Abstract
Blood tests are needed to aid in the early detection of pancreatic ductal adenocarcinoma (PDAC), and monitoring pancreatitis development into malignancy especially in high risk patients. This study exhibits efforts and progress toward developing such blood tests, using electrospray-mass spectrometry (MS) serum profiling to distinguish patients with early-stage PDAC or pancreatitis from each other and from controls. Identification of significant serum mass peak differences between these individuals was performed using t tests and “leave one out” cross validation. Serum mass peak distributions of control individuals were distinguished from those of patients with chronic pancreatitis or early-stage PDAC with P values <10−15, and patients with chronic pancreatitis were distinguished from those of patients with early-stage PDAC with a P value <10−12. Sera from 12 out of 12 patients with PDAC stages I, IIA and IIB were blindly validated from controls. Tandem MS/MS identified a cancer phenotype with elements of PDAC involved in early-stage PDAC/control discrimination. These studies indicate electrospray-MS mass profiling can detect serum changes in patients with pancreatitis or early-stage pancreatic cancer. Such technology has the potential to aid in early detection of pancreatic cancer, biomarker development, and in monitoring development of pancreatitis into PDAC.
Keywords: Pancreatic ductal adenocarcinoma, Serum mass profiling, Chronic pancreatitis, Electrospray mass spectrometry, Early detection, Cancer phenotype
Introduction
Early detection of pancreatic ductal adenocarcinoma (PDAC) is an important aspect of cancer treatment because early clinical stages (I, II) are easier to cure than later stages (III, IV) [1]. There is a need for robust, accurate and non-invasive detection methodology, e.g. from blood, for the early stages of pancreatic cancer [1,2]. Serum protein CA-19.9 is used to monitor existing pancreatic cancer but is not useful in diagnosis [3]. Multiple micro (mi) RNAs from plasma were shown to be indicators for pancreatic cancer, and mi-155 is possibly predictive for early-stage pancreatic neoplasia [4]. However there are still some discrepancies among micro RNA technologies [5,6]. A variety of serum biomarkers in an antibody-protein microarray format had positive results detecting late-stage pancreatic cancer and chronic pancreatitis (CP) [7]. Chronic pancreatitis is a significant risk factor for the development of pancreatic cancer [8,9]. One of the prominent mechanisms by which PDAC is hypothesized to develop, e.g., from pancreatitis to pancreatic cancer, is through cellular and genetic changes involving pancreatic intraepithelial neoplasias (PanINs) which can be found in chronic pancreatitis [10,11].
The profiling of bodily fluids using all-liquid electrospray ionization (ESI) mass spectrometry (MS) has the potential to distinguish differences between blood/sera of disease-free individuals and individuals with pathological conditions [12–15]. Serum mass profiling is useful in cancer diagnostics including pancreatic cancer, and in therapeutic development [14–17]. The underlying hypothesis is that sera contain ample numbers and kinds of peptides and other biomolecules (e.g., proteins, nucleic acids, glycoconjugates, lipids), and this complexity will vary between disease states [12–15]. The basis for some of this complexity involves exoprotease degradation of proteins [18] and cellular signaling mechanisms [19], and is hypothesized to reflect homeostatic as well as defense/stress mechanisms which change with physiological state [16–19]. Consequently, organs/tissues shed and/or secrete varying amounts and different kinds of biomolecules into the peripheral blood in response to different physiological conditions. All-liquid ESI-MS is possibly the simplest biomarker platform available, requiring only a serum dilution and injection into the mass spectrometer. Liquid MS analyzes disease-related phenotypic profiles in sera, as opposed to indirect genotypic/nucleic acid classifications. ESI-MS serum mass profiling examines potentially all biomolecules in sera, whereas other biomarker platforms (DNA, RNA, metabolomics, and various antibody methods) focus on a single component or small groups of similar components and can require a significant amount of preparation prior to analysis. To improve specificity in disease detection, the more biomolecules analyzed at once, the greater disease discriminatory powers of the platform [17,18]. Importantly, MS analysis meets the accuracy, robustness, and reproducibility guidelines for stringent clinical laboratory testing [20–23]. Standard statistical approaches, like those used in this study, are better suited than novel algorithms [20,23].
Previously, we utilized electrospray ionization mass spectrometry (ESI-MS) peaks to distinguish sera from early-stage ovarian, lung, and pancreatic cancer patients from healthy disease-free individuals [15–17,24–26]. In the present study, electrospray serum mass profiling is used to distinguish early-stage PDAC patients (stages I, IIA, IIB) from healthy individuals and from patients with chronic pancreatitis. Leave one out cross validation (LOOCV) of the mass peak data and randomization of cohort sera samples is used to check for and help ameliorate “over-fitting” of the mass peak data. “Hold out” databases are formed and used to validate blinded early-stage PDAC, CP, or control serum sets. Tandem MS/MS [27] is used to help identify peptides/proteins potentially involved in PDAC/control discrimination. Such straight-forward analyses, from an accessible body fluid like serum, holds promise for aiding in the diagnosis and disease monitoring and, in the future, understanding pancreatic carcino-genesis mechanisms as well as aiding in the development and analysis of therapeutic interventions for this deadly disease.
Materials and methods
Patients and clinical samples
Patient-related information concerning individuals with stage I, IIA, or IIB pancreatic cancer, chronic pancreatitis, as well as healthy control individuals, is listed in Table 1. Patient/serum samples are divided into three groups: complete databases, validation databases, and blind validation samples. Tumor pathological staging was according to the TNM staging system (tumor size, node involvement, metastasis presence) [28]. Tumor and pancreatitis pathology was determined at the Surgical Pathology Laboratories of the University of Oklahoma Health Sciences Center Hospital. Sera were obtained from patient peripheral blood at the University of Oklahoma Health Sciences Center, before treatments, according to standard procedures [29]. Blood and serum samples were also collected from healthy volunteers from the University community in identical fashion. Sera aliquots (100 μl) were frozen at −80 °C, and not reused after initial freezing and thawing. Histology and hematoxylin and eosin (H&E) staining of PDAC, CP, and control tissues were performed as described [25].
Table 1.
Patient groups and characteristics.
| Databases | Patient groups | Mixed age Mean (range) | Female age Mean (range) | Male age Mean (range) | Patient (N) (male/female) |
|---|---|---|---|---|---|
| Complete LOOCV databases (all patient samples) | Pancreatitis | 55.0 (39-80) | 52.8 (46-74) | 56.3 (39-80) | 14(9/5) |
| Pancreatic cancer stages I & IIA | 60.0 (42-73) | 66.6 (56-73) | 56.6 (42-68) | 9(6/3) | |
| Pancreatic cancer stage IIB | 68.2 (49-84) | 65.4 (49-76) | 71.3 (60-84) | 19(9/10) | |
| Pancreatic cancer stage IIA & IIB | 68.0 (49-84) | 66.5 (49-76) | 69.8 (58-84) | 23(11/12) | |
| Pancreatic cancer I, IIA & IIB | 65.5 (42-84) | 65.6 (49-76) | 65.4 (42-84) | 28 (15/13) | |
| Non-cancer (control)a | 55.9 (40-69) | 54.9 (40-63) | 56.8 (47-69) | 22 (12/10) | |
| Validation LOOCV databases (blind validation patient samples excluded) | Pancreatitis | 51.2(39-69) | 47.6 (46-49) | 53.0 (39-69) | 9(6/3) |
| Pancreatic cancer stage IIA & IIB | 70(56-84) | 70.2 (56-76) | 69.9 (58-84) | 13(8/5) | |
| Pancreatic cancer stage IIB | 70 8 (56-84) | 69.5 (56-76) | 71.5 (60-84) | 11 (7/4) | |
| Pancreatic cancer I, IIA & IIB | 65.9 (42-84) | 68.2 (56-76) | 64.6 (42-84) | 20 (13/7) | |
| Non-cancer (control) | 56.1 (40-69) | 54.0 (40-63) | 57.8 (47-69) | 14(8/6) | |
| Blind validation samples withheld from validation database groups | Pancreatitis | 62.0 (47-80) | 60.5 (47-74) | 63.0 (50-80) | 5 (3/2) |
| Pancreatic cancer stage IIA & IIB | 60.5 (39-76) | 60.7 (49-76) | 60.0 (39-74) | 10(3/7) | |
| Pancreatic cancer stage IIB | 64.6 (47-80) | 62.7 (49-76) | 70.5 (67-74) | 8 (2/6) | |
| Pancreatic cancer stage I, IIA & IIB | 59.9 (39-76) | 59.9 (49-76) | 60.0 (39-74) | 12(4/8) | |
| Non-cancer (control) | 55.6 (51-58) | 56.3 (51-58) | 55.0 (53-56) | 8 (4/4) | |
| Non-cancer (control) group 2b | 57.7 (41-75) | 55.9 (41-65) | 59.2 (47-69) | 18(10/8) | |
| Lung cancer | Lung cancer stage I | 64.2 (45-84) | 62.3 (45-84) | 65.0 (50-82) | 40 (28/12) |
Non-cancer (control)
non-cancer (control) group 2 are unique groups and do not have any members in common.
ESI-MS of sera from PDAC and CP patients and healthy controls
A serum aliquot from patients with PDAC, CP, or control individuals was diluted 1–300 into a solution of 50% methanol and 2% formic acid. The samples were loop injected (20 μl) into the nano source of an LCQ Advantage ion trap mass spectrometer (ThermoScientific), fitted with a 20 micron inner diameter (100 micron outer diameter) fused silica (Polymicro Technologies) tip at a flow rate of 0.5 μl/min using an Eldex MicroPro series 1000 pumping system [24]. High-resolution triplicate mass spectra were collected from disease and disease-free sera in random fashion per day. The spectra were sampled at an m/Z (mass divided by charge) resolution of two hundredths over an m/Z range of 400–2000. Positive ion mode spectra were collected over 30 min for each injection. Raw spectral data from the Advantage LCQ instrument were extracted using the manufacturer’s software “Qual Browser” version 1.4SR1. Spectral data were exported in a format providing rounded unit m/Z and intensity values. Data were then normalized to the highest m/Z sum intensity value in segments of 25 m/Z from 400 to 2000. MS spectral peak assignments were calculated as centroid m/Z¬ peak area values (valley to valley) using Mariner Data Explorer 4.0.0.1 software (Applied BioSystems). Centroid area is defined as the area of the peak calculated from its geometrical m/Z center. For tandem MS/MS mass peak identifications [27], 60 m/Z ions (peak range, 700–940 m/Z, identified by LOOCV analysis for discriminating non-cancer controls from patients with stage IIB pancreatic cancer) were screened at 12 m/Z ion intervals in 10 control and 10 PDAC serum samples in the ThermoScientific ion-trap MS instrument used in the cohort discriminations. Samples were diluted 1–390 in 50% methanol, 2% formic acid, 48% water, and analyzed at a flow rate of 0.20 microliters/min. Peak protein identifications were determined using SEQUEST Proteome Discoverer 1.0 (Thermo Scientific) using the “no cleavage” setting on a “Homo sapien” database created through the Discoverer software from a non-redundant database downloaded from NCBI on 07/08/2014. Serum samples on average contained 1.95 (range: 0–5) parent ions with significant differences of standard MS spectral data between the pre and post MS/MS scans of the 60 parental ions observed.
Statistical and quantitative analysis
m/Z peak area data were exported into Excel 2010, and triplicate peak areas at each m/Z value were averaged for each serum sample. Using a nested LOOCV protocol [30,31], individual m/Z peak areas of a “left out” serum sample were analyzed for significance against the “left in” database (e.g., remaining control versus PDAC samples) using t-tests (one-tailed, two sample unequal variance) [16,26]. LOOCV was performed by removing one patient or control serum m/Z peak area data set at a time from the total m/Z peak database for each class of sera samples, and then reforming the total MS peak database in the absence of that single sample m/Z data set. For each “left out” m/Z LOOCV peak area tested against the database of significant peaks, a value greater than the 50% “cutoff “ (see Fig. 1E) was assigned to a patient group descriptor like PDAC, and a value equal to or less than the ‘cutoff mean’ was assigned to the control or other group descriptor. This procedure was repeated for all sera samples in the control, PDAC, and pancreatitis cohorts. In addition, 50% of serum samples in the LOOCV analysis must select a particular m/Z peak to be deemed significant. A % of the total descriptor (e.g., PDAC) mass peak database was then plotted versus patient and control sample number (see Fig. 2A). Blinded sera validation was performed in similar LOOCV fashion against training validation subsets obtained from their respective retrospective databases from which the blinded sera samples were “left out”. Validation database construction was performed using the described LOOCV method with the complete exclusion of the blinded samples. Random sampling of groups/cohorts was obtained using the RAND function in Excel 2010.
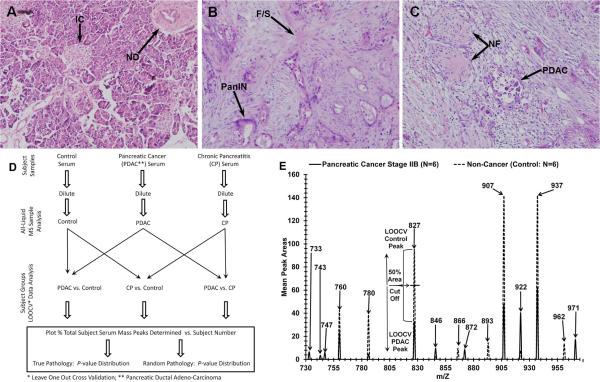
Fig. 1.
Histology of human pancreas tissue and electrospray MS m/Z serum peaks with analysis. Tissues are as follows: Control (Panel A), chronic pancreatitis (B), and (C) stage IIB pancreatic cancer. Hematoxylin and eosin (H&E) staining was performed as described in Materials and Methods. Demarcations and annotations are described in the Results. ESI-MS was performed as described in Materials and Methods. Panel D, flowchart of ESI-MS serum mass profiling. Panel E, electrospray MS methodology used to identify, quantify, and classify significant sera m/Z peaks into pancreatic cancer (PDAC) or non-cancer control descriptors; mass peak areas are averages from 6 individual serum samples per category.
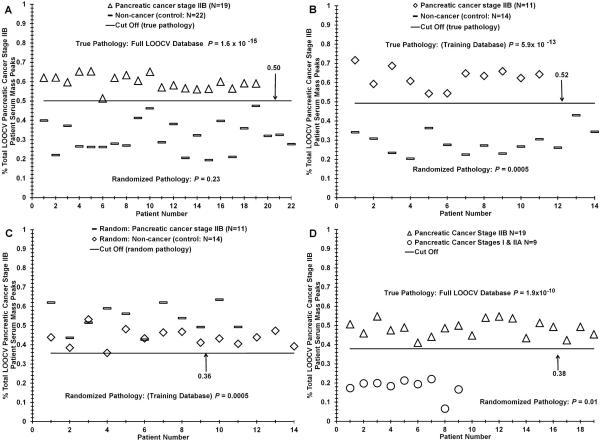
Fig. 2.
Distinguishing sera between early-stage PDAC patients and control individuals using ESI-MS mass profiling. Panel A, distribution difference between stage IIB PDAC patients versus sera of control individuals based on significant “% PDAC stage IIB patient serum peaks” using the mass peak analyses described in the Materials and Methods and Fig. 1D. Panel B, distribution analysis of the PDAC IIB versus control training database used in Table 3, panel I. Panel C, distribution analysis of the random p value obtained from panel B. Panel D, distinguishing PDAC stage I + IIA patient sera from stage IIB patient sera.
Test metrics
The diagnostic value of a test/procedure is defined by its sensitivity, specificity, predictive value, and efficiency [32,33]. Test sensitivity was determined from TP/(TP + FN) where TP was the number of true positives for disease presence, and FN was the number of false negatives for disease presence. Specificity was calculated from TN/(TN + FP) where TN is the number of true negatives and FP is the number of false positives. PDAC, CP disease, and control TP, FP, TN, and FN values were determined using cutoffs of the average “% patient serum peaks categorized” minus 2.2–4 standard deviations [SD] (Figs. 2–4). Receiver operator characteristic (ROC) curve analysis was performed as described previously [34].
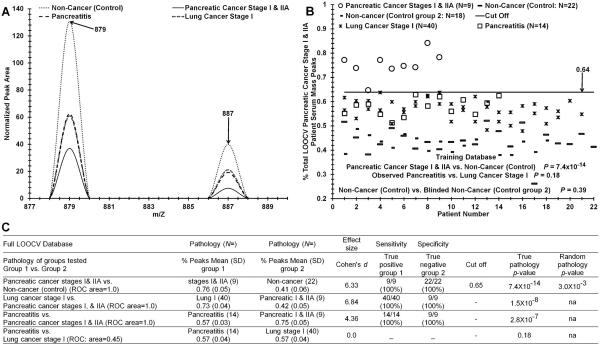
Fig. 4.
Discriminating sera from early-stage PDAC patients, CP patients, stage I lung cancer patients, and from control individuals. Panel A, mass peaks are averages from 5 individual serum samples per category. Panel B, distinguishing sera of PDAC stage I + IIA patients, CP patients, stage I lung cancer patients, and control individuals based on significant “% PDAC patient serum peaks”. Panel C, test metrics (described in Materials and Methods) for data in panel B.
Results
Serum mass peak profiling for distinguishing patients with early-stage pancreatic cancer
The histology in Fig. 1A–C illustrates the problem addressed, i.e., trying to distinguish, in a minimally-invasive manner, patients with early-stage pancreatic cancer (panel C, stage IIB), chronic pancreatitis (panel B), from control individuals (normal pancreas, panel A). A normal duct (ND) and Islet cells (IC), fibrotic scarring (F/S) and PanIN, or ductal adenocarcinoma (PDAC) and neuronal fibers (NF) are exhibited in panels A, B, and C respectively. Development of such a diagnostic tool would aid significantly in the monitoring, for example, of high-risk patients for PDAC. A hypothesis in the present study is that disease/organ insults exhibited in Fig. 1A–C can elicit systemic responses that can be detected with our serum ESI-MS methodology. Panel D is a flowchart depicting mass peak profiling of serum from control, PDAC, and CP subjects. Note this is an all-liquid process versus solid state MALDI/SELDI which has been subject to criticism (see Discussion). This procedure depicted here only requires a single sample dilution and is potentially the simplest biomolecule profiling platform currently available. LOOCV and group randomizations are employed to mitigate “over-fitting” (described below and in Discussion). Fig. 1E illustrates 15 of the significant ESI- MS mass peaks (range 730–960 m/Z, mass divided by charge) used to discriminate sera from patients with stage II PDAC (solid line) from control individuals. These significant (P < 0.05) peak area means differ as a group between 6 subject sera samples for each category (control, PDAC stage IIB). Prominent serum mass peak areas (higher value) from control individuals include m/Z 827, 907, and 937, and peaks 922 and 971 from PDAC patients. This m/Z region is only one of many analyzed (total range 400–2000 m/Z), and the large number of significant peak differences likely is contributing to the disease discrimination ability of this novel technology. This panel also exhibits our novel approach for categorization significant PDAC, PC, or control sera m/Z peaks used for construction of disease/control peak databases, as well as for “% disease peak” categorizations and “leave one out cross validation” (LOOCV) peak assignments in serum discrimination studies (see panel 1D and Figs. 2–4). LOOCV helps mitigate a phenomenon termed “over-fitting” which can result from assigning relatively large amounts of experimental data to two groups, e.g. pancreatic cancer or control [30,31]. LOOCV is performed by removing one at a time (“left out”) an MS m/Z peak data set for each serum sample in the database and then re-forming the rest of the samples in the “left in” database in the absence of that single sample MS m/Z “left out” data set. MS sera peak areas in a particular m/Z range (triplicate averaged) are identified as differing significantly between “left in” PDAC/PC patients and control individuals by comparing each of N pathology or control patient sera by t-tests. By convention for a particular m/Z peak, the highest mean is assigned that classifier, either disease or control (see control 827 m/Z peak in panel E). Then the “left in” database is tested for its ability to discriminate the new “blinded” sample MS m/Z data set that was “left out”. All the significant m/z peaks in an individual “left out” serum sample are then assigned a disease or control peak identifier by examining whether a particular “left out” peak area exceeds or falls below the 50% threshold area mean cutoff value necessary for a particular classifier label (e.g., 827 peak, Fig. 1E). That particular “left out” serum sample is then assigned a “% total subject serum peaks categorized” out of the total number of subject peaks in the complete “left in” database. This value is then plotted on the y-axis, e.g., in Figs. 2–4.
Fig. 2 illustrates our ability to distinguish patients with stage IIB pancreatic cancer from healthy control individuals. Panel A exhibits the % PDAC stage IIB patient serum MS peaks identified (y-axis) in individual patient sera spectra (diamonds) versus individual control sera spectra (circles, plotted against patient/control number (x-axis). For this analysis, a LOOCV range of 292–320 significant cancer m/Z peaks was employed (18–20% of total). The presence of “left out” PDAC patient m/Z peaks in a control serum sample is derived from those individual control peaks which happened to have an area value below the 50% threshold cutoff (Fig. 1E), which would give them the “cancer” designation. The P value for these two distribution differences is 1.6 × 10−15. Importantly, the P value for this binary comparison upon randomization of the two groups (control and stage IIB) increases substantially to 0.23 (also see Table 2). This is consistent with unique and real physiological differences in the sera of PDAC stage IIB patients versus controls using our test. The cutoff value for test metric analysis (true positives, false negatives, etc., Table 2) for this analysis is 0.50. Panel B exhibits the true pathology training distribution set used for PDAC stage IIB versus control blinded analysis (Table 3, panel 1) as well as the non-discriminatory nature of the 0.0005 randomization P value for this training set (C). Panel D ill-ustrates the mass peak analysis for distinguishing sera of PDAC stage IIB individuals versus sera of patients with stage I and IIA PDAC, plotted against % stage IIB patient serum peaks categorized (y axis). For this analysis, a LOOCV range of 136–157 significant m/Z peaks was employed (8–10% of total). The P value for the distribution difference in panel A was 1.9 × 10−10. The P value for this binary comparison upon randomization of the two groups (stage IIB and stage I/IIA) increases to 0.01 (also see Table 2). Of note, this panel D comparison is for no lymph node involvement (stage I + IIA) versus lymph node involvement (IIB) of the malignancy.
Table 2.
LOOCV test metrics.
| Pathology (N=) | Pathology (N=) | Effect size | Sensitivity | Specificity | ||||
|---|---|---|---|---|---|---|---|---|
|
I Full LOOCV series | ||||||||
| Pathology of groups tested Group 1 vs. Group 2 | % Peaks Mean (SD) Group 1 | % Peaks Mean (SD) Group 2 | Cohen's d | True positive group 1 | True negative group 2 | Cutoff | True pathology P-value | Random pathology P-value |
| Pancreatic cancer stage IIB vs. non-cancer (control) (ROC area = 1.0)a,c | Stage IIB (19) 0.60 (0.04) | Non-cancer (22) 0.32 (0.08) | 4.42 | 19/19 (100%) | 22/22 (100%) | 0.50 | 1.6 × 10−15 | 0.23 |
| Pancreatic cancer stages IIA & IIB vs. non-cancer (control) (ROC area = 1.0)c | Stages IIA & IIB (23) 0.58 (0.04) | Non-cancer (22) 0.31 (0.08) | 4.26 | 23/23 (100%) | 22/22 (100%) | 0.51 | 2.1 × 10−15 | 0.47 |
| Pancreatic cancer stages I, IIA & IIB vs. non-cancer (control) (ROC area = 0.99)c | Stages I, IIA & IIB (28) 0.54 (0.04) | Non-cancer (22) 0.30 (0.07) | 4.21 | 21/22 (95%) | 27/28 (96%) | 0.44 | 4.3 × 10−15 | 0.08 |
| Pancreatitis vs. non-cancer (control) (ROC area = 1.0)b,c | Pancreatitis (14) 0.68 (0.04) | Non-cancer (22) 0.38 (0.05) | 6.62 | 14/14 (100%) | 22/22 (100%) | 0.53 | 1.6 × 10−20 | 0.02 |
| Pancreatic cancer stages I& IIA vs. pancreatic cancer stage IIB (ROC area = 1.0)a | Stages I& IIA(9) 0.81 (0.05) | Stage IIB (19) 0.52 (0.04) | 6.41 | 9/9 (100%) | 19/19 (100%) | 0.37 | 1.9 × 10−10 | 0.01 |
| Pancreatitis vs pancreatic cancer stage IIB (ROC area = 1.0)b | Pancreatitis (14) 0.75 (0.04) | Stage IIB (19) 0.47 (0.05) | 6.18 | 14/14 (100%) | 19/19 (100%) | 0.63 | 1.5 × 10−18 | 8.5 × 10−3 |
| Pancreatitis vs pancreatic cancer stages I & IIA (ROC area = 1.0) | Pancreatitis (14) 0.60 (0.04) | Pancreatic I & IIA (9) 0.25 (0.04) | 7.73 | 14/14 (100%) | 9/9 (100%) | 0.65 | 1.3 × 10−12 | 4.8 × 10−4 |
| II Training LOOCV series for sample validation | ||||||||
|---|---|---|---|---|---|---|---|---|
| Pathology of groups tested Group 1 vs. Group 2 | % Peaks Mean (SD) Group 1 | % Peaks Mean (SD) Group 2 | Cohen's d | True positive group 1 | True negative group 2 | Cutoff | True pathology P-value | Random pathology P-value |
| Pancreatic cancer stage IIB vs. non-cancer (control) (ROC area = 1.0)a,c | Stage IIB (11) 0.64 (0.06) | Non-cancer (14) 0.30 (0.07) | 5.21 | 11/11 (100%) | 14/14 (100%) | 0.53 | 1.8 × 10−11 | 5 × 10−4 |
| Pancreatic cancer stages IIA & IIB vs. non-cancer (control) (ROC area = 1.0)c | Stages IIA & IIB (13) 0.65 (0.05) | Non-cancer (14) 0.31 (0.06) | 6.15 | 13/13 (100%) | 14/14 (100%) | 0.52 | 9.8 × 10−15 | 6 × 10−4 |
| Pancreatic cancer stages I, IIA & IIB vs non-cancer (control) (ROC area = 1.0)c | Stages I, IIA & IIB (16) 0.61 (0.05) | Non-cancer (14) 0.27 (0.07) | 5.58 | 16/16 (100%) | 14/14 (100%) | 0.45 | 5.2 × 10−15 | 2 × 10−5 |
| Pancreatitis vs non-cancer (control) (ROC area = 1.0)b,c | Pancreatitis (9) 0.79 (0.07) | Non-cancer (14) 0.33 (0.08) | 5.99 | 9/9 (100%) | 14/14 (100%) | 0.58 | 6.7 × 10−12 | 0.22 |
| Pancreatitis vs. pancreatic cancer stage IIB (ROC area = 1.0)b | Pancreatitis (9) 0.83 (0.06) | Stage IIB (11) 0.43 (0.06) | 6.66 | 9/9 (100%) | 11/11 (100%) | 0.65 | 1.8 × 10−11 | 0.03 |
Table 3.
Sample validation results and serum protein identifications.
| I | |||
|---|---|---|---|
| A: Samples validateda (N) | B: Validationa database (N) | C: Effect sizeb Cohen's d | D: % Correct True pathology |
| Pancreatic cancer stage IIB (N = 8) & non-cancer (control) (N = 8) | Pancreatic cancer stage IIB (N = 11) vs. non-cancer (control: N = 14) | 4.4 | 100% (16/16) |
| Pancreatic cancer stages IIA & IIB (N = 10) & non-cancer (control) (N = 8) | Pancreatic cancer stages IIA & IIB (N = 13) vs. non-cancer (control: N = 14) | 4.6 | 100% (18/18) |
| Pancreatic cancer stages I, IIA & IIB (N = 12) & non-cancer (control) (N = 8) | Pancreatic cancer stages I, IIA & IIB (N = 16) vs. non-cancer (control: N = 14) | 4.5 | 100% (20/20) |
| Pancreatitis(N = 5) & non-cancer (control) (N = 8) | Pancreatitis (N = 9) vs. non-cancer (control: N = 14) | 4.4 | 100% (13/13) |
| Pancreatitis (N = 5) & pancreatic cancer stage IIB (N = 8) | Pancreatitis (N = 9) vs. pancreatic cancer stage IIB (N = 11) | 4.0 | 100% (13/13) |
| II | |||||
|---|---|---|---|---|---|
| Protein identified | # Identified sequences Control [Cancerc] | Protein identified | # Identified sequences Control [Cancerc] | Protein identified | # Identified sequences Control [Cancerc] |
| Ig heavy chaind | 323 [268] | RYR2e | 39 [0] | SMARCAL1e | 7 [6] |
| Cytochrome c oxidased | 102 [103] | INSRd | 19[17] | TICRR | 8 [5] |
| NEBd | 40 [m] | PCLO | 26 [10] | ANKRD12e | 11 [1] |
| Ig light chaind | 105 [48] | TENM3 | 9 [19] | SUPV3L1e | 9 [3] |
| OBSCNd | 78 [35] | CSMD3e | 17 [10] | TMC2 | 12 [4] |
| TTNd | 38 [70] | CNTNAP4e | 17 [9] | WDR90 | 12 [0] |
| MUC16d | 33 [51] | MACF1e | 25 [1] | SSPO | 6 [5] |
| KMT2Ae | 12 [69] | WDFY3e | 15 [8] | BCL9Ld | 2 [9] |
| SYNE1e | 26 [55] | NSD1e | 14 [9] | NADHe | 6 [4] |
| BAI2e | 52 [22] | TRPM1e | 21 [2] | PITPNM3e | 9 [1] |
| CACNA1Ae | 22 [38] | VWA5B2e | 18 [3] | Cytochromed | 4 [5] |
| EIF4G1e | 18 [41] | RLTPR | 0 [21] | SLC35A2 | 9 [0] |
| MYCBP2 | 0 [51] | COL19A1 | 19 [0] | DNAH3 | 3 [4] |
| PLECd | 49 [2] | SHANK1e | 12 [7] | PLXNB1e | 2 [5] |
| SYNE2e | 0 [50] | TCRd | 6 [12] | SSTR5d | 3 [4] |
| ABCA1d | 30 [15] | BSN | 17 [0] | MUC19 | 3 [3] |
| DMDe | 23 [22] | DISP1d | 3 [13] | AMHe | 6 [0] |
| CEP164e | 21 [20] | ACACBe | 7 [8] | HLA-Bd | 3 [1] |
| NPNTe | 20 [21] | MUC2d | 14 [0] | MHCd | 4 [0] |
Blind samples are not members of the validation database against which they were tested;
observed between blinded sample groups.
Pancreatic cancer stage IIB
pancreatic cancer related
other cancer related, unique sequence segments range (3-273), 60 peaks (700-940)m/Z.
Distinguishing sera of CP patients from early-stage PDAC patients and healthy controls
Chronic pancreatitis is a risk factor for the development of pancreatic cancer [8,9]. Minimally invasive and inexpensive tools are needed to assist in pathological monitoring of this disorder and the potential transition of this disease into PDAC, especially among high-risk groups. Data in Fig. 3 indicate we are making progress in the development of such a tool involving ESI-MS analysis of patient serum. Panel A exhibits our ability to distinguish patients with chronic pancreatitis (N = 14) from healthy control individuals (N = 22).
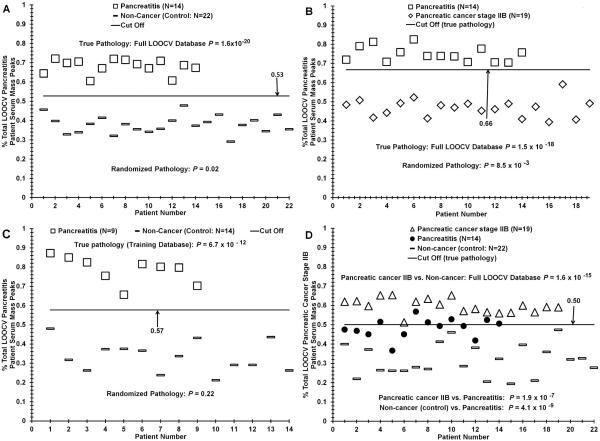
Fig. 3.
ESI-MS analysis discriminating sera from patients with pancreatitis from control individuals and from patients with early-stage PDAC. Panel A, serum sample distribution difference based on “% pancreatitis patient serum peaks” between CP patients and controls using the mass peak analyses described in Fig. 2 legend. Panel B, distinguishing CP patient serum from PDAC stage IIB patient serum. Panel C, distribution analysis of sera from CP patients versus control individuals for the training database used in validation experiments exhibited in Table 3, panel I. Panel D, distinguishing sera from patients with PDAC or CP, and control individuals.
The “% pancreatitis patient serum peaks” (y-axis) in individual patient sera spectra (triangles) are plotted versus individual control sera spectra (circles), plotted against patient/control number (x-axis). For this analysis, a LOOCV range of 224–252 significant cancer m/Z peaks was employed (14–16% of total). The P value for these two distribution differences is 1.6 × 10−20. The P value for this binary comparison upon randomization of the two groups (control and CP) increases substantially to 0.02 (Table 2). Panel B exhibits the mass peak sera data outcome for distinguishing patients with chronic pancreatitis from patients with stage IIB PDAC (N = 19). The “% pancreatitis patient serum peaks” (y-axis) in individual patient sera spectra (squares) are plotted versus patient PDAC stage IIB sera spectra (diamonds), plotted against patient number (x-axis). For this analysis, a LOOCV range of 176–198 significant cancer m/Z peaks was employed (11–12% of total). The P value for these two distribution differences is 1.5 × 10−18. The P value for this binary comparison upon randomization of the two groups (CP and stage IIB) increases substantially to 0.0085. This still low random p value did not yield patient/control discriminations (not shown as well as the 0.02 value in panel A), as exhibited by the 0.0005 random p value in Fig. 2C. Panel C, Fig. 3, exhibits the training database distribution used for the blinded pancreatitis analysis (Table 3, panel I). Panel D illustrates discrimination of patient serum from PDAC stage IIB, pancreatitis, and controls, with the pancreatitis distribution falling between the PDAC and controls.
Distribution differences between sera from stage I lung cancer patients and patients with stage I + IIA PDAC versus sera from control individuals
It is of interest to determine how sera from different cancer patients as well as other disease states separate using our serum mass profiling technology. How disease-specific is this methodology? Fig. 4 exhibits serum discrimination of cohorts of PDAC stage I + IIA patients versus control individuals; also included within these comparisons are sera analyses from pancreatitis patients and stage I lung cancer patients. Panel A exhibits two normalized and significant (P < 0.05) peak m/Z areas from 5 different sera samples from the 4 categories listed (control, PDAC stage I + IIA, CP, and lung cancer stage I). Note how the “normalized peak area” metric clusters the CP and stage I lung cancer sera mass peak areas together. Panel B illustrates the mass peak sera data for distinguishing patients with stage I + IIA PDAC (circles) from control individuals (dashes). For this analysis, a LOOCV range of 183–233 significant cancer m/Z peaks was employed (11–15% of total). The P value for these two distribution differences is 7.4 × 10−14. The P value for this binary comparison upon randomization of the two groups (control and stage I/IIA) increases to 0.003 (panel C). Also included in the panel B sera distribution data are samples from stage I lung cancer patients (Xs, N = 40) and pancreatitis patients (squares) plotted against the total peak database on the y-axis. Note how the “% PDAC patient serum peaks” metric clusters the CP and stage I lung cancer sera samples together. The P value for this CP versus stage I lung cancer sera peak distribution difference, exhibited in panel B, is 0.18 (panel C). It is noted that two independent sets of controls (small, large dashes, panel B) cluster together (p = 0.39).
Test metrics, blind validation, and serum peptide identifications
Table 2 exhibits the sera test metrics for the LOOCV patient ESI-MS peak distribution data exhibited in Figs. 2 and 3, using nomenclature from predictive value theory [32,33]. The pathological groups tested in binary fashion for these Figs. are listed in the far-left column. The “% MS peaks” means and their standard deviation (SD) are all well separated and have narrow SD boundaries for all the groups tested. The cut-off values to obtain the test metric data (false positives and false negatives) are exhibited next to the means and SDs. ROC area values for all analyzed binary group distribution differences are 1.0 where a 0.5 value would be random discrimination. The test sensitivity (measure of true positive rate) and test specificity (true negative rate) with the cohort numbers exhibited here are both 100%. Listed also in Table 2 are the P values for the true binary group distribution differences or the randomized mixed binary group distribution differences. Inherent physiological values in the original distribution differences are indicated by the very large increases in P values when the groups are randomized. In addition, a Cohen's d effect size is provided (http://www.danielsoper.com/statcalc3/calc.aspx?id=48) as another quantitative measure of the strength and reliability of the distribution differences; these large values indicate high reliability [35]. Table 3, panel I, gives the results of our blind sample analysis, with the list of sera samples tested and their group N values in the left column (A). The center column (B) exhibits the binary validation grouping used to test these corresponding blind samples (A). The column on the far right (D) exhibits the blind testing results/ percentages for testing column A against the column B binary test validation database. For this sample cohort pool, 5 out of 5 blind groups were 100% identified. Effect sizes for these distribution differences (described in Table 2) are reported as Cohens’ d values (C). All are large which is consistent with the 100% observed validation.
Panel II in Table 3 exhibits tandem MS/MS identified peptides (listed by their corresponding protein abbreviations) and their numbers of different identified sequences (parentheses and brackets) from discriminating sera sample m/Z mass peaks from 10 control individuals and 10 patients with stage IIb PDAC. A m/Z peak range of 700–940 m/Z was employed and only discriminating mass peaks (60 total, expanded to – or + 1 m/Z) from the control versus patient stage IIb sera discrimination (Fig. 2A) were screened in the MS/MS process. Unique peptide sequences observed per protein identified ranged from 3 to 273 (lower right to upper left, panel II), for a total of 57 peptide/proteins listed. This number represents 2% of the 2862 total peptides/proteins that were observed in the 1–273 unique sequence range (not shown). Medline searches (Ovid Technologies, Inc.) revealed 29% of the listed peptide/proteins have a previously identified PDAC relationship, e.g., mucin 16 (MUC16) [36] and dispatched (DISP1) which is a regulator in the hedgehog signaling pathway and is up-regulated in PDAC [37]. 51% of the peptides/proteins in Table 3, panel II have a cancer relationship (79% total cancer related). These results suggest we are discriminating controls from stage IIb patients using an overall cancer phenotype with known elements of PDAC.
Discussion
The early detection and prevention of pancreatic cancer are of utmost importance and major clinical and research interests [1,2,16,38]. The earlier this disease can be diagnosed, the earlier life-saving treatments can begin, thus increasing the survival rate of this cancer much above the present 4–5% [1,2]. Serum mass profiling is a promising technology for identifying potential biomarkers and their patterns relevant to the diagnosis, monitoring, understanding, and treating a variety of disease states and their progressions at the individual patient level [12,14–16,24–26]. An individual's serum mass profile is hypothesized to reflect, in part, homeostatic as well as defense/stress mechanisms which change with physiological state, e.g., with disease. Direct inputs from disease tissues are also possible. An underlying hypothesis of serum mass profiling is that the human organism is an exceedingly complex entity capable of complex responses to seemingly small bodily insults. We previously used this all-liquid technology to distinguish patients who had lung cancer, ovarian cancer, and pancreatic cancer from healthy control individuals [15,16,24–26,39]. We have also used this methodology to distinguish the earliest stages of PDAC development in a rodent model system [17]. In the present study, we demonstrate that all-liquid ESI-MS serum mass profiling can distinguish patients with early stage pancreatic cancer (stages I, IIA, IIB) from patients with pancreatitis, and from healthy control individuals.
Serum profiling of biomolecules associated with disease states like pancreatic cancer, by ESI-MS, has several advantages over other early detection platforms. For example, ESI-MS allows single step, direct injection analysis of a disease phenotype in blood without potential artifacts from in vitro manipulations involving extensive sample handling, and nucleic acid and enzyme amplifications. The most commonly used MS methodology for serum profiling is solid-state SELDI/MALDI (surface enhanced laser desorption ionization/matrix assisted laser desorption ionization)-MS [20,40,41]. The ESIMS approach used in the present study has potential advantages over SELDI/MALDI-MS, because of all-liquid handling, much reduced sample manipulation, absence of grid washings, no chemical additives, and no involvement of random crystallization processes, all which can lead to observational artifacts that might occur with SELDI [42]. To our knowledge our ESI-MS methodology presents a new paradigm to distinguish sera from individuals with and without early-stage pancreatic cancer, as well as pancreatitis. This paradigm involves comparing mass peaks at individual m/Z values which differed significantly between two distinct sera groups, e.g., from control or pancreatic disease (see Fig. 1D, E). During this process one serum sample, from either a control or a patient, was “left out” of the analysis (LOOCV). Subsequently, peaks which have greater areas among the disease group were by convention designated “disease” peaks. Peaks having greater areas among control samples were designated “control” peaks. All mass peaks from the “left out” serum samples were then tested against this “disease” and “control” mass peak LOOCV database in blinded fashion. We used an arbitrary 50% area cut-off value for each significant mass peak in the “disease” and “control” database that we used to assign a particular classification. We identified each “left out” peak as either a control peak (non-pancreatic disease) if that peak area value was equal to or below the 50% value, or a pancreatic disease peak if that particular m/Z peak area value was above the 50% value. In this way, we built up a “% PDAC patient serum MS peaks categorized” fraction for each sera sample (control or disease) out of the total number of group peaks in that pancreatic disease peak database. Therefore, a certain % of control individual serum peaks was assigned a pancreatic disease peak classification. We then plotted this % PDAC value versus sample number as exhibited in Figs. 2–4, and arrived at distributions for control and pancreatic disease which were distinguishable by t tests and ROC curves (Table 2).
With respect to our ability to discriminate sera samples from early-stage PDAC patients, CP patients, and control individuals, besides the leave one out cross validation (LOOCV) process we perform to validate these results, we also utilize t tests to describe the distribution difference probabilities for the null hypothesis that no discrimination is observed between the true binary group comparisons or the binary group comparisons where the two differing cohorts have been randomized together. For example, for the control (N of 22) versus PDAC stage IIB (N of 19) sera peak comparison (Fig. 2A), the t test P value for true binary comparison is 1.6 × 10−15 and the corresponding random group comparison P value is 0.23 (Table 2). These P values support strong discrimination in the true binary comparison and the null hypothesis (no discrimination) is true in the random comparison. This is indicative of unique and real physiological differences appearing in the sera of PDAC stage IIB patients versus controls using our test. Panel D in Fig. 2 is a comparison of the % stage IIB patient sera mass peaks categorized in PDAC stage IIB patients versus sera in stage I and IIA PDAC patients (N of 9). The P value for the true binary comparison is 1.9 × 10−10 and the P value for the randomized comparison is 0.01 (Table 2). Although the randomization is sending the null hypothesis metric in the correct direction (becoming much larger than the very low P value for the true binary comparison), this “random” P value (and others listed in Table 2) would still be considered significant for distinguishing two groups. However, the notion that such a “significant” p value could give actual true-pathology sample discriminations is discounted upon plotting “discriminations” (or lack thereof) of a much more “significant” random p value (0.0005, for the stage IIb/control training database comparison, Fig. 2C). The important PDAC stage I + IIA patient serum discrimination from control individual serum is a very low P value of 7.4 × 10−14 for the true binary comparison and another “significant” value of 0.003 for the random comparison (Table 2). We are looking at small physiological serum changes here (we refer to such changes as “quantum” or minimal changes) that are likely very difficult for any technology to distinguish in a minimally invasive bodily fluid like serum. These significant “random” p values, although non-discriminatory, suggest that a phenomenon termed “over-fitting” (random chance “significant” pairings due to large number of mass peaks analyzed) may be contributing some “significance” to this random distinction.
Chronic pancreatitis is considered a risk factor for developing PDAC, especially among individuals with a genetic predisposition toward CP where occurrence of PDAC can be as high as 40% [43]. Thus, minimally invasive and inexpensive tools would be of use in assisting the pathological monitoring of this disorder and its potential transition into PDAC. In order to develop such a test the analysis needs to discriminate patients with CP from patients with early-stage PDAC (I, IIA, IIB) and from control individuals. We report in this study that our ESI-MS serum mass profiling technology, with the cohorts utilized (Fig. 3A, Table 2), could distinguish a CP patient group (N = 14) from a control group (N = 22) with a P value of 1.6 × 10−20 for the true binary comparison and a value of 0.02 for the randomized binary comparison. Although this later value is still in the significant range, it is significantly increased from the specific true binary (CP versus control) distribution comparison. With respect to distinguishing sera of CP patients versus early-stage PDAC patients, we exhibit P values for the true binary comparisons of CP versus stage IIB or CP versus stage I + IIA of 1.5 × 10−18 (Fig. 3B) and 1.7 × 10−13 respectively; the random comparison for these two groupings is 0.0085 and 0.00013 respectively (Table 2). Although these later p values are still in the significant range, they are greatly increased over the specific binary pairings. And like Fig. 2C, no group discrimination is observed (not shown). To our knowledge no other laboratory group has published these types of fine (quantum) pancreatic disease distinctions using a readily available bodily fluid like serum. With respect to these cohort discriminations observed in Figs. 2 and 3 using serum ESI-MS analysis, we employed four forms of sample validation: LOOCV, ROC (receiver-operator characteristic) curve analysis, cohort randomization, and true blinded validation of a subset of group samples against the corresponding binary database lacking the blinded samples. The LOOCV and randomization results have been discussed. The ROC curve discriminations (between values of 0.5 [random] and 1.0 [non-random] were all 1.0 (Table 2).
Pancreatic cancer like lung cancer has a tobacco smoking risk factor, as much as 30% [8]. And smoking doubles the risk that individuals with hereditary chronic pancreatitis will develop PDAC [44]. We have previously utilized our ESI-MS serum mass profiling methodology to distinguish patients with early-stage lung cancer from control individuals, with tobacco smoking not confounding cancer discrimination in our ESI-MS analysis [26,39]. It was of interest to see in the present study if we could distinguish the sera of early-stage pancreatic cancer patients from early-stage lung cancer patients. From Fig. 4B using % PDAC patient serum peaks categorized as the metric, sera from stage I lung cancer patients were distinguished from sera from stage I + IIA PDAC patients with a distribution difference P value of 1.5 × 10−8 (Table 2). The corresponding random binary comparison P value was 0.4. Of note in Fig. 4 panel B is the grouping together of the pancreatitis cohort and the stage I lung cancer cohort, exhibiting a distribution difference P value between the two of 0.18. Could these observations possibly suggest a similar physiology/host response between pancreatitis and stage I lung cancer ?
Results of validation with true blinded cohort sample subsets are exhibited in Table 3, panel I. Although the blinded cohorts are somewhat small compared to a full prospective study, the % correct values for the blind PDAC stage IIB (N = 8), IIA + IIB (N = 10), stage I, IIA, IIB (N = 12) pancreatitis (N = 5), and non-cancer controls (N = 8) are all 100%. The test sensitivity (measure of false negative rate [type II error]) and specificity (false positive rate [type I error]) values for this study were all in the 95–100% range (Table 2). All these values portend well for this ESI-MS technology and for its use in prospective analysis of PDAC and pancreatitis patient groups. Such significant cohort groupings observed in Tables 2 and 3 and Figs. 2–4 pose the question, what physiological changes are possibly underlying these mass peak discriminations? These changes in serum mass profiles are in accord with the basic principles of serum profiling, namely, that disease and disease progression can be distinguished in steady state because these physiological differences cause measurable biomolecule changes in the peripheral blood due to host systemic responses, homeostasis and defense, as well as stress mechanisms. In addition, direct inputs from diseased tissues like tumors are also possible. At the m/Z data values being mined here, e.g., from 730 to 890 (Figs. 1D and 4A), these ranges likely encompass the lower mass peptide “serome” which result from differential host tissue/organ exoprotease activities and other cell/tissue signaling activities [18,19]. Since we are seeing fairly evident biomolecule changes as reflected in our serum profiles from apparent small physiological inputs, for example, distinguishing stage I + IIA PDAC (no lymph node involvement) and stage IIB PDAC (node involvement), one would need to hypothesize a mechanism to account for such apparent amplification of small signal(s) from minute starting inputs. Possibilities could involve “alarmin”-like molecules believed to be shed/secreted by cells which have been damaged/altered in some fashion which in turn bind to signal transduction pathway receptors to activate in a synergistic and cooperative manner more extensive innate defense/stress responses [19]. With respect to the biochemical changes associated with the ESI-MS serum discriminations involved in distinguishing control individuals from patients with stage IIb PDAC, an overall cancer phenotype was observed with biochemical elements of a PDAC response (Table 3, panel II). Specifically, peptides/proteins suggested to have roles in the physiological changes observed with these discriminations include an interesting trilogy of proteins previously shown to have roles in muscle structure and function (nebulin [NEB], obscurin [OBSCN], and titin [TTN}, Table 3, panel II), but with renewed interest in cancer biology [45]. Does this suggest non-muscle roles for these proteins for example in signal transduction and cell proliferation control? Is muscle degradation/regeneration associated with pancreatic cancer progression as a stress response? Amounts of NEB and TTN (i.e., their peptides) are relatively higher in sera from stage IIb PDAC patients versus controls and one (OBSCN) is relatively lower (Table 3, panel II). It is of interest that OBSCN peptides are lower in PDAC sera and that proteins may have a tumor suppressor function [45]. Panel II exhibits elevated levels of mucin 16/peptides in PDAC stage IIb patient sera, in line with previous PDAC observations [46,47]. Mucin 16 is a known serum biomarker for ovarian cancer (CA125), and was proposed previously to have a role in PDAC progression and metastasis through possible interaction with mesothelin [36]. This glycosylated protein has roles in tissue interactions and possible tumor protection from the immune system and chemotherapeutic drugs [47]. In summary, blood tests are needed to aid in the early detection of pancreatic ductal adenocarcinoma (PDAC). This study exhibits efforts and progress toward developing such blood tests, using a novel platform involving electrospray ionization-mass spectrometry (ESI-MS) serum mass profiling. This technology is able to distinguish patients with early-stage PDAC from control individuals and from individuals with chronic pancreatitis. Tandem MS of serum from early-stage PDAC patients, when compared with serum from control individuals, identified peptides/proteins characteristic of a cancer phenotype involved in these ESI-MS PDAC discriminations.
Acknowledgements
Authors acknowledge financial support from the Oklahoma Center for the Advancement of Science and Technology (OCAST Grant Number AR11-001), National Center for Research Resources and the National Institute of General Medical Sciences of the National Institutes of Health (Grant Number 8P20GM103447), Initiative for Minority Students: Bridges to the Baccalaureate Degree Award No: 5-R25-GM054938-10, Oklahoma Tobacco Research Center, and the University of Oklahoma Health Sciences Center Department of Surgery Research Fund.
Funding
This study was funded by the Oklahoma Center for the Advancement of Science and Technology, and the Department of Surgery, University of Oklahoma Health Sciences Center.
Footnotes
Conflict of interest
The authors declare no conflicts of interest.
References
- 1.American Cancer Society . Cancer Facts & Figures 2013. American Cancer Society; Atlanta: 2013. [Google Scholar]
- 2.Jones S, Zhang X, Parsons DW, Lin JD, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gattani AM, Mandeli J, Bruckner HW. Tumor markers in patients with pancreatic carcinoma. Cancer. 1996;78:57–62. doi: 10.1002/(SICI)1097-0142(19960701)78:1<57::AID-CNCR10>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 4.Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev. Res. (Phila.) 2009;2:807–813. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker M. MicroRNA profiling: separating signal from noise. Nat. Methods. 2010;7:687–692. doi: 10.1038/nmeth0910-687. [DOI] [PubMed] [Google Scholar]
- 6.Jayaprakash AD, Jabado O, Brown BD, Sachidanandam R. Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Res. 2011;39:e141. doi: 10.1093/nar/gkr693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wingren C, Sandström A, Segersvärd R, Carlsson A, Andersson R, Löhr M, et al. Identification of serum biomarker signatures associated with pancreatic cancer. Cancer Res. 2012;72:2481–2490. doi: 10.1158/0008-5472.CAN-11-2883. [DOI] [PubMed] [Google Scholar]
- 8.Malka D, Hammel P, Maire F, Rufat P, Madeira I, Pessione F, et al. Risk of pancreatic adenocarcinoma in chronic pancreatitis. Gut. 2002;51:849–852. doi: 10.1136/gut.51.6.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gukovsky I, Li N, Todoric J, Gukovskaya A, Karin M. Inflammation, autophagy, and obesity: common features in the pathogenesis of pancreatitis and pancreatic cancer. Gastroenterology. 2013;144:1199–1209. e4. doi: 10.1053/j.gastro.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hermanová M, Nenutil R, Kren L, Feit J, Pavlovský Z, Díte P. Proliferative activity in pancreatic intraepithelial neoplasias of chronic pancreatitis resection specimens: detection of a high-risk lesion. Neoplasma. 2004;51:400–404. [PubMed] [Google Scholar]
- 11.Löhr M, Klöppel G, Maisonneuve P, Lowenfels AB, Lüttges J. Frequency of K-ras mutations in pancreatic intraductal neoplasias associated with pancreatic ductal adenocarcinoma and chronic pancreatitis: a meta-analysis. Neoplasia. 2005;7:17–23. doi: 10.1593/neo.04445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richter R, Schulz-Knappe P, Schrader M, Ständker L, Jürgens M, Tammen H, et al. Composition of the peptide fraction in human blood plasma: database of circulating human peptides. J. Chromatogr. B. Biomed Sci. Appl. 1999;726:25–35. doi: 10.1016/s0378-4347(99)00012-2. [DOI] [PubMed] [Google Scholar]
- 13.Chambers G, Lawrie L, Cash P, Murray GI. Proteomics: a new approach to the study of disease. J. Pathol. 2000;192:280–288. doi: 10.1002/1096-9896(200011)192:3<280::AID-PATH748>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 14.Verma M, Wright GL, Jr, Hanash SM, Gopal-Srivastava R, Srivastava S. Proteomic approaches within the NCI early detection research network for the discovery and identification of cancer biomarkers. Ann. N. Y. Acad. Sci. 2001;945:103–115. doi: 10.1111/j.1749-6632.2001.tb03870.x. [DOI] [PubMed] [Google Scholar]
- 15.Hanas JS, Hocker JR, Cheung JY, Larabee JL, Lerner MR, Lightfoot SA, et al. Biomarker identification in human pancreatic cancer sera. Pancreas. 2008;36:61–69. doi: 10.1097/mpa.0b013e3180d0a738. [DOI] [PubMed] [Google Scholar]
- 16.Hocker JR, Lerner MR, Mitchell SL, Lightfoot SA, Lander TJ, Quillet AA, et al. Distinguishing early-stage pancreatic cancer patients from disease-free individuals using serum profiling. Cancer Invest. 2011;29:173–179. doi: 10.3109/07357907.2010.543214. [DOI] [PubMed] [Google Scholar]
- 17.Hocker JR, Mohammed A, Aston CE, Brewer M, Lightfoot SA, Rao CV, et al. Mass profiling of serum to distinguish mice with pancreatic cancer induced by a transgenic Kras mutation. Int. J. Cancer. 2013;133:2662–2671. doi: 10.1002/ijc.28285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villanueva J, Shaffer DR, Philip J, Chaparro CA, Erdjument-Bromage H, Olshen AB, et al. Differential exoprotease activities confer tumor-specific serum peptidome patterns. J. Clin. Invest. 2006;116:271–284. doi: 10.1172/JCI26022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bianchi ME. DAMPs, PAMPs, and alarmins: all we need to know about danger. J. Leukoc. Biol. 2007;81:1–5. doi: 10.1189/jlb.0306164. [DOI] [PubMed] [Google Scholar]
- 20.Semmes OJ, Feng Z, Adam BL, Banez LL, Bigbee WL, Campos D, et al. Evaluation of serum protein profiling by surface-enhanced laser esorption/ionization time-of- flight mass spectrometry for the detection of prostate cancer: I. Assessment of platform reproducibility. Clin. Chem. 2005;51:102–112. doi: 10.1373/clinchem.2004.038950. [DOI] [PubMed] [Google Scholar]
- 21.Hortin GL. Can mass spectrometric protein profiling meet desired standards of clinical laboratory practice? Clin. Chem. 2005;51:3–5. doi: 10.1373/clinchem.2004.043281. [DOI] [PubMed] [Google Scholar]
- 22.West-Nørager M, Bro R, Marini F, Høgdall EV, Høgdall CK, Nedergaard L, et al. Feasibility of serodiagnosis of ovarian cancer by mass spectrometry. Anal. Chem. 2009;81:1907–1913. doi: 10.1021/ac802293g. [DOI] [PubMed] [Google Scholar]
- 23.Sorace JM, Zhan M. A data review and re-assessment of ovarian cancer serum proteomic profiling. BMC Bioinformatics. 2003;4:24. doi: 10.1186/1471-2105-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hocker JR, Bishop EA, Lightfoot SA, Lerner MR, Peyton MD, Bracket DJ, et al. Serum profiling to distinguish early-stage and late-stage ovarian cancer patients from disease-free individuals. Cancer Invest. 2012;30:189–197. doi: 10.3109/07357907.2011.636115. [DOI] [PubMed] [Google Scholar]
- 25.Hocker JR, Peyton MD, Lerner MR, Lightfoot SA, Hanas RJ, Brackett DJ, et al. Distinguishing non-small cell lung adenocarcinoma patients from squamous cell carcinoma patients and from control individuals using serum profiling. Cancer Invest. 2012;30:180–188. doi: 10.3109/07357907.2011.633294. [DOI] [PubMed] [Google Scholar]
- 26.Hocker JR, Peyton MD, Lerner MR, Mitchell SL, Lightfoot SA, Lander TJ, et al. Serum discrimination of early-stage lung cancer patients using electrospray-ionization mass spectrometry. Lung Cancer. 2011;74:206–211. doi: 10.1016/j.lungcan.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 27.Larabee JL, Hocker JR, Cheung JY, Gallucci RM, Hanas JS. Serum profiling of rat dermal exposure to JP-8 fuel reveals an acute-phase response. Toxicol. Mech. Methods. 2008;18:41–51. doi: 10.1080/15376510701697072. [DOI] [PubMed] [Google Scholar]
- 28.Greene FL, Sobin LH. A worldwide approach to the TNM staging system: collaborative efforts of the AJCC and UICC. J. Surg. Oncol. 2009;99:269–272. doi: 10.1002/jso.21237. [DOI] [PubMed] [Google Scholar]
- 29.Tuck MK, Chan DW, Chia D, Godwin AK, Grizzle WE, Krueger KE, et al. Standard operating procedures for serum and plasma collection: Early Detection Research Network Consensus Statement Standard Operating Procedure Integration Working Group. J. Proteome Res. 2009;8:113–117. doi: 10.1021/pr800545q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ransohoff DR. Rules of evidence for cancer molecular-marker discovery and validation. Nat. Rev. Cancer. 2004;4:309–314. doi: 10.1038/nrc1322. [DOI] [PubMed] [Google Scholar]
- 31.Guan W, Zhou M, Hampton CY, Benigno BB, Walker LD, Gray A, et al. Ovarian cancer detection from metabolomic liquid chromatography/mass spectrometry data by support vector machines. BMC Bioinformatics. 2009;10:259–274. doi: 10.1186/1471-2105-10-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altman DG, Bland JM. Statistics notes: diagnostic tests 2: predictive values. BMJ. 1994;309:102. doi: 10.1136/bmj.309.6947.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altman DG, Bland JM. Diagnostic tests. 1: sensitivity and specificity. BMJ. 1994;308:1552. doi: 10.1136/bmj.308.6943.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Postier RG, Lerner MR, Lightfoot SA, Vannarath R, Lane MM, Hanas JS, et al. DNA ploidy and Markovian analysis of Neoplastic progression in experimental pancreatic cancer. J. Histochem. Cytochem. 2003;51:303–309. doi: 10.1177/002215540305100305. [DOI] [PubMed] [Google Scholar]
- 35.Cohen J. A power primer. Psychol. Bull. 1992;112:155–159. doi: 10.1037//0033-2909.112.1.155. [DOI] [PubMed] [Google Scholar]
- 36.Haridas D, Chakraborty S, Ponnusamy MP, Lakshmanan I, Rachagani S, Cruz E, et al. Pathobiological implications of MUC16 expression in pancreatic cancer. PLoS ONE. 2011;6:e26839. doi: 10.1371/journal.pone.0026839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martínez-Bosch N, Fernandez-Barrena MG, Moreno M, Ortiz-Zapater E, Munne-Collado J, Iglesias M, et al. Galectin-1 drives pancreatic carcinogenesis through stroma remodeling and hedgehog signaling activation. Cancer Res. 2014;74:3512–3524. doi: 10.1158/0008-5472.CAN-13-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohammed A, Janakiram NB, Li Q, Madka V, Ely M, Lightfoot S, et al. The epidermal growth factor receptor inhibitor gefitinib prevents the progression of pancreatic lesions to carcinoma in a conditional LSL-KrasG12D/+ transgenic mouse model. Cancer Prev. Res. (Phila.) 2011;3:1417–1426. doi: 10.1158/1940-6207.CAPR-10-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanas JS, Peyton MD, Lerner MR, Lightfoot SA, Deb SJ, Hanas RJ, et al. Distinguishing patients with stage I lung cancer versus control individuals using serum mass profiling. Cancer Invest. 2014;32:136–143. doi: 10.3109/07357907.2014.883528. [DOI] [PubMed] [Google Scholar]
- 40.Yu Y, Chen S, Wang LS, Chen WL, Guo WJ, Yan H, et al. Prediction of pancreatic cancer by serum biomarkers using surface-enhanced laser desorption/ionization-based decision tree classification. Oncology. 2005;68:79–86. doi: 10.1159/000084824. [DOI] [PubMed] [Google Scholar]
- 41.Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin. Chem. 2002;481:296–304. [PubMed] [Google Scholar]
- 42.McLerran D, Grizzle WE, Feng Z, Thompson IM, Bigbee WL, Cazares LH, et al. SELDI-TOF MS whole serum proteomic profiling with IMAC surface does not reliably detect prostate cancer. Clin. Chem. 2008;54:53–59. doi: 10.1373/clinchem.2007.091496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowenfels AB, Maisonneuve P, DiMagno EP, Elitsur Y, Gates LK, Jr, Perrault J, et al. Hereditary pancreatitis and the risk of pancreatic cancer. International Hereditary Pancreatitis Study Group. J. Natl. Cancer Inst. 1997;89:442–446. doi: 10.1093/jnci/89.6.442. [DOI] [PubMed] [Google Scholar]
- 44.Lowenfels AB, Maisonneuve P, Whitcomb DC, Lerch MM, DiMagno EP. Cigarette smoking as a risk factor for pancreatic cancer in patients with hereditary pancreatitis. JAMA. 2001;286:169–170. doi: 10.1001/jama.286.2.169. [DOI] [PubMed] [Google Scholar]
- 45.Balakrishnan A, Bleeker FE, Lamba S, Rodolfo M, Daniotti M, Scarpa A, et al. Cancer Res. 2007;67:3545–3550. doi: 10.1158/0008-5472.CAN-07-0065. [DOI] [PubMed] [Google Scholar]
- 46.Szwedziak K, Szymański D, Strzelczyk J. CA 125 concentration in portal blood as a predictor of resectability in pancreatic tumor. Contemp. Oncol. 2013;17:394–399. doi: 10.5114/wo.2013.35057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Streppel MM, Vincent A, Mukherjee R, Campbell NR, Chen S-H, Konstantopoulos K, et al. Mucin 16 (cancer antigen 125) expression in human tissues and cell lines and correlation with clinical outcome in adenocarcinomas of the pancreas, esophagus, stomach, and colon. Hum. Pathol. 2012;43:1755–1763. doi: 10.1016/j.humpath.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]