Abstract
Human mature erythrocytes are terminally differentiated cells that have lost their nuclei and organelles during development. Even though mature erythrocytes lack ribosomal and other large-sized RNAs, they still retain small-sized RNAs. We have recently shown that there are abundant and diverse species of microRNAs in mature erythrocytes through the use of several different techniques, including northern blot, miRNA microarray, and real-time PCR. Furthermore, fractionation and genomic analysis has revealed that erythrocyte microRNA expression is different from that of reticulocytes or leukocytes and that mature erythrocytes contribute the majority of microRNA expression in whole blood. Therefore, global analysis of microRNA expression in circulating erythrocytes has the potential to provide mechanistic insights into erythrocyte biology and erythrocyte-related disorders. Here, we have provided the detailed methods for isolating and characterizing the microRNAs from human mature erythrocytes to enable such researches into human diseases involving erythrocytes.
1. Introduction
Human erythrocytes are end products of a highly regulated differentiation process that involves the gradual loss of cellular organelles, a decline in nucleic acid content, and a step-wise acquisition of erythrocyte characteristics (1). The nuclei are extruded as they become differentiated into reticulocytes. Cytoplasmic RNA and translation activities, while still detectable in reticulocytes, fall below detection limit as they become mature erythrocytes (2). The prevailing view that mature erythrocytes lack most RNAs primarily comes from their inability to stain with RNA-binding dyes (e.g., acridine orange, methylene blue), the basis of these dyes to distinguish reticulocytes from mature erythrocytes in clinical setting (3). Given the potential limitations and biases of these methods, certain RNA species may not be detected. We have recently found that human mature erythrocytes, though largely lacking in ribosomal and large-sized RNA, possess diverse and abundant microRNAs (4). It is also important to note the recent discovery of microRNAs and proteins mediating microRNA function in platelets, another group of terminally differentiated anuclear blood cells (5). These erythrocyte microRNAs allow the application of DNA microarray technology to capture and extract the biological information to understand erythrocyte phenotypes during physiological and pathological adaptations in many human diseases affecting erythrocytes.
MicroRNAs are noncoding RNAs of 19–25 nt in size which mediate posttranscriptional regulation of target mRNAs through the formation of noncanonical base pairing with the 3′ UTR. MicroRNAs regulate a wide variety of biological processes (e.g., differentiation, apoptosis, and oncogenic transformation) (6). Many publications have highlighted the role of several microRNAs that have been implicated in the process of erythropoiesis, (7–16) as well as during in vitro erythroid differentiation (8, 17–19). Since erythroid cells lose their nuclei and active transcription during the reticulocyte stages of their development, posttranscriptional regulation of remaining mRNAs probably plays a very important role. Although microRNAs are likely to play a regulatory role in posttranscriptional regulation in erythroid cells, we have very limited information thus far. The genomic study of microRNAs in mature erythrocytes may provide a unique and accessible window to enhance our understanding of their regulatory roles in erythroid cells, both under normal circumstances and in pathological states, such as various anemic diseases (4) and polycythemia vera (14, 20). To facilitate the study of microRNAs in the human erythrocytes, we are providing the detailed methodology for the isolation and characterization of erythrocyte microRNAs.
2. Materials
Miltenyi MACS® Separators (autoMACS).
Miltenyi autoMACS® Columns, Miltenyi.
Miltenyi human CD71 microbeads, Miltenyi.
Blood samples (~20 mL) to be analyzed, collected from donors.
Human blood for the isolation of common reference RNAs used for microarrays.
Purecell Neo-leukoreduction filter (PALL Biomedical Products, East Hills, NY).
Phosphate-buffered saline (PBS), pH 7.4 (10× stock solution: 1.37 M NaCl, 27 mM KCl, 100 mM Na2HPO4, 18 mM KH2PO4).
Staining buffer: PBS with 2% of fetal bovine serum (FBS).
mirVana miRNA isolation kit (Ambion, Applied Biosystems, Foster City, CA).
Vac-Man® Laboratory Vacuum Manifold (Promega, WI).
miRCURY LNA™ microRNA Array (Exiqon, Denmark).
miRCURY LNA™ microRNA Hy3/Hy5 Power labeling kit (Exiqon, Denmark).
Qiagen RNeasy Mini Kit.
MAUI® SC mixer for microarray hybridization.
Wash Buffer A: For 1 L, 100 mL of 20× SSC, 20 mL 10% detergent, 880 mL water.
Wash Buffer B: For 1 L, 50 mL of 20× SSC, 950 mL water.
Wash Buffer C: For 1 L, 10 mL 20× SSC, 990 mL water.
-
20× SSC:
Dissolve 175.3 g of NaCl, 88.2 g of sodium citrate in 800 mL of distilled H2O.
Adjust the pH to 7.0 with a few drops of 1 M HCl.
Adjust the volume to 1 L with additional distilled H2O.
Sterilize by autoclaving.
GenePix 4000B microarray scanner (Axon, Fremont, CA).
3. Methods
We have divided our discussion of the methods into four sections: (Subheading 3.1) the isolation of human mature erythrocytes using leukoreduction filters and autoMACS®, (Subheading 3.2) the purification all RNAs larger than 10 nt using the microRNA isolation kits, (Subheading 3.3) the labeling of erythrocyte microRNA, and (Subheading 3.4) the profiling of erythrocyte microRNA species using spotted Exiqon microRNA microarrays. The conventional wisdom that mature erythrocytes lack RNA may be due to a technical issue during traditional RNA isolation procedures (e.g., Trizol or Qiagen RNeasy or other column-based RNA isolation kit), during which the majority of small-sized RNAs are lost. Therefore, we modified an isolation protocol using Ambion’s miRVana kit to capture all RNA larger than ten nucleotides (nt) for our analysis of erythrocytic microRNAs. To isolate the microRNA from mature erythrocytes, it is important to first remove other cell types in the human blood because the higher level of RNAs from contaminating cells has the potential to complicate the miRNA analysis. Finally, we label the erythrocyte microRNAs for interrogation by the microRNA microarrays. In the labeling procedures, we use a higher amount of ethanol in the Qiagen RNeasy kit to remove the unlabeled Cy-Dye while retaining the labeled microRNAs.
3.1. Purification of Mature Erythrocytes from Human Blood
Centrifuge whole blood sample and aspirate plasma and buffy coat layer for initial removal of plasma and leukocytes. Resuspend the cells in PBS (see Note 1).
Cut the outlet tubing of the Pall leukoreduction filter and insert into the filters with the pointed ends to allow the infusion of the whole blood from the top of the filter to allow filtration by simple gravity.
Pour the blood into the leukoreduction filter bag and then collect the filtrate which contains red blood cells. Use PBS to wash the filter, and collect filtrate into a 50-mL Falcon tube.
Wash the leukocyte-depleted blood with cold PBS and add 80 μL of CD71 (this protocol uses the presence/absence of CD71 to differentiate reticulocytes from erythrocytes) micro-beads to each 20 mL of blood sample (see Note 2). We mix the sample gently by inverting the tubes and incubating for 20 min on ice to allow the binding of the beads to the CD71+ reticulocytes.
Wash the cells by adding staining buffer to the top once and centrifuge at 805 × g in a table top centrifuge for 5 min without brake. Pipette off supernatant carefully and completely. The red cell pellet can be loose, therefore requiring extra caution.
Resuspend the cells in staining buffer again and bring the volume to less than 50% hematocrit in 25 or 50 mL of PBS. For each run on Miltenyi MACS® Separators, 25 mL is the highest capacity, so you may need two separate runs for some samples.
Apply the cell suspension onto the column, run negative selection (DEPLETES program) for mature RBC, collect both CD71+ and CD71− samples for the same patient samples. The cells in the CD71+ fraction will be in approximately 2 mL, while the CD71− fractions (the majority of the erythrocytes) will be in similar volume of starting sample volume.
After the completion of the separation, clean all the tubes and empty the waste, and then place new tubes and run sleep program of the autoMACS® Separator.
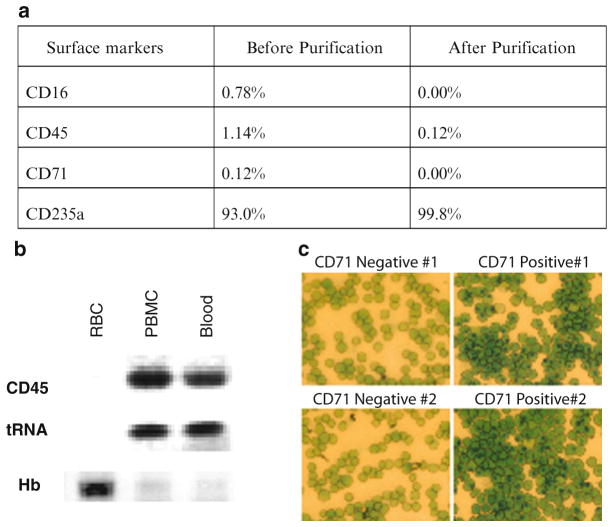
The purity of the resulting mature erythrocytes can be evaluated using flow cytometry (for the percentage of CD71+ cells, Fig. 1a), methylene blue stain (Fig. 1b), or RT-PCR for lineage-specific transcripts (Fig. 1c). Alternatively, the samples can be sent to a reticulocyte lab for similar determination using RNA-staining dye (e.g., acridine orange).
Fig. 1.
(a) Assessment of the purified mature erythrocytes was performed by examining the surface expression of indicated lineage markers on whole blood (before purification) and purified erythrocytes (after purification) with flow cytometry to estimate the percentage of cells with surface expression of CD16 (leukocyte marker), CD45 (leukocyte marker), CD71 (reticulocyte marker), and CD235a (mature erythrocytes marker). (b) RT-PCR assay to evaluate the cell purity based on the abundance of indicated transcripts (CD45, tRNA and beta-hemoglobin) in the RNA from purified erythrocytes (RBC), peripheral blood mononuclear cells (PBMC) and whole blood. (c) New methylene blue stain of the CD71− erythrocytes (left) and CD71+ reticulocytes (right) from two independent isolations.
3.2. Isolation of MicroRNAs from the Purified Human Mature Erythrocytes
Pool all the CD71 cells together and fill the tube up to the top with cold PBS and centrifuge 805 × g without brake to pellet the red cells.
After centrifugation, remove the supernatant carefully due to the looseness of the red cell pellets.
The CD71+ population can be lysed using the recommended 500 μL of lysis buffer (in the Ambion miRVana RNA isolating kit). However, for the CD71− population (mature erythrocytes), you will need to lyse the cell pellet using 5 mL of lysis buffer in a 50-mL Falcon tube due to the large cell number and amount of protein in red cells (see Note 3). It is important to ensure complete lysis of the red cell pellet by repeated pipetting and thorough vortexing (at least 60 s). Since we mostly follow the recommended sample amount for the CD71+ cells, we will emphasize the modifications we have made for CD71− population in the remaining protocol.
Add 50 μL (for the CD71+) and 500 μL (for the CD71−) homogenate additive in the Ambion miRVana RNA isolating kit to each sample, consistent with the one-tenth volume of the lysis buffer used in the previous step.
Leave the mixtures of lysed samples and homogenate additive on ice for 10 min.
Add acid phenol choloroform (5 mL) to the CD71− samples and vortex vigorously for 1–2 min to ensure thorough mixing and adequate protein extraction.
Separate the aqueous and organic phase by centrifuging the 50-mL Falcon tube for 5 min at 10,000 × g at room temperature with complete brake ON.
Collect the supernatant (aqueous) which contains RNA and transfer it to a fresh tube and repeat the phenol/chloroform extraction step until the supernatant becomes clear to indicate the completeness of the organic phase extraction.
Estimate the volume of the resulting supernatant and then add 1.25 volumes 100% ethanol and mix thoroughly by carefully inverting the tube several times.
The lysate/ethanol mixture from one sample will be passed through one or two RNA-binding filter cartridges provided by miRVana kits by vacuum manifold modified using the Promega Vacuum Manifold. The bottom of an Eppendorf tube is cut off and connected with the adaptor in the Vacuum Manifold through a 200 μL pipette tip sealed with parafilm to allow the vacuum. We pipette the sample continuously onto the membrane before the membrane becomes dry after the passing of the samples. (Note: It is important to ensure that the flow rate of the vacuum is not too high as this will decrease yields. Also, make sure to check the filters for small holes, which will also reduce yields.)
The RNA-binding filter cartridge with the erythrocyte RNAs will be removed from the vacuum manifold and placed in a fresh Eppendorf tube. The filter cartridge will be washed with 700 μL of the miRNA Wash Buffer A using a microcentrifuge as recommended in the kit.
Wash the filter cartridge with 500 μL Wash Buffer B/C twice.
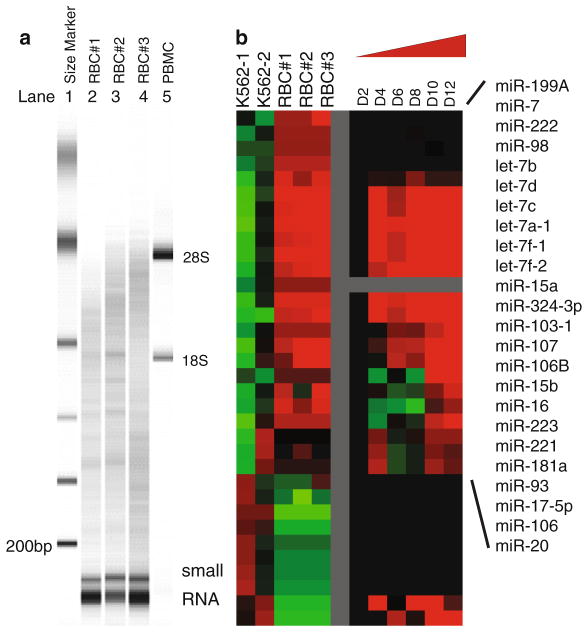
Elute RNA with 100 μL DEPC-water at 95°C (in two washes of 50 μL each to increase RNA yield) and measure the concentration via Nanodrop. The size distribution of the isolated RNAs can be assessed by Agilent Bioanalyzer (Fig. 2a) or RNA gels (see Note 4).
Fig. 2.
(a) The size distribution of RNAs of three independent erythrocyte samples (lanes 2–4 labeled RBC #1–3) and one PBMC sample (lane 5 labeled as PBMC) were determined with Agilent Bioanalyzer with the indicated size markers (lane 1 as indicated). (b) Left: the microRNA expression pattern obtained with microRNA microarrays of three mature erythrocyte samples was compared to that of two erythroleukemia K562 cell lines. Right: the expression of erythrocyte-specific microRNAs in the CD34+ erythroid progenitor cells at indicated stages of erythroid differentiation in a previously published study (14).
3.3. Profiling of Erythrocyte MicroRNAs Using MicroRNA Microarrays
Start with 5 μg of total RNA with the following sample sheet (Table 1).
Place all Exiqon labeling kit components on ice and thaw for 20 min. Add 29 μL of nuclease-free water to the supplied Cy-Dyes, mix by vortexing, and spin down. Keep the dyes covered since they are photosensitive.
Prepare the samples (table above) with total volume of RNA samples with water to 6 μL and prepare master mix for all samples and keep on ice until all the samples are ready.
Vortex and spin down, add 13 μL of premade master mix to each samples.
Use PCR machine to incubate all samples at 0°C for 1 h, followed by 15 min at 65°C, and then 4°C forever. But do not leave the completed samples on PCR machine for more than 30 min.
Place the labeling reaction in a Speedvac® and dry the sample to complete dryness or until the volume is less than 10 μL. It usually takes about 20 min.
Redissolve the dry sample in 5–10 μL nuclease-free water.
Add 350 μL buffer RLT (of the RNeasy Mini Kit from Qiagen) to the sample, and disrupt and homogenize immediately by vigorous vortexing.
Add 3.5 volumes of 100% ethanol (1,225 μL) and mix thoroughly by vortexing. Do not centrifuge. (Please note that here the large amount of ethanol is used to trap the small-sized RNAs.)
Pipette 700 μL of the sample, including any precipitate that may have formed, into an RNeasy Mini spin column placed in a 2-mL collection tube. Centrifuge for 15 s at ≥8,000 × g (≥10,000 rpm) and discard the flow through.
Repeat the loading of the samples onto the mini spin column until the whole sample has been pipetted into the spin column. Discard the flow-through each time.
Place the RNeasy Mini spin column into a new 2-mL collection tube. Pipette 500 μL buffer RPE into the spin column, and centrifuge for 15 s at ≥8,000 × g (≥10,000 rpm). Discard the flow-through.
Pipette another 500 μL buffer RPE (in the RNeasy kit) into the column and centrifuge for 15 s at ≥8,000 × g (≥10,000 rpm) again.
Centrifuge at full speed for 1 min finally.
Place the RNeasy Mini spin column into a 1.5-mL collection tube. Pipette 25 μL of RNase-free water directly onto the spin column membrane. Centrifuge for 1 min at ≥8,000 × g (≥10,000 rpm) to elute the labeled RNAs for the hybridization with microarrays.
Table 1.
Sample sheet for the assembly reagent for microRNA labeling for microarray experiments
| Amt Cy3 | Amt Cy5 | Cy3 Conc | Cy5 Conc | Cy3 | LBl | H2O | Cy5 | Lbl | H2O | Slides | Notes | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 5 | 5 | miRNA-EX_01_ | |||||||||
| 2 | 5 | 5 | miRNA-EX_01_ | |||||||||
| 3 | 5 | 5 | miRNA-EX_01_ | |||||||||
| 4 | 5 | 5 | miRNA-EX_01_ | |||||||||
| 5 | 5 | 5 | miRNA-EX_01_ | |||||||||
| 6 | 5 | 5 | miRNA-EX_01_ | |||||||||
| 7 | 5 | 5 | miRNA-EX_01_ | |||||||||
| 8 | 5 | 5 | miRNA-EX_01_ | |||||||||
| 9 | 5 | 5 | miRNA-EX_01_ | |||||||||
| 10 | 5 | 5 | miRNA-EX_01_ |
Cy3 erythrocyte RNA samples from individual patients, Cy5 common RNA reference isolated from one unit of blood, which will be used in all samples as normalization control (the dye choice for samples vs. reference is optional)
3.4. Microarray Hybridization
Add 33 μL of 2× Exiqon hybridization buffer to make the total volume 66 μL.
Heat the sample tubes to 95° for 3 min and then spin briefly before adding them to the array.
Apply the samples to slides in MAUI® SC mixer – need 61–62 μL to load and hybridize at 60°C using nonevaporation trays covering slides. The hybridization depends on the melting temperature of the probes used on the microRNA microarrays.
Use glass stain dishes with metal slide holders for all washes.
Remove mixer with Wash Buffer A at 60°C.
Rinse the slides for 2 min wash at 60°C with Wash Buffer A.
Rinse the slides for 10 s at room temperature with Wash Buffer B.
Wash the slides for 2 min at room temperature with fresh Wash Buffer B.
Wash the slides for 2 min at room temperature with Wash Buffer C.
Spin dry the slides in the racks at 200–450 g at the table top centrifuge for 3 min.
Store the slides in a dark box until ready to scan using the Axon GenePix 4000B microarray scanner. It would be best to scan the arrays within a few hours before the signals decay. The obtained microRNA expression can be used to derive a biological conclusion (Fig. 2b).
Acknowledgments
We thank the Duke microarray facility and members of the Chi lab for technical assistance and constructive feedback and the Telen lab for assistance with sample collection. This research was funded by NIH R21DK080994 and Roche Foundation for Anemia Research (RoFAR).
Footnotes
For blood samples of each patient, collect 20 μL of whole blood for CBC analysis to determine the percentage of reticulocytes before and after purification procedures.
During the removal of CD71+ reticulocytes for the blood cells with high level of reticulocytes, the amount of CD71 beads may be limiting. Therefore, it is important to increase the amount of beads used to label the CD71+ cells to ensure efficient and successful immunodepletion.
Given the large amount of protein in the mature red cells relative to the RNAs, it is often important to increase the volume as well as the repetition of the phenol extraction to ensure the successful separation of the aqueous phase for RNA purification.
When the RNA gel or Bioanalyzer are used to analyze RNAs, the intactness and the 28S/18S rRNA ratio are usually used to indicate the integrity and quality of RNA. Given the fact that large-sized RNAs are often degraded in the mature erythrocytes, it is usual to observe only small-sized RNAs with significant degradation of the large-sized RNAs.
References
- 1.Hoffman R, Benz EJB, Shattil SJ, Furie B, Cohen HJ, et al. Hematology: Basic Principles and Practice. Churchill Livingstone; Edinburgh: 2004. [Google Scholar]
- 2.Goh SH, Lee YT, Bouffard GG, Miller JL. Hembase: browser and genome portal for hematology and erythroid biology. Nucleic Acids Res. 2004;32:D572–D574. doi: 10.1093/nar/gkh129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linda G, Lee C-HCLAC. Thiazole orange: a new dye for reticulocyte analysis. Cytometry. 1986;7:508–517. doi: 10.1002/cyto.990070603. [DOI] [PubMed] [Google Scholar]
- 4.Chen SY, Wang Y, Telen MJ, Chi JT. The genomic analysis of erythrocyte microRNA expression in sickle cell diseases. PLoS One. 2008;3:e2360. doi: 10.1371/journal.pone.0002360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landry P, Plante I, Ouellet DL, Perron MP, Rousseau G, et al. Existence of a microRNA pathway in anucleate platelets. Nat Struct Mol Biol. 2009;16:961–966. doi: 10.1038/nsmb.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci U S A. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Q, Huang Z, Xue H, Jin C, Ju XL, et al. MicroRNA miR-24 inhibits erythropoiesis by targeting activin type I receptor ALK4. Blood. 2007;111:588–595. doi: 10.1182/blood-2007-05-092718. [DOI] [PubMed] [Google Scholar]
- 9.Yang GH, Wang F, Yu J, Wang XS, Yuan JY, et al. MicroRNAs are involved in erythroid differentiation control. J Cell Biochem. 2009;107:548–556. doi: 10.1002/jcb.22156. [DOI] [PubMed] [Google Scholar]
- 10.Felli N, Pedini F, Romania P, Biffoni M, Morsilli O, et al. MicroRNA 223-dependent expression of LMO2 regulates normal erythropoiesis. Haematologica. 2009;94:479–486. doi: 10.3324/haematol.2008.002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu YF, Du TT, Dong M, Zhu KY, Jing CB, et al. Mir-144 selectively regulates embryonic alpha-hemoglobin synthesis during primitive erythropoiesis. Blood. 2009;113:1340–1349. doi: 10.1182/blood-2008-08-174854. [DOI] [PubMed] [Google Scholar]
- 12.Pase L, Layton JE, Kloosterman WP, Carradice D, Waterhouse PM, et al. miR-451 regulates zebrafish erythroid maturation in vivo via its target gata2. Blood. 2009;113:1794–1804. doi: 10.1182/blood-2008-05-155812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dore LC, Amigo JD, Dos Santos CO, Zhang Z, Gai X, et al. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc Natl Acad Sci U S A. 2008;105:3333–3338. doi: 10.1073/pnas.0712312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruchova H, Yoon D, Agarwal AM, Mendell J, Prchal JT. Regulated expression of microRNAs in normal and polycythemia vera erythropoiesis. Exp Hematol. 2007;35:1657–1667. doi: 10.1016/j.exphem.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masaki S, Ohtsuka R, Abe Y, Muta K, Umemura T. Expression patterns of microRNAs 155 and 451 during normal human erythropoiesis. Biochem Biophys Res Commun. 2007;364:509–514. doi: 10.1016/j.bbrc.2007.10.077. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q, Huang Z, Xue H, Jin C, Ju XL, et al. MicroRNA miR-24 inhibits erythropoiesis by targeting activin type I receptor ALK4. Blood. 2008;111:588–595. doi: 10.1182/blood-2007-05-092718. [DOI] [PubMed] [Google Scholar]
- 17.Georgantas RW, 3rd, Hildreth R, Morisot S, Alder J, Liu CG, et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci U S A. 2007;104:2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhan M, Miller CP, Papayannopoulou T, Stamatoyannopoulos G, Song CZ. MicroRNA expression dynamics during murine and human erythroid differentiation. Exp Hematol. 2007;35:1015–1025. doi: 10.1016/j.exphem.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choong ML, Yang HH, McNiece I. MicroRNA expression profiling during human cord blood-derived CD34 cell erythropoiesis. Exp Hematol. 2007;35:551–564. doi: 10.1016/j.exphem.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Bruchova H, Yoon D, Agarwal AM, Swierczek S, Prchal JT. Erythropoiesis in poly-cythemia vera is hyper-proliferative and has accelerated maturation. Blood Cells Mol Dis. 2009;43:81–87. doi: 10.1016/j.bcmd.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]