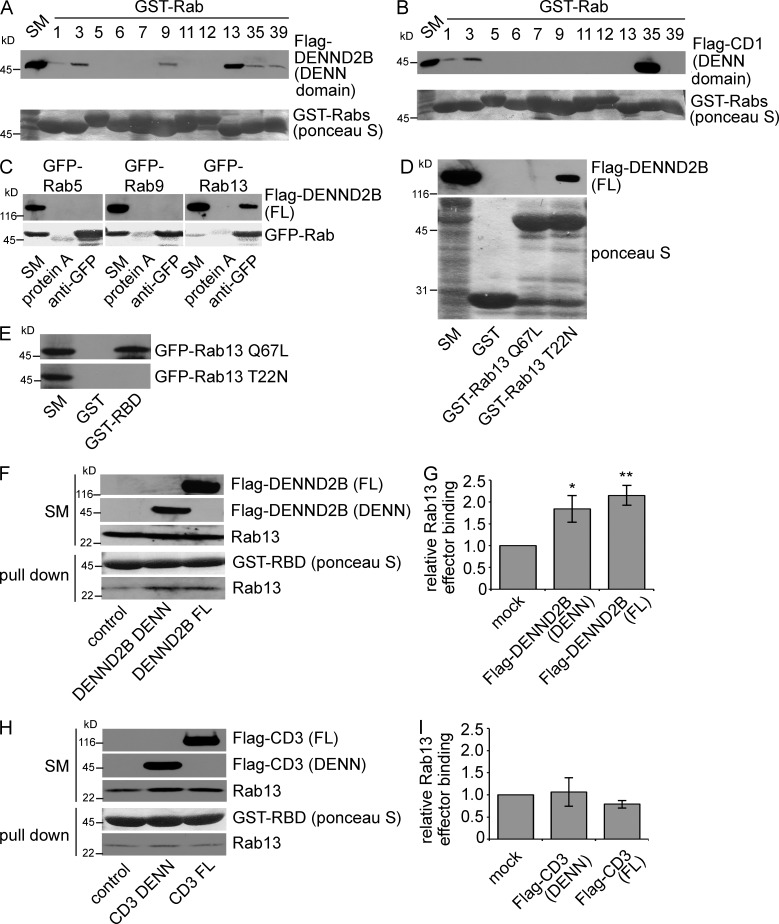
To examine a potential role for DENND2B in membrane trafficking via Rab activation, we used the DENND2B DENN domain in an affinity selection screen with a panel of GST-Rabs, including many of the proposed Rab substrates for DENN domain proteins. Flag-DENND2B DENN domain binds to Rab13 with minimal binding to other Rabs, including Rab9 (Figs. 1 A and S1 A). Notably, DENND2B does not bind to Rab8, which is highly conserved with Rab13 (Fig. S1 A). Consistent with previous studies (Allaire et al., 2010; Marat and McPherson, 2010), the DENN domain of connecdenn 1 binds strongly to Rab35 in similar assays (Fig. 1 B). In coimmunoprecipitation experiments, full-length Flag-DENND2B interacts with Rab13 but not Rab9 (Figs. 1 C and S1 B). Thus, DENND2B interacts preferentially with Rab13.
Figure 1.
DENND2B is a GEF for Rab13. (A and B) GST-Rabs, bound to glutathione beads, were incubated with HEK-293T cell lysates expressing the Flag-DENND2B DENN domain (A) or Flag–connecdenn 1 (CD1) DENN domain (B) in the presence of 5 mM EDTA. Ponceau S staining reveals the level of GST-Rabs, whereas specifically bound DENN domains were detected by blotting. Starting material (SM) equals 10% of the lysate used per condition. (C) Lysates from HEK-293T cells coexpressing Flag-DENND2B full length (FL) and GFP-Rabs were immunoprecipitated with the anti-GFP antibody in the presence of 5 mM EDTA, and the indicated proteins were detected by blotting. (D) GST-Rab13 Q67L, T22N, or GST alone, bound to glutathione beads, was incubated with HEK-293T cell lysates expressing Flag-DENND2B full length and analyzed as in A. (E) GST-RBD or GST alone, bound to glutathione beads, were incubated with HEK-293T cell lysates expressing GFP-Rab13 Q67L or T22N and analyzed as in A. (F–I) GST-RBD, bound to glutathione beads, was incubated with HEK-293T cell lysates expressing full-length Flag-DENND2B or DENN domain (F) or full-length Flag–connecdenn 3 (CD3) or DENN domain (H) and analyzed as in A. (G) Quantification of F (mean ± SEM in which n = 7 for mock and DENN domain and n = 6 for full length pooled from four independent experiments; one-way analysis of variance [ANOVA] with Dunnett’s post-test; *, P < 0.05; **, P < 0.01). (I) quantification of H (mean ± SEM in which n = 6 for mock and DENN domain and n = 5 for full length pooled from three independent experiments; statistical analysis used as in G).
To examine whether Rab13 has a nucleotide preference in its interaction with DENND2B, we performed binding experiments with GST-Rab13 T22N, a constitutively inactive GDP-bound mutant, and GST-Rab13 Q67L, a constitutively active GTP-bound mutant. Flag-DENND2B prefers Rab13 T22N (Fig. 1 D), a hallmark of GEFs. To verify the GEF activity of DENND2B toward Rab13, we developed a Rab13 activation assay using the Rab13-binding domain (RBD) of MICAL-L1 (Abou-Zeid et al., 2011). We confirmed that GST-RBD binds to GFP-Rab13 Q67L but not GFP-Rab13 T22N (Fig. 1 E). Overexpression of full-length, Flag-DENND2B, or its DENN domain results in an approximate twofold increase in binding of endogenous Rab13 to GST-RBD (Fig. 1, F and G). Thus, DENND2B is a GEF for Rab13. Previous studies reported that connecdenn 3 possesses weak GEF activity toward Rab13 in vitro (Yoshimura et al., 2010). However, overexpression of Flag–connecdenn 3, full-length or DENN domain, does not increase the amount of active Rab13 in cells (Fig. 1, H and I).
DENND2B is required for invasive behavior of epithelial cells
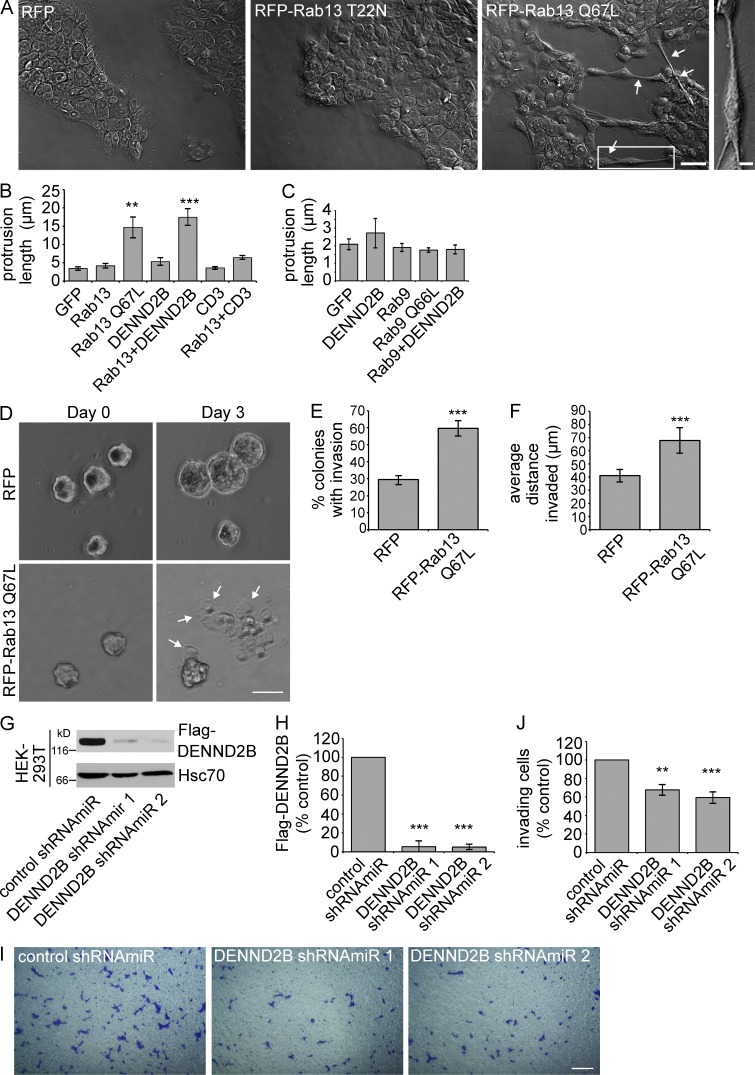
Scattering of epithelial cells induced by the tumor promoter 12-O-tetradecanoylphorbol-13-acetate (TPA) is a model for cell migration and invasion (Imamura et al., 1998), and knockdown of Rab13 decreases TPA-induced epithelial cell scattering (Kanda et al., 2008). We thus sought to examine the role of Rab13 and DENND2B in cell migration and invasion using MCF10A cells, a breast epithelial cell line expressing DENND2B (Fig. S2 A). When grown in a monolayer, MCF10A cells form adherent islands, with an epithelial-like morphology (Debnath et al., 2003). However, single cells are dynamic and will actively migrate toward each other to form such islands or will invade through a thin-layer Matrigel barrier toward high serum when plated in serum-free media (Pignatelli et al., 2012; Hu et al., 2014). Interestingly, when we overexpressed RFP-Rab13 Q67L in MCF10A cells (Fig. S2, B and C), we noticed that a percentage of the cells acquired a spindle-like morphology with disruption of the adherent island arrangement, and this was not observed with overexpression of RFP or RFP-Rab13 T22N (Figs. 2 A and S2 C). An obvious feature of the morphological change is the elongation of the cells and the appearance of plasma membrane protrusions (Fig. 2 A). Using protrusion length as a measure of morphological change, we find that expression of GFP-DENND2B or mCherry (mCh)-Rab13 alone are insufficient to induce a change in morphology, but their coexpression induces protrusions similar to mCh-Rab13 Q67L (Fig. 2, A and B). Coexpression of mCh-Rab13 with GFP–connecdenn 3 fails to induce membrane protrusions (Fig. 2 B). Moreover, expression of active mCh-Rab9 Q66L or coexpression of wild-type mCh-Rab9 and GFP-DENND2B also fails to induce membrane protrusions (Fig. 2 C). Therefore, activation of Rab13 by DENND2B mediates a morphological alteration of MCF10A cells.
Figure 2.
Rab13 and DENND2B promote epithelial cell invasion. (A) MCF10A cells were transduced with lentivirus driving expression of RFP, RFP-Rab13 T22N, or Q67L. Cells were fixed and imaged using DIC microscopy. Arrows indicate the presence of plasma membrane protrusions. Boxed region is magnified on the right. Bars: (low power view) 50 µm; (magnified view) 10 µm. (B) Protrusion length of transfected MCF10A cells was quantified (mean ± SEM from a minimum of three independent experiments measuring a minimum of 40 cells per condition per experiment; one-way ANOVA with Dunnett’s post-test; **, P < 0.01; ***, P < 0.001). (C) Transfected MCF10A cells were quantified as in B (mean ± SEM from three independent experiments measuring a minimum of 45 cells per condition per experiment). (D) Invasion of MCF10A spheroids transduced with RFP or RFP-Rab13 Q67L into collagen matrix. Bar, 50 µm. Arrows indicate invading cells. The images correspond to Video 1. (E and F) Quantification of the invasion from D (mean ± SD from three independent experiments measuring a minimum of 10 spheroids per condition per experiment; Student’s t test; ***, P < 0.001). (G) Flag-DENND2B expressing HEK-293T cells were transduced with control shRNAmiR or two different shRNAmiRs targeting DENND2B, and the indicated proteins were detected by blotting. (H) Quantification of Flag-DENND2B levels from G normalized to Hsc70 and expressed relative to the control shRNAmiR cells (mean ± SD from three independent experiments; statistical analysis used as in B). (I) MCF10A cells transduced with the indicated shRNAmiRs were plated on Matrigel-coated membranes for 14 h, and cells that invaded through the membranes were fixed and stained with crystal violet. Bar, 300 µm. (J) Quantification of MCF10A cell invasion from I normalized to control shRNAmiR cells (mean ± SEM from four independent experiments measuring 24 fields of view per condition per experiment; statistical analysis used as in B).
As the physiological relevance of the morphological change induced by Rab13/DENND2B overexpression is unclear, we next examined whether Rab13 influences the invasiveness of MCF10A cells. We first grew MCF10A cells expressing RFP or RFP-Rab13 Q67L on a Geltrex matrix to form multicell colonies, after which we overlaid them with collagen and performed time-lapse imaging for 3 d. Expression of active Rab13 causes increased cell invasion into the collagen (Fig. 2, D–F; and Video 1). We next examined the invasive behavior of MCF10A cells in a loss-of-function paradigm, in which we plated MCF10A cells on a thin layer of Matrigel on transwell membranes. Cells in serum-free media in the upper chamber migrate toward serum-containing media in the bottom chamber, a standard assay of cell invasiveness (Shaw, 2005). We used lentivirus to deliver two independent shRNAmiRs (Ritter et al., 2013), reducing DENND2B expression compared with a nontargeting shRNAmiR control (Fig. 2, G and H). Interestingly, knockdown of DENND2B significantly reduces the invasive capacity of MCF10A cells (Fig. 2, I and J) with no influence on total cell number or viability during the 14-h time course of the assay (Fig. S2, D and E). Together, these data indicate that DENND2B, through its activation of Rab13, is required for epithelial cell invasiveness. Whether or not this enhanced invasiveness correlates to a transformation of the cells is currently unknown.
DENND2B activates Rab13 selectively at the leading edge of migrating cells
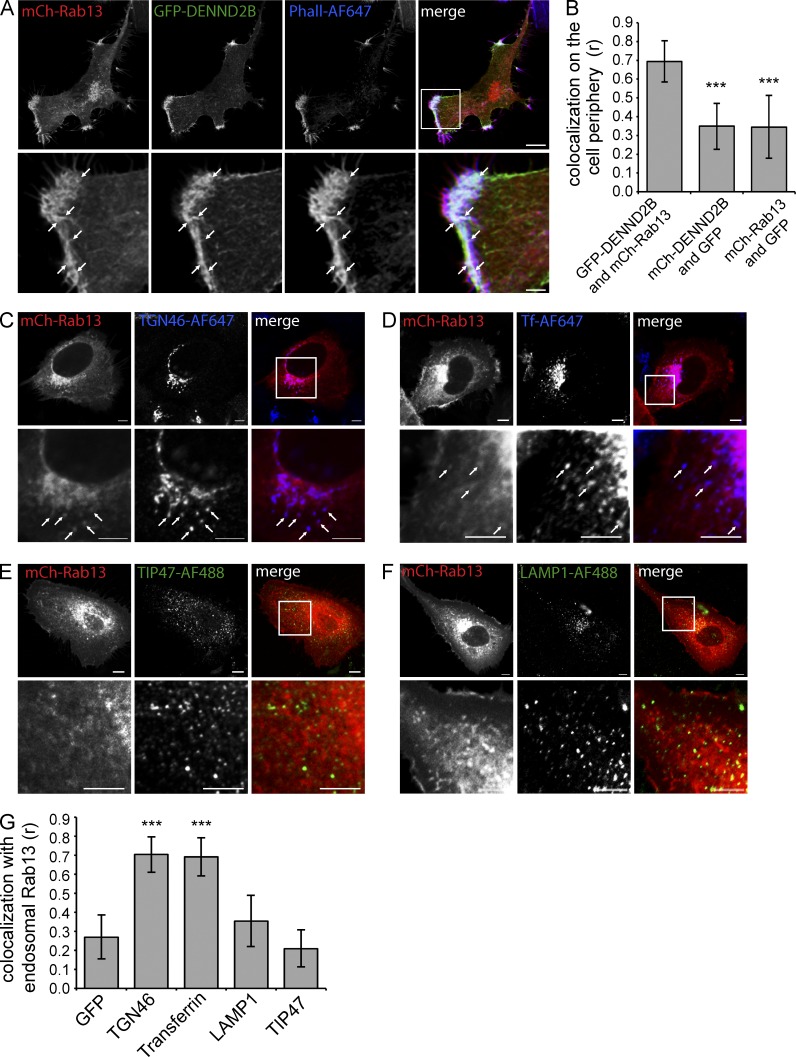
To gain insight into the mechanism of Rab13-mediated changes in cell morphology and invasion, we sought to identify the cellular site of Rab13 activation. GFP-DENND2B localizes predominantly to the cell periphery where it colocalizes with actin (Fig. 3 A). Rab13 also localizes to the cell periphery with additional pools in the perinuclear region and on cytoplasmic vesicles (Fig. 3, A–G). Interestingly, Rab13 and DENND2B colocalize predominantly at the actin-rich leading edge (Fig. 3, A and B).
Figure 3.
Rab13 colocalizes with DENND2B at the cell’s leading edge. (A) MCF10A cells were cotransfected with GFP-DENND2B and mCh-Rab13, fixed, and stained with Phalloidin–Alexa Fluor 647 (AF647). Bars: (low magnification) 10 µm; (high magnification) 2.5 µm. (B) Quantification of colocalization at the cell periphery using Pearson correlation coefficient (mean ± SD measuring 10 cells per condition pooled from two independent experiments; one-way ANOVA with Dunnett’s post-test; ***, P < 0.001). (C–F) MCF10A expressing mCh-Rab13 were fixed and labeled with TGN46-AF647 (C) internalized transferrin-AF647 (D), TIP47-AF488 (E), or LAMP-1–AF488 (F). Arrows point to colocalizing structures. Bars, 5 µm. (G) Quantification of colocalization from C–F as described in B. Boxed regions are magnified on the bottom.
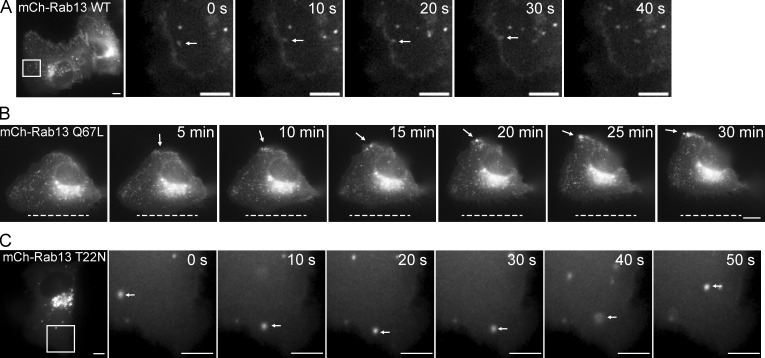
Consistent with a previous study (Nokes et al., 2008), the perinuclear pool of Rab13 partially colocalizes with TGN46, which at steady state is found prominently at the TGN, as well as with recycling endosomes labeled with internalized transferrin (Fig. 3, C, D, and G). There is little or no colocalization with TIP47 and LAMP-1, markers of late endosomes and late endosomes/lysosomes, respectively (Fig. 3, E–G). Live-cell imaging reveals that wild-type mCh-Rab13 travels on vesicles from the perinuclear region to the cell periphery, where a subset of vesicles appear to associate closely with the plasma membrane, leading to a pool of Rab13 on the cell surface (Fig. 4 A, Video 2, and Video 3). Vesicles are also seen to move away from the surface, and it is not currently known whether there is a net anterograde flow. mCh-Rab13 Q67L shows a similar trafficking behavior and also accumulates on the plasma membrane (Fig. 4 B and Video 4). Surprisingly, mCh-Rab13 T22N also travels on vesicles; however, it does not accumulate on the plasma membrane (Fig. 4 C and Video 5). We thus speculate that Rab13 travels on vesicles to the cell periphery where it encounters its GEF, DENND2B. This allows for localized activation and association with the plasma membrane.
Figure 4.
Active Rab13 accumulates at the cell’s leading edge. (A) MCF10A cells expressing wild-type (WT) mCh-Rab13 were imaged live. The arrows follow a vesicle that appears to fuse with the plasma membrane. The images correspond to Videos 2 and 3. (B) MCF10A cells expressing mCh-Rab13 Q67L were imaged live. The arrows indicate a site of vesicle accumulation. The dashed lines indicate the starting position of the lagging edge. The images correspond to Video 4. (C) MCF10A cells expressing mCh-Rab13 T22N were imaged live. The arrows follow a vesicle that fails to associate with the plasma membrane. The images correspond to Video 5. Bars, 10 µm. Boxed regions are magnified on the right.
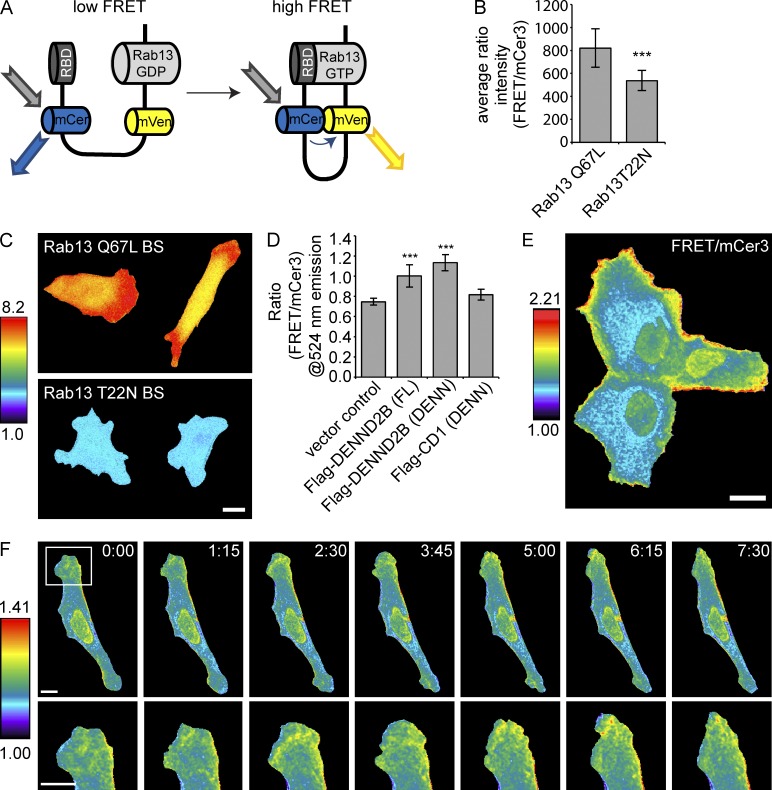
To directly test our model of localized Rab activation at the plasma membrane, we designed a genetically encoded, single-chain biosensor that uses FRET for temporal and spatial detection of Rab13 activation. The design of the biosensor is based on that previously reported for a RhoA biosensor (Pertz et al., 2006). From N to C terminus, the biosensor encodes the RBD of MICAL-L2, which binds specifically to active Rab13, monomeric Cerulean3 (mCer3) fluorescent protein, an unstructured linker, circularly permutated monomeric Venus (mVen) fluorescent protein and full-length Rab13 (Figs. 5 A and S3 A). This design maintains the correct C-terminal structure of the GTPase to preserve prenylation (Pertz et al., 2006). In principle, when Rab13 is activated it will bind the RBD, altering the mCer3 and mVen dipole-coupling state to modulate FRET (Fig. 5 A). We first tested and optimized the biosensor construction, measuring the emission spectra in live HEK-293 cells in suspension as described (Pertz et al., 2006). Inserting three repeats of an 18–amino acid linker (Whitlow et al., 1993) gives optimal differences in the FRET to mCer3 ratio of the Rab13 Q67L biosensor versus the Rab13 T22N form (Fig. S3 B), as does the circular permutation cp229 of mVen (Fig. S3 C; Nagai et al., 2004). The optimized Rab13 Q67L biosensor shows 53% higher FRET/mCer ratio intensity at 524 nm than does the Rab13 T22N biosensor (Figs. 5 B and S3 D), whereas the mean FRET efficiencies are 17% and 2.5%, respectively (Fig. S3, E and F). The enhanced efficiency of the Rab13 Q67L biosensor can also be visualized by ratio imaging in adherent mouse embryonic fibroblasts (MEFs; Fig. 5 C).
Figure 5.
Rab13 activation occurs at the leading edge of migrating cells. (A) Schematic of a Rab13 biosensor. (B) Mean emission ratio FRET/mCer3 of the optimized Q67L and T22N Rab13 biosensors in adherent MEFs using whole cell imaging (mean ± SD measuring 14 samples per condition pooled from two independent experiments; Student’s t test; ***, P < 0.001). (C) Ratiometric image (FRET/mCer3) of the optimized Q67L and T22N Rab13 biosensors (BS) in MEFs using whole cell imaging. (D) Fluorometric analysis of HEK-293T cells in suspension coexpressing Rab13 biosensor and RabGDI with empty vector, Flag-DENND2B full length (FL) or DENN domain, and Flag–connecdenn 1 (CD1) DENN domain (mean ± SD measuring six samples per conditions pooled from two independent experiments; one-way ANOVA with Dunnett’s post-test; ***, P < 0.001). (E) Ratiometric image (FRET/mCer3) of Rab13 wild-type biosensor in MCF10A cells. (F) Ratiometric image (FRET/mCer3) of Rab13 biosensor wild type in a randomly migrating MEF. The images correspond to Video 6. Time in minutes and seconds is indicated. Bars, 10 µm.
As binding of the biosensor to endogenous proteins (effectors or other Rabs) could conceivably influence the FRET signal, we tested whether the biosensor prefers intra- or intermolecular interactions. We created a Rab13-Q67L biosensor in which we replaced the RBD with the Cdc42/Rac1 interactive binding (CRIB) domain from p21-activated kinase (PAK; Moshfegh et al., 2014). The Rab13-Q67L-PAK-CRIB biosensor shows a lower FRET/mCer ratio compared with Rab13-Q67L-RBD, confirming that the PAK-CRIB domain does not interact with active Rab13 (Fig. S3 G). Next, using pull-down assays, we find that GST-RBD binds to the Rab13-Q67L-PAK-CRIB biosensor, which lacks intramolecular interactions, but not the Rab13-Q67L-RBD biosensor that has intramolecular interactions (Fig. S3 H). This demonstrates that the biosensor prefers intramolecular interactions and that other effectors or Rabs are unlikely to outcompete for binding to the biosensor.
Next, we confirmed that the wild-type biosensor shows the same distribution in MCF10A cells as wild-type Rab13 (Fig. S3 I). Finally, we coexpressed wild-type Rab13 biosensor with Flag-DENND2B, full-length or DENN domain, and the DENN domain of connecdenn 1 and observed that DENND2B expression enhances the FRET signal, whereas connecdenn 1 does not (Fig. 5 D).
With this characterization in hand, we expressed the biosensor in MCF10A cells and observed robust activation of wild-type Rab13, predominantly at the cell periphery (Fig. 5 E). Remarkably, we also observe Rab13 activation at the leading edge of randomly migrating cells (Fig. 5 F and Video 6), with waves of Rab13 activation at sites of plasma membrane protrusions (Video 6). As the ratiometric imaging measurements were made using wide-field microscopy, we tested the extent to which cell volume could influence these measurements using a previously developed model (Spiering et al., 2013). We determined that the distribution of the biosensor activity is significantly different from what would be obtained purely based on the volumetric variation (Fig. S3 J). Thus, the measured activation patterns are not influenced by the variability in cell thickness. These results confirm that Rab13 activation occurs at the cell periphery, where DENND2B and Rab13 colocalize, and further support a model in which local activation of Rab13 by DENND2B contributes to the dynamics of the plasma membrane and the development of a leading edge during cell migration and invasion.
DENND2B functions in a complex with the Rab13 effector MICAL-L2
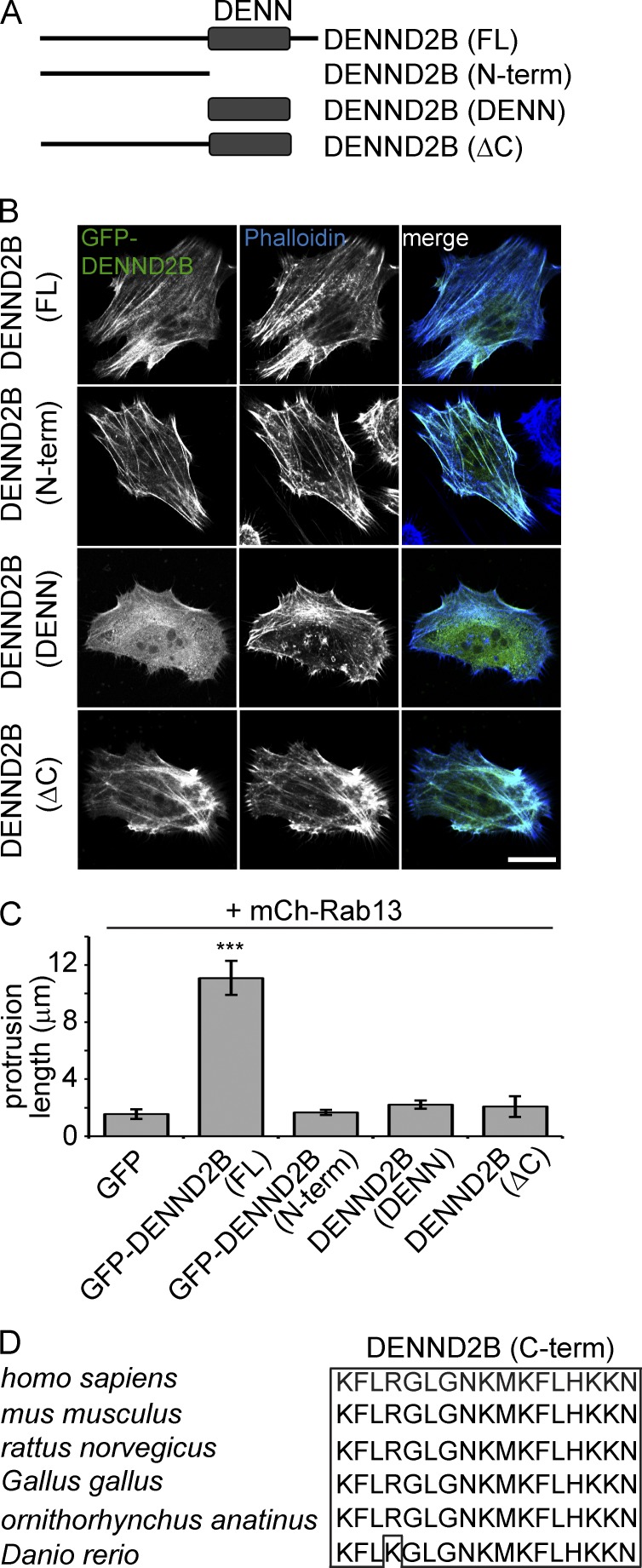
We next sought to identify determinants in DENND2B required for its localized activation of Rab13. We thus generated a series of DENND2B constructs (Fig. 6 A) that we tested for their ability to localize to actin (Fig. 6 B) and alter cell morphology when coexpressed with mCh-Rab13 (Fig. 6 C). To clearly distinguish DENND2B localized to actin, we examined the colocalization of DENND2B with actin stress fibers in HeLa cells (Fig. 6 B). As previously demonstrated (Figs. 2 B and 3 A), full-length DENND2B colocalizes with actin and induces membrane protrusions (Fig. 6, B and C). In contrast, the isolated DENN domain is predominantly soluble (Fig. 6 B) and does not induce membrane protrusions in the presence of Rab13 (Fig. 6 C), despite the fact it activates Rab13 in cells (Fig. 1, F and G; and Fig. 5 D). It is likely that the isolated DENN domain fails to alter cell morphology because Rab13 activation occurs away from the cell periphery. A DENND2B N-terminal construct (DENND2B [N-term]) also fails to induce morphological changes despite the fact it has a similar localization as the full-length protein (Fig. 6, B and C). This was expected as it lacks the DENN domain. Intriguingly, a DENND2B construct with deletion of the last 17 amino acids (DENND2B [ΔC]), which are C-terminal of the DENN domain, also fails to induce morphological alterations despite the fact that it contains the DENN domain and has a similar localization as full-length protein (Fig. 6, B and C). Thus, in addition to localized activation of Rab13 on actin, DENND2B must contain the evolutionarily conserved C-terminal 17–amino acid region (Fig. 6 D) to induce morphological changes required for invasive behavior.
Figure 6.
DENND2B C-terminal region is required for the morphological transformation of MCF10A cells. (A) Schematic of DENND2B constructs used in these experiments. (B) HeLa cells were transfected with the indicated GFP-DENND2B constructs, fixed, and processed for fluorescence for GFP and Phalloidin-AF647. Bar, 10 µm. (C) Protrusion length of MCF10A cells cotransfected with mCh-Rab13 and the indicated constructs was quantified (mean ± SEM from a minimum of three independent experiments measuring a minimum of 25 cells per condition per experiment; one-way ANOVA with Dunnett’s post-test; ***, P < 0.001). (D) Sequence of the 17 C-terminal residues of human DENND2B aligned to the indicated species. Identical residues are boxed. C-term, C terminus; FL, full length; N-term, N terminus.
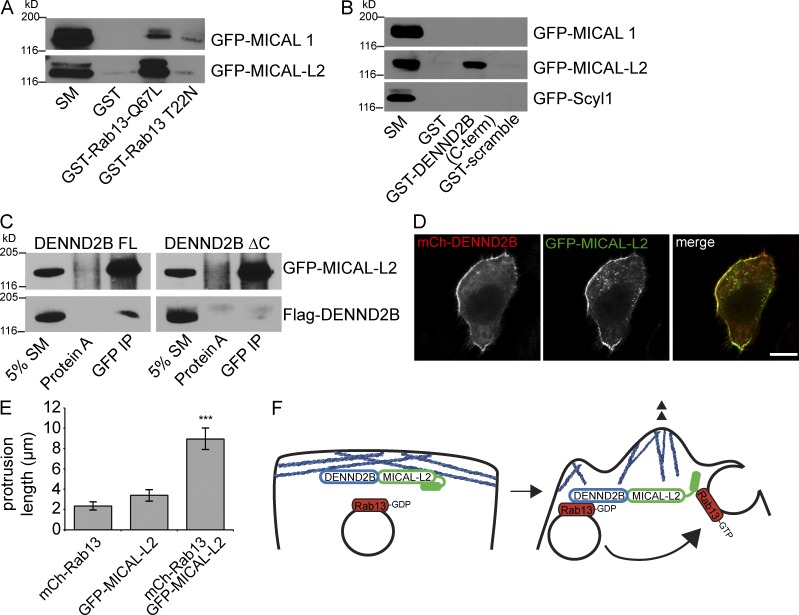
We hypothesized that the C-terminal region of DENND2B interacts with a protein required for DENND2B–Rab13-mediated alterations in cell morphology. MICAL-L2 is a Rab13 effector (Fig. 7 A; Nishimura and Sasaki, 2008), which upon binding Rab13 undergoes a conformational change that induces its association with the actin-binding filamin proteins, and both filamins and MICAL-L2 are required to induce actin changes causing membrane ruffling associated with cell migration (Baldassarre et al., 2009; Sakane et al., 2013). Moreover, like Rab13, MICAL-L2 is required for TPA-induced scattering of epithelial cells (Kanda et al., 2008). Remarkably, MICAL-L2, but not the related Rab13 effector MICAL 1, binds to the 17–amino acid C-terminal region of DENND2B (Fig. 7, A and B; Fukuda et al., 2008). Moreover, full-length DENND2B, but not DENND2B lacking the C-terminal region, coimmunoprecipitates with MICAL-L2 (Fig. 7 C). Finally, DENND2B and MICAL-L2 colocalize at the cell periphery (Fig. 7 D), and expression of MICAL-L2 in the presence of Rab13 induces similar morphological changes as those seen with expression of DENND2B (Fig. 7 E). Thus, DENND2B interacts with the Rab13 effector MICAL-L2, and this interaction is required for the morphological alterations observed in MCF10A cells.
Figure 7.
DENND2B C-terminal interacts with MICAL-L2. (A) GST-Rab13 Q67L or T22N, bound to glutathione beads, were incubated with HEK-293T cell lysates expressing GFP–MICAL 1 or GFP–MICAL-L2, and the indicated proteins were detected by blotting. Starting material (SM) equals 5% of the lysate used per condition. (B) GST-DENND2B C-terminal (C-term) region or GST coupled to a scrambled version of that sequence, bound to glutathione beads, were incubated with HEK-293T cell lysates expressing GFP–MICAL 1, GFP–MICAL-L2, or GFP-Scyl1, and the indicated proteins were detected by blotting. Starting material equals 2.5% of the lysate used per condition. (C) Lysates from HEK-293T cells coexpressing Flag-DENND2B full length (FL) or Flag-DENND2BΔC with GFP–MICAL-L2 were immunoprecipitated with the anti-GFP antibody, and the indicated proteins were detected by blotting. IP, immunoprecipitation. (D) MCF10A cells were transfected with mCh-DENND2B and GFP–MICAL-L2, fixed, and processed for fluorescence for GFP and mCh. Bar, 10 µm. (E) Protrusion length of transfected MCF10A cells was quantified (mean ± SEM of three independent experiments measuring a minimum of 40 cells per conditions per experiment; one-way ANOVA with Dunnett’s post-test; ***, P < 0.001). (F) Proposed model whereby vesicles carrying GDP-bound Rab13 encounter DENND2B in a complex with MICAL-L2 on cortical actin (blue lines). DENND2B activates Rab13, and the Rab13-GTP–positive vesicles fuse with the plasma membrane. Rab13-GTP also binds to and opens MICAL-L2, which then stimulates actin assembly for the formation of membrane ruffles.
We propose a model in which Rab13 is delivered on vesicles to the cell surface where it encounters DENND2B in a complex with MICAL-L2 (Fig. 7 F). This serves to ensure that MICAL-L2 is present at sites of Rab13 activation. Once activated, Rab13 stimulates MICAL-L2 to induce the formation of membrane ruffles while active Rab13 vesicles fuse with the plasma membrane to supply new membrane or deliver cargo, such as integrins (Nishikimi et al., 2014), required for migratory/invasive behavior. In this manner, DENND2B coordinates actin dynamics, with vesicle delivery contributing to the formation of a dynamic leading edge for cell migration and invasion.
Rab13 knockdown reduces cancer cell migration and invasion in vitro
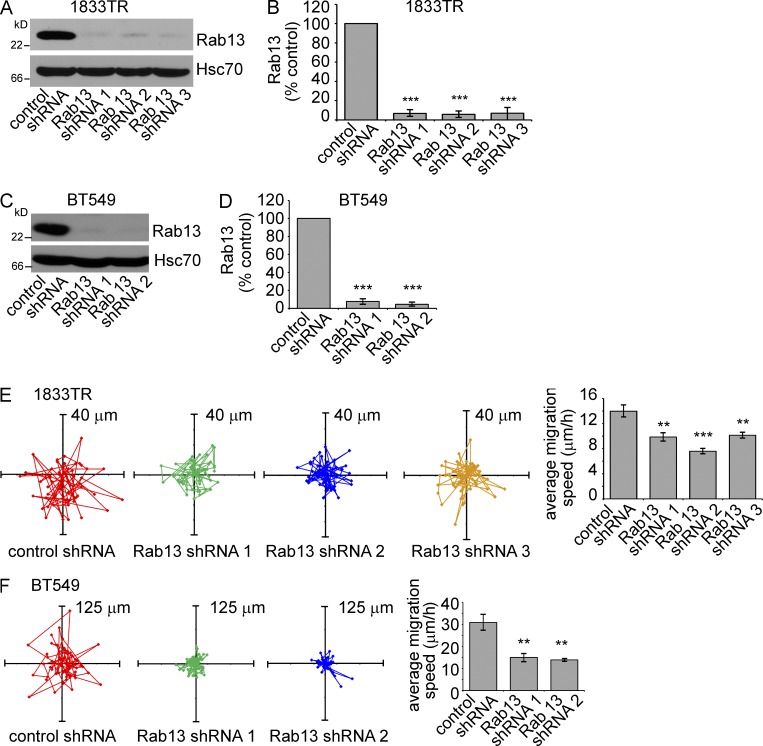
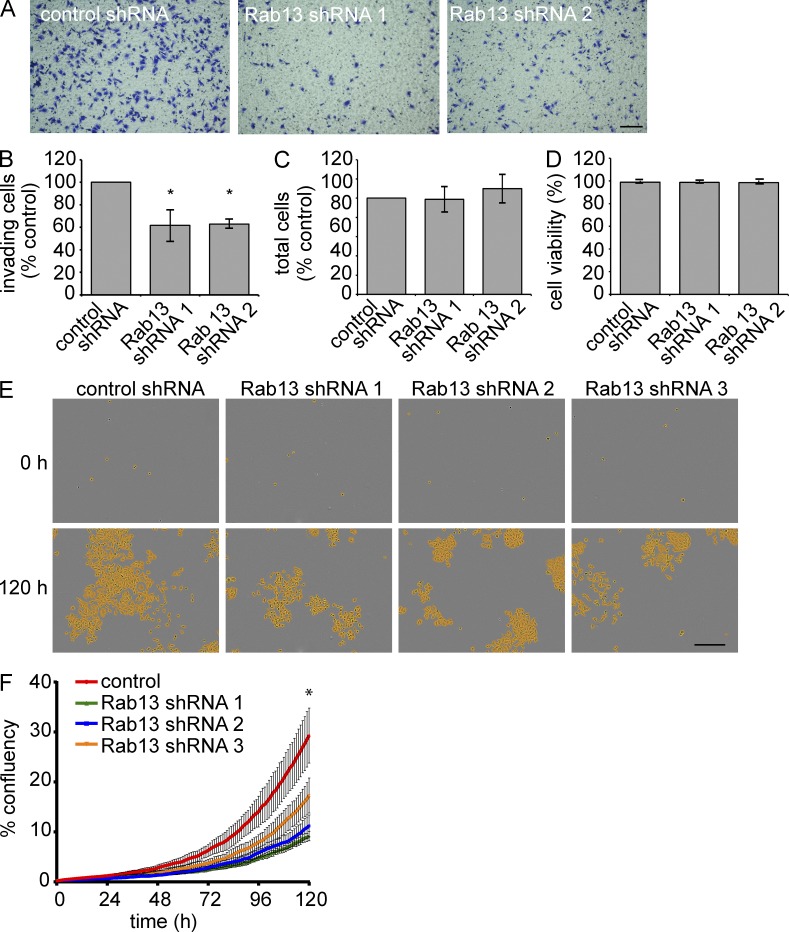
We next sought to examine the role of the Rab13 pathway in migration/invasion in a disease model and thus turned to MDA-MB-231-1833TR and BT549 breast cancer cells. We used lentivirus to deliver up to three independent shRNA sequences targeting Rab13, leading to efficient, stable Rab13 knockdown in both cell types (Fig. 8, A–D). Rab13 knockdown significantly reduces the migratory rate of both cell lines (Fig. 8, E and F). We then focused on the 1833TR cells as they have been selected for their robust metastatic capacity (Kang et al., 2003). Rab13 knockdown strongly decreases the ability of 1833TR cells to invade through Matrigel-coated membranes (Fig. 9, A and B), with no influence on cell number or viability over the time course of the experiment (Fig. 9, C and D). However, when measured over several days, Rab13 knockdown does significantly decrease cell proliferation (Fig. 9, E and F). Thus, in highly metastatic breast cancer cells, Rab13 is required for cell migration, invasion and proliferation, three key aspects of carcinoma progression.
Figure 8.
Rab13 knockdown reduces migration of breast cancer cells. (A) MDA-MB-231-1833TR cells were stably transduced with lentivirus driving expression of control shRNA or three different shRNAs targeting Rab13, and the indicated proteins were detected by blotting. (B) The levels of Rab13 from experiments as in A were normalized to Hsc70 and expressed relative to control cells (mean ± SD from four independent experiments; one-way ANOVA with Dunnett’s post-test; ***, P < 0.001). (C) As in A, but using BT549 cells. (D) As in B, but quantifying knockdown in C. (E) The migration pattern of single 1833TR cells expressing control or Rab13 targeting shRNAs. The mean migration speed of all cells measured is shown in the bar graph (mean ± SEM from three independent experiments measuring a minimum of 10 cells per condition per experiment; one-way ANOVA with Dunnett’s post-test; **, P < 0.01). (F) As in E, but with BT549 cells.
Figure 9.
Rab13 knockdown reduces invasiveness and growth of breast cancer cells. (A) 1833TR cells with stable expression of control or Rab13 targeting shRNAs were plated on Matrigel-coated membranes for 14 h and invading cells were fixed and stained with crystal violet. Bar, 300 µm. (B) Quantification of invading cells in A. normalized to control shRNA (mean ± SEM from four independent experiments measuring 24 fields of view per condition per experiment; one-way ANOVA with Dunnett’s post-test; *, P < 0.05). (C) 1833TR cells were plated on Matrigel-coated coverslips for 14 h and stained with Hoechst, and the number of cells was quantified (mean ± SEM from four independent experiments measuring 24 fields of view per condition per experiment, one-way ANOVA). (D) 1833TR cells were trypsinized and stained with Trypan blue, and the number of viable cells was quantified (mean ± SEM from five independent experiments measuring three wells per condition per experiment; statistical analysis used as in B). (E) 1833TR cells, plated at low density were imaged every hour for 120 h. Cells detected by imaging software are highlighted in yellow. Bar, 300 µm. (F) Quantification of cell confluency over time as in E. Graph shows a representative experiment (mean ± SD from three independent experiments measuring three wells per condition per experiment; two-way ANOVA; *, P < 0.05).
Rab13 knockdown reduces spread of cancer cells in vivo
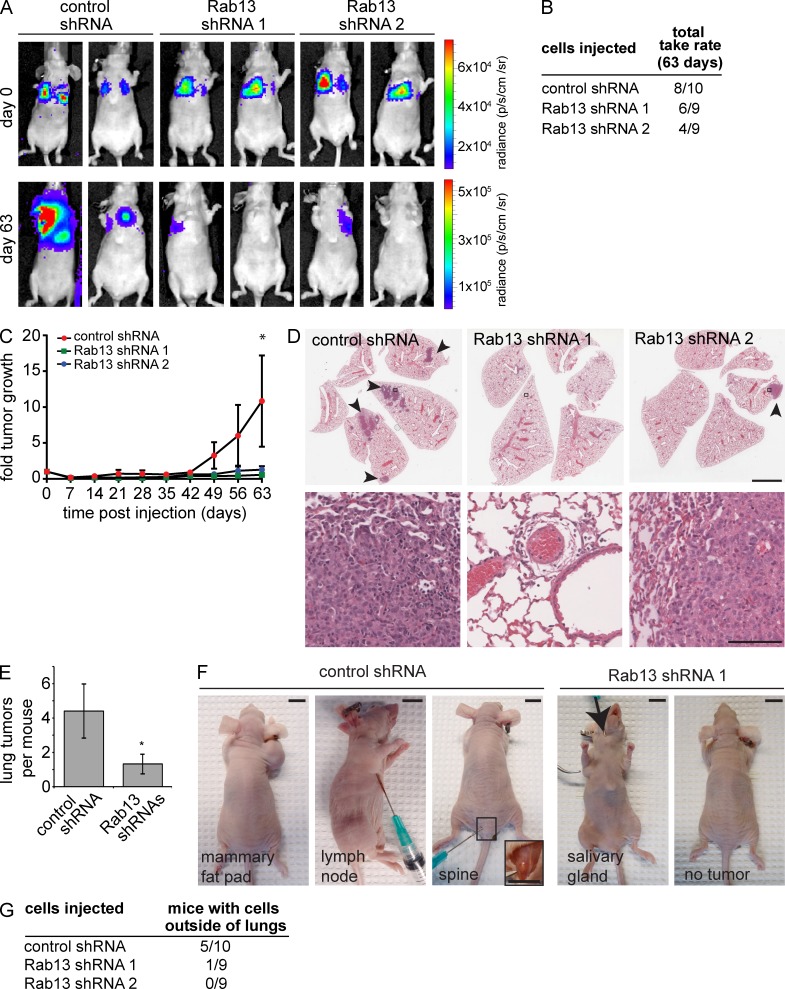
We next tested the involvement of Rab13 in invasive tumor growth in vivo. 1833TR cells with stable expression of control vector or Rab13-specific shRNA sequences (Fig. 8, A and B) were injected into the tail veins of nude mice as previously described (Knight et al., 2013). The cells also stably express a luciferase cassette allowing for analysis of tumor growth using in vivo bioluminescent imaging (Mourskaia et al., 2012). Mice were imaged shortly after injection (day 0) to confirm seeding of control and Rab13 shRNA cells in the lung microenvironment (Figs. 10 A and S4). Monitoring of tumor growth by luciferase activity 63 d after injection reveals higher rates of tumor take (80%) in mice injected with control cells compared with those injected with Rab13 shRNA cells (56%; Fig. 10, A and B; and Fig. S4). Among mice positive for tumors, injection with control cells generates significantly higher luciferase activity 63 d after injection than Rab13 shRNA cells, which reflects a greater tumor burden (Fig. 10, A and C; and Fig. S4). Consistently, histological analysis of lung sections show that mice injected with control cells have larger tumors and a significantly higher number of lung lesions per mouse relative to Rab13 shRNA cells (Fig. 10, D and E). Although lung lesions from both control and Rab13 shRNA cells demonstrate extravasation and proliferation external to blood vessels indicative of invasive growth, control cells also appeared in other tissues, including mammary fat pad, lymph nodes, and bone, reflecting the high metastatic capacity of the cell line (Figs. 10 F and S4). Remarkably, of those mice positive for lung tumor growth, 62% of mice injected with control cells exhibited cancer cells spread outside of the lungs compared with only 10% of mice injected with Rab13 knockdown cells (Figs. 10 G and S4). This demonstrates that Rab13 knockdown robustly diminishes the ability of aggressive tumor cells to migrate to new locations despite proliferation in the lung microenvironment. Coupled with the observation that RAB13 is amplified in multiple tumor types (Fig. S5; Cerami et al., 2012; Gao et al., 2013), our data suggest that Rab13 can promote tumor development and demonstrate that Rab13 knockdown diminishes the growth and spread of highly metastatic cancer cells in vivo.
Figure 10.
Rab13 knockdown reduces tumor growth and migration in vivo. (A) 1833TR cells stably expressing luciferase and either a control vector or shRNAs targeting Rab13 were injected into the tail veins of mice. The panel shows representative bioluminescence images of mice immediately after injection (day 0) and 63 d after injection. See Fig. S3 for all mice used in the study. (B) Tumor take rates of injected mice at day 63. (C) Total bioluminescence of luciferase-positive mice was quantified and normalized to day 0 (mean ± SEM from one independent experiment in which n = 9 for control mice and shRNA 2 and n = 8 for shRNA 1; two-way ANOVA with Tukey’s post-test; *, P < 0.05). (D) Representative hematoxylin and eosin staining of lung tissue collected from mice injected with control cells and Rab13 knockdown cells. Arrowheads indicate tumors. The bottom row corresponds to the areas indicated by the black boxes in the top row. Bars: (top row) 3 mm; (bottom row) 100 µm. (E) Quantification of tumor numbers in D (mean ± SEM from one independent experiment in which n = 10 and n = 18 for control and Rab13 shRNA, respectively; Student’s t test; *, P < 0.05). (F) Injected mice with visible tumors illustrate cell migration outside the lungs. A mouse with no detectable mass is shown for comparison. Bars, 1 cm. (G) The number of mice with a luciferase signal anywhere outside of the lung at day 63 was quantified.
Discussion
DENND2B is a Rab13 GEF
The identification of DENN domain proteins as a diverse family of Rab GEFs (Allaire et al., 2010; Yoshimura et al., 2010), coupled with the recognition that Rab-mediated membrane trafficking controls the key cellular phenotypes that drive cancer cell biology (Caswell and Norman, 2006, 2008; Goldenring, 2013; Mellman and Yarden, 2013), emphasizes the need to understand the cell physiological relationships between DENN domain proteins and their Rab substrates. Here, we present multiple lines of evidence that Rab13 is a physiologically relevant substrate for DENND2B. Although it is possible that DENND2B could activate Rab13 indirectly, this seems unlikely as Rab13 and DENND2B interact in pull-down and coimmunoprecipitation assays in a nucleotide-dependent manner. Moreover, coexpression of DENND2B and Rab13 cause the same morphological change of cells as that seen with an active Rab13 mutant, and knockdown of either protein decreases the invasive capacity of cells. A previous study reported that all members of the DENND2 family activate Rab9 and, consistently, knockdown of DENND2A or Rab9 results in enlarged late endosomes positive for LAMP-1 (Yoshimura et al., 2010). However, knockdown of DENND2B has no effect on the morphology of late endosomes, suggesting that it may act on a different Rab (Yoshimura et al., 2010). Yoshimura et al. (2010) note that although Rab9 is expressed in Drosophila, the DENND2 family is not, and therefore, a GEF other than DENND2 family members must be able to activate Rab9. In fact, DENND2B is absent from all invertebrates including Caenorhabditis elegans, the mollusk Lottia gigantea, and the eel-like invertebrate Branchiostoma floridae, a close relative of vertebrates. DENND2B is, however, present in vertebrates, including fish, and thus appears to have evolved early in the vertebrate lineage. Interestingly, Rab13 is also absent from invertebrates, including B. floridae, and appears to have diverged from its close homologue Rab8 (which is present throughout eukaryotes) in the vertebrate lineage (Klöpper et al., 2012). Thus, DENND2B may have evolved in response to the evolution of a new vertebrate Rab.
Localized activation of Rab13 at the plasma membrane leading edge
An intriguing finding of this study is the trafficking of inactive Rab13 on vesicles. The majority of Rabs are thought to associate with membranes upon activation (Goody et al., 2005) and to be extracted from membranes by GDP dissociation inhibitors (GDIs) once they switch to the inactive, GDP-bound form (Pfeffer et al., 1995). However, this does not seem to be the case for all Rabs (Allaire et al., 2010), and Rab13 is notable in that it resists membrane extraction by GDI (Marzesco et al., 1998). Only when the Rab13-labeled vesicles reach the plasma membrane is Rab13 activated by DENND2B, allowing for its association with the plasma membrane. Thus, the primary function of Rab13 appears to be in the fusion or association of TGN/endosome-derived transport vesicles with the plasma membrane. The use of the Rab13 biosensor has been paramount in elucidating this mechanism of Rab13 activation. Although this type of technology has been applied extensively to the study of GTPases, such as Ras, RhoA, and Cdc42 (Mochizuki et al., 2001; Nalbant et al., 2004; Pertz et al., 2006; Hanna et al., 2014; Moshfegh et al., 2014), to our knowledge only one other Rab biosensor has been reported (Kitano et al., 2008). Given that there are ∼70 mammalian Rabs regulating trafficking at nearly all cellular compartments, future development of Rab biosensors, based on the proof-of-principle concept presented here, will be extremely valuable in understanding how they function.
DENND2B functions in a complex with MICAL-L2
In addition to demonstrating that DENND2B is a GEF for Rab13, we discovered that DENND2B binds to the Rab13 effector MICAL-L2 and associates with MICAL-L2 at the cell periphery. This interaction would serve to concentrate MICAL-L2 at sites where Rab13 is being activated and is necessary for morphological alteration of cells. Such a positive GEF–effector feedback loop has been previously described. In yeast, Sec2p acts as a GEF for the small GTPase Sec4 and binds the Sec4 effector molecule Sec15 on the surface of secretory vesicles to facilitate tethering to the plasma membrane (Walch-Solimena et al., 1997; Guo et al., 1999; Medkova et al., 2006). In mammalian cells, Rab5 is activated by its GEF, Rabex-5, and recruits its effector Rabaptin-5. Rabaptin-5 not only binds to Rabex-5 but also enhances its GEF activity for Rab5 (Lippé et al., 2001). Similarly, Cdc42 is activated by its GEF, intersectin, and intersectin binds the Cdc42 effector N-WASP, which further enhances intersectin’s GEF activity (Hussain et al., 2001). It will thus be interesting to determine whether MICAL-L2 binding enhances the GEF activity of DENND2B.
DENND2B and Rab13 promote cancer phenotypes
A key finding of this study is that the DENND2B–Rab13 pathway promotes migration of cancer cells in vivo. During metastasis, epithelial cells exhibit loss of cell–cell contact. Previous studies have shown that knockdown of Rab13 or MICAL-L2 reduces levels of the tight junction proteins claudin-1 and occludin at the plasma membrane (Yamamura et al., 2008). Given the role of Rab13 in vesicle fusion, this could involve alterations in the delivery of these proteins to the cell surface. Rab13 could also modulate tight junctions indirectly through its effects on actin (Sakane et al., 2012). In fact, tight junction formation relies heavily on F-actin organization. For instance, the actin nucleator Arp2/3 is essential for the assembly of tight junctions, and Arp2/3 knockdown causes severe defects in tight junctions of developing embryonic epidermis (Zhou et al., 2013). This is not surprising considering that claudin-1 and occludin are linked to actin by zona occludin-1 (Fanning et al., 1998). Thus, the reported effects of Rab13 on tight junction formation could be explained by its regulation of cortical actin through MICAL-L2.
Metastasis also requires migration of cancer cells away from the primary tumor. Here, we demonstrate that Rab13 controls the migration of tumor cells from the lungs to secondary sites. Moreover, we show that Rab13 is activated at the leading edge of migrating cells where actin is being dynamically remodeled. This is consistent with the reported role of Rab13 and MICAL-L2 in cell scattering as inactive Rab13 or MICAL-L2 knockdown inhibit TPA-induced epithelial cell scattering (Kanda et al., 2008). Under normal conditions, MICAL-L2 is autoinhibited (Sakane et al., 2010). However, local activation of Rab13 by DENND2B at the leading edge relieves this inhibition, allowing for actin remodeling through interaction with the actin-binding protein filamin (Sakane et al., 2013). Expression of constitutively active mutants of MICAL-L2 induces the formation of membrane ruffles (Sakane et al., 2013), a process that also depends on filamins (Baldassarre et al., 2009). Thus, the association of DENND2B with MICAL-L2 would allow actin rearrangements to be coupled to new membrane fusion required for cell migration. Through this mechanism, amplification of RAB13 as seen in several cancer types would cause enhanced invasive capacity of cancerous cells.
The ability of DENND2B to promote phenotypes consistent with metastasis and tumorigenicity appears at odds with a previous study that it is a suppressor of tumorigenicity (Lichy et al., 1992). In fact, DENND2B was initially discovered as a suppressor of tumorigenicity by screening a cDNA expression library for genes that reduce tumorigenicity of HeLa cells when injected into nude mice (Lichy et al., 1992). However, in addition to the full-length form of DENND2B studied here, a shorter, 70-kD isoform (p70) exists that contains a truncated N-terminal region. Interestingly, p70 is produced from a distinct promoter, and only p70 levels, not full-length DENND2B, have been correlated to tumor suppression in cancer cell lines (Lichy et al., 1996). In fact, p70 was found to inhibit DENND2B signaling to restore contact-dependent growth (Hubbs et al., 1999; Majidi et al., 2000). We now demonstrate that DENND2B activates Rab13 at the cell surface to promote cell invasiveness and phenotypes consistent with tumorigenicity. Given that the N terminus is required for proper localization of DENND2B and that this site is absent from p70, perhaps p70 suppresses tumorigenicity by activating Rab13 away from the plasma membrane. This would be consistent with our finding that coexpression of Rab13 with the isolated DENN domain does not induce morphological changes of epithelial cells. It is possible that during cancer, when p70 is down-regulated, there would be an increase in localized Rab13 activation on cortical actin, resulting in increased metastatic capacity. It will be interesting in future studies to tease apart the functions of DENND2B isoforms in cancer in light of its role as a GEF for Rab13.
Rab GTPases are beginning to be recognized for their role in multiple types of cancer. It is now clear that Rab13 plays an important role in promoting tumorigenicity, but targeted therapies toward Rab13 may be difficult to achieve given the high degree of sequence homology across the Rab GTPase family. Characterization of Rab regulators, such as DENND2B, will provide a better mechanistic understanding of how Rabs contribute to disease and increase the potential for therapeutic targets.
Materials and methods
Antibodies
Mouse monoclonal Flag (M2) antibody was obtained from Sigma-Aldrich. Rabbit polyclonal GFP (A6455), AF647-conjugated Phalloidin, transferrin, and anti–sheep, and Alexa Fluor 488–conjugated anti–mouse and anti–rabbit were purchased from Invitrogen. Chicken polyclonal Rab13 and rabbit monoclonal LAMP-1 antibodies were obtained from Abcam. Rat monoclonal Hsc70 (1B5) antibody was obtained from Enzo Life Sciences. Rabbit polyclonal TIP47 antibody was purchased from Novus Biologicals. Sheep polyclonal TGN46 antibody was obtained from AbD Serotec. HRP-conjugated anti–mouse and anti–rabbit were obtained from Jackson ImmunoResearch Laboratories, Inc.
DNA constructs
GST-Rab1a, GST-Rab3a, GST-Rab5, GFP-Rab5, GST-Rab11a, GST-Rab13, GST-Rab35 in pGEX-6P1, Flag–connecdenn 1 full length, Flag–connecdenn 3 full length, and Flag–connecdenn 3 DENN domain in pCMVtag2B and GFP-Scyl1 in pEGFP-C2 were previously described (Burman et al., 2008; Allaire et al., 2010; Marat et al., 2012). GFP-Rab7 and GFP-Rab9 (canine) in pEGFP-C1 were a gift from F. Meunier (University of Queensland, Brisbane, Australia). GFP–MICAL 1, –MICAL-L1, and –MICAL-L2 in pEGFP-C3 were gifts from S. Caplan (University of Nebraska, Omaha, NE). His-RabGDI in pETH6 was a gift from W. Balch (Scripps Research Institute, San Diego, CA). Rab6b (GenBank accession no. BC036864), Rab12 (BC098407), Rab39 (BC028064), and DENND2B (BC036655) were obtained from GE Healthcare and cloned into pGEX-6P1 containing a GST tag. DENND2B C-terminal (amino acid 1,121–1,137) and scrambled control were generated by oligonucleotide annealing and subcloned into pGEX-4T1. Rab13 T22N, Rab13 Q67L, and Rab9 Q66L were produced using the megaprimer approach. These constructs along with MICAL-L1 RBD (691–858), DENND2B DENN domain (687–1,121), DENND2B N-term (1–686), and DENND2B ΔC (1–1,121) were cloned into pGEX-6P1 (under the tac promoter and induced with 500 µM isopropyl β-d-1-thiogalactopyranoside; Wisent Bioproducts), pEGFP-C2, mCh-C2, and/or pCMV-tag2B. Primers used to generate constructs can be found in Table S1. All constructs are of human sequences unless otherwise indicated. All constructs were verified by sequence analysis.
RNA extraction and RT-PCR
To obtain cellular RNA, 3 × 106 cells grown in standard cell culture conditions at roughly 80% confluence were washed with PBS and then processed with RNeasy kit (QIAGEN). To generate cDNA, 500 ng RNA was added to an Omniscript cDNA reaction (QIAGEN) according to the manufacturer’s instructions using random nonamer oligonucleotides (Sigma-Aldrich). Final PCR amplicons were generated using a standard PCR reaction with Taq DNA polymerase in a 20-µl final reaction volume, using 2 µl of the cDNA reaction as a template. Primers used for PCR reactions can be found in Table S1.
Lentivirus production
Target sequences for DENND2B were designed using the BLOCK-iT RNAi Designer (Invitrogen), subcloned first into the pcDNA6.2/GW-emerald GFP–miR cassette and subsequently into the pRRLsinPPT viral expression vector (Invitrogen; Ritter et al., 2013). Viral particles were produced in HEK-293T cells and concentrated as previously described (Thomas et al., 2009; Ritter et al., 2013). Nontargeting control shRNAmiR virus was previously described (Thomas et al., 2009). For Rab13 overexpression virus, Rab13 was subcloned first into pcDNA6.2/GW-RFP–miR cassette and subsequently into pRRLsinPPT. Rab13 shRNA 1 (TRCN0000381528), Rab13 shRNA 2 (TRCN0000382005), and Rab13 shRNA 3 (TRCN0000343510) in pLKO-TRC005 were obtained from the MISSION at The RNAi Consortium genome-wide shRNA collection (Sigma-Aldrich). Empty pLKO-TRC005 vector was used as a control. Viral particles were produced by coexpressing Rab13 shRNA or control with packaging plasmids pMD.G, RSV-Rev, and pCMVDR8.2 in HEK-293T cells using FuGENE (Promega). The media containing viral particles were collected and filtered with a 0.45-µm filter to remove cell debris. Sequences used to generate shRNA can be found in Table S1.
Protein knockdown/overexpression with lentivirus
For knockdown of DENND2B, MCF10A cells were plated the day of the transduction, virus was added at an MOI of 10, and the media were replaced with fresh culture media the following day. All experiments were performed 4–14 d after transduction. For overexpression of Rab13, virus was added to MCF10A cells at an MOI of 5, and the media were replaced with fresh culture media the following day. All experiments were performed within 5 d of transduction. For knockdown of Rab13, filtered culture media containing viral particles were added to MDA-MB-231-1833TR (gift from P. Siegel, McGill University, Montreal, Quebec, Canada) and BT549 cells in media supplemented with 8 µg/ml polybrene. 1 d after transduction, the media were replaced with fresh culture media. Starting 2 d after transduction, the cells were selected and maintained in media containing 1.5 µg/ml puromycin dihydrochloride (Sigma-Aldrich).
Biochemical assays
For Rab13 effector binding assays, HEK-293T cells, grown to 40% confluency in 15-cm dishes, were transfected with FLAG-DENN domain constructs. At 18 h after transfection, cells were lysed in 20 mM Hepes, pH 7.4, 100 mM NaCl, 10 mM MgCl2, 5% glycerol, 1% Triton-X 100, 0.5% deoxycholate, and 0.1% SDS, supplemented with protease inhibitors (0.83 mM benzamidine, 0.25 mM phenylmethylsulfonyl fluoride, 0.5 µg/ml aprotinin, and 0.5 µg/ml leupeptin). Cell lysates were incubated with 40 µg GST–MICAL-L1 RBD precoupled to glutathione–Sepharose beads for 2 h at 4°C and washed three times with the same buffer. Samples were eluted in SDS-PAGE sample buffer, resolved by SDS-PAGE, and processed for Western blotting. Various GST-Rab proteins were expressed in Escherichia coli BL21 and purified using standard procedures in Tris buffer (20 mM Tris, pH 7.4, 100 mM NaCl, and 1 mM dithiothreitol) supplemented with protease inhibitors. Biosensor binding assays were performed in Tris buffer containing 5 mM MgCl2 and protease inhibitors. HEK-293T cells, grown to 40% confluency in 15-cm dishes, were transfected with Flag-tagged or GFP-tagged constructs. At 18 h after transfection, cells were washed with PBS, scraped into Tris buffer supplemented with protease inhibitors, and sonicated, and Triton X-100 was added to 1% final concentration. After 15 min of incubation at 4°C, the lysates were centrifuged at 305,000 g for 15 min at 4°C. For GST pull-down experiments, supernatants were incubated with 40 µg GST fusion proteins precoupled to glutathione–Sepharose beads for 2 h at 4°C. For coimmunoprecipitation experiments, supernatants were incubated with protein A–Sepharose beads (GE Healthcare) and the anti-GFP antibody for 2 h at 4°C. Samples were washed three times with the same buffer, eluted in SDS-PAGE sample buffer, resolved by SDS-PAGE, and processed for Western blotting.
Invasion and cell viability assays
Invasion assays were performed in Boyden chambers using Transwell Permeable Inserts with 8.0-µm pores in 24-well plates (Corning). Matrigel (BD) was diluted to 0.25 µg/µl in serum-free media, and 50 µl was added to the top reservoir of the chamber using chilled pipette tips and allowed to solidify at 37°C for 1 h. MCF10A or MDA-MB-231-1833TR cells were grown to 100% confluence the day of the assay, trypsinized, and resuspended in serum-free media containing 0.1% BSA, and 2 × 104 cells in 100 µl were added to the top reservoir while 650 µl serum-containing media was added to the bottom chamber. The chambers were then placed at 37°C for 14 h. Cells from the top reservoir that did not invade were removed using a cotton swab. Invading cells were fixed in 4% PFA at room temperature for 15 min and stained in 0.2% crystal violet at room temperature for 15 min. Excess stain was removed by rinsing in water, and dried cells were imaged using a microscope (Axio Observer.Z1; Carl Zeiss) equipped with a Fluar 10× air objective (NA 0.5) with transmitted light and a digital camera (EOS Rebel XS f/18-55 mm-IS; Canon). Acquisition and analyses were performed using EOS Utility software version 2.8 (Cannon). The number of invaded cells per field of view was quantified. To measure viability, MCF10A or MDA-MB-231-1833TR cells were grown to 100% confluence for the day of the assay, trypsinized, and resuspended in serum-free media containing 0.1% BSA. Cells were stained with Trypan blue, and the number of viable cells was quantified on a hemocytometer. For Geltrex invasion assays, 20 µl Geltrex-reduced growth factor basement membrane (Life Technologies) was spread into each well of an 8-well chambered coverglass (Lab-Tek II; Thermo Fisher Scientific) that had been pretreated for 10 min with 100% ethanol. After 30 min at 37°C, 1.5 × 103 cells/well were plated in complete media containing 2% Geltrex and cultured for 4 d to form colonies. Media were removed, and colonies were covered with 100 µl of 2.5-mg/ml rat tail collagen I (Invitrogen) in DMEM, 0.7% NaHCO3, and 0.1 M Hepes and incubated 1 h at 37°C. 300 µl of complete media/well was added, and invasion proceeded for 72 h (37°C at 5% CO2). Images were collected using a microscope (Axiovert 200M; Carl Zeiss) equipped with a long-distance A-PLAN 20× Ph1 objective (NA 0.3) with transmitted light and a camera (AxioCam HRm; Carl Zeiss). Quantification was performed using MetaMorph software version 7.6.4 (Molecular Devices).
Proliferation and cell migration assays
For short-time proliferation assays, Matrigel was diluted to 0.25 µg/µl in serum-free media, and 300 µl was used to coat glass coverslips. Aliquots of MCF10A or MDA-MB-231-1833TR cells (2.4 × 105 cells in 600 µl serum-free media) were added and incubated for 14 h at 37°C. Cells were fixed with 4% PFA and stained with Hoechst. The total number of cells per field of view was quantified. For longer term proliferation assays, 500 MDA-MB-231-1833TR cells per well were plated in a 96-well plate and imaged every hour for 5 d using the kinetic live-cell imaging system (IncuCyte; Essen Biosciences). The cell confluency was determined using the IncuCyte software. Cell migration distances were tracked using MetaMorph software version 7.6.4.
Cellular imaging and protrusion assays
For live-cell imaging, MCF10A cells were plated at 60% confluence on poly-l-lysine–coated 35-mm No. 1.5 glass-bottom dishes (MatTek Corporation) in complete MCF10A media. Cells were transfected with DNA constructs using jetPRIME reagent and incubated for 12–14 h. Live-cell imaging was performed using a microscope (Axio Observer.Z1) equipped with a Plan Apochromat 40× oil objective (NA 1.4), Definite Focus system (Carl Zeiss), and a camera (AxioCam MR3; Carl Zeiss). Cells were kept at 37°C in 5% CO2 using Incubation System S (PeCon). GFP and mCh were imaged using 470- and 591-nm laser lines, respectively, from an illumination source (Colibri.2; Carl Zeiss). Acquisition and analyses were performed using ZEN 11.0 software (Carl Zeiss), whereas videos were made using ImageJ 1.43m (National Institutes of Health). MCF10A or HeLa cells were plated on poly-l-lysine–coated glass coverslips and transfected with DNA constructs using jetPRIME transfection reagent (Polyplus) according to the manufacturer’s instructions. For internalization of transferrin, transfected cells were serum starved for 1 h and incubated in serum-free media containing 25 µg/ml of labeled transferrin for 30 min. Cells were washed in PBS and fixed for 15 min in 3% PFA. After fixation, cells were permeabilized with 0.2% Triton X-100 in PBS and processed for immunofluorescence with the appropriate antibodies in PBS. Imaging was performed using a laser-scanning confocal microscope (LSM 710; Carl Zeiss) equipped with a Plan Apochromat 63× oil objective (NA 1.4; Carl Zeiss). Acquisition was performed using ZEN 11.0 software, and analysis was performed using ImageJ 1.43m. For differential interference contrast (DIC) microscopy, MCF10A cells were prepared on 35-mm No. 1.5 glass-bottom dishes. Imaging was performed using microscope (Axio Observer.Z1) equipped with a Plan Apochromat 20× oil objective (NA 0.8), DIC II 0.35/0.4 condenser module, and a camera (AxioCam MR3). Acquisition and analyses were performed using ZEN 11.0 software. For protrusion assays, healthy transfected cells were selected blind in the DAPI channel and imaged for GFP or mCh. The length of the longest protrusion was measured using ImageJ 1.43m. Pearson correlation coefficients were determined using the colocalization analysis plugin from McMaster Biophotonics Facility ImageJ for Microscopy bundle (Collins, 2007).
Rab13 biosensor, construction, and optimization
To make the Rab13 construct compatible with the vector system used to construct the biosensor (Pertz et al., 2006), the internal restriction sites NcoI and XhoI were silenced using site-directed mutagenesis. Primers used to generate Rab13 biosensors can be found in Table S1. The RBD of MICAL-L2 (aa 808–904) and Rab13 (wild type, Q67L, and T22N) were PCR amplified, digested, and ligated into the biosensor backbone system as shown in Fig. S3 A. Constitutively active Rab13 biosensor (Q67L mutant version) was modified to abrogate the binding of the integral RBD to Rab13 GTPase by exchanging the RBD with the CRIB domain of PAK 1, containing mutations to eliminate any GTPase binding effects (H83/86D and W103D mutations; Moshfegh et al., 2014). Point mutagenesis was performed using the PCR mutagenesis kit (QuikChange; Agilent Technologies). The intramolecular linker length between the mCer3 and mVen fluorescent proteins (Fig. S3 A) was optimized for the best FRET ratio difference between the constitutively active (Q67L) versus the dominant-negative (T22N) versions of the biosensor, by using cassettes of 18–amino acid flexible, structure-less, and protease-resistant linker encoding the sequence TSGSGKPGSGEGSTKGGS (Whitlow et al., 1993) in increments between 1 and 5 linker units. To further optimize the FRET dipole-coupling angles, circularly permutated Venus fluorescent proteins were tested on the 3-linker version of the biosensor. The circularly permutated mVen (cp49, 157, 173, 195, and 229; Nagai et al., 2004) fluorescent proteins were PCR amplified, digested with NotI and EcoRI, and ligated into position. For fluorometric measurements, the Rab13 biosensors were expressed in HEK-293T cells using Lipofectamine 2000 following the manufacturer’s protocols (Invitrogen). In situations in which RabGDI or DENND2B constructs were not used, an empty pCDNA3.1 vector was used as a DNA control. The DNA constructs were transfected with the cDNA concentration ratio of 1:4:4 for the biosensor/GDI/GEF. The procedure for the fluorometry followed the previously published protocols (Pertz et al., 2006). In brief, at 48 h after transfection, cells were briefly trypsinized and detached into cold PBS. The cell suspensions were transferred to a cuvette, and the fluorometric measurements were performed. Emission spectra between 450 and 600 nm were obtained upon 433-nm excitation of the cell suspensions in a spectrofluorometer (Fluorolog-MF2; Horiba Jobin-Yvon). The mock-transfected cell suspension was used to obtain the background fluorescence response, which was subtracted from the raw data. The emission maximum value at 475 nm was used to normalize the entire spectrum, and the changes in the FRET emission at 525 nm were quantified.
Acceptor photobleaching analysis
Constitutively active (Q67L) and dominant-negative (S22N) Rab13 biosensors were transiently transfected into HEK293T cells using the JetPRIME reagent following the manufacturer’s protocols. Cells were plated onto coverslips and imaged with an inverted microscope (IX81-ZDC; Olympus) equipped with an 40× oil immersion DIC objective lens (NA 1.3) and a charge-coupled device camera (CoolSNAP ESII; Roper Scientific), using the fluorescence excitation and emission optical configuration as used for imaging of the FRET biosensor MCF10A and MEF cell lines. The mCer3 fluorescence emission intensities were recorded from 10 random fields of view followed by additional 10 random fields of view but postirradiated through a filter (FF01-525/50-25; Semrock) for 2 min to photobleach the mcp229Venus, using the 100-W Hg Arc lamp as the light source. The flat-field–corrected and background-subtracted mCer3 images before and after photobleaching were analyzed for the mean per pixel gray levels, and the FRET efficiency limits were calculated using the following equation:
Live-cell imaging of the Rab13 biosensor
MCF10A cells and MEFs were plated on poly-l-lysine–coated coverslips and fibronectin-coated coverslips, respectively, and transfected with the Rab13 biosensor using JetPRIME. MCF10A were imaged after a 14-h transfection, and MEFs were imaged after an 8-h transfection. Imaging was performed in Ham’s F-12K without phenol red (Biosource) containing 2% fetal bovine serum and 5% horse serum for MEFs and MCF10A, respectively, in a closed chamber heated to 37°C. Activation of Rab13 was determined by monitoring the ratio of FRET between mCer3 to mVen and the donor mCer3 intensities for the Rab13 sensor (Pertz et al., 2006; Bravo-Cordero et al., 2011). Time-lapse sequences were acquired using an inverted microscope (IX81-ZDC) equipped with two internal beam splitters (T555LPXR for the internal prism replacement and T505LPXR for the FRET/CFP split in the external beam splitter attached to the left side camera port; Chroma Technology Corp.) to allow simultaneous acquisition of both FRET and mCer3 channels using two charge-coupled device cameras (CoolSNAP ESII) operating at 2 × 2 binning mode (Spiering et al., 2013). Images were acquired using a 60× Plan Apochromat N (1.45 NA) objective (Olympus) and MetaMorph version 7.1.5 software. The filter sets used were ET436/20X (excitation) and ET480/40M (emission) for mCer3 and ET436/20X (excitation) and ET535/30M (emission) for FRET (Chroma Technology Corp.). Cells were illuminated with a 100-W mercury lamp through 5–10% transmittance neutral density filters. mCer3 and FRET images were recorded simultaneously for 700–1,000-ms exposure times. Imaging and postacquisition processing were performed using MetaMorph version 7.8.8 software and Matlab R2012b (MathWorks) as previously described (Hodgson et al., 2006, 2008, 2010). In brief, camera noise image was subtracted from the raw mCer3 and FRET image stacks followed by a flat-field correction using a set of shading images acquired from cell-free fields of view under identical imaging conditions as the actual raw image data stacks. The flat-field correction corrects for the uneven illumination in the field of view, stemming primarily from the defocusing of the arc lamp source used. This is performed by dividing the camera noise–subtracted raw data images by the corresponding shading images of each channel. The camera noise–subtracted and flat field–corrected images are then convolved against a 3 × 3 median filter kernel (Matlab medfilt2.m) to eliminate the hot pixels. The program to perform the steps up to this point is available as scImage.m (Spiering et al., 2013). The camera noise–subtracted, flat field–corrected, and median-filtered image stacks were then processed for the background subtraction. Because of the flat-field correction, the cell-free regions within the field of view should have near uniform intensity values; thus, the cell-free background intensity values within a small region of interest is averaged and subtracted from the entire field of view and repeated for every time point of the time-lapse data stacks in both mCer3 and FRET sets. This process is performed using MetaMorph software using the statistical correction tool. The background-subtracted images are corrected for pixel-by-pixel matching to eliminate alignment-related artifacts stemming from small differences in the mounting angles of the two cameras (also including imperfections in the mounting angles of the dichromatic mirrors and emission filters in the light path) used during the simultaneous imaging. To do this, the misalignments in the mCer3 and FRET channels are characterized using a calibration slide containing multispectral beads (TetraSpeck microspheres; Invitrogen). The calibration images are used to determine the nonlinear coordinate transformation parameters, using Matlab programs tkMorph.m (Spiering et al., 2013) and the spot-detector (Thomann et al., 2002), which fits the Gaussian point-spread function to the fluorescent bead spots to determine the centroid location with subpixel accuracy. The coordinate transformation parameters obtained from this calibration steps are used to align the processed mCer3 and FRET image stacks, using an affine transformation (morph.m program in Matlab; Spiering et al., 2013). The coordinate-transformed mCer3 and FRET image stacks were intensity thresholded using the thresholding tool in the MetaMorph software to generate binary masks. The binary masks are then multiplied into the corresponding image stacks, which sets the regions outside of the cell to 0, thus eliminating the outside-of-cell noise during the final ratiometric processing. Before the ratiometric calculation, the binary-masked image stacks of mCer3 and FRET channels are globally optimized against each other at subpixel accuracy by an x-y translational image alignment tool based on the normalized cross-correlation method using the Matlab program regFC2.m (Shen et al., 2006; Spiering et al., 2013). This second level of image alignment after the nonlinear coordinate transformation procedure is necessary because the image cropping step after processing of the coordinate transformation may produce a slight x-y misalignment depending on the severity of the nonlinear correction applied. The ratio stack of images was obtained by dividing the aligned and masked FRET image by the masked mCer3 image. The time-lapse ratio image set was corrected for photobleaching by modeling the intensity decay in the ratio as a second order exponential function, accounting for the two photobleaching components as previously described (Hodgson et al., 2006). This assumption is based on the idea that the majority of GTPases in cytosol exists in complex with the GDI and thus is a passive component in the modulation of the ratio signals and that only a very small fraction of the total GTPase goes through activation cycles at any given time (Del Pozo et al., 2002). The whole-cell mean ratio values are calculated, and the double exponential fit is performed using a Matlab program bleachcorrectWhole.m (Shen et al., 2006; Spiering et al., 2013). The inverse of the resulting double exponential function is then multiplied at each time point of the ratio image stack to obtain the corrected ratio data. To test for the influence of cell volume on ratio measurements, line scans were made perpendicular to the cell edge in six cells from three independent experiments, and the following parameters were measured, averaged, and normalized arbitrarily to a position 12 µm away from the edge: (a) EGFP was nonspecifically expressed in the cytoplasm of parental MCF10A cells without biosensor to determine the mean cell volume distribution at the edge; (b) mCer3 intensities were measured in the biosensor expressing MCF10A cells to reflect the distribution of the Rab13 biosensor; and (c) ratio intensities of the biosensor measurements were taken in MCF10A cells expressing the wild-type Rab13 biosensor. The predicted FRET/mCer3 was calculated using the previously described calculation (Spiering et al., 2013):
The optical bleed through from the mCer3 to the FRET channel on our imaging system was assumed to be a constant and at 0.6:1.0 (FRET/mCer3). We calculated the theoretical distribution of the ratio values if it was based on the volume and the biosensor distribution alone. Where mCer3 intensity is experimentally determined, ρFRET and ρmCer3 describes the optical bleed through of 0.6 and 1.0, respectively, from our optical configuration, Ah is the volume as measured by the GFP distribution and averaged over 10 cells, and α-factor is a parameter to match the maximum ratio between the model predictions and the actual ratio.
Analysis of tumor growth and migration in vivo
Female nude mice (6–8 wk old; Taconic Farms, Inc.) were injected via the tail vein with 7.5 × 104 MDA-MB-231-1833TR cells (vector control, n = 10 mice; Rab13 shRNA 1, n = 9 mice; and Rab13 shRNA 2, n = 9 mice). Luciferase activity in the lungs was confirmed and measured immediately after injection using bioluminescence imaging, and mice were imaged once per week thereafter to monitor the development of metastases. For imaging, mice were anaesthetized with isoflurane and injected intraperitoneally with 50 µl of the luciferase substrate d-luciferin (PerkinElmer; 30 mg/ml in PBS), and bioluminescence was detected at 1-min intervals for 10 min using Xenogen IVIS 100 (Caliper Life Sciences, Inc.) in batches of five mice. All images to be compared were opened as a group using Living Image Version 4.2 software (PerkinElmer) to allow visualization on a uniform scale and analysis using the Measurement ROI tool to quantify all detectable regions of luciferase activity in the body of each mouse. The resulting sum total luminescence signal per mouse was normalized to the signal generated by the initial cell inoculum for that mouse as measured on the day of injection. Tumor outgrowth was then expressed as a fold change in luminescence signal. Mice negative for luciferase activity at day 63 were excluded from tumor outgrowth quantification. Similarly, two mice without reliable quantification as a result of oversaturation (one shRNA control and one Rab13 shRNA 1) were excluded from quantitative analysis. Tumor “take” was considered positive if luciferase activity was detectable anywhere in the animal at 63 d after injection. Mice were sacrificed 10 wk after injection. Metastases were confirmed and counted at necropsy and by histological analysis. Tissue samples for histology were fixed for 24 h in 10% formalin, embedded in paraffin, and sectioned at 5 µm. Sections were stained with hematoxylin and eosin and imaged using a slide scanner (Aperio-XT; Aperio Technologies). The number of lesions in the lungs was quantified blind. All animal experiments were performed in accordance with the guidelines of the McGill University Animal Ethics Committee and the Canadian Council on Animal Care as a protocol approved by the Facility Animal Care Committee (Protocol #5562).
Online supplemental material
Fig. S1 shows the interaction between DENND2B and Rab13 by GST pull-down and coimmunoprecipitation assays. Fig. S2 shows DENND2B levels in MCF10A cells, expression of RFP-Rab13 constructs in MCF10A cells, MCF10A morphology changes induced by RFP-Rab13 Q67L, and controls (cell number and cell viability) for invasion assays in MCF10A cells. Fig. S3 shows the optimization and validation of the Rab13 biosensor. Fig. S4 shows the bioluminescence images of all mice injected with cancer cells used in this study. Fig. S5 shows the alteration frequency of RAB13 across several cancer types. Table S1 contains all oligonucleotides used in this study. Video 1 shows MCF10A cells overexpressing Rab13 Q67L invading through a collagen matrix. Video 2 shows transport of wild-type mCh-Rab13 vesicles toward the leading edge of a cell. Video 3 shows wild-type mCh-Rab13 vesicles associating with active membrane ruffles. Video 4 shows transport of mCh-Rab13 Q67L vesicles toward the leading edge of a cell. Video 5 shows the failure of mCh-Rab13 T22N vesicles to accumulate on the plasma membrane despite trafficking from the perinuclear pool to the cell surface. Video 6 shows ratiometric imaging of the Rab13 biosensor revealing activation at growing protrusions. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201407068/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201407068.dv.
Supplementary Material
Acknowledgments
We thank Dr. Pietro De Camilli and Dr. Andrea L. Marat for comments on the manuscript, Dr. Jonathan M. Lee for helpful discussion, and Jacynthe Philie for excellent technical assistance. We thank Dr. William Balch for the RabGDI construct, Dr. Steve Caplan for the MICAL constructs, Dr. Fred Meunier for GFP-Rab7 and GFP-Rab9, and Dr. Peter Siegel for the MDA-MB-231-1833TR cell line.
This work was supported by grant MOP-62684 from the Canadian Institutes of Health Research (CIHR) to P.S. McPherson and National Institutes of Health grant GM093121 to L. Hodgson. M.S. Ioannou was supported by a Frederick Banting and Charles Best Canada Graduate Scholarship from CIHR. P.S. McPherson is a James McGill Professor.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AF647
- Alexa Fluor 647
- ANOVA
- analysis of variance
- CRIB
- Cdc42/Rac1 interactive binding
- DIC
- differential interference contrast
- FRET
- Förster resonance energy transfer
- GDI
- GDP dissociation inhibitor
- GEF
- guanine-nucleotide exchange factor
- mCer3
- monomeric Cerulean3
- mCh
- mCherry
- MEF
- mouse embryonic fibroblast
- mVen
- monomeric Venus
- PAK
- p21-activated kinase
- RBD
- Rab13-binding domain
- TPA
- 12-O-tetradecanoylphorbol-13-acetate
References
- Abou-Zeid N., Pandjaitan R., Sengmanivong L., David V., Le Pavec G., Salamero J., and Zahraoui A.. 2011. MICAL-like1 mediates epidermal growth factor receptor endocytosis. Mol. Biol. Cell. 22:3431–3441 10.1091/mbc.E11-01-0030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allaire P.D., Marat A.L., Dall’Armi C., Di Paolo G., McPherson P.S., and Ritter B.. 2010. The Connecdenn DENN domain: a GEF for Rab35 mediating cargo-specific exit from early endosomes. Mol. Cell. 37:370–382 10.1016/j.molcel.2009.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allaire P.D., Seyed Sadr M., Chaineau M., Seyed Sadr E., Konefal S., Fotouhi M., Maret D., Ritter B., Del Maestro R.F., and McPherson P.S.. 2013. Interplay between Rab35 and Arf6 controls cargo recycling to coordinate cell adhesion and migration. J. Cell Sci. 126:722–731 10.1242/jcs.112375 [DOI] [PubMed] [Google Scholar]
- Baldassarre M., Razinia Z., Burande C.F., Lamsoul I., Lutz P.G., and Calderwood D.A.. 2009. Filamins regulate cell spreading and initiation of cell migration. PLoS ONE. 4:e7830 10.1371/journal.pone.0007830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-Cordero J.J., Oser M., Chen X., Eddy R., Hodgson L., and Condeelis J.. 2011. A novel spatiotemporal RhoC activation pathway locally regulates cofilin activity at invadopodia. Curr. Biol. 21:635–644 10.1016/j.cub.2011.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burman J.L., Bourbonniere L., Philie J., Stroh T., Dejgaard S.Y., Presley J.F., and McPherson P.S.. 2008. Scyl1, mutated in a recessive form of spinocerebellar neurodegeneration, regulates COPI-mediated retrograde traffic. J. Biol. Chem. 283:22774–22786 10.1074/jbc.M801869200 [DOI] [PubMed] [Google Scholar]
- Caswell P.T., and Norman J.C.. 2006. Integrin trafficking and the control of cell migration. Traffic. 7:14–21 10.1111/j.1600-0854.2005.00362.x [DOI] [PubMed] [Google Scholar]
- Caswell P., and Norman J.. 2008. Endocytic transport of integrins during cell migration and invasion. Trends Cell Biol. 18:257–263 10.1016/j.tcb.2008.03.004 [DOI] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2:401–404 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins T.J.2007. ImageJ for microscopy. Biotechniques. 43(Suppl. 1):S25–S30 10.2144/000112517 [DOI] [PubMed] [Google Scholar]
- Debnath J., Muthuswamy S.K., and Brugge J.S.. 2003. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 30:256–268 10.1016/S1046-2023(03)00032-X [DOI] [PubMed] [Google Scholar]
- Del Pozo M.A., Kiosses W.B., Alderson N.B., Meller N., Hahn K.M., and Schwartz M.A.. 2002. Integrins regulate GTP-Rac localized effector interactions through dissociation of Rho-GDI. Nat. Cell Biol. 4:232–239 10.1038/ncb759 [DOI] [PubMed] [Google Scholar]
- Fanning A.S., Jameson B.J., Jesaitis L.A., and Anderson J.M.. 1998. The tight junction protein ZO-1 establishes a link between the transmembrane protein occludin and the actin cytoskeleton. J. Biol. Chem. 273:29745–29753 10.1074/jbc.273.45.29745 [DOI] [PubMed] [Google Scholar]
- Fukuda M., Kanno E., Ishibashi K., and Itoh T.. 2008. Large scale screening for novel rab effectors reveals unexpected broad Rab binding specificity. Mol. Cell. Proteomics. 7:1031–1042 10.1074/mcp.M700569-MCP200 [DOI] [PubMed] [Google Scholar]
- Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenring J.R.2013. A central role for vesicle trafficking in epithelial neoplasia: intracellular highways to carcinogenesis. Nat. Rev. Cancer. 13:813–820 10.1038/nrc3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goody R.S., Rak A., and Alexandrov K.. 2005. The structural and mechanistic basis for recycling of Rab proteins between membrane compartments. Cell. Mol. Life Sci. 62:1657–1670 10.1007/s00018-005-4486-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Roth D., Walch-Solimena C., and Novick P.. 1999. The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J. 18:1071–1080 10.1093/emboj/18.4.1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna S., Miskolci V., Cox D., and Hodgson L.. 2014. A new genetically encoded single-chain biosensor for Cdc42 based on FRET, useful for live-cell imaging. PLoS ONE. 9:e96469 10.1371/journal.pone.0096469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsay E., and Schekman R.. 2007. Avl9p, a member of a novel protein superfamily, functions in the late secretory pathway. Mol. Biol. Cell. 18:1203–1219 10.1091/mbc.E06-11-1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattula K., Furuhjelm J., Tikkanen J., Tanhuanpää K., Laakkonen P., and Peränen J.. 2006. Characterization of the Rab8-specific membrane traffic route linked to protrusion formation. J. Cell Sci. 119:4866–4877 10.1242/jcs.03275 [DOI] [PubMed] [Google Scholar]
- Hodgson L., Nalbant P., Shen F., and Hahn K.. 2006. Imaging and photobleach correction of Mero-CBD, sensor of endogenous Cdc42 activation. Methods Enzymol. 406:140–156 10.1016/S0076-6879(06)06012-5 [DOI] [PubMed] [Google Scholar]
- Hodgson L., Pertz O., and Hahn K.M.. 2008. Design and optimization of genetically encoded fluorescent biosensors: GTPase biosensors. Methods Cell Biol. 85:63–81 10.1016/S0091-679X(08)85004-2 [DOI] [PubMed] [Google Scholar]
- Hodgson L., Shen F., and Hahn K.. 2010. Biosensors for characterizing the dynamics of rho family GTPases in living cells. Curr. Protoc. Cell Biol. Chapter 14:Unit 14.11.1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P., Yang J., Hou Y., Zhang H., Zeng Z., Zhao L., Yu T., Tang X., Tu G., Cui X., and Liu M.. 2014. LncRNA expression signatures of twist-induced epithelial-to-mesenchymal transition in MCF10A cells. Cell. Signal. 26:83–93. [DOI] [PubMed] [Google Scholar]
- Hubbs A.E., Majidi M., and Lichy J.H.. 1999. Expression of an isoform of the novel signal transduction protein ST5 is linked to cell morphology. Oncogene. 18:2519–2525 10.1038/sj.onc.1202554 [DOI] [PubMed] [Google Scholar]
- Hussain N.K., Jenna S., Glogauer M., Quinn C.C., Wasiak S., Guipponi M., Antonarakis S.E., Kay B.K., Stossel T.P., Lamarche-Vane N., and McPherson P.S.. 2001. Endocytic protein intersectin-l regulates actin assembly via Cdc42 and N-WASP. Nat. Cell Biol. 3:927–932 10.1038/ncb1001-927 [DOI] [PubMed] [Google Scholar]
- Imamura H., Takaishi K., Nakano K., Kodama A., Oishi H., Shiozaki H., Monden M., Sasaki T., and Takai Y.. 1998. Rho and Rab small G proteins coordinately reorganize stress fibers and focal adhesions in MDCK cells. Mol. Biol. Cell. 9:2561–2575 10.1091/mbc.9.9.2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen K.R., Sørensen E., Brøndum K.K., Daugaard T.F., Thomsen R., and Nielsen A.L.. 2013. Direct RNA sequencing mediated identification of mRNA localized in protrusions of human MDA-MB-231 metastatic breast cancer cells. J. Mol. Signal. 8:9 10.1186/1750-2187-8-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda I., Nishimura N., Nakatsuji H., Yamamura R., Nakanishi H., and Sasaki T.. 2008. Involvement of Rab13 and JRAB/MICAL-L2 in epithelial cell scattering. Oncogene. 27:1687–1695 10.1038/sj.onc.1210812 [DOI] [PubMed] [Google Scholar]
- Kang Y., Siegel P.M., Shu W., Drobnjak M., Kakonen S.M., Cordón-Cardo C., Guise T.A., and Massagué J.. 2003. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 3:537–549 10.1016/S1535-6108(03)00132-6 [DOI] [PubMed] [Google Scholar]
- Kitano M., Nakaya M., Nakamura T., Nagata S., and Matsuda M.. 2008. Imaging of Rab5 activity identifies essential regulators for phagosome maturation. Nature. 453:241–245 10.1038/nature06857 [DOI] [PubMed] [Google Scholar]
- Klöpper T.H., Kienle N., Fasshauer D., and Munro S.. 2012. Untangling the evolution of Rab G proteins: implications of a comprehensive genomic analysis. BMC Biol. 10:71 10.1186/1741-7007-10-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight J.F., Lesurf R., Zhao H., Pinnaduwage D., Davis R.R., Saleh S.M., Zuo D., Naujokas M.A., Chughtai N., Herschkowitz J.I., et al. 2013. Met synergizes with p53 loss to induce mammary tumors that possess features of claudin-low breast cancer. Proc. Natl. Acad. Sci. USA. 110:E1301–E1310 10.1073/pnas.1210353110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine T.P., Daniels R.D., Gatta A.T., Wong L.H., and Hayes M.J.. 2013. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics. 29:499–503 10.1093/bioinformatics/bts725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levivier E., Goud B., Souchet M., Calmels T.P., Mornon J.P., and Callebaut I.. 2001. uDENN, DENN, and dDENN: indissociable domains in Rab and MAP kinases signaling pathways. Biochem. Biophys. Res. Commun. 287:688–695 10.1006/bbrc.2001.5652 [DOI] [PubMed] [Google Scholar]
- Li W., Li K., Zhao L., and Zou H.. 2014. Bioinformatics analysis reveals disturbance mechanism of MAPK signaling pathway and cell cycle in Glioblastoma multiforme. Gene. 547:346–350 10.1016/j.gene.2014.06.042 [DOI] [PubMed] [Google Scholar]
- Lichy J.H., Modi W.S., Seuanez H.N., and Howley P.M.. 1992. Identification of a human chromosome 11 gene which is differentially regulated in tumorigenic and nontumorigenic somatic cell hybrids of HeLa cells. Cell Growth Differ. 3:541–548. [PubMed] [Google Scholar]
- Lichy J.H., Majidi M., Elbaum J., and Tsai M.M.. 1996. Differential expression of the human ST5 gene in HeLa-fibroblast hybrid cell lines mediated by YY1: evidence that YY1 plays a part in tumor suppression. Nucleic Acids Res. 24:4700–4708 10.1093/nar/24.23.4700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippé R., Miaczynska M., Rybin V., Runge A., and Zerial M.. 2001. Functional synergy between Rab5 effector Rabaptin-5 and exchange factor Rabex-5 when physically associated in a complex. Mol. Biol. Cell. 12:2219–2228 10.1091/mbc.12.7.2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majidi M., Gutkind J.S., and Lichy J.H.. 2000. Deletion of the COOH terminus converts the ST5 p70 protein from an inhibitor of RAS signaling to an activator with transforming activity in NIH-3T3 cells. J. Biol. Chem. 275:6560–6565 10.1074/jbc.275.9.6560 [DOI] [PubMed] [Google Scholar]
- Marat A.L., and McPherson P.S.. 2010. The connecdenn family, Rab35 guanine nucleotide exchange factors interfacing with the clathrin machinery. J. Biol. Chem. 285:10627–10637 10.1074/jbc.M109.050930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marat A.L., Dokainish H., and McPherson P.S.. 2011. DENN domain proteins: regulators of Rab GTPases. J. Biol. Chem. 286:13791–13800 10.1074/jbc.R110.217067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marat A.L., Ioannou M.S., and McPherson P.S.. 2012. Connecdenn 3/DENND1C binds actin linking Rab35 activation to the actin cytoskeleton. Mol. Biol. Cell. 23:163–175 10.1091/mbc.E11-05-0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzesco A.M., Galli T., Louvard D., and Zahraoui A.. 1998. The rod cGMP phosphodiesterase delta subunit dissociates the small GTPase Rab13 from membranes. J. Biol. Chem. 273:22340–22345 10.1074/jbc.273.35.22340 [DOI] [PubMed] [Google Scholar]
- Medkova M., France Y.E., Coleman J., and Novick P.. 2006. The rab exchange factor Sec2p reversibly associates with the exocyst. Mol. Biol. Cell. 17:2757–2769 10.1091/mbc.E05-10-0917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I., and Yarden Y.. 2013. Endocytosis and cancer. Cold Spring Harb. Perspect. Biol. 5:a016949 10.1101/cshperspect.a016949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo W., Zhang J., Li X., Meng D., Gao Y., Yang S., Wan X., Zhou C., Guo F., Huang Y., et al. 2013. Identification of novel AR-targeted microRNAs mediating androgen signalling through critical pathways to regulate cell viability in prostate cancer. PLoS ONE. 8:e56592 10.1371/journal.pone.0056592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N., Yamashita S., Kurokawa K., Ohba Y., Nagai T., Miyawaki A., and Matsuda M.. 2001. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 411:1065–1068 10.1038/35082594 [DOI] [PubMed] [Google Scholar]
- Moshfegh Y., Bravo-Cordero J.J., Miskolci V., Condeelis J., and Hodgson L.. 2014. A Trio-Rac1-Pak1 signalling axis drives invadopodia disassembly. Nat. Cell Biol. 16:574–586 10.1038/ncb2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourskaia A.A., Amir E., Dong Z., Tiedemann K., Cory S., Omeroglu A., Bertos N., Ouellet V., Clemons M., Scheffer G.L., et al. 2012. ABCC5 supports osteoclast formation and promotes breast cancer metastasis to bone. Breast Cancer Res. 14:R149 10.1186/bcr3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T., Yamada S., Tominaga T., Ichikawa M., and Miyawaki A.. 2004. Expanded dynamic range of fluorescent indicators for Ca(2+) by circularly permuted yellow fluorescent proteins. Proc. Natl. Acad. Sci. USA. 101:10554–10559 10.1073/pnas.0400417101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbant P., Hodgson L., Kraynov V., Toutchkine A., and Hahn K.M.. 2004. Activation of endogenous Cdc42 visualized in living cells. Science. 305:1615–1619 10.1126/science.1100367 [DOI] [PubMed] [Google Scholar]
- Nishikimi A., Ishihara S., Ozawa M., Etoh K., Fukuda M., Kinashi T., and Katagiri K.. 2014. Rab13 acts downstream of the kinase Mst1 to deliver the integrin LFA-1 to the cell surface for lymphocyte trafficking. Sci. Signal. 7:ra72 10.1126/scisignal.2005199 [DOI] [PubMed] [Google Scholar]
- Nishimura N., and Sasaki T.. 2008. Identification and characterization of JRAB/MICAL-L2, a junctional Rab13-binding protein. Methods Enzymol. 438:141–153 10.1016/S0076-6879(07)38010-5 [DOI] [PubMed] [Google Scholar]
- Nokes R.L., Fields I.C., Collins R.N., and Fölsch H.. 2008. Rab13 regulates membrane trafficking between TGN and recycling endosomes in polarized epithelial cells. J. Cell Biol. 182:845–853 10.1083/jcb.200802176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nookala R.K., Langemeyer L., Pacitto A., Ochoa-Montaño B., Donaldson J.C., Blaszczyk B.K., Chirgadze D.Y., Barr F.A., Bazan J.F., and Blundell T.L.. 2012. Crystal structure of folliculin reveals a hidDENN function in genetically inherited renal cancer. Open Biol. 2:120071 10.1098/rsob.120071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshimura M., Kugoh H., Koi M., Shimizu M., Yamada H., Satoh H., and Barrett J.C.. 1990. Transfer of a normal human chromosome 11 suppresses tumorigenicity of some but not all tumor cell lines. J. Cell. Biochem. 42:135–142 10.1002/jcb.240420304 [DOI] [PubMed] [Google Scholar]
- Pertz O., Hodgson L., Klemke R.L., and Hahn K.M.. 2006. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 440:1069–1072 10.1038/nature04665 [DOI] [PubMed] [Google Scholar]
- Pfeffer S.R., Dirac-Svejstrup A.B., and Soldati T.. 1995. Rab GDP dissociation inhibitor: putting rab GTPases in the right place. J. Biol. Chem. 270:17057–17059 10.1074/jbc.270.29.17057 [DOI] [PubMed] [Google Scholar]
- Pignatelli J., Tumbarello D.A., Schmidt R.P., and Turner C.E.. 2012. Hic-5 promotes invadopodia formation and invasion during TGF-β–induced epithelial–mesenchymal transition. J. Cell Biol. 197:421–437 10.1083/jcb.201108143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A.J.2011. Life at the leading edge. Cell. 145:1012–1022 10.1016/j.cell.2011.06.010 [DOI] [PubMed] [Google Scholar]
- Ritter B., Murphy S., Dokainish H., Girard M., Gudheti M.V., Kozlov G., Halin M., Philie J., Jorgensen E.M., Gehring K., and McPherson P.S.. 2013. NECAP 1 regulates AP-2 interactions to control vesicle size, number, and cargo during clathrin-mediated endocytosis. PLoS Biol. 11:e1001670 10.1371/journal.pbio.1001670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakane A., Honda K., and Sasaki T.. 2010. Rab13 regulates neurite outgrowth in PC12 cells through its effector protein, JRAB/MICAL-L2. Mol. Cell. Biol. 30:1077–1087 10.1128/MCB.01067-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakane A., Abdallah A.A., Nakano K., Honda K., Ikeda W., Nishikawa Y., Matsumoto M., Matsushita N., Kitamura T., and Sasaki T.. 2012. Rab13 small G protein and junctional Rab13-binding protein (JRAB) orchestrate actin cytoskeletal organization during epithelial junctional development. J. Biol. Chem. 287:42455–42468 10.1074/jbc.M112.383653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakane A., Alamir Mahmoud Abdallah A., Nakano K., Honda K., Kitamura T., Imoto I., Matsushita N., and Sasaki T.. 2013. Junctional Rab13-binding protein (JRAB) regulates cell spreading via filamins. Genes Cells. 18:810–822 10.1111/gtc.12078 [DOI] [PubMed] [Google Scholar]
- Shaw L.M.2005. Tumor cell invasion assays. Methods Mol. Biol. 294:97–105. [DOI] [PubMed] [Google Scholar]
- Shen F., Hodgson L., Rabinovich A., Pertz O., Hahn K., and Price J.H.. 2006. Functional proteometrics for cell migration. Cytometry A. 69A:563–572 10.1002/cyto.a.20283 [DOI] [PubMed] [Google Scholar]
- Spiering D., Bravo-Cordero J.J., Moshfegh Y., Miskolci V., and Hodgson L.. 2013. Quantitative ratiometric imaging of FRET-biosensors in living cells. Methods Cell Biol. 114:593–609 10.1016/B978-0-12-407761-4.00025-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H.2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10:513–525 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- Thomann D., Rines D.R., Sorger P.K., and Danuser G.. 2002. Automatic fluorescent tag detection in 3D with super-resolution: application to the analysis of chromosome movement. J. Microsc. 208:49–64 10.1046/j.1365-2818.2002.01066.x [DOI] [PubMed] [Google Scholar]
- Thomas S., Ritter B., Verbich D., Sanson C., Bourbonnière L., McKinney R.A., and McPherson P.S.. 2009. Intersectin regulates dendritic spine development and somatodendritic endocytosis but not synaptic vesicle recycling in hippocampal neurons. J. Biol. Chem. 284:12410–12419 10.1074/jbc.M809746200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Solimena C., Collins R.N., and Novick P.J.. 1997. Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J. Cell Biol. 137:1495–1509 10.1083/jcb.137.7.1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlow M., Bell B.A., Feng S.L., Filpula D., Hardman K.D., Hubert S.L., Rollence M.L., Wood J.F., Schott M.E., Milenic D.E., et al. 1993. An improved linker for single-chain Fv with reduced aggregation and enhanced proteolytic stability. Protein Eng. 6:989–995 10.1093/protein/6.8.989 [DOI] [PubMed] [Google Scholar]
- Wu X., Bradley M.J., Cai Y., Kümmel D., De La Cruz E.M., Barr F.A., and Reinisch K.M.. 2011. Insights regarding guanine nucleotide exchange from the structure of a DENN-domain protein complexed with its Rab GTPase substrate. Proc. Natl. Acad. Sci. USA. 108:18672–18677 10.1073/pnas.1110415108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura R., Nishimura N., Nakatsuji H., Arase S., and Sasaki T.. 2008. The interaction of JRAB/MICAL-L2 with Rab8 and Rab13 coordinates the assembly of tight junctions and adherens junctions. Mol. Biol. Cell. 19:971–983 10.1091/mbc.E07-06-0551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S., Gerondopoulos A., Linford A., Rigden D.J., and Barr F.A.. 2010. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J. Cell Biol. 191:367–381 10.1083/jcb.201008051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Iyer L.M., He F., and Aravind L.. 2012. Discovery of novel DENN proteins: Implications for the evolution of eukaryotic intracellular membrane structures and human disease. Front. Genet. 3:283 10.3389/fgene.2012.00283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K., Muroyama A., Underwood J., Leylek R., Ray S., Soderling S.H., and Lechler T.. 2013. Actin-related protein2/3 complex regulates tight junctions and terminal differentiation to promote epidermal barrier formation. Proc. Natl. Acad. Sci. USA. 110:E3820–E3829 10.1073/pnas.1308419110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.