Interaction between K17 and hnRNP K regulates CXCR3 signaling in an RSK-dependent fashion to promote epithelial tumor cell growth and invasion.
Abstract
High levels of the intermediate filament keratin 17 (K17) correlate with a poor prognosis for several types of epithelial tumors. However, the causal relationship and underlying mechanisms remain undefined. A recent study suggested that K17 promotes skin tumorigenesis by fostering a specific type of inflammation. We report here that K17 interacts with the RNA-binding protein hnRNP K, which has also been implicated in cancer. K17 is required for the cytoplasmic localization of hnRNP K and for its role in regulating the expression of multiple pro-inflammatory mRNAs. Among these are the CXCR3 ligands CXCL9, CXCL10, and CXCL11, which together form a signaling axis with an established role in tumorigenesis. The K17–hnRNP K partnership is regulated by the ser/thr kinase RSK and required for CXCR3-dependent tumor cell growth and invasion. These findings functionally integrate K17, hnRNP K, and gene expression along with RSK and CXCR3 signaling in a keratinocyte-autonomous axis and provide a potential basis for their implication in tumorigenesis.
Introduction
The type I keratin 17 (K17) is rapidly and robustly up-regulated in various types of epithelial cells after tissue injury and other forms of insult (McGowan and Coulombe, 1998) and in several diseases including psoriasis and cancer (Nielsen et al., 2004; Ide et al., 2012; Wang et al., 2013), where it has been used as a diagnostic and prognostic marker (DePianto et al., 2010; Karantza, 2011). K17 serves multiple roles in a cell-autonomous fashion in vivo, including regulation of cell growth signaling (Kim et al., 2006), tumorigenesis (DePianto et al., 2010; Sankar et al., 2013), and cytokine expression including the CXCR3 ligands CXCL9, CXCL10, and CXCL11 (DePianto et al., 2010). In a mouse model of tumorigenesis involving constitutive Hedgehog signaling in the skin, genetic ablation of Krt17 delays tumor onset in correlation with reduced keratinocyte proliferation, reduced inflammation, and a polarization of the immune response from a T helper cell type 1 (Th1)- and Th17-dominated to a Th2-dominated character (DePianto et al., 2010). In particular, the Krt17 status was found to impact mRNA transcript levels for cytokines known to participate in the pathogenesis of human basal cell carcinoma, including the CXCR3 ligands CXCL9, CXCL10, and CXCL11 (Lo et al., 2010), in a keratinocyte-autonomous fashion (DePianto et al., 2010).
Cytokine expression and inflammation play key roles in the development of chronic inflammatory disease and in the progression and metastasis of tumors (Tauler and Mulshine, 2009; Elinav et al., 2013). CXCR3, in particular, plays an essential role in epidermal inflammation, proliferation, and skin tumorigenesis (Winkler et al., 2011). Increased CXCR3 expression occurs in additional types of tumors, and its elevated expression has been linked to a worse prognosis in melanoma, colon, and breast cancer (Fulton, 2009). In skin psoriasis, which is in part driven by inflammation and various types of immune effectors, expression of CXCR3 and its ligands are significantly elevated (Chen et al., 2010). K17 has also been shown to contribute to the pathogenesis of psoriasis (Jin and Wang, 2014). Accordingly, the emerging connection between K17 and the expression of CXCR3 ligands and other pro-inflammatory cytokines (Lo et al., 2010) may also represent a defining step in hyperproliferative and inflammatory disorders related to tumorigenesis. How a cytoskeletal protein such as K17 regulates cytokine expression during tumorigenesis and related processes is a wide-open issue.
hnRNP K is a member of the heterogeneous nuclear ribonucleoprotein (hnRNP) family of DNA/RNA-binding proteins that can impact all steps involved in gene expression, from de novo transcription to translation (Bomsztyk et al., 2004; Chaudhury et al., 2010). hnRNPs are among the most abundant proteins in the nucleus and are ubiquitously expressed in all tissue types (Chaudhury et al., 2010). Depending on context, hnRNP K (and other hnRNPs) can participate in the regulation of a broad array of genes including ones mediating inflammation. Further, hnRNP K has been found to be overexpressed in many cancers, where it enhances cell proliferation and transformation (Mandal et al., 2001; Gao et al., 2013), and its cytoplasmic accumulation has been correlated with tumor cell growth and metastasis (Inoue et al., 2007; Chen et al., 2009). Here, we report on a novel mechanism whereby a physical and functional partnership between K17 and hnRNP K regulates CXCR3 signaling in a RSK (p90 ribosomal protein S6 kinase)-dependent fashion to promote tumor epithelial cell growth and invasion.
Results
K17 interacts with hnRNP K and is required for its cytoplasmic localization
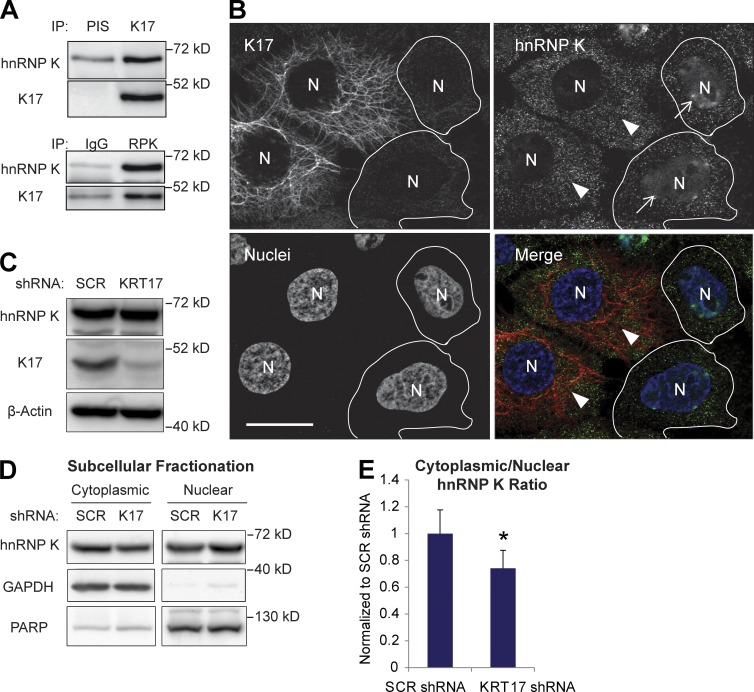
We combined immunoprecipitation (IP) with mass spectrometry (Chung et al., 2012) to identify K17-binding proteins with a demonstrated role in regulating gene expression. hnRNP K, a multifunctional protein that can impact all the steps involved in gene expression (Bomsztyk et al., 2004; Chaudhury et al., 2010) and plays a key role in tumorigenesis (Gao et al., 2013), was identified in this screen. The K17–hnRNP K interaction was confirmed by reciprocal co-IP in the A431 human epidermoid carcinoma cell line (Fig. 1 A). Another hnRNP protein, hnRNP A2/B1, did not co-IP with K17 (Fig. S1 A), which supports the specificity of the K17–hnRNP K interaction. hnRNP K also co-IPs with K5 (Fig. S1 B), a type II keratin binding partner for K17 (DePianto et al., 2010), and other keratins present in A431 cells (Fig. S1 C), which suggests that hnRNP K forms a complex with either keratin subunits or filaments. The keratin–hnRNP K interaction is markedly reduced upon KRT17 knockdown (Fig. S1, B and C), which otherwise does not impact keratin filament organization (not depicted). This suggests a specific requirement for K17 in the keratin–hnRNP K interaction.
Figure 1.
K17 binds to hnRNP K and regulates cytoplasmic localization of hnRNP K. (A) Co-IP of K17 and hnRNP K (RPK). IP was performed in A431 cells with anti-K17 or hnRNP K (RPK) antibody with preimmune serum (PIS) or IgG as a control. Immunoblotting was performed with antibodies against the indicated proteins. (B) Immunostaining of hnRNP K (green) and K17 (red) in A431 cells transduced with KRT17 shRNA. Nuclei (N) are shown in blue. Arrows indicate cells expressing KRT17 shRNA and arrowheads indicate cells (outlined) that did not become transduced with KRT17 shRNA. Bar, 10 µm. (C) Cell lysates from A431 cells stably expressing SCR or KRT17 shRNA were subjected to SDS-PAGE and immunoblotting was performed with the antibodies against the indicated proteins. (D) Subcellular fractionation of hnRNP K in A431 cells stably expressing SCR or KRT17 shRNA. Immunoblotting was performed with antibodies against the indicated proteins. PARP was used as a control for the nuclear fraction, whereas GAPDH was used for the cytoplasmic fraction. (E) Signal intensities of hnRNP K bands from D were quantitated and shown as the cytoplasmic-to-nuclear hnRNP K ratio. Data from three experimental repeats were normalized to A431 SCR shRNA cells and are represented as mean ± SEM (error bars). *, P < 0.03.
hnRNP K is predominantly nuclear in normal cells but also occurs in the cytoplasm of tumor cells, where it is purported to have a role in cancer progression and metastasis (Inoue et al., 2007; Zhou et al., 2010; Barboro et al., 2014). We surmised that K17, being a cytoskeletal protein, may play a role in regulating hnRNP K localization to the cytoplasm. Immunostaining for hnRNP K in A431 cells under mild detergent conditions (0.01% digitonin) reveals a modest but distinct signal in the cytoplasm (Fig. 1 B, arrowheads). The nuclear localization of hnRNP K is significantly enhanced in cells transduced with KRT17 shRNA (Fig. 1 B, arrows), whereas its total cellular levels are unaffected (Fig. 1 C). HeLa cervical cancer cells, which express endogenous K17 and hnRNP K, also showed decreased cytoplasmic localization of hnRNP K in K17-depleted cells (Fig. S1, D and E). Expression of a shRNA-resistant K17 in cells expressing KRT17 shRNA rescued decreased cytoplasmic and increased nuclear localization of hnRNP K in A431 cells, confirming the requirement of K17 on cytoplasmic localization of hnRNP K (Fig. S1 F). Subcellular fractionation of A431 cells confirmed the reduced cytoplasmic-to-nuclear ratio of hnRNP K in KRT17 shRNA-expressing cells compared with scrambled (SCR) shRNA control (Fig. 1, D and E). Virtually identical observations were made when comparing ear skin tissue from Krt17−/− Gli2tg/+ and Krt17+/+ Gli2tg/+ (Fig. S2, A and B), which show comparable levels of total hnRNP K protein (Fig. S2 C), and in primary cultures of newborn skin keratinocytes established from these sources (Fig. S2, D and E). These findings establish that K17 protein interacts with hnRNP K and enhances its cytoplasmic localization, raising the prospect that it may regulate its function in this setting.
hnRNP K binds to and regulates CXCR3 ligand transcripts
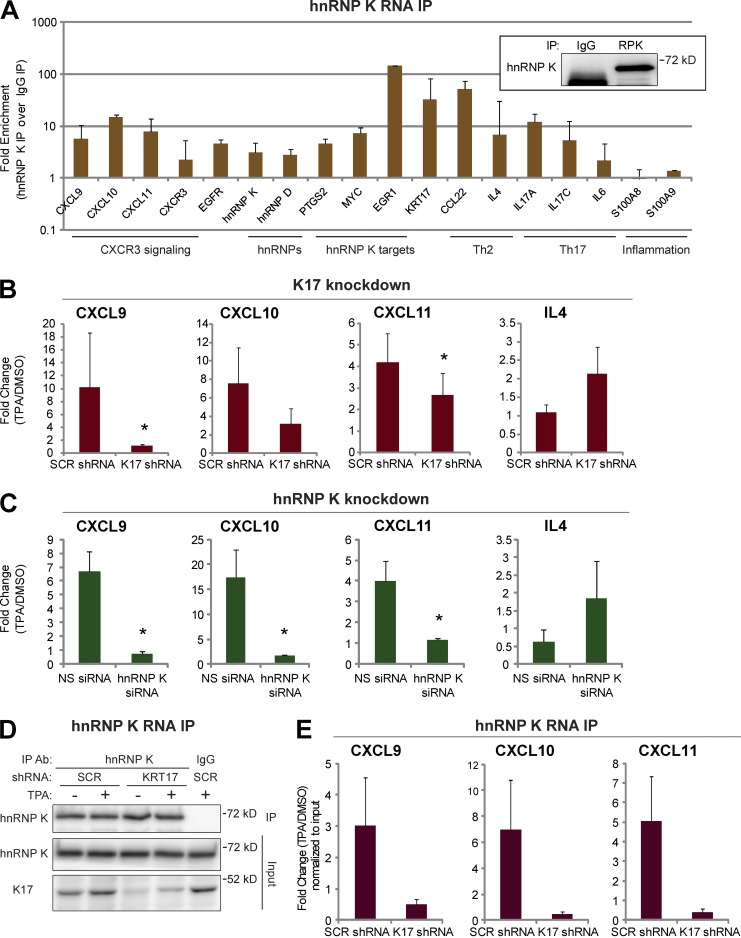
To probe whether hnRNP K binds mRNA transcripts shown to be regulated in a K17-dependent fashion, we next performed RNA IP (RIP) assays from A431 cells (Fig. 2 A) as well as Gli2tg/+ mouse skin keratinocytes in primary culture (Fig. S3 A). We isolated RNA from hnRNP K or control (IgG) IP and subjected it to quantitative RT-PCR (qRT-PCR) with target-specific primer sets. Consistent with the literature, the MYC (Evans et al., 2003), PTGS2 (Shanmugam et al., 2008), and EGR1 (Mikula and Bomsztyk, 2011) mRNAs are selectively enriched in hnRNP K IPs. Several pro-inflammatory mRNA transcripts, including CCL22, IL4, IL17A, IL17C, and KRT17, were newly identified as being highly enriched (fivefold or more) in hnRNP K IP samples (Fig. 2 A). Of particular interest, the CXCL9, CXCL10, and CXCL11 mRNAs, which are known to be regulated in a K17-dependent manner (DePianto et al., 2010), are highly enriched in hnRNP K IP samples from both mouse and human tumor keratinocytes.
Figure 2.
CXCR3 ligands are hnRNP K–bound and regulated in a K17-dependent manner. (A) RNA IP with anti–hnRNP K antibody or IgG control was performed in A431 cells. mRNA levels of the indicated genes from immunoprecipitates were measured using qRT-PCR. Fold enrichment normalized to IgG control is shown in a log scale. Data from five experimental repeats are represented as mean ± SEM (error bars). (inset) Immunoblotting showing hnRNP K (RPK) pull-down. IP with anti–hnRNP K (RPK) antibody or IgG was performed. Immunoblotting was performed with an hnRNP K antibody. (B) A431 cells stably expressing SCR or KRT17 shRNA were treated with 200 nM TPA or DMSO (vehicle control) for 1.5 h. mRNA levels of the indicated genes were measured using qRT-PCR. Data from nine experimental repeats were normalized to DMSO control and are represented as mean ± SEM (error bars). *, P < 0.05. (C) A431 cells transfected with NS or hnRNP K siRNA (RPK1) were treated with 200 nM TPA or DMSO (vehicle control) for 1.5 h. mRNA levels of the indicated genes were measured using qRT-PCR. Data from 10 experimental repeats were normalized to DMSO control and are represented as mean ± SEM (error bars). *, P < 0.04. (D) RNA IP with anti–hnRNP K antibody was performed in 200 nM TPA- or DMSO-treated (16 h) A431 cells stably expressing SCR or KRT17 shRNA. The Western blot shows the extent of hnRNP K pull-down. IP with anti–hnRNP K antibody or IgG was performed. Immunoblotting was performed with antibodies against the indicated proteins. (E) mRNA levels of the indicated genes from hnRNP K immunoprecipitates (D) were measured using qRT-PCR. Data from five experimental repeats were normalized to DMSO control and input, and are represented as mean ± SEM (error bars).
Treatment with 12-o-tetradecanoylphorbol-13-acetate (TPA) induces acute inflammation and up-regulates cytokine gene expression in a K17-dependent manner in mouse keratinocytes in vivo and in primary culture (DePianto et al., 2010). We find that TPA treatment of human A431 tumor keratinocytes enhances the physical interaction between K17 and hnRNP K (Fig. S3, B and C) and otherwise robustly induces the CXCL9, CXCL10, and CXCL11 mRNAs, and does so in a K17-dependent manner (Fig. 2 B). The expression of these transcripts under basal (unstimulated) conditions remained mostly unaffected by the K17 status (Fig. S3 D). Similar to Krt17−/− Gli2tg/+ mouse keratinocytes (DePianto et al., 2010), we find that induction of the Th2 cytokine IL4 mRNA was unimpeded in K17 knockdown cells, which supports an element of specificity between K17 and mRNA levels for CXCR3 ligands. We next modulated hnRNP K levels in the context of this paradigm, and investigated the associated impact on its bound transcripts. siRNA-mediated knockdown of hnRNP K (Fig. S4 A) results in decreased levels of CXCL9, CXCL10, and CXCL11 mRNAs upon TPA induction, whereas IL4 mRNA is increased (Fig. 2 C). Use of two additional siRNAs targeting different regions of HNRNPK confirms the requirement for normal levels of hnRNP K to maintain CXCL9, CXCL10, and CXCL11 mRNA levels in TPA-treated A431 cells (Fig. S4, B and C). Even under basal (unstimulated) conditions, the CXCL9, CXCL10, and CXCL11 mRNAs are down-regulated upon hnRNP K knockdown (Fig. S4 D). Together, these findings indicate that CXCL9, CXCL10, and CXCL11 transcripts are bona fide targets of hnRNP K, and that hnRNP K is required for their expression at normal levels.
K17 is required for hnRNP K–dependent regulation of CXCR3 ligands
We next investigated whether K17 and hnRNP K regulate CXCR3 ligand mRNAs in an interdependent fashion. We used RNA IP to assess the binding of hnRNP K to CXCL9, CXCL10, and CXCL11 transcripts in A431 cells with KRT17 knockdown (Fig. 2, D and E). Serum-deprived A431 cells stably expressing SCR or KRT17 shRNA were treated with DMSO or TPA, and hnRNP K IP was performed. Similar levels of hnRNP K were immunoprecipitated under all conditions, and CXCL9, CXCL10, and CXCL11 transcripts are robustly enriched in hnRNP K immunoprecipitates relative to IgG control, as expected (not depicted; see Fig. 2 A). In A431 cells expressing SCR shRNA (A431 SCR shRNA cells), TPA treatment enhances the levels of CXCL9, CXCL10, and CXCL11 mRNAs bound to hnRNP K. This phenomenon is markedly lessened in cells expressing KRT17 shRNA (A431 KRT17 shRNA cells; Fig. 2 E). Similar findings were obtained in A431 cells under basal unstimulated conditions (Fig. S4 E) as well as in primary cultures of newborn skin keratinocytes from HPV16tg mice (comparing K17+/+ with K17−/− substrains; unpublished data), which form tumors resembling human skin squamous cell carcinomas (Arbeit et al., 1994). hnRNP K thus requires K17 to bind the CXCL9, CXCL10, and CXCL11 mRNAs across several paradigms involving two species.
RSK regulates the K17–hnRNP K interaction and CXCR3 ligand expression
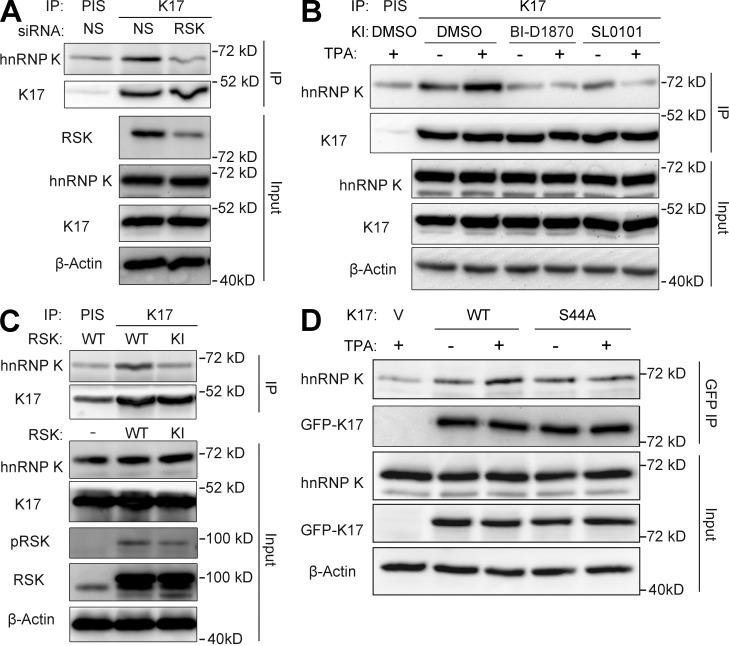
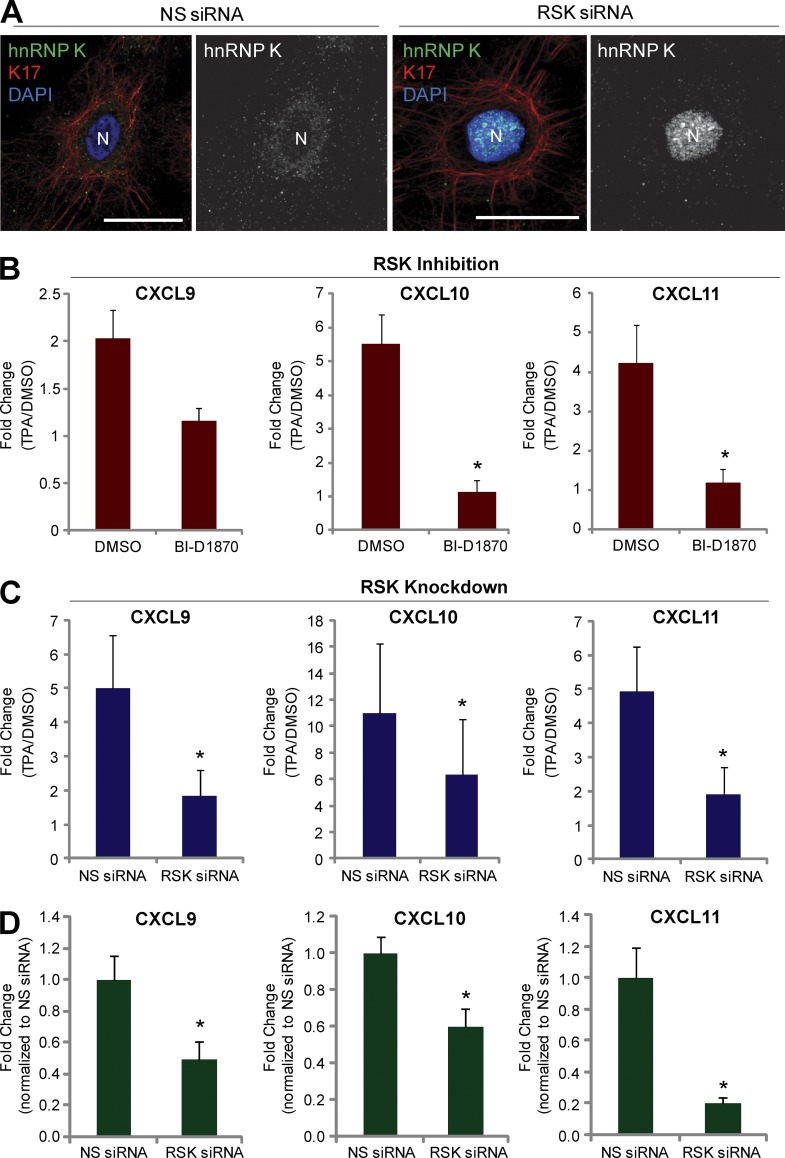
In addition to the aforementioned increase in the K17–hnRNP K interaction, TPA also activates RSK (Fig. S3 C), a regulator of various cellular processes including cell growth, proliferation, and motility (Anjum and Blenis, 2008). As we previously showed that K17 is phosphorylated by RSK (Pan et al., 2011), we next investigated whether RSK modulates the K17–hnRNP K interaction and CXCR3 ligand expression. siRNA-mediated RSK1 knockdown mitigated the physical interaction between hnRNP K and K17 (co-IP assays shown in Fig. 3 A). Further, treatment with the RSK inhibitors BI-D1870 or SL0101 before TPA induction (Fig. 3 B) or overexpression of a dominantly acting kinase-inactive (KI) RSK1 (Fig. 3 C) also hampered co-IP of K17 and hnRNP K in A431 cells, indicating a requirement for RSK kinase activity in the K17–hnRNP K interaction. Because RSK phosphorylates K17 on residue S44 (Pan et al., 2011), we tested the requirement of K17 phosphorylation by RSK on the K17–hnRNP K interaction. Plasmids encoding GFP-tagged wild-type (WT) or S44A K17 were transfected into A431 KRT17 shRNA cells. Exogenous K17 was then pulled down using anti-GFP–conjugated beads, and the extent of endogenous hnRNP K co-IP was measured. Similar to the data reported in Fig. S3 C, WT K17 showed enhanced complex formation with hnRNP K upon TPA treatment. In contrast, co-IP between hnRNP K–S44A K17 mutant did not increase after TPA stimulation, which suggests that S44 phosphorylation on K17 plays a role in the K17–hnRNP K interaction (Fig. 3 D). Given that S44 phosphorylation occurs concomitantly with other phosphorylation events on K17 (Pan et al., 2011), that the K17–hnRNPK complex features other keratins (Fig. S1 C), and that hnRNP K itself can be phosphorylated by multiple kinases (Bomsztyk et al., 2004; Chaudhury et al., 2010), other phospho-residues may be involved in regulating the K17–hnRNP K interaction. We next tested the requirement for RSK in the cytoplasmic localization of hnRNP K by performing immunofluorescence staining for hnRNP K in RSK1 siRNA-treated cells (Fig. 4 A). Compared with cells transfected with nonsilencing (NS) siRNA, cells transfected with RSK1 siRNA showed markedly increased nuclear hnRNP K staining, which indicates that RSK regulates the cytoplasmic/nuclear partitioning of hnRNP K. Finally, pretreatment with an RSK inhibitor (Fig. 4 B) or RSK knockdown resulted in decreased levels of CXCL9, CXCL10, and CXCL11 upon TPA induction (Fig. 4 C), even under basal unstimulated conditions (Fig. 4 D). Together, these findings establish that both the physical and functional interaction between K17 and hnRNP K and expression of CXCR3 ligands are subject to RSK-dependent regulation.
Figure 3.
RSK regulates the interaction between K17 and hnRNP K. (A) Co-IP of K17 and hnRNP K. IP with anti-K17 antibody or PIS as a control was performed in A431 cells transiently transfected with RSK or NS siRNA. Immunoprecipitates (IP) or inputs were subjected to SDS-PAGE, and immunoblotting was performed with antibodies against the indicated proteins. (B) Co-IP of K17 and hnRNP K. A431 cells pretreated with 2.5 µM BI-D 1870, 1.44 µM SL0101, or DMSO as a control for 2 h were treated with 200 nM TPA or DMSO (vehicle control) for 15 min. IP with anti-K17 antibody or PIS as a control was performed. Immunoprecipitates (IP) or inputs were subjected to SDS-PAGE and immunoblotting was performed with antibodies against the indicated proteins. (C) Co-IP of K17 and hnRNP K. IP with anti-K17 antibody or PIS as a control was performed in A431 cells transiently transfected with WT or KI-RSK. Immunoprecipitates (IP) or inputs were subjected to SDS-PAGE and immunoblotting was performed with antibodies against the indicated proteins. (D) Co-IP of K17 and hnRNP K. A431 KRT17 shRNA cells were transfected with pEGFP-C3 vector, K17 WT, or S44A mutant. Cells were treated with 200 nM TPA or DMSO (vehicle control) for 15 min, and IP with GFP-Trap_A was performed. Immunoprecipitates (IP) or inputs were subjected to SDS-PAGE and immunoblotting was performed with antibodies against the indicated proteins.
Figure 4.
RSK regulates hnRNP K localization as well as the expression of CXCR3 ligands. (A) Immunostaining of hnRNP K (green) and K17 (red) in A431 cells transfected with RSK or NS siRNA. Nuclei are shown in blue. Bars, 10 µm. (B) A431 cells pretreated with 2.5 µM BI-D 1870 or DMSO as a control for 2 h were treated with 200 nM TPA or DMSO (vehicle control) for 1.5 h. mRNA levels of the indicated genes were measured using qRT-PCR. Data from three experimental repeats were normalized to DMSO control and are represented as mean ± SEM (error bars). *, P < 0.05. (C and D) A431 cells transiently transfected with RSK or NS siRNA were treated with 200 nM TPA or DMSO (vehicle control) for 1.5 h (C) or grown in normal growth condition (D). mRNA levels of the indicated genes were measured using qRT-PCR. Data from three experimental repeats were normalized to DMSO control and are represented as mean ± SEM. *, P < 0.05. (D) mRNA levels for the indicated genes from A431 cells transiently transfected with RSK or NS siRNA under basal condition were measured using qRT-PCR. Data from three experimental repeats (n = 6) were normalized to NS siRNA control and are represented as mean ± SEM. *, P < 0.05.
K17 fosters the growth and invasiveness of tumor keratinocytes
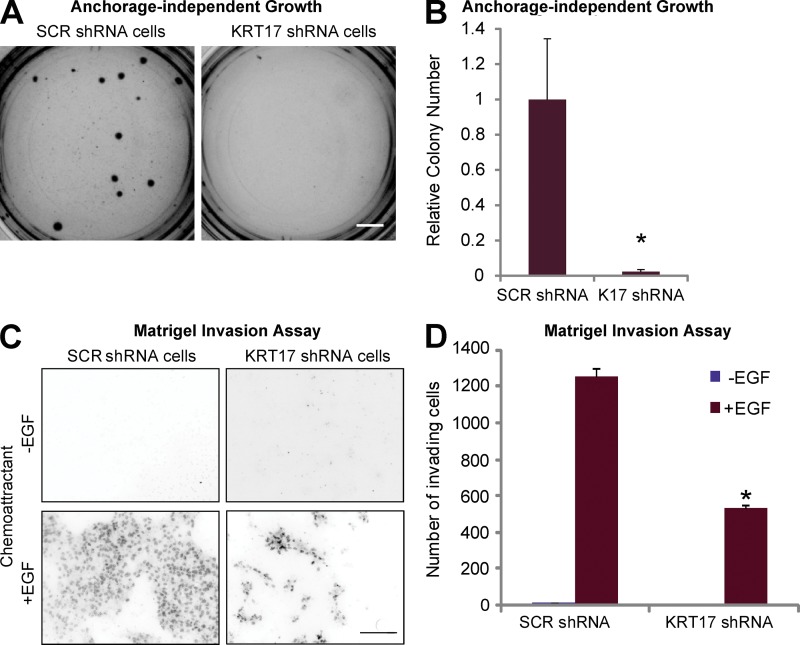
CXCR3 signaling (Winkler et al., 2011), K17 expression (DePianto et al., 2010), and cytoplasmic localization of hnRNP K (Inoue et al., 2007) have all been linked to tumor growth and/or invasiveness. We next investigated whether CXCR3 ligands play an active role in K17’s ability to promote a tumorigenic phenotype in two classical assays: cell growth in soft agar and Matrigel invasion. A431 SCR shRNA cells readily formed colonies after 3 wk of culture in soft agar, but this property is significantly lessened in A431 KRT17 shRNA cells (Fig. 5, A and B). Use of an additional shRNA (KRT17D shRNA) targeting KRT17 also showed decreased colony formation in the anchorage-independent growth assay, in a manner that is proportional to the extent of K17 down-regulation (Fig. S5 A), further supporting the requirement for K17 in the growth properties of tumor keratinocytes.
Figure 5.
K17 is required for tumor cell growth and cell invasion. (A) A431 cells stably expressing SCR or KRT17 shRNA were grown in soft agar, and phase-contrast images of colonies were taken after 3 wk. Bar, 1 cm. (B) Colony numbers from A were quantitated using ImageJ, and data from seven experimental repeats (each performed in triplicate) were normalized to SCR shRNA and are represented as mean ± SEM (error bars). *, P < 0.05. (C) A431 cells stably expressing SCR or KRT17 shRNA were subjected to a Matrigel invasion assay with or without 10 ng/ml EGF as a chemoattractant for 48 h. Nuclei of invaded cells were stained with propidium iodide and imaged under a fluorescent microscope. Bar, 100 µm. (D) The numbers of invaded cells from C were quantitated using ImageJ, and data from three experimental repeats (each performed in triplicate) are represented as mean ± SEM (error bars). *, P < 0.009.
In the Matrigel invasion assay (and in the presence of EGF), A431 KRT17 shRNA cells show significantly reduced invasion compared with the A431 SCR shRNA cells (Fig. 5, C and D). Expression of shRNA-resistant K17 rescues the invasion defect of KRT17 shRNA cells in the Matrigel invasion assay (Fig. S5 B). These findings confirm and extend a previous report that K17 is required for cell transformation and cellular adhesion (Sankar et al., 2013) and provide an ideal paradigm in which to test for, using conditioned medium, the involvement of secreted factors in these processes.
K17 and hnRNP K are required for the secretion of factors fostering the transformed phenotype
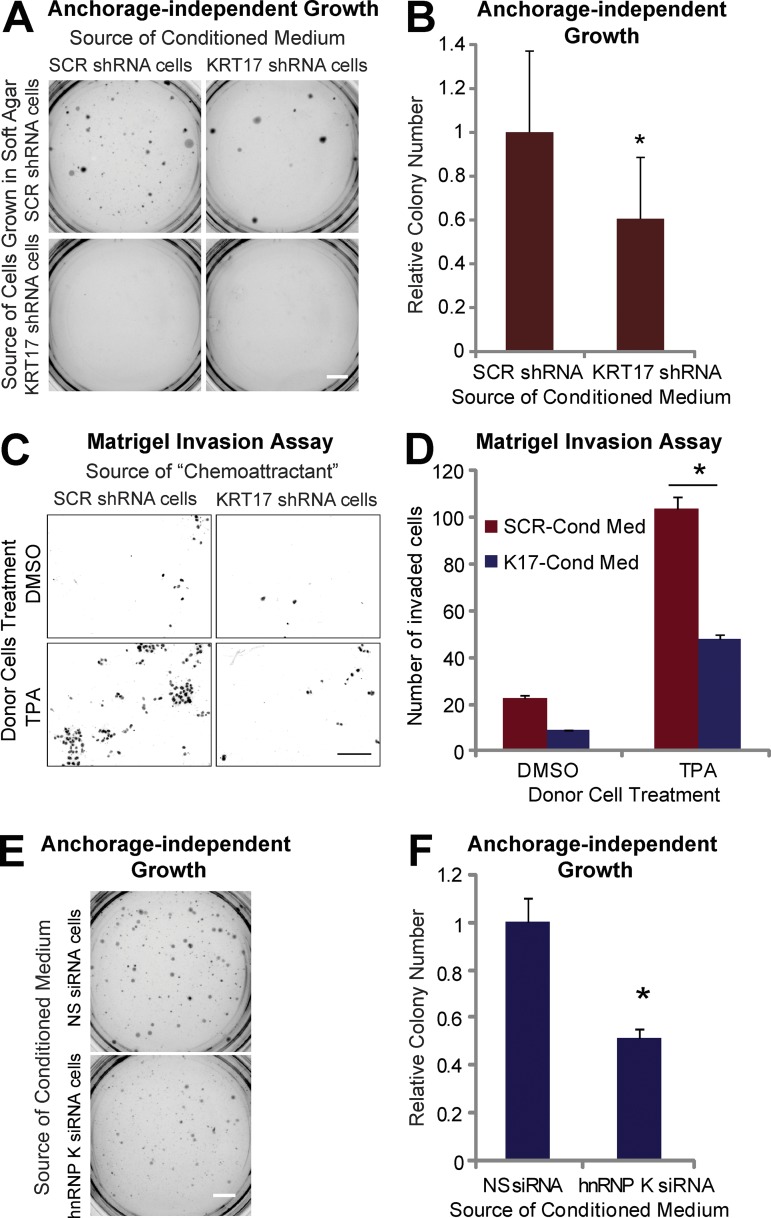
As CXCR3 ligands are secreted and can affect cell proliferation and invasion in a paracrine manner (Kawada et al., 2004; Groom and Luster, 2011; Winkler et al., 2011), conditioned medium from either SCR or KRT17 shRNA cells was used to feed A431 SCR or KRT17 shRNA cells cultured in soft agar, and colony formation was tested. A431 KRT17 shRNA cells did not form colonies in soft agar (see Fig. 5 A and Fig. 4 A), whereas A431 SCR shRNA cells grown in conditioned medium from SCR shRNA cells did (Fig. 6 A). In contrast, A431 SCR shRNA cells grown in conditioned medium from KRT17 shRNA cells showed a significantly smaller number of colonies (Fig. 6 B). The use of conditioned medium from cells transduced with a different shRNA against KRT17 (Fig. S5 C) or from cells with rescued K17 expression using shRNA-resistant KRT17 (Fig. S5 D) confirmed that K17, specifically, plays a significant role in the production of secreted factors that promote colony growth in this assay.
Figure 6.
K17 and hnRNP K are required for the secretion of factor(s) necessary for tumor cell growth. (A) Anchorage-independent growth assay using A431 cells stably expressing SCR or KRT17 shRNA in soft agar. Cells were grown in conditioned medium from A431 SCR or KRT17 shRNA cells. Bar, 1 cm. (B) Colony numbers of SCR shRNA cells grown in conditioned medium from SCR or KRT17 shRNA cells from A were quantitated using ImageJ and normalized to those grown in conditioned medium from A431 SCR shRNA cells. Data from nine experimental repeats (each performed in triplicate) are represented as mean ± SEM (error bars). *, P < 0.05. (C) Matrigel invasion assay of A431 SCR shRNA cells. Cells were allowed to invade using conditioned medium from TPA- or DMSO-treated A431 SCR or KRT17 shRNA cells as a chemoattractant for 48 h. Nuclei of invaded cells were stained with propidium iodide and imaged under a fluorescent microscope. Bar, 100 µm. (D) The number of invaded cells from C was quantitated using ImageJ and normalized to the cells exposed to conditioned medium from DMSO-treated A431 SCR shRNA cells. Data from three experimental repeats (each performed in triplicate) are represented as mean ± SEM (error bars). *, P < 0.01. (E) Anchorage-independent growth assay using A431 SCR shRNA cells in soft agar. Cells were grown in conditioned medium from A431 SCR shRNA cells transfected with NS (NS siRNA cells) or hnRNP K (RPK1; hnRNP K siRNA cells) siRNA. Bar, 1 cm. (F) Colony numbers from E were quantitated using ImageJ and normalized to cells grown in conditioned medium from cells expressing NS siRNA. Data from three experimental repeats (each performed in triplicate) are represented as mean ± SEM. *, P < 0.005.
We also used the Matrigel assay to monitor invasion of A431 SCR shRNA cells when conditioned medium from TPA- or DMSO-treated SCR or KRT17 shRNA cells (termed donor cells) was used as a chemoattractant (Fig. 6 C). Cells show limited invasion toward conditioned medium from DMSO-treated SCR shRNA cells, and enhanced invasion toward conditioned medium from TPA-treated A431 SCR shRNA cells. By comparison, reduced invasion is seen when using conditioned medium from DMSO- or TPA-treated KRT17 shRNA cells (Fig. 6 D), which suggests that K17 is required for the secretion of chemokines that are effective in this setting. These findings strengthen the notion that K17 is required for the production of secreted factors that promote the aberrant growth and invasiveness of tumor keratinocytes.
Because the expression of CXCR3 ligands depends on both hnRNP K and K17 (Fig. 2), the involvement of hnRNP K in the transformed phenotype was assessed next. Conditioned medium from A431 SCR shRNA cells expressing NS or hnRNP K siRNA was used to culture A431 SCR shRNA cells in soft agar, and colony formation was assessed. A431 keratinocytes grown in medium from hnRNP K siRNA-expressing cells show a markedly reduced number of colonies compared with cells grown in conditioned medium from NS siRNA-expressing cells, (Fig. 6, E and F), which suggests that hnRNP K is also required for the secretion of key factors regulating tumor cell growth.
CXCR3 signaling is required for K17-dependent tumor cell growth
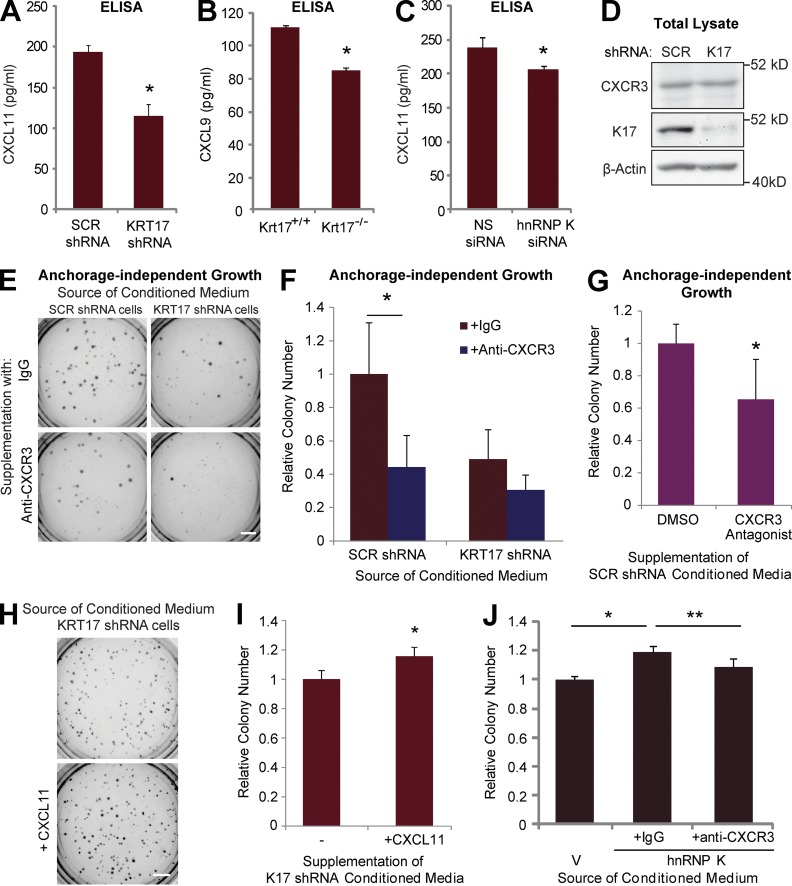
We next tested whether K17 plays a role in the secretion of CXCR3 ligands. Use of a CXCL11-specific ELISA confirmed the reduced CXCR3 ligand level in medium from KRT17 shRNA-expressing A431 cells (Fig. 7 A). Alternatively, ELISAs performed for CXCL8, IL-17F, and thymic stromal lymphopoietin (TSLP) showed no difference in SCR versus KRT17 shRNA-expressing cells (unpublished data), again supporting the specificity of the link between K17 and CXCR3 ligand expression. Reduced levels of another CXCR3 ligand protein, CXCL9, also occur in primary cultures of keratinocytes from ear skin lesions of K17−/− Gli2tg/+ mice (Fig. 7 B; note that the C57/Bl6 mouse strain harbors a premature stop codon in Cxcl11 [Groom and Luster, 2011], precluding the production of a functional protein). Reduced levels of secreted CXCL11 were also observed in A431 cells after hnRNP K knockdown (Fig. 7 C), which confirms the requirement of hnRNP K for full expression of CXCL11. Unlike its ligands, levels of CXCR3 receptor protein are the same in A431 SCR and KRT17 shRNA cells (Fig. 7 D). Finally, to assess whether CXCR3 signaling plays a role in the transformed phenotype, conditioned medium from A431 SCR or KRT17 shRNA cells was supplemented with anti-CXCR3 neutralizing antibody (or IgG control) and used to treat A431 SCR shRNA cells grown in soft agar. Adding control IgG to conditioned media harvested from SCR shRNA-expressing cells does not affect its ability to impact tumor cell growth, or the negative impact of medium obtained from KRT17 shRNA-expressing cells (Figs. 7, E and F). In contrast, adding the anti-CXCR3 antibody significantly reduces the ability of SCR shRNA-expressing cell-conditioned medium to stimulate tumor cell growth (Fig. 7, E and F). Use of a chemical inhibitor of CXCR3 also confirmed the requirement of CXCR3 signaling in colony growth (Fig. 7 G). Moreover, supplementation of conditioned medium from KRT17 shRNA cells with recombinant CXCL11 increased the ability of cells to form colonies, further highlighting the critical role of CXCR3 ligand downstream of K17 (Fig. 7, H and I). Conditioned medium from cells overexpressing hnRNP K also increased the colony formation compared with cells treated with conditioned medium from a vector control, and this increase was abrogated when anti-CXCR3 neutralizing antibody was supplemented (Fig. 7 J). Together, these findings establish that CXCR3 signaling plays a role in K17- and hnRNP K–dependent tumor cell growth.
Figure 7.
CXCR3 inhibition results in an attenuated transformed phenotype. (A) CXCL11 protein amounts in conditioned medium from A431 SCR or KRT17 shRNA cells were measured by ELISA. Data from three experimental repeats (each performed in 10 replicates) are represented as mean ± SEM (error bars). *, P < 0.016. (B) CXCL9 protein amounts in conditioned medium from primary keratinocytes isolated from ear lesions of P81 Krt17+/+ or Krt17−/− Gli2tg/+ mice were measured by ELISA. Data from four experimental repeats (each performed in triplicate) are represented as mean ± SEM (error bars). *, P < 0.04. (C) CXCL11 protein amounts in conditioned medium from A431 SCR shRNA cells transfected with either NS or hnRNP K siRNA (RPK1) were measured by ELISA. Data from three experimental repeats (each performed in 10 replicates) are represented as mean ± SEM (error bars). *, P < 0.04. (D) Immunoblotting performed on lysates from A431 cells stably expressing SCR or KRT17 shRNA with antibodies against the indicated proteins. (E) Soft agar colony assay using A431 SCR shRNA cells. Cells were grown in conditioned medium from A431 SCR or KRT17 shRNA cells, supplemented with anti-CXCR3 neutralizing antibody (0.5 ng/ml) or IgG control. Bar, 1 cm. (F) Colony numbers from E were quantitated using ImageJ and normalized to those grown in IgG-supplemented conditioned medium from A431 SCR shRNA cells. Data from four experimental repeats (each performed in triplicate) are represented as mean ± SEM (error bars). *, P < 0.03. (G) Soft agar colony assay using A431 SCR shRNA cells. Cells were grown in conditioned medium from A431 SCR shRNA cells, supplemented with DMSO or CXCR3 antagonist (10 µM). Colony numbers were quantitated using ImageJ and normalized to those grown in DMSO control. Data from three experimental repeats (each performed in triplicate) are represented as mean ± SEM (error bars). *, P < 0.05. (H) Soft agar colony assay using A431 SCR shRNA cells. Cells were grown in conditioned medium from A431 KRT17 shRNA cells, supplemented with or without 10 ng/ml recombinant CXCL11. Bar, 1 cm. (I) Colony numbers from H were quantitated using ImageJ and normalized to those grown without CXCL11. Data from five experimental repeats (each performed in triplicate) are represented as mean ± SEM (error bars). *, P < 0.02. (J) Soft agar colony assay using A431 SCR shRNA cells. Cells were grown in conditioned medium from A431 cells transfected with mCherry-C1 vector or hnRNP K, supplemented with anti-CXCR3 neutralizing antibody (0.5 ng/ml) or IgG control. Colony numbers were quantitated using ImageJ and normalized to those grown in conditioned medium from vector-expressing cells. Data from four experimental repeats (each performed in triplicate) are represented as mean ± SEM (error bars). *, P < 0.02; **, P < 0.05.
Discussion
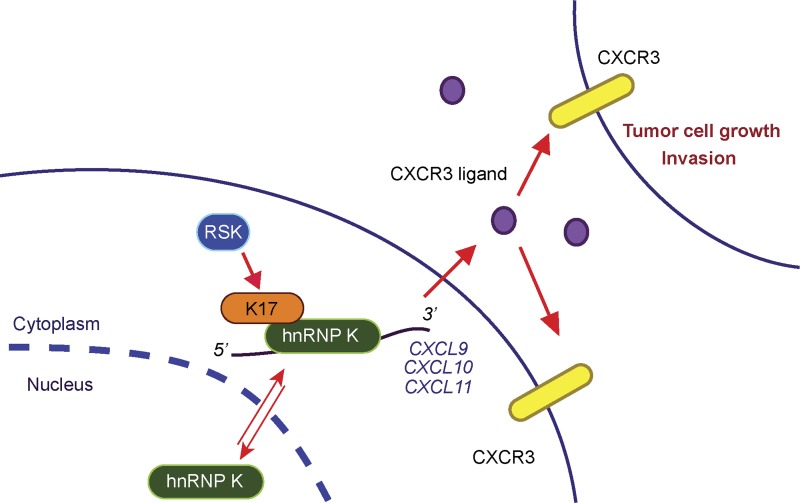
The contribution of cytoskeletal elements to regulating gene expression during normal tissue homeostasis and in disease processes such as cancer is increasingly recognized (Xu et al., 2009). This study reveals that K17’s ability to regulate pro-inflammatory and pro-tumorigenic gene expression involves its interaction with hnRNP K. A specific pathway, namely the CXCR3-dependent signaling axis, is shown to be subjected to hnRNP K regulation and to account for, at least in part, K17’s ability to promote inflammation, tumor cell growth, and invasiveness (Fig. 8). We establish a role for the RSK signaling kinase in regulating the interaction between K17 and hnRNP K, and expression of CXCR3 ligands. Collectively, the findings reported here expose, for the first time, an unanticipated link between RSK and inflammation through the regulation of CXCR3 ligand expression, a mechanism that may account for RSK’s established role in several cancers including skin cancer (Anjum and Blenis, 2008; Cho et al., 2012).
Figure 8.
A model depicting the significance of K17- and hnRNP K–dependent expression of CXCR3 ligands. K17 binds to hnRNP K and is required for its cytoplasmic localization. hnRNP K binds to and regulates mRNA transcripts for the CXCR3 ligands CXCL9, CXCL10, and CXCL11 in a K17-dependent manner. The K17–hnRNP K interaction and expression of CXCR3 ligands are both regulated by RSK activity. CXCR3 ligand secretion and CXCR3 signaling are required for K17- and hnRNP K–dependent enhancement of tumor cell growth and invasion.
The sharp sensitivity of Krt17 expression to pro-inflammatory and pro-tumorigenic signals, such as IFNγ (Vogel et al., 1995), is well-established (DePianto et al., 2010; Jin and Wang, 2014). The K17/hnRNP K/CXCR3 signaling axis exposed by our study implicates K17 in positive feedback loops that not only promote but also sustain a specific type of inflammation and immune response in chronic skin disease settings. CXCR3 ligand-expressing keratinocytes are poised to recruit CXCR3-positive cells into their local environment, which would then secrete soluble factors (e.g., IFNγ; Li et al., 2010) to induce additional CXCR3 ligand expression in keratinocytes (or other cell types in the proximal environment), triggering an amplification loop of inflammation that can act in autocrine and paracrine fashions (Lacotte et al., 2009). Our findings thus elevate the status of K17 from being an intriguing biomarker to a genuine effector in complex disease processes, and strengthen the emerging connection between “intra- and extra-cellular matrices” and the regulation of gene expression and tissue homeostasis (Xu et al., 2009). Future studies are needed to elucidate the exact mechanism of K17–hnRNP K interaction and identify the steps of gene expression control (e.g., is it transcriptional? posttranscriptional? translational?) at which hnRNP K and K17 together regulate CXCR3 ligand expression and function.
It appears likely, based on the literature, that K17-dependent activation of CXCR3 plays important roles in disease conditions other than basal cell carcinoma and, possibly, in normal tissue settings as well. Expression of CXCR3 and its ligands is markedly elevated in psoriatic skin (Chen et al., 2010), in which K17 contributes to disease pathogenesis (Jin and Wang, 2014). CXCR3 and its ligand CXCL11 are both required for wound repair in healthy skin (Yates et al., 2011), a physiological setting in which K17 expression is robustly induced (McGowan and Coulombe, 1998). Finally, ectodysplasin (Eda) stimulates CXCL10 and CXCL11 expression in developing mouse ectoderm and this, along with CXCR3 activation, contributes to the patterning and morphogenesis of hair follicles (Lefebvre et al., 2012), yet another setting in which K17 is prominently expressed (McGowan and Coulombe, 1998). Whether K17 is properly specified to impact gene expression in an hnRNP K–dependent fashion, as described here, in these additional settings represents an issue worth examining.
The idea of intermediate filament–dependent regulation of gene expression is consistent with the tissue-, differentiation-, and context-specific nature of their distribution, and has been around for decades (Traub, 1995). The interdependence of hnRNP K and K17 in regulating CXCR3 ligand expression suggests that intermediate filaments and hnRNPs may coordinate in additional settings for the purpose of regulating gene expression. For instance, hnRNPs interact with vimentin (Inoue et al., 2005), a type III intermediate filament protein that is up-regulated in various cancers (Satelli and Li, 2011), though the associated significance has not been defined. hnRNP K also regulates the mRNA encoding the neurofilament-M subunit, and both are required for normal development and growth of neuronal axons (Liu and Szaro, 2011). Our findings significantly extend these previous efforts, and provide a blueprint for investigating the extent of the functional partnership between hnRNPs and IF proteins, and its relevance to biological processes.
Materials and methods
Mouse models
Keratin 17–null (Krt17−/−) mice (C57BL/6 strain; see McGowan et al., 2002) were crossed to transgenic mice overexpressing mouse Gli2 under the control of the bovine Krt5 promoter (Gli2tg/+; C57BL/6 strain; see Grachtchouk et al., 2000) to create Krt17−/−; Gli2tg/+ mice, and relevant controls, as described previously (see DePianto et al., 2010). Genotyping was performed for the Krt17 locus and Gli2 transgene using PCR as described previously (DePianto et al., 2010).
Plasmids and siRNAs
Plasmid pKH3-avian RSK1 (chicken RPS6KA1 cDNA in the CMV promoter–containing pKH3 vector backbone; plasmid #8998; Addgene) and pKH3-avian RSK1 K112R/K464R (kinase-dead RSK1 coding sequence in the same vector backbone; plasmid #8981; Addgene) were gifts from J. Blenis (Harvard Medical School, Boston, MA). Cloning of mouse Krt17 cDNA into pEGFP-C3 vector backbone (CMV promoter; Takara Bio Inc.) to generate pEGFP-C3 K17 WT and subsequent PCR mutagenesis for pEGFP-C3 K17 S44A (in which Ser 44 is mutated to Ala) have been described previously (Pan et al., 2011). An shRNA-resistant K17 sequence was generated using a QuikChange II XL Site-Directed Mutagenesis kit (Agilent Technologies) and PCR using the oligonucleotide primers listed in Table 1. The GFP and GFP-K17 coding sequences were cloned into pENTR1A (Invitrogen) using PCR with the primers listed in Table 1 and also cloned into pLenti CMV/TO hygro (featuring a CMV promoter; Addgene) using LR Clonase (Invitrogen). hnRNP K cDNA was cloned out of pCMV6-AC HNRNPK (CMV promoter; OriGene) with PCR using the primers listed in Table 1 and cloned into mCherry-C1 (CMV promoter; Takara Bio Inc.).
Table 1.
Oligonucleotide primers used in PCR reactions for cloning indicated genes into overexpression plasmids
| Gene | Direction | Oligonucleotide primer sequence |
| shRNA-resistant K17a | Forward | 5′-GAGAATCTCAAGGAAGAACTAGCGTATCTGAAGAAGAACCACGAG-3′ |
| Reverse | 5′-CTCGTGGTTCTTCTTCAGATACGCTAGTTCTTCCTTGAGATTCTC-3′ | |
| GFP-K17b | Forward | 5′-GACTGGATCCAGCATGGTGAGCAAGGGCGAGG-3′ (BamHI restriction site underlined) |
| Reverse | 5′-GCTGGGTCTAGATATCTCGAGTGTTAACGGGTGGTCTGGTGC-3′ (XbaI restriction site underlined) | |
| GFPc | Reverse | 5′-GGTCTAGATTTACTTGTACAGCTCGTCCATGCCGAG-3′ (XbaI restriction site underlined) |
| hnRNP Kd | Forward | 5′-CAGTCGACGATGGAAACTGAACAG-3′ (SalI restriction site underlined) |
| Reverse | 5′-ATCCCGGGCTTAGAATCCTTCAAC-3′ (XmaI restriction site underlined) |
Used with a QuikChange II XL Site-Directed Mutagenesis kit.
Includes a BamHI or XbaI restriction site for ligation into pENTR1A plasmid (Invitrogen).
Used forward primer of GFP-K17b for PCR reaction.
Includes a SalI or XmaI restriction site for ligation into mCherry-C1 plasmid (Takara Bio Inc.).
To silence KRT17 expression, shRNA targeting the KRT17 cDNA at positions 387 (KRT17C) or 411 (KRT17D) was devised as described previously (Chung et al., 2012). An NS shRNA has also been described previously (Chung et al., 2012). A SCR shRNA was designed by scrambling the KRT17 targeting sequence at position 387 (Table 2) and cloned in pSuper.retro.puro vector (OligoEngine). To silence HNRNPK expression, siRNAs targeting the 3′-UTR of HNRNPK were devised. The following Dicer-substrate RNA duplex (IDT) were used to target HNRNPK or used as a NS siRNA control (Table 3). ON-TARGET plus SMARTpool RSK1 siRNAs from GE Healthcare (Table 4) has been described previously (Pan et al., 2011).
Table 2.
Oligonucleotide primer sets used to generate SCR shRNA control for KRT17 shRNA
| shRNA | Direction | shRNA sequence (SCR sequence underlined) |
| SCR shRNA | Forward | 5′-GATCCCCGCGATGTGGCGAACTGACACGTTCAAGAGACGTGTCAGTTCGCCACATCGCTTTTTA-3′ |
| Reverse | 5′-AGCTTAAAAAGCGATGTGGCGAACTGACAACGTCTCTTGAACGTGTCAGTTCGCCACATCGCGGG-3′ |
Table 3.
Dicer-substrate RNA duplex (IDT) used to target HNRNPK or used as a nonsilencing siRNA control
| siRNA | Strand | siRNA sequence |
| hnRNP K siRNA (RPK1) | Antisense strand | 5′-rCrCrUrArUrGrArArGrCrArGrArGrGrArArUrGrUrUrGrGrCrUrUrU-3′ |
| Sense strand | 5′-rArGrCrCrArArCrArUrUrCrCrUrCrUrGrCrUrUrCrArUrAGG-3′ | |
| hnRNP K siRNA (RPK2) | Antisense strand | 5′-rGrArUrGrCrCrArGrGrGrArCrArUrGrUrGrGrArCrUrArUrUrGrUrU-3′ |
| Sense strand | 5′-rCrArArUrArGrUrCrCrArCrArUrGrUrCrCrCrUrGrGrCrATC-3′ | |
| hnRNP K siRNA (RPK3) | Antisense strand | 5′-rArArGrArGrCrArUrUrArCrArUrUrCrUrGrCrArCrArUrGrCrUrC-3′ |
| Sense strand | 5′-rGrCrArGrUrGrUrGrCrArGrArArUrGrUrArArUrGrCrUrCTT-3′ | |
| NS siRNA | Antisense strand | 5′-rGrUrCrUrArArCrArGrCrCrGrUrUrArCrArCrArCrCrUrUrArGrArU-3′ |
| Sense strand | 5′-rCrUrArArGrGrUrGrUrGrUrArArCrGrGrCrUrGrUrUrArGAC-3′ |
Table 4.
Sequences for ON-TARGET plus SMARTpool RSK1 siRNAs from GE Healthcare
| Gene | siRNA sequence |
| RPS6KA1 | 5′-AGGGCAAGCUCUAUCUUAU-3′ |
| 5′-GAACGGACUCCUCAUGACA-3′ | |
| 5′-ACACGAUGCUGGCAGGAUA-3′ | |
| 5′-CAAAGGAGGUCAUGUUUAC-3′ |
Cell culture and TPA stimulation
A431 or HeLa (ATCC) cells were grown in Dulbecco’s modified essential medium (Invitrogen) containing 10% FBS (Atlanta Biologicals), 100 units/ml penicillin, and 100 µg/ml streptomycin (Invitrogen) at 37°C in 5% CO2. For cells stably expressing KRT17 or SCR shRNA, the medium was supplemented with 0.5 µg/ml puromycin. Primary cultures of skin keratinocytes from 2-d-old mouse littermate pups that are either heterozygous (Krt17+/−) or homozygous (Krt17−/−) for a Krt17 null allele (McGowan et al., 2002) were isolated and cultured as described previously (Chung et al., 2012). Primary cultures of skin keratinocytes from ear lesions of age-matched Gli2tg/+ mice that are either WT (Krt17+/+), heterozygous (Krt17+/−), or homozygous (Krt17−/−) for the Krt17 allele were isolated by mincing ears into fine pieces and subjecting them to 0.25% Trypsin overnight. After vigorous vortexing, free keratinocytes were isolated as described previously (Chung et al., 2012) and cultured in Cnt-57 medium (CELLnTEC) before harvesting. For TPA stimulation, cells previously incubated for 18 h in 0.1% FBS-containing medium were treated with either DMSO (vehicle control) or 200 nM TPA for the indicated time periods.
Antibodies and other reagents
The following antibodies were obtained from commercial sources: mouse monoclonal (mAb) anti-GAPDH (1D4) and anti–hnRNP K (3C2) were from Santa Cruz Biotechnology, Inc.; rabbit polyclonal (pAb) anti-PARP (9542), pAb anti-RSK1 (9333), and pAb phospho-RSK (9341) were from Cell Signaling Technology; pAb anti-mCherry was from BioVision; mAb anti–β-actin (clone AC-15) and pAb anti-K18 (C-terminal) were from Sigma-Aldrich; mAb anti-K5 (AF138) and pAb anti-K14 (AF-64) were from Covance; pAb anti–hnRNP K (EP943Y), rabbit monoclonal anti-K13 (EPR3671), and chicken polyclonal K8 (ab107115) were from Abcam; and mAb anti–human CXCR3 (MAB160) neutralizing antibody was from R&D Systems.
Peptides NH2-C-S-S-R-E-Q-V-H-Q-T-T-R-COOH, NH2-C-S-S-T-I-K-Y-T-T-COOH, and NH2-C-S-T-S-F-S-Q-S-Q-S-Q-S-S-R-D-COOH were used for the production of an antiserum to rabbit pAb anti-K17, anti-K6, and anti-K16 (McGowan and Coulombe, 1998; Bernot et al., 2002). mAb anti-hnRNP A2/B1 (4C2) was a gift from M. Matunis (Johns Hopkins University, Baltimore, MD). Normal mouse IgG (sc-2025) and normal rabbit IgG (sc-2027) were obtained from Santa Cruz Biotechnology, Inc. GFP-Trap_A was from ChromoTek. Recombinant human CXCL11 protein was obtained from Reprokine. RSK inhibitors BI-D1870 and SL0101were obtained from Symansis and EMD Millipore, respectively. All other chemicals (including TPA) were from Sigma-Aldrich unless noted otherwise.
Transfection
Overexpression plasmids were transiently transfected using FuGENE HD transfection reagent (Promega) according to the manufacturer’s protocol. siRNAs were transiently transfected using Lipofectamine RNAiMAX transfection reagent (Life Technologies) using the manufacturer’s protocol.
Retroviral and lentiviral infections
Retroviral supernatants were generated by calcium phosphate–mediated cotransfection of the shRNA plasmids and the packaging plasmids into the packaging cell line Phoenix as described previously (Chung et al., 2012). Lentiviral supernatants were generated using the pLenti plasmids as described previously (Wan et al., 2007). Retroviral or lentiviral supernatants, collected 24 h after transfection, were used to infect subconfluent cells in three sequential 4-h incubations in the presence of 4 µg/ml polybrene (Sigma-Aldrich). Transductants were selected in puromycin (0.5 µg/ml) and/or hygromycin (0.25 µg/ml), beginning 48 h after infection.
RNA harvest, cDNA synthesis, and qRT-PCR
RNA was harvested using QIAshredder and RNeasy kit (both from QIAGEN) or NucleoSpin RNA II kit (Takara Bio Inc.) according to the manufacturers’ protocols. 1.0 µg RNA was reverse-transcribed with the iScript cDNA Synthesis kit (Bio-Rad Laboratories) using the manufacturer’s protocol. qRT-PCR was performed on the first strand cDNA with the primers listed in Table 5 and Sso Advanced SYBR Green supermix (Bio-Rad Laboratories) using the CFX96 qRT-PCR apparatus (Bio-Rad Laboratories). The following program was used for all qRT-PCR reactions: denaturation step at 95°C for 10 min, 40 cycles of PCR (denaturation at 95°C for 10 s, annealing and elongation at 60°C for 30 s). Amplification curves were read with the CFX Manager (Bio-Rad Laboratories, Inc.) using the comparative cycle threshold method. Relative quantifications or fold changes of the target mRNAs were calculated after normalization of cycle thresholds with respect to at least two reference genes between ACTB, RPS18, and GAPDH levels.
Table 5.
Oligonucleotide primer sets used for qRT-PCR assays
| Gene | Direction | Primer sequence (5′ to 3′) |
| KRT17 | Forward | GGTGGGTGGTGAGATCAATGT |
| Reverse | CGCGGTTCAGTTCCTCTGTC | |
| CXCL9 | Forward | GTAGTGAGAAAGGGTCGCTGT |
| Reverse | AGGGCTTGGGGCAAATTGTT | |
| CXCL10 | Forward | GTGGCATTCAAGGAGTACCTC |
| Reverse | TGATGGCCTTCGATTCTGGATT | |
| CXCL11 | Forward | GACGCTGTCTTTGCATAGGC |
| Reverse | GGATTTAGGCATCGTTGTCCTTT | |
| CXCR3 | Forward | CCACCTAGCTGTAGCAGACAC |
| Reverse | AGGGCTCCTGCGTAGAAGTT | |
| IL17A | Forward | TCCCACGAAATCCAGGATGC |
| Reverse | TGTTCAGGTTGACCATCACAGT | |
| IL6 | Forward | AATTCGGTACATCCTCGACGG |
| Reverse | TTGGAAGGTTCAGGTTGTTTTCT | |
| IL4 | Forward | CGGCAACTTTGTCCACGGA |
| Reverse | TCTGTTACGGTCAACTCGGTG | |
| S100A8 | Forward | ATGCCGTCTACAGGGATGAC |
| Reverse | ACACTCGGTCTCTAGCAATTTCT | |
| S100A9 | Forward | GGTCATAGAACACATCATGGAGG |
| Reverse | GGCCTGGCTTATGGTGGTG | |
| HNRNPK | Forward | GCAGGAGGAATTATTGGGGTC |
| Reverse | TGCACTCTACAACCCTATCGG | |
| HNRNPD | Forward | GCGTGGGTTCTGCTTTATTACC |
| Reverse | TTGCTGATATTGTTCCTTCGACA | |
| MYC | Forward | GGCTCCTGGCAAAAGGTCA |
| Reverse | AGTTGTGCTGATGTGTGGAGA | |
| PTGS2 | Forward | CCAGTATAAGTGCGATTGTACCC |
| Reverse | TCAAAAATTCCGGTGTTGAGCA | |
| CCL22 | Forward | ATCGCCTACAGACTGCACTC |
| Reverse | GACGGTAACGGACGTAATCAC | |
| EGR1 | Forward | GGTCAGTGGCCTAGTGAGC |
| Reverse | GTGCCGCTGAGTAAATGGGA | |
| IL17C | Forward | CCACACTGCTACTCGGCTG |
| Reverse | CACACGGTATCTCCAGGGTGA | |
| EGFR | Forward | AGGCACGAGTAACAAGCTCAC |
| Reverse | ATGAGGACATAACCAGCCACC | |
| ACTB | Forward | CATGTACGTTGCTATCCAGGC |
| Reverse | CTCCTTAATGTCACGCACGAT | |
| GAPDH | Forward | AAGGTGAAGGTCGGAGTCAAC |
| Reverse | GGGGTCATTGATGGCAACAATA | |
| RPS18 | Forward | GCGGCGGAAAATAGCCTTTG |
| Reverse | GATCACACGTTCCACCTCATC |
RNA IP
RNA IP was performed and RNA was harvested from normally grown cells or serum-deprived cells treated with DMSO or 200 nM TPA overnight using an RIP assay kit and pAb anti–hnRNP K (both from MBL International) according to the manufacturer’s protocol. Normal rabbit IgG (MBL International) was used as a negative control. After cell lysis (in accordance with the MBL protocol), equal aliquots of lysate were taken for quality control by Western blotting and RNA isolation followed by qRT-PCR. RNA harvested was reverse transcribed, and qRT-PCR was performed as described in the previous section. Every experiment was performed at least four times.
Co-IP, protein gel electrophoresis, and immunoblotting
Cells were washed with PBS, and cell lysates were prepared in cold Triton lysis buffer (1% Triton X-100; 40 mm Hepes, pH 7.5; 120 mm sodium chloride; 1 mm EDTA; 1 mm phenylmethylsulfonyl fluoride; 10 mm sodium pyrophosphate; 1 µg/ml each of chymostatin, leupeptin, and pepstatin; 10 µg/ml each of aprotinin and benzamidine; 2 µg/ml antipain; 1 mm sodium orthovanadate; and 50 mm sodium fluoride) for Western blotting, cold Triton lysis buffer supplemented with 2% Empigen (BD) for anti-K17, K5, or GFP IP, or cold NP-40 lysis buffer (0.25% NP-40; 50 mM Tris, pH 8.0; 100 mM sodium chloride; 1 mm phenylmethylsulfonyl fluoride; 10 mm sodium pyrophosphate; 1 µg/ml each of chymostatin, leupeptin, and pepstatin; 10 µg/ml each of aprotinin and benzamidine; 2 µg/ml antipain; 1 mm sodium orthovanadate; and 50 mm sodium fluoride) for anti–hnRNP K IP (3C2). Protein concentration was determined using the Bio-Rad protein assay (Bio-Rad Laboratories) with bovine serum albumin (Thermo Fisher Scientific) as a standard. For IP, aliquots of cell lysate were incubated with the indicated antibody, preimmune serum, or IgG control, and immune complexes were captured using the TrueBlot anti–rabbit Ig IP beads (Rockland Immunochemicals Inc.) or Protein G Sepharose (GE Healthcare). For immunoblotting, protein concentration was determined using the Bio-Rad protein assay, and cell lysates were prepared in Laemmli SDS-PAGE sample buffer. Aliquots of protein lysate were resolved by SDS-PAGE, transferred to nitrocellulose membranes (0.45 µm; Bio-Rad Laboratories, Inc.), and immunoblotted with the indicated antibodies followed by HRP-conjugated goat anti–mouse or anti–rabbit IgG or rabbit anti–chicken IgY (Sigma-Aldrich) and SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) or Amersham ECL Select Western Blotting Detection Reagent (GE Healthcare). Signals were detected using the FluorChem Q imaging system (ProteinSimple). For Western blot signal quantitation, the Alphaview software (ProteinSimple) was used.
Biochemical subcellular fractionation
Subcellular fractionation was performed as described previously (Wan et al., 2011). Cells were washed in PBS and resuspended for 5 min at 4°C in ice-cold NP-40 buffer (10 mM Hepes, pH 7.9; 10 mM KCl; 1.5 mM MgCl2; 0.1 mM EDTA; 0.5 mM dithiothreitol; 0.4% NP-40; 1 mm phenylmethylsulfonyl fluoride; 10 mm sodium pyrophosphate; 1 µg/ml each of chymostatin, leupeptin, and pepstatin; 10 µg/ml each of aprotinin and benzamidine; 2 µg/ml antipain; 1 mm sodium orthovanadate; and 50 mm sodium fluoride). Lysates were centrifuged for 3 min at 500 g and 4°C, and supernatants were collected as cytosolic fractions. Pellets were incubated for 10 min at 4°C in a high-salt buffer (20 mM Hepes, pH 7.9; 420 mM NaCl; 1.5 mM MgCl2; 25% [vol/vol] glycerol; 0.5 mM PMSF; 0.2 mM EDTA; 0.5 mM dithiothreitol; 1 mm phenylmethylsulfonyl fluoride; 10 mm sodium pyrophosphate; 1 µg/ml each of chymostatin, leupeptin, and pepstatin; 10 µg/ml each of aprotinin and benzamidine; 2 µg/ml antipain; 1 mm sodium orthovanadate; and 50 mm sodium fluoride). Supernatants were collected as nuclear fractions after centrifugation for 10 min at 13,800 g and 4°C.
Immunofluorescence staining
For immunostaining of cells in culture, cells grown on glass coverslips (VWR) were washed in PBS, fixed in 4% paraformaldehyde in PBS, and permeabilized in 0.01% digitonin or 0.1% Triton X-100. For ear lesions from P80 Krt17+/+ or Krt17−/− Gli2tg/+ mice, tissues were fresh-frozen in Tissue-Tek O.C.T. (Sakura) and sections were cut at 5 µm. Samples were blocked in 5% normal goat serum (NGS) in PBS for at least 1 h before staining with primary antibodies diluted in blocking buffer for 16 h followed by Alexa Fluor 488– or Alexa Fluor 594–conjugated goat anti–mouse or goat anti–rabbit secondary antibodies (Invitrogen) for 1 h. 1 µg/ml of Hoechst 33342 was used to stain the nuclei, and coverslips were mounted on microscope slides with mounting media containing 1,4-diaza-bicyclo[2.2.2]octane (Electron Microscopy Sciences). Fluorescence images were obtained using the Zen Pro 2012 software (Carl Zeiss) with an Axio Observer.Z1 fluorescence microscope (Carl Zeiss) equipped with an AxioCam MRm camera (Carl Zeiss) at 37°C under a 63× Plan-Apochromat oil immersion lens (Carl Zeiss). The images were processed using Photoshop software (Adobe) to split and pseudo-color individual channels.
ELISA
2,500 cells were plated on each well of a 96-well tissue culture plate (BD) with at least 10 replicates per condition. The medium was left unchanged starting from 48 h after plating or 16 h after transfection. After 48 h, the medium was collected and CXCL9, CXCL11, CXCL8, IL-17F, and TSLP levels were determined using mouse CXCL9/MIG Duoset, human CXCL11/I-TAC Duoset, human CXCL8/IL-8 Duoset, human IL-17F Duoset, and human TSLP Duoset (all from R&D Systems) according to the manufacturer’s protocol. Every experiment was performed at least three times, each with at least three biological replicate samples.
Anchorage-independent growth in soft agar
100,000 cells were plated per well in a 6-well plate in 1 ml of 0.3% agarose in media on top of a 2-ml bottom layer of 0.5% agarose in media. Cells were fed every 3 d with either normal or conditioned media. Phase-contrast images were obtained using the FluorChem Q imaging system, and light intensity was adjusted to specifically show colonies in dark shades. Images were then processed using the ImageJ software (National Institutes of Health) to quantify colony numbers. Each experiment contained at least three replicates per condition, and every experiment was performed at least three times.
Transwell cell invasion assay
Transwell chambers with 8-µm pores (Corning) were coated with 20 µl Matrigel (BD) followed by blocking with 0.1% FBS-containing medium for 2 h. 100,000 serum-deprived cells were plated in the top chamber for 4 h to allow attachment. Either 0.1% FBS-containing medium with or without 10 ng/ml EGF or conditioned medium were then added to the bottom chamber. After 48 h, cells were washed, fixed in methanol at –20°C, and stained with propidium iodide. A cotton swab was used to remove cells from the top chamber. Fluorescence images of migrated cells were taken, and fluorescence intensity was adjusted to specifically show cell nuclei in white. Images were processed using the ImageJ software to quantify the number of cells migrated. Each experiment contained at least three replicates per condition, and every experiment was performed at least three times.
Conditioned medium
50,000 cells were plated on each well of a six-well tissue culture plate (Thermo Fisher Scientific). The culture medium was left unchanged starting from 16 h after plating or transfection. After 48–96 h, the medium was collected, filtered through a 0.45 µm PVDF syringe filter (EMD Millipore), and used fresh to feed cells in soft agar. For the experiments reported in Fig. 6, conditioned medium was supplemented with either 0.5 ng/ml of anti-CXCR3 antibody or IgG control, or 10 µM of CXCR3 antagonist (EMD Millipore) or DMSO control before feeding cells. To assays for invasiveness, cells were serum-deprived in 0.1% FBS-containing medium for 16 h and placed in 0.1% FBS-containing medium with DMSO or 200 nM TPA for 48 h. Conditioned medium was collected, filtered, and used fresh.
Graphs and statistics
All graphs in the manuscript are shown as mean ± SEM. For comparisons between two datasets, a Student’s t test (tails = 2, type = 1) was used, and statistically significant p-values are indicated in the figures and figure legends.
Online supplemental material
Fig. S1 shows the specificity of the K17–hnRNP K interaction by testing interactions between hnRNPs and keratins, and confirms K17-dependent hnRNP K localization in additional settings. Similar to what was observed in human cancer cell lines in Fig. 1, Fig. S2 demonstrates that K17 regulates the cytoplasmic localization of hnRNP K in keratinocytes from Gli2tg/+ mice. Fig. S3 reports on a survey of hnRNP K–bound transcripts in mouse keratinocytes and TPA-induced RSK phosphorylation along with its impact on hnRNP K’s interaction with K17. In Fig. S4, dependency on hnRNP K for the expression of CXCR3 ligands is shown with three different siRNAs targeting HNRNPK along with the binding of hnRNP K to various transcripts in A431 cells with KRT17 knockdown. Fig. S5 validates the requirement of K17 for the growth and invasiveness of tumor cells with a series of experiments using an additional KRT17 shRNA and shRNA-resistant K17. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201408026/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201408026.dv.
Supplementary Material
Acknowledgments
We thank members of the Coulombe laboratory and Dr. Janis Taube for advice and support. We thank Drs. Ryan Hobbs, Yen Wu, Stéphane Lajoie, Fengyi Wan, Luis Garza, Michael Matunis, and Carole Parent for helpful discussions and advice.
This work was supported by grants AR44232 and CA160255 to P.A. Coulombe from the National Institutes of Health. B.M. Chung was supported in part by training grant T32CA009110 from the National Cancer Institute.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- IP
- immunoprecipitation
- NS
- nonsilencing
- qRT-PCR
- quantitative RT-PCR
- SCR
- scrambled
- TPA
- 12-o-tetradecanoylphorbol-13-acetate
- TSLP
- thymic stromal lymphopoietin
- WT
- wild type
References
- Anjum R., and Blenis J.. 2008. The RSK family of kinases: emerging roles in cellular signalling. Nat. Rev. Mol. Cell Biol. 9:747–758 10.1038/nrm2509 [DOI] [PubMed] [Google Scholar]
- Arbeit J.M., Münger K., Howley P.M., and Hanahan D.. 1994. Progressive squamous epithelial neoplasia in K14-human papillomavirus type 16 transgenic mice. J. Virol. 68:4358–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboro P., Ferrari N., and Balbi C.. 2014. Emerging roles of heterogeneous nuclear ribonucleoprotein K (hnRNP K) in cancer progression. Cancer Lett. 352:152–159 10.1016/j.canlet.2014.06.019 [DOI] [PubMed] [Google Scholar]
- Bernot K.M., Coulombe P.A., and McGowan K.M.. 2002. Keratin 16 expression defines a subset of epithelial cells during skin morphogenesis and the hair cycle. J. Invest. Dermatol. 119:1137–1149 10.1046/j.1523-1747.2002.19518.x [DOI] [PubMed] [Google Scholar]
- Bomsztyk K., Denisenko O., and Ostrowski J.. 2004. hnRNP K: one protein multiple processes. BioEssays. 26:629–638 10.1002/bies.20048 [DOI] [PubMed] [Google Scholar]
- Chaudhury A., Chander P., and Howe P.H.. 2010. Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: Focus on hnRNP E1’s multifunctional regulatory roles. RNA. 16:1449–1462 10.1261/rna.2254110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.C., Liu H.P., Li H.P., Hsueh C., Yu J.S., Liang C.L., and Chang Y.S.. 2009. Thymidine phosphorylase mRNA stability and protein levels are increased through ERK-mediated cytoplasmic accumulation of hnRNP K in nasopharyngeal carcinoma cells. Oncogene. 28:1904–1915 10.1038/onc.2009.55 [DOI] [PubMed] [Google Scholar]
- Chen S.C., de Groot M., Kinsley D., Laverty M., McClanahan T., Arreaza M., Gustafson E.L., Teunissen M.B., de Rie M.A., Fine J.S., and Kraan M.. 2010. Expression of chemokine receptor CXCR3 by lymphocytes and plasmacytoid dendritic cells in human psoriatic lesions. Arch. Dermatol. Res. 302:113–123 10.1007/s00403-009-0966-2 [DOI] [PubMed] [Google Scholar]
- Cho Y.Y., Lee M.H., Lee C.J., Yao K., Lee H.S., Bode A.M., and Dong Z.. 2012. RSK2 as a key regulator in human skin cancer. Carcinogenesis. 33:2529–2537 10.1093/carcin/bgs271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung B.M., Murray C.I., Van Eyk J.E., and Coulombe P.A.. 2012. Identification of novel interaction between annexin A2 and keratin 17: evidence for reciprocal regulation. J. Biol. Chem. 287:7573–7581 10.1074/jbc.M111.301549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePianto D., Kerns M.L., Dlugosz A.A., and Coulombe P.A.. 2010. Keratin 17 promotes epithelial proliferation and tumor growth by polarizing the immune response in skin. Nat. Genet. 42:910–914 10.1038/ng.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E., Nowarski R., Thaiss C.A., Hu B., Jin C., and Flavell R.A.. 2013. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat. Rev. Cancer. 13:759–771 10.1038/nrc3611 [DOI] [PubMed] [Google Scholar]
- Evans J.R., Mitchell S.A., Spriggs K.A., Ostrowski J., Bomsztyk K., Ostarek D., and Willis A.E.. 2003. Members of the poly (rC) binding protein family stimulate the activity of the c-myc internal ribosome entry segment in vitro and in vivo. Oncogene. 22:8012–8020 10.1038/sj.onc.1206645 [DOI] [PubMed] [Google Scholar]
- Fulton A.M.2009. The chemokine receptors CXCR4 and CXCR3 in cancer. Curr. Oncol. Rep. 11:125–131 10.1007/s11912-009-0019-1 [DOI] [PubMed] [Google Scholar]
- Gao R., Yu Y., Inoue A., Widodo N., Kaul S.C., and Wadhwa R.. 2013. Heterogeneous nuclear ribonucleoprotein K (hnRNP-K) promotes tumor metastasis by induction of genes involved in extracellular matrix, cell movement, and angiogenesis. J. Biol. Chem. 288:15046–15056 10.1074/jbc.M113.466136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grachtchouk M., Mo R., Yu S., Zhang X., Sasaki H., Hui C.C., and Dlugosz A.A.. 2000. Basal cell carcinomas in mice overexpressing Gli2 in skin. Nat. Genet. 24:216–217 10.1038/73417 [DOI] [PubMed] [Google Scholar]
- Groom J.R., and Luster A.D.. 2011. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol. Cell Biol. 89:207–215 10.1038/icb.2010.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide M., Kato T., Ogata K., Mochiki E., Kuwano H., and Oyama T.. 2012. Keratin 17 expression correlates with tumor progression and poor prognosis in gastric adenocarcinoma. Ann. Surg. Oncol. 19:3506–3514 10.1245/s10434-012-2437-9 [DOI] [PubMed] [Google Scholar]
- Inoue A., Watanabe T., Tominaga K., Tsugawa K., Nishio K., Takahashi K.P., and Kaneda K.. 2005. Association of hnRNP S1 proteins with vimentin intermediate filaments in migrating cells. J. Cell Sci. 118:2303–2311 10.1242/jcs.02345 [DOI] [PubMed] [Google Scholar]
- Inoue A., Sawata S.Y., Taira K., and Wadhwa R.. 2007. Loss-of-function screening by randomized intracellular antibodies: identification of hnRNP-K as a potential target for metastasis. Proc. Natl. Acad. Sci. USA. 104:8983–8988 10.1073/pnas.0607595104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L., and Wang G.. 2014. Keratin 17: a critical player in the pathogenesis of psoriasis. Med. Res. Rev. 34:438–454 10.1002/med.21291 [DOI] [PubMed] [Google Scholar]
- Karantza V.2011. Keratins in health and cancer: more than mere epithelial cell markers. Oncogene. 30:127–138 10.1038/onc.2010.456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawada K., Sonoshita M., Sakashita H., Takabayashi A., Yamaoka Y., Manabe T., Inaba K., Minato N., Oshima M., and Taketo M.M.. 2004. Pivotal role of CXCR3 in melanoma cell metastasis to lymph nodes. Cancer Res. 64:4010–4017 10.1158/0008-5472.CAN-03-1757 [DOI] [PubMed] [Google Scholar]
- Kim S., Wong P., and Coulombe P.A.. 2006. A keratin cytoskeletal protein regulates protein synthesis and epithelial cell growth. Nature. 441:362–365 10.1038/nature04659 [DOI] [PubMed] [Google Scholar]
- Lacotte S., Brun S., Muller S., and Dumortier H.. 2009. CXCR3, inflammation, and autoimmune diseases. Ann. N. Y. Acad. Sci. 1173:310–317 10.1111/j.1749-6632.2009.04813.x [DOI] [PubMed] [Google Scholar]
- Lefebvre S., Fliniaux I., Schneider P., and Mikkola M.L.. 2012. Identification of ectodysplasin target genes reveals the involvement of chemokines in hair development. J. Invest. Dermatol. 132:1094–1102 10.1038/jid.2011.453 [DOI] [PubMed] [Google Scholar]
- Li B., Xu W., Xu L., Jiang Z., Wen Z., Li K., and Xiong S.. 2010. I-TAC is a dominant chemokine in controlling skin intragraft inflammation via recruiting CXCR3+ cells into the graft. Cell. Immunol. 260:83–91 10.1016/j.cellimm.2009.09.004 [DOI] [PubMed] [Google Scholar]
- Liu Y., and Szaro B.G.. 2011. hnRNP K post-transcriptionally co-regulates multiple cytoskeletal genes needed for axonogenesis. Development. 138:3079–3090 10.1242/dev.066993 [DOI] [PubMed] [Google Scholar]
- Lo B.K., Yu M., Zloty D., Cowan B., Shapiro J., and McElwee K.J.. 2010. CXCR3/ligands are significantly involved in the tumorigenesis of basal cell carcinomas. Am. J. Pathol. 176:2435–2446 10.2353/ajpath.2010.081059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M., Vadlamudi R., Nguyen D., Wang R.A., Costa L., Bagheri-Yarmand R., Mendelsohn J., and Kumar R.. 2001. Growth factors regulate heterogeneous nuclear ribonucleoprotein K expression and function. J. Biol. Chem. 276:9699–9704 10.1074/jbc.M008514200 [DOI] [PubMed] [Google Scholar]
- McGowan K.M., and Coulombe P.A.. 1998. Onset of keratin 17 expression coincides with the definition of major epithelial lineages during skin development. J. Cell Biol. 143:469–486 10.1083/jcb.143.2.469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan K.M., Tong X., Colucci-Guyon E., Langa F., Babinet C., and Coulombe P.A.. 2002. Keratin 17 null mice exhibit age- and strain-dependent alopecia. Genes Dev. 16:1412–1422 10.1101/gad.979502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikula M., and Bomsztyk K.. 2011. Direct recruitment of ERK cascade components to inducible genes is regulated by heterogeneous nuclear ribonucleoprotein (hnRNP) K. J. Biol. Chem. 286:9763–9775 10.1074/jbc.M110.213330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen T.O., Hsu F.D., Jensen K., Cheang M., Karaca G., Hu Z., Hernandez-Boussard T., Livasy C., Cowan D., Dressler L., et al. 2004. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin. Cancer Res. 10:5367–5374 10.1158/1078-0432.CCR-04-0220 [DOI] [PubMed] [Google Scholar]
- Pan X., Kane L.A., Van Eyk J.E., and Coulombe P.A.. 2011. Type I keratin 17 protein is phosphorylated on serine 44 by p90 ribosomal protein S6 kinase 1 (RSK1) in a growth- and stress-dependent fashion. J. Biol. Chem. 286:42403–42413 10.1074/jbc.M111.302042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankar S., Tanner J.M., Bell R., Chaturvedi A., Randall R.L., Beckerle M.C., and Lessnick S.L.. 2013. A novel role for keratin 17 in coordinating oncogenic transformation and cellular adhesion in Ewing sarcoma. Mol. Cell. Biol. 33:4448–4460 10.1128/MCB.00241-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satelli A., and Li S.. 2011. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell. Mol. Life Sci. 68:3033–3046 10.1007/s00018-011-0735-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam N., Reddy M.A., and Natarajan R.. 2008. Distinct roles of heterogeneous nuclear ribonuclear protein K and microRNA-16 in cyclooxygenase-2 RNA stability induced by S100b, a ligand of the receptor for advanced glycation end products. J. Biol. Chem. 283:36221–36233 10.1074/jbc.M806322200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauler J., and Mulshine J.L.. 2009. Lung cancer and inflammation: interaction of chemokines and hnRNPs. Curr. Opin. Pharmacol. 9:384–388 10.1016/j.coph.2009.06.004 [DOI] [PubMed] [Google Scholar]
- Traub P.1995. Intermediate filaments and gene regulation. Physiol. Chem. Phys. Med. NMR. 27:377–400. [PubMed] [Google Scholar]
- Vogel U., Denecke B., Troyanovsky S.M., Leube R.E., and Böttger E.C.. 1995. Transcriptional activation of psoriasis-associated cytokeratin K17 by interferon-γ. Analysis of gamma-interferon activation sites. Eur. J. Biochem. 227:143–149 10.1111/j.1432-1033.1995.tb20370.x [DOI] [PubMed] [Google Scholar]
- Wan F., Anderson D.E., Barnitz R.A., Snow A., Bidere N., Zheng L., Hegde V., Lam L.T., Staudt L.M., Levens D., et al. 2007. Ribosomal protein S3: a KH domain subunit in NF-κB complexes that mediates selective gene regulation. Cell. 131:927–939 10.1016/j.cell.2007.10.009 [DOI] [PubMed] [Google Scholar]
- Wan F., Weaver A., Gao X., Bern M., Hardwidge P.R., and Lenardo M.J.. 2011. IKKβ phosphorylation regulates RPS3 nuclear translocation and NF-κB function during infection with Escherichia coli strain O157:H7. Nat. Immunol. 12:335–343 10.1038/ni.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.F., Lang H.Y., Yuan J., Wang J., Wang R., Zhang X.H., Zhang J., Zhao T., Li Y.R., Liu J.Y., et al. 2013. Overexpression of keratin 17 is associated with poor prognosis in epithelial ovarian cancer. Tumour Biol. 34:1685–1689 10.1007/s13277-013-0703-5 [DOI] [PubMed] [Google Scholar]
- Winkler A.E., Brotman J.J., Pittman M.E., Judd N.P., Lewis J.S. Jr, Schreiber R.D., and Uppaluri R.. 2011. CXCR3 enhances a T-cell-dependent epidermal proliferative response and promotes skin tumorigenesis. Cancer Res. 71:5707–5716 10.1158/0008-5472.CAN-11-0907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Boudreau A., and Bissell M.J.. 2009. Tissue architecture and function: dynamic reciprocity via extra- and intra-cellular matrices. Cancer Metastasis Rev. 28:167–176 10.1007/s10555-008-9178-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates C.C., Bodnar R., and Wells A.. 2011. Matrix control of scarring. Cell. Mol. Life Sci. 68:1871–1881 10.1007/s00018-011-0663-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., Shanas R., Nelson M.A., Bhattacharyya A., and Shi J.. 2010. Increased expression of the heterogeneous nuclear ribonucleoprotein K in pancreatic cancer and its association with the mutant p53. Int. J. Cancer. 126:395–404 10.1002/ijc.24744 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.