Abstract
Imbalanced chromosomal content, or aneuploidy, strongly affects the physiology of eukaryotic cells. The consequences of these effects are frequently detrimental, in particular in Metazoans. In humans, aneuploidy has been causatively linked to pathological conditions such as spontaneous abortions, trisomy syndromes and cancer. However, only in recent years have we witnessed an unraveling of the complex phenotypes that are caused by aneuploidy. Importantly, it has become apparent that aneuploidy evokes global and uniform changes that cannot be explained by the altered expression of the specific genes located on aneuploid chromosomes. Recent discoveries show that aneuploidy negatively affects protein folding; in particular, the functions of the molecular chaperone Heat Shock Protein 90 (HSP90) and the upstream regulator of heat shock-induced transcription, Heat Shock Factor 1 (HSF1), are impaired. Here we discuss the possible causes and consequences of this impairment and propose that the protein folding stress instigated by aneuploidy may be a common feature of conditions as variable as cancer and trisomy syndromes.
Keywords: aneuploidy, cancer, HSF1, HSP90, protein folding, proteostasis, trisomy
Equal segregation of chromosomes into daughter cells during cell division is essential for the proper proliferation of any organism. Errors in chromosome segregation lead to unbalanced chromosome numbers, so called aneuploidy. Aneuploidy is a deviation from the norm, and includes the gain and loss of one or a few chromosomes as well as massively aberrant karyotypes with chromosomal rearrangements and highly variable chromosome numbers. The effects of aneuploidy on physiology are determined by the altered expression of genes encoded on aneuploid chromosomes.1-3 Thus, the phenotypes of aneuploid cells are often variable, yet almost always detrimental, at least in Metazoans. Consequently, aneuploidy is the main cause of spontaneous abortions in humans and the rare aneuploid survivors suffer from multiple developmental defects. Paradoxically, however, aneuploidy is also a hallmark of cancer and promotes malignancy. But what exactly are the consequences of aneuploidy? What are the molecular mechanisms which underlie its detrimental phenotypes?
In recent years, the use of techniques that allow de novo generation of aneuploid cells of diverse karyotypes, coupled with systematic ‘omics’ analyses have facilitated significant advances in our understanding of the effects of whole chromosomal aneuploidy on cellular physiology (e.g.4-7). It is clear that imbalanced karytopes lead to chromosome- and thus gene-specific phenotypes.1,8 However, a novel view which has emerged is that aneuploidy also evokes specific global and uniform changes, largely conserved from yeast to man, that cannot be attributed simply to the expression changes of the specific genes located on aneuploid chromosomes. These changes go to the heart of cellular physiology and include detrimental effects on proliferation and genome stability, global changes in gene expression, as well as a rewiring of cellular metabolism.4,5,9,10 Elucidating the molecular basis for these complex phenomena is critical for a deeper understanding of trisomy syndromes as well as cancer, with which aneuploidy is tightly linked.
An important feature of the global and uniform changes elicited by aneuploidy is the effect that it exerts on protein homeostasis or proteostasis. Aneuploid cells accumulate ubiquitin- or Heat Shock Protein 104 (Hsp104)-positive inclusions,5,11,12 indicative of defective protein folding or insufficient clearance (or both), manifest higher levels of protein degradation,5,11,12 and exhibit sensitivity to conditions or drugs that impair proteostasis.1,11 Compelling evidence suggests that the primary reason for these phenomena is the gene expression from supernumerary chromosomes,2 but the molecular mechanisms involved are not well understood. Moreover, it is unknown whether these disturbances in proteostasis underlie some of the complex phenotypes observed in aneuploid cells.
Aneuploid Human Cells Exhibit Impaired HSF1 and HSP90 Function
To better understand the effects of aneuploidy on proteostasis in human cells, we employed luciferase-based constructs that serve as highly sensitive sensors of protein folding capacity.13 Tri- and tetrasomic aneuploid cells exhibited a pronounced impairment in their capacity to refold these sensors after heat shock and were also compromised in their ability to fold the proteins in the presence of a HSP90 inhibitor. These results indicated that protein folding stress and impaired function of the HSP90 molecular chaperone may be characteristic features of human aneuploid cells.12 Consistent with a specific defect in HSP90 function, aneuploid human cells were more sensitive to chemical inhibition of HSP90 than cognate diploids but did not display general sensitivity to other conditions or drugs that impair protein folding, such as HSC70/HSP70 inhibition or heat shock. A similar sensitivity was previously also observed in yeast1 and murine aneuploids,11 suggesting that diminished HSP90 capacity represents an evolutionarily conserved consequence of aneuploidy.
Interestingly, we observed that the ability of aneuploid cells to induce the expression of Heat Shock Proteins (HSPs) as part of the HSF1-mediated Heat Shock Response (HSR) is compromised. Thus, aneuploid cells exhibit consistently lower mRNA and protein levels of HSP90 and other HSF1-induced molecular chaperones as well as lower protein levels of HSF1 itself. This led us to hypothesize that inadequate activity of HSF1, the master regulator of chaperone transcription, may underlie the protein folding problems in human aneuploid cells. In fact, aneuploid cell lines with endogenous overexpression of HSF1, achieved by transfer of chromosome 8 which harbors the HSF1 coding sequence, were spared the aneuploidy-induced defects in HSP90 function, as evidenced by their folding of the mutant luciferase sensor and lack of sensitivity to chemical inhibition of HSP90. Likewise, ectopic overexpression of a constitutively active HSF1 allele in other aneuploids rescued the protein folding defect and protected against HSP90 inhibition.
A salient, but as yet poorly understood characteristic of aneuploid cells is a complex, genome-wide and conserved dysregulation of the transcriptome and proteome.5,6,14 HSF1 and HSP90 play critical and broad roles in regulating transcription and protein abundance and function, respectively.15,16 Thus, we hypothesized that there is a link between the aneuploidy-induced defects in the function of these proteins and the patterns of mRNA and protein abundance observed in aneuploid cells. In fact, we observed striking similarities between the changes in the transcriptome of aneuploid cells and the transcriptional changes identified in a cancer cell line in which HSF1 was depleted.17 Moreover, analysis of the proteome revealed that the abundance of HSP90 interacting proteins was significantly lower in 3 out of 4 aneuploid cell lines tested. Further, pathway analysis revealed a pronounced overlap between proteome changes in aneuploid cells and proteome changes occurring in response to HSP90 inhibition.18 In particular, we observed a marked overlap between downregulated pathways, suggesting the possibility that the inhibition of DNA and RNA metabolism, chromatin remodeling and cell cycle-related pathways might be directly or indirectly caused by a defect in HSP90. Taken together, our analysis suggests that the characteristic changes in mRNA and protein abundance as well as pathway activity in aneuploid cells are at least partially due to impaired protein folding capacity.
The Causes of the Protein Folding Defect in Human Aneuploid Cells
The causes of the protein folding defect in aneuploid cells and how aneuploidy impairs HSF1 function and specifically HSP90 remain unclear. What is certain is that the defects are due to the gene expression from the aneuploid chromosomes, as aneuploid yeast harboring transcriptionally silent Yeast Artificial Chromosomes (YACs) do not exhibit the impairment.2 How can the imbalanced gene expression in aneuploid cells lead to defects in protein folding? The findings made thus far are consistent with a model in which aneuploid cells experience low-level but chronic protein folding stress. Since the protein folding capacity of eukaryotic cells is tightly matched to the load of proteins that must be folded,19 the enhanced and imbalanced protein production in aneuploid cells is sufficient to trigger a genome-wide competition for protein folding factors. This, in turn, leads to the misfolding and degradation of a significant set of proteins. The chronic proteotoxic stress which ensues may inhibit the activation of the heat shock response in a manner which is reminiscent of observations in cellular models of neurodegenerative disease,20-22 where it is thought that the chronic expression of misfolded proteins impairs proteostasis capacity by titrating away key elements of the network. Thus, we propose that human aneuploid cells are subject to a vicious cycle of proteotoxic stress, in which impaired protein folding is constantly exacerbated by inhibition of HSF1 activity (Fig. 1). A similar concept has been invoked to explain the relationship between aneuploidy and chromosomal instability,23 and we therefore suggest that several of the phenotypes of aneuploid cells may be self-reinforcing.
Figure 1.
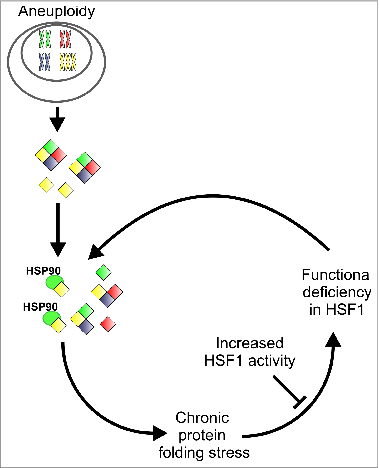
Effects of aneuploidy on cellular protein folding. The elevated levels of genes located on supernumerary chromosomes leads to imbalanced expression, which triggers a competition for protein folding factors, in particular among HSP90 clients, and leads to persistent protein folding stress. This sustained protein misfolding may then inhibit the activity of HSF1, resulting in a vicious cycle in which chronic protein misfolding is continuously aggravated by defective HSF1 activity.
We propose that the chronic protein folding stress experienced by aneuploid cells may be most clearly discerned in the expression changes of proteins encoded on supernumerary chromosomes. While the majority of gene products encoded on supernumerary chromosomes are present in aneuploid cells at levels expected based on gene copy number (e.g., approximately 1.5-fold elevated compared to diploid for a gene encoded on a trisomic chromosome), the expression of certain classes of proteins is attenuated or "compensated" posttranscriptionally.5,7,24,25 The first class of proteins subject to this compensation are members of protein complexes.5,24,25 It is noteworthy in this regard that protein complex members when present in stoichiometric excess relative to their binding partners likely rely upon chaperones to remain soluble and to be protected from degradation. Additionally, it has been proposed that HSP90 specifically may play a prominent role in the assembly of multi-molecular protein complexes.26 Reduced expression of a second class of proteins, kinases, was described in human aneuploid cells.5 Intriguingly, a large number of protein kinases are dependent on HSP90 function in order to fold and function properly.27 We previously proposed28 that the decreased abundance of these protein classes stems from the insufficient protein folding capacity triggered by imbalanced gene expression (Fig. 2). Consistent with such a scenario, it was recently demonstrated in aneuploid yeast that the downregulation of compensated proteins occurs posttranslationally and is mediated by the proteasome and autophagy.25 Taken together, we propose that the enhanced protein production due to the presence of extra chromosomes might titrate away chaperones, thus leading to the chronic misfolding and degradation of a subset of the cellular proteome.
Figure 2.
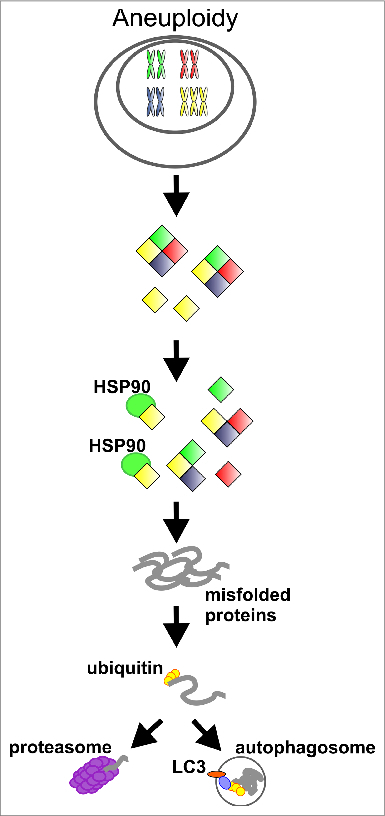
Proposed model for protein compensation in aneuploid cells. The elevated levels of genes located on supernumerary chromosomes leads to imbalanced expression and to competition for protein folding factors, in particular among HSP90 clients, such as protein kinases, subunits of multimolecular complexes and probably others. This competition results in chronic protein misfolding and degradation of proteins with exaggerated needs for the protein folding machinery.
Protein Folding Stress and the Phenotypes of Aneuploid Cells
The prominent and pervasive protein folding defect in aneuploid cells raises the tantalizing possibility that other hallmarks of aneuploidy may be a result of impaired protein folding capacity. A potential link between the disturbed proteostasis of aneuploid cells and their defects in proliferation is particularly intriguing. There is now a large body of evidence implicating HSF1 and HSP90 as important facilitators of cellular proliferation, particularly in cancer cells. It is particularly noteworthy that through its chaperone activity HSP90 regulates either directly or indirectly a large number of proteins with important roles in promoting G1 to S transition, such as Cyclin D and Cyclin E as well as Cyclin-dependent kinase 4 (CDK4), Cyclin-dependent kinase 6 (CDK6) and Cyclin-dependent kinase 2 (CDK2).29 In fact, levels of HSP90 protein peak at the G1/S boundary in a HSF1-dependent manner,30 indicating that both these factors are involved in promoting G1 to S transition and chemical inhibition of HSP90 most commonly leads to a cell cycle arrest at G1/S.29 It has long been recognized that aneuploidy often impairs proliferation in yeast as well as Metazoans, and although this has been linked with defects in progression through the G1 and S phases of the cell cycle,5,31 the molecular basis for this impairment is not understood. We anticipate that future studies will resolve if and how the impaired protein folding capacity of aneuploid cells is linked with their defective cell cycle progression.
HSP90 also chaperones a large number of proteins with crucial roles in maintaining the integrity of the genome.32 For example, HSP90 is an essential regulator of DNA polymerase eta activity during translesion synthesis at stalled replication forks, promotes stabilization and correct localization of the repair factors Fanconi anemia, complementation group A (FANCA) and breast cancer 2/Fanconi anemia, complementation group D (FANCD1/BRCA2),33 and its activity is required for stabilization of Mis12 complexes at kinetochores, thus contributing to efficient microtubule-kinetochore attachments.34 Further, inhibition of HSP90 itself leads to increased sensitivity to DNA damaging agents and to aneuploidy in yeast.35 Recently, the first direct evidence emerged that aneuploidy promotes genomic instability and additional whole chromosomal aneuploidy by elevating the levels of chromosome gain and loss and of mitotic recombination.9,10,23 However, the mechanisms behind this have so far remained elusive. It is tempting to speculate that an underlying reason for the genomic instability of aneuploid cells is defective protein folding.
Aneuploidy, Cancer and HSF1
It is now firmly established that HSF1 plays a critical role in promoting many of the hallmarks of malignant cells.36 High expression of HSF1 has been reported in a number of cancers,15 and elevated levels of nuclear HSF1 are an independent predictor of poor outcome in breast cancer,37 in hepatocellular carcinoma,38 and in endometrial carcinoma.39 Additionally, loss of HSF1 suppresses tumor formation in several murine models.40 It is likely that HSF1 exerts its tumor-promoting functions by stimulating the expression of molecular chaperones that ameliorate the cell stress which appears to be an unavoidable part of tumorigenesis.41 Through induction of HSP90 expression, HSF1 promotes the stabilization of numerous tumor-promoting HSP90 clients such as macrophage migration inhibitory factor (MIF), protein kinase B (PKB) and HSF1 itself42 as well as a plethora of factors involved in promoting cell proliferation such as cyclin-dependent kinases,43-45 and unstable oncogenes.46 In fact, several studies have shown that HSF1 is necessary for promoting and maintaining cancer cell proliferation,40,47 as well as supporting additional critical facets of tumor biology, such as glucose metabolism and signal transduction.36,40 Further, the transcriptional program of HSF1 is altered in tumor samples in comparison to non-transformed cells and includes many genes that support critical oncogenic processes.15 Finally, HSF1 even facilitates carcinogenesis in a non-cell-autonomous manner by rewiring the transcriptome of cancer-associated fibroblasts to promote and support malignant cells.48
How can our finding that HSF1 activity is impaired in aneuploid cells be reconciled with the fact that both aneuploidy and heightened activity of HSF1 are frequently observed in cancer? The answer to this question may be illuminated by the observation that the parental colorectal cancer cell line HCT116, used as a parental cell line for our studies on aneuploidy, is characterized by pre-existing segmental aneuploidy of the long arm of chromosome 8 where the HSF1 locus is located. Upon chromosome transfer-induced aneuploidization protein folding is impaired in these cells, indicating that de novo induction of aneuploidy always exerts a strong negative effect on HSF1 activity, irrespective of prior levels of HSF1 protein. This negative effect can only be overridden by additional augmentation of HSF1 levels. Thus, our results suggest that, in addition to the already described functions of HSF1 in tumorigenesis, an important reason for why HSF1 activity is elevated in cancer might be to protect against the deleterious effects of aneuploidy on proteostasis. Indeed, although almost ubiquitous in cancer, there is strong evidence that, through its inhibitory effects on proliferation and spontaneous immortalization, aneuploidy also acts as a tumor suppressor.4,49,50 We propose that one explanation for this is the protein folding defect instigated by aneuploidy. As a consequence, proteotoxic stress represents a hurdle that aneuploid cells must negotiate on their way to malignancy or in chromosomally unstable tumors, a chronic problem that must be efficiently counteracted. Intriguingly, it has been shown that loss-of-function of the deubiquitinating enzyme Ubp6, a protein proposed to negatively regulate proteasomal degradation, can suppress the formation of cytoplasmic protein inclusions in aneuploid yeast.2 Taken together with our observations, this suggests that cells can alleviate the detrimental effects of aneuploidy on proteostasis by either augmenting protein folding capacity or increasing protein degradation (Fig. 3), and further indicates that adaptation to aneuploidy might partially explain the heavy reliance of cancer cells on HSF1 function and on proteasomal degradation.
Figure 3.
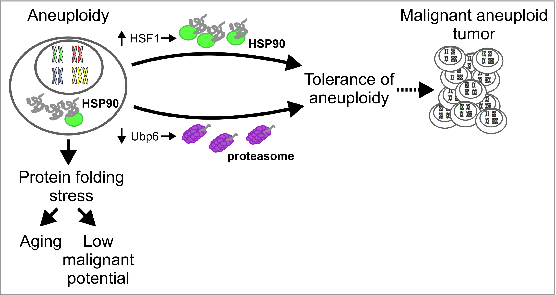
Potential paths to ameliorating the effects of aneuploidy on proteostasis. The impaired protein folding capacity of aneuploid cells represents a barrier to tumorigenesis and may result in cellular aging. Cells may overcome these detrimental effects by either improving protein folding capacity or by enhancing protein degradation, thus elevating their proliferative capacity and potentially giving rise to malignant aneuploid tumors.
Aneuploidy and Protein Folding Stress in Trisomy Syndromes
The observations summarized above may offer a new way of understanding trisomy syndromes, such as Down syndrome and it will be important to determine whether chronic proteotoxic stress partially underlies the deleterious phenotypes seen in these conditions. Further, although trisomy is always detrimental in humans, we speculate that one reason that certain trisomies are not compatible with life or even prenatal development is due to a high burden of proteotoxic stress. Conversely, embryos that survive and develop may either be subject to relatively mild proteotoxic stress or have found a way to alleviate it. Consistent with such a notion, chromosome 8, which harbors the HSF1 gene is the largest somatic chromosome whose trisomy is tolerated in post-natal development,51,52 and chromosome 21, trisomy of which represents the most common viable aneuploid karyotype in humans, is the smallest and most gene-poor somatic human chromosome.
Conclusions/Outlook
Recent research on aneuploidy has facilitated an ever greater understanding of the effects of imbalanced chromosome numbers on cellular physiology. It now appears clear that impaired function of the proteostasis network and protein folding stress represent evolutionarily conserved hallmarks of aneuploid cells. The two major challenges to be negotiated are a more intricate dissection of the molecular basis for the protein folding stress in aneuploid cells as well as a delineation of the relationship between defects in protein folding and other deleterious phenotypes of aneuploidy. It is to be hoped that progress on both of these questions may lead to a clearer grasp of the pathology of trisomy syndromes. Indeed, our evolving understanding of the effects of aneuploidy on proteostasis make a compelling case for investigating the status of the proteostasis network in cells isolated from people with trisomy syndromes. It has long been known that aneuploidy is a recurring feature of cancer cells. However, as the effects of aneuploidy on cellular physiology have remained obscure, so too has the role of aneuploidy in tumorigenesis been enigmatic. The insights gained thus far together with a deeper appreciation of the molecular mechanisms involved should lead to a fuller understanding of carcinogenesis.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Boris Pfander, Lisa Vincenz and the members of the Maintenance of Genome Stability Group for their helpful comments on the manuscript and figures.
Funding
This work was supported by the Max Planck Society, the Center for Integrated Protein Science, Munich, and by a grant from Deutsche Forschungsgemeinschaft to Z.S.
References
- 1. Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 2007; 317:916-24; PMID:17702937; http://dx.doi.org/ 10.1126/science.1142210 [DOI] [PubMed] [Google Scholar]
- 2. Oromendia AB, Dodgson SE, Amon A. Aneuploidy causes proteotoxic stress in yeast. Genes Dev 2012; 26:2696-708; PMID:23222101; http://dx.doi.org/ 10.1101/gad.207407.1122,12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jiang J, Jing Y, Cost GJ, Chiang JC, Kolpa HJ, Cotton AM, Carone DM, Carone BR, Shivak DA, Guschin DY, et al. Translating dosage compensation to trisomy 21. Nature 2013; 500:296-300; PMID:23863942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 2008; 322:703-9; PMID:18974345; http://dx.doi.org/ 10.1126/science.1160058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchova Z. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol 2012; 8:608; PMID:22968442; http://dx.doi.org/ 10.1038/msb.2012.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sheltzer JM, Torres EM, Dunham MJ, Amon A. Transcriptional consequences of aneuploidy. Proc Natl Acad Sci U S A 2012; 109:12644-9; PMID:22802626; http://dx.doi.org/ 10.1073/pnas.1209227109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 2010; 468:321-5; PMID:20962780; http://dx.doi.org/ 10.1038/nature09529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rancati G, Pavelka N, Fleharty B, Noll A, Trimble R, Walton K, Perera A, Staehling-Hampton K, Seidel CW, Li R. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell 2008; 135:879-93; PMID:19041751; http://dx.doi.org/ 10.1016/j.cell.2008.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, Humpton TJ, Brito IL, Hiraoka Y, Niwa O, Amon A. Aneuploidy drives genomic instability in yeast. Science 2011; 333:1026-30; PMID:21852501; http://dx.doi.org/ 10.1126/science.1206412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu J, Pavelka N, Bradford WD, Rancati G, Li R. Karyotypic determinants of chromosome instability in aneuploid budding yeast. PLoS Genet 2012; 8:e1002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tang YC, Williams BR, Siegel JJ, Amon A. Identification of aneuploidy-selective antiproliferation compounds. Cell 2011; 144:499-512; PMID:21315436; http://dx.doi.org/ 10.1016/j.cell.2011.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Donnelly N, Passerini V, Durrbaum M, Stingele S, Storchova Z. HSF1 deficiency and impaired HSP90-dependent protein folding are hallmarks of aneuploid human cells. EMBO J 2014; 33:2374-87; PMID:25205676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gupta R, Kasturi P, Bracher A, Loew C, Zheng M, Villella A, Garza D, Hartl FU, Raychaudhuri S. Firefly luciferase mutants as sensors of proteome stress. Nat Methods 2011; 8:879-84; PMID:21892152; http://dx.doi.org/ 10.1038/nmeth.1697 [DOI] [PubMed] [Google Scholar]
- 14. Durrbaum M, Kuznetsova AY, Passerini V, Stingele S, Stoehr G, Storchova Z. Unique features of the transcriptional response to model aneuploidy in human cells. BMC Genomics 2014; 15:139; PMID:24548329; http://dx.doi.org/ 10.1186/1471-2164-15-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mendillo ML, Santagata S, Koeva M, Bell GW, Hu R, Tamimi RM, Fraenkel E, Ince TA, Whitesell L, Lindquist S. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell 2012; 150:549-62; PMID:22863008; http://dx.doi.org/ 10.1016/j.cell.2012.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer 2005; 5:761-72; PMID:16175177; http://dx.doi.org/ 10.1038/nrc1716 [DOI] [PubMed] [Google Scholar]
- 17. Chuma M, Sakamoto N, Nakai A, Hige S, Nakanishi M, Natsuizaka M, Suda G, Sho T, Hatanaka K, Matsuno Y, et al. Heat shock factor 1 accelerates hepatocellular carcinoma development by activating nuclear factor-kappaB/mitogen-activated protein kinase. Carcinogenesis 2014; 35:272-81; PMID:24130164; http://dx.doi.org/ 10.1093/carcin/bgt343 [DOI] [PubMed] [Google Scholar]
- 18. Sharma K, Vabulas RM, Macek B, Pinkert S, Cox J, Mann M, Hartl FU. Quantitative proteomics reveals that Hsp90 inhibition preferentially targets kinases and the DNA damage response. Mol Cell Proteomics 2012; 11:M111 014654; PMID:22167270; http://dx.doi.org/ 10.1074/mcp.O111.015149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gidalevitz T, Prahlad V, Morimoto RI. The stress of protein misfolding: from single cells to multicellular organisms. Cold Spring Harb Perspect Biol 2011; 3:pii: a009704; PMID:21536706; http://dx.doi.org/ 10.1101/cshperspect.a009704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 2011; 144:67-78; PMID:21215370; http://dx.doi.org/ 10.1016/j.cell.2010.11.050 [DOI] [PubMed] [Google Scholar]
- 21. Chafekar SM, Duennwald ML. Impaired heat shock response in cells expressing full-length polyglutamine-expanded huntingtin. PloS One 2012; 7:e37929; PMID:22649566; http://dx.doi.org/ 10.1371/journal.pone.0037929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hipp MS, Park SH, Hartl FU. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol 2014; 24:506-14; PMID:24946960; http://dx.doi.org/ 10.1016/j.tcb.2014.05.003 [DOI] [PubMed] [Google Scholar]
- 23. Potapova TA, Zhu J, Li R. Aneuploidy and chromosomal instability: a vicious cycle driving cellular evolution and cancer genome chaos. Cancer Metastasis Rev 2013; 32:377-89; PMID:23709119; http://dx.doi.org/ 10.1007/s10555-013-9436-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A. Identification of aneuploidy-tolerating mutations. Cell 2010; 143:71-83; PMID:20850176; http://dx.doi.org/ 10.1016/j.cell.2010.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dephoure N, Hwang S, O'Sullivan C, Dodgson SE, Gygi SP, Amon A, Torres EM. Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. eLife 2014; 3:e03023; PMID:25073701; http://dx.doi.org/ 10.7554/eLife.03023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Makhnevych T, Houry WA. The role of Hsp90 in protein complex assembly. Biochim Biophys Acta 2012; 1823:674-82; PMID:21945180; http://dx.doi.org/ 10.1016/j.bbamcr.2011.09.001 [DOI] [PubMed] [Google Scholar]
- 27. Caplan AJ, Mandal AK, Theodoraki MA. Molecular chaperones and protein kinase quality control. Trends Cell Biol 2007; 17:87-92; PMID:17184992; http://dx.doi.org/ 10.1016/j.tcb.2006.12.002 [DOI] [PubMed] [Google Scholar]
- 28. Donnelly N, Storchova Z. Dynamic karyotype, dynamic proteome: buffering the effects of aneuploidy. Biochim Biophys Acta 2014; 1843:473-81; PMID:24295790; http://dx.doi.org/ 10.1016/j.bbamcr.2013.11.017 [DOI] [PubMed] [Google Scholar]
- 29. Burrows F, Zhang H, Kamal A. Hsp90 activation and cell cycle regulation. Cell Cycle 2004; 3:1530-6; PMID:15539946; http://dx.doi.org/ 10.4161/cc.3.12.1277 [DOI] [PubMed] [Google Scholar]
- 30. Nakai A, Ishikawa T. Cell cycle transition under stress conditions controlled by vertebrate heat shock factors. EMBO J 2001; 20:2885-95; PMID:11387221; http://dx.doi.org/ 10.1093/emboj/20.11.2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thorburn RR, Gonzalez C, Brar GA, Christen S, Carlile TM, Ingolia NT, Sauer U, Weissman JS, Amon A. Aneuploid yeast strains exhibit defects in cell growth and passage through START. Mol Biol Cell 2013; 24:1274-89; PMID:23468524; http://dx.doi.org/ 10.1091/mbc.E12-07-0520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kaplan KB, Li R. A prescription for 'stress'–the role of Hsp90 in genome stability and cellular adaptation. Trends Cell Biol 2012; 22:576-83; PMID:22959309; http://dx.doi.org/ 10.1016/j.tcb.2012.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stecklein SR, Kumaraswamy E, Behbod F, Wang W, Chaguturu V, Harlan-Williams LM, Jensen RA. BRCA1 and HSP90 cooperate in homologous and non-homologous DNA double-strand-break repair and G2/M checkpoint activation. Proc Natl Acad Sci U S A 2012; 109:13650-5; PMID:22869732; http://dx.doi.org/ 10.1073/pnas.1203326109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Davies AE, Kaplan KB. Hsp90-Sgt1 and Skp1 target human Mis12 complexes to ensure efficient formation of kinetochore-microtubule binding sites. J Cell Biol 2010; 189:261-74; PMID:20404110; http://dx.doi.org/ 10.1083/jcb.200910036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen G, Bradford WD, Seidel CW, Li R. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature 2012; 482:246-50; PMID:22286062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ciocca DR, Arrigo AP, Calderwood SK. Heat shock proteins and heat shock factor 1 in carcinogenesis and tumor development: an update. Arch Toxicol 2013; 87:19-48; PMID:22885793; http://dx.doi.org/ 10.1007/s00204-012-0918-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Santagata S, Hu R, Lin NU, Mendillo ML, Collins LC, Hankinson SE, Schnitt SJ, Whitesell L, Tamimi RM, Lindquist S, et al. High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc Natl Acad Sci U S A 2011; 108:18378-83; PMID:22042860; http://dx.doi.org/ 10.1073/pnas.1115031108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fang F, Chang R, Yang L. Heat shock factor 1 promotes invasion and metastasis of hepatocellular carcinoma in vitro and in vivo. Cancer 2012; 118:1782-94; PMID:22009757; http://dx.doi.org/ 10.1002/cncr.26482 [DOI] [PubMed] [Google Scholar]
- 39. Engerud H, Tangen IL, Berg A, Kusonmano K, Halle MK, Oyan AM, Kalland KH, Stefansson I, Trovik J, Salvesen HB, et al. High level of HSF1 associates with aggressive endometrial carcinoma and suggests potential for HSP90 inhibitors. Br J Cancer 2014; 111:78-84; PMID:24853175; http://dx.doi.org/ 10.1038/bjc.2014.262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell 2007; 130:1005-18; PMID:17889646; http://dx.doi.org/ 10.1016/j.cell.2007.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 2009; 136:823-37; PMID:19269363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schulz R, Streller F, Scheel AH, Ruschoff J, Reinert MC, Dobbelstein M, Marchenko ND, Moll UM. HER2/ErbB2 activates HSF1 and thereby controls HSP90 clients including MIF in HER2-overexpressing breast cancer. Cell Death Dis 2014; 5:e980; PMID:24384723; http://dx.doi.org/ 10.1038/cddis.2013.508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Prince T, Sun L, Matts RL. Cdk2: a genuine protein kinase client of Hsp90 and Cdc37. Biochemistry 2005; 44:15287-95; PMID:16285732; http://dx.doi.org/ 10.1021/bi051423m [DOI] [PubMed] [Google Scholar]
- 44. Stepanova L, Leng X, Parker SB, Harper JW. Mammalian p50Cdc37 is a protein kinase-targeting subunit of Hsp90 that binds and stabilizes Cdk4. Genes Dev 1996; 10:1491-502; PMID:8666233; http://dx.doi.org/ 10.1101/gad.10.12.1491 [DOI] [PubMed] [Google Scholar]
- 45. Mahony D, Parry DA, Lees E. Active cdk6 complexes are predominantly nuclear and represent only a minority of the cdk6 in T cells. Oncogene 1998; 16:603-11; PMID:9482106; http://dx.doi.org/ 10.1038/sj.onc.1201570 [DOI] [PubMed] [Google Scholar]
- 46. Blagosklonny MV. Hsp-90-associated oncoproteins: multiple targets of geldanamycin and its analogs. Leukemia 2002; 16:455-62; PMID:11960322; http://dx.doi.org/ 10.1038/sj.leu.2402415 [DOI] [PubMed] [Google Scholar]
- 47. Meng L, Gabai VL, Sherman MY. Heat-shock transcription factor HSF1 has a critical role in human epidermal growth factor receptor-2-induced cellular transformation and tumorigenesis. Oncogene 2010; 29:5204-13; PMID:20622894; http://dx.doi.org/ 10.1038/onc.2010.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Scherz-Shouval R, Santagata S, Mendillo ML, Sholl LM, Ben-Aharon I, Beck AH, Dias-Santagata D, Koeva M, Stemmer SM, Whitesell L, et al. The reprogramming of tumor stroma by HSF1 is a potent enabler of malignancy. Cell 2014; 158:564-78; PMID:25083868; http://dx.doi.org/ 10.1016/j.cell.2014.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol 2010; 188:369-81; PMID:20123995; http://dx.doi.org/ 10.1083/jcb.200905057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 2007; 11:25-36; PMID:17189716; http://dx.doi.org/ 10.1016/j.ccr.2006.12.003 [DOI] [PubMed] [Google Scholar]
- 51. Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010; 463:899-905; PMID:20164920; http://dx.doi.org/ 10.1038/nature08822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ganmore I, Smooha G, Izraeli S. Constitutional aneuploidy and cancer predisposition. Hum Mol Genet 2009; 18:R84-93; PMID:19297405; http://dx.doi.org/ 10.1093/hmg/ddp084 [DOI] [PMC free article] [PubMed] [Google Scholar]


