SUMMARY
Chromosomes form three-dimensional structures that are critical to the regulation of cellular and genetic processes. Here, we present a study of global chromatin interaction patterns in Arabidopsis thaliana. Our genome-wide approach confirmed interactions that were previously observed by other methods as well as uncovered previously unknown long-range interactions such as those among small heterochromatic regions embedded in euchromatic arms. We also found that interactions are correlated with various epigenetic marks that are localized in active or silenced chromatin. Arabidopsis chromosomes do not contain large local interactive domains that resemble the topological domains described in animals, but instead contain relatively small interactive regions scattered around the genome that contain H3K27me3 or H3K9me2. We generated interaction maps in mutants that are defective in specific epigenetic pathways and found altered interaction patterns that correlate with changes in the epigenome. These analyses provide further insights into molecular mechanisms of epigenetic regulation of the genome.
INTRODUCTION
Spatial organization of the genome and higher order chromosome structures can be studied by a series of chromosome conformation capture (3C)-based approaches (Dekker et al., 2013). Eukaryotic chromosomes are organized in three dimensions inside the nucleus (Dixon et al., 2012; Gibcus and Dekker, 2013; Jin et al., 2013; Lieberman-Aiden et al., 2009; Sexton et al., 2012). Folding of chromosomes leads different regions of chromatin to interact with each other, which often bears important functional significance such as maintaining genome integrity, compartmentalizing silent chromatin, regulating gene expression, regulating DNA replication, and forming highly interactive local domains (Dixon et al., 2012; Jin et al., 2013; Sexton et al., 2012; Zhang et al., 2012). Chromatin interaction studies have uncovered looping interactions in genes, such as those between enhancers and CTCF-binding sites with their target promoters, as well as interactions between centromeres, telomeres, early origins of replication, and chromosomal breakpoints (Crevillen et al., 2013; Dekker et al., 2013; Dixon et al., 2012; Duan et al., 2010; Jin et al., 2013; Li et al., 2012; Sanyal et al., 2012; Shen et al., 2012). Highly expressed and transcriptionally related genes have also been shown to interact with each other (Dekker et al., 2013; Gibcus and Dekker, 2013; Osborne et al., 2004; Tanizawa et al., 2010). Interestingly, mammalian chromosomes often have subchromosomal compartments preferentially enriched for either gene-rich transcriptionally active or gene-poor transcriptionally silent chromatin regions (Gibcus and Dekker, 2013; Lieberman-Aiden et al., 2009).
Transcriptionally active and silent chromatin regions are distinguished by distinct epigenetic marks such as DNA methylation and particular histone modifications (Feng and Jacobsen, 2011). For example, in Arabidopsis, silent heterochromatin is marked by histone H3 lysine 9 di-methylation (H3K9me2) and H3K27 mono-methylation (H3K27me1) (Bernatavichute et al., 2008; Jacob et al., 2009), whereas H3K4 tri-methylation (H3K4me3) marks actively-transcribed regions, and is absent from heterochromatin (Zhang et al., 2009). Constitutively highly expressed protein-coding genes are also modified by DNA methylation occurring in the CG dinucleotide context within the transcribed region (gene body methylation) (Feng et al., 2010; Zemach et al., 2010). On the other hand, H3K27me3 (mediated by Polycomb pathway) marks silent genes and genes with tissue-specific expression patterns, and is highly anticorrelated with gene body methylation (Zhang et al., 2007). DNA methylation in non-CG sequence contexts (CHG and CHH, where H = A, C, or T) is often found on repetitive DNA, such as transposable elements (“TEs”) that form heterochromatin (Feng and Jacobsen, 2011). These observations raise the possibility that epigenetic marks might influence the architecture and interaction patterns of the chromosomes. This idea is consistent with a recent study that employed a circular chromosome conformation capture (4C) technique in Arabidopsis and found that interacting regions tend to have similar epigenetic landscapes (Grob et al., 2013).
Chromosomes from mammals, Drosophila, and bacteria form large local interactive domains, termed “topological”, “physical”, and “chromosome interaction” domains, respectively (Dixon et al., 2012; Le et al., 2013; Sexton et al., 2012). Although the sizes of these domains vary in different organisms, it is common in eukaryotes (mammals and Drosophila) that these domains correlate extensively with active or repressive epigenetic modifications (Dixon et al., 2012; Sexton et al., 2012). Furthermore, the boundaries of topological and physical domains are often marked by the insulator binding protein CTCF, as well as other factors such as actively transcribed genes or certain types of TEs (Dixon et al., 2012; Sexton et al., 2012). So far, it remains unclear whether chromosomes of the model plant Arabidopsis also contain well-defined regular domain structures or whether they have unique folding and interaction patterns.
In this study, we adapted genome-wide Hi-C (Lieberman-Aiden et al., 2009) to study chromatin interactions in Arabidopsis. Our datasets in wild type and various epigenetic mutants provide a framework for further understanding of the nuclear organization of plant genomes and the relationship between epigenetic marks and chromosome architectures.
RESULTS AND DISCUSSION
High Resolution Maps of Chromatin Interaction in Arabidopsis
We previously developed a Hi-C protocol suitable for analysis of the genome of Arabidopsis thaliana (L.) Heynh (Moissiard et al., 2012). Using this protocol we generated Hi-C libraries for wild-type Arabidopsis, as well as from a suite of mutants defective in gene silencing, DNA methylation, or specific histone modifications. We obtained ~41 to 66 million usable paired-end reads from each library, which, given the relatively small size of the Arabidopsis genome (125 million bases) (Arabidopsis Genome, 2000) and the dynamic smoothing process we applied, permits analysis of interaction at high resolution. Sequencing data were first processed using previously described analysis filters and pipelines (Lieberman-Aiden et al., 2009; Moissiard et al., 2012) with modifications, and then plotted in two dimensional matrices to present interaction tendency between any two locations in the Arabidopsis genome (Figure 1A and Data S1) (see Supplemental Experimental Procedures online). We also generated one-dimensional plots depicting the general relationship between interaction tendency and genomic distance (Figure S1A).
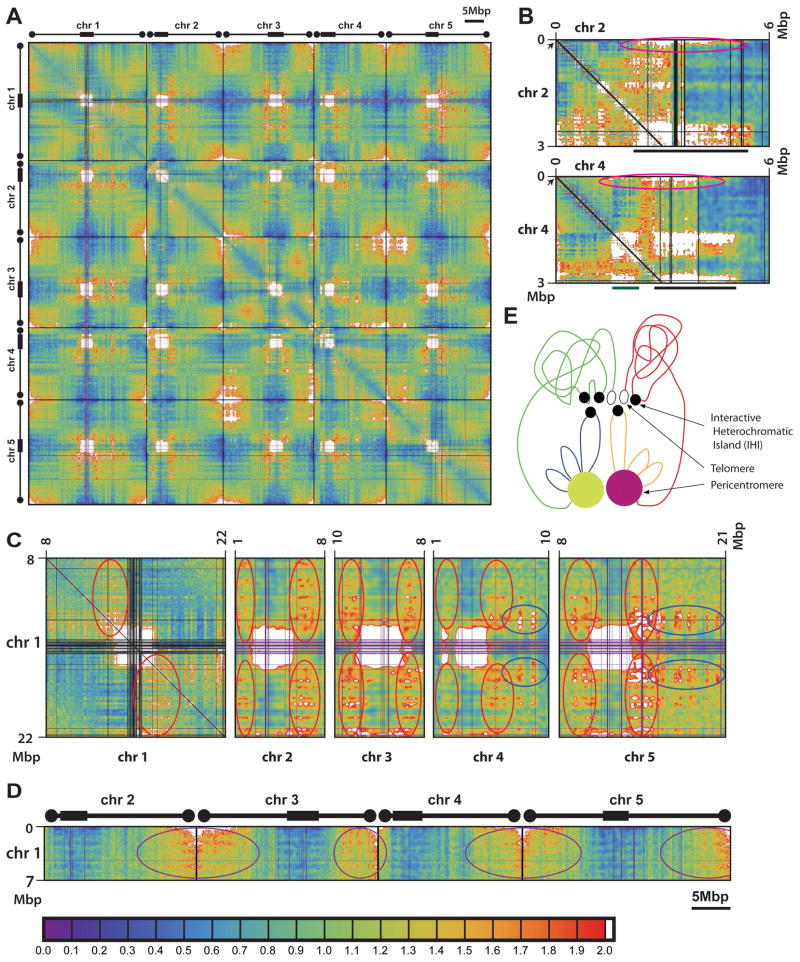
Figure 1. Chromatin interaction patterns in Arabidopsis.
(A) Two dimensional interaction map of wild type. The five Arabidopsis chromosomes are shown from left to right and top to bottom. Chromosomes are separated by thin black bars. Thick boxes mark approximate positions of the pericentromeric regions, and circles mark approximate positions of the telomeric regions. Blue to green to yellow to orange to red is weak to strong interaction tendency, with white for very strong interaction beyond a threshold. Light gray indicates areas withheld from analysis due to, e.g., problematic 50-mer mapping.
(B) Selected detail of wild type interaction map. Chromosomal coordinates are labeled on the top and left sides. Pericentromeres of both chromosomes and the knob of chromosome 4 are labeled by black and green bars at bottom, respectively. Locations of the NORs are indicated by black arrows. Pink circles indicate interaction of NORs with pericentromeres and the knob of chromosome 4.
(C) Selected detail of wild type interaction map illustrating the interaction between proximal euchromatic arms and peripheral areas of pericentromeric heterochromatin. Red circles indicate interaction of the arms of chromosome 1 with pericentromeres of all five chromosomes. Blue circles indicate interaction of the arms of chromosomes 4 and 5 with pericentromere of chromosome 1.
(D) Selected detail of wild type interaction map illustrating the interaction of a distal euchromatic arm of chromosome 1 with the distal euchromatic arms of chromosomes 2–5. Purple circles indicate areas of strong interaction.
(E) Working model of typical major chromatin interaction patterns in Arabidopsis. The diagram depicts the pericentromeres of chromosomes 3 (yellow) and 5 (pink), along with the long arm of chromosome 3 (proximal chromatins in blue and distal chromatins in green) and the short arm of chromosome 5 (proximal chromatins in orange and distal chromatins in red). IHIs are indicated by closed black circles, and the telomeres are indicated by open black circles. See text for details.
Color bar for in panels (A) to (D) is shown at the bottom of the figure. See Supplemental Experimental Procedures online for details.
General Patterns of Chromatin Interaction Exhibited by the Arabidopsis Genome
A prominent feature of the wild-type Arabidopsis genome is that it is generally not segmented into large local adjacent interactive domains as has been previously described in several other organisms (Figure 1A) (Dixon et al., 2012; Le et al., 2013; Sexton et al., 2012), although a small number of interactive domains do exist (see later sections). To ensure this difference was not due to differences in our analysis methods, we applied our pipeline to a previously published mouse Hi-C dataset (Dixon et al., 2012) and readily observed topological domains that have been described (Figure S1B). The boundaries of topological domains are highly correlated with the binding of insulator protein CTCF (Dixon et al., 2012; Sexton et al., 2012), suggesting that CTCF may play a central role in defining the borders of topological domains. The lack of CTCF in plants (Heger et al., 2012) may help explain the lack of regular large topological domains, and suggests that nuclear architecture of plants is significantly different from that of animals.
The strongest interactions we observed were exhibited by the blocks of pericentromeric heterochromatin, both among sequences within the same pericentromere and between sequences of different pericentromeres (Figure 1A) (Moissiard et al., 2012). This is consistent with previous studies showing that Arabidopsis has well-defined chromocenters formed by pericentromeric heterochromatin, these being visible by light microscopy, and that homologous pericentromeric regions frequently associate with each other in DNA-FISH assays (Fransz et al., 2002; Schubert et al., 2012). We detect interaction between all pairs of heterologous pericentromeres (Figure 1A). Also, consistent with a previous 4C study in Arabidopisis (Grob et al., 2013), the interaction of pericentromeres includes the heterochromatic knob on the short arm of chromosome 4, which is close to the pericentromere (Figure 1A) (Arabidopsis Genome, 2000). The knob is composed of heterochromatin regions that were originally derived from the pericentromere, and therefore the interaction of the knob with pericentromeres is analogous to the interactions between the pericentromeres. The region between the knob and the pericentromere is not included in these strong interactions, indicating the presence of a euchromatic loop (Figure 1A), consistent with earlier FISH studies (Fransz et al., 2002). Moreover, pericentromeres have very little interaction with regions outside of the pericentromeres or the knob, except for the Nucleolar Organizing Regions (“NORs”; see below) (Figure 1A) (Moissiard et al., 2012).
We also detected strong interactions among telomeres (Figure 1A) (Moissiard et al., 2012), supported by previous DNA-FISH assays (Schubert et al., 2012), and consistent with the “telomere bouquet model” in which telomeres cluster around the nucleolus (Harper et al., 2004; Scherthan, 2007). Figures 1A and S1A shows that telomere interactions take place among all telomeres of all chromosomes, suggesting that all telomeres cluster together randomly. The telomeres on the short arms of chromosomes 2 and 4 take part in these interactions, but interact more weakly than the other telomeres (Figure 1A). The short arms of chromosomes 2 and 4 contain the 45S rDNA containing NORs, which are very close to the telomeres (Arabidopsis Genome, 2000) (Fransz et al., 2002). By DNA-FISH, the two NORs not only interact with each other, but also colocalize with the chromocenters of chromosomes 2 and 4 (Fransz et al., 2002), as seen with Hi-C (Figure 1A/B). The strong association of the NORs and chromocenters likely prevents the adjacent telomeres from strongly interacting with other telomeres. While not seen in Figure 1A (due to genomic distance-related data modeling; see Supplemental Experimental Procedures online), the strong interaction between the two telomeres of each single chromosome is clearly evident both from the general relationship between interactivity and genomic distance (Figure S1A) and two dimensional plots in which the average distance-related interactivity has not been removed (Figure S1C). The interactive domains at the ends of chromosomes extend much further than the telomere territories themselves (Figure 1A), in line with previous findings showing that distal regions of chromosome arms interact more frequently than regions close to the centromeres (Grob et al., 2013; Schubert et al., 2012).
A previous DNA-FISH-based model of Arabidopsis nuclear architecture hypothesized that regions of the euchromatic arms associate with pericentromeric heterochromatin, with euchromatic loops emanating from the chromocenters (Fransz et al., 2002). Our data support this model by revealing that there are regions in the euchromatic arms that frequently interact with the peripheral areas of pericentromeric heterochromatin, both intra- and inter-chromosomally (Figures 1A and 1C). However, we found that the euchromatic regions that participate in these interactions are those in the roughly one half of the chromosome arm that is closest to the pericentromere (Figures 1A and 1C). We also found that for a given chromosome, euchromatic-pericentromeric interactions are higher between sequences on a given euchromatic arm with the half of the pericentromere that it is adjacent to (Figure 1C chromosome 1). This preference might contribute to the observation of a generally higher level of interaction between the chromatin regions on one side of a centromere with other chromatin on that same side, as opposed to interaction with chromatin regions on the opposite sides of the centromere (Figure S1C), which is supported by FISH studies showing that the two euchromatic arms of the same chromosome tend not to intermingle (Berr and Schubert, 2007). Interestingly, these same regions tend to interact with the areas adjacent to the pericentromeres on all of the other chromosomes as well, and in this case they interact with chromatin regions on both sides of the centromere (Figure 1C chromosomes 2–5). This could be explained by the observation that all of the pericentromeric regions interact strongly (Figure 1A), which would bring these euchromatic sequences in closer proximity to each other.
In sharp contrast to the interactions of the proximal half of the euchromatic arms, the distal half of the arms preferentially interact with the distal half of the other chromosome arms (Figures 1A, 1D and S1C), and sharp boundaries between these two halves of the arms are frequently apparent (e.g., arms of chromosome 1 with those of chromosome 3 in Figure 1A). These distal interactions mirror, and may be in part driven by, the telomere interactions described above.
The Arabidopsis genome thus shows a unique nuclear organization that is different from that of animal genomes previously studied, showing a lack of regular large topological domains but prominent interaction patterns involving pericentromeres, telomeres, and very large areas of the euchromatic arms partitioned into a centromere proximal region that interacts with itself and the pericentromeric regions, and a distal region that interacts with itself and the telomeric regions (Figure 1E). This is further supported by a hierarchical clustering analysis (Figure S1D).
Interactive Heterochromatic Islands Formed by Multiple Loci in Arabidopsis
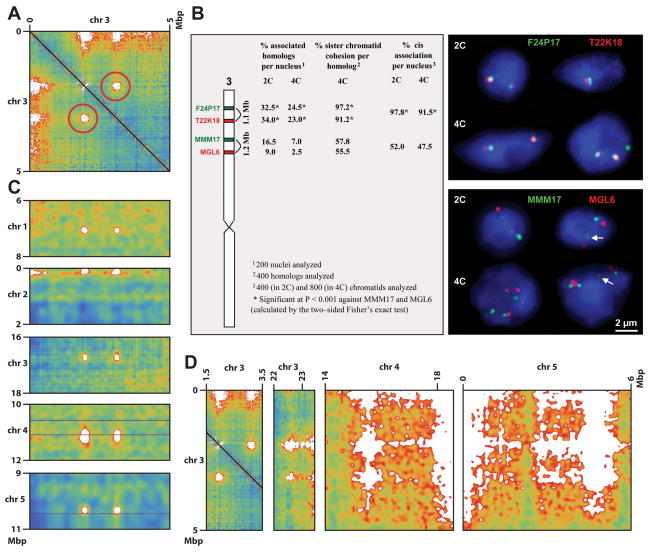
Inspection of the interaction maps revealed a number of high intensity off-diagonal punctate signals, indicating strong interactions between loci far apart in the primary DNA sequence (Figure 1A). Prominent examples include two loci near the beginning of chromosome 3, which can also be readily detected by 3C analyses (Figure 2A and S2A, and Table S1). DNA-FISH further confirmed a high frequency of interaction between these two loci (Figure 2B and Table S1B). We found that regions participating in these interactions all contain small patches of heterochromatin (average size ~7 Kbp) that exist in the otherwise euchromatic arms. They are marked, for instance, by H3K9me2 and contain numerous TE-related repetitive sequences, but are flanked by expressed protein-coding genes (Figure S2B–E). We therefore term these structures “Interactive Heterochromatic Islands”, or “IHIs”. The euchromatic arms also contain many additional similar patches of heterochromatin, some larger in size, but do not show these long-range interactions. While the IHI interacting regions all contained these heterochromatin patches, the interacting region is much larger than the patches themselves, ranging from 200 to 1,600 Kbp (Table S1A). It is clear, though, that the peaks of highest interaction intensity overlap small heterochromatin patches (sharp H3K9me2 peaks) within the IHIs (Figure S2B–F), suggesting that the interactions of IHIs are possibly mediated through the heterochromatin patches. Nonetheless, Figure S2F also suggests that heterochromatin patches alone are not sufficient to cause chromatin interaction, since there are peaks of H3K9me2 within the IHIs that do not overlap with peaks of interaction. Intriguingly, all IHIs showed interaction with all other IHIs (Figures 2A, 2C, and 2D). Because our Hi-C data is generated from large collections of cells, it is not possible to tell if all IHIs are clustered together simultaneously, or whether only a fraction of these interactions exist at any given moment in a given single cell. Curiously, although IHIs have heterochromatic features similar to sequences in pericentromeric heterochromatin, they do not interact with pericentromeric regions. Instead, we found that IHIs frequently interact with telomeric and subtelomeric regions (Figures 1A and 2A/D). In addition, the broad regions surrounding IHIs show a higher level of interchromosomal interactions, especially for those in the distal half of the euchromatic arms (Figures 1A and 2D). Although the function of IHI interactions is not known, it is possible they strengthen the interactions of larger domains, such as the telomere-containing distal halves of the euchromatic chromosomes (Figure 1E). It is interesting that the dynamics and participants of the IHI interaction network are altered in several epigenetic mutants (see below), suggesting that epigenetic marks are in part regulating these interactions. Together, these findings suggest that a novel network of interactions takes place among a small set of epigenetically silent regions and the telomeres, forming a previously unappreciated level of complexity of interaction within the Arabidopsis nucleus.
Figure 2. Interactive Heterochromatic Islands (IHIs) in Arabidopsis.
(A) Two loci on chromosome 3 interact strongly with each other. Chromosomal coordinates are labeled on the top and left sides. Red circles indicate the interaction (interaction maps are symmetric about their main diagonals and so it appears twice).
(B) DNA-FISH analysis of the two loci on chromosome 3 from panel (A). The left subpanel diagrams chromosome 3 and the approximate positions of BACs used, along with a statistical summary of the results (*P < 0.001 via two-sided Fisher’s exact test) from 200 nuclei. Here, “homologs” refer to the same regions on homologous chromosomes, and “cis association” refers to the association between different regions on the same chromosome. Representative images of 2C and 4C nuclei show different configurations of associated and/or separated chromatin segments of the frequently associated BACs F24P17 and T22K18 (upper panel) in comparison to configurations in nuclei labeled by BACs MMM17 and MGL6 as a control (lower panel). Chromatin elongation in 2C and 4C nuclei (indicated by white arrows) and sister chromatid separation in 4C may lead to more than two or four signals per nucleus in 2C and 4C nuclei, respectively.
(C) Interaction of the two loci on chromosome 3 from panel (A) with five other loci, one from each chromosome.
(D) Interaction of the two loci on chromosome 3 from panel (A) with loci toward the beginning of chromosome 5 and the ends of chromosomes 3 and 4.
Color scales in panels (A), (C), and (D) are the same as in Figure 1A.
Comparisons of Genomic Features and Chromatin Interactions
We assessed connections between chromatin interaction patterns and various genomic datasets, including histone modifications (ChIP-Seq and ChIP-Chip), DNA methylation (BS-Seq), and RNA abundance (RNA-Seq) (all data utilized are displayed at http://genomes.mcdb.ucla.edu/AthBSseq/ and were produced from similar tissues as the Hi-C data) (Bernatavichute et al., 2008; Jacob et al., 2009; Stroud et al., 2014; Stroud et al., 2013; Zhang et al., 2009; Zhang et al., 2007). We permuted 2.5 Kbp-binned copies of the Arabidopsis genome based on bin average signal intensity for each genomic dataset, and then rendered the interaction map in each new order. Thus, regions with the highest signal intensity for a given genomic feature (e.g., H3K9me2) are clustered to the top left of each permuted map, and those with low intensity toward the bottom right (Figures 3 and S3, and Data S1). This allows for easy visualization of increased or decreased chromatin interactions in regions enriched or depleted for particular genomic features.
Figure 3. Genomic features and chromatin interactions.
(A) Two dimensional interaction map of wild type, except with chromosomal positions (2.5 Kbp bins) permuted within each chromosome based on intensity of H3K9me2 signals. Panels on the left and bottom indicate the chromosomal positions of the loci in the map. Panels on the right and top indicate the H3K9me2 modification level of the corresponding loci. Regions with the highest and lowest H3K9me2 levels are clustered toward the top left corner and bottom right corner, respectively, of each permuted map. Red indicates higher interaction and green indicates lower interaction than average.
(B) Two dimensional interaction map of wild type as in (A), except with chromosomal positions permuted based on intensity of H3K4me3 signals.
Color bar is shown at the bottom of the figure. See Supplemental Experimental Procedures online for details.
Most repressive epigenetic marks, including DNA methylation, H3K9me2, and H3K27me1, strongly colocalized with the highest levels of chromatin interactions (upper left corners in Figures 3A and S3C, and Data S1). This is consistent with our finding that the strongest chromatin interactions exist among and between pericentromeric regions that are enriched in these marks (Figures 1A, 3A, and S3C, and Data S1). We also observed lower interaction between the heavily DNA, H3K9 di-, or H3K27 mono-methylated regions with the rest of each chromosome (Figures 3A and S3C, and Data S1). This likely reflects the association of pericentromeric regions with themselves and avoidance of the rest of the genome (Figure 1A). We also found that if we exclude previously defined pericentromeric regions (Bernatavichute et al., 2008) from the permutation analysis, there is no clear association of chromatin interaction strength with either highly or lowly H3K9m2 modified regions (Figure S3D), suggesting that the effect observed in Figure 3A is primarily the result of pericentromere interactions.
Marks of active chromatin, such as H3K4 mono-, di-, and tri-methylation showed anti-localization with the brightest Hi-C signals (Figure 3B and Data S1), suggesting that transcriptional active regions do not show unusually high interaction among themselves. Consistently, these marks almost exclusively exist outside of the pericentromeric regions that show the strongest interactions in the genome (Figure 3B and Data S1). We also examined the relationship between mRNA-Seq-derived gene expression levels and chromatin interactivity. Notably, we did not see a correlation of chromatin interactivity with genes of the highest level of RNA-Seq expression, even when pericentromeric regions were excluded (Figure S3E), suggesting a lack of clustering of the most actively transcribed genes generally. This result is consistent with our recent observation that plant RNA Polymerase II, while excluded from the heterochromatic regions, exhibits a homogeneous distribution pattern within the euchromatic regions of interphase nuclei (Schubert, In press).
Local Interactive Domains Correlated with Certain H3K27me3 and H3K9me2 Regions
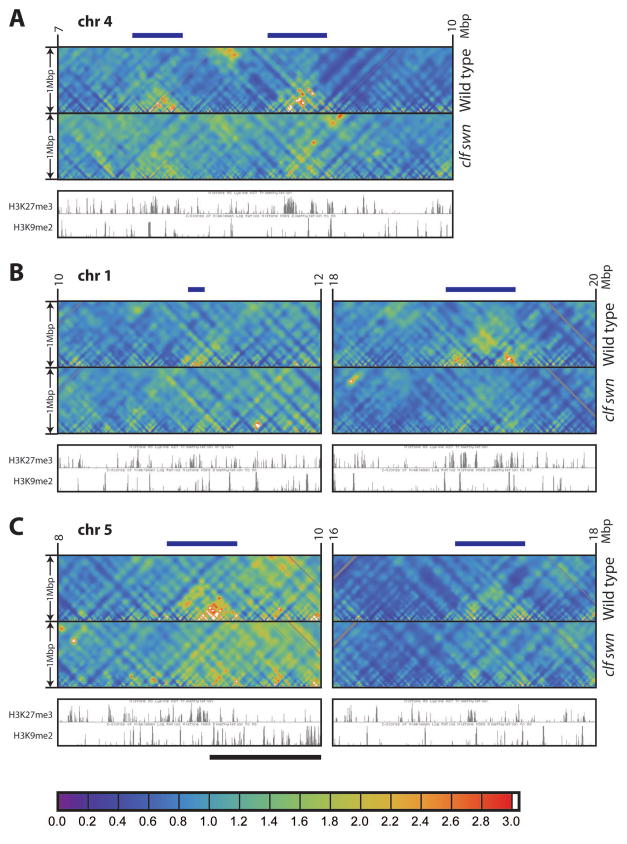
To investigate interactive domains on a short to medium distance scale (up to a few megabasepairs), we extracted strips of the interaction maps near the main diagonal and rendered them as detailed local interaction maps (see Figure S4A for chromosome 3 of wild type, and Data S1 for a complete set). From these, we identified a small number of interactive domains scattered throughout the genome, which are correlated with certain regions marked with either of the two histone modifications, H3K27me3 or H3K9me2.
H3K27me3 is a silencing mark of the Polycomb system and is present on about 17% of protein-coding genes (Turck et al., 2007; Zhang et al., 2007). As opposed to the frequent clustering of Polycomb-regulated genes known in animals (e.g., Hox gene clusters) (Schwartz et al., 2006; Tolhuis et al., 2006), H3K27me3 regions are more evenly scattered throughout the Arabidopsis genome and are generally restricted to the transcribed regions of single genes, with only a few H3K27me3 regions clustered together, especially in the case of tandemly repeated homologous genes (Turck et al., 2007; Zhang et al., 2007). Recent work (Rosa et al., 2013) has shown that the two FLOWERING LOCUS C (FLC, a well-known target of the Polycomb/H3K27me3 pathway) alleles on homologous chromosomes cluster together, suggesting long-range interactions. Furthermore, in Drosophila, Polycomb/H3K27me3 target regions have been shown to interact with each other (Sexton et al., 2012). We identified two heavily H3K27me3-marked regions on chromosome 4 of wild type, lying within ~1 Mbp of each other, which show strong interaction within the domains (Figure 4A wild type), as well as a number of other similar regions (Figure 4B/C wild type). These regions correspond to unusually large clusters of adjacent H3K27me3-marked genes (Figures 4 and S4B). On the other hand, we did not observe a higher chromatin interactivity for H3K27me3-marked regions generally in the genome (Figure S3F and Data S1). Therefore, the H3K27me3 interactive domains are apparently limited to a small number of sites in the genome, where H3K27me3-marked genes are clustered.
Figure 4. Local interactive domains in Arabidopsis.
Local interaction maps of a selected 3 Mbp region in chromosome 4 (A) and 2 Mbp regions in chromosomes 1 (B) and 5 (C) from wild type and clf swn double mutant, to a distance of 1 Mbp (see y axis). Chromosomal coordinates are labeled on top. H3K27me3 and H3K9me2 tracks shown are from wild type on the UCSC Genome Browser. The local interactive domains are labeled by dark blue bars on tops of plots. The left subpanel of (C) is close to pericentromeric heterochromatin, and thus a large interactive domain overlapping with strong H3K9me2 signals appears in the right half (black bar).
Color bar (same as Figure S1D) is shown at the bottom of the figure. See Supplemental Experimental Procedures online for details.
See also Figure S4, and Data S1 and S6.
As discussed above, pericentromeric regions marked by H3K9me2 interact extensively within and among each other (Figure 3A), readily visualized in local interaction maps as large domains near the centromeres of each chromosome (Figure S4A/C and Data S1). In addition, we observed a number of small patches of H3K9me2 outside of the pericentromeric regions (Bernatavichute et al., 2008) that show a high level of interactivity within the domain (Figure S4A/C). All of the most prominent H3K9me2-containing small local interactive domains correspond to the IHIs described above; however, not all IHIs form locally interacting domains of this type (Figures 2 and S4A/C, and Table S1A). Indeed, the IHIs form stronger interactions among each other than they do with other areas of chromatin in the vicinity of the IHI (Figures 1 and 2). This suggests that IHIs, unlike pericentromeric regions, do not necessarily form strong local interactive domains, but instead have a relatively high level of interactivity with other IHIs and the telomere regions.
Epigenetic Mutants Show Altered Chromatin Interaction Patterns
We performed Hi-C in a number of mutants affecting epigenetic regulation, including the curly leaf (clf) swinger (swn) double mutant, arabidopsis thaliana microrchidia 6 (atmorc6), morpheus’ molecule 1 (mom1), decrease in dna methylation 1 (ddm1), methyltransferase 1 (met1), chromomethylase 3 (cmt3), and the su(var)3–9 homolog 4 (suvh4) suvh5 suvh6 triple mutant. We generated two dimensional interaction maps (Figures 5A and S5, and Data S2–S5), local interaction maps (Data S6), and histograms and two dimensional plots of interaction differences between each mutant and wild type (Figures 6 and S6, and Data S2 to S5). Histograms suggest that the overall differences between mutants and wild type are readily detectable and are visibly larger than the difference between two wild type replicates (Figure S6B).
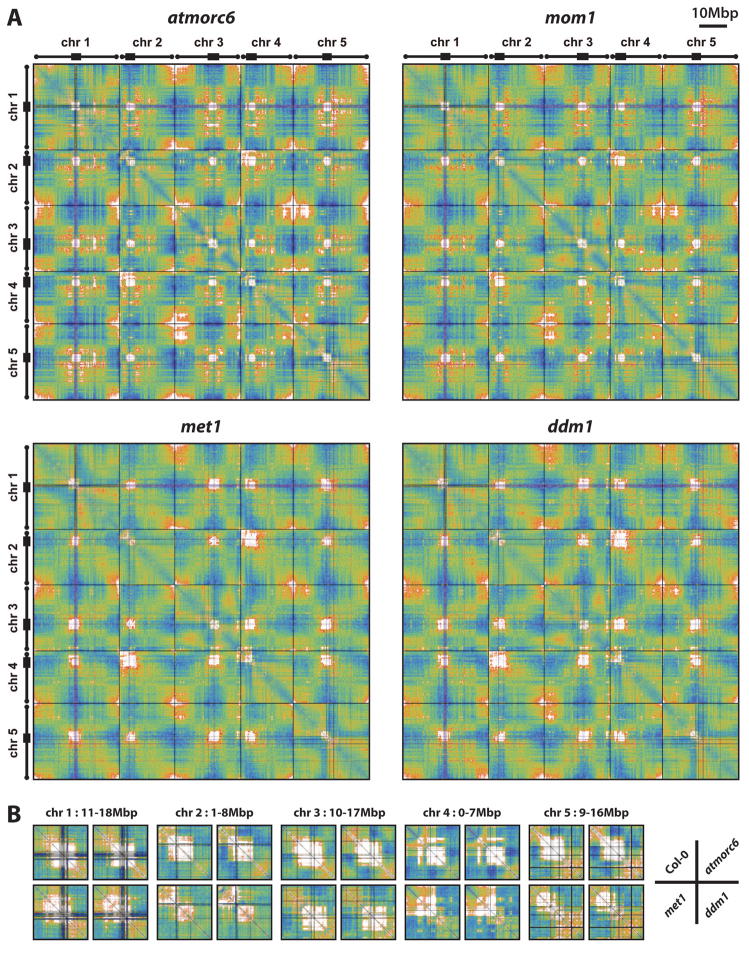
Figure 5. Chromatin interaction patterns in mutants affecting epigenetic processes.
(A) Two dimensional interaction maps of atmorc6, mom1, met1, and ddm1 generated in the same way as Figure 1A.
(B) Detail of selected pericentromeric interactions across wild type and mutants. The region selected for each comparison is indicated on top.
Color scales are the same as in Figure 1A.
See also Figure S5, and Data S2–S5.
Figure 6. Comparison of interaction patterns across mutants and wild type.
Chromatin interaction patterns of atmorc6, mom1, met1, and ddm1 vs. wild type. Colors show difference of mutant relative to mean of wild type and mutant (“percent difference”); black is no change and brightest red (mutant higher) and brightest green (mutant lower) are 100%.
Chromosome labels and gray are as in Figure 1A.
Color bar is shown at the bottom of the figure. See Supplemental Experimental Procedures online for details.
See also Figure S6, and Data S2–S5.
CLF and SWN are two enhancer of zeste (Ez) homologs that control H3K27me3, and the clf swn double mutant lacks virtually all H3K27me3 (Lafos et al., 2011). We analyzed the local interactive domains consisting of clustered H3K27me3 genes described above, and found that the interaction within these domains was dramatically reduced or eliminated in the double mutant (Figure 4, and Data S1 and S6). Hence, H3K27me3 may act directly or indirectly to regulate the interactivity of these domains. Although the functions of the clustering of these H3K27me3-marked genes and their higher level of local interactivity are unknown, the change in the behavior of these domains in clf swn suggests regulation of these interactions at an epigenetic level.
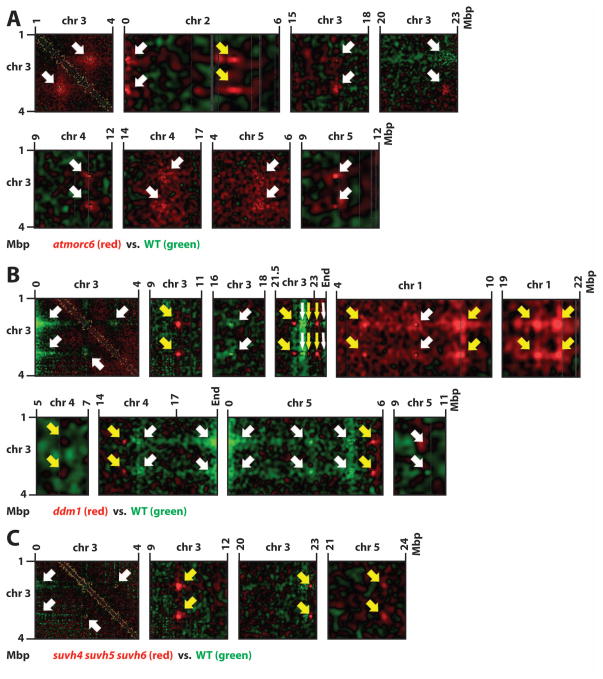
AtMORC6 is an ATPase that is required for heterochromatin condensation, and we have previously shown a low-resolution Hi-C comparison of wild type vs. atmorc6 (Moissiard et al., 2012). We repeated Hi-C analysis of atmorc6 at higher coverage and resolution. As we showed previously, atmorc6 exhibits decreased association of pericentromeric heterochromatin with itself as well as elevated interaction between pericentromeric heterochromatin and euchromatic arms (Figure 6), consistent with decondensed chromocenters and de-repressed transposon expression in atmorc6 (Moissiard et al., 2012). Interestingly, the atmorc6 mutant clearly shows enhanced interaction among telomeres when compared to wild type, and the interactive chromatin regions at the ends of chromosomes extend much further from the telomere territories into euchromatic arms than in wild type (Figures 1A, 5A, and 6). In addition, atmorc6 affects the above-mentioned interactions involving the distal and proximal halves of the euchromatic chromosome arms, such that the proximal interaction is enhanced and the distal interaction is reduced in the atmorc6 mutant (Figure 6). Thus, AtMORC6 appears to regulate large-scale nuclear organization, and shifts the balance of interaction of euchromatic arms with themselves and with pericentromeres. Moreover, AtMORC6 affects the interactions of the IHIs: compared to wild type, atmorc6 shows enhanced interaction among IHIs (Figure 7A and Data S3). This is particularly interesting in light of the fact that both IHIs and pericentromeric regions are characterized by high levels of H3K9me2 and transposon sequences, yet atmorc6 reduces the interaction among pericentromeres while increasing the interactions among IHIs (Figures 6 and 7A).
Figure 7. Dynamics of IHIs in mutants.
Details of plots in the style of Figure 6 (with mutant higher/lower red/green) showing changes in IHI interaction in atmorc1 (A), ddm1 (B), and suvh4 suvh5 suvh6 (C). Chromosomal coordinates are labeled on top and left sides. White arrows indicate IHIs that are also found in wild type (see Figure 2A/C/D). Yellow arrows indicate IHIs only found in the mutants.
Color scales are the same as in Figure 6.
See also Figure S7, Table S1, and Data S3–S5.
Similar to AtMORC6, MOM1 is another Arabidopsis gene that represses genes and TEs without altering DNA methylation (Amedeo et al., 2000). Interestingly, several loss-of-function mom1 alleles were recovered from the same genetic screen that identified atmorc6 (see Supplemental Experimental Procedures online), again implicating a functional resemblance of the MOM1 and AtMORC6 genes. We found very little difference in the pattern of chromatin interaction of mom1 vs. wild type, and we did not observe a decrease in interaction within and between pericentromeric regions (Figure 6 and Data S3). These results are consistent with previous DNA-FISH observations showing that mom1 does not show decondensed chromocenters (Probst et al., 2003), and suggest that, although MOM1 and AtMORC6 share some similarities, they likely employ different mechanisms in regulating transcriptional gene silencing.
Since chromatin interaction is positively correlated with repressive epigenetic marks (see above), we sought to examine whether reducing DNA methylation and H3K9me2 levels would impact chromatin interaction in the regions with these marks. We therefore performed Hi-C analysis in met1, ddm1, cmt3, and suvh4 suvh5 suvh6 triple mutants. The two dimensional interaction maps and comparative maps revealed striking changes in chromatin interaction patterns, particularly in met1 and ddm1 (Figures 5A and 6, and Data S4), two mutants that reduce DNA methylation in all sequence contexts (Stroud et al., 2013). First, both met1 and ddm1 resemble atmorc6 in having less interaction of pericentromeric regions with themselves and more interaction of pericentromeres with euchromatic chromosome arms. However, the degree of change displayed by each of met1 and ddm1 is even higher than atmorc6. This is consistent with previous microscopic studies showing that met1 and ddm1 mutants have decondensed chromocenters, and that DDM1 and MET1 are both required for the repression of a larger number of genes and TEs than AtMORC6 (Moissiard et al., 2012; Probst et al., 2003; Soppe et al., 2002; Stroud et al., 2012). The most drastic changes in pericentromere association take place on chromosomes 2 and 4; while areas of pericentromeric heterochromatin on these that interact with NORs still interact with NORs in the mutants, other areas of the pericentromeres show very little interaction with adjacent areas, especially for chromosome 4 (Figure 5B). We also observed a loss of interactivity of internal regions of chromocenters, such as on chromosome 5, suggesting the formation of loops that no longer participate in the main block of pericentromeric interactions (Figure 5B). These results are consistent with previous work showing that the visible chromocenters are smaller in met1 and ddm1 mutants (Soppe et al., 2002). The effects of met1 and ddm1 on the pericentromeric regions are very different from the effect of the atmorc6 mutant, which shows decreased interaction for all pericentromeric regions, but maintains the same regions in the generally still-strongly-interacting pericentromeres (Figure 5B). This is also consistent with the different morphology of the chromocenters; atmorc6 mutants show larger and more diffused chromocenters, while met1 and ddm1 show smaller but still very punctate chromocenters (Moissiard et al., 2012; Soppe et al., 2002). We also observed another unique and striking phenotype in both met1 and ddm1 — a shift in the locations of the chromatin regions that participate in strong interactions within pericentromeric regions — suggesting that the regions that fold into the chromocenters have changed in these mutants (Figure 5B). This phenomenon is possibly due to a combinatorial effect of reduction of DNA methylation in pericentromeres of met1 and ddm1 (Stroud et al., 2013) together with changes in the histone methylation landscapes in the pericentromeres of these mutants (Deleris et al., 2012; Mathieu et al., 2005). We also observed an interesting behavior of IHIs in met1 and ddm1, with the IHIs defined in wild type showing slightly decreased mutual interaction in met1 and ddm1. However, this slight decrease is perhaps the result of recruitment of an increased number of participating IHI loci in the mutants (Figure 7B, Table S1A/C, and Data S4). The newly-appearing IHI loci resemble the previously described loci in wild type in that they are centered on small patches of heterochromatin in the otherwise euchromatic arms (Figure S7A–D). A comparison of wild type and mutant expression profiles did not reveal a clear correlation between reactivation of transposons and recruitment of new IHI loci in the mutants (Figure S7E); therefore, the mechanism by which the new IHIs get recruited is unknown. Together, these results demonstrate dramatic alteration of chromatin interactions in the met1 and ddm1 mutants, suggesting that DNA methylation is a major epigenetic determinant of the natural nuclear architecture of chromatin.
SUVH4, SUVH5, and SUVH6 are histone methyltransferases for H3K9 (Feng and Jacobsen, 2011). The suvh4 suvh5 suvh6 triple mutant exhibits extensive loss of non-CG methylation (Stroud et al., 2014; Stroud et al., 2013) and has been shown to have somewhat decondensed chromocenters (Rajakumara et al., 2011). Consistent with these data, we observed an atmorc6-like pericentromeric interaction pattern in suvh4 suvh5 suvh6 (Figure S6C) (Stroud et al., 2014). However, unlike the met1 and ddm1 mutants, suvh4 suvh5 suvh6 did not shift the regions interacting with pericentromeric regions (Figure S5), suggesting that suvh4 suvh5 suvh6 generates a less severe nuclear organization change than met1 and ddm1. We also observed that all of the IHI interactions still occurred in suvh4 suvh5 suvh6, while three new IHI loci also appeared, two of which correspond to the same ectopic IHI loci in met1 and ddm1 (Figure 7C, Table S1A/C, and Data S5). ChIP-Seq (Stroud et al., 2014) suggests that the majority of IHI loci, including the ones that ectopically interact in suvh4 suvh5 suvh6, have lost detectable H3K9me2 signal in suvh4 suvh5 suvh6 (Figures S7F). Collectively, the findings from met1, ddm1, and suvh4 suvh5 suvh6 suggest that, despite the IHIs being enriched in DNA methylation and H3K9me2, the interactions among IHI loci are not dependent on DNA methylation or H3K9me2 marks, and that, in fact, additional loci are recruited to IHIs when these marks are reduced.
CMT3 mediates CHG methylation (Stroud et al., 2013), but loss of CMT3 does not lead to chromocenter decondensation (Moissiard et al., 2012). Consistently, we did not observe atmorc6-like chromatin interaction patterns in cmt3 mutant, and cmt3 also lacks the dramatic chromocenter interaction alterations observed in met1 and ddm1 (Figures S5 and S6C, and Data S5). In fact, the difference in Hi-C maps between cmt3 and wild type is quite minimal — not much bigger than the difference between wild type replicates (Figure S6A–C). This result suggests that CHG methylation alone plays a relatively minor role in regulating chromatin interaction.
Collectively, our investigations of the chromatin interaction patterns in various epigenetic mutants indicate that loss of DNA methylation and histone H3K9 methylation affect chromatin interaction, leading to losses of interaction among pericentromeric regions and gains in interaction among IHIs. Further, H3K27me3 is also important in regulating interactions within large domains consisting of adjacent H3K27me3-marked genes.
CONCLUSION
Our Hi-C analyses show that Arabidopsis chromosomes interact extensively through their pericentromeric regions, as well as through two domains of the euchromatic arms, one consisting of the proximal half that also interacts with pericentromere adjacent regions, and one consisting of the distal half that also interacts with telomeres (see model in Figure 1E). A number of Interactive Heterochromatic Islands (IHIs) show strong long-range interactions with each other, which are also associated with telomeric regions. In addition, special regions in the euchromatic arms that are either H3K9me2- or H3K27me3-modified form local interactive hotspots. On the other hand, we do not observe strong interactions among highly expressed genes as has been observed in animals. Mutants that affect various repressive epigenetic processes exhibit altered chromosome architectures that are related to the effect of these mutations on heterochromatin condensation or their effects on the maintenance of histone or DNA methylation. These results reveal the complexity of chromatin interactions within the Arabidopsis nucleus and will form the basis of future studies on the regulatory mechanisms underlying chromosome folding in plants.
EXPERIMENTAL PROCEDURES
Additional details of experimental and analysis methods can be found in the Supplemental Experimental Procedures.
Hi-C Library Construction and Sequencing
Hi-C libraries compatible with Illumina sequencing were generated as described previously (Moissiard et al., 2012). The libraries were sequenced on HiSeq 2000 DNA sequencers obtaining paired-end 50- or 51-nucleotide reads following manufacturer instructions (Illumina).
Hi-C Data Analysis
The analysis pipeline includes a “dynamic smoothing” algorithm to reduce statistical noise in low coverage regions, but retain high resolution in high coverage regions. Provision was also made for two aspects of Hi-C signals that can lead to confounding effects. The first phenomenon is that some regions of the genome have much higher or lower counts than average due to issues such as density of HindIII sites, the mapability of 50-mers, the fidelity of the reference genome, and any biases introduced by library amplification or sequencing procedures. The second relates to the fact that interactions on the same chromosome correlate strongly with genomic distance. For instance, interactions between two areas of chromatin are naturally higher if those areas have only a short distance between them (Figure S1A). Raw counts are separated into components consisting of mapability effect, distance effect, and the remaining signal. This procedure allowed for higher sensitivity in detecting subtle interactions occurring both within and between chromosomes.
Supplementary Material
HIGHLIGHTS.
Chromatins interact through pericentromeres, telomeres, and adjacent regions
Interactive Heterochromatic Islands (IHIs) show strong long-range interactions
Interactions are correlated with epigenetic marks in active or silenced chromatins
Mutants in repressive epigenetic pathways alter chromatin interaction patterns
Acknowledgments
We thank M. Akhavan for Illumina sequencing, H. Stroud and G. Moissiard for assistance with plant work, H. Stroud, G. Moissiard, S. Bischof, D. Husmann, C. Hale, and J. Goodrich for seeds, Y. Zhan and J. Dekker for advice on Hi-C library construction, M. Kühne for technical assistance, and I. Schubert, P. Fransz, and members of the Jacobsen laboratory for supportive discussions and comments on the manuscript. Illumina sequencing was performed at the UCLA BSCRC BioSequencing Core Facility. This work was supported by grants from U.S. National Institutes of Health to S.E.J. (GM60398). S.F. was a Special Fellow of the Leukemia & Lymphoma Society. S.E.J. is an investigator of the Howard Hughes Medical Institute.
Footnotes
ACCESSION NUMBER
Sequencing data were deposited in the NCBI Sequence Read Archive (SRA) as accession SRP043612.
Supplemental information includes seven supplemental figures, one supplemental table, six supplemental datasets, supplemental experimental procedures, and supplemental references, and can be found with this article online at http://#######.
AUTHOR CONTRIBUTIONS
S.F., S.J.C., M.P., and S.E.J. designed the project; S.F. and V.S. performed experiments; S.J.C. conceived, implemented, and ran the analysis pipeline; S.J.C., J.Z., and S.E.J. analyzed the data; and S.F., S.J.C., and S.E.J. wrote the manuscript.
References
- Amedeo P, Habu Y, Afsar K, Mittelsten Scheid O, Paszkowski J. Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature. 2000;405:203–206. doi: 10.1038/35012108. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome I. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE. Genomewide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS One. 2008;3:e3156. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A, Schubert I. Interphase chromosome arrangement in Arabidopsis thaliana is similar in differentiated and meristematic tissues and shows a transient mirror symmetry after nuclear division. Genetics. 2007;176:853–863. doi: 10.1534/genetics.107.073270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crevillen P, Sonmez C, Wu Z, Dean C. A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J. 2013;32:140–148. doi: 10.1038/emboj.2012.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris A, Stroud H, Bernatavichute Y, Johnson E, Klein G, Schubert D, Jacobsen SE. Loss of the DNA Methyltransferase MET1 Induces H3K9 Hypermethylation at PcG Target Genes and Redistribution of H3K27 Trimethylation to Transposons in Arabidopsis thaliana. PLoS Genet. 2012;8:e1003062. doi: 10.1371/journal.pgen.1003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS. A three-dimensional model of the yeast genome. Nature. 2010;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Jacobsen SE. Epigenetic modifications in plants: an evolutionary perspective. Curr Opin Plant Biol. 2011;14:179–186. doi: 10.1016/j.pbi.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P, De Jong JH, Lysak M, Castiglione MR, Schubert I. Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc Natl Acad Sci U S A. 2002;99:14584–14589. doi: 10.1073/pnas.212325299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013;49:773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob S, Schmid MW, Luedtke NW, Wicker T, Grossniklaus U. Characterization of chromosomal architecture in Arabidopsis by chromosome conformation capture. Genome Biol. 2013;14:R129. doi: 10.1186/gb-2013-14-11-r129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper L, Golubovskaya I, Cande WZ. A bouquet of chromosomes. J Cell Sci. 2004;117:4025–4032. doi: 10.1242/jcs.01363. [DOI] [PubMed] [Google Scholar]
- Heger P, Marin B, Bartkuhn M, Schierenberg E, Wiehe T. The chromatin insulator CTCF and the emergence of metazoan diversity. Proc Natl Acad Sci U S A. 2012;109:17507–17512. doi: 10.1073/pnas.1111941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y, Feng S, LeBlanc CA, Bernatavichute YV, Stroud H, Cokus S, Johnson LM, Pellegrini M, Jacobsen SE, Michaels SD. ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat Struct Mol Biol. 2009;16:763–768. doi: 10.1038/nsmb.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013 doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafos M, Kroll P, Hohenstatt ML, Thorpe FL, Clarenz O, Schubert D. Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet. 2011;7:e1002040. doi: 10.1371/journal.pgen.1002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TB, Imakaev MV, Mirny LA, Laub MT. High-Resolution Mapping of the Spatial Organization of a Bacterial Chromosome. Science. 2013 doi: 10.1126/science.1242059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O, Probst AV, Paszkowski J. Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. Embo J. 2005;24:2783–2791. doi: 10.1038/sj.emboj.7600743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissiard G, Cokus SJ, Cary J, Feng S, Billi AC, Stroud H, Husmann D, Zhan Y, Lajoie BR, McCord RP, et al. MORC family ATPases required for heterochromatin condensation and gene silencing. Science. 2012;336:1448–1451. doi: 10.1126/science.1221472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Probst AV, Fransz PF, Paszkowski J, Scheid OM. Two means of transcriptional reactivation within heterochromatin. Plant J. 2003;33:743–749. doi: 10.1046/j.1365-313x.2003.01667.x. [DOI] [PubMed] [Google Scholar]
- Rajakumara E, Law JA, Simanshu DK, Voigt P, Johnson LM, Reinberg D, Patel DJ, Jacobsen SE. A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo. Genes Dev. 2011;25:137–152. doi: 10.1101/gad.1980311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa S, De Lucia F, Mylne JS, Zhu D, Ohmido N, Pendle A, Kato N, Shaw P, Dean C. Physical clustering of FLC alleles during Polycomb-mediated epigenetic silencing in vernalization. Genes Dev. 2013;27:1845–1850. doi: 10.1101/gad.221713.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H. Telomere attachment and clustering during meiosis. Cell Mol Life Sci. 2007;64:117–124. doi: 10.1007/s00018-006-6463-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert V. RNA polymerase II forms transcription networks in rye and Arabidopsis nuclei and its amount increases with endopolyploidy. Cytogenet Genome Res. doi: 10.1159/000365233. In press. [DOI] [PubMed] [Google Scholar]
- Schubert V, Berr A, Meister A. Interphase chromatin organisation in Arabidopsis nuclei: constraints versus randomness. Chromosoma. 2012;121:369–387. doi: 10.1007/s00412-012-0367-8. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe WJ, Jasencakova Z, Houben A, Kakutani T, Meister A, Huang MS, Jacobsen SE, Schubert I, Fransz PF. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 2002;21:6549–6559. doi: 10.1093/emboj/cdf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Do T, Du J, Zhong X, Feng S, Johnson L, Patel DJ, Jacobsen SE. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol. 2014;21:64–72. doi: 10.1038/nsmb.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152:352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Hale CJ, Feng S, Caro E, Jacob Y, Michaels SD, Jacobsen SE. DNA methyltransferases are required to induce heterochromatic re-replication in Arabidopsis. PLoS Genet. 2012;8:e1002808. doi: 10.1371/journal.pgen.1002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizawa H, Iwasaki O, Tanaka A, Capizzi JR, Wickramasinghe P, Lee M, Fu Z, Noma K. Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucleic Acids Res. 2010;38:8164–8177. doi: 10.1093/nar/gkq955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, van Steensel B, van Lohuizen M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet. 2006;38:694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, Buisine N, Gagnot S, Martienssen RA, Coupland G, Colot V. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 2007;3:e86. doi: 10.1371/journal.pgen.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009;10:R62. doi: 10.1186/gb-2009-10-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE. Whole-Genome Analysis of Histone H3 Lysine 27 Trimethylation in Arabidopsis. PLoS Biology. 2007;5(5):e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.