SUMMARY
Regulatory T cells (Treg cells) are required for immune homeostasis. Chromatin remodeling is essential for establishing diverse cellular identities, but how the epigenetic program in Treg cells is maintained throughout the dynamic activation process remains unclear. Here we have shown that CD28 co-stimulation, an extracellular cue intrinsically required for Treg cell maintenance, induced the chromatin-modifying enzyme, Ezh2. Treg-specific ablation of Ezh2 resulted in spontaneous autoimmunity with reduced Foxp3+ cells in non-lymphoid tissues and impaired resolution of experimental autoimmune encephalomyelitis. Utilizing a model designed to selectively deplete wild-type Treg cells in adult mice co-populated with Ezh2-deficient Treg cells, Ezh2-deficient cells were destabilized and failed to prevent autoimmunity. After activation, the transcriptome of Ezh2-deficient Treg cells was disrupted, with altered expression of Treg cell lineage genes in a pattern similar to Foxp3-deficient Treg cells. These studies reveal a critical role for Ezh2 in the maintenance of Treg cell identity during cellular activation.
INTRODUCTION
Regulatory T cells (Treg cells) are a subset of T lymphocytes that suppress auto-reactive effector T cells and are essential for immune homeostasis. Treg cell maintenance is critical because their loss leads to the rapid onset of fatal autoimmunity (Kim et al., 2007). CD28 signaling is essential for the generation and maintenance of Treg cells (Tai et al., 2005; Tang et al., 2003), which, in the case of CD28-deficient NOD mice, leads to exacerbated autoimmunity due to disrupted Treg cell homeostasis (Lenschow et al., 1996; Salomon et al., 2000). While CD28 signaling contributes to Treg cell identity via multiple mechanisms, including induction of Foxp3 itself, our previous studies indicated that CD28 signals also regulate enzymes that control chromatin structure (Martínez-Llordella et al., 2013). Chromatin-mediated support of Treg cell identity might be especially important in the context of inflamed tissues where activated Treg cells must preserve their core gene-expression program in the face of a complex milieu of extracellular cues.
The epigenetic regulator Enhancer of Zeste Homolog 2 (Ezh2) functions primarily within the multi-subunit polycomb repressive complex 2 (PRC2) and catalyzes the tri-methylation of lysine 27 on the exposed N-terminal tail of histone H3 (H3K27me3) (Margueron and Reinberg, 2011). H3K27me3 recruits protein complexes involved in chromatin compaction and is associated with inactive genes (Spivakov and Fisher, 2007). Ezh2 and H3K27me3-marked histones have been shown to be critical for proper B and T cell lineage development (Mandal et al., 2011; Raaphorst et al., 2001; Su et al., 2003; Su et al., 2005), cytokine gene regulation in distinct T helper cell subsets (Chang and Aune, 2007; Jacob et al., 2008; Koyanagi et al., 2005), and T helper-1 (Th1) versus Th2 cell polarization in vitro (Tumes et al., 2013). By comparison, Treg cells have a distinct H3K27me3 landscape compared to naive or polarized CD4+ T helper cells (Wei et al., 2009). Furthermore, Ezh2 can directly control Foxp3 expression (Xiong et al., 2012) and, during inflammatory responses, Ezh2 is recruited by Foxp3 to repress key genes in Treg cells (Arvey et al., 2014). However, genetic ablation of Ezh2 does not disrupt induced Treg cell generation in vitro (Tumes et al., 2013; Zhang et al., 2014). Therefore, the importance of Ezh2 to Treg cell stability and function, especially in naturally arising Treg cells in vivo, is unresolved.
Here we have shown that Ezh2 is induced after CD28-mediated activation and stabilizes the Treg cell transcriptional program. Mice with Ezh2 deficiency targeted specifically to Foxp3-expressing cells succumbed to autoimmunity and were incapable of resolving an induced, acute form of autoimmune disease. Activated Ezh2-deficient Treg cells showed selective destabilization of Treg cell signature genes and a pronounced induction of genes normally repressed in Treg cells after activation. The effect of Ezh2 deletion in activated Treg cells was most prominent in non-lymphoid tissue sites where the frequency of Foxp3+ cells and the stability of Foxp3 expression were reduced. Thus, Ezh2 is critical for proper Treg cell function by supporting Foxp3-driven gene expression patterns following cellular activation.
RESULTS
CD28-Dependent Induction of Ezh2 in T Regulatory Cells
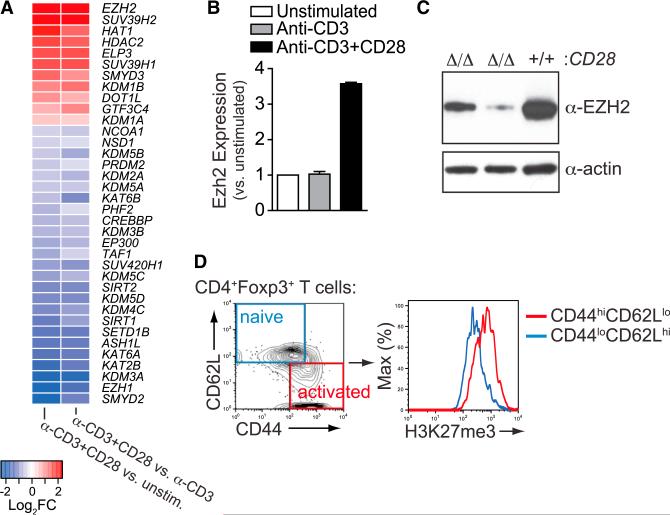
A survey of all differentially expressed histone acetyltransferase, methyltransferase, and demethylase genes upon activation of human naive CD4+ T cells (Martínez-Llordella et al., 2013) revealed that Ezh2, which encodes an H3K27 methyltransferase, was the most highly induced CD28-dependent chromatin modifier (Figure 1A). Blockade of CD28 co-stimulation, either using CD28- or B7-deficient mice, confirmed the CD28 co-stimulation-dependent induction of Ezh2 protein in mouse T cells (Figures S1A and S1B). CD28 co-stimulation also induced Ezh2 mRNA and protein in murine Treg cells (Figures 1B and 1C). Furthermore, there was concordance between reduced Ezh2 expression and reduced enzymatic activity in activated CD28-deficient Treg cells, based on deposition of tri-methylated Lys27 histone H3 (H3K27me3) marks by quantitative flow cytometric analysis (Figure S1C). The specificity of the anti-H3K27me3 antibody, and lack of redundancy with Ezh1 in activated Treg cells, was confirmed by H3K27me3 staining of Ezh2-deficient Treg cells (Figures S1D–S1G). Finally, activated CD44hiCD62Llo Treg cells had more H3K27me3 marks than resting CD44loCD62Lhi cells in vivo (Figure 1D). Thus, CD28-mediated co-stimulation in Treg cells was required to induce Ezh2 expression and activity.
Figure 1. CD28-Dependent Induction of Ezh2 in T Regulatory Cells.
(A) Microarray analysis to reveal CD28-dependent genes by comparison of human naive CD4+ T cells that were unstimulated, stimulated with anti-CD3, or stimulated with anti-CD3 and anti-CD28 antibodies for 24 hr. Heat map of known histone lysine demethylases, methyltransfereses, acetyltransferases, and deacetylases that were significantly differentially expressed by comparing anti-CD3+CD28-stimulated versus unstimulated and anti-CD3+ CD28-stimulated versus anti-CD3 stimulated (36 out of all 9,824 DEGs, FDR < 0.05) are plotted.
(B) Ezh2 expression in sorted mouse Treg cells 24 hr after stimulation with anti-CD3 or anti-CD3 and anti-CD28 antibody-coated beads relative to unstimulated Treg cells (n = 2 per condition).
(C) Ezh2 Western blot analysis of CD28-deficient (Δ/Δ) or WT (+/+) mouse Treg cells (from Foxp3-cre;CD28fl/fl mice) 36 hr after stimulation with anti-CD3 and anti-CD28 antibody-coated beads.
(D) H3K27me3 staining in naive versus activated Treg cells in vivo by gating populations based on CD44 and CD62L expression.
Data are mean ± SEM and representative of at least three experiments unless noted otherwise. See also Figure S1.
Phenotypic Analysis Does Not Reveal Defects in Ezh2-Deficient Treg Cells
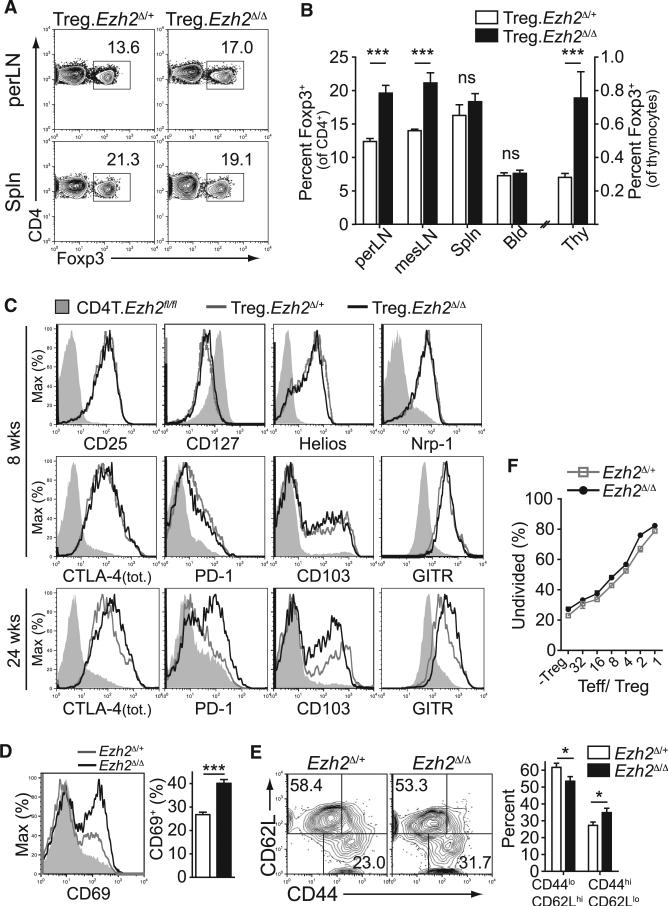
To directly investigate the role of Ezh2 in Treg cells in vivo, we generated mice with partial, Foxp3-GFP-hcre;Ezh2fl/+ (termed Treg.Ezh2Δ/+ mice), or complete deletion of Ezh2 in Treg cells, Foxp3-GFP-hcre;Ezh2fl/fl (termed Treg.Ezh2Δ/Δ mice) (Su et al., 2003; Zhou et al., 2008). The frequency of Foxp3+ Treg cells (as a percent of total CD4+ T cells) was increased in lymph nodes and thymi of Treg.Ezh2Δ/Δ mice but unchanged in the blood and spleen (Figures 2A and 2B). Phenotypic analysis by flow cytometry showed no differences in the expression of many important Treg-associated proteins (Figure 2C), although markers of Treg cell activation (e.g., CTLA-4, PD-1, CD103, GITR) were increased in Treg cells of older Treg.Ezh2Δ/Δ mice. In addition, Ezh2Δ/Δ Treg cells from Treg.Ezh2Δ/Δ mice had higher frequencies of CD69+ cells and greater proportions of CD44hiCD62Llo cells at all time points examined as compared to Treg cells from Treg.Ezh2Δ/+ littermates (Figures 2D and 2E). Finally, CD62Lhi Ezh2Δ/Δ Treg cells maintained normal suppressive capacity in vitro (Figure 2F). Thus, Treg cells from Treg.Ezh2Δ/Δ mice are not reduced in frequency, are phenotypically normal and are capable of being activated and functioning properly. The increased frequency and activation of Treg cells in Treg.Ezh2Δ/Δ mice might result from increased demand for Treg cell mediated tolerance, a phenomenon commonly described in mice with dysfunctional or depleted Treg cells (Pierson et al., 2013; Zhou et al., 2008).
Figure 2. Ezh2-Deficient Treg Cells Are Unaltered in Lymphoid Tissues.
(A) Representative flow cytometric analysis of the frequencies of Treg cells (as percentage of gated CD4+ cells) in peripheral lymph nodes and spleen.
(B) Quantification of data obtained as in (A) for all lymphoid tissues (perLN, inguinal and axillary; mesLN, mesenteric; Spln, spleen; Bld, blood; Thy, thymus).
(C) Phenotypic analysis of Treg cells (solid lines) from perLNs of indicated mice or CD4+Foxp3− cells from Treg.Ezh2Δ/Δ mice (shaded, CD4T. Ezh2fl/fl) by staining for indicated proteins at 8 or 24 weeks of age.
(D and E) Flow cytometric analysis of Treg cells for CD69 (D) or CD44 versus CD62L expression (E) and quantification of results from mice analyzed between 8 and 17 weeks of age.
(F) In vitro Treg cell suppression assay monitoring the percentage of undivided CD4+ T cells (Teff) by dilution of CTV three days after co-culture with indicated ratio of Teff/ Treg (-Treg, no Treg cells). Mean of triplicate assays from one of two experiments shown.
Data are mean ± SEM and representative of the analysis of at least three mice per genotype and time point unless noted otherwise.
Uncontrolled T Cell Activation and Tissue Infiltration in Treg.Ezh2Δ/Δ Mice
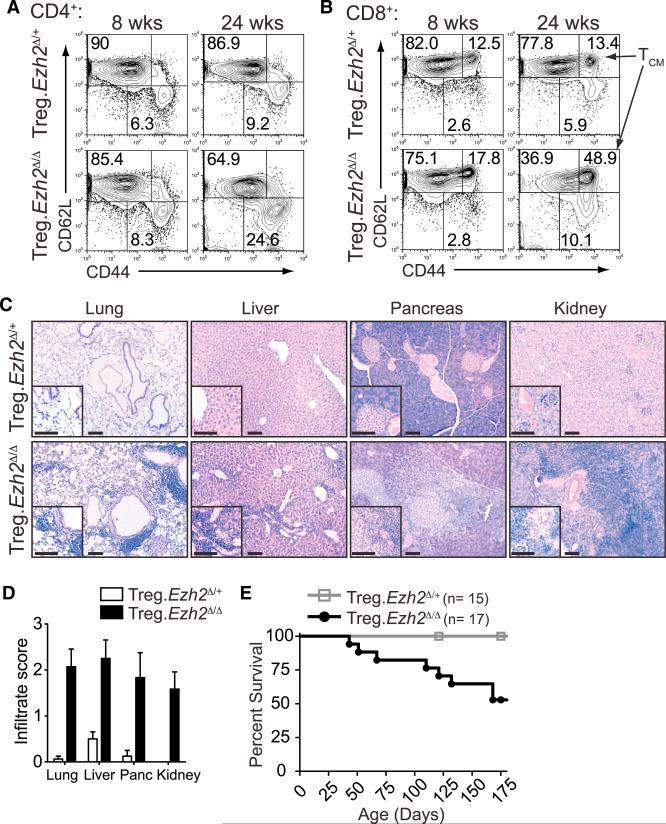
To investigate whether Ezh2 deletion in Treg cells disrupts Treg function in vivo, we followed Treg.Ezh2Δ/+ and Treg.Ezh2Δ/Δ mice over time. Young Treg.Ezh2Δ/Δ mice (6 to 8 weeks old) had increased lymph node cellularity compared to Treg.Ezh2Δ/+ littermates (Figures S2A and S2B). However, the proportions of activated CD44hiCD62Llo CD4+ and CD8+ T cells, as well as CD44hiCD62Lhi CD8+ T cells (TCM), were increased most noticeably in older Treg.Ezh2Δ/Δ mice (Figures 3A and 3B). In addition, unlike Treg.Ezh2Δ/+ mice, Treg.Ezh2Δ/Δ mice displayed various symptoms of autoimmunity, such as reduced weight, hair loss, scaly tails, and swelling around their eyes and ears (Figures S2C and S2D). Examination of non-lymphoid tissues, including the lung, liver, pancreas, and kidney, revealed large infiltrates of mononuclear immune cells, especially in peri-vascular areas (Figures 3C and 3D). Treg.Ezh2Δ/Δ mice had markedly reduced life spans, eventually succumbing to the uncontrolled inflammation observed in all tissues examined (Figure 3E). Notably, we observed similar defects in immune tolerance utilizing a different Foxp3YFP-cre allele to delete Ezh2 specifically in Treg cells (data not shown) (Rubtsov et al., 2008). This suggests that although Ezh2-deficient Treg cells in Treg.Ezh2Δ/Δ mice are present at normal to increased frequencies, appear grossly phenotypically normal, and are functional in vitro, they are unable to properly maintain immune homeostasis.
Figure 3. Ezh2 Deletion in Treg Cells Disrupts Immune Homeostasis.
(A and B) CD44 versus CD62L staining in CD4+−Foxp3− cells (A) and CD8+ cells (B) from peripheral lymph nodes at 8 or 24 weeks of age.
(C) Representative H&E stained sections of lung, liver, pancreas, and kidney from age-matched Treg.Ezh2Δ/+ and Treg.Ezh2Δ/Δ mice (17 weeks pictured). Scale bar represents 100 mM.
(D) Quantification of histological analysis from (C) of mice aged 12–18 weeks old (n = 7–8 per genotype). 0–3 score: 0, no mononuclear infiltration; 3 high degree of mononuclear infiltration (score 3 depicted in (C) for Treg.Ezh2Δ/Δ mice).
(E) Survival of Treg.Ezh2Δ/+ (n = 15) and Treg.Ezh2Δ/Δ (n = 17) mice.
Data are mean ± SEM and representative of at least three mice per genotype and age unless noted otherwise. See also Figure S2.
Impaired Immune Tolerance by Ezh2-Deficient Treg Cells in Adult Mice
To determine whether mice with Ezh2-deficient Treg cells were prone to autoimmunity due to an intrinsic defect in Treg cell function, we utilized female mice heterozygous for the Foxp3YFP-cre genetically targeted allele. Due to the presence of Foxp3 on the X chromosome and X-inactivation, Foxp3YFP-cre/Foxp3WT;Ezh2fl/fl females are chimeric, harboring both wild-type and Ezh2-deficient Treg cells. These females did not exhibit increased T cell activation or any autoimmune symptoms despite harboring equal frequencies of Ezh2-deficient Treg cells as mice with Ezh2Δ/+ Treg cells (Figures S2E and S2F, and data not shown). Thus, the disease in Treg.Ezh2Δ/Δ mice is suppressed by the function of wild- type Treg cells, supporting a hypothesis that Ezh2-deficient Treg cells are intrinsically defective.
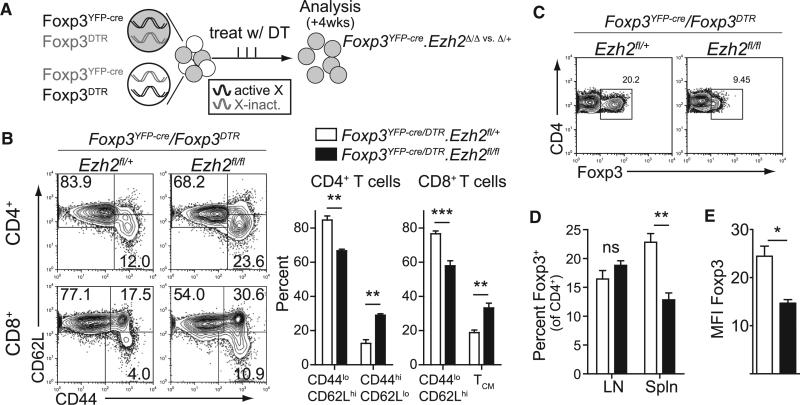
Next we generated female mice that carried the Foxp3YFP-cre and a Foxp3DTR allele, which expresses the diphtheria toxin receptor (DTR) under the control of the endogenous Foxp3 promoter (Kim et al., 2007). By breeding Foxp3YFP-cre/Foxp3DTR females with Ezh2fl alleles, we could selectively delete the wild-type Treg cells expressing the Foxp3DTR allele by treatment with diphtheria toxin (DT). This setting allowed us to acutely challenge Ezh2Δ/Δ Treg cells to maintain immune homeostasis in adult mice (Figures 4A and S3A). As seen in Figure 4B, acute depletion of wild-type Treg cells in mice harboring Ezh2Δ/Δ Treg cells, as compared to mice with Ezh2Δ/+ Treg cells, led to increased activation of both CD4+ and CD8+ T cells in lymph nodes and the development of more severe pathology in these mice (Figures S3B and S3C). In addition, the Ezh2Δ/Δ Treg cells were reduced in frequency in the spleen and had a significant reduction in the expression of Foxp3 (Figures 4C–4E and S3D). Therefore, Ezh2-deficient Treg cells could not prevent the activation of CD4+ and CD8+ T cells in vivo and this was associated with a defect in Treg cell stability in the activated Treg cell pool.
Figure 4. Acutely Challenged Ezh2-Deficient Treg Cells Are Unstable and Do Not Control T Cell Activation.
(A) Experimental model to selectively deplete wild-type Treg cells to challenge Ezh2Δ/Δ Treg cells to maintain immune homeostasis.
(B) Analysis of CD4+ and CD8+ T cell activation in lymph nodes by CD44 and CD62L expression after four weeks of DT treatment (33/week) of Foxp3YFP-cre/ Foxp3DTR;Ezh2fl/+ or Foxp3YFP-cre/Foxp3DTR;Ezh2fl/fl mice.
(C and D) Representative flow cytometric analysis of the percentage of Foxp3+ Treg cells (of CD4+ cells) in spleens of mice treated with DT (C) and quantification of the percentage of Foxp3+ cells in the spleens or lymph nodes (D).
(E) MFI of Foxp3 expression by antibody staining of splenocytes of DT treated mice.
Data are mean ± SEM and representative of at least three mice per genotype from three independent experiments. See also Figure S3.
Reduced Frequency and Increased Destabilization of Treg Cells in Non-lymphoid Tissues of Treg.Ezh2Δ/Δ Mice
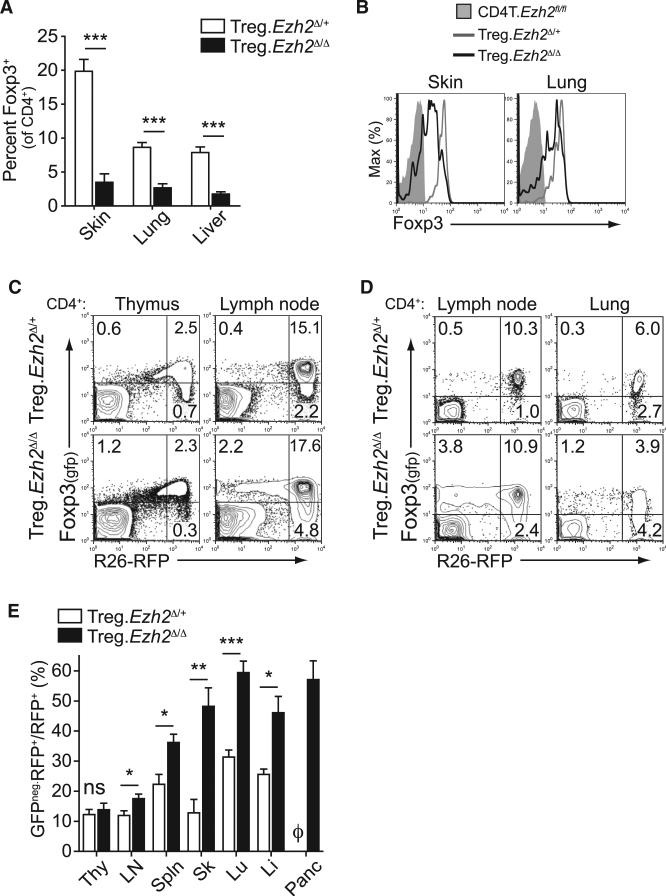
We next examined Treg cells in non-lymphoid tissues. There were massive reductions in the frequencies of Treg cells in aged Treg.Ezh2Δ/Δ mice in all tissues examined (Figures 5A, S4A, and S4B). Ezh2Δ/Δ Treg cells also exhibited lower Foxp3 expression in tissues based on Foxp3 reporter expression and anti-FoxP3 antibody staining (Figures 5B and S4C), suggesting that the autoimmune symptoms in Treg.Ezh2Δ/Δ were related to a profound deficiency of Treg cells in the non-lymphoid tissues.
Figure 5. Reduced Frequency and Increased Destabilization of Ezh2-Deficient Treg Cells in Tissues.
(A) Quantification of the frequency of Treg cells (as percentage of Foxp3+ cells of CD4+ T cells) from indicated tissues of age-matched mice 18 to 30 weeks old.
(B) Representative Foxp3-GFP reporter expression by flow cytometry of CD4+CD25+ populations from the skin and lung of indicated mice.
(C and D) Representative lineage tracing of Treg cells in Treg.Ezh2Δ/+ and Treg.Ezh2Δ/Δ mice by flow cytometry of thymus and lymph nodes (C) or lymph nodes and lung (D). Cells depicted are gated CD4+. Mice depicted were 17 to 18 weeks old.
(E) Quantification of the percentage of GFPneg.RFP+ cells (of all CD4+RFP+ cells) across indicated tissues of Treg.Ezh2Δ/+ and Treg.Ezh2Δ/Δ mice. φ, no T cells detected.
Data are mean ± SEM and representative of at least three mice per genotype from at least three independent experiments. See also Figure S4.
The origin and fate of the Treg cell lineage with Ezh2-deficiency was examined utilizing a lineage-tracing strategy by breeding mice with an R26LSL-RFP allele to mice with the Foxp3-GFP-hcre allele (Zhou et al., 2009). This allowed the tracking of newly generated Treg cells (CD4+GFP+RFPneg cells), stable Treg cells (CD4+GFP+RFP+ cells), and cells that did not maintain Foxp3 expression (CD4+ GFPnegRFP+ cells). Comparison of Treg.Ezh2Δ/+ and Treg.Ezh2Δ/Δ mice revealed an increased proportion of GFP+RFPneg cells in the thymus and lymph nodes of Treg.Ezh2Δ/Δ mice, suggesting that Treg.Ezh2Δ/Δ mice generate more new Treg cells (Figure 5C). The GFP+RFPneg cells were CD25+, Nrp-1+, and more naive (CD44lo CD62Lhi) than GFP+RFP+ cells, which suggests committed Treg cells of thymic origin (Figures S4D and S4E) (Miyao et al., 2012; Weiss et al., 2012; Yadav et al., 2012). In addition, Treg.Ezh2Δ/Δ mice had increased proportions of GFPnegRFP+ cells across all tissues analyzed (except for the thymus), indi cating that Ezh2Δ/Δ Treg cells had a reduced capacity to maintain the stable Treg cell pool (CD4+GFP+RFP+ cells) (Figures 5C–5E). Increased proportions of GFPnegRFP+ cells were also observed in healthy Foxp3YFP-cre/Foxp3WT;Ezh2fl/fl females, indicating that an impaired ability to maintain the Foxp3+ cell pool was intrinsic to Ezh2Δ/Δ Treg cells and not just a byproduct of the increased inflammation in Treg.Ezh2Δ/Δ mice (Figure S4F).
The highest percentages of GFPnegRFP+ cells were found in non-lymphoid tissues (Figure 5E), where Treg cells appear highly activated and showed decreased Foxp3 expression. While these lineage-tracing experiments cannot distinguish whether Ezh2-deficient Treg cells became destabilized early (GFP+RFPneg cells) or late (GFP+RFP+ cells) after Foxp3 is expressed, recombination of all the alleles, including both Ezh2fl alleles and the R26LSL-RFP allele, would require the Foxp3-cre allele to be sufficiently expressed (Miyao et al., 2012; Rubtsov et al., 2008). In addition, the increased proportion of Foxp3 negative cells with Ezh2-deficiency was restricted to the CD44hiCD62Llo population with no significant increase in GFPnegRFP+ cells in the CD44loCD62Lhi population (Figures S4G and S4H), suggesting that Ezh2, which is induced in Treg cells after activation, promotes the maintenance of Foxp3 expression during Treg cell responses.
Ezh2-Deficient Treg Cells Exhibit Altered Gene-Expression Patterns after Activation
Gene-expression analysis (RNaseq) of CD62Lhi or CD62Llo Ezh2Δ/+ and Ezh2Δ/Δ Treg cells (CD4+CD25+YFP+RFP+ cells) sorted from lymph nodes and spleens of Foxp3YFP-cre/Foxp3WT;Ezh2fl/+ or Foxp3YFP-cre/Foxp3WT;Ezh2fl/fl mice was performed to investigate the underlying defects in Ezh2Δ/Δ Treg cells following activation and/or differentiation (Figures S5A–5C). Foxp3+ cells from chimeric female mice were used for this analysis to identify alterations that precede Foxp3 protein loss in mice that do not exhibit any inflammation-driven abnormalities.
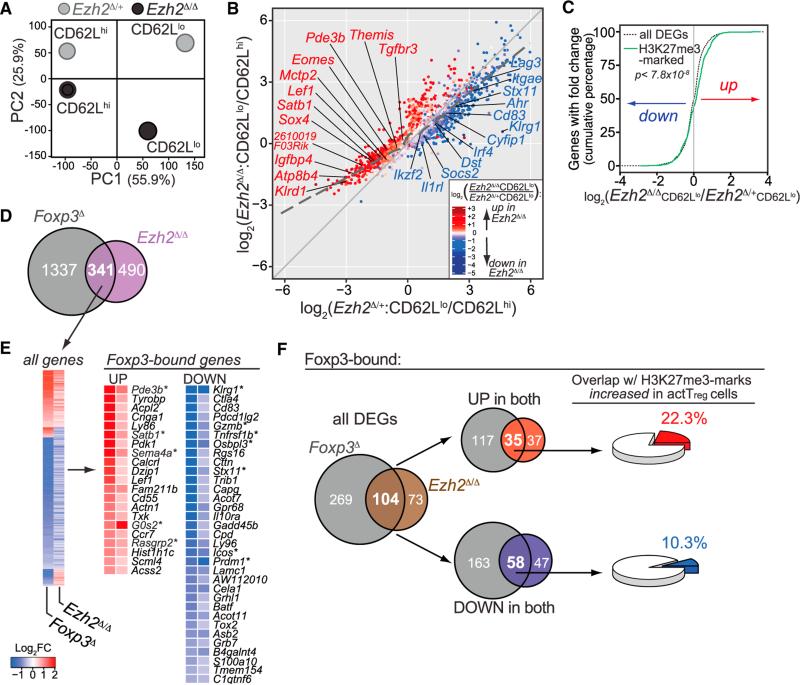
Principal component analysis revealed that the greatest differences in gene expression patterns between Ezh2Δ/+ and Ezh2Δ/Δ Treg cells occurred in the CD62Llo populations (Figure 6A), consistent with the hypothesis that defects in Ezh2Δ/Δ Treg cells are manifested after activation. Next, we compared the differentially expressed genes (DEGs) between CD62Llo and CD62Lhi populations in Ezh2Δ/+ compared to Ezh2Δ/Δ Treg cells to determine whether there were global differences in the pattern of genes induced or repressed after activation (Figure 6B). Ezh2Δ/Δ Treg cells had dramatically increased expression of genes normally repressed in CD62Llo cells (lower left of plot) as compared to a modest reduction in the expression of genes normally induced in CD62Llo cells (upper right of plot). To determine whether these changes in global gene repression after activation resulted from decreased Ezh2-mediated H3K27me3 deposition, we utilized the recently reported H3K27me3 ChIP-seq dataset by Arvey et al. (2014) to identify genes associated with increased deposition of H3K27me3 marks after Treg cell activation. As shown in Figure 6C, genes associated with increasing H3K27me3 marks in activated Treg cells were specifically enriched in the upregulated (or de-repressed) DEGs in Ezh2Δ/Δ CD62Llo Treg cells (p < 7.8 × 10−8, K-S test). Furthermore, only the cumulative distribution of the upregulated genes marked by H3K27me3 differed significantly (K-S test) from all DEGs (p < 1.95 × 10−9), whereas for H3K27me3-marked, downregulated genes, there was no significant difference (p < 0.13). Thus, Ezh2, via H3K27me3 deposition, is required for the repressive gene program in Treg cells after activation.
Figure 6. Ezh2 Is Required in Activated Treg Cells to Stabilize the Foxp3-Driven Treg Program.
(A) Principal-component analysis of the transcriptomes of sorted CD62Lhi and CD62Llo YFP+ Treg cells from lymph nodes and spleens of female Foxp3YFP-cre/ Foxp3WT;Ezh2fl/+ (Ezh2Δ/+) and Foxp3YFP-cre/Foxp3WT;Ezh2fl/fl (Ezh2Δ/Δ) mice. The contribution of each principal component to the total variance in the data is shown on each axes (%). Each point represents the combined average of three biological samples except for CD62Lhi Ezh2Δ/+, which is from two samples.
(B) Comparison of all differentially expressed genes (DEGs = 3097, FDR < 0.05) between CD62Lhi and CD62Llo Treg cells in the absence of Ezh2 (Ezh2Δ/Δ, y axis) or presence of Ezh2 (Ezh2Δ/+, x axis). Dashed line represents the trend of all genes differentially regulated between the Ezh2Δ/Δ and Ezh2Δ/+ datasets. Data points are colored by significant differences in gene expression between the CD62Llo populations of Ezh2Δ/Δ versus Ezh2Δ/+ Treg cells: red, increased expression; blue, reduced expression. Differentially expressed “Treg cell signature” genes are labeled and colored with respect to expression change.
(C) Cumulative distribution of fold changes in all DEGs (black dotted line, 3097 genes) or DEGs with increased H3K27me3 marks in activated Treg cells (green line, 465 genes) between CD62Llo Ezh2Δ/Δ and Ezh2Δ/+ Treg cells, p < 7.8 × 10−8 using the two-sample Kolmogorov-Smirnov (K-S) statistical test.
(D) Venn diagram of DEGs shared between Foxp3D Treg cells (Foxp3gfpkoTreg versus WT Treg: 1,678 genes) and activated Ezh2Δ/Δ Treg cells (CD62Llo Ezh2Δ/Δ versus CD62Llo Ezh2Δ/+ Treg cells: 831 genes).
(E) Clustering of all genes up or down (341 DEGs, FDR < 0.05) between Foxp3D and Ezh2Δ/Δ data sets (left) and expression of Foxp3-bound genes that are up- or downregulated ≥0.5 log2 fold change (right). *, genes also identified by GSEA of Ezh2Δ/Δ Treg cells compared to Treg versus Tconv immunological signatures.
(F) Left to right: Venn diagram depicting Foxp3-bound DEGs in-common between Foxp3D and Ezh2Δ/Δ Treg cells (left); overlap in Foxp3-bound DEGs either up-(top) or downregulated (bottom) in both datasets (middle); percentage of shared up- (top) or downregulated Foxp3-bound DEGs (bottom) in Foxp3D and Ezh2Δ/Δ that are associated with increased H3K27me3 marks in activated versus resting WT Treg cells (right).
See also Figure S5.
CD62Llo Ezh2-Deficient Treg Cells Are Similar to Foxp3-Deficient Treg Cells
Analysis of the genes whose expression was dysregulated in the absence of Ezh2 revealed a large number of “Treg cell signature genes” (Figure 6B, red and blue text) (Hill et al., 2007). Comparison of differentially expressed genes (DEGs) between CD62Llo populations of Ezh2Δ/Δ versus Ezh2Δ/+ Treg cells (termed Ezh2Δ/Δ) from our dataset to DEGs between “Foxp3gfpko” (cells with gfp inserted into the Foxp3 locus blocking Foxp3 protein expression) versus wild-type Treg cells (termed Foxp3D) from Gavin et al. dataset (2007) revealed that Ezh2Δ/Δ Treg cells had global defects in the Foxp3-mediated gene-expression program. Forty-one percent of all DEGs in Ezh2Δ/Δ Treg cells were also differentially expressed in Foxp3D Treg cells (Figure 6D). Furthermore, the qualitative changes in the expression patterns of the shared genes were similar (Figure 6E). Gene set enrichment analysis showed that the expression pattern of Ezh2Δ/Δ Treg cells more closely resembled conventional CD4+ T than Treg cells (Figure S5D). Finally, as shown in Figure 6E, many of the overlapping genes were direct Foxp3 targets based on published data of chromatin immunoprecipitation for Foxp3 (Samstein et al., 2012). In fact, when limiting the DEGs from the two datasets to only Foxp3-bound genes, 58.8% of all DEGs in Ezh2Δ/Δ Treg cells overlapped with DEGs from Foxp3D Treg cells, thus dramatically enriching the overlap in these two datasets (Figures 6F and S5E). These overlapping DEGs were represented among both up- and downregulated genes in both datasets. However, the upregulated (or de-repressed) genes in Foxp3D and Ezh2Δ/Δ Treg cells were enriched two-fold in H3K27me3 marks that are increased upon Treg cell activation (22.3%, red from Arvey et al. [2014]) versus the coordinately downregulated genes (10.3%, blue) (Figures 6F and S5F). These results suggest an essential role for Ezh2 in promoting the Foxp3-dependent Treg cell program during activation and are consistent with the model proposed by Arvey et al. (2014) that a number of Foxp3-bound genes are repressed by Ezh2-mediated H3K27 tri-methylation in activated Treg cells. There was also an overlap of Foxp3-bound and downregulated genes in Foxp3D and Ezh2Δ/Δ Treg cells (i.e., genes weakly induced in Ezh2Δ/Δ Treg cells upon activation) that were less enriched in activation-associated H3K27me3 marks (Figures 6F). This may suggest that Ezh2 promotes the expression of a subset of genes bound by Foxp3, possibly in a PRC2-independent manner, as described in the context of cancer (Xu et al., 2012), although downregulation of these genes could also be an indirect result of Ezh2 deficiency in these cells.
Inability of Ezh2-Deficient Treg Cells to Control Remission in Experimental Autoimmune Encephalomyelitis
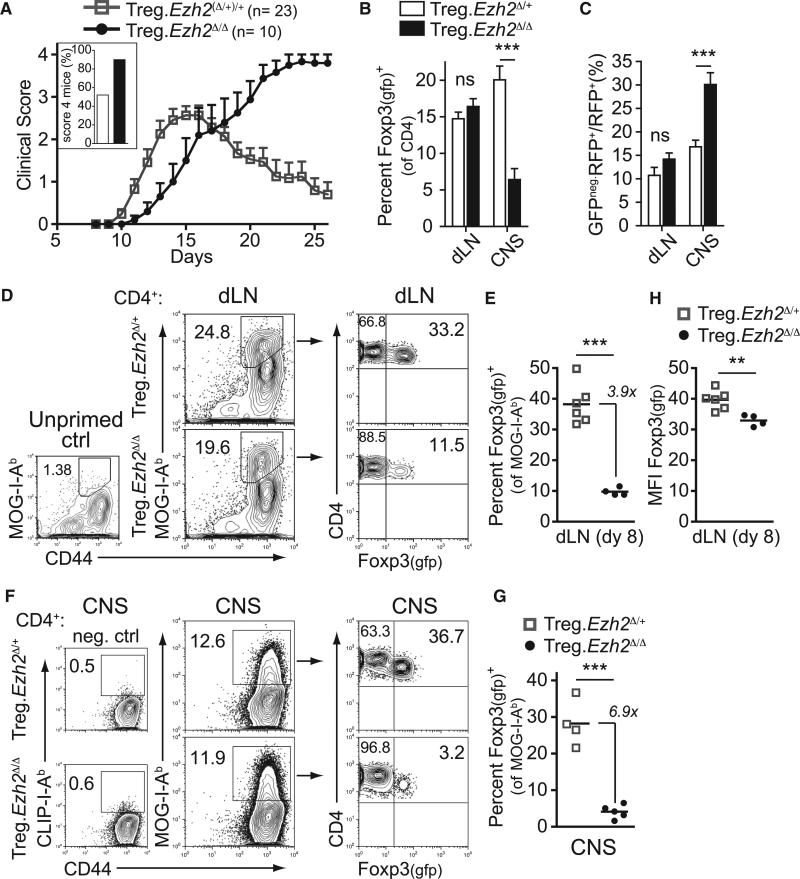
To determine the acute clinical impact of Ezh2-deficiency in Treg cells, we examined Ezh2Δ/Δ Treg cell phenotypes in experimental autoimmune encephalomyelitis (EAE), an inducible mouse model of multiple sclerosis. As seen in Figure 7A, immunization of Treg.Ezh2Δ/+ (or Ezh2+/+) mice with the neural antigen myelin oligodendrocyte glycoprotein (MOG35–55 peptide), resulted in moderate disease, with 52% of the mice achieving the highest clinical disease score of 4. In contrast, 90% of Treg.Ezh2Δ/Δ mice exhibited the most severe form of the disease. Furthermore, while all Treg.Ezh2Δ/+ mice ultimately regained hind limb mobility, none of the Treg.Ezh2Δ/Δ mice recovered from the disease. There was a reduction in Treg cell frequency, as well as an increased percentage of GFPnegRFP+ cells, specifically in the CNS tissues of Treg.Ezh2Δ/Δ mice (Figures 7B and 7C). These results recapitulated the phenotypes observed spontaneously in Treg.Ezh2Δ/Δ mice. Importantly, Ezh2Δ/Δ Treg cells displayed the same defective phenotypes in a competitive environment with wild-type Treg cells in Foxp3YFP-cre/Foxp3WT female mice with EAE (Figures S6A and S6B). However, female Foxp3YFP-cre/Foxp3WT;Ezh2fl/fl mice recovered from the disease (Figure S6C), strongly suggesting that Ezh2 deficiency renders Treg cells intrinsically defective in regulating tissue inflammation.
Figure 7. Mice with Ezh2-Deficient Treg Cells Do Not Resolve EAE-Induced Tissue Inflammation.
(A) Plot of clinical score versus time (days) after induction of EAE in indicated mice. Inset shows percentage of mice with score 4 disease. Mean cumulative data from three experiments.
(B and C) Percentage of Foxp3+ cells (of CD4+ T cells, B) or GFPneg.RFP+ cells (of CD4+RFP+ cells, C) in draining lymph nodes (dLN) or central nervous system tissues (CNS) at peak disease (18–27 days after EAE induction). Data pooled from three independent experiments.
(D) Representative analysis of CD4+ T cells from dLN enriched and stained with MOG38–49-I-Ab tetramers from unprimed control mice (left plot) or mice primed with MOG35–55+CFA eight days earlier (middle plots) and the fraction of Foxp3-gfp+ cells of the total MOG-I-Ab+ cells from dLN (right plots).
(E) Quantification of the percentage of Foxp3-gfp+ cells of all MOG-I-Ab+ cells in enriched dLN from (D).
(F) Representative analysis of CD4+ T cells from CNS stained with CD44 and CLIP-I-Ab (neg. ctrl, left plots) or MOG38–49-I-Ab tetramers (middle plots) and the fraction of Foxp3-gfp+ cells of the total MOG-I-Ab+ cells from CNS (right plots).
(G) Quantification of the percentage of Foxp3-gfp+ cells of all MOG-I-Ab+ cells in CNS.
(H) MFI of Foxp3 expression by GFP reporter in dLN of indicated mice 8 days after EAE induction.
Data are mean ± SEM and representative of at least three mice per genotype from at least three independent experiments. See also Figures S6 and S7.
The EAE system allowed us to follow newly activated, antigen-specific Treg cells over time utilizing I-Ab tetramers loaded with MOG38–49 peptide. MOG-specific Ezh2Δ/Δ Treg cells were readily detectable in the draining lymph node 8 days after EAE induction, indicative of an expansion of these cells, which are nearly undetectable in unprimed mice (Figure 7D) (Bailey-Bucktrout et al., 2013). However, the frequency of MOG-specific Treg cells (as a fraction of all MOG-specific CD4+ T cells) was reduced as compared to controls (Figures 7D and 7E), especially in the CNS tissues at the peak of disease (Figures 7F and 7G). To directly assess whether Ezh2Δ/Δ Treg cells were activated and proliferated normally in response to antigenic stimulation in vivo, we labeled 2D2 T cell receptor (TCR) transgenic (MOG35–55-specific) Ezh2Δ/+ and Ezh2Δ/Δ Treg cells with CFSE and co-transferred the cells into MOG-primed wild-type mice (Bettelli et al., 2003). Four days after transfer, both Ezh2Δ/+ and Ezh2Δ/Δ MOG-specific Treg cells had divided to a similar extent (Figure S7A). Similarly, the activation and proliferation of Ezh2Δ/Δ Treg cells in vitro was intact (Figure S7B); however, Ezh2Δ/Δ Treg cells did exhibit increased apoptosis following several rounds of division (Figures S7C and S7D). Finally, the newly activated MOG-specific Treg cells exhibited reduced Foxp3 expression (Figure 7H), suggesting that Ezh2Δ/Δ Treg cells are destabilized early after activation, driving reduced frequencies of Treg cells in tissues. These results support the conclusion that deletion of Ezh2 in Treg cells leads to a selective loss of antigen-specific activated cells at the site of inflammation leading to uncontrolled autoimmunity.
DISCUSSION
While the role of the polycomb genes in maintaining cellular fate decisions during development is established, their role in maintaining the identity of mature, differentiated cells after cellular activation is less clear. In this study, we identified Ezh2 as an epigenetic modifier induced by CD28 co-stimulation that is necessary for the maintenance of Treg cell identity after activation. Mice with Ezh2-deficient Treg cells failed to maintain immune tolerance, developed multi-organ autoimmunity, and were incapable of resolving inflammation in CNS tissues upon acute induction of autoimmunity (EAE). These mice exhibited reductions in Treg cells specifically in the non-lymphoid tissues, suggesting that Ezh2-deficiency selectively disrupts the activated Treg cell pool. Comparative gene-expression analysis of activated (CD62Llo) versus quiescent (CD62Lhi) Treg cells from healthy mice confirmed that the activated Ezh2-deficient Treg cells had aberrant expression of the Foxp3-dependent gene-expression program, including many Treg cell signature genes that were direct targets of Foxp3 and were de-repressed. Furthermore, Ezh2-deficient Treg cells, despite expressing Foxp3, had a gene-expression pattern that resembled Foxp3-deficient Treg cells and exhibited lineage instability after activation. These results reveal an important role for CD28-induced epigenetic control in the differentiation and maintenance of responding Treg cells and highlight the importance of chromatin regulation in the maintenance of immune homeostasis particularly in non-lymphoid tissues.
Epigenetic mechanisms that alter chromatin organization are thought to control, or at least reinforce, the differentiation and maintenance of polarized T helper cell subsets (Ansel et al., 2003; Wilson et al., 2009). Deletion of Ezh2 in CD4+ T cells was recently shown to promote the capacity of in vitro differentiated T helper subsets to convert to opposing phenotypes; however, a role for this chromatin modifier in Treg cells was not elucidated (Tumes et al., 2013). Treg cells harbor a unique constellation of epigenetic histone modifications compared to other T cell subsets (Wei et al., 2009), yet the molecules and mechanisms that regulate these distinctions are less clear. Recent studies indicate that the unique epigenetic state in Treg cells might be independent of their lineage-specifying transcription factor Foxp3 and depend on signals from the T cell receptor and co-stimulatory pathways to alter the chromatin accessibility of key Treg-cell-expressed genes (Ohkura et al., 2012; Samstein et al., 2012). Hence, it is noteworthy that we identified Ezh2 as a chromatin modifier dependent on CD28 co-stimulation.
Ezh2-deficient Treg cells emigrating from the thymus appeared phenotypically normal and functional and were even increased in Treg.Ezh2Δ/Δ mice. Importantly, Ezh2 was only deleted subsequent to Foxp3 expression in this model, thus confining our analysis to the effects of Ezh2 deletion in established Treg cells and excluding its role in the development of the Treg cell lineage. In this context, Ezh2-deficient Treg cells exhibited altered transcription and destabilization only after they were activated. During CD4+ T cell activation, cellular proliferation is accompanied by coordinated changes in chromatin structure that are linked to the differentiation of divergent types of T helper cells (Agarwal and Rao, 1998; Bird et al., 1998; Grogan et al., 2001). Treg cells exit the thymus in a relatively naive state and upon peripheral activation, differentiate to tailor their suppressive activity to the particular inflammatory environment they will control (Campbell and Koch, 2011; Darrasse-Jèze et al., 2009; Fisson et al., 2003; Rosenblum et al., 2011). A recent study indicated that TCR stimulation alone is insufficient to drive the differentiation of Treg cells from a naive-like to effector-like state and that additional inflammatory stimuli are required, presumably to induce the expression of co-stimulatory molecules like B7-1 and B7-2 on antigen-presenting dendritic cells that provide additional signals with TCR stimulation (Smigiel et al., 2014). Therefore, a stepwise connection between CD28-dependent activation in lymphoid tissues, differentiation into effector-like Treg cells, and ultimately the migration and function of these Treg cells in non-lymphoid tissues seems probable.
From this perspective, the paucity of Ezh2-deficient Treg cells specifically in non-lymphoid tissues would be expected if these Treg cells were destabilized after activation, before their migration to target tissues. Indeed, induction of EAE in Treg.Ezh2Δ/Δ mice generated greatly reduced Treg frequencies in the CNS and, as a result, these mice never resolved the inflammation in the CNS. However, the induction of disease in Treg.Ezh2Δ/Δ mice was not accelerated, likely because Ezh2-deficient Treg cells were activated and proliferated normally at the earliest time points after priming (and were suppressive in vitro) but showed signs of Foxp3 destabilization and Treg cell loss at later time points. Together, these data indicate that Ezh2-deficient Treg cells become defective subsequent to activation and this is consistent with our gene-expression analysis showing transcriptional divergence specifically in the activated (CD62Llo) Treg cell pool. The data also suggest that different subsets of Treg cells might be specialized for controlling different stages of disease, with naive Treg cells being most important to the control of initial priming events in lymph nodes and activated effector Treg cells being most important to the resolution of inflammation at the site of tissue destruction.
We found that Ezh2 functions largely repress genes after cellular activation in Treg cells. This is consistent with the known epigenetic activity of Ezh2 but also might suggest a specific role for gene repression upon activation in Treg cells. Indeed, Foxp3 directly binds many genes exhibiting increased expression in activated Ezh2-deficient Treg cells. Several studies have reported that Foxp3 acts predominantly as a repressor upon activation of Treg cells (Arvey et al., 2014; Marson et al., 2007; Morikawa et al., 2014). This might serve to preserve the Tregspecific gene expression program by dampening the induction of genes normally upregulated during T cell activation. Foxp3 might exert this repression by recruiting the Ezh2-containing polycomb repressive complex to key targets during activation, because Foxp3 repressed genes are associated with H3K27me3 deposition and reduced chromatin accessibility (Arvey et al., 2014). By genetically ablating Ezh2 in Treg cells in this study, we have functionally validated the importance of Ezh2 activity in activated Treg cell stability and function. We hypothesize that Ezh2, in collaboration with Foxp3, is responsible for controlling an entire network of genes essential for the Treg cell transcriptional program during activation and that the dysregulation of many genes ultimately drives the phenotypes observed in Ezh2-deficient Treg cells. Among this set of co-regulated genes that are upregulated in Ezh2-deficient Treg cells and normally enriched in H3K27me3 marks with activation are several genes that are known to antagonize Treg cell function (e.g., Pde3b, Tcf7, Lef1) (Gavin et al., 2007; Keerthivasan et al., 2014). Importantly, while our conclusions from the comparison of H3K27me3 marks in wild-type Treg cells to de-repressed genes in Ezh2-deficient Treg cells are consistent with a direct role for Ezh2-mediated H3K27me3 marks supporting a Treg cell gene-expression program, only an assessment of the changes to H3K27me3 marks in Ezh2-deficient cells would provide direct evidence of this mechanism. Thus, we cannot rule out that indirect effects of Ezh2 deficiency, unrelated to its role in H3K27 tri-methylation specifically in Treg cells, might contribute to the phenotypes observed.
In this study, we have demonstrated that the epigenetic regulator, Ezh2, can coordinate cellular activation and the maintenance of cellular identity. Ezh2 bridges these processes by directly responding to extracellular cues that drive proliferation (CD28 co-stimulation) and functioning with lineage specifying transcription factors (Foxp3) to reinforce the cells’ transcriptional program. In the context of the adaptive immune system, where the activation and expansion of subsets of cells with unique specificities is essential, epigenetic regulation of the fidelity of cell identity is paramount. With the recent development of several drugs that target specific chromatin modifiers, new opportunities might exist to modulate the cellular identity of immune cells, namely Treg cells, for therapeutic benefit.
EXPERIMENTAL PROCEDURES
Mice
All mice used were bred onto a C57BL/6 background a minimum of five generations. All mouse experiments (or cells from mice of given genotypes) used comparisons between littermates or age-matched control mice. CD28fl mice harbor loxP sites flanking the extracellular and transmembrane domains of Cd28 (exons 2–3) (Zhang et al., 2013). Foxp3-GFP-hcre and Foxp3YFP-cre mice express Cre recombinase in Foxp3+ cells utilizing distinct technologies (Rubtsov et al., 2008; Zhou et al., 2008). Ezh2fl mice harbor loxP sites flanking exons 16–19 encoding the SET domain (Su et al., 2003). For diphtheria toxin treatments, mice were treated with a dose of 50 mg/kg three times per week, every other day. For EAE studies, disease was induced as described previously (Bailey-Bucktrout et al., 2013) and clinical disease was scored by ascending hind-limb paralysis as follows: 1, paralysis of tail; 2, hind limb weakness; 3, paralysis of one hind limb; 4, paralysis of both hind-limbs. Mice were monitored at least 6 days for recovery after reaching score 4 disease. All experiments were done according to the Institutional Animal Care and Use Committee guidelines of the University of California, San Francisco.
Lymphocyte Isolation
Cells from lymphoid organs were prepared by mechanical disruption between frosted slides or from non-lymphoid organs by digestion in RPMI supplemented with HEPES and 20 μg/ml DNase I (Roche) with these modifications: lungs and liver were prepared by mincing and digesting the tissues for 1 hr at 37°C with 125 U/ml Collagenase D (Roche); whole pancreas was prepared by digesting for 30 min at 37°C with 0.8 mg/ml Collagenase P (Roche) followed by mincing; skin was prepared from the trunk skin between limbs, removing hair and subcutaneous fat, and digesting for 45 min at 37°C with 2 mg/ml Collagenase XI (Sigma) and 0.5 mg/ml Hyaluronidase (Sigma). All suspensions were passaged over 40 μm filters before cell staining or activation. Isolation of lymphocytes from the spinal cord and cerebellum (CNS) of mice with EAE was done as described (Bailey-Bucktrout et al., 2013).
CD4 Enrichment, Cell Sorting, and Flow Cytometry
CD4+ T cells were enriched for western analyses by negative selection using EasySep magnetic bead kit (STEMCELL Technologies) prior to activation (with simultaneous depletion of CD25+ cells) or 24 hr after mixed lymphocyte activation. All other purified populations (confirmed ≥95%) were obtained by sorting single cells using MoFlo (Beckman Coulter) or FACSAria (BD-Biosciences) machines. Utilizing Foxp3-driven GFP (or YFP) expression in combination with antibody staining for CD8α (53–6.7), CD4 (GK1.5), CD25 (PC61), and CD62L (MEL-14), naive CD4+ T cells (CD4+CD62L+CD8− GFP°CD25− ) and Treg cells (CD4+GFP+CD25+CD62L+/− CD8− ) were sorted. When available, the Cre-activated R26LSL-RFP reporter was also used. Live cells were stained with antibodies to extracellular antigens for 20–30 min on ice and then fixed and permeabilized (eBiosciences) for staining of intracellular antigens. Flow cytometry was performed on LSR II machines (BD Biosciences) and analyzed with Flowjo software (Tree Star).
MHCII Tetramer Staining
Draining lymph nodes (inguinal, brachial, and axillary) or CNS-infiltrating cells from mice immunized with MOG+CFA were stained with MOG38–49-I-Ab-APC tetramer (or CLIP-I-Ab-APC, CNS only) for 2 hr at room temperature, followed by staining with other antibodies on ice. For dLN cells, tetramer-positive cells were first enriched using anti-APC microbeads and MACS LS separation columns (Miltenyi Biotec). Tetramers provided by the National Institutes of Health Tetramer Core.
In Vitro Stimulation and Proliferation
Cells were activated with anti-CD3 and anti-CD28 coated beads (Dynabeads Mouse T-Activator CD3/CD28, Invitrogen) at a ratio of 1:1 (cell:bead), beads coated selectively with anti-CD3 or anti-CD3 and anti-CD28 (T Cell Activation/Expansion Kit mouse, Miltenyi Biotec), or 1 μg/ml soluble antibodies against CD3 (clone 145-2C11) ±CD28 (clone PV-1). Cells were kept at a concentration of 106 cells/ml in RPMI medium supplemented with 10% FBS, non-essential amino acids, sodium pyruvate, L-glutamine, HEPES, and β-ME, and cells cultured more than 24 hr were supplemented with 200–2,000 IU/ml recombinant human IL-2 (Chiron Corp). To monitor proliferation, we labeled sorted cells with CFSE or Cell Trace Violet (CTV, Invitrogen). In vitro Treg cell suppression was performed by labeling naive CD4+ T cells with CTV in the presence of 1 μg/ml anti-CD3 antibody (clone 145-2C11), irradiated splenocytes (from TCRα-deficient mice), and different ratios of Treg cells. The percentage of undivided cells (no dilution of CTV) was analyzed 3 days later.
RNA Isolation and qPCR
RNA was isolated by TRIzol homogenization and extraction (Invitrogen). For qPCR of Ezh2 transcripts, RNA was converted to cDNA using SuperScript III First-Strand Synthesis (Invitrogen), and measured using Taqman probe Mm00468464 (Life Technologies) with Taqman Fast Universal mix or the primers 5′-GCAGGGACTGAAACTGGGGGAG (forward) and 5′- CAGCACCACTCCACTCCACATTC (reverse) with Fast SYBR PCR master mix (Applied Biosystems). Samples were run in duplicate or triplicate on the Applied Biosystems 7500/7900 Fast Real-Time PCR System and normalized to HPRT or 18S RNA.
Western Blotting
Cells were lysed at a concentration ~106 cells/50 μl of RIPA lysis buffer (50 mM Tris-HCl pH7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.5% DOC, 1 mM DTT) with protease inhibitor cocktail (Sigma) and PhosSTOP (Roche), separated on a NuPAGE Novex 4%–12% Bis-Tris Gel (Invitrogen), transferred to PVDF membranes, stained with anti-Ezh2 monoclonal antibody (D2C9, Cell Signaling Technology) or anti-Actin polyclonal antibody (Sigma) followed by anti–rabbit IgG-HRP (GE Healthcare), and detected with SuperSignal West Femto Substrate (Thermo Scientfic) on film (Kodak).
Histology
Tissues fixed in 10% formalin overnight, preserved in 70% EtOH, and embedded in paraffin were cut and sections stained with hematoxylin and eosin (H&E). Infiltrate scoring was performed by blinded assessment of at least three sections of each tissue per mouse.
Transcriptome Analysis and Bioinformatics
RNA extracted from sorted populations Ezh2Δ/Δ CD62Lhi, Ezh2Δ/Δ CD62Llo, Ezh2Δ/+ CD62Lhi, and Ezh2Δ/+ CD62Llo Treg cells (n = 3 biological samples/ group, except Ezh2Δ/+ CD62Lhi, n = 2) underwent RNA sequencing (RNaseq) at the UCSF Sandler Asthma Basic Research Center Functional Genomics Core using the Illumina HiSeq SE™ 50bp platform. Detailed methods for RNaseq, microarray, GSEA, and genome-wide binding site analyses are provided in the Supplemental Information.
Statistics and Experimental Design
Statistical tests used for bioinformatics analysis were described in methods or legends. p values from unpaired two-tailed Student’s t tests were used for all other statistical comparisons between two groups and data were displayed as the mean ± SEM. p values are denoted in figures by: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Supplementary Material
Highlights.
Ezh2 is induced after activation in a CD28-dependent manner
Treg-specific ablation of Ezh2 in mice leads to spontaneous autoimmunity
Activation drives lineage instability and loss of Ezh2-deficient Treg cells in vivo
Activated Ezh2-deficient Treg cells transcriptionally resemble Foxp3-deficient cells
ACKNOWLEDGMENTS
We thank D. Erle, A. Barczak, R. Barbeau, and J. Pollack of the UCSF Sandler Center Functional Genomics Core for assistance with RNAseq data; M. Lee, V. Nguyen, N. Lescano, and J. Paw for assisting with flow cytometry; K. Fasano for preparation of tissues for histological analyses; N. Ali and M. Rosenblum for assistance with harvesting cells from mouse skin; and A. Abbas, M.S. Anderson, F. van Gool, S.A. Villalta, and A.G. DuPage for critical reading of this manuscript. This work was supported by NIH grants R01 AI046643 and UM1 AI-12-059. G.C. was supported by a JDRF fellowship. M.D. was supported by the Helen Hay Whitney Foundation and NIH T32 grant to UCSF (A1007334-23A1). J.A.B. is the A.W. and Mary Margaret Clausen Distinguished Professor in Metabolism and Endocrinology.
Footnotes
AUTHOR CONTRIBUTIONS
M.D. and J.A.B. designed the study; M.D. performed all experiments with assistance from J.Q., W.L.R., M.M.M., D.H., R.Z., and L.T.; G.C. performed bioinformatic analyses; A.M. provided conceptual advice; M.D. and J.A.B. wrote the manuscript.
ACCESSION NUMBERS
The GEO accession number for the RNA-seq data reported in this paper is GSE58998.
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures, one table, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.immuni.2015.01.007.
REFERENCES
- Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat. Immunol. 2003;4:616–623. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- Arvey A, van der Veeken J, Samstein RM, Feng Y, Stamatoyannopoulos JA, Rudensky AY. Inflammation-induced repression of chromatin bound by the transcription factor Foxp3 in regulatory T cells. Nat. Immunol. 2014;15:580–587. doi: 10.1038/ni.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Bucktrout SL, Martinez-Llordella M, Zhou X, Anthony B, Rosenthal W, Luche H, Fehling HJ, Bluestone JA. Self-antigen-driven activation induces instability of regulatory T cells during an inflammatory autoimmune response. Immunity. 2013;39:949–962. doi: 10.1016/j.immuni.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E, Pagany M, Weiner HL, Linington C, Sobel RA, Kuchroo VK. Myelin oligodendrocyte glycoprotein-specific T cell receptor transgenic mice develop spontaneous autoimmune optic neuritis. J. Exp. Med. 2003;197:1073–1081. doi: 10.1084/jem.20021603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird JJ, Brown DR, Mullen AC, Moskowitz NH, Mahowald MA, Sider JR, Gajewski TF, Wang CR, Reiner SL. Helper T cell differentiation is controlled by the cell cycle. Immunity. 1998;9:229–237. doi: 10.1016/s1074-7613(00)80605-6. [DOI] [PubMed] [Google Scholar]
- Campbell DJ, Koch MA. Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nat. Rev. Immunol. 2011;11:119–130. doi: 10.1038/nri2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Aune TM. Dynamic changes in histone-methylation ‘marks’ across the locus encoding interferon-gamma during the differentiation of T helper type 2 cells. Nat. Immunol. 2007;8:723–731. doi: 10.1038/ni1473. [DOI] [PubMed] [Google Scholar]
- Darrasse-Jèze G, Deroubaix S, Mouquet H, Victora GD, Eisenreich T, Yao KH, Masilamani RF, Dustin ML, Rudensky A, Liu K, Nussenzweig MC. Feedback control of regulatory T cell homeostasis by dendritic cells in vivo. J. Exp. Med. 2009;206:1853–1862. doi: 10.1084/jem.20090746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisson S, Darrasse-Jèze G, Litvinova E, Septier F, Klatzmann D, Liblau R, Salomon BL. Continuous activation of autoreactive CD4+ CD25+ regulatory T cells in the steady state. J. Exp. Med. 2003;198:737–746. doi: 10.1084/jem.20030686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, Rudensky AY. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–775. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- Grogan JL, Mohrs M, Harmon B, Lacy DA, Sedat JW, Locksley RM. Early transcription and silencing of cytokine genes underlie polarization of T helper cell subsets. Immunity. 2001;14:205–215. doi: 10.1016/s1074-7613(01)00103-0. [DOI] [PubMed] [Google Scholar]
- Hill JA, Feuerer M, Tash K, Haxhinasto S, Perez J, Melamed R, Mathis D, Benoist C. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27:786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Jacob E, Hod-Dvorai R, Schif-Zuck S, Avni O. Unconventional association of the polycomb group proteins with cytokine genes in differentiated T helper cells. J. Biol. Chem. 2008;283:13471–13481. doi: 10.1074/jbc.M709886200. [DOI] [PubMed] [Google Scholar]
- Keerthivasan S, Aghajani K, Dose M, Molinero L, Khan MW, Venkateswaran V, Weber C, Emmanuel AO, Sun T, Bentrem DJ, et al. beta-Catenin promotes colitis and colon cancer through imprinting of proinflammatory properties in T cells. Sci Transl Med. 2014;6:225ra228. doi: 10.1126/scitranslmed.3007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 2007;8:191–197. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- Koyanagi M, Baguet A, Martens J, Margueron R, Jenuwein T, Bix M. EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in Th1 cells. J. Biol. Chem. 2005;280:31470–31477. doi: 10.1074/jbc.M504766200. [DOI] [PubMed] [Google Scholar]
- Lenschow DJ, Herold KC, Rhee L, Patel B, Koons A, Qin HY, Fuchs E, Singh B, Thompson CB, Bluestone JA. CD28/B7 regulation of Th1 and Th2 subsets in the development of autoimmune diabetes. Immunity. 1996;5:285–293. doi: 10.1016/s1074-7613(00)80323-4. [DOI] [PubMed] [Google Scholar]
- Mandal M, Powers SE, Maienschein-Cline M, Bartom ET, Hamel KM, Kee BL, Dinner AR, Clark MR. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat. Immunol. 2011;12:1212–1220. doi: 10.1038/ni.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Kretschmer K, Frampton GM, Jacobsen ES, Polansky JK, MacIsaac KD, Levine SS, Fraenkel E, von Boehmer H, Young RA. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445:931–935. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Llordella M, Esensten JH, Bailey-Bucktrout SL, Lipsky RH, Marini A, Chen J, Mughal M, Mattson MP, Taub DD, Bluestone JA. CD28-inducible transcription factor DEC1 is required for efficient autoreactive CD4+ T cell response. J. Exp. Med. 2013;210:1603–1619. doi: 10.1084/jem.20122387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao T, Floess S, Setoguchi R, Luche H, Fehling HJ, Waldmann H, Huehn J, Hori S. Plasticity of Foxp3(+) T cells reflects promiscuous Foxp3 expression in conventional T cells but not reprogramming of regulatory T cells. Immunity. 2012;36:262–275. doi: 10.1016/j.immuni.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Morikawa H, Ohkura N, Vandenbon A, Itoh M, Nagao-Sato S, Kawaji H, Lassmann T, Carninci P, Hayashizaki Y, Forrest AR, et al. FANTOM Consortium Differential roles of epigenetic changes and Foxp3 expression in regulatory T cell-specific transcriptional regulation. Proc. Natl. Acad. Sci. USA. 2014;111:5289–5294. doi: 10.1073/pnas.1312717110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura N, Hamaguchi M, Morikawa H, Sugimura K, Tanaka A, Ito Y, Osaki M, Tanaka Y, Yamashita R, Nakano N, et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 2012;37:785–799. doi: 10.1016/j.immuni.2012.09.010. [DOI] [PubMed] [Google Scholar]
- Pierson W, Cauwe B, Policheni A, Schlenner SM, Franckaert D, Berges J, Humblet-Baron S, Schönefeldt S, Herold MJ, Hildeman D, et al. Antiapoptotic Mcl-1 is critical for the survival and niche-filling capacity of Foxp3+ regulatory T cells. Nat. Immunol. 2013;14:959–965. doi: 10.1038/ni.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaphorst FM, Otte AP, van Kemenade FJ, Blokzijl T, Fieret E, Hamer KM, Satijn DPE, Meijer CJLM. Distinct BMI-1 and EZH2 expression patterns in thymocytes and mature T cells suggest a role for Polycomb genes in human T cell differentiation. J. Immunol. 2001;166:5925–5934. doi: 10.4049/jimmunol.166.10.5925. [DOI] [PubMed] [Google Scholar]
- Rosenblum MD, Gratz IK, Paw JS, Lee K, Marshak-Rothstein A, Abbas AK. Response to self antigen imprints regulatory memory in tissues. Nature. 2011;480:538–542. doi: 10.1038/nature10664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X, Treuting P, Siewe L, Roers A, Henderson WR, Jr., et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- Salomon B, Lenschow DJ, Rhee L, Ashourian N, Singh B, Sharpe A, Bluestone JA. B7/CD28 costimulation is essential for the homeostasis of the CD4+CD25+ immunoregulatory T cells that control autoimmune diabetes. Immunity. 2000;12:431–440. doi: 10.1016/s1074-7613(00)80195-8. [DOI] [PubMed] [Google Scholar]
- Samstein RM, Arvey A, Josefowicz SZ, Peng X, Reynolds A, Sandstrom R, Neph S, Sabo P, Kim JM, Liao W, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151:153–166. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smigiel KS, Richards E, Srivastava S, Thomas KR, Dudda JC, Klonowski KD, Campbell DJ. CCR7 provides localized access to IL-2 and defines homeostatically distinct regulatory T cell subsets. J. Exp. Med. 2014;211:121–136. doi: 10.1084/jem.20131142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nat. Rev. Genet. 2007;8:263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- Su I-H, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat. Immunol. 2003;4:124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- Su I-H, Dobenecker M-W, Dickinson E, Oser M, Basavaraj A, Marqueron R, Viale A, Reinberg D, Wülfing C, Tarakhovsky A. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 2005;121:425–436. doi: 10.1016/j.cell.2005.02.029. [DOI] [PubMed] [Google Scholar]
- Tai X, Cowan M, Feigenbaum L, Singer A. CD28 costimulation of developing thymocytes induces Foxp3 expression and regulatory T cell differentiation independently of interleukin 2. Nat. Immunol. 2005;6:152–162. doi: 10.1038/ni1160. [DOI] [PubMed] [Google Scholar]
- Tang Q, Henriksen KJ, Boden EK, Tooley AJ, Ye J, Subudhi SK, Zheng XX, Strom TB, Bluestone JA. Cutting edge: CD28 controls peripheral homeostasis of CD4+CD25+ regulatory T cells. J. Immunol. 2003;171:3348–3352. doi: 10.4049/jimmunol.171.7.3348. [DOI] [PubMed] [Google Scholar]
- Tumes DJ, Onodera A, Suzuki A, Shinoda K, Endo Y, Iwamura C, Hosokawa H, Koseki H, Tokoyoda K, Suzuki Y, et al. The poly-comb protein Ezh2 regulates differentiation and plasticity of CD4(+) T helper type 1 and type 2 cells. Immunity. 2013;39:819–832. doi: 10.1016/j.immuni.2013.09.012. [DOI] [PubMed] [Google Scholar]
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh T-Y, Watford WT, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss JM, Bilate AM, Gobert M, Ding Y, Curotto de Lafaille MA, Parkhurst CN, Xiong H, Dolpady J, Frey AB, Ruocco MG, et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. The Journal of experimental medicine. 2012;209:1723–1742. S1721. doi: 10.1084/jem.20120914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat. Rev. Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Khanna S, Grzenda AL, Sarmento OF, Svingen PA, Lomberk GA, Urrutia RA, Faubion WA., Jr. Polycomb antagonizes p300/CREB-binding protein-associated factor to silence FOXP3 in a Kruppel-like factor-dependent manner. J. Biol. Chem. 2012;287:34372–34385. doi: 10.1074/jbc.M111.325332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, Wu X, Stack EC, Loda M, Liu T, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav M, Louvet C, Davini D, Gardner JM, Martinez-Llordella M, Bailey-Bucktrout S, Anthony BA, Sverdrup FM, Head R, Kuster DJ, et al. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. The Journal of experimental medicine. 2012;209:1713–1722. S1711–1719. doi: 10.1084/jem.20120822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Huynh A, Whitcher G, Chang J, Maltzman JS, Turka LA. An obligate cell-intrinsic function for CD28 in Tregs. J. Clin. Invest. 2013;123:580–593. doi: 10.1172/JCI65013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kinkel S, Maksimovic J, Bandala-Sanchez E, Tanzer MC, Naselli G, Zhang JG, Zhan Y, Lew AM, Silke J, et al. The poly-comb repressive complex 2 governs life and death of peripheral T cells. Blood. 2014;124:737–749. doi: 10.1182/blood-2013-12-544106. [DOI] [PubMed] [Google Scholar]
- Zhou X, Jeker LT, Fife BT, Zhu S, Anderson MS, McManus MT, Bluestone JA. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J. Exp. Med. 2008;205:1983–1991. doi: 10.1084/jem.20080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Bailey-Bucktrout SL, Jeker LT, Penaranda C, Martínez-Llordella M, Ashby M, Nakayama M, Rosenthal W, Bluestone JA. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat. Immunol. 2009;10:1000–1007. doi: 10.1038/ni.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.