Highlights
-
•
HRV-A was associated with more severe ILI symptoms in adults, but not in children.
-
•
Adults with HRV-A had prolonged duration of viral shedding.
-
•
Adults had longer HRV shedding than children.
Keywords: Rhinovirus, HRV genotypes, Viral shedding, Military
Abstract
Background
human rhinovirus (HRV) is a major cause of influenza-like illness (ILI) in adults and children. Differences in disease severity by HRV species have been described among hospitalized patients with underlying illness. Less is known about the clinical and virologic characteristics of HRV infection among otherwise healthy populations, particularly adults.
Objectives
to characterize molecular epidemiology of HRV and association between HRV species and clinical presentation and viral shedding.
Study design
observational, prospective, facility-based study of ILI was conducted from February 2010 to April 2012. Collection of nasopharyngeal specimens, patient symptoms, and clinical information occurred on days 0, 3, 7, and 28. Patients recorded symptom severity daily for the first 7 days of illness in a symptom diary. HRV was identified by RT-PCR and genotyped for species determination. Cases who were co-infected with other viral respiratory pathogens were excluded from the analysis. We evaluated the associations between HRV species, clinical severity, and patterns of viral shedding.
Results
eighty-four HRV cases were identified and their isolates genotyped. Of these, 62 (74%) were >18 years. Fifty-four were HRV-A, 11HRV-B, and 19HRV-C. HRV-C infection was more common among children than adults (59% vs. 10%, P < 0.001). Among adults, HRV-A was associated with higher severity of upper respiratory symptoms compared to HRV-B (P = 0.02), but no such association was found in children. In addition, adults shed HRV-A significantly longer than HRV-C (P trend = 0.01).
Conclusions
among otherwise healthy adults with HRV infection, we observed species-specific differences in respiratory symptom severity and duration of viral shedding.
1. Background
Human rhinovirus (HRV)—the most prevalent respiratory virus—causes up to half of common colds [1], [2] and imposes a significant economic burden [3]. HRV has also been associated with bronchiolitis [4], pneumonia [5], and exacerbation of breathing difficulties in populations with underlying respiratory conditions, including asthma, cystic fibrosis, and chronic obstructive pulmonary disease (COPD) [6], [7], [8]. Prior to the era of molecular testing, rhinovirus was considered to be a relatively mild pathogen of questionable importance. However, with the ability to more readily identify HRV infection and species—including the newly identified species (HRV-C)—evidence is emerging that severity of HRV may be dependent upon the species and/or serotype.
The three species of HRV (A–C) comprise a large group of genetically diverse viruses with more than 150 serotypes [9], [10]. HRV-A and C are associated with more severe clinical manifestations in children [11], [12], [13] and adults [14]. In particular, HRV-C is associated with more severe illness in young children, particularly those with asthma [12], [15], [16] or cystic fibrosis [17]. However, less is known about the epidemiology and clinical characteristics of HRV infection in otherwise healthy populations, especially adults without underlying illness. Moreover, little is known about the persistence of HRV shedding, which may influence duration of symptoms, clinical course, and infectiousness.
2. Objectives
The objective of the study was to understand the full spectrum of HRV disease and species-specific differences in symptom severity, clinical course, and viral shedding among patients with HRV infection in a longitudinal study of influenza-like illness (ILI) among otherwise healthy individuals.
3. Study design
3.1. Overview of the ARIC study
Established in July 2009, the Acute Respiratory Infection Consortium (ARIC) is a multi-site, multi-disciplinary clinical research network for the study of ILI among otherwise healthy military personnel and beneficiaries. At the core of the ARIC is the natural history study, an observational, longitudinal cohort study to determine the etiology, epidemiology, and clinical characteristics of ILI among patients presenting for care at five US-based military treatment facilities (Table 1 ).
Table 1.
Characteristics of HRV patients by species.
| Among rhinovirus positive (n = 84) |
||||||||
|---|---|---|---|---|---|---|---|---|
| Total | HRV-A (n = 54) |
HRV-B (n = 11) |
HRV-C (n = 19) |
Pa | ||||
| N | N | (%) | N | (%) | N | (%) | ||
| Age (years) | ||||||||
| 0–17 | 22 | 9 | (40.9) | 0 | (0.0) | 13 | (59.1) | <0.01 |
| 18–65 | 62 | 45 | (72.6) | 11 | (17.7) | 6 | (9.7) | |
| Sex | ||||||||
| Male | 48 | 31 | (64.6) | 6 | (12.5) | 11 | (22.9) | 1.00 |
| Female | 36 | 23 | (63.9) | 5 | (13.9) | 8 | (22.2) | |
| Study siteb | ||||||||
| SAMHS, San Antonio, TX | 3 | 2 | (66.7) | 0 | (0.0) | 1 | (33.3) | 0.71 |
| NMCSD, San Diego, CA | 13 | 6 | (46.1) | 3 | (23.1) | 4 | (30.8) | |
| NMCP, Portsmouth, VA | 64 | 43 | (67.2) | 8 | (12.5) | 13 | (20.3) | |
| MAMC, Tacoma, WA | 4 | 3 | (75.0) | 0 | (0.0) | 1 | (25.0) | |
| Ethnicity | ||||||||
| Caucasian | 57 | 39 | (68.4) | 6 | (10.5) | 12 | (21.1) | 0.09 |
| African American | 17 | 6 | (35.3) | 4 | (23.5) | 7 | (41.2) | |
| Asian | 4 | 4 | (100) | 0 | (0.0) | 0 | (0.0) | |
| Other | 6 | 5 | (83.3) | 1 | (16.7) | 0 | (0.0) | |
| Current smoker (only for patients >13 years of age)c | ||||||||
| Yes | 17 | 10 | (22.2) | 4 | (36.4) | 3 | (50.0) | 0.21 |
| No | 45 | 35 | (77.8) | 7 | (63.6) | 3 | (50.0) | |
| N/Ac or missing | 22 | 9 | 0 | 13 | ||||
| Smoker in the household | ||||||||
| Yes | 28 | 19 | (67.9) | 3 | (10.7) | 6 | (21.4) | 0.87 |
| No | 49 | 32 | (65.3) | 4 | (8.2) | 13 | (26.5) | |
| Missing | 3 | 4 | 0 | |||||
| Children attending daycare | ||||||||
| Yes | 7 | 3 | (42.9) | 0 | (0.0) | 4 | (57.1) | 1.00 |
| No | 13 | 6 | (46.2) | 0 | (0.0) | 7 | (53.8) | |
| N/Ad or missing | 64 | 45 | 11 | 8 | ||||
| Household member attending daycare | ||||||||
| Yes | 24 | 16 | (66.7) | 2 | (8.3) | 6 | (25.0) | 1.00 |
| No | 51 | 34 | (66.7) | 5 | (9.8) | 12 | (23.5) | |
| Missing | 9 | 4 | 4 | 1 | ||||
| Hospitalization | ||||||||
| Yes | 4 | 2 | (50.0) | 0 | (0.0) | 2 | (50.0) | 0.30 |
| No | 80 | 52 | (65.0) | 11 | (13.7) | 17 | (21.3) | |
P-value of exact test.
SAMHS: San Antonio Military Health System, TX, NMCSD: Naval Medical Center San Diego, CA, NMCP: Naval Medical Center Portsmouth, VA, MAMC: Madigan Army Medical Center, Tacoma, WA.
This question was not available to children under 13 years of age.
This question was not available to adults and was excluded from the analysis.
3.2. Patient population and procedures
From February 2010 to April 2012, we recruited patients aged 0–65 years who presented within 72 h of ILI symptom onset. ILI was defined by fever (temperature ≥ 100.4 °F) accompanied by one of the following respiratory symptoms: cough, sputum production, shortness of breath, chest pain, and/or sore throat. Both inpatient and outpatient subjects were eligible for participation. Patients with the following co-morbidities were excluded: type 1 or 2 diabetes, immunodeficiency, COPD, cystic fibrosis, severe asthma, chronic neuromuscular disease, chronic cardiac disease, or chronic renal disease.
At enrollment, demographic information and clinical symptoms were collected by interview. A nasopharyngeal specimen was collected using a nylon flocked swab (Copan Diagnostics, Corona, CA). Participants returned at days 3 ± 1, 7 ± 2 and 28 ± 7 for collection of clinical symptoms and a nasopharyngeal swab at each visit.
Written informed consent was obtained at enrollment. The study was approved by the Infectious Disease Institutional Review Board of the Uniformed Services University of the Health Sciences (IDCRP-045).
In the current study, we performed retrospective analysis on a sub-population that participated in a prospective study on ILI. We included HRV-positive cases with sufficient nasal swab specimens for HRV genotyping and excluded cases with co-detection of other respiratory viruses (see details in Section 3.4) from February 2010 to April 2012.
3.3. Clinical characteristics and severity measures
The presence and severity of clinical symptoms were recorded at each study visit either by self-report or, in subjects <4 years, by observation using a standardized four-point scale (0: none; 1: mild; 2: moderate; and 3: severe). Symptom grading was explained by research personnel. In addition, patients or their guardians completed symptom diaries for 7 days since ILI onset using the same four-point severity scale. Patients also reported number of days with reduced activity. Potential lower respiratory tract involvement was inferred by abnormality of chest radiograph exam (if done) and physical exam performed by health attendants, including the presence of crackles, egophany, decreased breath sounds, inspiratory wheezing, expiratory wheezing, stridor, rhonchi, accessory muscle breath, dullness to percussion, nasal flaring and grunting. Clinical severity was characterized by composite scores (sum score) of symptoms in four categories: upper respiratory (earache, runny nose, sore throat and sneezing), lower respiratory (cough, breathing difficulty, hoarseness and chest pain), systemic illness (muscle ache, fatigue, headache and chills) and total severity (the above 12 symptoms). The measure was modified from severity scores published by Hayden et al. [18] and was only applied to adults because the measurements were originally designed and verified among adult ILI patients.
3.4. Detection of respiratory virus and HRV genotyping
Nasopharyngeal swabs were placed immediately into viral transport media, frozen at either −70° or −80 °C, and shipped to the Naval Health Research Center (San Diego, CA). HRV was identified using real-time reverse transcription polymerase chain reaction (rtRT-PCR) as previously reported [19]. The presence of other viral respiratory pathogens (i.e., influenza virus, adenovirus (type A–F), respiratory syncytial virus, coronavirus (HKU1, NL63, OC43, and 229E), parainfluenza virus (type 1–4), human metapneumovirus and bocavirus) was assessed by multiplex assays (xTAG Respiratory Viral Panel, Luminex, Austin, TX or PLEX-ID Viral IC Spectrum, Abbott, Chicago, IL). We excluded cases with co-detection from the analysis to avoid possible interference in clinical severity from non-HRV respiratory viruses.
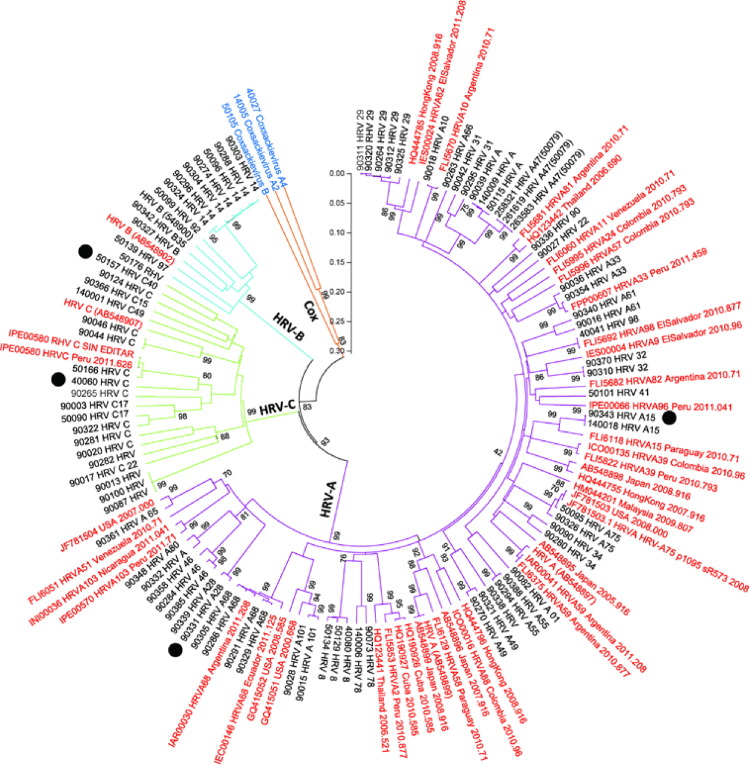
HRV sequencing was targeted toward nucleotide 165–1079 of the VP4/VP2 coding region of the genomic RNA [20], [21], the most commonly studied region of HRV. Genotyping and serotyping was performed by aligning RNA sequences of our samples with standard HRV sequences from the GenBank database and previous reports (Fig. 2 ). Nucleotide sequences from our study samples are available at GenBank (accession number: KF957884-KF957967 and KF957971). Detailed methods of HRV detection, sequencing and genotyping, in addition to standard sequences used in genotyping analysis, are available in the Appendix.
Fig. 2.
Phylogenetic tree of partial HRV VP4/VP2 RNA sequences from 84 study samples. Prototype strains of HRV-A–C from GenBank were included for comparison (Clustal X version 2.0.1). The tree was constructed using the neighbor-joining method in MEGA software (version 5). The statistical significance of the tree topology was tested by bootstrapping (1000 replicas). Pairwise distances between and within the genotypes at the nucleotide level were calculated with Kimura 2 parameters and with Poisson correction at the amino acid level with MEGA software. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Black dot: hospitalized case, sequence name in red: standard sequence, Cox: coxsackievirus.
3.5. Statistical analysis
We first described differences in demographics, geographic location, and potential risk factors by HRV species. Second, we assessed species-specific differences in severity of clinical symptoms (individual and composite), and hospitalization. Symptom data from children were analyzed separately. We performed chi-square tests (exact test, as appropriate) to examine the association between categorical variables. We performed Kruskal–Wallis tests to examine species-specific differences in individual symptom score and composite scores. We then compared symptoms with P value of Kruskal–Wallis tests lower than 0.05 within each HRV species pair (e.g., HRV-A vs. HRV-B) using Wilcoxon rank-sum tests and false discovery rate (FDR) to adjust for multiple comparisons. Last, we compared the pattern of serial HRV detection by performing Cochran–Armitage Trend tests. Detection of HRV at consecutive visits implied ongoing viral shedding during that time period. A two-sided P value lower than 0.05 was considered statistically significant. Analyses were performed using SAS software, Version 9.3 (SAS Institute, Cary, North Carolina).
4. Results
4.1. Detection of HRV species
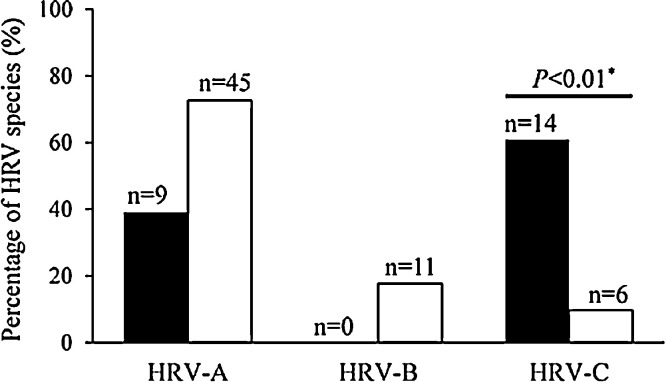
Between February 2010 and April 2012, a total of 160HRV-positive cases were identified. Seventy-six cases were not genotyped due to insufficient nasal swab specimens (n = 64) and/or co-detection with other respiratory viral pathogens (n = 12). The remaining 84HRV-confirmed cases were genotyped, including 62 (74%) adults, 48 (57%) males, 54 (64%) active duty military members. Among the 84 cases, 54 (64%) had HRV-A, 11 (13%) had HRV-B, and 19 (23%) had HRV-C. HRV-C was detected more frequently among children (13/22, 59%) than adults (6/62, 10%; P < 0.01, Fig. 1 ). The distribution of HRV species did not differ by other demographic characteristics (Table 1). The phylogenetic tree revealed significant genetic diversity in the HRV VP4/VP2 regions (Fig. 2). Forty-four serotypes were identified. There was no genetic correlation or cluster of HRV serotypes among hospitalized cases (Fig. 2), children attending day care centers, and/or adults living in dorms.
Fig. 1.
Proportion of HRV species, by age group.
Black bar: children, white bar: adults.
Note: *P-value of chi-square test on the proportion of patients with HRV-C infection between children and adults.
4.2. HRV species and clinical severity
All cases reported or were observed for symptom severity at enrollment. Adult patients with HRV-A and HRV-C had higher composite scores of lower respiratory and total symptoms compared to those with HRV-B, although the differences were not statistically significant (Table 2 ). No other differences in clinical severity, including individual symptoms, abnormal chest X-ray findings, abnormal physical exams and duration of reduced activity, were observed at enrollment among either adults or children (Table 2). A total of four patients (5%, one child and three adults) were hospitalized: two with HRV-A and two with HRV-C. All four patients survived.
Table 2.
HRV species and clinical symptoms and severity by age group.
| Children (age <18 years old) (n = 22) |
Adults (age ≥18 years old) (n = 62) |
||||||
|---|---|---|---|---|---|---|---|
| HRV-A (n = 9) |
HRV-C (n = 13) |
Pa | HRV-A (n = 45) |
HRV-B (n = 11) |
HRV-C (n = 6) |
Pb | |
| Med N (%)d | Med N (%)d | Med N (%)d | Med N (%)d | Med N (%)d | |||
| Upper respiratory symptoms | |||||||
| Sneezing | 0, 4 (44.4) | 1, 9 (69.2) | 0.59 | 1, 34 (75.6) | 1, 7 (63.6) | 1.5, 4 (66.7) | 0.85 |
| Sore throatc | 0, 3 (37.5) | 0, 1 (10.0) | 0.22 | 2, 41 (91.1) | 2, 9 (81.8) | 3, 6 (100) | 0.08 |
| Earachec | 0, 1 (11.1) | 0, 1 (10.0) | 0.87 | 1, 24 (53.3) | 0, 3 (27.3) | 1, 4 (66.7) | 0.17 |
| Runny nose | 2, 9 (100) | 2, 11 (84.6) | 0.86 | 2, 41 (91.1) | 3, 10 (90.9) | 2, 5 (83.3) | 0.85 |
| Cough | 2, 9 (100) | 2, 13 (100) | 1.00 | 2, 44 (97.8) | 1, 10 (90.9) | 2, 6 (100) | 0.17 |
| Hoarseness | 0, 4 (44.4) | 0, 2 (15.4) | 0.12 | 1, 35 (77.8) | 1, 8 (72.7) | 2, 5 (83.3) | 0.81 |
| Lower respiratory symptoms | |||||||
| Shortness of breath | 0, 1 (11.1) | 0, 2 (15.4) | 0.74 | 1, 30 (66.7) | 0, 4 (36.4) | 0, 2 (33.3) | 0.12 |
| Chest painc | 0, 0 (0.0) | 0, 0 (0.0) | NA | 0, 22 (48.9) | 0, 5 (45.5) | 0, 2 (33.3) | 0.73 |
| Gastroenteritis symptoms | |||||||
| Decreased appetite | 2, 7 (77.8) | 2, 12 (92.3) | 0.19 | 2, 30 (66.7) | 2, 7 (63.6) | 2, 6 (100) | 0.47 |
| Vomiting | 0, 1 (11.1) | 0, 3 (23.1) | 0.46 | 0, 9 (20.0) | 0, 3 (27.3) | 0, 0 (0.0) | 0.43 |
| Diarrhea | 0, 1 (11.1) | 0, 1 (7.7) | 0.84 | 0, 11 (24.4) | 0, 5 (45.5) | 0, 2 (33.3) | 0.57 |
| Abdominal painc | 0, 1 (11.1) | 0, 0 (0.0) | 0.26 | 0, 13 (28.9) | 0, 5 (45.5) | 1.5, 4 (66.7) | 0.06 |
| Systemic symptoms | |||||||
| Headachec | 0, 2 (22.2) | 0, 0 (0.0) | 0.10 | 2, 36 (80.0) | 2, 9 (81.8) | 2.5, 6 (100) | 0.33 |
| Red eyes | 0, 4 (44.4) | 0, 6 (46.2) | 0.82 | 0, 21 (46.7) | 1, 6 (54.5) | 1.5, 4 (66.7) | 0.51 |
| Chills | 0, 0 (0.0) | 0, 1 (7.7) | 0.41 | 1, 37 (82.2) | 1, 9 (81.8) | 2, 4 (66.7) | 0.68 |
| Nauseac | 0, 0 (0.0) | 0, 1 (7.7) | 0.37 | 0, 21 (46.7) | 1, 7 (63.6) | 1, 4 (66.7) | 0.53 |
| Joint painc | 0, 0 (0.0) | 0, 0 (0.0) | NA | 2, 34 (75.6) | 2, 10 (90.9) | 2, 5 (83.3) | 0.09 |
| Muscle achesc | 0, 0 (0.0) | 0, 0 (0.0) | NA | 2, 35 (77.8) | 2, 9 (81.8) | 2, 4 (66.7) | 0.75 |
| Fatigue | 2, 6 (66.7) | 2, 9 (69.2) | 1.00 | 2, 43 (95.6) | 2, 10 (90.9) | 2, 5 (83.3) | 0.83 |
| Dizzinessc | 0, 0 (0.0) | 0, 0 (0.0) | NA | 0, 21 (46.7) | 0, 5 (45.5) | 1, 4 (66.7) | 0.61 |
| Potential lower respiratory tract involvement | |||||||
| Presence of self-reported LRT symptoms | 1 (11.1) | 2 (14.3) | 1.00f | 33 (73.3) | 6 (54.5) | 3 (50.0) | 0.32f |
| Abnormal chest X-ray or PE finding | 1 (16.7) | 4 (30.8) | 0.48f | 5 (15.6) | 0 (0.0) | 0 (0.0) | 0.74f |
| Did not have PE | 3 | 1 | 13 | 2 | 2 | ||
| Fever (>100.4 °F) | 8 (88.9) | 13 (92.9) | 1.00f | 11 (24.4) | 2 (18.2) | 2 (33.3) | 0.79f |
| Hospitalizationd | 0 (0.0) | 1 (7.1) | 0.41f | 2 (4.4) | 0 (0.0) | 1 (16.7) | 0.30f |
| Composite severity score (median (range), only available for adult ILI patients) | |||||||
| Upper respiratory symptoms | 6 (6, 8) | 6 (3, 7) | 7 (6, 10) | 0.15 | |||
| Lower respiratory symptoms | 5 (3, 7) | 4 (2, 6) | 4.5 (3, 7) | 0.42 | |||
| Systemic symptoms | 6 (5, 9) | 7 (4, 9) | 7.5 (5, 9) | 0.89 | |||
| Total symptoms | 18 (14, 22) | 15 (13, 18) | 18 (16, 22) | 0.41 | |||
| Days of reduced activitye | 2 (2–3) | 2 (2–2) | 0.69 | 2 (1–3) | 2 (1–2) | 2 (1–3) | 0.56 |
NA: not available.
P-value of chi-square test comparing score of HRV-B with HRV-A/C in adults was 0.02.
P-value of Wilcoxon rank-sum test comparing symptom scores and days of reduced activity between children with HRV-A and HRV-C.
P-value of Kruskal–Wallis test comparing symptom scores and days of reduced activity across three HRV species in adults.
One child with HRV-A and three with HRV-C did not respond to this question.
Med represents median severity score of the designated symptom. Number and proportion of patients reporting the designated symptom and hospitalization in each category.
Average of three respiratory symptoms (sore throat, cough and shortness of breath) and systemic symptoms (chills, muscle ache, headache, fatigue, and dizziness) with their standard deviations in parentheses.
P-value of Fisher’s exact test comparing the proportion of patients with the outcome across HRV species.
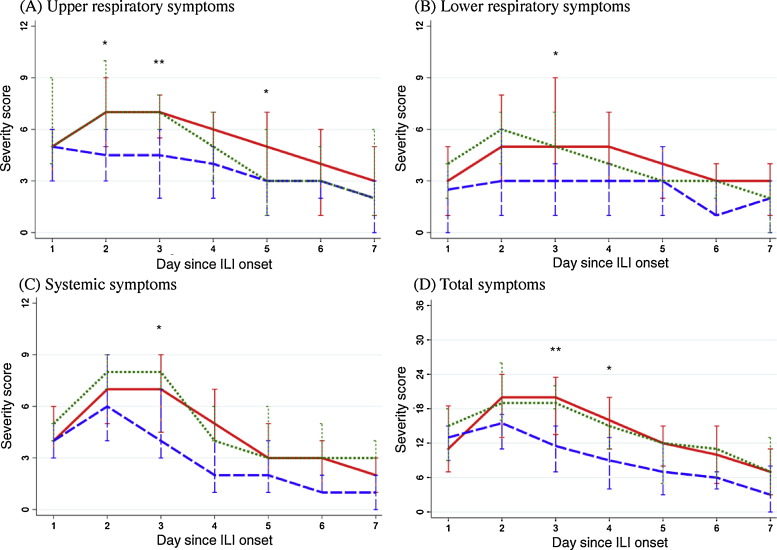
The majority (89%) of adults reported daily symptom scores through study day 7. Daily composite scores peaked on the second day of illness and decreased thereafter, regardless of HRV genotype and symptom category (Fig. 3 ). Patients with HRV-A and HRV-C tended to report higher composite scores compared to those with HRV-B. Species-specific differences were detected on day 3 of illness (upper respiratory symptoms, P = 0.02; total symptoms, P = 0.03, Fig. 3). Pair-wise comparison among three HRV genotypes on day 3 showed that patients with HRV-A had higher scores than HRV-B (median score of HRV-A vs. HRV-B: 7 vs. 4.5, P = 0.02 for upper respiratory; 20 vs. 11.5, P = 0.04 for total symptom scores with FDR adjustment), while no significant differences were found between those with HRV-B and HRV-C, presumably due to small numbers.
Fig. 3.
Median and interquartile range of composite symptom scores among adults, by HRV genotypes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Red solid line: HRV-A; blue dashed line: HRV-B; green dotted line: HRV-C.
**P ≤ 0.05, *P > 0.05 & P ≤ 0.1 for comparison across three HRV genotypes.
4.3. HRV species and persistent viral shedding
Fifty (60%) of the 84 patients completing all four study visits were included in the analysis of viral shedding. Eleven (22%) were positive for HRV on day 0 only, 25 (50%) on days 0 and 3, 12 (24%) on days 0, 3, and 7. Only two (4%) were positive for HRV at all four visits (Table 3 ).
Table 3.
Patterns of viral shedding among HRV cases who completed 4 study visit, by species and age group.
| 0–17 years of age (n = 12) |
18–65 years of age (n = 38) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HRV detection at consecutive visits | HRV-A | HRV-C | Total |
Pa HRV-A vs. -C |
HRV-A | HRV-B | HRV-C | Total |
Pb HRV-A vs. -B |
Pb HRV-A vs. -C |
Pb HRV-B vs. -C |
Pc adult vs. child | |
| Day 0 | 3 | 4 | 7 | 0.39 | 1 | 1 | 2 | 4 | 0.35 | 0.01 | 0.25 | <0.01 | |
| Day 0 and 3 | 2 | 2 | 4 | 16 | 3 | 2 | 21 | ||||||
| Day 0, 3, and 7 | 0 | 0 | 0 | 9 | 3 | 0 | 12 | ||||||
| Day 0, 3, 7 and 28 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | ||||||
| Total | 5 | 7 | 12 | 27 | 7 | 4 | 38 | ||||||
Note: numbers of HRV detection in group 3 (Day 0, 3, and 7) and group 4 (Day 0, 3, 7, and 28) were combined in the Cochran–Armitage Trend test given the small number.
P-value of Cochran–Armitage Trend test comparing HRV shedding pattern at 4 study visits between HRV-A and HRV-C among children.
P-value of Cochran–Armitage Trend test comparing HRV shedding pattern at 4 study visits between designated HRV species among adults with FDR adjustment.
P-value of Cochran–Armitage Trend test comparing HRV shedding pattern (regardless of HRV species) at 4 study visits between children and adults.
Prolonged detection of HRV was more common among adults than children. Only four adults (11%) had detection of HRV solely at enrollment, in contrast to 7 (59%) children; thirteen adults (13/38, 34%) had evidence of HRV shedding on or after day 7, while only one child (1/12, 8%) had evidence of HRV shedding on or after day 7 (P trend < 0.01, by visit category, Table 3). When examined by species, HRV-C was only detectable at day 0 and 3, while HRV-A and HRV-B was detected on day 7 and later (HRV-A vs. HRV-C, P trend = 0.01 with FDR adjustment). We did not find different shedding patterns between children with HRV-A and HRV-C—the majority of them were tested positive only on enrollment or on enrollment and day 3.
5. Discussion
Our study of the epidemiology and clinical characteristics of HRV infection among healthy adults and children without underlying medical conditions differs from previous reports that described infection among patients with asthma, COPD, or cystic fibrosis. We excluded cases of HRV in whom other respiratory viral pathogens were detected. The prospective, longitudinal design of the study allowed us to evaluate both clinical and virologic (i.e., shedding) characteristics of HRV infection in these patients and to minimize bias of patient-recalled symptoms using a standardized severity score.
The prevalence of HRV species differed by age. HRV-C infection was more common in children than adults, while the majority of adults were infected with HRV-A. Other groups have described high prevalence of HRV-C in children, regardless of clinical status [11], [12], [22]. However, information on the frequency of HRV-C detection in adults with respiratory illness is less consistent, ranging from 4.8% to 30% [7], [14], [23], [24], [25], regardless of patients’ comorbid status. It is unclear why such variation existed in adult population. While our data, along with others’, suggest age differences in the prevalence of HRV species, there are as yet no data explaining reasons of these differences. It may be that a longer history of exposure and disease in adults versus children may account for the diversity of clinical presentation and the patterns of viral shedding [26], [27], [28].
We found adult patients with HRV-A had higher composite severity scores compared to those with HRV-B in the early (i.e., day 3 of illness) stages of illness. Similar trends were also found among HRV-C adult patients, although the finding was not statistically significant. In addition, all four hospitalized cases had either HRV-A or HRV-C. The findings were in accordance with the literature—HRV-A and/or HRV-C are associated with more severe manifestations, including severe upper respiratory illness and pneumonia [14], [29]. While symptoms related to HRV infection are generally mild among patients without underlying health conditions, such species-specific variation implies differences in viral pathogenesis and/or host immunity. We were unable to confirm such differences using clinical data collected at enrollment. Differences in the timing of presentation and/or enrollment may have contributed to the result. Patients were enrolled within 72 h after illness, but this relied upon their initial presentation to the facility. Even within such a short period, subtle differences in the severity of clinical symptoms may have been masked.
We did not find that HRV-C or HRV-A was more severe in children, unlike previous reports [12], [13], [24], [30], [31]. Since we restricted enrollment to children without underlying conditions, it is possible that HRV-C is not more severe than other HRV species in this group. This finding adds context to the literature regarding HRV-C in children, suggesting that the observed higher severity of some HRV infections may be confined to specific risk groups.
Persistent viral shedding may indicate different levels of virulence, host immune response and infectiousness [32], [33]. In sharp contradistinction to influenza infection where children shed virus longer than adults [34], [35], [36], our data suggest that adults shed HRV longer than children. In addition, we observed adults to shed HRV-A longer than HRV-C. Longer shedding of HRV-A among adults may be correlated to more severe clinical presentation with such species. Further studies are needed to understand interaction of host response and viral virulence. To the best of our knowledge, this is the first study to describe species-specific and age-specific differences in the patterns of viral shedding during natural HRV infection.
Our study has several limitations. First, the small sample size, especially among children (n = 23), may have precluded us from identifying an association between HRV species and clinical outcomes. Second, the study population was comprised of individuals who presented to healthcare facilities with ILI. This may not reflect the epidemiology of HRV infection among individuals in the community who do not seek medical care. In addition, the majority of HRV cases in this study were from a single site. This may limit the generalizability of our results to different geographical regions. On the other hand, our study population also had a relatively small proportion of hospitalized patients and may not be fully representative of severe cases of HRV infection. Third, symptom diaries were not completed by all patients. It is possible that some participants suspended use of their diaries when their symptoms subsided. To address this, we performed a sensitivity analysis using data only from participants who completed the diary per protocol (n = 42). The analysis confirmed the results that adult patients with HRV-A had higher composite severity scores than those with HRV-B (data not shown). Finally, shedding of virus was measured by detection of RNA, not using cell culture. The fastidious nature of HRV has led to the use of PCR as the main tool for detection, so we feel this is acceptable, though it is only inferential of infectious particles. In addition, the duration of shedding was measured by consecutive detection of HRV in nasal swabs while genotyping was performed for only the baseline specimen. It is possible that the HRV detected at subsequent visits could have been new infections.
In conclusion, this study lends new insight into the role of HRV with respect to species-specific patterns of clinical symptom patterns and viral shedding in otherwise healthy children and adults with ILI. Our findings may inform development of strategies for prevention of HRV disease and transmission. Moreover, studies are needed to understand host-pathogen interactions across HRV species in different age groups, which will ultimately lead to improved clinical management of severe HRV infection.
Funding
The study was funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, under Inter-Agency Agreement Y1-AI-5072, and the Armed Forces Health Surveillance Center, Global Emerging Infections Surveillance and Response System.
Competing interests
None to report.
Ethical approval
The study was approved by the Infectious Disease Institutional Review Board of the Uniformed Services University of the Health Sciences (IDCRP-045).
Key points
HRV species A and C were associated with more severe respiratory and systemic symptoms in adults, but not in children. Adults with HRV-A had prolonged duration of viral shedding.
Disclaimer
The views expressed are those of the author(s) and do not necessarily reflect the official views of the Uniformed Services University of the Health Sciences, the US Navy, the US Army, the US Air Force, or the US Department of Defense.
Acknowledgements
We acknowledge the contributions of the ARIC team of clinical research coordinators, clinical site managers, data managers and administrative support personnel to the success of this project. We are grateful to Vidal Felices and Victoria Espejo for their assistance in HRV genotyping and sequencing.
Appendix.
Details regarding HRV detection, genotyping and phylogenetic analysis
HRV detection
Human rhinovirus was screened from clinical samples using real-time reverse transcription-PCR (RT-PCR). Rhinovirus primers and probe from [37]: F: 5′ CPX GCC ZGC GTG GC 3′; R: 5′ GAA ACA CGG ACA CCC AAA GTA 3′; P: 5′ FAM-TCC TCC GGC CCC TGA ATG YGG C-BHQ1 3′ target the viral 5′ noncoding region. This was part of a multiplex assay, created in house to generate a research use only test. Amplification was performed with an ABI 7500 with reverse transcription at 50° for 30 min; activation at 95° for 10 min; followed by 40 cycles of amplification at 95° for 15 s and 55° for 30 s. Reactions were carried out in 20-μl reaction volumes with 2.5-μl Nuclease-free water, 3.0-μl Primer/probe mix, 2.0-μl PATH-ID Real Time Multiplex Enzyme, 12.5-μl PATH-ID Real time Multiplex 2X Mix. Primer mix uses a 0.8-μM concentration of rhinovirus primers and a 0.2-μM concentration of rhinovirus probe.
HRV genotyping and phylogenetic analysis
DNA Extraction and PCR
We extracted and amplified viral RNA from 140 μl of the viral transport media using a viral RNA kit (QIAamp, Qiagen®). This was performed in 22 μl of reaction mixture consisting of 2.2 μl of nuclease-free water, 3 μl of MgSO4, (5.0 X), 15 μl of 2x reaction mix (Super Script III One-Step RT-PCR System) with platinum (Taq High Fidelity kit), 0.6 μl 20 μM concentrations of the sense and antisense primers, 0.6 μl Enzyme Mix and 8 μl of template. HRV and coxsackievirus detection was performed by semi-nested reverse transcription-PCR (RT-PCR) targeting nucleotides 165–1079 of the genomic RNA. We used P1-1 HRV (CAAGCACTTCTGTYWCCCC) and 9565_R HRV (GCATCNGGYARYTTCCACCACCAICC) [38], [39], [40]. The amplification was carried out in a thermocycler 7700 (Applied Biosystems). Cycling conditions included a reverse transcription step at 50 °C for 30 min and 95 °C for 15 min followed by 45 PCR cycles: 94 °C for 30 s; 55 °C for 30 s; 72 °C for 90 s; and final incubation for 72 °C for 10 min. The amplified products of 915 bp were analyzed by electrophoresis on an agarose 2% gel. PCR products were purified with Centri-Seps Columns (Princeton Separations, Inc.). Purified products were directly used for sequencing of viral nucleic acids from clinical specimens.
Sequencing and phylogenetic analyses
The VP4/VP2 coding region of all HRV positive samples obtained were sequenced and included in the following phylogenetic analyses. This region was selected for sequencing because VP4/VP2 is the most commonly studied region of HRV. For direct sequencing of viral nucleic acids from clinical specimens, gene fragments were amplified and sequenced with the use of a Big Dye terminator cycle sequencing kit (version 3.1, Applied Biosystems) and internal primers Generic F HRV (AGCCTGCGTGGCKGCC) and NCR2HRV (ACTACTTTGGGTGTCCGTGTTTC) on a Genetic Analyzer system (version 3130xL, Applied Biosystems). Nucleotide sequences of PCR products (n = 441 nt) were analyzed by sequencing using Sequencher and BioEdit (version 7.0.0, Isis Pharmaceuticals, Inc.) software and then aligned with the CLUSTAL X version 2.0.1 software to compare with HRV sequences from the GenBank database. Phylogenetic trees were constructed by the neighbor-joining method in MEGA software (version 5). The statistical significance of the tree topology was tested by bootstrapping (1000 replicas). Pairwise distances between and within the genotypes at the nucleotide level were calculated with Kimura 2 parameters and with Poisson correction at the amino acid level with MEGA software. Nucleotide sequences from our study samples are available at GenBank (accession number: KF957884-KF957967 and KF957971).
Accession number of standard HRV sequences used in the study
| Reference sequence | GenBank accession # | Reference |
|---|---|---|
| FPP00607_HRVA33_Peru_2011.459 | [41] | |
| FLI5996_HRVA57_Colombia_2010.793 | [41] | |
| IES00004_HRVA9_ElSalvador_2010.96 | JX129407.1 | [41] |
| FLI6060_HRVA11_Venezuela_2010.71 | [41] | |
| FLI5995_HRVA24_Colombia_2010.793 | [41] | |
| FLI5682_HRVA82_Argentina_2010.71 | [41] | |
| IPE00066_HRVA96_Peru_2011.041 | [41] | |
| FLI5670_HRVA10_Argentina_2010.71 | JX129395.1 | [41] |
| FLI5681_HRVA81_Argentina_2010.71 | [41] | |
| HQ123442_Thailand_2006.690 | HQ123442 | [42] |
| AB548895_Japan_2005.916 | AB548895.1 | [43] |
| HM044201_Malaysia_2009.807 | HM044201 | [44] |
| IAR00041_HRVA59_Argentina_2011.208 | [41] | |
| FLI6375_HRVA59_Argentina_2010.877 | JX129393.1 | [41] |
| HRV_A_ (AB548897) | AB548897 | [43] |
| JF781503_USA_2008.000 | JF781503 | [45] |
| FLI5692_HRVA98_ElSalvador_2010.877 | JX129408 | [41] |
| HQ444785_HongKong_2008.916 | HQ444785 | [46] |
| IES00024_HRVA62_ElSalvador_2011.208 | [41] | |
| AB548898_Japan_2008.916 | AB548898 | [43] |
| HQ444755_HongKong_2007.916 | HQ444755 | [46] |
| ICO00135_HRVA39_Colombia_2010.96 | [41] | |
| FLI5822_HRVA39_Peru_2010.793 | [41] | |
| FLI6118_HRVA15_Paraguay_2010.71 | [41] | |
| HQ444796_HongKong_2008.916 | HQ444796 | [46] |
| ICO00016_HRVA88_Colombia_2010.96 | [41] | |
| AB548896_Japan_2007.916 | AB548896 | [43] |
| FLI6129_HRVA58_Paraguay_2010.71 | [41] | |
| AB548899_Japan_2008.916 | AB548899 | [43] |
| HQ190927_Cuba_2010.585 | HQ190927 | [47] |
| FLI5853_HRVA12_Peru_2010.877 | [41] | |
| HQ190926_Cuba_2010.585 | HQ190926 | [47] |
| HQ123441_Thailand_2006.521 | HQ123441 | [42] |
| IEC00146_HRVA68_Ecuador_2011.125 | JX129404.1 | [41] |
| IAR00030_HRVA68_Argentina_2011.208 | [41] | |
| JF781504_USA_2007.000 | JF781504 | [41] |
| FLI6051_HRVA51_Venezuela_2010.71 | JX129434.1 | [41] |
| IPE00570_HRVA103_Peru_2011.71 | [41] | |
| INI00036_HRVA103_Nicaragua_2011.041 | [41] | |
| GQ415052_USA_2008.585 | GQ415052 | [48] |
| GQ415051_USA_2000.668 | GQ415051 | [48] |
| HRV_B_ (AB548900) | AB548900 | [43] |
| HRV_B_ (AB548902) | AB548902 | [43] |
| HRV_C_ (AB548907) | AB548907 | [43] |
| IPE00580_HRVC_Peru_2011.626 | [41] |
References
- 1.Makela M.J., Puhakka T., Ruuskanen O. Viruses and bacteria in the etiology of the common cold. J. Clin. Microbiol. 1998;36:539–542. doi: 10.1128/jcm.36.2.539-542.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monto A.S. Epidemiology of viral respiratory infections. Am. J. Med. 2002;112(Suppl. 6A):4S–12S. doi: 10.1016/s0002-9343(01)01058-0. [DOI] [PubMed] [Google Scholar]
- 3.Fendrick A.M., Monto A.S., Nightengale B., Sarnes M. The economic burden of non-influenza-related viral respiratory tract infection in the United States. Arch. Intern. Med. 2003;163:487–494. doi: 10.1001/archinte.163.4.487. [DOI] [PubMed] [Google Scholar]
- 4.Henquell C., Mirand A., Deusebis A.L. Prospective genotyping of human rhinoviruses in children and adults during the winter of 2009–2010. J. Clin. Virol. 2012;53:280–284. doi: 10.1016/j.jcv.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Lieberman D., Shimoni A., Shemer-Avni Y., Keren-Naos A., Shtainberg R., Lieberman D. Respiratory viruses in adults with community-acquired pneumonia. Chest. 2010;138:811–816. doi: 10.1378/chest.09-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denlinger L.C., Sorkness R.L., Lee W.M. Lower airway rhinovirus burden and the seasonal risk of asthma exacerbation. Am. J. Respir. Crit. Care Med. 2011;184:1007–1014. doi: 10.1164/rccm.201103-0585OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flight W.G., Bright-Thomas R.J., Tilston P. Incidence and clinical impact of respiratory viruses in adults with cystic fibrosis. Thorax. 2013 doi: 10.1136/thoraxjnl-2013-204000. [DOI] [PubMed] [Google Scholar]
- 8.McManus T.E., Marley A.M., Baxter N. Respiratory viral infection in exacerbations of COPD. Respir. Med. 2008;102:1575–1580. doi: 10.1016/j.rmed.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palmenberg A.C., Spiro D., Kuzmickas R. Sequencing and analyses of all known human rhinovirus genomes reveal structure and evolution. Science. 2009;324:55–59. doi: 10.1126/science.1165557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bochkov Y.A., Gern J.E. Clinical and molecular features of human rhinovirus C. Microbes Infect. 2012;14:485–494. doi: 10.1016/j.micinf.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee W.M., Lemanske R.F., Jr., Evans M.D. Human rhinovirus species and season of infection determine illness severity. Am. J. Respir. Crit. Care Med. 2012;186:886–891. doi: 10.1164/rccm.201202-0330OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwane M.K., Prill M.M., Lu X. Human rhinovirus species associated with hospitalizations for acute respiratory illness in young US children. J. Infect. Dis. 2011;204:1702–1710. doi: 10.1093/infdis/jir634. [DOI] [PubMed] [Google Scholar]
- 13.Linsuwanon P., Payungporn S., Samransamruajkit R. High prevalence of human rhinovirus C infection in Thai children with acute lower respiratory tract disease. J. Infect. 2009;59:115–121. doi: 10.1016/j.jinf.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiang Z., Gonzalez R., Wang Z. Human rhinoviruses in Chinese adults with acute respiratory tract infection. J. Infect. 2010;61:289–298. doi: 10.1016/j.jinf.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calvo C., Casas I., Garcia-Garcia M.L. Role of rhinovirus C respiratory infections in sick and healthy children in Spain. Pediatr. Infect. Dis. J. 2010;29:717–720. doi: 10.1097/INF.0b013e3181d7a708. [DOI] [PubMed] [Google Scholar]
- 16.Mak R.K., Tse L.Y., Lam W.Y., Wong G.W., Chan P.K., Leung T.F. Clinical spectrum of human rhinovirus infections in hospitalized Hong Kong children. Pediatr. Infect. Dis. J. 2011;30:749–753. doi: 10.1097/INF.0b013e31821b8c71. [DOI] [PubMed] [Google Scholar]
- 17.de Almeida M.B., Zerbinati R.M., Tateno A.F., et al. Rhinovirus C. and respiratory exacerbations in children with cystic fibrosis. Emerging Infect. Dis. 2010;16:996–999. doi: 10.3201/eid1606.100063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayden F.G., Fritz R., Lobo M.C., Alvord W., Strober W., Straus S.E. Local and systemic cytokine responses during experimental human influenza A virus infection: relation to symptom formation and host defense. J. Clin. Invest. 1998;101:643–649. doi: 10.1172/JCI1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu X., Holloway B., Dare R.K. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J. Clin. Microbiol. 2008;46:533–539. doi: 10.1128/JCM.01739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang T., Wang W., Bessaud M. Evidence of recombination and genetic diversity in human rhinoviruses in children with acute respiratory infection. PLoS One. 2009;4:e6355. doi: 10.1371/journal.pone.0006355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savolainen C., Blomqvist S., Mulders M.N., Hovi T. Genetic clustering of all 102 human rhinovirus prototype strains: serotype 87 is close to human enterovirus 70. J. Gen. Virol. 2002;83:333–340. doi: 10.1099/0022-1317-83-2-333. [DOI] [PubMed] [Google Scholar]
- 22.Lauinger I.L., Bible J.M., Halligan E.P. Patient characteristics and severity of human rhinovirus infections in children. J. Clin. Virol. 2013;58:216–220. doi: 10.1016/j.jcv.2013.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fry A.M., Lu X., Olsen S.J. Human rhinovirus infections in rural Thailand: epidemiological evidence for rhinovirus as both pathogen and bystander. PLoS One. 2011;6:e17780. doi: 10.1371/journal.pone.0017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau S.K., Yip C.C., Lung D.C. Detection of human rhinovirus C in fecal samples of children with gastroenteritis. J. Clin. Virol. 2012;53:290–296. doi: 10.1016/j.jcv.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe A., Carraro E., Kamikawa J., Leal E., Granato C., Bellei N. Rhinovirus species and their clinical presentation among different risk groups of non-hospitalized patients. J. Med. Virol. 2010;82:2110–2115. doi: 10.1002/jmv.21914. [DOI] [PubMed] [Google Scholar]
- 26.Bochkov Y.A., Palmenberg A.C., Lee W.M. Molecular modeling, organ culture and reverse genetics for a newly identified human rhinovirus C. Nat. Med. 2011;17:627–632. doi: 10.1038/nm.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McErlean P., Shackelton L.A., Andrews E. Distinguishing molecular features and clinical characteristics of a putative new rhinovirus species, human rhinovirus C (HRVC) PLoS One. 2008;3:e1847. doi: 10.1371/journal.pone.0001847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wark P.A., Grissell T., Davies B., See H., Gibson P.G. Diversity in the bronchial epithelial cell response to infection with different rhinovirus strains. Respirology. 2009;14:180–186. doi: 10.1111/j.1440-1843.2009.01480.x. [DOI] [PubMed] [Google Scholar]
- 29.Lau S.K., Yip C.C., Lin A.W. Clinical and molecular epidemiology of human rhinovirus C in children and adults in Hong Kong reveals a possible distinct human rhinovirus C subgroup. J. Infect. Dis. 2009;200:1096–1103. doi: 10.1086/605697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller E.K., Khuri-Bulos N., Williams J.V. Human rhinovirus C associated with wheezing in hospitalised children in the middle east. J. Clin. Virol. 2009;46:85–89. doi: 10.1016/j.jcv.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller E.K., Edwards K.M., Weinberg G.A. A novel group of rhinoviruses is associated with asthma hospitalizations. J. Allergy Clin. Immunol. 2009;123:98–104. doi: 10.1016/j.jaci.2008.10.007. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lau L.L., Ip D.K., Nishiura H. Heterogeneity in viral shedding among individuals with medically attended influenza A virus infection. J. Infect. Dis. 2013;207:1281–1285. doi: 10.1093/infdis/jit034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walsh E.E., Peterson D.R., Kalkanoglu A.E., Lee F.E., Falsey A.R. Viral shedding and immune responses to respiratory syncytial virus infection in older adults. J. Infect. Dis. 2013;207:1424–1432. doi: 10.1093/infdis/jit038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chartrand C., Leeflang M.M., Minion J., Brewer T., Pai M. Accuracy of rapid influenza diagnostic tests: a meta-analysis. Ann. Intern. Med. 2012;156:500–511. doi: 10.7326/0003-4819-156-7-201204030-00403. [DOI] [PubMed] [Google Scholar]
- 35.Fiore A.E., Fry A., Shay D. Antiviral agents for the treatment and chemoprophylaxis of influenza—recommendations of the Advisory Committee on Immunization Practices (ACIP) MMWR Recomm. Rep. 2011;60:1–24. [PubMed] [Google Scholar]
- 36.Suess T., Remschmidt C., Schink S.B. Comparison of shedding characteristics of seasonal influenza virus (sub) types and influenza A(H1N1) pdm09; Germany, 2007–2011. PLoS One. 2012;7:e51653. doi: 10.1371/journal.pone.0051653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu X., Holloway B., Dare R.K. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J. Clin. Microb. 2008;46:533–539. doi: 10.1128/JCM.01739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang T., Wang W., Bessaud M. Evidence of recombination and genetic diversity in human rhinoviruses in children with acute respiratory infection. PLoS One. 2009;4:e6355. doi: 10.1371/journal.pone.0006355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Savolainen C., Blomqvist S., Mulders M.N., Hovi T. Genetic clustering of all 102 human rhinovirus prototype strains: serotype 87 is close to human enterovirus 70. J. Gen. Virol. 2002;83:333–340. doi: 10.1099/0022-1317-83-2-333. [DOI] [PubMed] [Google Scholar]
- 40.Laine P., Savolainen C., Blomqvist S., Hovi T. Phylogenetic analysis of human rhinovirus capsid protein VP1 and 2A protease coding sequences confirms shared genus-like relationships with human enteroviruses. J. Gen. Virol. 2005;86:697–706. doi: 10.1099/vir.0.80445-0. [DOI] [PubMed] [Google Scholar]
- 41.Garcia J., Espejo V., Nelson M. Human rhinoviruses and enteroviruses in influenza-like illness in Latin America. Virol. J. 2013;10:30. doi: 10.1186/1743-422X-10-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Linsuwanon P., Payungporn S., Suwannakarn K., Chieochansin T., Theamboonlers A., Poovorawan Y. Complete coding sequence characterization and comparative analysis of the putative novel human rhinovirus (HRV) species C and B. Virol. J. 2011;8:5. doi: 10.1186/1743-422X-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakamura M., Itokazu K., Taira K. Detection and phylogenetic analysis of human rhinoviruses in Okinawa, Japan. Jpn. J. Infect. Dis. 2010;63:221–223. [PubMed] [Google Scholar]
- 44.Etemadi M.R., Othman N., Savolainen-Kopra C., Sekawi Z., Wahab N., Sann L.M. Biodiversity and clinico-demographic characteristics of human rhinoviruses from hospitalized children with acute lower respiratory tract infections in Malaysia. J. Clin. Virol. 2013;58:671–677. doi: 10.1016/j.jcv.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.S.B. Liggett, A. Godinez, E. Hine, et al. Genome sequencing of human rhinovirus strains (unpublished).
- 46.Mak R.K., Tse L.Y., Lam W.Y., Wong G.W., Chan P.K., Leung T.F. Clinical spectrum of human rhinovirus infections in hospitalized Hong Kong children. Pediatr. Infect. Dis. J. 2011;30:749–753. doi: 10.1097/INF.0b013e31821b8c71. [DOI] [PubMed] [Google Scholar]
- 47.A. Pinon, B.Acosta, O. Valdes, et al. First molecular study of human rhinoviruses in Cuba: detection of the newly human rhinovirus C. (unpublished).
- 48.Rathe J.A., Liu X., Tallon L.J., Gern J.E., Liggett S.B. Full-genome sequence and analysis of a novel human rhinovirus strain within a divergent HRV-A clade. Arch. Virol. 2010;155:83–87. doi: 10.1007/s00705-009-0549-8. [DOI] [PMC free article] [PubMed] [Google Scholar]