Abstract
Background
A qualitative highly predictive urinary test for polyomavirus nephropathy (PVN) is the PV-Haufen test. This article evaluates whether a quantitative PV-Haufen analysis, that is, the number of PV-Haufen shed per milliliter urine, predicts PVN disease grades and the severity of intrarenal PV replication.
Methods
Polyomavirus-Haufen were counted in 40 urine samples from patients with biopsy-proven definitive PVN. The number of PV-Haufen was correlated with both histologic PVN disease grades 1 to 3 and the number of SV40-T–expressing cells as indicators of intrarenal PV replication in corresponding renal allograft biopsies (manual counts and automated morphometry). Findings from quantitative PV-Haufen analyses were compared to conventional laboratory test results, that is, BK viremia (quantitative polymerase chain reaction [PCR]) and BK viruria (quantitative PCR and decoy cell counts).
Results
Polyomavirus-Haufen counts showed excellent correlation (α0.77–0.86) with the severity of intrarenal PV replication and disease grades. In particular, low PV-Haufen numbers strongly correlated with early PVN grade 1 and minimal intrarenal expression of SV40-T antigen (P < 0.001). In comparison, BK viremia and viruria levels by PCR showed only modest correlations with histologic SV40-T expression (α0.40–0.49) and no significant correlation with disease grades or minimal intrarenal PV replication. No correlations were seen with urinary decoy cell counts. In contrast to conventional quantitative PCR assays or decoy cell counts, quantitative urinary PV-Haufen testing accurately reflects the severity of PV replication, tissue injury, and PVN disease grades.
Conclusions
Quantitative PV-Haufen testing is a novel noninvasive approach to patient management for the diagnosis and prediction of PVN disease grades and monitoring of disease course during therapy.
Quantitative testing of polyoma virus within shed renal tubule cells in the urine is superior to conventional BKV urinary or blood quantitation to define both the presence and degree of polyoma nephropathy. Supplemental digital content is available in the text.
Polyomavirus nephropathy (PVN) is the most important viral infection in renal allografts. The incidence of PVN in kidney transplant recipients ranges from 4% to 12%.1-4 ABO-incompatible grafts seem to be at increased risk with a prevalence of 18%.5 Graft failure because of PVN occurs in approximately 20% to 30% with a higher prevalence of 50% in cases of sclerosed PVN disease grade 3 (data collected by the Banff working group on polyomavirus nephropathy). Specific anti-PV therapy is not available, and outcome largely depends on an early diagnosis of PVN with limited renal injury that tends to respond favorably to therapeutic intervention (mainly based on the reduction of immunosuppression).
Polyomavirus nephropathy is defined morphologically by the histologic demonstration of polyomavirus (PV) replication in the renal parenchyma, and a renal biopsy is required to render a definitive diagnosis.6-9 However, establishing a diagnosis of PVN in a kidney biopsy can be challenging. Deterioration of allograft function and a rise in serum creatinine levels does not always accompany early PVN, resulting in a possible delay of a diagnostic renal biopsy.6,8
Patient management after kidney transplantation includes risk assessment for potential PVN with quantitative polymerase chain reaction (PCR) assays for BK virus loads in plasma or urine or quantitation of urinary decoy cells in urine cytology specimens. These assays have been the mainstays for both clinical care and intervention.2,10-19 They are based on the observation that all patients with PVN show systemic signs of PV replication with viremia or viruria, and the assumption that plasma PCR levels for BK virus reflect intrarenal PV replication. However, only a minority of patients with PV viremia or viruria ultimately develops PVN, underscoring a well-established fact: latency establishing DNA viruses, such as PV, can be reactivated without causing manifest disease.8,18,20-24 Moreover, in immunocompromised patients, BK virus–associated disease with viremia can also be seen in the bladder (hemorrhagic cystitis) or in the salivary glands (human immunodeficiency virus–associated salivary gland sclerosis), typically lacking concurrent PVN.25-28 Thus, positive PCR-based BK viremia or viruria test results or the presence of decoy cells in the urine indicate viral activation and an increased risk for PVN, but the tests cannot reliably predict the actual presence of intrarenal viral disease, that is, definitive PVN. In the setting of PVN, an ideal biomarker would not only indicate the presence or absence of intrarenal disease but also provide information on the severity of virally induced tissue injury.
We recently described a noninvasive biomarker to accurately predict PVN in fixed voided urine samples, that is, the PV-Haufen test. We showed that a qualitative analysis, that is, PV-Haufen present or absent, marked intrarenal disease with positive and negative predictive values exceeding 95%.20,29 The PV-Haufen test is based on a novel concept, that is, the detection of dense, cast-like PV aggregates in fixed voided urine samples. Polyomavirus-Haufen form in virally injured renal tubules subsequent to PV release from infected tubular epithelial cells. They are then flushed out of the diseased kidneys and can be detected in the urine. In our previous publication, we reported that PV-Haufen were exclusively found in patients with biopsy-proven definitive PVN and were never seen in 139 control patients with varying degrees of BK viremia and viruria and absence of PVN. In patients with a definitive biopsy-proven diagnosis of PVN who were followed longitudinally, PV-Haufen shedding went from negative to positive at the time of initial biopsy diagnosis. Polyomavirus-Haufen testing subsequently remained positive during persistent viral nephropathy. At the time of disease resolution, PV-Haufen testing turned from positive to negative and remained negative thereafter during long-term follow-up. Thus, the qualitative detection of urinary PV-Haufen, that is, present or absent, is disease-specific and represents a novel clinical biomarker for definitive PVN.20,30
Currently, it is undetermined whether a quantitative urinary PV-Haufen analysis, that is, recording the number of PV-Haufen shed per milliliter urine, could provide additional clinical information on the severity of intrarenal disease. It is also undetermined how a quantitative urinary PV-Haufen assay might compare to conventional mainly PCR-based laboratory screening tests for PVN.
We address these questions in our current study that expands on our previous qualitative PV-Haufen test findings by now focusing on quantitative testing. Because PV-Haufen shedding is only found in patients with biopsy-proven definitive PVN, we hypothesize that the degree of urinary PV-Haufen shedding reflects the severity of virally induced kidney injury. The aim of our current study is to correlate traditional screening assays and quantitative PV-Haufen test results with the severity of intrarenal PV replication or PV burden and disease grades in patients with biopsy-proven PVN. Which test is most accurate?
RESULTS
Correlation of Quantitative Screening Test Results With the Severity of Intrarenal PV Replication
The severity of intrarenal PV replication was determined by using an immunohistochemical stain for the SV40-T antigen and manually counting the number of positive staining nuclei, the number of positive staining tubular cross-sections, or alternatively performing (automated) morphometry. Irrespective of manual or automated analysis, the number of urinary PV-Haufen most accurately reflected the severity of intrarenal PV replication–SV40-T expression with excellent correlation coefficients between 0.77 and 0.86 (Table 1 and Figure 1). Of note, correlations were tightest for comparisons based on the manual counts rather than the (automated) morphometric deconvolution analyses using the Aperio system. Plasma and urine PCR readings (BK viremia and viruria) showed a modest correlation with intrarenal PV replication (correlation coefficients between 0.40 and 0.49). No correlation was seen between the number of decoy cells in the urine and the SV40-T expression in the corresponding biopsy samples.
TABLE 1.
Correlation of quantitative screening test results with the severity of intrarenal polyomavirus replication/ viral burden in cases of PVN
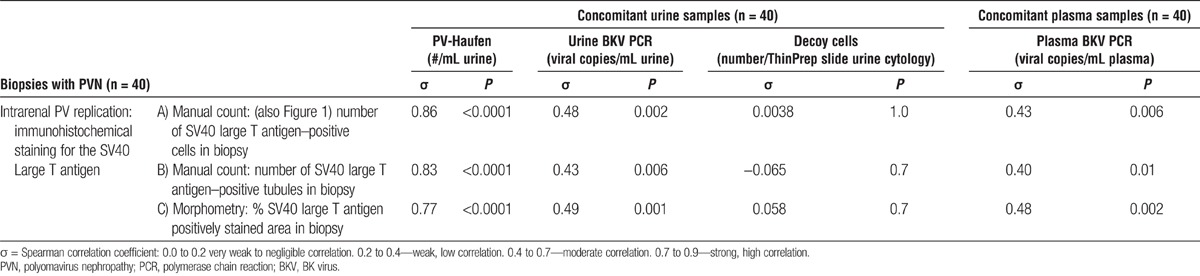
FIGURE 1.
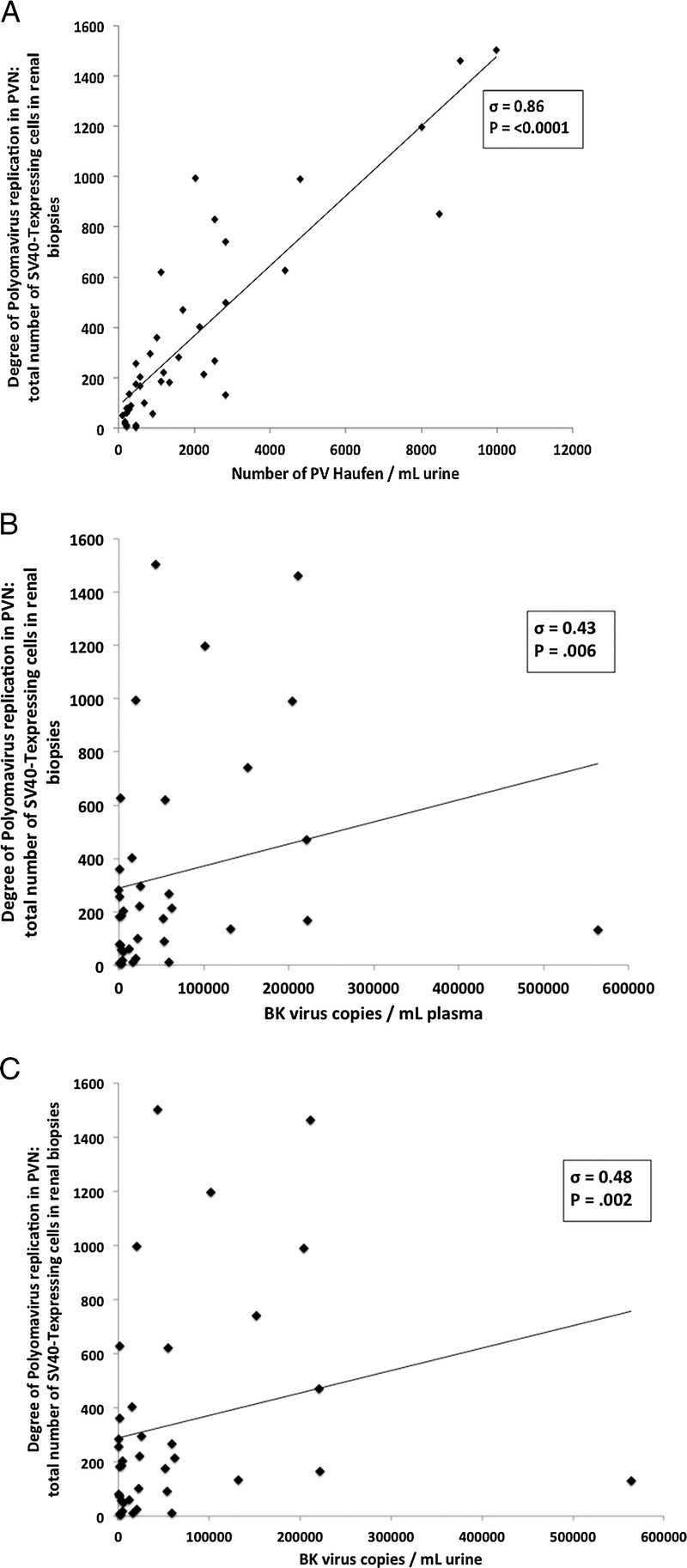
Correlation of quantitative screening test results with intrarenal PV replication, that is, the number of SV40-T–expressing tubular epithelial cells in a biopsy sample with PVN. A, Urinary PV-Haufen; (B) Plasma PCR for BK virus; and (C) Urine PCR for BK virus. Scatterplots with regression lines are depicted. PVN, polyomavirus nephropathy; PCR, polymerase chain reaction.
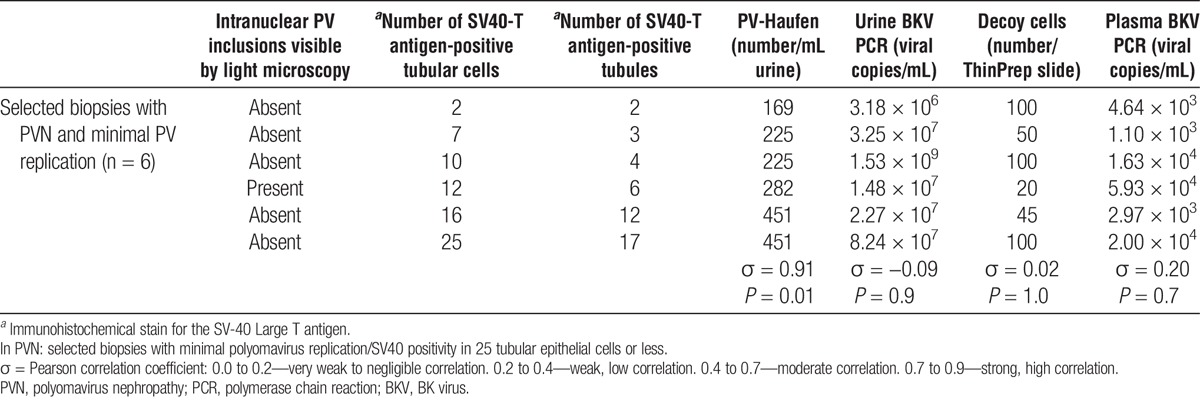
For correlation studies of a subgroup of PVN cases with minimal PV burden, six biopsies with 25 or less intrarenal SV40-T antigen–expressing cells were selected. Five of these six biopsies lacked intranuclear PV inclusion bodies by light microscopy further supporting an early and limited PVN disease stage in these cases. All six cases showed low urinary PV-Haufen numbers that tightly correlated with the low intrarenal PV burden (σ = 0.91; P = 0.01). Lowest PV-Haufen counts were found in one case with only two intrarenal SV40-T antigen-expressing cells (Table 2). The PV-Haufen counts increased in increments in the remaining five cases parallel to increased intrarenal PV load levels. In contrast, in this cohort, no significant correlations were found between all other screening assays and minimal intrarenal PV replication: plasma PCR σ = 0.20, P = 0.7; urine PCR σ = −0.09, P = 0.9; and decoy cell shedding σ = 0.02, P = 1.0.
TABLE 2.
Correlation of quantitative screening test results with intrarenal polyomavirus replication or viral burden
Correlation of Screening Tests With PVN Disease Grades
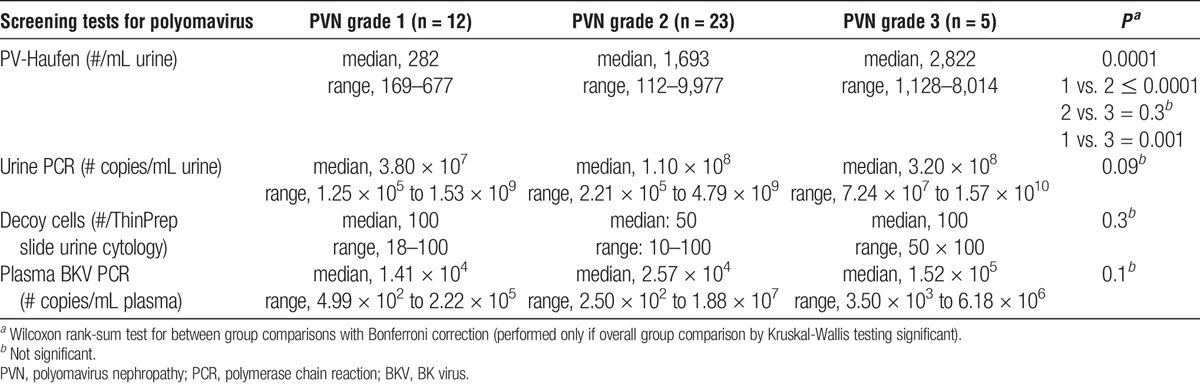
Low numbers of urinary PV-Haufen were statistically significantly associated with early or mild PVN disease grade 1 in 12 of the 12 cases with a median PV-Haufen count of 282/mL urine and a maximum count of 677 PV-Haufen/mL urine seen in one patient. Polyomavirus nephropathy disease grade 2 showed median PV-Haufen counts significantly higher at 1693/mL (range, 112–9977/mL urine; P =< 0.0001). On an individual case basis, PV-Haufen counts showed only little overlap between PVN disease grades 1 and 2 (Table 3 and Figure 1S, A, SDC, http://links.lww.com/TP/B46). Polyomavirus nephropathy disease grade 3 demonstrated median PV-Haufen counts of 2822/mL, which differed significantly from PVN disease grade 1 (1 vs. 3 P = 0.002). In comparison, the degree of BK viremia and BK viruria by PCR testing and the degree of decoy cell shedding by urine cytology were not significantly associated with PVN disease grades 1–3 (Table 3).
TABLE 3.
Quantitative screening test results by PVN disease grade
DISCUSSION
Urinary PV-Haufen are tight three-dimensional cast-like viral aggregates that are easily detected in fixed urine samples by negative staining electron microscopy (EM).20,29 In a previous study, we showed that the qualitative detection of PV-Haufen, that is, present or absent, in a fixed urine specimen predicted PVN with positive and negative predictive values greater than 95%.20
In the current study, we extended our analysis to quantitative urinary PV-Haufen testing. We correlated the number of PV-Haufen in voided urine samples with the severity of PVN and compared the data to standard PV screening assays. This comprehensive comparative analysis showed that quantitative urinary PV-Haufen testing most accurately reflected the severity of intrarenal PV replication and virally induced renal injury. Few PV-Haufen in urine samples were associated with limited intrarenal PV replication, including one case with only two intrarenal SV40-T–expressing cells. Counts increased in increments parallel to the increase of intrarenal PV replication and high numbers of urinary PV-Haufen reflected marked virally induced tubular injury, a high intrarenal PV burden (correlation coefficient, 0.86).
Quantitative PV-Haufen testing also reflected PVN disease grades as proposed by the Banff Working Group on PVN.31 Early mild PVN disease grade 1 was reflected by low numbers of urinary PV-Haufen that differed significantly from much higher counts in the florid PVN disease grade 2, and the late and sclerosed PVN disease grade 3.
How can the striking correlation between the severity of virally induced tissue injury in PVN and the numbers of urinary PV-Haufen shed into the urine be explained? The formation of PV-Haufen appears to be similar to the formation of other intratubular casts such as red or white blood cell casts. Polyomavirus-Haufen form within virally injured renal tubules containing high concentrations of uromodulin (Tamm-Horsfall protein) that has previously been shown to facilitate aggregation of influenza, mumps, and Newcastle viruses.32 High concentrations of Tamm-Horsfall protein facilitate the aggregation of PV in vitro (personal observation; manuscript in preparation).30 Because PV-Haufen are flushed with the urine out of tubules and into the bladder, they can serve as specific urinary biomarkers for intrarenal PVN.
In contrast, quantitative PCR assays to detect BK virus in plasma and urine only showed modest correlation with the severity of intrarenal PV replication or PV burden (correlation coefficients, 0.43 to 0.48). No significant correlations were found between quantitative PCR test results and PVN disease grades or limited PVN with only minimal intrarenal disease. No significant correlations were seen with urinary decoy cell counts. These results could be explained by potential PV activation in sites other than the kidneys, such as the urothelium or salivary glands.25-28
In daily practice, we do not recommend using quantitative urinary PV-Haufen testing as a mass screening tool. Rather, it should be used as a targeted test to separate patients with asymptomatic transient BK viral activation from those with intrarenal disease, that is, definitive PVN. The test provides specific diagnostic evidence of intrarenal PVN and gives prognostic information on the severity of virally induced tissue injury. It is particularly suited for the diagnosis of PVN in early disease grade 1 with minimal intrarenal disease. Quantitative urinary PV-Haufen testing could be used to monitor PVN progression or regression during persistent disease.
Urinary PV-Haufen testing is performed using negative staining EM, a well-established, easily performed technique specifically developed for the identification and quantitation of viruses in various body fluids.33 It has been in routine use in virology since the early 1960s and is the method of choice for viral studies at major virology centers, such as The Centers for Disease Control in Atlanta, GA. The incorporation of urinary PV-Haufen testing only requires “thinking outside the box.” The turnaround time from receipt of a voided urine sample to reporting of results is approximately 3 hr with costs lower or comparable to PCR-based assays.
Although this single center study has limitations because of relatively few cases analyzed, we nevertheless think that quantitative urinary PV-Haufen testing will help to better define, diagnose, and manage PVN.
In conclusion, urinary PV-Haufen are accurate biomarkers of PVN and the number of PV-Haufen shed in fixed voided urine samples accurately reflects the severity of intrarenal disease. Quantitative PV-Haufen testing allows for an early diagnosis of PVN and monitoring of changing intrarenal viral load levels during therapeutic interventions such as in the setting of new clinical drug trials.
MATERIALS AND METHODS
Based on the availability of concomitant blood, urine, and renal biopsy samples, cases of definitive PVN were selected from the archives of The University of North Carolina at Chapel Hill (total n = 40 sample sets). The study protocol was approved by the institutional review board. Per definition, all diagnostic renal allograft biopsies showed evidence of PVN.6-8 Histologic material was evaluated by two pathologists (V.N., H.K.S.) at a multiheaded scope and scoring results based on consensus (including the count of SV40-T–positive cells and tubules) were used for statistical analysis. Urine samples collected on the day of biopsy were analyzed by conventional urine cytology (ThinPrep; Hologic Incorporated, Bedford, MA) for decoy cells, negative staining EM for the detection and quantitation of PV-Haufen, and quantitative PCR for BKV gene sequences. For negative staining EM, urine was fixed in a 1:1 ratio of 4% paraformaldehyde with subsequent EM grid preparation using previously published methods. This protocol does not require any embedding procedures and thin sectioning is not needed (SDC, Materials and Methods, http://links.lww.com/TP/B46).20,29,33 Plasma samples collected on the day of biopsy were analyzed by quantitative PCR for BKV gene sequences.
Biopsy Samples and Diagnosis of PVN
All renal allograft biopsies fulfilled the Banff ’97 criteria for tissue adequacy and were evaluated according to standard guidelines.34 The PVN classification into disease grades 1 to 3 followed proposals made by the 2013 BANFF working group on the classification of Polyomavirus Nephropathy.31 Polyomavirus nephropathy grades (1 = early/mild; 2 = florid; 3 = late/sclerosed) were defined based on the severity of PV replication or degree of staining for the SV40-T antigen in combination with the Banff “ci” interstitial fibrosis score. Twelve of 40 cases were diagnosed as early or mild PVN grade 1 (Figure 2S, A and D, SDC, http://links.lww.com/TP/B46), 23/40 as florid PVN grade 2 (Figure 2S, B and E, SDC, http://links.lww.com/TP/B46) and 5 of 40 as late sclerosed PVN grade 3 (Figure 2S, C and F, SDC, http://links.lww.com/TP/B46).
Quantitative Analysis of the SV40-T Antigen Immunostain in Renal Biopsies
Intrarenal polyomavirus replication was assessed by immunohistochemistry with a monoclonal mouse antibody directed against the SV40-T antigen following previously published protocols (Clone 416, Calbiochem, San Diego, CA) (Figure 2S, D, E, and F, SDC, http://links.lww.com/TP/B46).35 Expression levels of the SV40-T antigen, that is, the number of epithelial cell nuclei expressing the T antigen, were quantitatively analyzed by manual count and also by (automated) morphometry using the Aperio system (Aperio, Vista, CA).
Manual Counts
(a) The total number of tubular epithelial cell nuclei expressing the SV40-T antigen and (b) the total number of stained tubular cross-sections lined by one or more epithelial cells expressing the SV-40 large T antigen were recorded.
Morphometric Analysis
The percentage area of a renal biopsy demonstrating positive expression with the SV40-T antigen was accomplished as follows:
Aperio Color Deconvolution Software version 10.0 was applied to 40× virtual images in “svs” format scanned on the Aperio CS scanner (Aperio, Vista, CA). The total biopsy area was determined using the total stained area of all cellular components read using a channel optimized for blue nuclear counterstain. To evaluate the % of SV40-T–positive staining in the total biopsy area (TS1), the total immunopositive area including both specific intranuclear staining and nonspecific background staining was determined using the DAB channel as specified by Aperio (% DAB). To calculate the specific intranuclear SV-40 large T antigen signals, the weak positive threshold (TSDAB) was set to eliminate presumed nonspecific background staining as, for example, seen in the cytoplasm. The final % of SV-40–positive staining (%IMMUNOPOSITIVE) was calculated as: 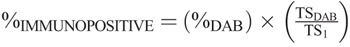 .
.
Negative Staining EM for the Detection and Quantitation of PV Haufen
PV-Haufen Definition
“Haufen” (after a German word for “cluster” or “stack”) were defined as discrete three-dimensional, cast-like, dense polyomavirus aggregates in voided urine samples analyzed by negative staining EM (Figure 2). Polyomavirus casts containing six or more polyomavirus virions were classified as “PV-Haufen” following our previously published approach.20,29
FIGURE 2.
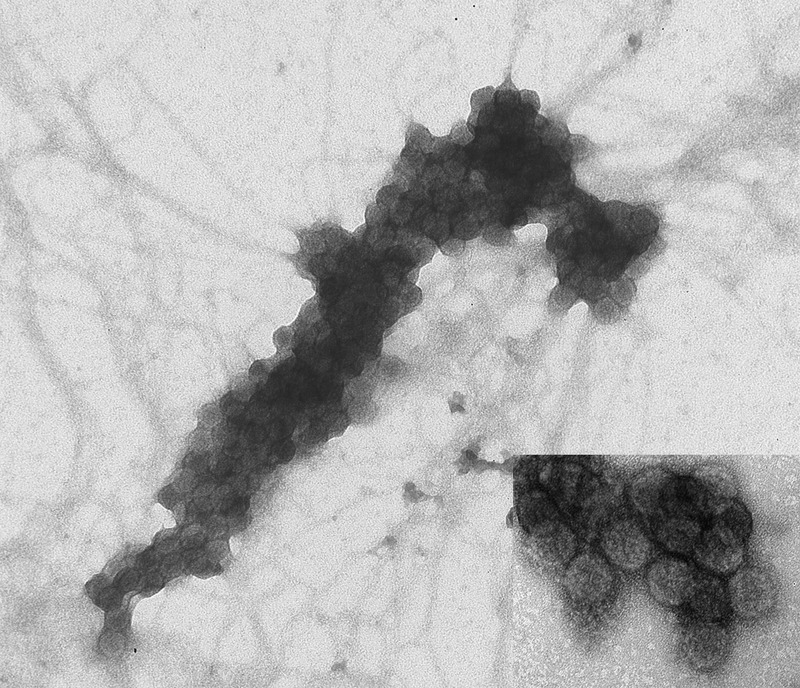
Negative staining electron microscopy depicting a characteristic PV-Haufen in a voided urine sample. Haufen are tight three dimensional viral casts composed of six or more individual PV virions. Note the large size of this PV-Haufen and the diagnostic viral capsid substructure and uniformity allowing easy identification by electron microscopy (inset) (uranyl acetate stain: ×100,000 magnification). PV, polyomavirus.
Negative Staining EM and PV-Haufen Quantitation
Negative staining EM was performed using previously published standard procedures with a two-step methodology of urine clarification and urine concentration (SDC, Materials and Methods, http://links.lww.com/TP/B46 and Table 1S, SDC, http://links.lww.com/TP/B46).20,29,33 One EM grid was prepared for each voided urine sample. Haufen quantitation was performed according to established previously published methodologies specifically designed for the quantitation of viruses in body fluids.33,36 Briefly, PV-Haufen in the voided urine sample adhered to the EM grid by electrostatic binding properties (Figure 3S, SDC, http://links.lww.com/TP/B46). Electron microscopy grids were examined on a transmission electron microscope (LEO EM-910, LEO Electron Microscopy, Thornwood, NY, accelerating voltage, 80–100 kV). A total of 25 randomly selected grid fields were viewed at 50,000× magnification and PV-Haufen were counted in these 25 grid fields at 80,000–100,000× magnification. The examination time was, on average, 20 to 30 min per grid. The total number of PV-Haufen per mL urine was then calculated using the following formula:

(methodology used by the Centers for Disease Control per Dr. Charles Humphrey, Chief, Infectious Disease Pathology).33,36
Quantitative PCR
Plasma and urine BK polyomavirus loads were determined by a real-time quantitative PCR assay using the ABI PRISM 7900HT Sequence Detection System with well characterized probes and primers specific for BK virus as previously published.15,20 Real-time detection of PCR products was achieved with a fluorescent hydrolysis (Taqman) probe. Primers and probes were purchased from TIB Molbiol LLC, Adelphia, NJ. Quantitative linearity of the assay extended from 10 to 109 measured copies of BKV equivalents correlating with a dynamic linear range of 250 to 2.5 × 1010 BKV copies/mL urine or plasma. Detectable BKV DNA below the lower limit of linearity (<250 BKV copies/mL) was classified as a “low-positive” result.
Urine Cytology for Decoy Cell Quantitation
All urine samples were processed for cytology using standard liquid-based procedures (Hologic Incorporated, Bedford, MA) and stained by Papanicolaou method. Decoy cells were identified and quantified as reported previously.11
Statistical Analysis
Statistical analyses were performed with standard programs (Microsoft Excel and Stata 12.1; Stata Corp, College Station, TX). The number of urinary PV-Haufen, number of decoy cells, urine and plasma BKV DNA load levels were correlated with: (a) the severity of intrarenal PV replication/SV40-T staining using Pearson’s correlation coefficient and Spearman’s correlation coefficient and, (b) PVN disease grades 1 to 3 using Wilcoxon rank-sum testing, and Kruskal-Wallis testing with ties. Wilcoxon rank-sum testing for pairwise comparisons with Bonferroni correction was performed only if overall group comparison by Kruskal-Wallis testing was significant. All testing was performed with two-sided α = 0.05.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the expert technical assistance provided by Bruna Brylawski, MS. The authors also thank the University of North Carolina Hospitals Cytology Laboratory and the transplant unit coordinators for help with the collection of study samples.
Footnotes
The authors declare no funding or conflicts of interest.
All authors participated in designing the study, performing the experiments, and preparing the article.
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s website (www.transplantjournal.com).
REFERENCES
- 1. Benavides CA, Pollard VB, Mauiyyedi S, et al. BK virus-associated nephropathy in sirolimus-treated renal transplant patients: incidence, course, and clinical outcomes. Transplantation 2007; 84: 83. [DOI] [PubMed] [Google Scholar]
- 2. Hirsch HH, Randhawa P. BK polyomavirus in solid organ transplantation. Am J Transplant 2013; 13 (Suppl 4): 179. [DOI] [PubMed] [Google Scholar]
- 3. van Aalderen MC, Heutinck KM, Huisman C, et al. BK virus infection in transplant recipients: clinical manifestations, treatment options and the immune response. Neth J Med 2012; 70: 172. [PubMed] [Google Scholar]
- 4. Ranzi AD, Prolla JC, Keitel E, et al. The role of urine cytology for ‘decoy cells’ as a screening tool in renal transplant recipients. Acta Cytol 2012; 56: 543. [DOI] [PubMed] [Google Scholar]
- 5. Sharif A, Alachkar N, Bagnasco S, et al. Incidence and outcomes of BK virus allograft nephropathy among ABO- and HLA-incompatible kidney transplant recipients. Clin J Am Soc Nephrol 2012; 7: 1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nickeleit V. The Pathology of Kidney Transplantation. In: Ruiz P, ed. Transplantation Pathology. 1st ed New York, NY: Cambridge University Press; 2009: 83. [Google Scholar]
- 7. Colvin RB, Nickeleit V. Renal Transplant Pathology. In: Jennette JC, Olson JL, Schwartz MM, eds. Heptinstall’s Pathology of the Kidney. 6th ed Philadelphia, PA: Lippincott, Williams & Wilkins; 2007: 1441. [Google Scholar]
- 8. Nickeleit V, Mihatsch MJ. Polyomavirus nephropathy in native kidneys and renal allografts: an update on an escalating threat. Transpl Int 2006; 19: 960. [DOI] [PubMed] [Google Scholar]
- 9. Hirsch HH, Babel N, Comoli P, et al. European perspective on human polyomavirus infection, replication and disease in solid organ transplantation. Clin Microbiol Infect 2014; 20 (Suppl 7): 74. [DOI] [PubMed] [Google Scholar]
- 10. Chakera A, Dyar OJ, Hughes E, et al. Detection of polyomavirus BK reactivation after renal transplantation using an intensive decoy cell surveillance program is cost-effective. Transplantation 2011; 92: 1018. [DOI] [PubMed] [Google Scholar]
- 11. Singh HK, Bubendorf L, Mihatsch MJ, et al. Urine cytology findings of polyomavirus infections. In: Ahsan N, ed. Polyomaviruses and Human Diseases. 1st ed Georgetown, TX: Springer Science+Business Media, Landes Bioscience/Eurekah.com; 2006: 201. [DOI] [PubMed] [Google Scholar]
- 12. Nickeleit V, Steiger J, Mihatsch MJ. Re: noninvasive diagnosis of BK virus nephritis by measurement of messenger RNA for BK virus VP1. Transplantation 2003; 75: 2160. [DOI] [PubMed] [Google Scholar]
- 13. Nickeleit V, Klimkait T, Binet IF, et al. Testing for polyomavirus type BK DNA in plasma to identify renal-allograft recipients with viral nephropathy. N Engl J Med 2000; 342: 1309. [DOI] [PubMed] [Google Scholar]
- 14. Hirsch HH, Drachenberg CB, Steiger J, et al. Polyomavirus associated nephropathy in renal transplantation: critical issues of screening and management. In: Ahsan N, ed. Polyomaviruses and Human Diseases. 1st ed New York, NY: Springer Science+Business Media, Landes Bioscience /Eurekah.com; 2006: 160. [DOI] [PubMed] [Google Scholar]
- 15. Randhawa P, Ho A, Shapiro R, et al. Correlates of quantitative measurement of BK polyomavirus (BKV) DNA with clinical course of BKV infection in renal transplant patients. J Clin Microbiol 2004; 42: 1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thamboo TP, Jeffery KJ, Friend PJ, et al. Urine cytology screening for polyoma virus infection following renal transplantation: the Oxford experience. J Clin Pathol 2007; 60: 927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hirsch HH, Brennan DC, Drachenberg CB, et al. Polyomavirus-associated nephropathy in renal transplantation: interdisciplinary analyses and recommendations. Transplantation 2005; 79: 1277. [DOI] [PubMed] [Google Scholar]
- 18. Hirsch HH, Knowles W, Dickenmann M, et al. Prospective study of polyomavirus type BK replication and nephropathy in renal-transplant recipients. N Engl J Med 2002; 347: 488. [DOI] [PubMed] [Google Scholar]
- 19. Nickeleit V, True K, Detwiler R, et al. Risk assessment for polyomavirus nephropathy using urine cytology and the detection of decoy cells: cheap and efficient. Transplantation 2012; 94: e42. [DOI] [PubMed] [Google Scholar]
- 20. Singh H, Andreoni KA, Madden V, et al. Presence of urinary Haufen accurately predicts polyomavirus nephropathy. J Am Soc Nephrol 2009; 20: 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hirsch HH. Polyomavirus BK nephropathy: a (re-)emerging complication in renal transplantation. Am J Transplant 2002; 2: 25. [DOI] [PubMed] [Google Scholar]
- 22. Grinde B. Herpesviruses: latency and reactivation—viral strategies and host response. J Oral Microbiol 2013; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fishman JA. Overview: cytomegalovirus and the herpesviruses in transplantation. Am J Transplant 2013; 13 (Suppl 3): 1. [DOI] [PubMed] [Google Scholar]
- 24. Egli A, Binggeli S, Bodaghi S, et al. Cytomegalovirus and polyomavirus BK posttransplant. Nephrol Dial Transplant 2007; 22 (Suppl 8): viii72. [DOI] [PubMed] [Google Scholar]
- 25. Burger-Calderon R, Madden V, Hallett RA, et al. Replication of oral BK virus in human salivary gland cells. J Virol 2014; 88: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jeffers LK, Madden V, Webster-Cyriaque J. BK virus has tropism for human salivary gland cells in vitro: implications for transmission. Virology 2009; 394: 183. [DOI] [PubMed] [Google Scholar]
- 27. Jeffers L, Webster-Cyriaque JY. Viruses and salivary gland disease (SGD): lessons from HIV SGD. Adv Dent Res 2011; 23: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Erard V, Kim HW, Corey L, et al. BK DNA viral load in plasma: evidence for an association with hemorrhagic cystitis in allogeneic hematopoietic cell transplant recipients. Blood 2005; 106: 1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Singh HK, Donna Thompson B, Nickeleit V. Viral Haufen are urinary biomarkers of polyomavirus nephropathy: new diagnostic strategies utilizing negative staining electron microscopy. Ultrastruct Pathol 2009; 33: 222. [DOI] [PubMed] [Google Scholar]
- 30. Nickeleit V, Brylawski B, Rivier L, et al. Urinary polyomavirus-Haufen shedding in mouse and man: a proof-of-concept study for a non-invasive urine biomarker for polyomavirus nephropathy. Lab Invest 2013; 93: 390A. [Google Scholar]
- 31. Haas M, Sis B, Racusen LC, et al. Banff 2013 meeting report: inclusion of c4d-negative antibody-mediated rejection and antibody-associated arterial lesions. Am J Transplant 2014; 14: 272. [DOI] [PubMed] [Google Scholar]
- 32. Tamm I, Bugher JC, Horsfall FL. Ultracentrifugation studies of a urinary mucoprotein which reacts with various viruses. J Biol Chem 1955; 212: 125. [PubMed] [Google Scholar]
- 33. Hayat MA, Miller SE. Negative Staining. New York, NY: McGraw-Hill Publishing Company; 1990. [Google Scholar]
- 34. Racusen LC, Solez K, Colvin RB, et al. The Banff 97 working classification of renal allograft pathology. Kidney Int 1999; 55: 713. [DOI] [PubMed] [Google Scholar]
- 35. Nickeleit V, Hirsch HH, Binet IF, et al. Polyomavirus infection of renal allograft recipients: from latent infection to manifest disease. J Am Soc Nephrol 1999; 10: 1080. [DOI] [PubMed] [Google Scholar]
- 36. Miller SE. Virus particle counting by electron microscopy. In: Griffith J, ed. Electron Microscopy in Biology. New York, NY: Wiley; 1982: 306. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


