Abstract
To design appropriate antiretroviral therapy regimens and avoid the emergence of human immunodeficiency virus (HIV)-1 variants with reduced susceptibility to antiretroviral drugs, genotypic drug-resistance testing (HIV genotyping) is strongly recommended. To monitor the quality of HIV genotyping in Japan, we performed an external quality assessment (EQA), named the Japanese external quality assessment program, to standardize HIV genotyping (JEQS). To accurately evaluate the quality of HIV genotyping, we employed as reference material (RM) a well-characterized sample, in vitro transcribed RNA (trRNA) that includes the HIV gag–pol sequence, and created a JEQS2010 panel consisting of three single variant and three mixed trRNA samples. All 11 participating laboratories showed high concordance rates (>96%) for the single variant samples. Eight laboratories also showed good rates of detecting minor variants, but three laboratories failed to detect the variants comprising one-half of the sample. These three laboratories used a common primer that had four internal mismatches to the minor trRNA clone. This program showed the usefulness of trRNA as RM, the high quality of HIV genotyping, and extensive interlaboratory variation in the ability to detect minor variants. These results suggest that improving the quality of HIV genotyping in Japan requires regularly implementing the EQA program and improving the HIV genotyping protocol in each laboratory.
Introduction
Standardizing clinical laboratory testing is necessary for maintaining the technical excellence of tests and providing identical quality medical services everywhere the test is offered.1 Clinical tests using genetic analysis have benefited from great technological advances over the past two decades, making such tests more important and indispensable for precise diagnosis and detailed understanding of diseases. As these advances have occurred so quickly and dynamically, many genetic tests such as polymerase chain reaction (PCR) and sequencing have not yet been standardized. Exceptions are tests for hepatitis B virus, hepatitis C virus (HCV), and quantification of HIV-1 plasma viral copy,2,3 where FDA-approved commercial kits are available, but other tests are mostly designed in each laboratory as in house protocols, and have not been standardized.
One important genetic test that has not been standardized in the field of HIV/AIDS diagnosis is HIV-1 drug-resistance genotypic testing (HIV genotyping).4 For HIV-infected individuals, prognosis and quality of life have dramatically improved with antiretroviral therapy (ART) that combines at least three antiretroviral (ARV) drugs. However, to gain the maximum benefit of ART and avoid the risk of incomplete viral suppression, the possibility of drug-resistant HIV-1 must be considered when selecting an appropriate ARV regimen.5 To determine drug resistance and select the optimal ARV regimen, patient plasma samples must be tested for HIV genotype, and the target genome of ARVs must be sequenced. This testing applies not only for selecting salvage regimens for virological failure cases, but also for newly infected/diagnosed individuals as transmission of antiretroviral-resistant HIV has been reported worldwide.6–11
HIV genotyping is a relatively complicated test compared to other clinical laboratory tests, as it comprises many processes such as extracting RNA from plasma, converting RNA to DNA, PCR amplification, DNA sequencing, editing sequences, and interpreting drug resistance. As no clinically approved HIV genotypic test kits are available in Japan, all laboratories in Japan carry out HIV genotyping according to their original protocols.12
To maintain the quality level of each in house genotypic test, HIV genotyping must be standardized. An important qualification protocol in standardizing clinical tests is external quality assessment (EQA). Several EQA studies of HIV genotyping have used HIV-seropositive plasma samples as reference material (RM).13–16 Though using clinical samples is ideal to evaluate the quality of the whole genotyping protocol, this usage has several difficulties. First, the HIV-1 population in plasma has wide genetic diversity, resulting in so-called quasispecies and making it difficult to establish a concrete reference sequence for scoring the quality of genotypic results from each laboratory. Reference sequences used in previous EQA studies13–16 to evaluate the sequence quality of each laboratory were generated as consensus sequences based on sequences reported by laboratories participating in the EQA. Second, as seropositive plasma must be drawn from HIV-infected patients, the amounts are limited, making it difficult to supply the same plasma lot to multiple laboratories. Thus, using patient plasma samples may restrict the number of laboratories able to join the EQA for the same assessment.
The problems of accurately sequencing and having sufficient amounts of plasma samples might be resolved by using artificial RM, for example, plasmids and recombinant viruses, as used in previous EQA studies.17–19 We employed in vitro transcribed RNA (trRNA) that includes the gag–pol sequence as an alternative RM for our HIV genotyping EQA study, named the Japanese external quality assessment program to standardize HIV genotyping (JEQS). To monitor the quality of HIV genotyping in Japan and to evaluate the quality and suitability of trRNA samples as RM for HIV genotyping, we sent a JEQS2010 panel to 11 laboratories that participated in the JEQS2010 program.
Materials and Methods
Laboratories participating in the JEQS2010 program
To monitor and maintain the quality of HIV genotyping in Japan, the JEQS program was established in 2003. By 2010, this program had 11 participating Japanese laboratories: two medical laboratories in hospitals, two commercial laboratories, and seven institutes of public health. Of these, three laboratories were accredited under the International Organization for Standardization (ISO) 15189, indicating that laboratory tests met international standards. This study was approved by the Institutional Ethical Review Committee of the National Hospital Organization Nagoya Medical Center.
Construction of plasmids and in vitro transcription
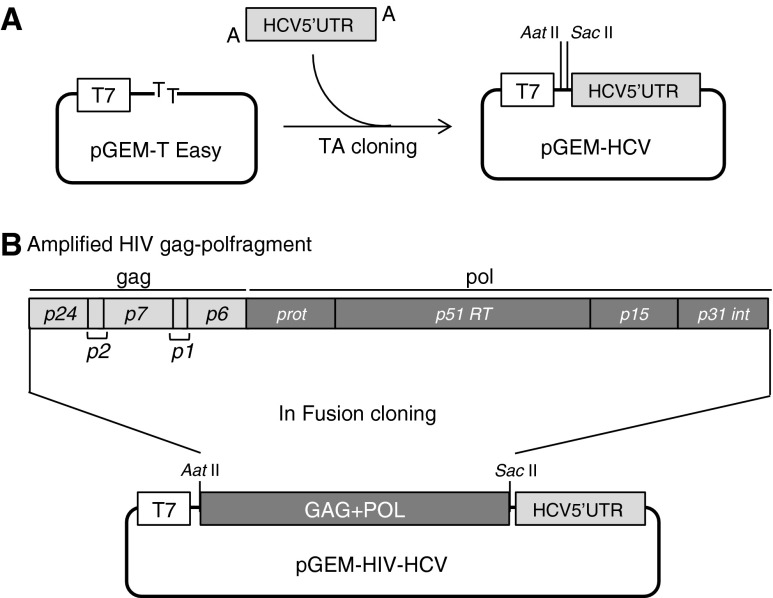
To quantify the amount of trRNA, we constructed a plasmid vector with an HIV-1-unrelated sequence, the 5′-UTR gene of HCV. Briefly, the HCV 5′-UTR gene (321 bps) was amplified from serum with HCV subtype 1b using primers HCV 5′-UTR STD01F and HCV 5′-UTR STD01R (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/aid). RT-PCR was set up using the One-Step RNA PCR kit (TAKARA, Inc., Shiga, Japan) according to the manual. Cycling conditions were 1 cycle at 50°C for 30 min and 1 cycle at 94°C for 2 min, followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s. This fragment was ligated into pGEM-T Easy vector using the TA cloning system (Promega Co, Madison, WI) according to the manual. After cloning, this vector named pGEM-HCV was linearized by digestion with AatII and SacII (Fig. 1A). The HIV-1 gag–pol gene was ligated into linearized pGEM-HCV (Fig. 1B). Briefly, the HIV-1 gag–pol gene (2,836 bps) was amplified from plasma HIV-1 using outer primers HIV p24 STD01F, HIV int STD01R and inner primers HIV p24 STD03F, HIV int STD04R (Supplementary Table S1).
FIG. 1.
Construction of plasmid for HIV-1 gag–pol RNA transcription. (A) An amplified hepatitis C virus (HCV) 5′-UTR fragment (nt 19–339, the accession number of the reference sequence is AB049088) is ligated into the TA cloning site of pGEM-T Easy vector. (B) The amplified HIV gag–pol fragment (nt 1,480–4,315, reference strain is HXB2, K03455) is ligated into pGEM-HCV, followed by HIV gag–pol RNA transcription by T7 RNA polymerase.
RT-PCR was set up using the One-Step RNA PCR kit. Cycling conditions were 1 cycle at 50°C for 30 min and 1 cycle at 94°C for 2 min, followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 4 min. Nested PCR was set up using KOD-Plus ver2 (TOYOBO Co., Ltd, Osaka, Japan) according to the manual. Cycling conditions were 1 cycle at 94°C for 2 min, followed by 35 cycles of denaturation at 98°C for 10 s, annealing at 55°C for 30 s, and extension at 68°C for 3 min. This amplified fragment was ligated into linearized pGEM-HCV using the In-Fusion Advantage PCR Cloning Kit (Clontech Laboratories, Inc., Mountain View, CA) according to the manual. After checking the correct sequences of the cloned plasmids, which were linearized by the restriction enzyme MluI, RNA was transcribed in vitro using the RiboMAX Large Scale RNA Production System T7 (Promega) according to the manual. Transcribed RNA was purified using the RNeasy Mini kit (QIAGEN, Hilden, Germany) according to the manual for RNA cleanup. The sequences of each trRNA were verified by three different sequencing protocols, including the JEQS working group recommended method, and used for EQA program panel samples. The complete HIV sequences inserted in the plasmids are summarized in Supplementary Fig. S1.
JEQS2010 panel
Six coded samples were prepared for the JEQS2010 panel (Table 1). Three samples, JEQS2010-014, -015, and -016 (described as #14, #15, and #16), comprised a single trRNA variant, i.e., sample #14 included multiple drug-resistance mutations (DRM) and sample #15 was identical to sample #16 with low copy number. The other three samples, JEQS2010-026, -028, and -029 (described as #26, #28, and #29), were trRNA mixtures of wild-type nucleotides and DRM, adjusted based on the amount of trRNA containing 30%, 10%, and 50% DRM, respectively. The amount of trRNA was measured with the COBAS AmpliPrep/COBAS TaqMan HCV test (Roche Diagnostics GmbH, Mannheim, Germany) by a laboratory accredited under ISO15189, and the concentration was adjusted by dilution with RNase-free water containing 50 ng/ml Escherichia coli tRNA. Each sample was divided into RNase-/DNase-free sterile tubes, lyophilized, and stored at −30°C until shipping. Prior to distributing the JEQS2010 panel, we confirmed the amplification of lyophilized trRNA kept for 12 weeks at −30°C (data not shown). The JEQS2010 panel was distributed on dry ice (−30°C) to participating laboratories.
Table 1.
Details of the JEQS2010 Panel
| DRM of PI | DRM of RTI | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample number | Subtypea | HIV RNA (log cp/ml) | Type | Major | Minor | Others | NRTIs | NNRTIs | Others |
| 014 | B | 3.07 | Single | D30N, M46I, I54V, V82A, N88D | L10F, L33I, A71V, T74S | I13V, L19I, A22V, E35D, M36I, R41K, I62V, L63P, Q92K, I93L | M41L, L74I, V118I, M184V, T215Y | None | V35K, T39A, K43Q, K122E, I135T, I142T, Q207E, R211K |
| 015 | B | 3.34 | Single | None | None | T12S, I15V, L63P | None | None | K13R, K122E, T200I |
| 016 | B | 2.83 | Single | None | None | T12S, I15V, L63P | None | None | K13R, K122E, T200I |
| 026 | B | 4.70 | 30% mix | (M46ML, V82VT, L90LM)b | (A71AV, T74TS)b | (L10LH, I15VI, K20KR, E35ED, M36MI, N37NE, K43KR, R57RK, D60ED, I62IV, L63P, I64IV, I72IV, Q92EQ, I93IL)b | (D67DN, K70KR, V118VI, M184MV, L210LF, T215TF, K219KE)b | None | (V35VT, T39TA, V60IV, K64KR, K122KE, I135TI, D123DE, M164VM, Q174H, D177DE, Q197QK, T200IA, I202IV, Q207E, R211KR, F214FL)b |
| 028 | B | 4.82 | 10% mix | ||||||
| 029 | B | 5.09 | 50% mix | ||||||
Subtype was identified by the NCBI genotyping tool.
First amino acid harbored in the majority, second one in the minority (e.g., M46ML, the majority is “M” and the minority is “L”).
DRM, drug-resistance mutation; PI, protease inhibitor; RTI, reverse transcriptase inhibitor; NRTIs, nucleoside analogue reverse transcriptase inhibitors; NNRTIs, nonnucleoside analogue reverse transcriptase inhibitors; Single, single variant.
HIV genotyping
HIV genotyping was carried out according to each participating laboratory's standard protocols. These laboratories' reagents and other technical information are shown in Supplementary Table S2. Laboratories electronically reported the edited DNA sequences in which protease (PR) and reverse transcriptase (RT) are 297 bp and 720 bp, respectively. In addition, laboratories filled out a questionnaire regarding their technologies and standard protocols.
Evaluation and data analysis
The quality of genotyping protocols was evaluated by three factors: (1) the concordance rate between the entire submitted nucleotide sequences and original trRNA clone sequences, (2) the sensitivity of detecting a minor variant with 30% abundance, and (3) the identification rate of the nucleotide sequence of a variant comprising one-half of the sample.
The concordance rate was evaluated using all samples. Sensitivity was evaluated using samples #26 (30% minority) and #28 (10% minority). The identification rate was evaluated using sample #29. The criteria for evaluations are summarized in Supplementary Table S3, with examples. Here, the ambiguity codes defined by the International Union of Biochemistry and Molecular Biology were interpreted to indicate mixtures. “Concordance” was defined as complete agreement (e.g., A is A, R is R) and partial concordance (e.g., A is R, R is M) was regarded as “discordance.” For sensitivity of detecting minority nucleotides (R: G>A), we evaluated the detection of minority nucleotides either alone (A) or in a mixture including (A) (e.g., R, M) as “detected,” but nucleotides alone (G) as “not detected.” As for the identification of nucleotide mixtures, only concordant patterns (R is R) were recognized as “identified” and others as “not identified.”
Results
Laboratories show high concordance rates in entire nucleotide sequence analysis of single variant samples and lower concordance rates for mixed samples
The quality of HIV genotyping was evaluated by examining the concordance rates between reported and reference sequences. All participating laboratories successfully analyzed all six samples regardless of their HIV RNA copy number and reported entire nucleotide sequences. For the single variant samples #14, #15, and #16, concordance rates were high, on average 99.7%, 99.9%, and 99.9%, respectively, independent of DRM presence (Fig. 2A). However, laboratory ID-3 had the lowest scores for all three samples. This laboratory reported more ambiguity code “N”s representing A/G/C/T in its results than the other 10 laboratories; 33, 14, and 3 “N”s were reported in samples #14, #15, and #26, respectively. Since “N” was regarded as “discordance,” too much ambiguity lowered the scores of ID-3. Besides reporting more “N”s, ID-3 reported several partial concordances, which also affected their scores. As the RMs were single variant samples, the genotyping quality of ID-3 needs to be discussed and improved.
FIG. 2.
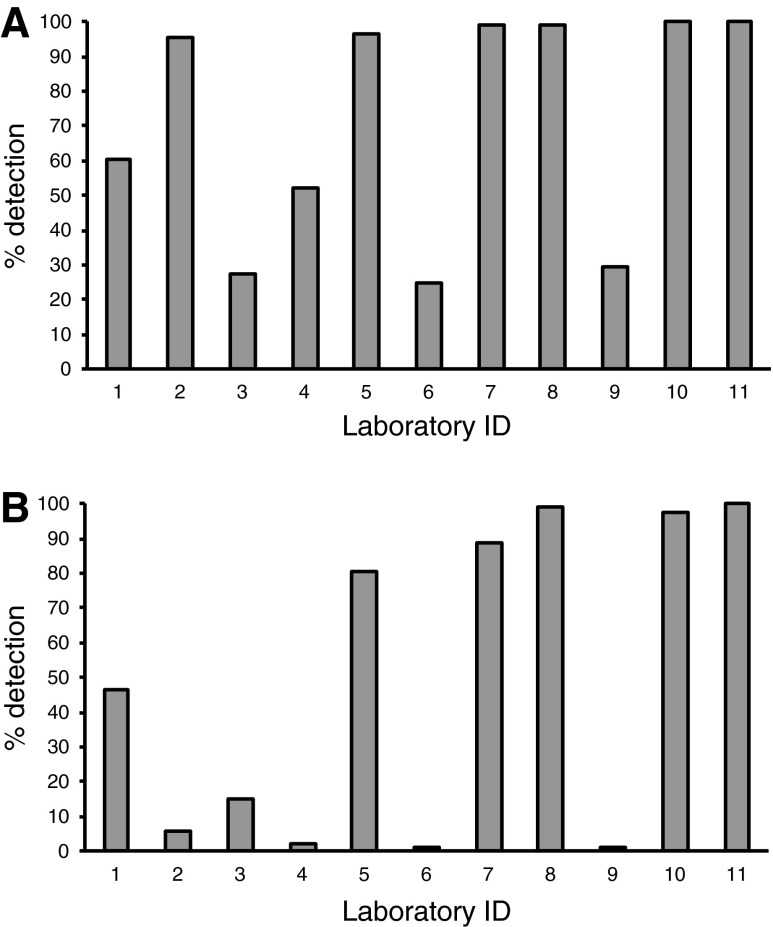
Concordance rates over the entire nucleotide sequence. (A) Samples #14, #15, and #16 consist of a single variant transcribed RNA (trRNA). Concordance rates reflect complete agreement between reported sequences and reference. (B) Samples #26 and #28 consist of two trRNAs containing a minor variant with 30% and 10% abundance, respectively. Sample #29 contains a variant comprising one-half of the sample.
In contrast to the high concordance rate in single variant samples, the concordance rates were lower in mixed samples. The average concordance rates for samples #26, #28, and #29 were 97.5%, 95.5%, and 98.2%, respectively (Fig. 2B). Almost all “discordances” were classified as “partial concordances,” except for five “complete discordances.” Of these, three were “N” in sample #26 reported from laboratory ID-3. Similar results were obtained in the analysis of concordance rates at 68 DRM positions as documented by the International AIDS Society, USA or the Stanford University HIV drug-resistance database (Supplementary Fig. S2). These results suggest high concordance rates for analysis of single variant samples, but not for mixed samples, with interlaboratory variations in concordance rates.
Detection rates of minor variants
To evaluate the ability of laboratories to detect minor variants, we examined their rates of detecting the nucleotide sequences of minority trRNA clones in samples #26 and #28, which were adjusted to contain 30% and 10% minority trRNA clones, respectively, of the total sample trRNA clones. Both samples had 88 discordant positions in nucleotide sequences, i.e., 26 and 62 positions in PR and RT, respectively (Supplementary Table S4). The average detection rates of minor variants in samples #26 (Fig. 3A) and #28 (Fig. 3B) were 71.3% and 48.9%, respectively, and median rates were 95.5% (range, 25.0–100) and 46.6% (range, 1.1–100), respectively. Thus, nearly half of these discordant positions were not detected in sample #28, suggesting that a 10% minority trRNA clone is difficult to detect consistently by population-based sequence methods. Detection rates for sample #26 were high in the majority of laboratories, except for laboratories ID-3, ID-6, and ID-9 (Fig. 3A). These results indicate extensive interlaboratory variations in the ability to detect minor variants.
FIG. 3.
Detection rate of the minor variant. (A) Detection rate of the nucleotide sequence of the minority trRNA clone in sample #26 (30% minority). (B) Detection rate of the nucleotide sequence of the minority trRNA clone in sample #28 (10% minority). Both samples have 88 discordant positions; theoretically 26 positions are in protease (PR) and 62 positions are in reverse transcriptase (RT).
Identification rates of the nucleotide sequence of the variant comprising one-half of the sample
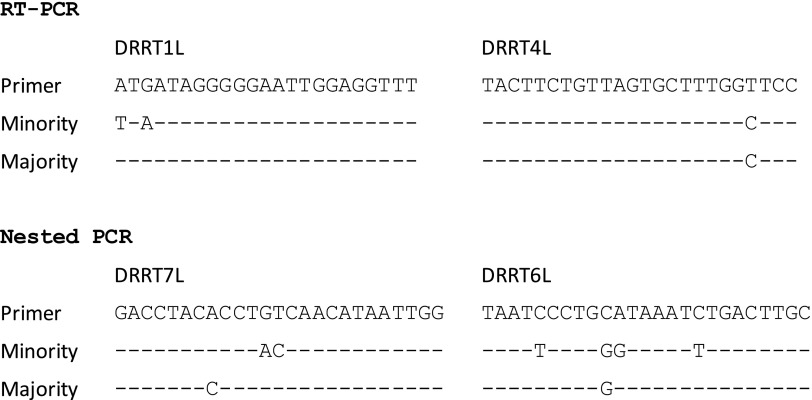
Sample #29 included a variant comprising one-half of the sample; therefore, we expected good identification rates of 88 discordant positions in nucleotide sequences (Supplementary Table S4). Indeed, 8 of 11 participating laboratories achieved over 90% identification rates, with an average rate of 77.3% among all 11 laboratories (Table 2). The identification rate for the PR region was better (93.4%) than that for the RT region (70.5%). Although eight laboratories achieved high identification rates in the PR and RT regions, three (ID-3, ID-6, and ID-9) failed to identify almost all discordant positions in the RT region. For these laboratories, the average identification rate in RT (1.6%) was significantly lower than that of the PR region (82.1%) (p=0.046 by Wilcoxon signed-rank test). These laboratories reported nucleotide sequences identical to that of the major variant of samples #26 and #28 (data not shown), clearly showing that these three laboratories specifically amplified only the major variant. To clarify the reasons for this specific amplification, we reviewed their genotyping protocols and found that all three laboratories used the same primer sets, DRRT1L/DRRT4L and DRRT7L/DRRT6L, for RT amplification. Comparison of the primer and trRNA sequences showed four internal mismatches to the minority trRNA clone in DRRT6L (Fig. 4), and these mismatches appear to have caused amplification failure of the minority trRNA clone.
Table 2.
Identification Rate of Correct Bases in a Sample with an Equal Mix
| Identification rate of mixture (%) | |||
|---|---|---|---|
| Laboratory ID | PR | RT | PR+RT |
| 1 | 100.0 | 90.3 | 93.2 |
| 2 | 100.0 | 100.0 | 100.0 |
| 3 | 65.4 | 4.8 | 22.7 |
| 4 | 92.3 | 98.4 | 96.6 |
| 5 | 100.0 | 100.0 | 100.0 |
| 6 | 84.6 | 0.0 | 25.0 |
| 7 | 100.0 | 93.5 | 95.5 |
| 8 | 96.2 | 98.4 | 97.7 |
| 9 | 96.2 | 0.0 | 28.4 |
| 10 | 92.3 | 90.3 | 90.9 |
| 11 | 100.0 | 100.0 | 100.0 |
PR, protease; RT, reverse transcriptase.
FIG. 4.
Sequence comparison between primers and trRNA in a mixed sample. Primer set DRRT1L and DRRT4L is for reverse transcriptase polymerase chain reaction (RT-PCR) and DRRT7L and DRRT6L for nested PCR. Identical bases are represented by dashes (-).
Identification rates of the amino acid sequences in individual codons
To investigate whether certain positions are particularly difficult to identify, we evaluated the identification rate of amino acid sequences for each codon. No differences in rates were observed in single variant samples #14, #15, and #16, whereas differences in rates were observed in mixed samples #26, #29, and #29 at codons with mixtures. Generally, the rates at codons with mixtures were low and varied, especially PR codon 37 and RT codon 200, which had two of three nucleotides in a codon as mixtures, demonstrating significantly lower identification rates. These findings suggest difficulty in accurately identifying amino acid sequences in codons with mixtures (Supplementary Fig. S3).
Discussion
This is the first report of an HIV-genotyping EQA study in Japan. Our evaluation of the quality of genotyping protocols of 11 participating laboratories showed that most of them scored high on sequence concordance, had good sensitivity in detecting a minor variant with 30% abundance, and accurately identified the nucleotide sequence of the variant comprising one-half of the sample. In this EQA study, our panel samples did not comprise seropositive plasma, but trRNA. Since our trRNAs are artificially constructed samples, their exact sequences were known; thus, we expected to precisely and critically evaluate the concordance rate and identification/detection sensitivity rates of participating laboratories' in house genotyping protocols and skills.
The greatest advantages of trRNA are that we can avoid drawing large amounts of blood from patients to prepare sample panels for EQA and have flexibility in inserting drug-resistance mutations that we wish to evaluate in EQA. Another advantage is that trRNAs are not infectious materials, allowing samples to be easily distributed to participating laboratories without any biohazard restrictions, as in Japan and many countries adopting the international model agreements of the United Nations Committee of Experts on the Transport of Dangerous Goods. On the other hand, the disadvantages of using trRNA are that samples prepared as single variant or mixed clonal templates may be limited by their narrow genetic diversity depending on the number of clones blended, not simulating the quasispecies of seropositive samples, and the process of RNA extraction cannot be evaluated by trRNA.
We chose to score participating laboratories on their rates of detecting a minor variant with 30% abundance and identifying the nucleotide sequence of a variant comprising one-half of the sample as important factors in the quality of their HIV-genotyping protocols and skills because previous EQA studies reported more discordance in mixed-sample analysis.13,14 Therefore, we tested not only single variant samples, but also three mixed samples. Indeed, 10%, 30%, and 50% mixtures were prepared because population sequencing is recognized to detect minor variants above 20% frequency.20 Thus, good detection/identification rates were expected in samples with 30% and 50% mixtures, but not with a 10% mixture. In fact, 8 of 11 laboratories scored high in detecting/identifying sequences in 30% and 50% mixture samples, but the detection rate was lower and varied with the 10% mixture sample (Fig. 3). Thus, we confirmed that the threshold of minority detection sensitivity of in house genotyping should be set between 10% and 30%.
We initially expected that all laboratories would detect the minor variant with 30% abundance and the variant comprising one-half of the sample. However, three laboratories (ID-3, ID-6, and ID-9) failed to detect almost all nucleotide sequences of the minority trRNA clone (Fig. 3). This failure appeared to be clearly due to nucleotide mismatches between the minority trRNA clone and the primers used by the three laboratories, suggesting PCR failure of the minority trRNA in these three laboratories. Nevertheless, this failure does not imply that their primers were misdesigned or inappropriate. We need to consider that the test results might have been affected in an extreme manner by the panel samples' narrow genetic diversity, only two trRNA clones.14,21 Although the failure might have been due to the mixed sample not having quasispecies, biased amplification due to primer sequences, namely “primer selection,” may happen in any protocol and sample unless a single primer pair is used for target amplification. Indeed, similar results were reported using clinical samples.13 Furthermore, the number of mismatches was correlated with PCR product yield despite the initial copy number input,22,23 suggesting the difficulty of avoiding biased amplification of quasispecies by PCR.
Therefore, multiple rounds of EQA are necessary, with careful evaluation followed by improvement of genotyping protocols to maintain genotyping quality. Furthermore, using trRNA, which allows more flexibility in designing mutations, may contribute to clarifying the mechanism of nonamplifiable samples that are sometimes observed among clinical samples, even those with high viral copy numbers. Making such panel samples and distributing them to participating laboratories may improve the quality of each laboratory's genotyping. Considering the advantages and limitations of clinical and artificial samples, we propose that EQA test panels should evaluate HIV genotyping using both clinical and artificial samples.
The importance of regular participation in HIV genotyping EQA programs was indicated in the European HIV Drug Resistance Guidelines.24 Although EQA programs have been implemented worldwide,15 region-specific factors, such as HIV-1 subtypes and transmitted drug-resistant variants, may affect EQA design. Therefore, in addition to managing our local EQA program, we encourage our members to participate in other EQA programs. Indeed, one laboratory in the JEQS2010 EQA participated in the EQA organized by NRL (National Reference Laboratory) Australia and received a good evaluation.13
The JEQS2010 program is the first step toward standardizing HIV genotyping in Japan. Accurately detecting minor variants and identifying the nucleotide sequence of the variant comprising one-half of the sample are critical not only for clinical testing, but also for basic science, surveillance of transmitted drug-resistant HIV-1, and detection of superinfections.25–28 More sensitive techniques, such as ultradeep pyrosequencing, can detect minor variants at less than 1% frequency, and are thus more useful than population-based sequencing for detecting minor variants with drug-resistance mutations.29,30 However, this technique is not yet popular in Japanese clinical laboratories; therefore, current standardization of HIV genotyping should be based on population-based sequencing.
Supplementary Material
Acknowledgments
We appreciate the cooperation and support of the participating laboratories and members of the JEQS2010 program and the Japanese Drug Resistance HIV-1 Surveillance Network: Haruyo Mori, Rumi Minami, Masayasu Oie, Kazue Uchida, Kenji Sadamasu, Mami Nagashima, Makiko Kondo, Atsushi Ajisawa, Takeshi Fujii, Teruhisa Fujii, Yuko Fujikawa, Jiro Fujita, Katsuyuki Fukutake, Tetsushi Goto, Motohiro Hamaguchi, Syuji Hanabusa, Naoki Hasegawa, Jun Hayashi, Tsunefusa Hayashida, Shigemi Hitomi, Masahide Horiba, Takeshi Iketani, Yoshiaki Ishigatsubo, Toshihiro Ito, Ichiro Itoda, Aikichi Iwamoto, Shinya Iwamuro, Akira Kaneda, Akiro Kimura, Ichiro Koga, Yoko Kojima, Mitsuru Konishi, Takeshi Kuwahara, Syuzo Matsushita, Motoo Matsuura, Hideaki Nagai, Masayoshi Negishi, Masako Nishizawa, Masaaki Noda, Shinichi Oka, Yasuo Ota, Chiho Otani, Hiroko Sagara, Norihiro Sato, Takeyuki Sato, Takuma Shirasaka, Koji Sudo, Akira Tachikawa, Masashi Taki, Sadahiro Tamashima, Yoshinari Tanabe, Yasuto Tanaka, Haruki Taniguchi, Masao Tateyama, Masanori Tei, Takahiro Tsuchiya, Atsuhisa Ueda, Mikio Ueda, Takamsa Ueno, Makoto Utsumi, Masahiro Yamamoto, Akira Yamanaka, Kunio Yano, Yoshiyuki Yokomaku, and Mihoko Yotsumoto. We also thank Claire Baldwin for her help in preparing the manuscript.
This study was supported by a Grant-in-Aid for AIDS research from the Ministry of Health, Labor, and Welfare of Japan (H22-AIDS-004).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Plebani M: Evaluating laboratory diagnostic tests and translational research. Clin Chem Lab Med 2010;48:983–988 [DOI] [PubMed] [Google Scholar]
- 2.Segondy M, Izopet J, Pellegrin I, et al. : Comparison of the QUANTIPLEX HIV-1 RNA 2.0 assay with the AMPLICOR HIV-1 MONITOR 1.0 assay for quantitation of levels of human immunodeficiency virus type 1 RNA in plasma of patients receiving stavudine-didanosine combination therapy. J Clin Microbiol 1998;36:3392–3395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pawlotsky JM, Bouvier-Alias M, Hezode C, et al. : Standardization of hepatitis C virus RNA quantification. Hepatology 2000;32:654–659 [DOI] [PubMed] [Google Scholar]
- 4.Shafer RW: Genotypic testing for human immunodeficiency virus type 1 drug resistance. Clin Microbiol Rev 2002;15:247–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geretti AM: Clinical implications of HIV drug resistance to nucleoside and nucleotide reverse transcriptase inhibitors. AIDS Rev 2006;8:210–220 [PubMed] [Google Scholar]
- 6.Gatanaga H, Ibe S, Matsuda M, et al. : Drug-resistant HIV-1 prevalence in patients newly diagnosed with HIV/AIDS in Japan. Antiviral Res 2007;75:75–82 [DOI] [PubMed] [Google Scholar]
- 7.Hattori J, Shiino T, Gatanaga H, et al. : Trends in transmitted drug-resistant HIV-1 and demographic characteristics of newly diagnosed patients: Nationwide surveillance from 2003 to 2008 in Japan. Antiviral Res 2010;88:72–79 [DOI] [PubMed] [Google Scholar]
- 8.Hogg RS, Bangsberg DR, Lima VD, et al. : Emergence of drug resistance is associated with an increased risk of death among patients first starting HAART. PLoS Med 2006;3:e356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Little SJ, Frost SD, Wong JK, et al. : Persistence of transmitted drug resistance among subjects with primary human immunodeficiency virus infection. J Virol 2008;82:5510–5518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vo TT, Ledergerber B, Keiser O, et al. : Durability and outcome of initial antiretroviral treatments received during 2000–2005 by patients in the Swiss HIV Cohort Study. J Infect Dis 2008;197:1685–1694 [DOI] [PubMed] [Google Scholar]
- 11.Huang HY, Daar ES, Sax PE, et al. : The prevalence of transmitted antiretroviral drug resistance in treatment-naive patients and factors influencing first-line treatment regimen selection. HIV Med 2008;9:285–293 [DOI] [PubMed] [Google Scholar]
- 12.Fujisaki S, Fujisaki S, Ibe S, et al. : Performance and quality assurance of genotypic drug-resistance testing for human immunodeficiency virus type 1 in Japan. Jpn J Infect Dis 2007;60:113–117 [PubMed] [Google Scholar]
- 13.Land S, Cunningham P, Zhou J, et al. : TREAT Asia Quality Assessment Scheme (TAQAS) to standardize the outcome of HIV genotypic resistance testing in a group of Asian laboratories. J Virol Methods 2009;159:185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sayer DC, Land S, Gizzarelli L, et al. : Quality assessment program for genotypic antiretroviral testing improves detection of drug resistance mutations. J Clin Microbiol 2003;41:227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pandit A, Mackay WG, Steel C, et al. : HIV-1 drug resistance genotyping quality assessment: Results of the ENVA7 Genotyping Proficiency Programme. J Clin Virol 2008;43:401–406 [DOI] [PubMed] [Google Scholar]
- 16.Descamps D, Delaugerre C, Masquelier B, et al. : Repeated HIV-1 resistance genotyping external quality assessments improve virology laboratory performance. J Med Virol 2006;78:153–160 [DOI] [PubMed] [Google Scholar]
- 17.Schuurman R, Demeter L, Reichelderfer P, et al. : Worldwide evaluation of DNA sequencing approaches for identification of drug resistance mutations in the human immunodeficiency virus type 1 reverse transcriptase. J Clin Microbiol 1999;37:2291–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schuurman R, Brambilla D, de Groot T, et al. : Underestimation of HIV type 1 drug resistance mutations: Results from the ENVA-2 genotyping proficiency program. AIDS Res Hum Retroviruses 2002;18:243–248 [DOI] [PubMed] [Google Scholar]
- 19.Neuwald P, Funelas M, DelCarmen J, et al. : Results of the 2001 AcroMetrix HIV-1 resistance proficiency program. J Clin Virol 2002;25(Suppl 3):S55–63 [DOI] [PubMed] [Google Scholar]
- 20.Vandamme A, Houyez F, Bànhegyi D, et al. : Laboratory guidelines for the practical use of HIV drug resistance tests in patient follow-up. Antivir Ther 2001;6:21–39 [PubMed] [Google Scholar]
- 21.Huang DD, Eshleman SH, Brambilla DJ, et al. : Evaluation of the editing process in human immunodeficiency virus type 1 genotyping. J Clin Microbiol 2003;41:3265–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christopherson C, Sninsky J, and Kwok S: The effects of internal primer-template mismatches on RT-PCR: HIV-1 model studies. Nucleic Acids Res 1997;25:654–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwok S, Kellogg DE, McKinney N, et al. : Effects of primer-template mismatches on the polymerase chain reaction: Human immunodeficiency virus type 1 model studies. Nucleic Acids Res 1990;18:999–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vandamme AM, Camacho RJ, Ceccherini-Silberstein F, et al. : European Recommendations for the Clinical Use of HIV Drug Resistance Testing: 2011 Update. Aids Rev 2011;13:77–108 [PubMed] [Google Scholar]
- 25.van der Kuyl AC. and Cornelissen M: Identifying HIV-1 dual infections. Retrovirology 2007;4:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Redd AD, Mullis CE, Serwadda D, et al. : The rates of HIV superinfection and primary HIV incidence in a general population in Rakai, Uganda. J Infect Dis 2012;206:267–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cornelissen M, Jurriaans S, Kozaczynska K, et al. : Routine HIV-1 genotyping as a tool to identify dual infections. AIDS 2007;21:807–811 [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Lerma JG, Nidtha S, Blumoff K, et al. : Increased ability for selection of zidovudine resistance in a distinct class of wild-type HIV-1 from drug-naive persons. Proc Natl Acad Sci USA 2001;98:13907–13912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunn DT, Coughlin K, and Cane PA: Genotypic resistance testing in routine clinical care. Curr Opin HIV AIDS 2011;6:251–257 [DOI] [PubMed] [Google Scholar]
- 30.Fisher R, van Zyl GU, Travers SA, et al. : Deep sequencing reveals minor protease resistance mutations in patients failing a protease inhibitor regimen. J Virol 2012;86:6231–6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.