A previously uncharacterized protein plays a role in cellulose deposition during the course of seed coat epidermal cell differentiation.
Abstract
Differentiation of the maternally derived seed coat epidermal cells into mucilage secretory cells is a common adaptation in angiosperms. Recent studies identified cellulose as an important component of seed mucilage in various species. Cellulose is deposited as a set of rays that radiate from the seed upon mucilage extrusion, serving to anchor the pectic component of seed mucilage to the seed surface. Using transcriptome data encompassing the course of seed development, we identified COBRA-LIKE2 (COBL2), a member of the glycosylphosphatidylinositol-anchored COBRA-LIKE gene family in Arabidopsis (Arabidopsis thaliana), as coexpressed with other genes involved in cellulose deposition in mucilage secretory cells. Disruption of the COBL2 gene results in substantial reduction in the rays of cellulose present in seed mucilage, along with an increased solubility of the pectic component of the mucilage. Light birefringence demonstrates a substantial decrease in crystalline cellulose deposition into the cellulosic rays of the cobl2 mutants. Moreover, crystalline cellulose deposition into the radial cell walls and the columella appears substantially compromised, as demonstrated by scanning electron microscopy and in situ quantification of light birefringence. Overall, the cobl2 mutants display about 40% reduction in whole-seed crystalline cellulose content compared with the wild type. These data establish that COBL2 plays a role in the deposition of crystalline cellulose into various secondary cell wall structures during seed coat epidermal cell differentiation.
In angiosperms, the seed coat (also called the testa), derived from the maternal ovule integuments, provides a boundary separating the embryo from the external environment (Haughn and Chaudhury, 2005; North et al., 2010). The seed coat plays a major role in protecting the next-generation embryo throughout development, desiccation and dormancy, seed dispersal, and the first stages of germination in the new habitat. In various plant species, including Arabidopsis (Arabidopsis thaliana), the seed coat epidermal cells accumulate a large amount of pectinaceous mucilage. Upon seed imbibition, the mucilage is extruded, forming a gel-like capsule surrounding the seed in a process known as myxospermy (Western et al., 2000). Seed mucilage is composed primarily of pectins, highly hygroscopic polysaccharides, with rhamnogalacturonan I serving as the main component; yet other polysaccharides can also be found in smaller quantities, including homogalacturonan, arabinans, galactans, xyloglucan, glucomannans, and cellulose (Arsovski et al., 2010; Western, 2012; Yu et al., 2014). The function of seed mucilage has not been definitively established, but potential roles include seed hydration during the first stages of seed germination, a role in seed dispersal through its effect on buoyancy, protection from predation by insects, and/or interactions with microorganisms (Engelbrecht and García-Fayos, 2012; Western, 2012; Yang et al., 2013; Saez-Aguayo et al., 2014).
The process of seed coat epidermal cell differentiation (also called mucilage secretory cells [MSCs]) has been well characterized in Arabidopsis. In summary, following a phase of cell expansion, there is a massive induction of pectin biosynthesis and polar secretion from the Golgi to the apoplast, leading to the formation of a cytoplasmic column (Fig. 1). Large quantities of secondary cell wall material are then deposited into the radial cell wall and throughout the cytoplasmic column, generating a volcano-shaped structure known as the columella. During the final stages of seed maturation, the cell undergoes apoptosis and seed/mucilage desiccation (Western et al., 2000). The sequential induction of cell wall-related metabolic pathways over the course of MSC differentiation makes it an attractive system for the study of plant cell wall polymer biosynthesis and modification (Arsovski et al., 2010; Haughn and Western, 2012; Kunieda et al., 2013; Saez-Aguayo et al., 2013; Voiniciuc et al., 2013; Griffiths et al., 2014).
Figure 1.
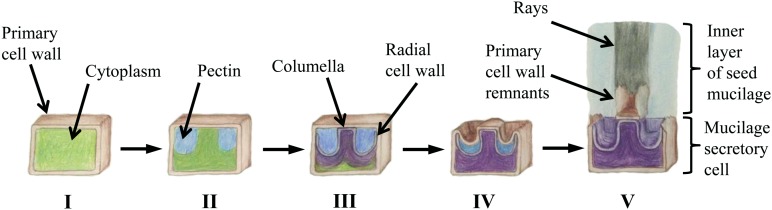
Schematic representation of seed epidermal cell differentiation and mucilage extrusion. The process of seed epidermal cell differentiation (also called MSCs) goes through a phase of cell expansion (I); massive biosynthesis and secretion of pectins from the Golgi to the apoplast leading to the formation of a cytoplasmic column (II); secondary cell wall deposition into the radial cell wall and throughout the cytoplasmic column, generating a volcano-shaped columella (III); and desiccation and apoptosis of the mature seed and mucilage (IV), as described previously by Western et al. (2000). Upon seed hydration, the hygroscopic pectic component of seed mucilage expands rapidly, breaking the outer cell wall and encapsulating the seed; a set of cellulosic rays anchor the pectic component of seed mucilage to the seed surface (V).
Upon hydration of the mature seed, the mucilage expands rapidly, rupturing the outer cell wall and encapsulating the seed (Western et al., 2000; Haughn and Chaudhury, 2005). Once released, seed coat mucilage is organized in two distinct domains: an adherent, inner layer, tightly attached to the seed surface; and a nonadherent, outer, diffuse layer. Both layers are composed primarily of the pectin rhamnogalacturonan I, with the adherent layer also containing other pectins and minor amounts of hemicellulose and cellulose (Western et al., 2000; Penfield et al., 2001; Willats et al., 2001; Macquet et al., 2007; Young et al., 2008). A number of previous studies provide data supporting a role for cellulose in seed coat mucilage: (1) Calcofluor, Congo Red, and Pontamine Fast Scarlet S4B stainings of seed mucilage are consistent with the presence of cellulose in the set of rays deposited across the inner layer of seed coat mucilage (Windsor et al., 2000; Willats et al., 2001; Macquet et al., 2007; Harpaz-Saad et al., 2011; Mendu et al., 2011a); (2) fluorescently labeled cellulose-binding modules identify the presence of crystalline and amorphous cellulose (Blake et al., 2006; Young et al., 2008; Dagel et al., 2011; Sullivan et al., 2011); (3) pectolytic enzymes could not degrade the set of rays radiating from the seed unless combined with cellulase treatment (Macquet et al., 2007); and (4) most recently, genetic studies confirmed the cellulosic composition of the rays of seed coat mucilage, identifying CELLULOSE SYNTHASE5 (CESA5) as an essential player in cellulose deposition in this context (Harpaz-Saad et al., 2011; Mendu et al., 2011a; Sullivan et al., 2011). Altogether, these data demonstrate the essential structural role of cellulose in anchoring the pectic component of seed coat mucilage to the seed surface.
Cellulose microfibrils, the primary load-bearing elements of plant cell walls, are synthesized at the plasma membrane by cellulose synthase complexes (Somerville et al., 2004; Baskin, 2005; Somerville, 2006). The glycosyl transferase, CELLULOSE SYNTHASE A (CESA), acts as the catalytic subunit of the cellulose synthase complex that produces paracrystalline chains of β(1→4)-linked Glc molecules. Multiple β(1→4)-linked Glc chains are then assembled into crystalline cellulose microfibrils, aligned according to cell function and shape requirements (Brown, 1985). The cellulose microfibrils are cross-linked by hemicelluloses and embedded in a matrix of pectins. During cell expansion, additional primary cell wall cellulose is deposited, dictating the extent and orientation of cell growth. Secondary cell wall is synthesized following the completion of cell expansion as part of the process of cell differentiation. Genetic and biochemical studies demonstrate that, of the 10 CESA genes encoded by the Arabidopsis genome, CESA1, CESA3, and CESA6 are involved in the biosynthesis of primary cell walls, shown to appear at a 1:1:1 molar ratio (Desprez et al., 2007; Persson et al., 2007; Gonneau et al., 2014). While CESA1 and CESA3 are essential components of the cellulose synthase complex involved in primary cell wall deposition, CESA2, CESA5, CESA6, and CESA9 act in a partially redundant manner, with predominant roles in different developmental contexts. CESA4, CESA7, and CESA8 are involved in secondary cell wall deposition during xylem cell differentiation (Arioli et al., 1998; Beeckman et al., 2002; Gillmor et al., 2002; Taylor et al., 2003; Desprez et al., 2007; Persson et al., 2007). In addition, recent findings demonstrate a partially redundant role for CESA2, CESA5, and CESA9 in secondary cell wall deposition during the course of MSC differentiation, suggesting that a broader perspective should be adopted with regard to the role of CESAs in various developmental contexts and demonstrating the utility of MSCs for the study of cellulose biosynthesis (Stork et al., 2010; Mendu et al., 2011a, 2011b).
In order to identify additional elements involved in the emerging mechanism of cellulose microfibril deposition in MSCs, we employed tissue-specific coexpression analysis covering the course of seed development. This seed-specific coexpression analysis led to the identification of COBRA-LIKE2 (COBL2) as a novel regulator of crystalline cellulose deposition. COBL2 is a member of the plant-specific, COBRA-LIKE (COBL) gene family composed of 12 genes in Arabidopsis. The putative COBL gene products share three common domains: an N-terminal cellulose-binding domain, a Cys-rich CCVS domain, and a C-terminal hydrophobic domain for the attachment of a glycosylphosphatidylinositol (GPI) anchor (Roudier et al., 2002; Brady et al., 2007). In both plant and animal systems, the presence of a GPI anchor is often associated with polar secretion of the protein to distinct areas of the plasma membrane. In certain cases, following secretion, the GPI anchor is cleaved, releasing the polypeptide, which then can act as a soluble signal molecule (Rodriguez-Boulan and Powell, 1992; Matter and Mellman, 1994; Friedrichson and Kurzchalia, 1998; Varma and Mayor, 1998). The mechanism of COBL protein function has yet to be elucidated. However, various COBL family members were shown previously to be required for anisotropic growth in different developmental contexts. COBRA, the founder member of the family, was first identified in a genetic screen for mutants with perturbations in root elongation. In wild-type seedlings, root elongation occurs through the deposition of cellulose microfibrils transverse to the longitudinal axis, yielding turgor-driven anisotropic cell expansion. Mutations in cobra (cob) result in reduced levels of crystalline cellulose and disorganized deposition of cellulose microfibrils at the root elongation zone, leading to short and swollen roots (Benfey et al., 1993; Hauser et al., 1995; Schindelman et al., 2001; Roudier et al., 2005; Sorek et al., 2014). Other characterized COBL family members have been assigned similar functions in other contexts: COBL9 in root hair elongation (Parker et al., 2000; Ringli et al., 2005; Jones et al., 2006), COBL10 and COBL11 in pollen tube elongation through the transmitting tract (Li et al., 2013), and COBL4, initially identified through its coexpression with CESA4, CESA7, and CESA8 (as well as its orthologs from rice [Oryza sativa] and maize [Zea mays]), in secondary cell wall deposition in xylem cells (Li et al., 2003; Brown et al., 2005; Persson et al., 2005; Ching et al., 2006; Sato et al., 2010b; Dai et al., 2011; Liu et al., 2013). Of the 12 COBL family members, seven remain uncharacterized. Despite accumulating evidence of the profound roles of various COBL proteins in crystalline cellulose deposition, their mechanism of action remains largely unknown. In this work, we assign a central role for the previously uncharacterized COBL2 in the mechanism of cellulose deposition during the course of seed coat epidermal cell differentiation in Arabidopsis.
RESULTS
Organ-Specific Coexpression Analysis Suggests That COBL2 Is Involved in Cellulose Deposition in the Arabidopsis Seed Coat
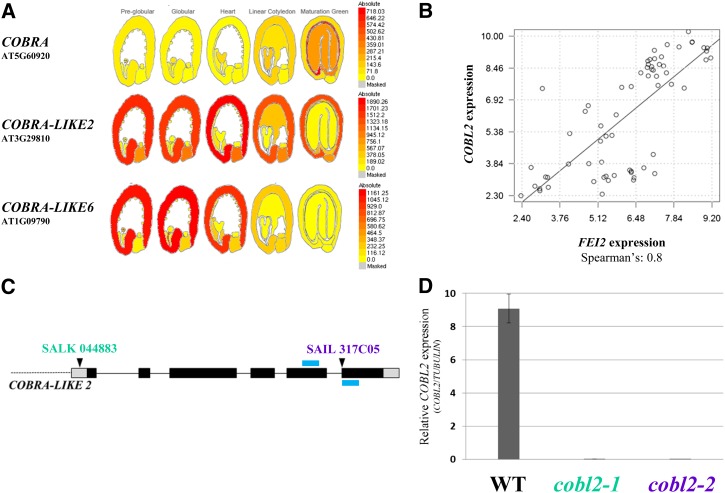
Cellulose, deposited into a set of rays radiating from the Arabidopsis seed upon hydration, anchors the pectic component of seed mucilage to the imbibed seed. CESA5, the FEI2 receptor-like kinase (FEI2), and the extracellular arabinogalactan protein, SALT OVERLY SENSITIVE5 (SOS5), were identified as important players in seed mucilage ray formation (Harpaz-Saad et al., 2011; Mendu et al., 2011a; Sullivan et al., 2011). Two complementary approaches were employed to further investigate the mechanism of cellulose deposition into the rays of seed coat mucilage: (1) investigation of the expression pattern of genes known to be involved in cellulose deposition (or their homologs); and (2) identification of the genes that are coexpressed with FEI2 and/or CESA5. SOS5 was not included in this analysis, due to its absence from the ATH1 Affymetrix chip. Both approaches used the publicly available gene expression profiling data set of laser-captured microdissected seeds (divided into seven subtissues), sampled throughout seed development, in five predefined developmental stages (Le et al., 2010). One of the gene families examined was COBL, members of which were shown previously to affect crystalline cellulose content and the orientation of cellulose microfibril deposition (Schindelman et al., 2001; Roudier et al., 2005). The expression pattern of COB, the founder member, did not support a role in cellulose deposition in MSCs. However, the expression pattern of other COBL genes, over the course of seed development (as observed using the Bio-Array Resource eFP browser), suggests that COBL2 and COBL6 may play roles in this process (Fig. 2A; Supplemental Fig. S1A; Winter et al., 2007; Le et al., 2010). Both genes exhibit a seed coat-specific induction of expression during the course of seed development and have not been assigned functions so far. Generally, these data are in agreement with the expression pattern of COBL2 as demonstrated by the Haughn laboratory data set following gene expression profiling in Arabidopsis seed coats during development (Dean et al., 2011). While COBL2 is expressed throughout seed coat differentiation, COBL6 is expressed specifically during the early stages of seed development in a pattern resembling that described previously for CESA2 (Fig. 2A; Supplemental Fig. S1, A and C; Harpaz-Saad et al., 2011). These data identify COBL2 and COBL6 as candidates for possible roles in cellulose deposition during seed development.
Figure 2.
Expression analysis of COBL2 during the course of seed development. A, Expression pattern of representative members of the COBL gene family through the course of seed development as depicted by the Bio-Array Resource eFP browser (Winter et al., 2007) based on the data set generated by Le et al. (2010) following gene expression in laser-capture microdissected seeds. B, Coexpression relationship between FEI2 and COBL2 as demonstrated by Spearman correlation coefficient calculated according to their expression patterns in the Le et al. (2010) gene expression data set. C, Schematic representation of the COBL2 gene as annotated by The Arabidopsis Information Resource, pinpointing the location of the T-DNA insertion in both alleles examined, named cobl2-1 and cobl2-2. The blue bars represent the locations of diagnostic primers. Alleles are indicated above the insertion lines. D, Quantitative RT-PCR analysis of the effect of the cobl2-1 and cobl2-2 mutations on COBL2 gene expression in developing seeds compared with the wild type (WT), performed with COBL2- or TUBULIN-specific primers, with TUBULIN serving as a reference gene.
In the second approach, Spearman and Pearson rank correlation coefficients were used to evaluate the expression correlation between the baits (FEI2 and CESA5) and each of the genes represented on the ATH1 Affymetrix chip during the course of seed development. Data set-specific coexpression analysis with FEI2 as a bait assigned a Spearman rank correlation coefficient value of 0.8 (and Pearson correlation of 0.77) between FEI2 and COBL2, implying significant correlation (Fig. 2B). The previously described role of various COBL family members in cellulose deposition in different developmental contexts (Brady et al., 2007) prompted us to further explore the role of COBL2 in seed mucilage. Examination of the correlation between FEI2 and other COBL family members using the Spearman correlation coefficient clearly demonstrates that the highest and only significant correlation in the seed development-specific context exists between FEI2 and COBL2 (Fig. 2B; Supplemental Fig. S1B). This context-specific coexpression analysis suggested that COBL2 is a candidate to be involved in cellulose deposition in seed coat epidermal cells.
COBL2 Gene Expression
To test the role of COBL2 in seed coat mucilage, two independent transfer DNA (T-DNA) insertion alleles were examined, cobl2-1 (SALK_044883), with an insertion in the COBL2 promoter sequence, and cobl2-2 (SAIL_317_C05), with an insertion in the last exon of the COBL2 gene (Fig. 2C; Alonso et al., 2003). Allelism tests between the cobl2 T-DNA insertion lines and the cob-1 mutant, serving as a negative control, demonstrate that cobl2-1 and cobl2-2 are indeed allelic (Supplemental Fig. S2). Phenotypic analysis (i.e. seed mucilage ray formation) was assessed in the F2 of these crosses (due to the maternal origin of the MSCs), confirming the allelic nature of the cobl2 insertion mutants (Supplemental Fig. S2). No seed coat-related phenotypes were observed for cobl6 mutants in the three different T-DNA insertion lines examined using the Calcofluor stain (Supplemental Fig. S3). The expression of COBL2 in the insertion mutants was analyzed using developing seeds of the wild type, cobl2-1, and cobl2-2 examined by quantitative reverse transcription (RT)-PCR conducted using TUBULIN as a reference gene. Both cobl2-1 and cobl2-2 appear to be null alleles, as neither had any detectable COBL2 transcript in developing seeds (Fig. 2D).
In order to further characterize the expression pattern of COBL2 during the course of seed development, RT-PCR was conducted on seeds at 4, 6, 8, 10, 12, and 14 DPA. As predicted from the expression profile of COBL2 in the Goldberg and Harada data set (Fig. 2A; Le et al., 2010), COBL2 was found to be expressed throughout seed development (Fig. 3A). The transgenic lines previously described by Brady et al. (2007) were examined, in developing seeds, for the expression of either GFP or GUS driven by the COBL2 promoter. While GUS staining of developing seeds at 12 DPA effectively demonstrated expression in endosperm/embryo, GUS expression does not appear to localize to the seed coat, contrary to the predicted COBL2 gene expression (Supplemental Fig. S4). Additionally, GFP fluorescence could not be observed in the developing seeds. To further study COBL2 expression in seeds, seed coat and embryos of wild-type seeds harvested 8, 10, and 12 DPA were separated and examined by RT-PCR for the expression of COBL2 in the different subseed contexts. GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE C SUBUNIT (GAPC) and TUBULIN served as reference genes, with NAC-RELATED SEED MORPHOLOGY1(NARS1) and PROTODERMAL FACTOR1 (PDF1) serving as seed coat- and embryo-specific markers, respectively (Kunieda et al., 2013). COBL2 gene expression was detected in the seed coat fraction and, interestingly, at similar levels also in the embryo fraction (Fig. 3B). The discrepancy between the GUS staining results and the COBL2 expression data by the two independent microarray experiments described above (Le et al., 2010; Dean et al., 2011) and the RT-PCR results obtained from seed coats versus embryos (Fig. 3B) might be attributed to the possible absence of regulatory elements required for proper in planta COBL2 expression or, alternatively, to posttranscriptional regulation mechanisms affecting COBL2 transcript levels. Previous work demonstrated that COBL2 promoter-driven GUS expression is observed in various developmental contexts, including the ovule, seed funiculus, embryonic root tip, root tip columella cells, and at lateral root emergence sites (Brady et al., 2007). Therefore, the cobl2 mutants were examined for additional phenotypes. Screening for perturbations in root development, seedlings were grown on medium with no additional Suc versus medium supplemented with 4.5% (w/v) Suc and observed for root elongation, root tip swelling, number and length of lateral roots, and root hair morphology. However, no significant phenotypes were evident (Supplemental Fig. S5). Additionally, the effect of the cobl2 alleles on the elongation and swelling of etiolated seedlings was examined, but no significant phenotypes were observed in this developmental context (Supplemental Fig. S6). Expression profiling of COBL2 by RT-PCR demonstrated that the highest level of expression is observed in siliques (Fig. 3C). Overall, these results confirm that COBL2 is expressed in the seed coat during the course of seed development and suggest that it may serve additional functions in other developmental contexts.
Figure 3.
Expression analysis of COBL2. A, Expression of COBL2 throughout seed development, using COBL2- and TUBULIN-specific primers, with TUBULIN serving as a reference gene. B, RT-PCR analysis of COBL2 expression in seed coat versus embryo at representative time points during the course of seed development. NARS1 serves as a seed coat-specific marker (Kunieda et al., 2013), PDF1 serves as an embryo-specific marker, and TUBULIN and GAPC serve as reference genes (Kunieda et al., 2013). C, COBL2 gene expression in different developmental contexts, performed with COBL2- or GAPC-specific primers, with GAPC serving as a reference gene.
cobl2 Mutants Display Malformation of the Rays Deposited across the Adherent Layer of Seed Mucilage
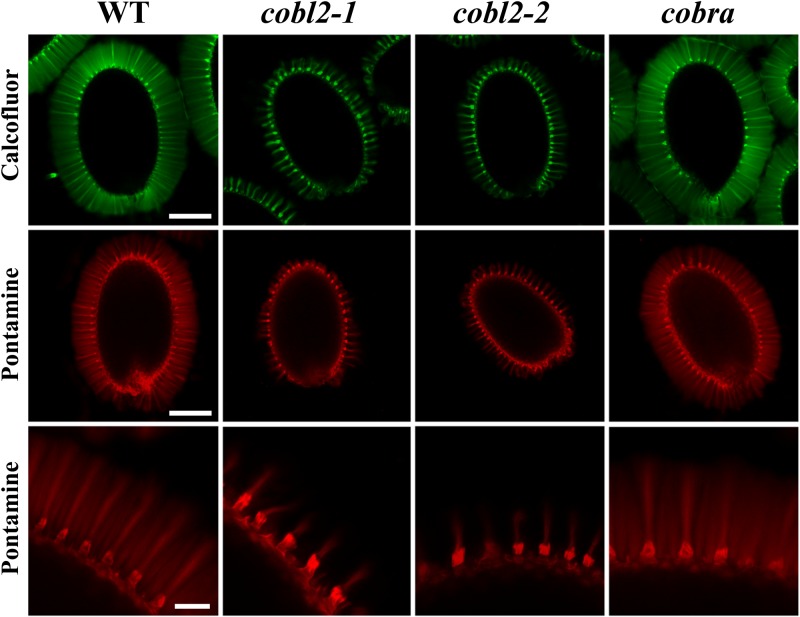
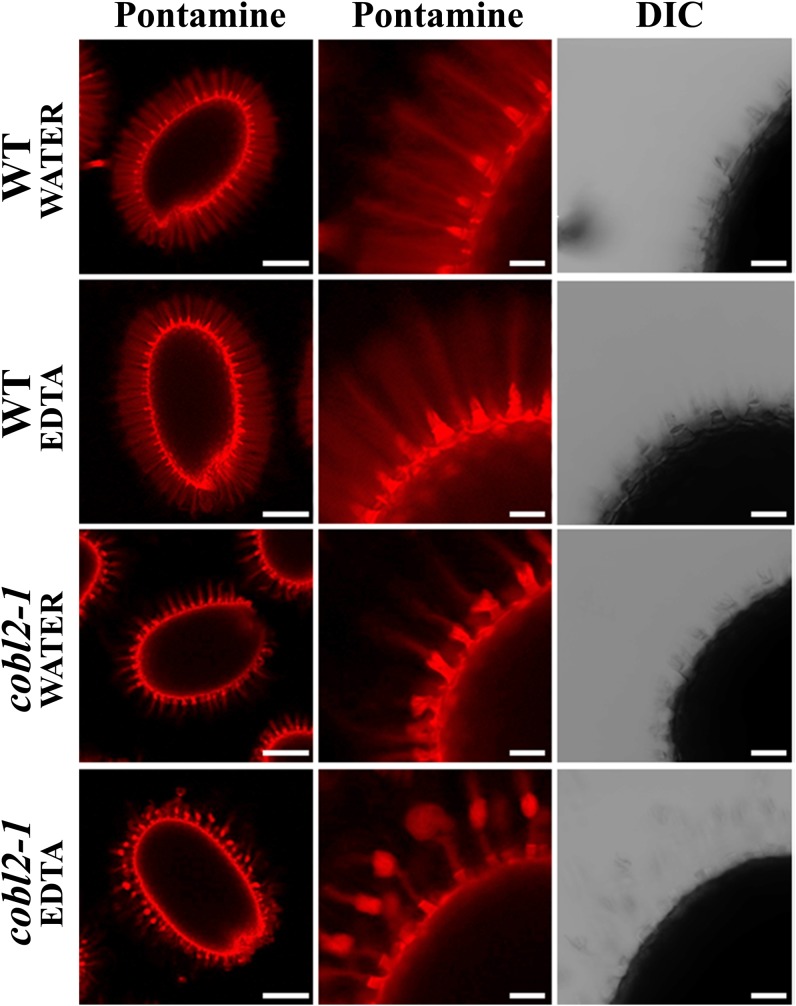
In order to investigate the role of COBL2 in cellulose deposition in seed mucilage, and in particular in the formation of the cellulosic rays, seed coat cellulosic structures were visualized by staining with Calcofluor White, which stains cellulose and other β-glucans, and Pontamine Fast Scarlet S4B, a cellulose-specific dye (Willats et al., 2001; Anderson et al., 2010; Wallace and Anderson, 2012). In wild-type seeds, both Calcofluor and Pontamine stain the columella, the remnants of the primary cell wall attached to the columella tips upon mucilage extrusion, and a set of organized rays extending from the tip of the columella at the seed surface and across the adherent layer of seed mucilage (Figs. 1 and 4). Although both stains label the columella and remnants of the primary cell wall in the cobl2-1 and cobl2-2 mutants, the rays appear substantially reduced and malformed compared with the wild type, suggesting a role for COBL2 in seed mucilage ray formation (Fig. 4). cob-1 seeds were indistinguishable from the wild-type seeds in this analysis. While COB plays a central role in cellulose deposition during root elongation, it does not appear to play a role in the developing seed coat, consistent with its expression pattern. Interestingly, when the Pontamine staining was conducted following treatment with the divalent cation chelator EDTA, detachment of the outer cell wall remnants from the tips of the columella was observed in cobl2 mutant seeds, similar to that described previously for fei2, sos5, and cesa5 mutants (Fig. 5; Harpaz-Saad et al., 2011). Cation chelators modulate the properties of pectin mucilage, presumably due to their effect on pectin cross-linking through Ca2+ and other divalent ions. The effect of cation chelators on the attachment of the outer cell wall remnants to the columella following mucilage extrusion may suggest a role for pectin-cellulose interactions in the adhesion of those two structures. Altogether, the data identify COBL2 as a novel player in cellulose deposition into the set of rays radiating from the seed upon mucilage extrusion.
Figure 4.
cobl2 mutant seeds exhibit reduced rays of cellulose observed in wild-type seed coat mucilage. Calcofluor staining for β-glucans (top row) and Pontamine staining for cellulose (middle and bottom rows) are shown following gentle shaking in water of the wild type (WT) and various mutants as indicated. Whole-seed views (top and middle rows) and closeups (bottom row) are shown. Both Calcofluor and Pontamine stain label the columella, outer cell wall remnants, and rays deposited across the inner, adherent layer of mucilage in the wild type. Bars = 0.25 mm (top and middle rows) and 0.05 mm (bottom row).
Figure 5.
Effects of cation chelators on seed mucilage morphology. Pontamine staining is shown following water or cation chelator treatment with gentle shaking of the wild type (WT) or various mutants as indicated. Whole-seed views (left column) and closeups of Pontamine-stained seeds (middle and right columns) are shown. Note the detachment of the outer cell wall remnants from the tips of the columella in cobl2 mutant seeds following treatment with the cation chelator EDTA. DIC, Differential interference contrast. Bars = 0.25 mm (left column) and 0.05 mm (middle and right columns).
cobl2 Mutants Display Increased Solubility of the Pectic Component of Seed Coat Mucilage
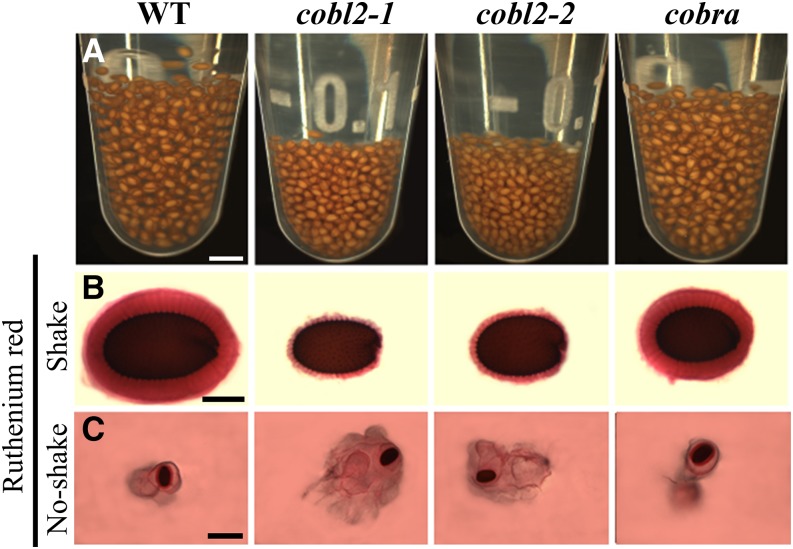
Recent studies have defined a new class of seed mucilage mutants in which perturbation of cellulose deposition into the rays radiating from the seed leads to increased solubility of the pectic component of seed mucilage. Malformed rays, as described previously for cesa5, fei2, sos5, and cellulose synthase-like A2 (csla2) mutants, are associated with malpartitioning of seed mucilage pectins, leading to a reduced inner, adherent layer alongside an expanded outer, nonadherent layer (Harpaz-Saad et al., 2011; Mendu et al., 2011a; Sullivan et al., 2011; Griffiths et al., 2014; Yu et al., 2014). In order to examine the effect of the malformed rays in the cobl2 seeds on the structure of the pectic component of seed mucilage, the relative levels of adherent and nonadherent mucilage in the mutant seeds were examined (Fig. 6). Upon imbibition, following mucilage extrusion, the wild-type seeds are separated by gaps, formed due to the clear, adherent layer of seed coat mucilage that remains attached to the seed surface. Both cobl2 mutant alleles occupy a substantially smaller volume, compared with the wild-type seeds (Fig. 6A), which was associated with greatly reduced gaps between individual seeds, suggesting a reduced adherent layer of seed mucilage in the mutants (Fig. 6A).
Figure 6.
cobl2 mutants display an increased solubility of the pectinaceous component of seed mucilage. A, Ten milligrams of seeds of the wild type (WT) and indicated mutants was immersed in water overnight at room temperature and occasionally shaken. B, Staining of seeds with Ruthenium Red for pectins, with gentle shaking, following pretreatment with EDTA. Note that only the adherent mucilage layer stays attached following shaking. C, Staining of seeds with Ruthenium Red with no shaking, demonstrating both adherent and nonadherent layers of seed mucilage. Bars = 2 mm (A), 0.25 mm (B), and 1 mm (C).
To further examine the adherent layer of seed mucilage, seeds were stained with the cationic dye Ruthenium Red, which marks acidic pectins and has been used extensively for visualization of the pectic component of seed mucilage (Hanke and Northcote, 1975; Western et al., 2000; Willats et al., 2001). The Ruthenium Red staining was conducted in conjunction with mild shaking to remove the nonadherent layer of mucilage. Both alleles of cobl2 exhibited a substantially reduced adherent layer as compared with seeds of both the wild type and the cob-1 mutant (Fig. 6B). This result confirms that the depletion of the adherent layer of seed coat mucilage in the cobl2 mutants, as observed under Ruthenium Red staining, is likely the cause of the reduced gaps between the seeds evident in the hydration assay (Fig. 6, A and B). In contrast, when the nonadherent layer of cobl2-1 and cobl2-2 seed coat mucilage was visualized via Ruthenium Red staining in the absence of shaking, it appeared substantially expanded in the mutant lines compared with both wild-type and cob-1 mutant seeds (Fig. 6C). The increased solubility of the pectic component of seed mucilage observed for cobl2 resembles that described previously for cesa5, fei2, and sos5 mutants, further confirming the structural role of seed mucilage cellulosic rays in the adhesion of the pectic component of seed mucilage to the seed surface.
COBL2 Is Required for Crystalline Cellulose Deposition into the Rays
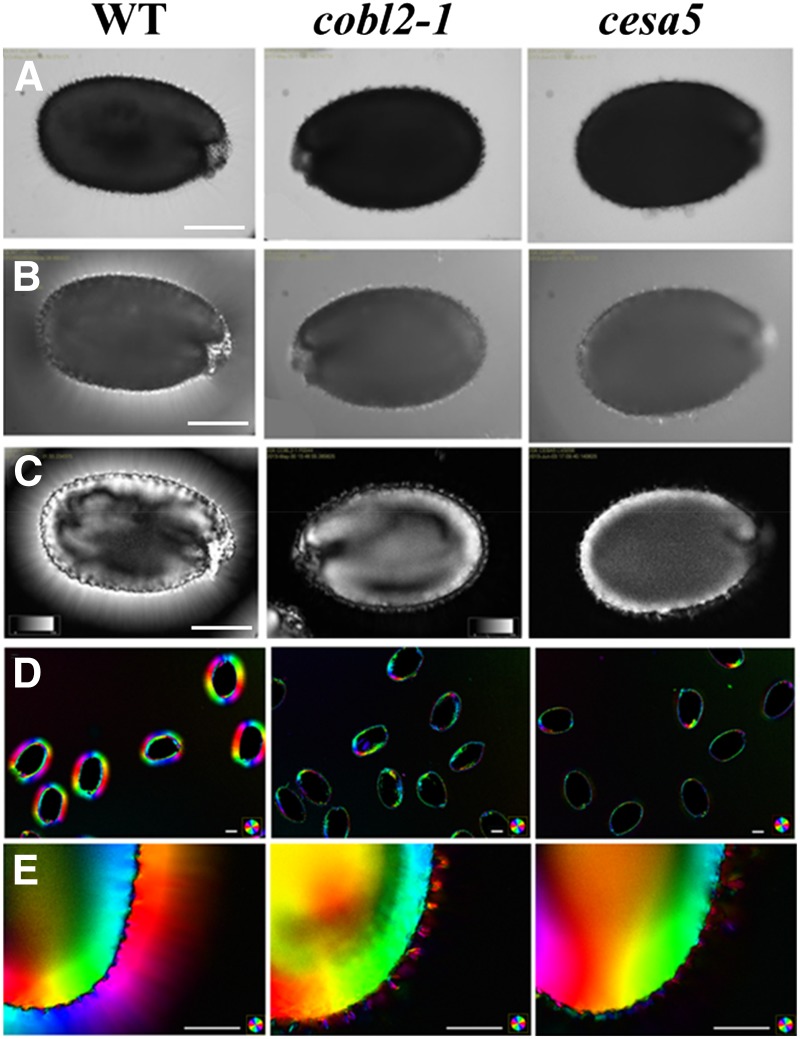
The mechanism by which the COBL genes affect cellulose deposition remains unclear. In other developmental contexts, COBL gene products have been shown to affect crystalline cellulose deposition (Schindelman et al., 2001; Li et al., 2003; Dai et al., 2011). Schindelman and colleagues elegantly employed polarized light microscopy to demonstrate a substantial reduction in crystalline cellulose deposition at the elongation zone of cob-1 mutant roots under restrictive conditions, a phenotype associated with the perturbation in anisotropic cell expansion (Schindelman et al., 2001; Roudier et al., 2005). Polarized light microscopy was also used to visualize crystalline cellulose deposition into the rays radiating from the seed upon mucilage extrusion (Sullivan et al., 2011; Yu et al., 2014). Under polarized light, the amount of light retardance observed correlates with the amount and average degree of alignment of crystalline cellulose microfibrils in the light path. A polarized light microscope system equipped with an automated liquid crystal compensator (LC-PolScope; Abrio Imaging System) was utilized to further investigate the role of COBL2 in cellulose deposition in seed mucilage (Schmidt, 1924; Oldenbourg, 1996). Use of the LC-PolScope is an informative method for in situ determination of cellulose microfibril orientation and crystalline cellulose content through its intrinsic ability to retard polarized light birefringence (Eder et al., 2010; Abraham and Elbaum, 2013). The LC-PolScope system enables high-resolution, noninvasive, in situ visualization and quantification of the effects of biological structures on polarized light due to their intrinsic anisotropic properties. Therefore, it is devoid of prelabeling treatments that may affect the examined tissue. Following imbibition, there was no observable difference in whole seeds of the cobl2-1 mutant when examined in bright light as compared with wild-type seeds (Fig. 7A). Under polarized light, wild-type seeds appeared surrounded by a halo, reflecting the cellulose rays radiating from the seed that cause a retardance of the polarized light. Moreover, the columella and remnants of the primary cell wall remaining following mucilage extrusion also can be identified at the seed surface due to their similar ability to retard polarized light (Fig. 7, B and C; Supplemental Fig. S6). While the columella and the remnants of the primary cell wall could be observed in the cobl2 seeds, the halo of cellulosic rays appears substantially reduced, resembling the phenotype of cesa5 seeds (Fig. 7; Supplemental Fig. S6). The similarity in phenotype between the cobl2 and cesa5 mutants suggests that COBL2 functions in the deposition of crystalline cellulose into the rays radiating from the seed upon imbibition.
Figure 7.
Polarized light birefringence demonstrates severe reduction in crystalline cellulose deposition into the rays of cobl2 mutant seeds, resembling the cesa5 seed phenotype. In situ visualization of crystalline cellulose deposition in MSCs of imbibed seeds was investigated by LC-PolScope for their ability to retard polarized light birefringence. The phenotypes of imbibed seeds of the indicated genotypes are shown by bright light (A), polarized light (B), and liquid crystal polarized light microscopy (LC-PolScope; C). The orientation of crystalline cellulose microfibril deposition is demonstrated by the angle of the slow optical axis per pixel and visualized by color-code index (D and E). WT, Wild type. Bars = 0.25 mm (A–C) and 0.1 mm (D and E).
The LC-PolScope system enables further analysis of the orientation of crystalline cellulose microfibril deposition by calculation of the orientation of the slow axis of the birefringent cellulose microfibrils per pixel. These data demonstrate that the crystalline cellulose microfibrils in the rays are deposited perpendicular to the seed surface (Fig. 7, D and E; Supplemental Fig. S6). Interestingly, while both Calcofluor and Pontamine stains reveal reduced rays in the cobl2 mutants, crystalline cellulose visualization by polarized light birefringence demonstrates a nearly complete lack of rays. This disparity suggests either a decreased crystallinity of the malformed rays remaining in the cobl2 and cesa5 mutant backgrounds or a limited sensitivity of the LC-PolScope visualization as compared with the cellulose stains (Figs. 3 and 6). In order to address this question, Pontamine-stained seeds were observed by both LC-PolScope and fluorescence microscopy (Supplemental Fig. S7). When the sensitivity of polarized light birefringence was set according to the optimal conditions for wild-type ray visualization, as in Figure 4, the residual rays of the mutants seen with the Pontamine stain were almost completely absent in the polarized light image. Increasing the sensitivity of the LC-PolScope led to saturated visualization of the rays in the wild type but did enable visualization of the residual rays evident in the cobl2 mutant background, which perfectly colocalizes with the Pontamine stain images (Supplemental Fig. S7). Interestingly, when stains such as Calcofluor or Pontamine are used for cellulose visualization, the rays of the wild-type seeds tend to stain to a lower extent than those of the mutants under the same conditions. This might be attributed to the increased accessibility of the rays to the stain as a result of the increased solubility of the pectinaceous component of seed coat mucilage in the mutants. In contrast, LC-PolScope imaging visualization based on the intrinsic properties of the visualized substance directly correlates with the amount of crystalline cellulose, unhindered by properties of stain diffusion, highlighting another distinct advantage of the LC-PolScope for the visualization of crystalline cellulose.
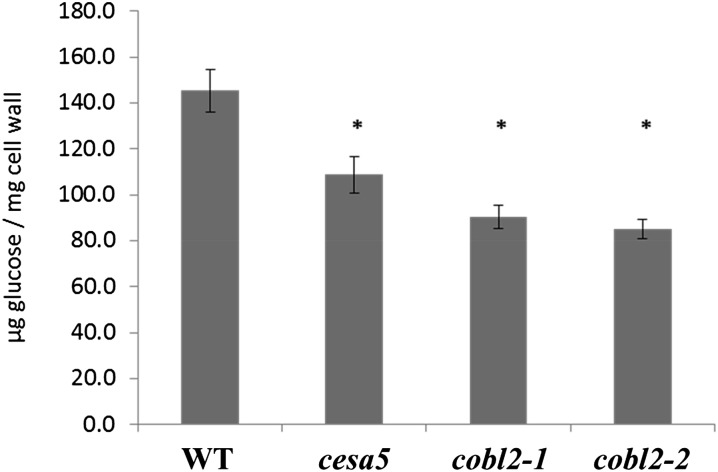
As a complementary approach aimed at the characterization of the role of COBL2 in crystalline cellulose deposition in seeds, we measured the crystalline cellulose content of mature, dry Arabidopsis seeds using the Updegraff assay. Seeds of the cesa5 mutant served as a control, as it has been shown previously to exhibit a reduction in whole-seed crystalline cellulose. Our results indicate a 25% reduction in crystalline cellulose content in seeds of the cesa5 mutant, consistent with previous results (Fig. 7; Sullivan et al., 2011). Disruption of COBL2 results in a significant reduction (approximately 40%) in crystalline cellulose levels in seeds of both cobl2 mutant alleles (cobl2-1 and cobl2-2; Fig. 8). As discussed previously, such a decrease in crystalline cellulose content cannot be attributed solely to a reduction of crystalline cellulose deposition in the rays (Sullivan et al., 2011), suggesting that COBL2 must play additional roles in cellulose deposition during seed development. These results demonstrate that COBL2 is essential for crystalline cellulose deposition in the seed. Specifically, COBL2 functions in crystalline cellulose deposition into the rays radiating from the Arabidopsis seed coat upon mucilage extrusion and is predicted to play additional roles in other developmental contexts in the seed as well.
Figure 8.
Dry seeds of cobl2 mutants display reduced amounts of crystalline cellulose as compared with the wild type. Whole-seed crystalline cellulose content was measured by Updegraff assay. Error bars represent sd, and asterisks indicate significant differences compared with the wild type (WT) when using the Mann-Whitney U test (P < 0.05; n ≥ 9).
COBL2 Is Required for Cellulose Deposition in the Radial Cell Wall and the Columella of MSCs
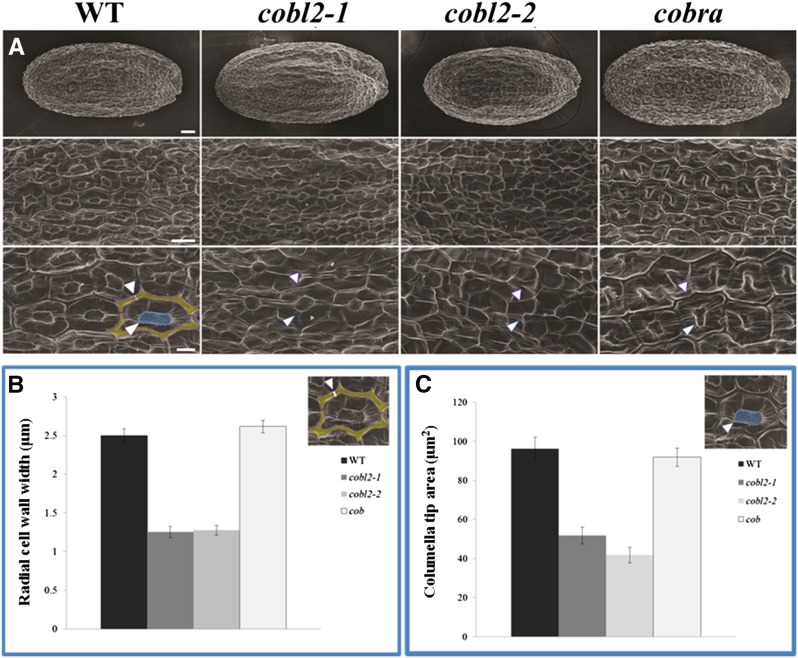
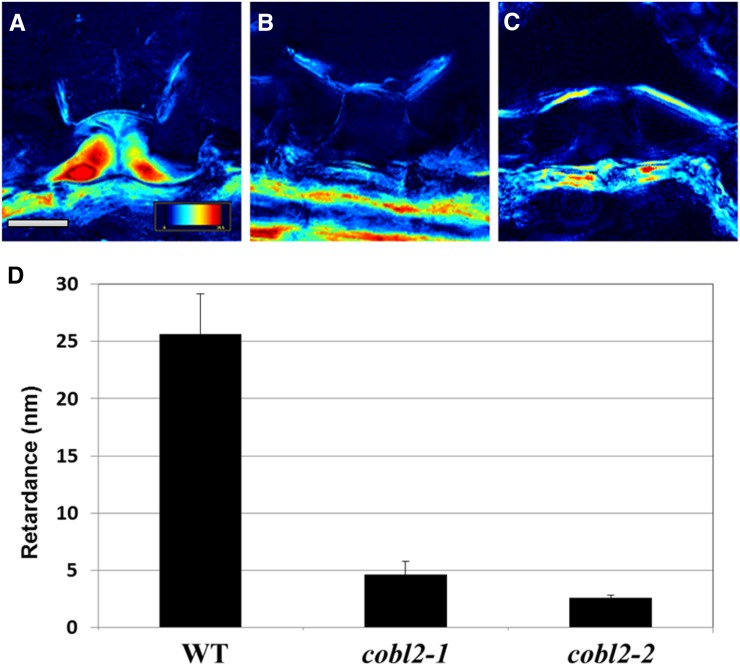
The significant reduction in crystalline cellulose in cobl2 mutant seeds prompted us to further investigate a role for COBL2 in other contexts in the seed. Previous studies have assigned roles for different members of the CESA gene family in cellulose biosynthesis in various contexts throughout seed development. In the Arabidopsis seed coat, CESA5 plays a nonredundant role in the formation of seed coat rays, yet it acts redundantly with CESA2 and CESA9 in cellulose deposition during radial cell wall thickening and the formation of the columella (Harpaz-Saad et al., 2011; Mendu et al., 2011a; Sullivan et al., 2011). In order to further examine the role of COBL2 during cellulose deposition in MSCs, scanning electron microscopy was conducted (Stork et al., 2010; Mendu et al., 2011a). Scanning electron microscopy imaging of dry seeds of cobl2 mutants revealed substantially thinner radial cell walls and reduced columella size compared with those observed in wild-type seeds (Fig. 9). Quantification of radial cell wall width and columella tip area revealed an approximately 50% reduction of both parameters in the two cobl2 alleles examined compared with wild-type and cob-1 mutant seeds (Fig. 9, B and C). To further confirm these results, 10-μm cryosections of imbibed seeds were examined by the LC-PolScope system (Fig. 10; Supplemental Fig. S8). The retardance of polarized light by the columella of both cobl2 alleles was reduced significantly compared with wild-type seeds, indicating that COBL2 plays an additional role in the deposition of crystalline cellulose into the cytoplasm of MSCs, giving rise to the columella (Fig. 10). Together, these data suggest a central role for COBL2 in multiple aspects of secondary cell wall deposition in MSCs, including ray formation, radial cell wall thickening, and the formation of the columella.
Figure 9.
cobl2 mutant seeds display alterations in MSC secondary cell wall structures like the columella and the radial cell wall. A, Scanning electron microscopy images of mature seeds of the indicated genotypes. The radial cell wall is artificially stained in yellow and the columella tip in blue. Bars = 25 mm (top row), 25 μm (middle row), and 10 μm (bottom row). B and C, Quantification of MSC radial cell wall width (B) and columella tip area (C) in the indicated genotypes. For each genotype, 150 epidermal cells from 10 seeds were measured. Error bars indicate se. WT, Wild type.
Figure 10.
The retardance of polarized light birefringence demonstrates significant reduction in crystalline cellulose deposition into the columella of cobl2 mutant seeds. Dry seeds of the indicated genotypes were cryosectioned and observed by LC-PolScope for in situ visualization of crystalline cellulose deposition in MSCs. A to C, Representative images of the retardance of polarized light birefringence by the columella of seed epidermal cells as visualized by LC-PolScope for the wild type (A), cobl2-1 (B), and cobl2-2 (C). The color-code index refers to the extent of polarized light retardance: a high degree of retardance is indicative of high levels of crystalline cellulose in the light path (as designated in red), and a low degree of retardance is indicative of a lack of crystalline cellulose in the light path (as designated in black). Bars = 5 μm. D, Quantification of the retardance of polarized light birefringence by the columella of epidermal cells of seeds from the indicated genotypes. For each genotype, five epidermal cells from three seeds were measured. Error bars indicate se. WT, Wild type.
DISCUSSION
Recent studies demonstrate multiple roles for cellulose deposition throughout the course of seed coat epidermal cell differentiation. Various CESAs were shown to play roles in the deposition of cellulose (1) into the set of cellulosic rays radiating from the seed upon mucilage extrusion and (2) into the secondary cell wall structures known as the columella and the radial cell wall (Stork et al., 2010; Harpaz-Saad et al., 2011; Mendu et al., 2011a; Sullivan et al., 2011). However, other components of the machinery employed in cellulose deposition in this developmental context remain largely uncharacterized. Here, we assign a novel function to COBL2, demonstrating its role in crystalline cellulose deposition in seed coat epidermal cells. Mutations in cobl2 lead to crystalline cellulose ray malformation associated with increased solubilization of seed mucilage pectins. These results identify the cobl2 mutants as part of an emerging class of seed coat mucilage mutants (including the previously described cesa5, fei2, sos5, csla2, and mum5) in which perturbation in mucilage cellulosic ray formation is associated with a redistribution of seed mucilage pectins (Harpaz-Saad et al., 2011; Mendu et al., 2011a; Sullivan et al., 2011; Yu et al., 2014). In a similar manner to CESA5, COBL2 affects cellulose deposition into the rays, the columella, and the radial cell wall. However, unlike CESA5, the effect of COBL2 on the columella and radial cell walls appears nonredundant, as a single cobl2 mutation significantly impacts the morphology of these secondary cell wall structures. Various results suggest that COBL2 acts through a direct effect on the deposition of crystalline cellulose microfibrils: (1) the high similarity in phenotype between the reduced rays observed for cobl2 and cesa5 mutants by both Pontamine stain and polarized light microscopy; (2) the significantly reduced retardance of polarized light by the columella of cobl2 mutant seeds as compared with the wild type; and (3) the reduction of approximately 40% in total seed crystalline cellulose content compared with the wild type. Therefore, COBL2 is essential for crystalline cellulose deposition into specialized secondary cell wall structures of seed epidermal cells, such as the set of rays radiating from the seed across the adherent layer of seed mucilage, the columella, and the radial cell wall. Additionally, although LC-PolScope visualization of whole-seed cryosections for cobl2-1, compared with the wild type and cesa5, did not indicate significant variance in crystalline cellulose content in other cell layers in the seed, the approximately 40% reduction in crystalline cellulose quantity measured in whole seeds and the RT-PCR results suggesting COBL2 expression during embryo development may indicate an even more extensive role for COBL2 in this context and require further study (Fig. 3; Supplemental Fig. S9).
COBL2 is a member of the COBL gene family encoding extracellular, GPI-anchored proteins. Other COBL gene products were shown previously to be required either for oriented crystalline cellulose deposition into the primary cell wall during cell expansion (e.g. COBRA in the root, COBL9 in root hairs, and COBL10 and COBL11 in pollen tube elongation) or for the assembly of crystalline cellulose microfibrils during the deposition of the secondary cell wall (such as COBL4 during the formation of the vascular system; Parker et al., 2000; Schindelman et al., 2001; Brown et al., 2005; Persson et al., 2005; Li et al., 2013). While the COBL gene products do not appear to function as part of the core machinery employed in the synthesis of the β(1→4)-linked Glc chains, accumulating evidence suggests that they may play a central role in the assembly of multiple β(1→4)-linked Glc chains into crystalline cellulose microfibrils (Schindelman et al., 2001; Liu et al., 2013; Sorek et al., 2014). Previous studies demonstrate that loss-of-function mutations in various COBL family members, like COBRA, COBL4, and several of the COBL4 orthologs, result in reduced levels of crystalline cellulose (Schindelman et al., 2001; Li et al., 2003; Brown et al., 2005; Ching et al., 2006; Liu et al., 2013). In this study, mutations in cobl2 led to reduced levels of crystalline cellulose, as demonstrated by both in situ light birefringence and whole-seed cellulose quantification. Nevertheless, deciphering the mechanism by which COBL2 affects crystalline cellulose deposition will require further investigation. Liu et al. (2013) showed that BRITTLE CULM1 (BC1), the rice ortholog of Arabidopsis COBL4, preferentially interacts with crystalline cellulose through conserved aromatic residues located at the N-terminal carbohydrate-binding module conserved across different members of the COBL family. The reduced crystallinity index in the bc1 mutant suggests a reduced ratio of crystalline to amorphous cellulose that can be complemented and even increased by the expression of the wild-type BC1 gene in the bc1 and wild-type backgrounds, respectively (Sato et al., 2010a; Liu et al., 2013). Additionally, Sorek et al. (2014) recently demonstrated that COBRA binds individual β(1→4)-linked glucan chains with higher affinity than crystalline cellulose. Solid-state NMR analysis of cob mutant cell walls indicated that cellulose microfibril crystallinity is reduced and more reducing ends are available as compared with the wild type. These results further support a role for the COBL gene products in the assembly of multiple β(1→4)-linked Glc chains into crystalline cellulose. Further studies will be required in order to understand the mechanism of COBL protein function and its interaction with other components involved in cellulose synthesis.
In this work, tissue-specific coexpression analysis was used to identify COBL2 as a candidate player in cellulose deposition in the Arabidopsis seed epidermis. Gene coexpression analysis is a powerful tool for the prediction of gene function and the discovery of biological pathways. Many genes involved in the biosynthesis of cell wall components, such as cellulose, hemicellulose, and lignin, have been identified using coexpression analyses (for review, see Ruprecht and Persson, 2012). Here, we hypothesized that, in certain cases, global coexpression analyses encompassing all available data sets may overlook tissue-specific coexpression relationships. Evolution of gene families by subfunctionalization leads to the development of similar yet specialized functions for different members in varying developmental contexts. In the case of cellulose biosynthesis, it was shown previously that different CESAs tend to heterodimerize to generate an array of cellulose synthase complexes with specialized functions in different developmental contexts (Desprez et al., 2007; Mendu et al., 2011a). Members of the receptor-like kinase superfamily to which FEI2 is related tend to heterodimerize with various receptor-like kinases in a context-specific manner as well (Russinova et al., 2004; Wang et al., 2005). In fact, seed-specific coexpression analysis with FEI2 led to the identification of the role of COBL2 in crystalline cellulose deposition in seed epidermal cells, a relationship entirely overlooked by available, global coexpression analysis methods. Various Web-based tools used for global coexpression analysis with FEI2 as bait, including ATTED-II, GeneCAT, and ACT (Manfield et al., 2006; Obayashi et al., 2007; Mutwil et al., 2008), failed to identify significant gene expression correlation between FEI2 and COBL2. These results suggest that commonly used global coexpression analysis methods may overlook tissue- or context-dependent coexpression relationships. The massive induction of cellulose deposition in a highly defined spatial localization (specifically localized to the epidermal layer of the seed coat) makes it an attractive developmental context for the study of cellulose metabolism through a tissue-specific coexpression analysis. This work provides evidence that organ- or physiology-specific coexpression analysis may uncover meaningful coexpression relationships overlooked by the commonly used global coexpression analysis methods. Therefore, we suggest that some biologically relevant transcriptional relationships will be revealed only under specific experimental conditions or in tissue-specific data sets.
The data presented further establish the role of crystalline cellulose in the adhesion of the pectinaceous component of seed mucilage to the seed surface. However, further studies are required in order to uncover the specialized machinery employed in cellulose deposition in this developmental context. Additionally, characterization of the interactions behind the cellulose-pectin network responsible for seed mucilage adhesion requires further investigation. Recent work by Saez-Aguayo et al. (2014) recently described the floating mucilage-releasing accessions, natural Arabidopsis variants with perturbation in ray formation associated with increased pectin solubility. These may serve as valuable tools for further study of the mechanism employed in seed mucilage ray formation and the cellulose-pectin interactions affecting pectin solubility. The development of new tools for correlative microscopy that facilitate the in situ study of cell wall composition will be valuable for future studies as well.
MATERIALS AND METHODS
Plant Material
The Columbia ecotype of Arabidopsis (Arabidopsis thaliana) was used in this study. The cobl2 (cobl2-1, SALK_044883; cobl2-2, SAIL_317_C05) alleles and cob-1 (Schindelman et al., 2001), cobl6 (cobl6-1, SAIL_144_C06; cobl6-2, SAIL_315_A08; cobl6-3, SALK_033122), and cesa5 (SALK_118491) mutants were obtained from the Arabidopsis Biological Resource Center. For growth in soil, plants were grown at 22°C in 75 µE of constant light. For growth in vitro, seeds were surface sterilized, cold treated at 4°C for 4 d in the dark on 1× Murashige and Skoog medium, and grown at 22°C in 100 µE of constant light.
Expression Analysis
For gene expression analysis, flowers were marked at anthesis and harvested 4, 6, 8, 10, 12, and 14 DPA. For 4, 6, and 8 DPA, whole siliques were harvested, while for 10, 12, and 14 DPA, developing seeds were collected from the siliques. Total RNA was extracted using the Plant/Fungi Total RNA Extraction kit (Norgen Biotek) with additional DNase treatment (Qiagen). First-strand synthesis of complementary DNA (cDNA) from RNA template (2 µg) was accomplished with SuperScript II Reverse Transcriptase (Invitrogen) and conducted according to the manufacturer’s instructions. For the amplification of PCR products from cDNA of the indicated genotypes (wild type, cobl2-1, and cobl2-2), the following exon-exon primers were used with TUBULIN serving as the reference gene: COBL2 CDS 3′, forward primer 5′-GGTTGCCATCACAAACTTTAAC-3′ and reverse primer 5′-CATGGCCGTGTCATTTATGTT-3′; and TUBULIN CDS, forward primer 5′-AAACTCACTACCCCCAGCTTTG-3′ and reverse primer 5′-GAGAGGAGCAAAACCAACCA-3′. For the analysis of COBL2 expression in seed coat and embryo, GAPC served as an additional general reference gene and NARSI and PDFI as seed coat- and embryo-specific genes. Primers were as follows: GAPC CDS, forward primer 5′-GATTCGGAAGAATTGGTCGTTT-3′ and reverse primer 5′-CTTCAAGTGAGCTGCAGCCTT-3′ (Voiniciuc et al., 2013); NARSI CDS, forward primer 5′-CCCTACCGACGAAGAGCTTGTTG-3′ and reverse primer 5′-CAATGGTGGAGTCTCTTCCATCATG-3′ (Kunieda et al., 2013); and PDFI CDS, forward primer 5′-ACTCCGGTTGTTGTGACTCC-3′ and reverse primer 5′-GTGCCTTCACGGTAGAGAGC-3′ (Kunieda et al., 2013). Quantitative real-time PCR was prepared with the above primers using Absolute Blue qPCR SYBR Green ROX Mix (Thermo Scientific) according to the manufacturer’s instructions (Bustin, 2000). Primer sensitivity and efficiency were analyzed on a dilution series of cDNA with reference sample (7 µg). The quantitative PCR and analysis were processed by Rotor-Gene 6000 (Corbett Research).
Seed Staining and Microscopy
For seed coat visualization, seeds were prehydrated with either water or 50 mm EDTA for 90 min, washed with water, and then stained as follows. The Ruthenium Red stain for pectins was prepared as described (Willats et al., 2001), and hydrated seeds were incubated in 0.01% (w/v) Ruthenium Red (Sigma) for 90 min at room temperature. The Calcofluor stain was conducted as described (Willats et al., 2001) using 25 µg mL−1 Fluorescent Brightener 28 (Sigma) for 20 min at room temperature. In both cases, seeds were washed for 16 h in water and then photographed. Pontamine staining was conducted as described (Anderson et al., 2010) using 0.01% (w/v) Pontamine Fast Scarlet S4B stain (Sigma) for 30 min following pretreatment as described above. The seeds were then destained with water for 4 h before examination. Ruthenium Red stain without shaking was prepared by dropping the seeds onto 12-well plates containing 0.0025% (w/v) Ruthenium Red in water for 90 min before imaging.
Observations were conducted with either a Leica compound dissection microscope or the Zeiss LSM710 confocal microscope equipped with a 405-nm laser diode for Calcofluor and a 561-nm laser for Pontamine. In order to determine the volume occupied by the seeds, 10 mg of dry seeds was hydrated with water overnight with occasional rough shaking.
Polarized Light Microscopy (LC-PolScope)
Sample birefringence was investigated using the LC-PolScope image-processing system (CRi) mounted on a microscope (Nikon Eclipse 80i) equipped with Plan Fluor 10×/0.3 OFN25 DIC L/N1, Plan Fluor 20×/0.5 OFN25 DIC N2, Plan Fluor 40×/0.75 OFN25 DIC M/N2, and Fluor 60×/100w DIC H/N2 ∞/0 WD 2.0 objectives. The system includes a computer-controlled universal compensator composed of two liquid crystal variable retarders. Images were taken by a cooled CCD camera at high optical resolution.
For whole-seed imaging, the complete seeds were imbibed in water for 1 h and mounted in water on a glass slide. For section imaging, the wet seeds were embedded in tissue-freezing medium, frozen, and sectioned at 10 μm thickness (Leica CM1850 Cryostat). The sections were mounted on glass slides, wetted, and sealed to prevent water evaporation. Retardance values were extracted manually from the LC-PolScope map in retardance mode using the Abrio software tools (CRi) from the columella of five epidermal cells from each seed section. The average retardance values were calculated for three different seed sections from each mutant line.
Scanning Electron Microscopy
Mature, dry seeds were mounted on aluminum stubs (Electron Microscopy Sciences), sputter coated with 40 nm of gold-palladium, and viewed using a Hitachi S-4700 field emission scanning electron microscope. Every genotype was examined in four independent groups of roughly 20 seeds each.
Crystalline Cellulose Quantification
Dry seeds of Arabidopsis of the various genotypes (33.2–33.9 mg) were frozen in liquid nitrogen and ground to a fine powder using three small metal beads in 2-mL plastic tubes (Retsch Ball Mill; two times at 25 Hz for 2.5 min). The ground material was washed two times with 70% (v/v) ethanol (1.5 mL) by vortexing, pelleting the wall residue (21,000g for 10 min), and discarding the supernatant. This was followed by three washes with a 1:1 (v/v) methanol:chloroform solution (1.5 mL per wash) using the same pelleting conditions as described above. The pellet was dried in a speed vacuum centrifuge at 60°C for 15 min. This pellet (1 mg) was treated with 1.5 mL of Updegraff reagent (acetic acid:nitric acid:water, 8:1:2 [v/v]; Updegraff, 1969) at 100°C for 30 min. Samples were allowed to cool at room temperature and spun down at 21,000g for 10 min, resulting in a pellet consisting primarily of crystalline cellulose. The supernatant was removed, and the pellet was washed four times with 1.5 mL of acetone and pelleted at 21,000g for 10 min each time. The samples were dried at room temperature followed by 15 min of drying under vacuum. The crystalline cellulose was then hydrolyzed with 175 µL of 72% (w/w) sulfuric acid for 1 h at room temperature under 300 rpm agitation. After hydrolysis, water was added (425 μL), and the samples were spun down at 21,000g for 30 s. The remaining solution was used for Glc determination using the anthrone assay (Laurentin and Edwards, 2003).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression analysis of the COBL gene family during seed development.
Supplemental Figure S2. Allelism tests between cobl2-1 and cobl2-2 demonstrate that both carry a mutation in the same gene.
Supplemental Figure S3. Mutants in cobl6 did not display seed mucilage phenotypes.
Supplemental Figure S4. Expression of COBL2 promoter-driven GUS in seeds.
Supplemental Figure S5. cobl2 mutants display no root elongation phenotype.
Supplemental Figure S6. Etiolated seedlings of cobl2 mutants are indistinguishable from the wild type.
Supplemental Figure S7. Both cobl2 mutant alleles display reduced crystalline cellulose levels in the rays of seed mucilage.
Supplemental Figure S8. Comparison of seed mucilage ray visualization by LC-PolScope and Pontamine staining.
Supplemental Figure S9. Crystalline cellulose in whole-seed sections of cobl2-1 as compared with the wild type.
Supplementary Material
Acknowledgments
We thank Tony Perdue (University of North Carolina) for assistance with confocal microscopy, Vicktoria Madden (University of North Carolina Microscopy Service Unit) for help with scanning electron microscopy, and the Harpaz-Saad laboratory and Kieber laboratory personnel for helpful discussions.
Glossary
- GPI
glycosylphosphatidylinositol
- MSC
mucilage secretory cell
- T-DNA
transfer DNA
- RT
reverse transcription
- cDNA
complementary DNA
Footnotes
This work was supported by the National Science Foundation (grant no. IOS 0624377 to J.J.K.), the Israel Science Foundation (I-CORE grant no. 757/12), and a Marie Curie Career Integration Grant (grant no. 618826 to S.H.-S).
This article is dedicated to the memory of Eran Barack.
Articles can be viewed without a subscription.
References
- Abraham Y, Elbaum R (2013) Quantification of microfibril angle in secondary cell walls at subcellular resolution by means of polarized light microscopy. New Phytol 197: 1012–1019 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anderson CT, Carroll A, Akhmetova L, Somerville C (2010) Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol 152: 787–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R, et al. (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279: 717–720 [DOI] [PubMed] [Google Scholar]
- Arsovski AA, Haughn GW, Western TL (2010) Seed coat mucilage cells of Arabidopsis thaliana as a model for plant cell wall research. Plant Signal Behav 5: 796–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin TI. (2005) Anisotropic expansion of the plant cell wall. Annu Rev Cell Dev Biol 21: 203–222 [DOI] [PubMed] [Google Scholar]
- Beeckman T, Przemeck GK, Stamatiou G, Lau R, Terryn N, De Rycke R, Inzé D, Berleth T (2002) Genetic complexity of cellulose synthase A gene function in Arabidopsis embryogenesis. Plant Physiol 130: 1883–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA (1993) Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119: 57–70 [DOI] [PubMed] [Google Scholar]
- Blake AW, McCartney L, Flint JE, Bolam DN, Boraston AB, Gilbert HJ, Knox JP (2006) Understanding the biological rationale for the diversity of cellulose-directed carbohydrate-binding modules in prokaryotic enzymes. J Biol Chem 281: 29321–29329 [DOI] [PubMed] [Google Scholar]
- Brady SM, Song S, Dhugga KS, Rafalski JA, Benfey PN (2007) Combining expression and comparative evolutionary analysis: the COBRA gene family. Plant Physiol 143: 172–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, Zeef LA, Ellis J, Goodacre R, Turner SR (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RM., Jr (1985) Cellulose microfibril assembly and orientation: recent developments. J Cell Sci Suppl 2: 13–32 [DOI] [PubMed] [Google Scholar]
- Bustin SA. (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25: 169–193 [DOI] [PubMed] [Google Scholar]
- Ching A, Dhugga KS, Appenzeller L, Meeley R, Bourett TM, Howard RJ, Rafalski A (2006) Brittle stalk 2 encodes a putative glycosylphosphatidylinositol-anchored protein that affects mechanical strength of maize tissues by altering the composition and structure of secondary cell walls. Planta 224: 1174–1184 [DOI] [PubMed] [Google Scholar]
- Dagel DJ, Liu YS, Zhong L, Luo Y, Himmel ME, Xu Q, Zeng Y, Ding SY, Smith S (2011) In situ imaging of single carbohydrate-binding modules on cellulose microfibrils. J Phys Chem B 115: 635–641 [DOI] [PubMed] [Google Scholar]
- Dai X, You C, Chen G, Li X, Zhang Q, Wu C (2011) OsBC1L4 encodes a COBRA-like protein that affects cellulose synthesis in rice. Plant Mol Biol 75: 333–345 [DOI] [PubMed] [Google Scholar]
- Dean G, Cao Y, Xiang D, Provart NJ, Ramsay L, Ahad A, White R, Selvaraj G, Datla R, Haughn G (2011) Analysis of gene expression patterns during seed coat development in Arabidopsis. Mol Plant 4: 1074–1091 [DOI] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder M, Lütz-Meindl U, Weiss IM (2010) Non-invasive LC-PolScope imaging of biominerals and cell wall anisotropy changes. Protoplasma 246: 49–64 [DOI] [PubMed] [Google Scholar]
- Engelbrecht M, García-Fayos P (2012) Mucilage secretion by seeds doubles the chance to escape removal by ants. Plant Ecol 213: 1167–1175 [Google Scholar]
- Friedrichson T, Kurzchalia TV (1998) Microdomains of GPI-anchored proteins in living cells revealed by crosslinking. Nature 394: 802–805 [DOI] [PubMed] [Google Scholar]
- Gillmor CS, Poindexter P, Lorieau J, Palcic MM, Somerville C (2002) Alpha-glucosidase I is required for cellulose biosynthesis and morphogenesis in Arabidopsis. J Cell Biol 156: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonneau M, Desprez T, Guillot A, Vernhettes S, Höfte H (2014) Catalytic subunit stoichiometry within the cellulose synthase complex. Plant Physiol 166: 1709–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths JA, Tsai AYL, Xue H, Voiniciuc C, Šola K, Seifert GJ, Mansfield SD, Haughn GW (2014) SALT-OVERLY SENSITIVE5 mediates Arabidopsis seed coat mucilage adherence and organization through pectins. Plant Physiol 165: 991–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke DE, Northcote DH (1975) Molecular visualization of pectin and DNA by ruthenium red. Biopolymers 14: 1–17 [DOI] [PubMed] [Google Scholar]
- Harpaz-Saad S, McFarlane HE, Xu S, Divi UK, Forward B, Western TL, Kieber JJ (2011) Cellulose synthesis via the FEI2 RLK/SOS5 pathway and cellulose synthase 5 is required for the structure of seed coat mucilage in Arabidopsis. Plant J 68: 941–953 [DOI] [PubMed] [Google Scholar]
- Haughn G, Chaudhury A (2005) Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci 10: 472–477 [DOI] [PubMed] [Google Scholar]
- Haughn GW, Western TL (2012) Arabidopsis seed coat mucilage is a specialized cell wall that can be used as a model for genetic analysis of plant cell wall structure and function. Front Plant Sci 3: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser MT, Morikami A, Benfey PN (1995) Conditional root expansion mutants of Arabidopsis. Development 121: 1237–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MA, Raymond MJ, Smirnoff N (2006) Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J 45: 83–100 [DOI] [PubMed] [Google Scholar]
- Kunieda T, Shimada T, Kondo M, Nishimura M, Nishitani K, Hara-Nishimura I (2013) Spatiotemporal secretion of PEROXIDASE36 is required for seed coat mucilage extrusion in Arabidopsis. Plant Cell 25: 1355–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurentin A, Edwards CA (2003) A microtiter modification of the anthrone-sulfuric acid colorimetric assay for glucose-based carbohydrates. Anal Biochem 315: 143–145 [DOI] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Ge FR, Xu M, Zhao XY, Huang GQ, Zhou LZ, Wang JG, Kombrink A, McCormick S, Zhang XS, et al. (2013) Arabidopsis COBRA-LIKE 10, a GPI-anchored protein, mediates directional growth of pollen tubes. Plant J 74: 486–497 [DOI] [PubMed] [Google Scholar]
- Li Y, Qian Q, Zhou Y, Yan M, Sun L, Zhang M, Fu Z, Wang Y, Han B, Pang X, et al. (2003) BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 15: 2020–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Shang-Guan K, Zhang B, Liu X, Yan M, Zhang L, Shi Y, Zhang M, Qian Q, Li J, et al. (2013) Brittle Culm1, a COBRA-like protein, functions in cellulose assembly through binding cellulose microfibrils. PLoS Genet 9: e1003704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macquet A, Ralet MC, Kronenberger J, Marion-Poll A, North HM (2007) In situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant Cell Physiol 48: 984–999 [DOI] [PubMed] [Google Scholar]
- Manfield IW, Jen CH, Pinney JW, Michalopoulos I, Bradford JR, Gilmartin PM, Westhead DR (2006) Arabidopsis Co-expression Tool (ACT): web server tools for microarray-based gene expression analysis. Nucleic Acids Res 34: W504–W509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter K, Mellman I (1994) Mechanisms of cell polarity: sorting and transport in epithelial cells. Curr Opin Cell Biol 6: 545–554 [DOI] [PubMed] [Google Scholar]
- Mendu V, Griffiths JS, Persson S, Stork J, Downie AB, Voiniciuc C, Haughn GW, DeBolt S (2011a) Subfunctionalization of cellulose synthases in seed coat epidermal cells mediates secondary radial wall synthesis and mucilage attachment. Plant Physiol 157: 441–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendu V, Stork J, Harris D, DeBolt S (2011b) Cellulose synthesis in two secondary cell wall processes in a single cell type. Plant Signal Behav 6: 1638–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M, Obro J, Willats WG, Persson S (2008) GeneCAT: novel webtools that combine BLAST and co-expression analyses. Nucleic Acids Res 36: W320–W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North H, Baud S, Debeaujon I, Dubos C, Dubreucq B, Grappin P, Jullien M, Lepiniec L, Marion-Poll A, Miquel M, et al. (2010) Arabidopsis seed secrets unravelled after a decade of genetic and omics-driven research. Plant J 61: 971–981 [DOI] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, Shibata D, Saito K, Ohta H (2007) ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res 35: D863–D869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenbourg R. (1996) A new view on polarization microscopy. Nature 381: 811–812 [DOI] [PubMed] [Google Scholar]
- Parker JS, Cavell AC, Dolan L, Roberts K, Grierson CS (2000) Genetic interactions during root hair morphogenesis in Arabidopsis. Plant Cell 12: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Meissner RC, Shoue DA, Carpita NC, Bevan MW (2001) MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 13: 2777–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR (2007) Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci USA 104: 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Wei H, Milne J, Page GP, Somerville CR (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA 102: 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringli C, Baumberger N, Keller B (2005) The Arabidopsis root hair mutants der2-der9 are affected at different stages of root hair development. Plant Cell Physiol 46: 1046–1053 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Boulan E, Powell SK (1992) Polarity of epithelial and neuronal cells. Annu Rev Cell Biol 8: 395–427 [DOI] [PubMed] [Google Scholar]
- Roudier F, Fernandez AG, Fujita M, Himmelspach R, Borner GHH, Schindelman G, Song S, Baskin TI, Dupree P, Wasteneys GO, et al. (2005) COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 17: 1749–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F, Schindelman G, DeSalle R, Benfey PN (2002) The COBRA family of putative GPI-anchored proteins in Arabidopsis: a new fellowship in expansion. Plant Physiol 130: 538–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprecht C, Persson S (2012) Co-expression of cell-wall related genes: new tools and insights. Front Plant Sci 3: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russinova E, Borst JW, Kwaaitaal M, Caño-Delgado A, Yin Y, Chory J, de Vries SC (2004) Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16: 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Aguayo S, Ralet MC, Berger A, Botran L, Ropartz D, Marion-Poll A, North HM (2013) PECTIN METHYLESTERASE INHIBITOR6 promotes Arabidopsis mucilage release by limiting methylesterification of homogalacturonan in seed coat epidermal cells. Plant Cell 25: 308–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Aguayo S, Rondeau-Mouro C, Macquet A, Kronholm I, Ralet MC, Berger A, Sallé C, Poulain D, Granier F, Botran L, et al. (2014) Local evolution of seed flotation in Arabidopsis. PLoS Genet 10: e1004221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Ito S, Fujii T, Suzuki R, Takenouchi S, Nakaba S, Funada R, Sano Y, Kajita S, Kitano H, et al. (2010a) The carbohydrate-binding module (CBM)-like sequence is crucial for rice CWA1/BC1 function in proper assembly of secondary cell wall materials. Plant Signal Behav 5: 1433–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Suzuki R, Nishikubo N, Takenouchi S, Ito S, Nakano Y, Nakaba S, Sano Y, Funada R, Kajita S, et al. (2010b) Isolation of a novel cell wall architecture mutant of rice with defective Arabidopsis COBL4 ortholog BC1 required for regulated deposition of secondary cell wall components. Planta 232: 257–270 [DOI] [PubMed] [Google Scholar]
- Schindelman G, Morikami A, Jung J, Baskin TI, Carpita NC, Derbyshire P, McCann MC, Benfey PN (2001) COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev 15: 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt WJ. (1924) Die Bausteine des Tierkörpers in Polarisiertem Lichte. Cohen Verlag, Bonn [Google Scholar]
- Somerville C. (2006) Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol 22: 53–78 [DOI] [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab T, et al. (2004) Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211 [DOI] [PubMed] [Google Scholar]
- Sorek N, Sorek H, Kijac A, Szemenyei HJ, Bauer S, Hématy K, Wemmer DE, Somerville CR (2014) The Arabidopsis COBRA protein facilitates cellulose crystallization at the plasma membrane. J Biol Chem 289: 34911–34920 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Stork J, Harris D, Griffiths J, Williams B, Beisson F, Li-Beisson Y, Mendu V, Haughn G, Debolt S (2010) CELLULOSE SYNTHASE9 serves a nonredundant role in secondary cell wall synthesis in Arabidopsis epidermal testa cells. Plant Physiol 153: 580–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan S, Ralet MC, Berger A, Diatloff E, Bischoff V, Gonneau M, Marion-Poll A, North HM (2011) CESA5 is required for the synthesis of cellulose with a role in structuring the adherent mucilage of Arabidopsis seeds. Plant Physiol 156: 1725–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100: 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegraff DM. (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32: 420–424 [DOI] [PubMed] [Google Scholar]
- Varma R, Mayor S (1998) GPI-anchored proteins are organized in submicron domains at the cell surface. Nature 394: 798–801 [DOI] [PubMed] [Google Scholar]
- Voiniciuc C, Dean GH, Griffiths JS, Kirchsteiger K, Hwang YT, Gillett A, Dow G, Western TL, Estelle M, Haughn GW (2013) Flying saucer1 is a transmembrane RING E3 ubiquitin ligase that regulates the degree of pectin methylesterification in Arabidopsis seed mucilage. Plant Cell 25: 944–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace IS, Anderson CT (2012) Small molecule probes for plant cell wall polysaccharide imaging. Front Plant Sci 3: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Goshe MB, Soderblom EJ, Phinney BS, Kuchar JA, Li J, Asami T, Yoshida S, Huber SC, Clouse SD (2005) Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17: 1685–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western TL. (2012) The sticky tale of seed coat mucilages: production, genetics, and role in seed germination and dispersal. Seed Sci Res 22: 1–25 [Google Scholar]
- Western TL, Skinner DJ, Haughn GW (2000) Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol 122: 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats WGT, McCartney L, Knox JP (2001) In-situ analysis of pectic polysaccharides in seed mucilage and at the root surface of Arabidopsis thaliana. Planta 213: 37–44 [DOI] [PubMed] [Google Scholar]
- Windsor JB, Symonds VV, Mendenhall J, Lloyd AM (2000) Arabidopsis seed coat development: morphological differentiation of the outer integument. Plant J 22: 483–493 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Baskin CC, Baskin JM, Gao R, Yang F, Wei L, Li L, He H, Huang Z (2013) Hydrated mucilage reduces post-dispersal seed removal of a sand desert shrub by ants in a semiarid ecosystem. Oecologia 173: 1451–1458 [DOI] [PubMed] [Google Scholar]
- Young RE, McFarlane HE, Hahn MG, Western TL, Haughn GW, Samuels AL (2008) Analysis of the Golgi apparatus in Arabidopsis seed coat cells during polarized secretion of pectin-rich mucilage. Plant Cell 20: 1623–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Shi D, Li J, Kong Y, Yu Y, Chai G, Hu R, Wang J, Hahn MG, Zhou G (2014) CELLULOSE SYNTHASE-LIKE A2, a glucomannan synthase, is involved in maintaining adherent mucilage structure in Arabidopsis seed. Plant Physiol 164: 1842–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.