SUMMARY
Mass-tag cell barcoding (MCB) labels individual cell samples with unique combinatorial barcodes, after which they are pooled for processing and measurement as a single multiplexed sample. The MCB method eliminates variability between samples in antibody staining and instrument sensitivity, reduces antibody consumption, and shortens instrument measurement time. Here, we present an optimized MCB protocol with several improvements over previously described methods. The use of palladium-based labeling reagents expands the number of measurement channels available for mass cytometry and reduces interference with lanthanide-based antibody measurement. An error-detecting combinatorial barcoding scheme allows cell doublets to be identified and removed from the analysis. A debarcoding algorithm that is single cell-based rather than population-based improves the accuracy and efficiency of sample deconvolution. This debarcoding algorithm has been packaged into software that allows rapid and unbiased sample deconvolution. The MCB procedure takes 3–4 h, not including sample acquisition time of ~1 h per million cells.
INTRODUCTION
Barcode multiplexing
As a general approach, pooled sample analysis has been used to improve efficiency and comparability for a diverse range of biological assays, from micro-sphere-based ELISA1 to high-throughput DNA sequencing2,3. For these applications, assay-specific identifiers such as fluorochrome combinations or oligonucleotide sequences are used as barcodes to uniquely label each sample, and the barcoded samples are pooled together for processing and measurement. Multiplexing in this manner eliminates sample-to-sample assay variability, increases assay throughput, and reduces reagent consumption. After pooled measurement, the uniquely identifiable barcodes are used to recover the individual samples for further analysis.
This multiplexing strategy was adapted to flow cytometry by the fluorescent cell barcoding (FCB) technique, which uses unique combinations of cell-reactive fluorophores to covalently label cell samples before pooled antibody staining and flow cytometry analysis4. Mass cytometry, a recently developed variation of flow cytometry, uses rare earth metal isotopes instead of fluorophores as detection reagents, allowing over 40 simultaneous antibody-based measurements at the single cell level5. The principles of FCB were extended to mass cytometry by the mass-tag cell barcoding (MCB) technique, which uses cell-reactive metal chelators to covalently label cell samples with combinatorial barcodes6. Both FCB and MCB use a single antibody cocktail to stain all samples simultaneously within a single tube, ensuring that all samples are exposed to the same antibody concentration at the same cell density. This uniform antibody exposure removes tube-to-tube variability from the assay, and is especially important when antibodies are used at non-saturating concentrations, as is often the case with mass cytometry because antibody concentrations must be titrated low enough to prevent ion detector saturation.
Analysis of multiplexed samples offers additional benefits that are specific to mass cytometry. The ion detection sensitivity of a mass cytometer will drift during instrument use and vary after each maintenance, and while this effect can be mitigated by normalization using bead standards7, measuring samples after pooling further reduces inter-sample variability. Additionally, the sample introduction loop of a mass cytometer is a potential source of carryover between samples, but the possibility of sample cross-contamination is bypassed by MCB because the samples are individually labeled with a unique barcode. Further improvements to MCB described here include 1) palladium-based cell labeling reagents, 2) a combinatorial doublet-filtering scheme, and 3) an improved barcode deconvolution algorithm implemented as a software application, all of which markedly improve the quality of mass cytometry data. In addition, we have recently modified the MCB protocol to allow barcode staining prior to methanol permeabilization, allowing methanol-sensitive surface markers to be assessed in combination with MCB multiplexing8. This modified protocol relies on transient permeabilization with saponin, and is included here as an alternate procedure.
Palladium-based MCB reagents
Lanthanide-based MCB reagents perform well as cell labeling reagents6, but their utility for mass cytometry analysis is limited in two ways. First, lanthanides are employed as antibody tags for mass cytometry, and therefore their use as MCB reagents reduces the number of antibody-based measurement parameters available. Second, MCB lanthanide reagents may interfere with other measurement channels due to isotopic impurity of enriched lanthanide isotopes and to mass spectrometry effects including the “+1 effect” which is due to the time-of-flight (TOF) mass trace distribution being skewed toward larger mass and overlapping with the next mass unit integration window and the “+16 effect” of oxidation during ionization in the plasma ion source (ICP). These contaminating effects are typically minor but become more pronounced when a high intensity MCB signal spills over into a low intensity antibody signal.
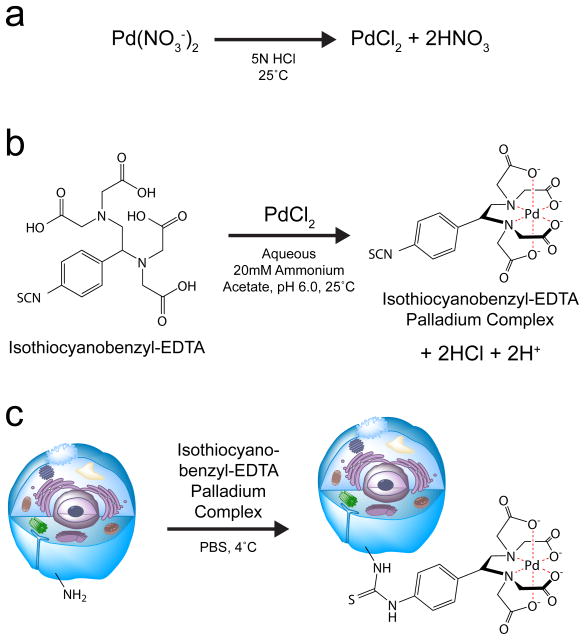
To prevent the reduction of available antibody measurement channels, and to avoid measurement channel cross talk, palladium was identified as a potential mass-tag for MCB. Since palladium is not compatible with the DTPA-based polymer used for antibody labeling, it is not used for mass cytometry antibody measurements9. Palladium has six stable isotopes with masses of 102, 104, 105, 106, 108, and 110 u, which fall well below the 139–176 u mass range of the lanthanides, and therefore its use in MCB reagents does not impinge upon lanthanide-based antibody detection. Pd110 does however overlap with the Cd110 present in quantum-dot conjugated antibodies, which are therefore not recommended for use in combination with the palladium-based MCB protocol. All six palladium isotopes were obtained in their 2+ charge state as nitrate salts, and dissolved in 5N HCl (Fig. 1a). Addition of the bifunctional molecule isothiocyano-benzyl-EDTA yielded a palladium-chelate (Fig. 1b) which may be subsequently used to covalently label cells (Fig. 1c).
Figure 1. Isothiocyanobenzyl-EDTA(palladium) MCB cell labeling reagent.
(a) Palladium nitrate is converted to palladium chloride after dissolving in 5N Hydrochloric Acid. (b) Palladium chelation by isothiocyanobenzyl-EDTA. (c) Cell labeling by the isothiocyanobenzyl-EDTA(palladium)chelate.
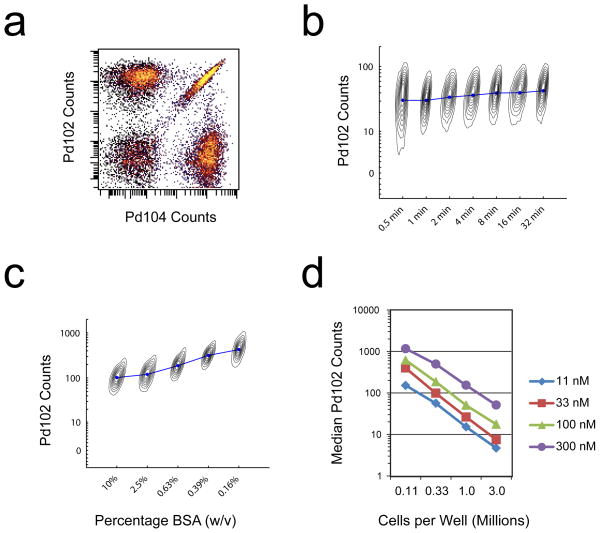
For MCB multiplexing, a binary labeling strategy is used where each sample is either positive or negative for each of the six palladium isotopes (Fig. 2a), and unique combinations of these positive and negative labels are used as a sample-identifying barcodes. The isothiocyanobenzyl-EDTA(palladium) chelates label cells rapidly in PBS, reaching completion between 0.5 to 1 minute at 4°C (Fig. 2b). Care must be taken to wash excess FBS or BSA away from the samples before MCB labeling, because nucleophile-containing proteins will compete for isothiocyanate reactivity and decrease cell labeling intensity (Fig. 2c). Additionally, care must be taken to quantify the number of cells in each sample to be labeled, because the isothiocyanobenzyl-EDTA(palladium) labeling reaction reaches completion rapidly in a stoichiometric manner, and therefore is highly sensitive to differences in cell number (Fig. 2d). While the EDTA-palladium dissociation rate is permissible for metal exchange between chelating groups on the time scale of days, in practice this effect and any resulting “cross-contamination” between labeled cell samples is observed to be negligible.
Figure 2. MCB cell labeling by the isothiocyanobenzyl-EDTA(palladium) chelate.
(a) Binary MCB labeling of PFA-fixed, methanol-permeabilized cells. (b) One million PFA-fixed, methanol-permeabilized cells were incubated with 100 nM Isothiocyanobenzyl-EDTA(Palladium) at 4 °C for the indicated times. Median intensities are shown as connected blue circles, and are overlaid on individual contour plots for each sample with Ir-intercalator along their hidden x axes. (c) One million PFA-fixed, methanol-permeabilized cells were mixed with the indicated concentrations of BSA before incubation with 300 nM Isothiocyanobenzyl-EDTA(Palladium) at 4 °C for 30 minutes. Median intensities are shown as connected blue circles, and are overlaid on individual contour plots for each sample with Ir-intercalator along their hidden x axes. (d) PFA-fixed, methanol-permeabilized cells were incubated at the indicated cell densities with the indicated Isothiocyanobenzyl-EDTA(Palladium) concentrations at 4 °C for 30 minutes.
Doublet-filtering barcode scheme
In mass cytometry, physical or coincident doublets will result in the false interpretation of two or more cells as a single cell. Physical doublets occur when multiple cells are physically attached due to some form of cell-cell interaction or to incomplete enzymatic separation and detachment of adherent cells. Coincident doublets occur when physically separate cells pass through the instrument in too quick succession to be identified as separate events by the mass cytometer’s cell detection software. Cell doublets will confound any single cell analysis, including algorithms such as SPADE10, visNE11, and Citrus12, and are especially problematic for the investigation of rare or uncharacterized cell types.
To improve doublet removal from mass cytometry datasets, a doublet-identifying barcode scheme was developed. Error-correcting codes such as Hamming codes have been used in barcode design for error detection and correction in multiplexed high-throughput sequencing13. This strategy relies on redundancy in the sample barcode: if one measurement is incorrect, the error is detected and may even be corrected depending on the level of barcode redundancy. Here, an n-choose-k barcoding scheme was chosen as the minimally redundant code that allows doublet identification and removal while maximizing the number of unique combinatorial identifiers, where n-choose-k equals n!/(k!(n−k)!).
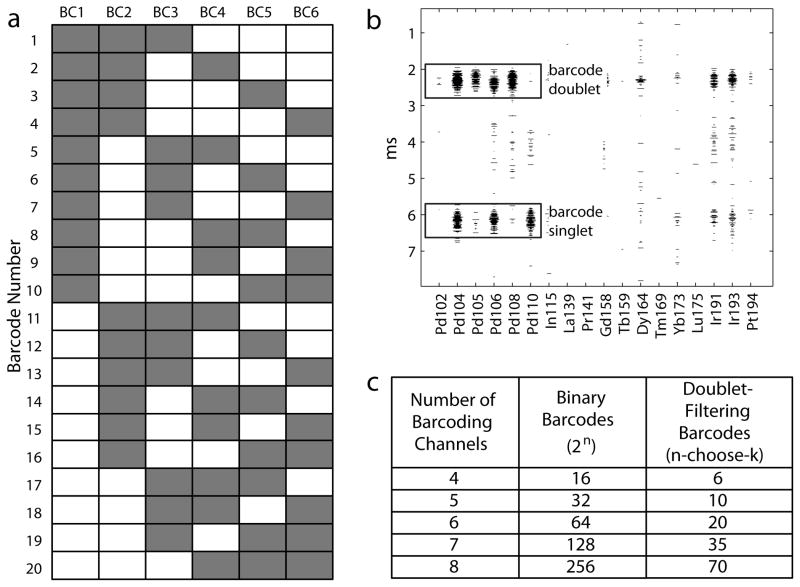
With six palladium isotopes available for MCB labeling, a 6-choose-3 barcoding scheme was used, in which each of the 20 individual barcodes are positive for exactly 3 of the 6 possible palladium MCB reagents (Fig. 3a). In this scheme, the combination of any two barcodes resulting from a cell doublet yields an ‘illegal’ barcode with at least four barcode channels that are positive, i.e., one that cannot belong to a single cell event (Fig. 3b). This scheme cannot detect coincident doublets between two cells from the same sample, but every other combination can be detected and removed. By this reasoning, 95% of the coincident doublets may be removed when using the 6-palladium 20-sample scheme, because for any single cell within a coincident doublet, the other cell within the doublet will have a different barcode 19 out of 20 times. However, in practice the actual doublet removal rate may be less than 95% depending on the number of physical doublets present in the samples.
Figure 3. Doublet-filtering MCB scheme.
(a) The 6-choose-3 doublet-filtering barcode scheme. Each well is positive (gray) and negative (white) for exactly 3 out of the 6 MCB reagents. (b) Examples of a barcode singlet (3 positive barcode channels) and a barcode doublet (>3 positive barcode channels) as seen in the time-of-flight spectra used to visualize cells while acquiring data at the instrument. (c) Maximum number of available barcodes as a function of number of barcode channels for both doublet-filtering n-choose-k schemes and for the non-redundant 2n binary scheme.
One tradeoff with this doublet-filtering scheme is a reduced number of cell samples that can be multiplexed by MCB. For every n metals used for barcoding, only n-choose-k combinations are available, which is maximized when k=floor(n/2), rather than the 2n that are available with a non-redundant binary scheme (Fig. 3c). With six palladium MCB reagents, the n-choose-k scheme results in 20 possible samples rather than 64. An appropriate barcode scheme must be chosen for each experiment based on 1) the number of measurement channels available for barcoding, 2) the desired number of samples to be multiplexed, and 3) the importance of doublet removal for sample analysis.
Single cell deconvolution algorithm
Traditionally, individual samples have been recovered from FCB and MCB datasets using Boolean combinations of manually drawn gates, but this method is not ideal because cell events that fall outside these gates must be discarded. This problem is exacerbated, and cell yield is made even lower, when there is variability in barcode staining intensity between pooled samples. This variability can be caused either by differences in cell number between samples without appropriate adjustment of MCB reagent (Fig. 2d), or by differences in cell size within samples (Fig. S1), and this variability leads to systematic depletion of certain cell types or samples from the dataset during deconvolution. To address this problem, an alternative deconvolution algorithm was developed that treats each cell individually instead of using gates to demarcate populations of cells. This de-convolution strategy is termed single cell debarcoding (SCD).
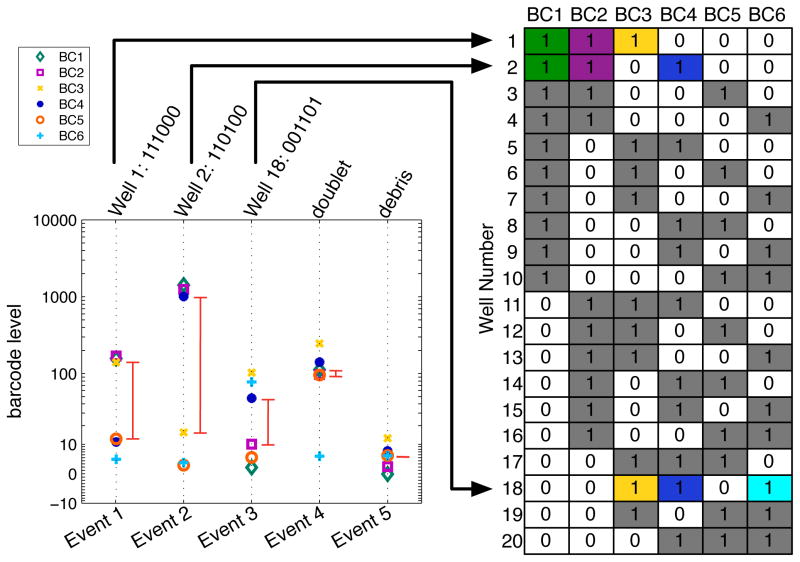
The SCD algorithm first rescales and then sorts the barcode intensities for each cell, in order to identify the largest barcode separation between adjacent barcode levels. This barcode separation is used as a boundary to define which barcode channels are ‘positive’ and ‘negative’ for each individual cell (Fig. 4). Cell events are assigned to a sample when they contain 1) a barcode separation larger than a threshold value, and 2) positive barcode channels corresponding to a combination used in the barcode scheme. Cell events remain unassigned if they contain 1) a barcode separation lower than the defined threshold, such as low signal debris, or 2) positive barcode channels that do not correspond to a barcoded population, such as doublets. When an n-choose-k doublet-filtering barcode scheme is used, the SCD algorithm uses the k highest and n-k lowest barcode channels instead of the largest barcode separation to assign positive and negative barcode values, and the barcode separation threshold is applied to the difference between the kth and (k−1)th highest normalized barcode intensities. Once preliminary barcodes have been assigned, outliers are filtered out by applying a Mahalanobis distance threshold to each barcode population, which takes into account the covariance of the barcode populations. Finally, each barcode population is output to a corresponding FCS file, and the cells that were discarded by the algorithm are output to a separate FCS file.
Figure 4. Single cell barcode deconvolution.
Five events from a 6-choose-3 MCB-multiplexed FCS file are shown in single cell format displayed on a vertical dashed line. Events 1–3 correspond to barcode singlets as indicated by the barcode key, Event 4 is a barcode doublet, and Event 5 is classified as debris. The red line segments indicate ‘barcode separation’ assuming the 6-choose-3 scheme, which is always set as the distance between the 3rd and 4th highest barcode intensities. Without this assumption, the last two events would have larger barcode separations but would still be discarded because their barcodes would not match any in the 20-sample scheme.
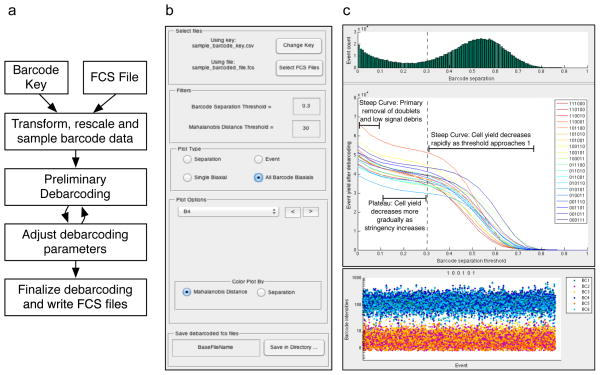
To facilitate deconvolution of barcoded datasets, the SCD algorithm was implemented as a standalone MATLAB application. By using the MATLAB Compiler Runtime (MCR), this application does not require a MATLAB installation or license. A flowchart describing the debarcoding workflow is shown in Figure 5a. The inputs are an FCS file containing a barcoded dataset, and a spreadsheet in CSV format that defines the barcoding scheme, referred to as the ‘Barcode Key’ (Fig. 5a). After selection of the input FCS file and Barcode Key in the control panel of the GUI (Fig. 5b), a preliminary round of barcode assignment is performed for a range of barcode separation thresholds. A histogram of cells binned by barcode separation, and a plot of the number of total events yielded for each barcoded sample as a function of the separation threshold, are then displayed in the top right panel of the GUI (Fig. 5c). This view of yield vs. separation threshold, as well as a single-cell view for each resulting barcode population (Fig. 5d) and biaxial scatter plots (Fig. 5e), aid the choice of deconvolution parameters that can favor either barcode stringency or cell yield. In most cases, there is no tradeoff necessary between stringency and yield, and high stringency settings may be used without substantially lowering cell yield. In certain exceptional cases with large differences in cell number, cell size, or even debris between samples, the MCB staining may not be uniform and the researcher may adjust the parameters according to his or her desire for barcode stringency vs. cell yield. A valuable internal control for the MCB protocol that provides an estimation of the deconvolution error rate is to leave a single well empty, for example only use 19 cell samples with the 20 sample 6-choose-3 MCB scheme. This allows for an estimation of incorrect sample assignment if any cells are assigned to the empty well after sample deconvolution, and is useful whether Boolean gating or the single cell deconvolution algorithm is used.
Figure 5. Single cell debarcoding software.
Mouse splenocytes were harvested from 20 individual mice, and treated with benzonase to minimize cell aggregates. 1.5 × 106 cells from each sample were MCB labeled and then pooled for mass cytometry processing. The pooled sample was blocked with anti-CD16/32, and then stained with an antibody cocktail including anti-CD4 and anti-CD8, followed by mass cytometry measurement.
(a) A flowchart of the single-cell debarcoding process. (b) The menu of the single-cell debarcoder. The lower plot portion dynamically changes depending on the plot type selected. (c) The analysis window which is used to guide selection of the barcode separation threshold parameter. The distribution of barcode separations is shown by green bars, and the resulting cell yields for each of the 20 unique populations after debarcoding are displayed as a function of this barcode separation threshold. The dashed line indicates a separation threshold value that best balances barcode assignment stringency and cell yield. (d) The ‘Event’ plot shows all cell events assigned to barcode 100101, with each cell event represented as a vertical line on which the 6 MCB reagent intensities are plotted, as in Figure 4. (e) The ‘All BC Biaxials’ plot type colored by Mahalanobis distance for barcode 100101 with the chosen parameters. All animal studies were performed in accordance with the investigators’ protocols approved by the Stanford University institutional animal care and use committee.
Doublet identification and removal
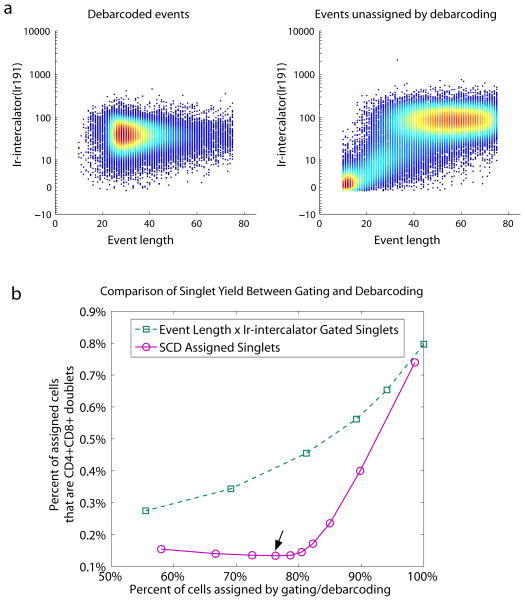
Unlike fluorescence-based flow cytometry, mass cytometry does not have scatter measurements to identify cell doublets for removal. Previous attempts to eliminate cell doublets from mass cytometry datasets used gating to remove events that are high for both Ir-Intercalator staining intensity and the number of time-of-flight scans per cell (event length). This gating strategy is imperfect for doublet removal because the doublet and singlet populations substantially overlap on the Ir-Intercalator × event length scatter plot as revealed by n-choose-k barcoding (Fig. 6a, Fig. S2a). Additionally, this gating strategy may result in the systematic removal of specific cell types, because event length is dependent on the total amount of metal labeling each cell, which in turn is cell type- and staining panel-dependent. When the stringency of the Ir-intercalator × event length gate matches the stringency of the SCD separation threshold, i.e. the two methods produced the same cell yield, the SCD algorithm consistently filtered out more doublets than the ‘singlet’ gate (Fig. 6b). The ‘singlet’ gates used for this analysis are shown in Figure S3; to achieve a similar range of yields, the SCD separation threshold was varied from 0 to 0.7, with the Mahalanobis distance threshold set to 30. In addition to its application for mass cytometry as presented here, this doublet-identifying barcode scheme may be applied to other single cell analysis methods, and will be especially useful when experimental analysis requires high-confidence discrimination between unique cells and removal of cell doublets.
Figure 6. Doublet removal with the 6-choose-3 MCB scheme and single cell de-barcoding.
(a) Biaxial plot of event length × Ir-intercalator of events that were assigned a barcode, and of events that were left unassigned. (b) Percent of cells assigned by gating (green squares) or debarcoding (purple circles) versus percent of assigned cells that are CD4+CD8+ doublets. The different yields were acquired by variable event length × Ir-intercalator gates (green squares) or debarcoding threshold stringency (purple circles). The arrow indicates the debarcoding parameters used in Figures 5 and 6a.
Comparison of SCD to Boolean gating
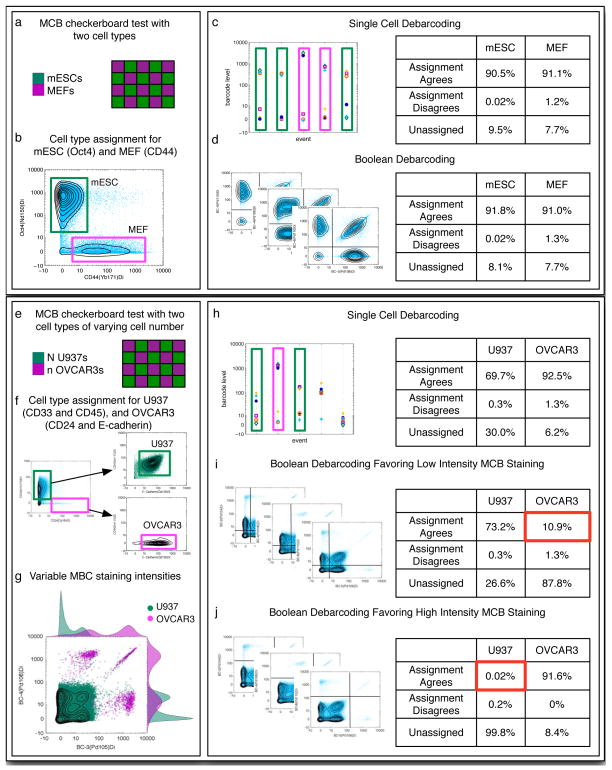
While Boolean gating has been used successfully for MCB deconvolution6, it relies on distinct populations with consistent positive and negative MCB labeling intensities across all samples and cell types. If high-intensity, low-background staining is achieved consistently in all samples, Boolean gating and SCD perform essentially the same (Fig. 7a–d). Consistent MCB labeling can, however, be difficult to achieve for several reasons, such as if cell number varies unexpectedly, if BSA washout is incomplete, or if one or more samples contain significant amounts of debris. An important attribute of the SCD algorithm is that it does not depend on uniform MCB labeling across samples or even within samples, and it performs well on sub-optimally MCB-multiplexed samples (Fig. 7e–h) where de-convolution by Boolean gating fails (Fig. 7i,j). Because the SCD algorithm assigns samples by identifying the highest MCB measurements from each cell individually rather than on a population basis, it works for multiplexed samples with a continuous distribution of MCB labeling intensity as well as for multiplexed samples with well-separated bimodal distributions.
Figure 7. Single cell deconvolution vs. Boolean gating for MCB samples with and without large differences in MCB labeling intensity.
(a) PFA-fixed, Methanol-permeabilized MEF and mESC cells were aliquoted into a 2 ml 96-well plate in a 20-sample checkerboard pattern at 0.2 × 106 and 0.5 × 106 cells per well, respectively. The cells were incubated with MCB reagents in a 6-choose-3 combinatorial scheme at 300 nM Isothiocyanobenzyl-EDTA(palladium). (b) After MCB labeling and pooling of the checkerboard-arranged samples, the MEF-specific antibody against CD44 and the mESC-specific antibody against Oct4 were used to differentiate between the two cell types. (c) Single-cell debarcoding and (d) boolean gate debarcoding produce similar cell yields and accuracies.
(e) PFA-fixed, Methanol-permeabilized U937 and OVCAR-3 cells were aliquoted into a 2 ml 96-well plate in a 20-sample checkerboard pattern at 30,000 and 100,000 cells per well, respectively. A large percentage of the OVCAR-3 cells were lost during the PBS wash steps before MCB labeling, which resulted in unusually high MCB staining intensity for these samples. The cells were incubated with MCB reagents in a 6-choose-3 combinatorial scheme at 30 nM Isothiocya-nobenzyl-EDTA(palladium). (f) After MCB labeling and pooling of the checkerboard-arranged samples, U937-specific antibodies against CD33 and CD45 and OVCAR-3-specific antibodies against CD24 and E-cadherin were used to differentiate between the two cell types. (g) Gating based on CD33, CD45, CD24, and E-Cadherin reveals the difference in MCB-labeling intensity between the U937 and OVCAR-3 cells. (h) Single-cell debarcoding successfully recovers both the U937 and OVCAR-3 populations. (i) Boolean gates bisecting the populations at a low MCB intensity primarily recovers U937 cells. The low percentage of recovered OVCAR-3 cells is highlighted in red. (j) Boolean gates bisecting the populations at a high MCB intensity primarily recovers OVCAR-3 cells. The low percentage of recovered U937 cells is highlighted in red.
It may not be possible to achieve equal levels of barcode staining across all wells when comparing multiple patient samples or different tissue types, yet it is precisely these situations in which the benefits of barcoding, in particular uniform antibody staining, will have the greatest benefit on the quality of the data. Performing barcode deconvolution with the single-cell rather than population-based method overcomes the challenges associated with variable barcode staining levels. Additionally, in contrast to the variability of manually chosen gate boundaries, the results of the single-cell method are solely determined by the chosen distance parameters, and therefore the deconvolution is reproducible.
MATERIALS
REAGENTS
Isotopically purified Palladium Nitrate. Pd102, Pd104, Pd105, Pd106, Pd108, and Pd110 Nitrates are commercially available as a custom order from Trace Sciences International. Palladium isotopes should be obtained with as high purity as possible, because debarcoding performance correlates positively with reagent purity, although in theory the MCB protocol would function reasonably well with only 60% pure reagents. The purities of the palladium reagents used in this study were Pd102 >91%, Pd104 >96%, Pd106 >99%, Pd108 >99%, Pd110 >99%. ! CAUTION Palladium Nitrate is an oxidizer. Keep away from heat and avoid contact with skin and eyes.
Hydrochloric Acid (Fisher Scientific # A466) ! CAUTION Hydrochloric acid is corrosive. Avoid contact with skin and eyes.
Sodium Hydroxide (Sigma S-8045) ! CAUTION Sodium Hydroxide is corrosive. Avoid contact with skin and eyes.
Sodium Phosphate Dibasic Heptahydrate (Sigma # S9390)
Potassium Phosphate Monobasic Anhydrous (Sigma # P0662)
Potassium Chloride (Fisher Scientific # P330)
Sodium Chloride (Fisher Scientific # S271)
Ammonium Acetate (Sigma # A7330)
1.5 ml Eppendorf tubes (Fisher Scientific # 14-222-155)
DMSO (Sigma # D2650)
Saponin (Sigma # S-7900)
Methanol (Fisher Scientific # A412-4) ! CAUTION Methanol is flammable. Keep away from heat. Avoid contact with skin and eyes. Avoid inhalation.
Bovine Serum Albumin (BSA) Fraction V (Sigma # A-2153)
Sodium Azide (Sigma # S-8032) ! CAUTION Sodium azide is highly toxic. Avoid contact with skin, eyes, and clothing. Fatal if swallowed.
16% Paraformaldehyde (PFA) EM Grade (Electron Microscopy Sciences # 15710) ! CAUTION PFA is an irritant. Avoid contact with skin and eyes. Avoid inhalation.
Cluster Tubes, 1.2ml Polypropylene (Fisher Scientific #07-200-319, # 07-200-317)
96-Well Deep Well Microplates Polypropylene 2.0ml Nonsterile (VWR # 40002-012)
96-Well PCR plates (Fisher Scientific # 14230237)
96-Well foil covers (Fisher Scientific # 14-222-342)
96-Well aluminum block (BioExpress # R-2027-S)
Isothiocyanobenzyl-EDTA (Dojindo # M030-10)
18G Needles (BD # 305195)
Ziplock Quart-size Freezer Bags (VWR # 82027-628)
Drierite Dessicant (VWR # 22891-050)
EQUIPMENT
P20, P200, and P1200 Multichannel Pipets (Rainin # L8-20, # L8-200, # L8-1200)
Vortex Mixer (GeneMate # S-3200-1)
Table-top Microfuge (5424, Eppendorf)
Table-top 96-well Format Refrigerated Centrifuge (Allegra X-22R, Beckman Coulter)
96-Well Aspirator (VP Scientific # VP 177A-1)
Lyophilizer (FreeZone 4.5, Labconco)
Mass Cytometer (CyTOF, DVS Sciences)
Aluminum 96-well Block (GeneMate # R-2027-S)
REAGENT SETUP
5N Hydrochloric Acid
Dilute concentrated Hydrochloric Acid solution to 5N in ddH2O and store indefinitely at room temperature (20–25 °C).
5N Sodium Hydroxide
Dissolve Sodium Hydroxide to 5N in ddH2O and store indefinitely at room temperature.
10× PBS
Dissolve 320 g Sodium Chloride (1.37 M), 8 g Potassium Chloride (27 mM), 46 g Sodium Phosphate Dibasic Heptahydrate (43 mM), 8 g Potassium Phosphate Monobasic Anhydrous (15 mM) in 3 L ddH2O, adjust to pH 7.4 with 5N Sodium Hydroxide, then bring final volume to 4 L and filter using a 0.8 micron filter. Store up to 5 years at room temperature.
PBS
Dilute 10× PBS 1:10 in ddH2O and store up to 5 years at room temperature.
PBS-S
Dissolve solid saponin powder in PBS to 0.02% w/v and store up to 6 months at room temperature.
2× Fixation Solution
Dilute 16% PFA 1:5 (to 3.2%) into PBS and store at 4 °C shielded from light for up to 2 weeks.
Cell Staining Medium (CSM)
Dissolve BSA to 0.5% (w/v) and NaN3 to 0.02% (w/v) in PBS. Store up to 6 months at 4 °C.
Ammonium Acetate Buffer
Dissolve Ammonium Acetate to 20 mM in ddH2O, and then confirm that solution is pH 6.0. Store up to 1 year at 4 °C.
10× Palladium Solution
Dissolve isotopically pure palladium nitrate to 100 mM in 5N HCl, giving a dark brown transparent solution. Store indefinitely at room temperature.
Intercalator Solution
Dilute 16% PFA 1:10 (to 1.6%) into PBS, and then add Ir-Intercalator diluted 1:10,000. Store at 4 °C shielded from light for up to 2 weeks.
PROCEDURE
MCB Labeling and Pooled Mass Cytometry Analysis ● TIMING ~ 3 hours plus ~ 1 hour for every million mass cytometry cell event measurements
-
1|
Bring PBS and a 2 ml 96-well plate to 4 °C on ice.
-
2|
Transfer cell samples from −80 °C storage to the bench.
-
PFA-fixed cells
Thaw on ice or in a cold water bath
-
PFA-fixed/methanol-permeabilized cells
Store on bench in dry ice
-
-
3|
Transfer cell samples to pre-labeled tubes. For large numbers of samples, it is efficient to use cluster tubes in 96-well format, or a 2 ml 96-well plate. Subsequent steps of this procedure will be written for 20 samples in a 2 ml 96-well plate.
-
PFA-fixed cells
Vortex cells to resuspend, then pipet a volume of the cell suspension that contains 500,000 cells (counted in Box 1 Step 9) into each well.
-
PFA-fixed/methanol-permeabilized cells
Add 1.2 ml CSM to each well. NOTE: CSM helps prevent cell loss in subsequent wash steps.
Vortex cells to resuspend, then pipet a volume of the cell suspension that contains 500,000 cells (counted in Box 1 Step 9) into each well. Pipet up and down 5 times or vortex to mix. NOTE: Methanol can cause BSA in the CSM to precipitate. This typically happens if methanol is greater than 50% of the total solution. Also, if the methanol and CSM are not completely mixed, then BSA will precipitate at the CSM/Methanol interface. The BSA precipitate does not appear to have an adverse effect on cell labeling or antibody staining. It dissolves and washes away with subsequent washes.
▲ CRITICAL STEP Equalizing the cell number for each sample ensures equal MCB labeling intensity, which is important for optimal barcode deconvolution. If samples cannot be equalized for cell number because some have too few cells, then adjust the concentration of MCB combinatorial mix added to the wells with fewer cells in Step 10.
? TROUBLESHOOTING
-
-
4|
Fill each well to the top with CSM, then centrifuge cells at 600 g for 5 minutes at 4 °C and aspirate supernatant.
-
5|
Wash cells and prepare for the MCB labeling reaction.
-
PFA-fixed cells
Wash cells one time with 4 °C PBS, centrifuging at 600 g for 5 minutes at 4 °C.
Wash cells one time with 4°C PBS-S (with 0.02% saponin), centrifuging at 600 g for 5 minutes at 4 °C, leaving residual volume of ~100 ul.
Aliquot 1.0 ml ice-cold PBS-S into 20 wells of the pre-chilled 2 ml 96-well plate corresponding to the wells of the MCB combinatorial plate.
-
PFA-fixed/methanol-permeabilized cells
Wash cells two times with 4 °C PBS, centrifuging at 600 g for 5 minutes at 4 °C, leaving residual volume of ~100 ul.
Aliquot 1.0 ml ice-cold PBS into 20 wells of the pre-chilled 2 ml 96-well plate corresponding to the wells of the MCB combinatorial plate.
? TROUBLESHOOTING
-
-
6|
Vortex or pipet to resuspend cell pellet in residual volume and store at 4 °C on ice.
-
7|
Thaw a 100× combinatorial MCB plate. An aluminum 96-well block may be used to increase the thaw speed.
-
8|
If any wells have an unequal number of cells: Adjust the concentration of each well corresponding to cell samples that are not cell number-equalized with fresh DMSO.
-
9|
Transfer 10 μl of MCB reagent into the 4 °C PBS-S or PBS-containing wells of the 2 ml 96-well plate with a P20 multichannel pipet. Mix by pipetting up and down 3 times with a P1000 multichannel pipet set to 0.9 ml, and then transfer 0.9ml of the ice-cold MCB mix to the resuspended cell samples. Mix cells and MCB reagent immediately by pipetting up and down 5–10 times to ensure uniform mixing and incubate for 15 min at room temperature.
▲ CRITICAL STEP The isothiocyanate group hydrolyzes rapidly in PBS, so work as quickly as possible to move MCB-containing PBS to the cell sample. Also ensure that the PBS is at 4°C when first mixed to slow isothiocyanate hydrolysis. NOTE: The volume of MCB reagent added to each well may be adjusted to accommodate changes in cell number. The MCB reagent volume may be approximately scaled 1:1 with cell number (Figure 2d), although for best results a new titration should be performed with the same number of cells per well as the final experiment.
? TROUBLESHOOTING
-
10|
Fill each well to the top with CSM, and then centrifuge at 600 g for 5 minutes at 4 °C and aspirate supernatant.
-
11|
Wash with CSM two additional times, and then pool cell samples into a single tube for antibody stain. Depending on sample number, residual volumes, and desired wash volumes, it may be necessary to pool in a 15ml or 50ml conical polypropylene tube before transferring to a smaller volume tube for antibody staining.
? TROUBLESHOOTING
-
12|
Add antibody staining cocktail to cells and incubate according to previously optimized staining protocol.
-
PFA-fixed cells
Perform surface marker antibody stain on MCB multiplexed sample.
If desired, permeabilize MCB multiplexed sample with methanol (Box 1 Step 8B), wash cells into CSM, and perform intracellular antibody stain.
-
PFA-fixed/methanol-permeabilized cells
-
Perform intracellular and methanol-resistant surface antibody stain on MCB multiplexed sample.
? TROUBLESHOOTING
-
-
-
13|
After antibody stain, wash cells 2× with CSM, and then incubate 15 minutes at room temperature with Intercalator Solution.
▲ CRITICAL STEP The Ir-Intercalator acts as a cell-identifying reagent during CyTOF analysis. The 1.6% PFA in the Intercalator Solution fixes the cell samples to prevent loss of metal and antibody when they are suspended in ddH2O at Step 16 before mass cytometry analysis. ■ PAUSE POINT Intercalating cell samples may be stored for up to 1 month at 4 °C.
-
14|
Wash cells 1× with CSM, 2× with ddH2O, and then analyze on a mass cytometer, collecting all pooled samples as a single FCS file.
Box 1. Preparation of Fixed, Permeabilized Cells ● TIMING ~ 1 hour.
REAGENTS
Cell culture medium (cell type-appropriate)
Cell dissociation reagent (cell type-appropriate, such as: PBS-EDTA/Versene (Life Technologies # 15040-066), Trypsin (Life Technologies # 25200-056), Try-pLE (Life Technologies # 12605-010), Accutase (Innovative Cell Technologies # AT 104), Collagenase (Life Technologies # 17100-017) (only required for adherent cell types and solid tissue samples)
Nylon Mesh Cell Strainer: 40 um (Corning # 352340), 70 um (Corning # 352350), or 100 um (Corning # 352360) (only required for adherent cell types and solid tissue samples)
Fixation Solution, see Reagent Setup in the main text
Cell Staining Medium (CSM), see Reagent Setup in the main text
DMSO (Sigma # D2650)
Methanol (Fisher Scientific # A412-4) ! CAUTION Methanol is flammable. Keep away from heat. Avoid contact with skin and eyes. Avoid inhalation.
EQUIPMENT
Table-top refrigerated swinging-bucket centrifuge (Allegra 6R, Beckman Coulter)
Haemocytometer (Hausser Scientific # 3200)
Vortex Mixer (GeneMate # S-3200-1)
-
For each cell sample to be multiplexed, prepare a single cell suspension concentrated at 1x108 cells/ml or less. Previously established protocols that are optimized for the cell type of interest should be followed, but here we provide generalized instructions. Follow option A for suspension cell lines and blood samples, option B for adherent cell lines, or option C for solid tissue samples.
-
Suspension cell lines or blood samples
Centrifuge cell suspension at 300g for 5 min, room temperature.
Aspirate or decant the supernatant.
Resuspend cell pellet in appropriate culture medium.
-
Adherent cell lines
Detach cells from culture plate by incubation with an appropriate dissociation reagent according to cell line-optimized protocol.
Pipet up and down with P1000 tip or plastic transfer pipet to break up cell clumps and achieve single cell suspension.
Transfer 10 ul to microscope slide or haemocytometer to confirm single cell suspension. Repeat previous step until single cell suspension is achieved.
Filter with nylon mesh (40 um, 70 um, or 100 um as appropriate) to remove any remaining clumps if necessary.
-
Solid tissue samples
Homogenize tissue sample with tissue-optimized mechanichal/enzymatic treatment.
Transfer 10 ul to microscope slide or haemocytometer to confirm single cell suspension. Repeat previous step until single cell suspension is achieved.
Filter with nylon mesh (40 um, 70 um, or 100 um as appropriate) to remove any remaining clumps if necessary.
-
Dilute all cell suspensions with culture medium to a uniform volume and transfer to an appropriate container for PFA fixation. If all cell samples to be multiplexed are available simultaneously, then dilute each sample to 0.5 ml and transfer to a 2 ml 96-well plate to facilitate multichannel pipetting in subsequent steps. If samples are not available simultaneously, the perform PFA fixation in sealable individual containers such as 50 ml conical tubes, 15 ml conical tubes, or 4 ml capped FACS tubes and scale all volumes appropriately.
Dilute 2× Fixation Solution 2-fold into the cell suspension for a final concentration of 1.6% PFA. Mix by pipetting up and down or vortexing, and then incubate at room temperature for 10 min. For a 0.5 ml cell suspension in a 2 ml 96-well plate, add 0.5 ml 2× Fixation Solution.
Slow the PFA-fixation reactions by diluting 2-fold with ice-cold CSM. This step can help equalize the sample fixation times by staggering CSM addition with the same timing as PFA addition. For a 1 ml PFA-fixation reaction in a 2 ml 96-well plate, add 1 ml CSM.
Centrifuge PFA-fixed cells at 600g for 5 min, 4 °C.
Aspirate or decant the supernatant, leaving enough residual volume to resuspend cells into a single cell suspension.
Vortex or pipet up and down to completely resuspend cell pellet in the residual volume and bring cell container to 4 °C on ice.
-
After PFA fixation, cells may either be permeabilized with methanol or frozen as is to allow saponin-mediated MCB staining8. If antibody staining will include any methanol-sensitive surface markers, cells should be frozen as is. If antibody staining will be only for intracellular markers and methanol-insensitive surface markers, cells should be methanol permeabilized.
-
Freeze PFA-fixed cells as is
Wash cells 1× with CSM.
Resuspend in CSM containing 10% DMSO.
-
Remove 10 μl and pipet onto a haemocytometer to determine cell density.
▲ CRITICAL STEP Cell density must be determined for each sample because MCB labeling intensity is dependent on cell number.
Freeze cells at −80 °C.
-
Methanol-permeabilize cells
Aspirate or decant the supernatant, leaving enough residual volume to resuspend cells into a single cell suspension.
Vortex or pipet up and down to completely resuspend cell pellet in the residual volume and bring cell container to 4 °C on ice.
-
Add 4°C methanol to the resuspended cell pellet, so that the final concentration of methanol is greater than 80%.
▲ CRITICAL STEP If cell pellet is not completely resuspended when methanol is added, cells will clump together and be lost. Adding methanol while vortexing the tube of cells works well, but take care to not let the methanol suspension overflow while vortexing.
-
Remove 20 μl cell/methanol suspension and diluting into 180 μl CSM in a 1.1 ml cluster tube. Vortex to mix and then pipet 10 μl onto a haemocytometer to determine cell density. If cell density is too low to accurately determine, scale up the volumes of methanol and CSM, centrifuge 5 min 4 °C 600g, aspirate the supernatant, and resuspend to an accurately determined smaller volume and transfer 10 μl to a haemocytometer to determine cell density.
▲ CRITICAL STEP Cell density must be determined for each sample because MCB labeling intensity is dependent on cell number.
-
■ PAUSE POINT Store PFA-fixed and PFA-fixed/methanol-permeabilized samples at −80 °C. PFA-fixed samples frozen in CSM/DMSO may be stored at −80 °C for 2 years or longer with little to no epitope degradation. PFA-fixed/methanol-permeabilized samples may be stored at −80 °C for 12 months or longer with little to no epitope degradation. Epitope behavior during methanol storage is not uniform, but in general epitopes are stable in methanol at −80 °C (years), −20 °C (weeks), 4 °C (overnight) or room temperature (minutes). Because cell samples in methanol are most stable at −80 °C, dry ice may be used for immediate storage of methanol-permeabilized cell samples before transfer to a −80 °C freezer.
Sample Deconvolution by Single Cell Debarcoder ● TIMING ~ 30 minutes
The debarcoding software, a detailed manual covering its installation and usage, and a sample dataset is available at https://github.com/nolanlab/single-cell-debarcoder/releases/latest.
-
15|
Modify sample names in the barcoding key CSV template file with text editing software or Microsoft Excel, and save modified version.
-
16|
Open the Single Cell Debarcoder and when prompted by the dialog box, select the correct saved barcode key.
-
17|
Click the ‘Select FCS File’ button in the ‘Select files’ panel and choose the FCS file containing the pooled samples. When the preliminary debarcoding is done, which may take several seconds depending on the size of the FCS file, a plot of the cell counts of the barcode populations will appear. To determine which barcode population corresponds to a particular line on the plot, click on that line and the label from the barcode key will appear.
-
18|
Enter a ‘separation threshold’ in the input box in the ‘Filters’ panel. This is a number between 0 and 1 that defines the minimum distance after normalization between the ‘positive’ and ‘negative’ barcode channels that is required for a cell to be assigned to a barcode channel. The goal is to filter out uncertain barcode assignments but still retain sufficiently large barcode populations; a suggested starting point is a value just below which the cell count of the barcode populations dramatically decreases (Fig. 5c). A barcode well that has been left blank can provide a guidance as to where to set the threshold, and also provides an estimate of false assignment rate. In a well stained sample, the false assignment rate calculated from cells assigned to wells left blank is typically <0.5%.
-
19|
Evaluate the barcode separation threshold by browsing through the ‘Event’ plots of different barcode populations. It may be useful to use the ‘zoom’ and ‘pan’ tools. Adjust the threshold if necessary, settling on the smallest value for which all the populations consist of cells in which positive and negative barcodes are sufficiently separated.
-
20|
Check the barcode deconvolution with biaxial plots of the barcode channels. To filter outliers from the populations, decrease the Mahalanobis threshold (Fig. 5d). This process of adjusting the separation threshold and the Mahalanobis threshold may be iterative until the appropriate balance between cell number and deconvolution confidence is reached. Examples of choosing the parameters can be found in the manual.
-
21|
Create a separate FCS file for each barcode population by pressing the ‘Save Debarcoded Files’ button, which will give a prompt to select the folder in which to save the files. By default the names of the wells as entered in the barcode key are appended to the original file name, but that may be adjusted prior to saving by editing the base file name next to the ‘Save Debarcoded Files’ button.
TIMING
Box 1, preparation of fixed, permeabilized cells: ~ 1 hour
Box 2, preparation of palladium MCB reagents: overnight
Box 3, titration of palladium MCB reagents: 4–6 hours
Box 4, preparation of 1000× and 100× combinatorial MCB plates: ~ 2 hours
Box 5, MCB combinatorial plate validation: ~2 hours
Steps 1–13, MCB labeling and antibody stain: ~ 3 hours
Step 14, mass cytometry measurement: ~ 1 hour for every million cells
Steps 15–21, sample deconvolution by single cell debarcoder ~ 15 minutes
Box 2. Preparation of Palladium MCB Cell Labeling Reagents ● TIMING Overnight.
REAGENTS
Isothiocyanobenzyl-EDTA (Dojindo # M030-10)
1.5 ml Eppendorf tubes (Fisher Scientific # 14-222-155)
10× Pd102, Pd104, Pd105, Pd106, Pd108, and Pd110 solutions, see Reagent Setup in the main text
20 mM Ammonium Acetate pH 6.0, see Reagent Setup in the main text
Liquid Nitrogen or Dry Ice
18G Needles (BD # 305195)
DMSO (Sigma # D2650)
Ziplock Quart-size Freezer Bags (VWR # 82027-628)
Drierite Dessicant (VWR # 22891-050)
EQUIPMENT
Vortex Mixer (GeneMate # S-3200-1)
Table-top Microfuge (5424, Eppendorf)
Lyophilizer (FreeZone 4.5, Labconco)
Remove isothiocyanobenzyl-EDTA powder from −20 °C freezer and warm to room temperature before opening to prevent condensation (water-sensitive).
For each palladium isotope MCB reagent to be prepared, tare a balance with a 1.5 ml polypropylene tube, dispense approximately 1.0 mg of isothiocyanoben-zyl-EDTA, then weigh the tube to 0.1 mg accuracy to determine the precise amount of reagent dispensed. Use this mass in the next step.
-
Calculate the volume x of 10mM palladium solution required to create a 2:1 isothiocyanobenzyl-EDTA:palladium molar ratio using the following formula, where a is the mass of isothiocyanobenzyl-EDTA in the tube from Step 2 and 439.33 mg/mmol is the molecular weight of isothiocyanobenzyl-EDTA:
The isothiocyanobenzyl-EDTA chelator is used in excess to ensure complete palladium chelation and to remove all free palladium from the final reagent, because the free metal will react with the cells in a reversible manner that is inappropriate for cell barcoding.
Prepare the volume of 10mM palladium solution calculated in the previous step by diluting 10× palladium solution 1:10 in ammonium acetate buffer, and then add this entire volume to the isothiocyanobenzyl-EDTA and vortex to mix. Multiple rounds of vortexing and spinning in a table top microfuge may be necessary to completely dissolve the isothiocyanobenzyl-EDTA solid, but it should dissolve completely within 15 seconds. While the ammonium acetate is insufficient to buffer the HCl, we have observed no benefit from increasing (or decreasing) the ammonium acetate buffer concentration.
Snap-freeze the isothiocyanobenzyl-EDTA:palladium mixture in liquid nitrogen or an ethanol-dry ice bath. Be sure to keep the tubes upright during freezing so that the frozen mixture will be at the bottom of the tube.
Prepare a vented cap for each tube by piercing the cap of an extra 1.5 ml tube with an 18G or similar needle, and then cutting off the cap. Warm the hinges of the frozen tubes with your fingers to prevent breaking, and then open the tubes and insert the vented caps, leaving the original caps attached. Work quickly to prevent thawing, and return tubes to dry ice after capping to ensure that they are thoroughly frozen before lyophilization.
Lyophilize overnight to yield a brown/orange film.
Dissolve the lyophilized isothiocyanobenzyl-EDTA:palladium in DMSO to 10 mM, using the same volume as calculated in Box 2 Step 3. Note that this solution is 20 mM isothiocyanobenzyl-EDTA, with approximately half of the EDTA chelating groups occupied by palladium.
Aliquot the 10 mM MCB reagent DMSO solutions and store at −80 °C with dessicant to protect from moisture. For example, place aliquots in a 9 × 9 freezer box, and then place this box in a Ziploc bag containing dessicant. Remove as much air as possible from the Ziploc bag before sealing.
■ PAUSE POINT 10 mM isothiocyanobenzyl-EDTA:palladium DMSO stocks may be stored up to 1 year at −80 °C with dessicant. Cell labeling activity may decrease with extended storage or with multiple freeze-thaw cycles.
Box 3. Titration of Palladium MCB Reagents ● TIMING 4–6 hours.
REAGENTS
Bulk PFA-fixed or PFA-fixed/Methanol-permeabilized cell sample, see Box 1 for reagent preparation
PBS, see Reagent Setup in main text
CSM, see Reagent Setup in main text
96-Well Deep Well Microplates Polypropylene 2.0ml Nonsterile (VWR # 40002-012)
10× Palladium Solutions, see Reagent Setup in main text
Intercalator Solution, see Reagent Setup in main text
EQUIPMENT
Table-top 96-well Format Refrigerated Centrifuge (Allegra X-22R, Beckman Coulter)
96-Well Aspirator (VP Scientific # VP 177A-1)
P20 and P1200 Multichannel Pipets (Rainin # L8-20, # L8-1200)
Titrate each MCB reagent on bulk cell samples to identify the optimal concentration for cell labeling. Typically the optimal concentration is between 50 nM and 1 uM for PFA-fixed/Methanol-permeabilized cells, depending on the number of cells per labeling reaction, the amount of residual BSA remaining after PBS wash, and the extent of isothiocyanate hydrolysis in the MCB reagent DMSO stock. PFA-fixed cells that have not been methanol permeabilized typically require 3-fold more barcoding reagent for optimal staining.
Perform each measurement in triplicate to ensure accurate titration. For a 5 point titration of 6 MCB reagents in triplicate, 90 cell labeling reactions will be performed. 96-well format eases pipetting and sample handling. While this protocol is written for 0.5x106 cells per well here, MCB multiplexing can be performed on higher cell numbers, at least up to 5x106 cells per well. When varying cell number, it is critical to titrate the barcoding reagent concentration for the number of cells (and fixation method) that will be used in the final experiment (Figure 2d).
Prepare a sufficient number of PFA-fixed, methanol-permeabilized cells in bulk to perform the MCB reagent titration, as described in Box 1. Any cell type may be used for this bulk sample, but suspension cell lines such U937 are preferred for ease of passage and collection.
Bring PBS and two 2 ml 96-well plates to 4 °C on ice. For one of the 2 ml 96-well plates, add 1 ml PBS to each well that will be used for titration, using the plate layout shown in Figure S4a. These wells will be used to prepare the MCB reagent DMSO stock dilutions before adding to the cells.
Transfer bulk, PFA-fixed, methanol-permeabilized cells from −80 °C to the bench on dry ice.
Vortex cells to mix, and then transfer a volume containing 0.5 × 106 cells per titration point into at least twice that volume of CSM. For a 5 point titration of 6 MCB reagents in triplicate, use 4.5 × 107 cells. Note that these intensity vs. concentration curves are specific for the number of cells per well (500,000 in this protocol), because MCB intensity varies with cell density as described in Fig. 2D. Adjust the number of bulk cells used in this MCB reagent titration to best match the expected cell densities of final experimental samples.
Centrifuge cells at 600 g for 5 minutes at 4°C, then aspirate supernatant.
Resuspend cell pellet to a concentration of 0.5 × 106 cells / ml (1.0 ml CSM per titration point) and aliquot 1.0 ml per well into the empty, chilled 96-well plate in the wells that will be used for the MCB labeling titration, using the plate layout shown in Figure S5a.
-
Wash the cell-containing 96-well plate 2× with PBS, centrifuging at 600 g for 5 minutes at 4 °C for each step and leaving a residual volume of 100 ul.
▲ CRITICAL STEP Residual BSA competes with cells for MCB reagent. Failure to adequately wash away BSA results in decreased and inconsistent MCB labeling intensity.
Thaw 10 mM MCB DMSO stocks, and dilute each to 1.1 uM with fresh DMSO. Perform a 1:3 serial dilution on this 100× working solution in a 96-well PCR plate by mixing 10 ul with 20 ul fresh DMSO at every step. Starting with a 1.1 uM 100× working solution yields final titration concentrations of 1,000 nM, 333 nM, 111 nM, 37 nM, and 12 nM. Perform the serial dilution in triplicate, using the plate layout shown in Figure S5a.
-
Pipet 10 ul of each 100× MCB working solution with a multichannel P20 pipet into the 4 °C PBS-containing 2 ml 96-well plate, mix by pipetting up and down 5 times with a multichannel P1000 pipet set to 900 ul, and then transfer 900 ul to the 2 ml 96-well plate containing resuspended PBS-washed cells. Mix immediately by pipetting up and down 5 times to ensure uniform mixing and incubate at 4 °C for 15 minutes.
▲ CRITICAL STEP The isothiocyanate group is unstable in PBS due to hydrolysis, so work as quickly as possible to move MCB-containing PBS to the cell sample. Ensure that the PBS used for dilution is 4 °C instead of room temperature to further slow isothiocyanate hydrolysis.
Add 0.5 ml CSM to each well of the MCB cell labeling reaction, and then centrifuge at 600 g for 5 minutes at 4 °C and aspirate supernatant.
Wash with CSM two additional times, and resuspend in 0.1 ml CSM.
-
Pool the wells as shown in Figure S4b, resulting in 3 tubes for each concentration point containing all MCB reagents.
▲ CRITICAL STEP In addition to reducing sample acquisition time, pooling each of the MCB reagent-labeled titration point samples into a single tube at this step allows for detection of any cell-to-cell transfer of the MCB label during the incubation in Box 2 Step 14. Improperly prepared MCB reagent may exchange readily between cells, hindering its use as a barcoding reagent.
Centrifuge cells for 5 minutes at 600g and 4 °C, then aspirate supernatant leaving residual volume of 100 ul.
Resuspend cell pellet in residual volume and incubate at room temperature for 60 minutes. This step simulates an antibody stain incubation and is necessary to test for cell-to-cell transfer of the MCB reagent.
-
Wash cells 1× with CSM, then incubate 15 minutes at room temperature with 1:10000 Ir-Intercalator in 1.6% PFA-containing PBS.
■ PAUSE POINT Intercalating cell samples may be stored for up to 1 month at 4 °C
-
Wash cells 1× with CSM, 2× with water, and then analyze on a mass cytometer. Run the samples in ascending order of MCB reagent concentration, and stop measuring samples before the MCB signal reaches the mass cytometer detection limit, which is approximately 10,000 counts per cell for any measurement channel. The maximum MCB concentration sample measured will usually be 111 nM or 333 nM for the titration described in this procedure. The higher MCB cell labeling concentrations in the titration are included in case the MCB reagent stock has lower than expected cell labeling activity.
! CAUTION Measuring samples with MCB staining intensity above the mass cytometer’s detection limit may substantially shorten the lifespan of the instrument’s ion detector.
Using the measured median intensities for each MCB reagent, and including only titration points that fall within the linear range of measurement (excluding points with a median intensity less than 10 or greater than 5,000 counts per cell), derive a curve to determine the cell counts observed vs. nM for each of the MCB reagents. This value will be used to determine the appropriate concentrations of each MCB reagent to use during preparation of the combinatorial MCB plates in Box 4.
BOX 4. Preparation of Combinatorial MCB Plates ● TIMING ~ 2 hours.
REAGENTS
10 mM MCB Reagent DMSO Stocks, see Box 2 for reagent preparation
DMSO (Sigma # D2650)
96-Well PCR plates (Fisher Scientific # 14230237)
96-Well foil covers (Fisher Scientific # 14-222-342)
Dry Ice
Ziplock Quart-size Freezer Bags (VWR # 82027-628)
Drierite Dessicant (VWR # 22891-050)
EQUIPMENT
P20 and P1200 Multichannel Pipets (Rainin # L8-20, # L8-1200)
Aluminum 96-well Block (GeneMate # R-2027-S)
The combinatorial MCB plates are used as a 100× working solution, but they are initially prepared at 1000× in order to conserve −80 °C freezer space, and to allow for dilution to different concentrations for use with different numbers of cells or to compensate for lost activity during extended storage. If freezer space is not limiting and the number of cells to multiplex will not change substantially, then the combinatorial MCB plates may be initially prepared at 100× concentration for storage and direct use.
Plan the MCB combinatorial scheme in advance using a spreadsheet program such as Microsoft Excel, and print out the layout to use as reference while pipetting each MCB reagent during Box 4 Step 3. Figure S5 shows the 6 pipetting layouts required for a 6-isotope 20-sample combinatorial MCB plate, and the subsequent steps are written for this scheme.
Thaw the 10 mM MCB reagent DMSO stocks, and dilute each MCB reagent with fresh DMSO to produce 660ul at 3000× the concentration required for optimal cell labeling, as determined by reagent titration in Box 3. Optimal cell labeling intensity is approximately 500 counts per cell, although this number may vary from instrument to instrument depending on ion detector sensitivity. The optimal cell labeling intensity should be determined for each instrument as approximately 5-fold below the level at which the +1 time-of-flight effect, isotopic impurity, and event length elongation become problematic due to high palladium signal intensity. If the 3000× concentration is chosen to be 3 mM, then mix 198 ul of the 10 mM MCB stock with 462 ul fresh DMSO.
-
Pipet an equal volume of each DMSO stock into the appropriate wells of a 96-well PCR plate following the printed key to create the 1000× combinatorial MCB master mix. For the 6-metal 20-sample doublet-filtering scheme that contains exactly 3 MCB reagents per well, pipet 60 ul of each MCB 3000× DMSO stock per well for a final volume of 180 ul in each well. We typically use the middle wells C4-F9 of the 96-well plate in a 5 × 4 grid so that the final working plates are better protected from moisture when sealed for long-term storage at −80 °C.
▲ CRITICAL STEP Visually inspect the PCR plate after all 3000× DMSO stocks have been pipetted, and check that every well contains the same volume of DMSO solution. If a well was missed or pipetted twice by mistake, the volume will be noticeably different: either 120 ul or 240 ul instead of the desired 180 ul. Take extra care to ensure correct pipetting at this step, because any mistake will result in unusable 1000× plates that must be discarded.
Inspect the 96-well PCR plate for bubbles present at the bottom of the wells. Remove any bubbles present with a P200 pipet, and then thoroughly mix each well of the 1000× combinatorial MCB master mix by pipetting up and down 10–20 times with a P200 multichannel pipet set to 120 ul.
Replicate by aliquoting the 1000× combinatorial MCB master mix plate into multiple 96-well PCR plates. For example, aliquot 18 ul from each well to new plates using a multichannel P20 pipet or pipetting robot.
-
Seal each aliquoted PCR plate with foil adhesive, bring to −80 °C with dry ice, and store in Ziploc bags with as much air removed as possible and with dessicant at −80 °C to protect from moisture.
■ PAUSE POINT 1000× combinatorial MCB plates may be stored up to 1 year at −80 °C with dessicant. Cell labeling activity may decrease with extended storage or with multiple freeze-thaw cycles.
▲ CRITICAL STEP Work as rapidly as possible to minimize DMSO absorbing water from the air, which can hydrolyze the MCB reagent isothiocyanate group.
Thaw one 1000× combinatorial MCB plate containing 18ul MCB reagent per well. An aluminum 96-well block may be used to increase the thaw speed.
Dilute each well 1:10 by adding 162 ul fresh DMSO to each well with a multi-channel pipet or pipetting robot.
Mix each well of 100× combinatorial MCB master mix by pipetting up and down 10–20 times with a P200 multichannel pipet set to 120 ul.
Aliquot the 100× combinatorial MCB master mix plate into multiple 96-well PCR plates. For example, aliquot 12 ul from each well to new plates using a multichannel pipet or pipetting robot.
-
Seal each aliquoted PCR plate with foil adhesive, bring to −80 °C with dry ice, and store in Ziploc bags with as much air removed as possible and with dessicant at −80 °C to protect from moisture.
■ PAUSE POINT 100× combinatorial MCB plates may be stored up to 1 year at −80 °C with dessicant. Cell labeling activity may decrease with extended storage or with multiple freeze-thaw cycles.
▲ CRITICAL STEP Work as rapidly as possible to minimize DMSO absorbing water from the air, which can hydrolyze the MCB reagent isothiocyanate group.
BOX 5. MCB Combinatorial Plate Validation ● TIMING ~ 2 hours.
REAGENTS
Bulk PFA-fixed, Methanol-permeabilized cells, see Box 1 for reagent preparation
PBS, see Reagent Setup in main text
CSM, see Reagent Setup in main text
96-Well Deep Well Microplates Polypropylene 2.0ml Nonsterile (VWR # 40002-012)
100× Combinatorial MCB Plates, see Box 4 for reagent preparation
Intercalator Solution, see Reagent Setup in main text
EQUIPMENT
Table-top 96-well Format Refrigerated Centrifuge (Allegra X-22R, Beckman Coulter)
96-Well Aspirator (VP Scientific # VP 177A-1)
P20, P200, and P1200 Multichannel Pipets (Rainin # L8-20, # L8-200, # L8-1200)
Aluminum 96-well Block (GeneMate # R-2027-S)
Before using the MCB combinatorial plates for experimental samples, it is useful to first test them on bulk PFA-fixed, methanol-permeabilized cells. The purpose of this test is two-fold: 1) to confirm that the MCB cell labeling intensity from the 100× combinatorial plate is at an appropriate level, and 2) to confirm there are no errors in sample assignment due to pipetting errors from Box 4 Step 3. The fidelity of sample assignment is tested with multiple cell sample layout patterns such as checkerboard, striped, and random arrangements to ensure that each of the 20 samples is correctly assigned.
Prepare a sufficient number of PFA-fixed, methanol-permeabilized cells in bulk to perform the MCB reagent titration, as described in Box 3. Any cell type may be used for this bulk sample, but suspension cell lines such U937 are preferred for ease of passage and collection.
Aliquot 1 × 106 cells per well into 20 wells of a 2 ml 96-well plate, corresponding to the 20 positions of the combinatorial MCB plate to be tested.
Label the 20 wells of bulk PFA-fixed methanol-permeabilized cells with the 100× combinatorial MCB plate to be tested, following the steps described in the Procedure section of the main text, Steps 5–12.
Arrange P200 Pipet tips in empty pipet tip boxes into the 8 patterns shown in Figure S6.
Using a P200 Multichannel pipet set to 150 ul, use the pipets arranged in Box 5 Step 4 and pool MCB-labeled cells into 8 groups corresponding to the 8 patterns shown in Figure S6. Use a 70 ml reservoir to pool each group before transferring to a 50 ml conical tube.
Centrifuge 50 ml conical tubes for 5 minutes at 600g and 4 °C, then aspirate supernatant.
Resuspend the 8 pooled samples in CSM, and transfer each sample to a labeled 4 ml FACS tube.
Centrifuge 4 ml FACS tubes for 5 minutes at 600g and 4 °C, then aspirate supernatant.
Resuspend cell pellet in residual volume and incubate at room temperature for 60 minutes. This step simulates the antibody stain and is necessary to test for cell-to-cell transfer of the MCB reagent.
For each of the 8 pooled samples, intercalate, measure by mass cytometer, and debarcode following the steps described in the Procedure section of the main text, Steps 15–23, and confirm that only the expected MCB combinations are present in each pooled sample.
TROUBLESHOOTING
| Step | Problem | Possible Reason | Solution |
|---|---|---|---|
| 3 | Uneven MCB staining between multiplexed samples | Unequal cell numbers input into the MCB staining reaction | Recount cell density for each cell sample and adjust volumes added for multiplexing to equalize cell number |
| If cell number cannot be equalized because some samples have too few cells, then decrease the MCB reagent cocktail concentration for these samples with fresh DMSO. MCB staining intensity is approximately linear in relation to cell number, so for 5-fold fewer cells, dilute the MCB reagent cocktail 5-fold with fresh DMSO. | |||
| Samples with large amounts of cellular debris will display reduced MCB labeling, because the debris behaves as additional cells and soaks up MCB reagent, reducing the amount available to label the cells of interest | If possible, re-prepare sample and minimize debris generation with gentler mechanical trituration | ||
| Debris may be removed from cell samples by gradient density centrifugation | |||
| Scale up the amount of MCB reagent added to high debris samples, or reduce the cell number added for multiplexing | |||
| Lower-than expected antibody staining intensity | Exceedingly high MCB staining intensity will reduce observed antibody staining intensity by ion-detector saturation/shutoff and by ion suppression | Adjust the cell number and MCB reagent volume so that MCB staining intensity is between 200 and 5000 counts per cell | |
| 5 | Lower-than expected MCB staining intensity | Quenching of MCB cell labeling by residual BSA | Increase the number and/or stringency of PBS washes before MCB cell labeling |
| 9 | Lower-than expected MCB staining intensity | MCB reagent has been hydrolyzed by multiple freeze-thaws, dessicant failure, or storage at elevated temperatures | Test additional 100× MCB combinatorial plates diluted from the same 1000× stock plate on bulk cells. If these also look bad, dilute additional 1000× MCB combinatorial plates prepared from the same batch and re-test with bulk cells. If 1000× stock looks bad, test the individual MCB reagent stocks on bulk cells. Discard and reprepare MCB combinatorial plates and reagents as necessary. |
| 11 | Lower-than expected cell yield (number of events collected) | Cell loss during sample handling and pooling, caused by loss of residual liquid and by cells sticking to the plastic of sample wells and pipet tips | During sample pooling, add an additional wash step for every well to collect residual cells. |
| Minimize the plastic surface area experienced by the cells by using a single pipet tip to transfer all samples, and mixing cells by pipetting rather than vortexing | |||
| 12 | Lower-than expected antibody staining intensity | Increased cell number relative to unpooled samples resulting in high cell density | Increase the antibody staining volume so that cell density less than or equal to 50 million cells per ml |
ANTICIPATED RESULTS
The MCB Protocol allows multiple samples to be pooled for antibody staining and mass cytometry analysis. Using the 6-palladium MCB reagents and the doublet-filtering n-choose-k barcoding scheme allows 20 sample multiplexing. After pooled sample processing and measurement on the mass cytometer, a single FCS file is produced for each multiplexed cell sample. The 20 individual samples are recovered from this pooled FCS file by the debarcoding software, which produces 20 FCS files plus an additional FCS file that contains the cell events that were unassigned by the deconvolution algorithm.
The MCB protocol and software are flexible enough to allow for additional MCB reagents and barcode schemes. If a researcher desires to multiplex more than 20 cell samples, a non-redundant barcoding scheme may be used at the expense of doublet filtering, or with additional MCB reagents using lanthanides chelates 6 but at the expense of additional antibody measurement channels. The procedure described here is designed to enable a researcher to barcode and antibody stain four groups of 20 samples in 3–4 hours. The attributes of MCB described in this report will enhance the quality of mass cytometry data.
Supplementary Material
Figure S1 | Comparison of MCB staining intensity between cell lines of different cell sizes that were pooled for MCB labeling in the same tube. BC2 (Pd104) staining intensity of OVCAR-3 and U937 cells from five barcoded samples, gated for cell type by surface marker expression as in Figure 7f. Debarcoding was performed independently of cell type gating.
Figure S2 | Time-of-flight traces of palladium barcodes of singlets and doublets with overlapping event lengths and Ir-intercalator intensities. (a) Example cell events are indicated on the event length × Ir-Intercalator biaxial plot. Event 1 is identified as a barcode singlet, but of similar length and Ir-intercalator intensity as event 4, which has been identified as a barcode doublet. Similarly, event 2 is of similar length and Ir-Intercalator intensity as event 3. (b) The per-spectrum traces of the barcoding channels from the IMD files confirm the barcode and single/doublet assignments.
Figure S3 | Gates used for Figure 6b. (a) Singlet gates of increasing stringency and their percent yields. (b) The percent of CD4+CD8+ cells within each of the singlet gates shown in (a).
Figure S4 | 96-well plate layout for MCB reagent titration in triplicate.
(a) Serial dilution layout for the 6 Palladium MCB reagents. (b) Wells to pool before for mass cytometry measurement.
Figure S5 | Plate layout for 6-choose-3 MCB combinatorial doublet-filtering scheme.
(a) MCB reagent combinations to use for a 20 sample 6-choose-3 combinatorial doublet-filtering scheme. (b) Mapping the 20 samples to a 5 × 4 grid. (c) Pipetting guide for each of the 6 Palladium MCB reagents into the 5 × 4 grid.
Figure S6 | Pooled sample groups for 20-sample MCB combinatorial plate testing and validation.
Wells to pool for 8 pooled sample groups that will be used to validate the sample assignment and correct orientation of the tested 100× MCB combinatorial plate.
Acknowledgments
We thank Angelica Trejo and Astraea Jager for mass cytometry quality control and instrument maintenance. We thank Erin Simonds and Peter Krutzik for helpful discussions. This work was supported by grants from the US National Institutes of Health (U19 AI057229, U54CA149145, N01-HV-00242, 1U19AI100627, 5R01AI07372405, R01CA184968, 1 R33 CA183654), NIH-Baylor Research Institute (41000411217), NIH-Northrop Grumman Corp.(7500108142), CIRM (DR1-01477), Department of Defense (OC110674), European Commission ( Health.2010.1.2-1), Food and Drug Administration (HHSF223201210194C), Bill and Melinda Gates Foundation (OPP 1017093), Alliance for Lupus Research, Entertainment Industry Foundation (NWCRA grant), and the Rachford and Carlota A. Harris Endowed Professorship to G.P.N.
Footnotes
AUTHOR CONTRIBUTIONS STATEMENTS
E.R.Z. developed the palladium MCB reagents and labeling protocol, implemented the doublet-filtering barcode scheme, developed the single-cell debarcoding algorithm, designed and performed the experiments, analyzed the data, and wrote the manuscript; R.F. developed the single-cell debarcoding algorithm, designed and implemented the debarcoding software, designed the experiments, analyzed the data, and wrote the manuscript; G.K.B. developed the palladium MCB reagents and labeling protocol, developed the saponin-mediated MCB labeling, and wrote the manuscript; E.D.A. developed the doublet-filtering barcode scheme; S.K. assisted with developing barcode deconvolution methods; V.D.G. performed experiments; C.G.L. performed experiments; Z.B. performed experiments and wrote the manuscript, M.H.S. performed experiments; B.B. developed the palladium MCB reagents and labeling protocol; W.J.F. designed the experiments and wrote the manuscript; D.P. developed the doublet-filtering barcode scheme and wrote the manuscript; G.P.N. designed the experiments and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests (see the HTML version of this article for details). G.P.N. has personal financial interest in the company Fluidigm, the manufacturer of the mass cytometer used in this manuscript. R.F. has been a paid consultant for the company DVS Sciences, the original manufacturer of the mass cytometer which has since merged with Fluidigm. G.K.B. has been a paid consultant for both DVS Sciences and Fluidigm.
References
- 1.Fulton RJ, McDade RL, Smith PL, Kienker LJ, Kettman JR. Advanced multiplexed analysis with the FlowMetrixTM system. Clin Chem. 1997;43:1749–1756. [PubMed] [Google Scholar]
- 2.Parameswaran P, et al. A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucleic Acids Res. 2007;35:e130. doi: 10.1093/nar/gkm760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer M, Stenzel U, Myles S, Prüfer K, Hofreiter M. Targeted high-throughput sequencing of tagged nucleic acid samples. Nucleic Acids Res. 2007;35:e97. doi: 10.1093/nar/gkm566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krutzik PO, Nolan GP. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat Methods. 2006;3:361–368. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- 5.Bandura DR, et al. Mass Cytometry: Technique for Real Time Single Cell Multitarget Immunoassay Based on Inductively Coupled Plasma Time-of-Flight Mass Spectrometry. Anal Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 6.Bodenmiller B, et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat Biotechnol. 2012;30:858–67. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finck R, et al. Normalization of mass cytometry data with bead standards. Cytometry A. 2013;83:483–94. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behbehani GK, et al. Transient partial permeabilization with saponin enables cellular barcoding prior to surface marker staining. Cytometry A. 2014 doi: 10.1002/cyto.a.22573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majonis D, Ornatsky O, Kinach R, Winnik MA. Curious Results with Palladium- and Platinum-Carrying Polymers in Mass Cytometry Bioassays and an Unexpected Application as a Dead Cell Stain. Biomacromolecules. 2011;12:3997–4010. doi: 10.1021/bm201011t. [DOI] [PubMed] [Google Scholar]
- 10.Qiu P, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amir ED, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruggner RV, Bodenmiller B, Dill DL, Tibshirani RJ, Nolan GP. Automated identification of stratifying signatures in cellular subpopulations. Proc Natl Acad Sci. 2014;111:E2770–7. doi: 10.1073/pnas.1408792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 | Comparison of MCB staining intensity between cell lines of different cell sizes that were pooled for MCB labeling in the same tube. BC2 (Pd104) staining intensity of OVCAR-3 and U937 cells from five barcoded samples, gated for cell type by surface marker expression as in Figure 7f. Debarcoding was performed independently of cell type gating.
Figure S2 | Time-of-flight traces of palladium barcodes of singlets and doublets with overlapping event lengths and Ir-intercalator intensities. (a) Example cell events are indicated on the event length × Ir-Intercalator biaxial plot. Event 1 is identified as a barcode singlet, but of similar length and Ir-intercalator intensity as event 4, which has been identified as a barcode doublet. Similarly, event 2 is of similar length and Ir-Intercalator intensity as event 3. (b) The per-spectrum traces of the barcoding channels from the IMD files confirm the barcode and single/doublet assignments.
Figure S3 | Gates used for Figure 6b. (a) Singlet gates of increasing stringency and their percent yields. (b) The percent of CD4+CD8+ cells within each of the singlet gates shown in (a).
Figure S4 | 96-well plate layout for MCB reagent titration in triplicate.
(a) Serial dilution layout for the 6 Palladium MCB reagents. (b) Wells to pool before for mass cytometry measurement.
Figure S5 | Plate layout for 6-choose-3 MCB combinatorial doublet-filtering scheme.
(a) MCB reagent combinations to use for a 20 sample 6-choose-3 combinatorial doublet-filtering scheme. (b) Mapping the 20 samples to a 5 × 4 grid. (c) Pipetting guide for each of the 6 Palladium MCB reagents into the 5 × 4 grid.
Figure S6 | Pooled sample groups for 20-sample MCB combinatorial plate testing and validation.
Wells to pool for 8 pooled sample groups that will be used to validate the sample assignment and correct orientation of the tested 100× MCB combinatorial plate.