Abstract
Tropomodulins (Tmods) interact with tropomyosins (TMs) via two TM-binding sites and cap the pointed ends of TM-coated actin filaments. To study the functional interplay between TM binding and TM-actin filament capping by Tmods, we introduced disabling mutations into the first, second, or both TM-binding sites of full-length Tmod1 (Tmod1-L27G, Tmod1-I131D, and Tmod1-L27G/I131D, respectively) and full-length Tmod3 (Tmod3-L29G, Tmod3-L134D, and Tmod3-L29G/L134D, respectively). Tmod1 and Tmod3 showed somewhat different TM-binding site utilization, but nearly all TM binding was abolished in Tmod1-L27G/I131D and Tmod3-L29G/L134D. Disruption of Tmod-TM binding had a modest effect on Tmod1's ability and no effect on Tmod3's ability to stabilize TM-actin pointed ends against latrunculin A-induced depolymerization. However, disruption of Tmod-TM binding did significantly impair the ability of Tmod3 to reduce elongation rates at pointed ends with α/βTM, albeit less so with TM5NM1, and not at all with TM5b. For Tmod1, disruption of Tmod-TM binding only slightly impaired its ability to reduce elongation rates with α/βTM and TM5NM1, but not at all with TM5b. Thus, Tmod-TM binding has a greater influence on Tmods’ ability to inhibit subunit association as compared to dissociation from TM-actin pointed ends, particularly for α/βTM, with Tmod3's activity being more dependent on TM binding than Tmod1's activity. Nevertheless, disruption of Tmod1-TM binding precluded Tmod1 targeting to thin filament pointed ends in cardiac myocytes, suggesting that the functional effects of Tmod-TM binding on TM-coated actin filament capping can be significantly modulated by the in vivo conformation of the pointed end or other factors in the intracellular environment.
INTRODUCTION
The tropomodulin (Tmod) family of tropomyosin (TM)-binding and actin filament pointed-end capping proteins includes four mammalian isoforms (Tmod1-4), which are differentially expressed in a tissue-specific manner and play roles in regulating actin filament architecture in diverse cell types (Gokhin and Fowler 2011b; Yamashiro et al. 2012). Tmods consist of an N-terminal TM/Pointed-End Actin Capping (TM-Cap) domain, containing two TM-binding sites and an actin binding site that confer TM-dependent actin pointed-end capping, followed by a C-terminal Leucine-Rich Repeat/Pointed-End Actin Capping (LRR-Cap) domain containing five leucine-rich repeat motifs and a TM-independent actin pointed-end capping site (Fig. 1A) (Kostyukova 2008a; Kostyukova 2008b; Yamashiro et al. 2012). All vertebrate Tmods tightly cap TM-actin pointed ends, with Kd's in the nanomolar to picomolar range, but Tmod capping of TM-free actin pointed ends is weaker, with Kd's of ~0.1-0.2 μM (Almenar-Queralt et al. 1999; Weber et al. 1994; Weber et al. 1999; Yamashiro et al. 2014). These properties also appear to extend to invertebrate Tmods, given that C. elegans Tmod caps TM-actin pointed ends more strongly than it caps TM-free actin pointed ends (Yamashiro et al. 2008).
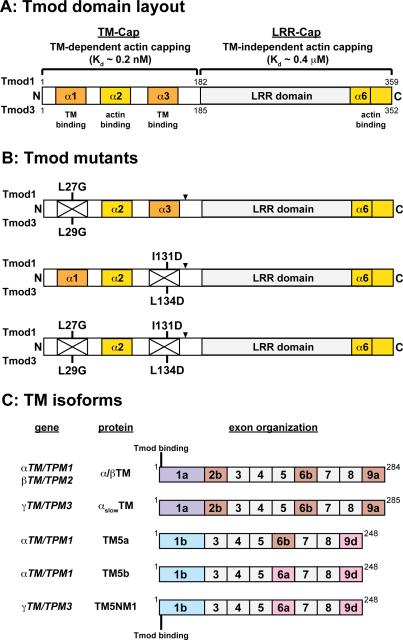
Figure 1. Schematics of the molecular layouts of the Tmod and TM isoforms used in this study.
(A) Structural and functional domains, TM-binding properties, and actin-capping properties of mammalian Tmod1 and Tmod3. Note that the TM-binding sites contain residues outside of their associated α-helical regions (Kostyukova 2008a; Kostyukova 2008b). (B) Mutant Tmod proteins used in this study. Point mutations disrupting the first and second TM-binding sites are labeled. In some experiments, Tmod1 was FLAG-tagged between theα3-helix and LRR domain (arrowheads). (C) Wild-type TM proteins (α/βTM, αslowTM, TM5a, TM5b, and TM5NM1) used in this study and the location of the Tmod-binding site on TMs.
Due to their expression in multiple cell types, Tmod1 and Tmod3 are the mammalian Tmod isoforms whose cellular functions have been studied in greatest detail (Yamashiro et al. 2012). Tmod1 predominantly functions in terminally differentiated, post-mitotic cells. For example, Tmod1 cooperates with α/βTM to stabilize actin filaments during myofibril assembly and adherens junction formation during cardiac morphogenesis in mice (Fritz-Six et al. 2003; McKeown et al. 2014; McKeown et al. 2008). Pointed-end capping and TM-binding by Tmod1 also regulate actin dynamics to control the precise lengths of the long α/βTM-coated actin filaments in mature cardiac myofibrils (Gregorio et al. 1995; Littlefield et al. 2001; Mudry et al. 2003; Sussman et al. 1998), as well as the short TM5b/TM5NM1-coated actin filaments in the spectrin-actin membrane skeleton in red blood cells (Moyer et al. 2010). By contrast, Tmod3 appears to predominantly function in more dynamic cellular contexts. These contexts include the leading lamellipodia of migrating endothelial cells, where Tmod3 acts as negative regulator of actin filament assembly and cell migration rates (Fischer et al. 2003), and proliferating erythroblasts prior to enucleation, where Tmod3 regulates terminal erythroid differentiation, erythroblast-macrophage island formation, cell cycle progression, and enucleation (Sui et al. 2014). Nevertheless, in the presence of TMs, Tmod3 can impart TM-actin filament stability in a manner similar to Tmod1, as shown in studies of TM5b/TM5NM1-coated actin filaments associated with the lateral membranes of polarized epithelial cells (Temm-Grove et al. 1998; Weber et al. 2007) as well as the TM4/TM5NM1-coated filaments linked to the sarcoplasmic reticulum of skeletal muscle fibers (Gokhin and Fowler 2011a; Vlahovich et al. 2009; Vlahovich et al. 2008). These studies raise the question of whether Tmod1 vs. Tmod3 interactions with TM isoforms drive these Tmods’ differential associations with diverse actin filament structures.
TMs are α-helical coiled-coil proteins that bind along the sides of actin filaments and protect filaments from depolymerization at their pointed ends (Broschat 1990; Broschat et al. 1989). Mammalian TMs exhibit even more dramatic isoform diversity than Tmods, with >40 unique protein products arising from alternative splicing of four genes (αTM/TPM1, βTM/TPM2, γTM/TPM3, and δTM/TPM4) (Gunning et al. 2008; Gunning et al. 2005). The TM-Cap domain of Tmods binds to sites at the N-termini of TMs (Fowler 1987; Fowler 1990; Greenfield and Fowler 2002; Greenfield et al. 2005; Gunning et al. 2008; Gunning et al. 2005; Kostyukova et al. 2007; Vera et al. 2000), and Tmod and TM isoforms exhibit complex selectivity, with different combinations of Tmod and TM isoforms showing distinct binding preferences and affinities (Babcock and Fowler 1994; Gokhin et al. 2010; Kostyukova 2008a; Kostyukova 2008b; Sussman and Fowler 1992; Uversky et al. 2011; Yamashiro et al. 2014). As described above, Tmods cap TM-coated actin filaments with >1000-fold greater affinity than TM-free actin filaments (Almenar-Queralt et al. 1999; Weber et al. 1994; Weber et al. 1999; Yamashiro et al. 2014), but there is relatively limited data comparing Tmod isoforms’ pointed-end capping activities for filaments coated with different TM isoforms (Yamashiro et al. 2014).
The first direct evidence of particular Tmod-TM isoform combinations influencing the pointed-end capping functionality of Tmods was a study showing that a truncated Tmod1 protein lacking Tmod1's 15 C-terminal residues (Tmod11-344) can cap TM5a-coated actin filaments several-fold more tightly than skeletal muscle α/βTM-coated filaments (Kostyukova and Hitchcock-DeGregori 2004)—a result consistent with stronger binding of Tmod1 to TM5a than to α/βTM (Greenfield and Fowler 2002; Kostyukova et al. 2007; Sussman and Fowler 1992; Uversky et al. 2011). It has also been observed that five-fold higher concentrations of TM5NM1, as compared to TM5a, are required to achieve high-affinity Tmod11-344-mediated capping of TM-coated actin filaments, while three-fold higher concentrations of TM5NM1 are required as compared to α/βTM (Kostyukova et al. 2007). Recently, it was shown that full-length Tmod1 and Tmod3 have similar pointed-end capping activities in the presence of α/βTM, TM5b, and TM5NM1, but Tmod3 is a weaker cap than Tmod1 when actin filaments are coated with recombinant αTM2 (Yamashiro et al. 2014). Because recombinant αTM2 is unacetylated at its N-terminus, binds F-actin weakly, and has a disabled Tmod-binding site (Greenfield and Fowler 2002; Hitchcock-DeGregori and Heald 1987; Urbancikova and Hitchcock-DeGregori 1994), we hypothesized that differences in pointed-end capping by Tmod1 vs. Tmod3 in the presence of native TMs could be revealed by experimentally attenuating the avidity of Tmod-TM binding.
Here, we sought to dissect the relationship between isoform-specific TM binding and TM-coated actin filament pointed-end capping by full-length Tmod1 and Tmod3. We studied this by disabling the TM-binding abilities of Tmod1 and Tmod3 via introducing mutations in the α-helices corresponding to the first, second, or both TM-binding sites of full-length Tmod1 (Tmod1-L27G, Tmod1-I131D, and Tmod1-L27G/I131D, respectively) and full-length Tmod3 (Tmod3-L29G, Tmod3-L134D, and Tmod3-L29G/L134D, respectively), which can serve as useful tools to test the significance of Tmod-TM binding in Tmods’ TM-actin pointed-end capping activity. To date, the functional effects of these amino acid substitutions have only been characterized for Tmod1. Both Tmod1-L27G and Tmod1-L27E exhibit impaired TM binding activity, but these mutants retain partial pointed-end capping activity (Greenfield et al. 2005; Kostyukova et al. 2005). Moreover, Tmod1-L27E exhibits somewhat reduced targeting to thin filament pointed ends in cardiac myocytes, Tmod1-I131D exhibits more severely reduced targeting, and Tmod1-L27G/I131D exhibits virtually no targeting (Tsukada et al. 2011). However, since TM-binding-site-disrupting point mutations were introduced in Tmod1 and never Tmod3, the possibility of differential functional effects of TM-binding site disruptions in Tmod1 vs. Tmod3 on pointed-end capping remains unexplored.
We found that disabling TM binding by simultaneous disruption of the first and second TM-binding sites significantly impaired Tmod3's ability to reduce pointed-end elongation rates only when the actin filaments were coated with α/βTM, with a more modest effect observed for TM5NM1, and no effect observed for TM5b. The ability of Tmod1 to reduce pointed-end elongation rates was even less sensitive to disruption of the first and second TM-binding sites, with only modest impairment observed for α/βTM and TM5NM1, and no impairment observed for TM5b. Surprisingly, in contrast to its effects on pointed-end elongation, disruption of TM-binding sites had only a modest effect on Tmod1's and no effect on Tmod3's ability to reduce pointed end depolymerization of TM-coated actin filaments from latrunculin A (LatA)-induced disassembly, for all TM isoforms examined. Collectively, these observations indicate that Tmod-TM interactions in the context of the actin filament pointed end are substantially different from Tmod-TM interactions in the absence of actin. Furthermore, the lack of sensitivity to Tmod-TM binding observed in the LatA depolymerization assays may reflect a greater reliance of actin filament pointed end stability on TM-actin interactions than on Tmod-TM interactions. Despite the relatively benign effects of disruptions in the TM-binding sites on Tmod1's ability to cap the pointed ends of α/βTM-coated actin filaments in vitro, we confirmed that disrupting Tmod-TM binding dramatically hinders Tmod1 targeting to thin filament pointed ends in cultured cardiac myocytes (Tsukada et al. 2011). Our findings point toward finely tuned interactions between TM isoforms and full-length Tmods, whose functional effects may be modulated by as-yet-unidentified factors in the intracellular environment.
RESULTS AND DISCUSSION
Tmod1 and Tmod3 differentially utilize their two TM-binding sites when interacting with TM isoforms
Previous studies with Tmod1 fragments and peptides have suggested that long, exon 1a-containing TMs predominantly utilize the first TM-binding site on Tmod1, while short, exon 1b-containing TMs predominantly utilize the second TM-binding site, thus contributing to selective TM-Tmod isoform recognition (Babcock and Fowler 1994; Kostyukova et al. 2007; Uversky et al. 2011). The TM-binding activity of Tmod1 can be perturbed by introducing disabling mutations into the first TM-binding site (Tmod1-L27G), the second TM-binding site (Tmod1-L134D), or both TM-binding sites (Tmod1-L27G/I131D) (Greenfield et al. 2005; Kostyukova et al. 2006; Kostyukova et al. 2005). To investigate TM-binding site utilization of Tmod1 and Tmod3 with various TMs, we prepared recombinant full-length Tmod proteins with mutations in the first, second, or both TM-binding sites: wild-type Tmod1, Tmod1-L27G, Tmod1-I131D, or Tmod1-L27G/I131D; and wild-type Tmod3, Tmod3-L29G, Tmod3-L134D, or Tmod3-L29G/L134D (Fig. 1A,B). We then compared Tmod-TM binding using a variety of representative sarcomeric and cytoskeletal TMs. Skeletal muscle TMs (containing exon 1a) were recombinant human αslowTM (γTM/TPM3 gene) prepared in baculovirus vectors (Akkari et al. 2002) and native rabbit skeletal muscle α/βTM (αTM/TPM1 and βTM/TPM2 genes) prepared from acetone powder. (Note that the N-terminal Tmod-binding sequences of αTM and βTM are identical (Gokhin et al. 2010), and that all muscle TMs used in our assays are acetylated at their N-termini.) Nonmuscle short TMs (containing exon 1b) were TM5a and TM5b (both from the αTM/TPM1 gene, and thus with identical N-termini) and TM5NM1 (γTM/TPM3 gene) (Fig. 1C).
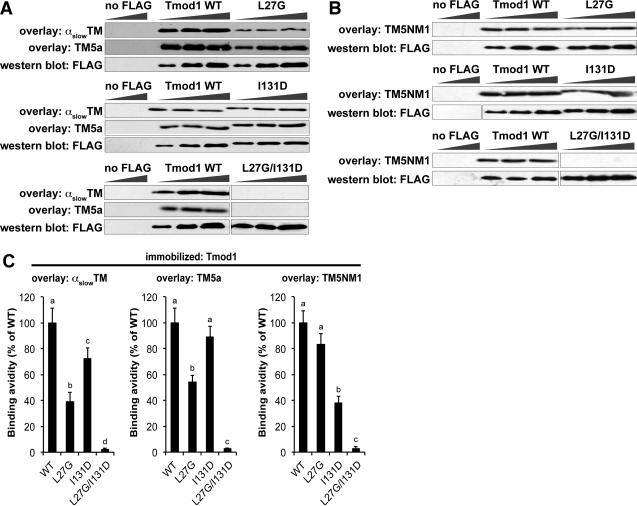
To evaluate the relative binding avidities of various TM isoforms to the two Tmods and their mutants, we performed blot overlay assays, where blots with immobilized Tmods and Tmod mutants were incubated with each TM, followed by antibodies to detect the bound TM and subsequent densitometric quantification of binding avidities (Gokhin et al. 2010; Ilkovski et al. 2008). This method reliably detects relative differences in Tmod-TM binding, as confirmed by dot blot overlay assays using native proteins in the absence of detergents (Yamashiro et al. 2014). The amount of TM bound was normalized to the level of Tmod or mutant protein by either western blotting for the Tmods on a duplicate blot or Coomassie blue staining on a duplicate gel (Gokhin et al. 2010). In our first series of experiments, we prepared FLAG-tagged Tmod1 and Tmod1 mutants, and then incubated the FLAG-Tmod1 blots with skeletal muscle αslowTM, TM5a, or TM5NM1. We observed that skeletal muscle αslowTM bound more weakly to the FLAG-Tmod1-L27G mutant than to the FLAG-Tmod1-I131D mutant (Fig. 2), suggesting that skeletal muscle αslowTM preferentially utilizes Tmod1's first TM-binding site. However, contrary to our expectations, the short TM5a also bound more weakly to the FLAG-Tmod1-L27G mutant than to the FLAG-Tmod1-I131D mutant (Fig. 2), indicating that, like skeletal muscle αslowTM, short TM5a also relies preferentially on Tmod1's first TM-binding site. On the other hand, TM5NM1 binding to Tmod1 was more affected by the I131D mutation in Tmod1's second TM-binding site (Fig. 2). Tmod1's first and second TM-binding sites appear to confer all of Tmod1's TM-binding activity in this assay, since binding of all three TMs to the double mutant (FLAG-Tmod1-L27G/I131D) was nearly undetectable (Fig. 2).
Figure 2. Tmod1 preferentially utilizes its first TM-binding site to interact with αslowTM and TM5a, but its second TM-binding site to interact with TM5NM1.
(A-C) Increasing amounts of FLAG-tagged Tmod1 fusion proteins (WT, L27G, I131D, or L27G/I131D) or mock-transfected cell lysates (“no FLAG”) were separated by SDS-PAGE, transferred to nitrocellulose, and overlaid with (A) skeletal muscle αslowTM, (B) TM5a, or (C) TM5NM1. Bound TMs were detected using anti-TM antibodies. Parallel blots were probed with an anti-FLAG antibody to show loading. (D) Binding avidities were quantified using densitometry by normalizing band intensities from the overlays to the corresponding band on the FLAG blot. Data shown are mean ± S.D. of three independent experiments. Different letters above bars within a single graph reflect significantly different values (p<0.05), as determined by one-way ANOVA with post hoc Fisher's PLSD tests.
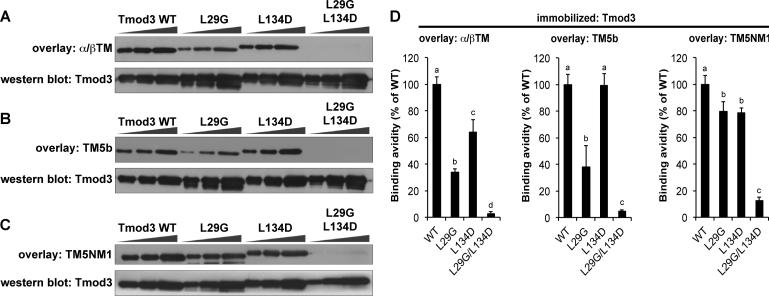
Next, we tested TM binding to Tmod3 and Tmod3 mutants using similar TM blot overlay procedures, incubating the blots with native skeletal muscle α/βTM, TM5b, or TM5NM1. Similar to Tmod1, mutation of the first TM-binding site (Tmod3-L29G) reduced skeletal muscle α/βTM and TM5b binding to Tmod3 more severely than mutation of the second TM-binding site (Tmod3-L134D) (Fig. 3). However, unlike Tmod1, TM5b binding to Tmod3 was not affected by mutating the second TM-binding site (Tmod3-L134D), suggesting that this site is less important for Tmod3's interaction with TM5b (Fig. 3). Also unlike Tmod1, where mutation of the second site significantly reduced binding of TM5NM1 (Fig. 2), TM5NM1 binding to Tmod3 was not affected by mutating either the first or second binding site on its own (Fig. 3). Nevertheless, the binding of all three TMs to Tmod3-L29G/L134D was negligible (Fig. 3), similar to the results with Tmod1-L27G/I131D (Fig. 2). Thus, TM isoforms utilize the first and second TM-binding sites on Tmod1 and Tmod3 to different extents, but, together, the two binding sites appear to be necessary for TM-binding by both Tmod isoforms for all TMs.
Figure 3. Tmod3 preferentially utilizes its first TM-binding site to interact with skeletal muscle α/βTM and TM5b, but not TM5NM1.
(A-C) Increasing amounts of Tmod3 proteins (WT, L29G, L134D, or L29G/L134D; 100 ng, 200 ng, and 400 ng from the left) were separated by SDS-PAGE, transferred to nitrocellulose, and overlaid with (A) skeletal muscle α/βTM, (B) TM5b, or (C) TM5NM1. Bound TMs were detected using anti-TM antibodies. Parallel Coomassie-stained gels show loading. (D) Binding avidities were densitometrically quantified by normalizing band intensities from the overlays to the corresponding Coomassie-stained bands. Data shown are mean ± S.D. of three independent experiments. Different letters above bars within a single graph reflect significantly different values (p<0.05), as determined by one-way ANOVA with post hoc Fisher's PLSD tests.
TM-binding-disabled Tmod mutants are competent to inhibit pointed-end depolymerization from TM-coated actin filaments
TMs bind cooperatively along the lengths of actin filaments and stabilize filaments against depolymerization by reducing the rate of actin subunit dissociation from pointed ends (Broschat 1990; Broschat et al. 1989; Hitchcock-DeGregori 2008; Tobacman 2008; Weber et al. 1999). Tmods cap the pointed ends of α/βTM-coated actin filaments with high affinity and enhance the ability of TMs to reduce actin subunit dissociation from pointed ends (Almenar-Queralt et al. 1999; Kostyukova and Hitchcock-DeGregori 2004; Weber et al. 1994; Weber et al. 1999). Therefore, we hypothesized that the relative abilities of Tmods or their mutants to stabilize TM-actin filament pointed ends may depend on the relative strengths of Tmods binding to TMs. To explore this idea, we tested the abilities of Tmod1 and Tmod3 to inhibit LatA-induced actin depolymerization from the pointed ends of TM-coated actin filaments. LatA is a drug that sequesters actin monomers that dissociate from filament ends at steady state, thereby depleting the monomer pool available for polymerization; the reduced extent of monomer association thus leads to net filament depolymerization (Coue et al. 1987; Morton et al. 2000; Spector et al. 1989; Yamashiro et al. 2008). To specifically measure the extent of pointed-end depolymerization, CapZ was used to cap the actin filament barbed ends, and the actin filaments were then pre-incubated with TMs along with 20 nM wild-type Tmod1, Tmod3, or their mutants before addition of LatA to induce depolymerization. The extent of F-actin depolymerization after incubation for 1.5 h was then measured as the percentage of total actin in the supernatant after ultracentrifugation.
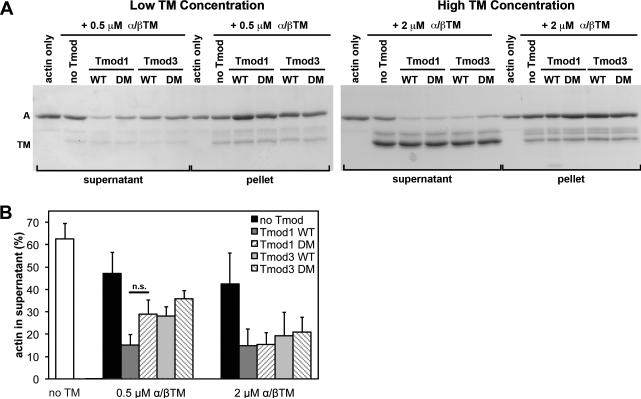
In the absence of Tmod and TM, ~60% of F-actin had depolymerized, and was present in the supernatant fraction after 1.5 h in the presence of LatA (Fig. 4-6). As expected (Broschat 1990; Broschat et al. 1989), addition of 0.5 or 2 μM TM to 20 nM CapZ-capped filaments slightly decreased the amount of actin in the supernatant to ~40-50%, consistent with a modest degree of F-actin stabilization, for both α/βTM (Fig. 4) and TM5b (Fig. 5). Addition of 0.5 μM TM5NM1 had little to no effect on F-actin stability, with ~60% of the F-actin remaining in the supernatant (i.e., no improvement over F-actin alone) (Fig. 6), consistent with the weaker binding of TM5NM1 to F-actin (Yamashiro et al. 2014). However, increasing the TM5NM1 concentration to 2 μM did impart a modest degree of F-actin stabilization (Fig. 6), similar to α/βTM (Fig. 4) and TM5b (Fig. 5). As expected (Yamashiro et al. 2008), the further addition of wild-type Tmod1 led to a several-fold decrease in actin in the supernatant, down to ~10-20%, also regardless of whether α/βTM (Fig. 4), TM5b (Fig. 5), or TM5NM1 (Fig. 6) was used. Because Tmod1 had no effect on the amount of actin in the supernatant in the absence of TM (Supplementary Fig. 1), these data confirm that Tmod1 inhibits actin pointed-end depolymerization by a TM-dependent pointed-end capping mechanism (Weber et al. 1994; Weber et al. 1999). The ability of Tmod1 to inhibit LatA-induced F-actin depolymerization was similar for all three TMs tested and was not significantly different at 0.5 μM (subsaturating) or 2 μM (saturating) TM concentrations in the presence of 2 μM total actin (20 nM CapZ-capped filaments) (Fig. 4-6).
Figure 4. Effects of wild-type and TM-binding-disabled mutant Tmod proteins on LatA-induced depolymerization from the pointed ends of CapZ-capped actin filaments coated with α/βTM.
(A) CapZ-capped actin filaments (CapZ:actin ratio of 1:100) were incubated with skeletal muscle α/βTM then mixed with wild-type Tmod1, wild-type Tmod3, Tmod1-L27G/I131D (double-mutant; DM), or Tmod3-L29G/L134D (double-mutant; DM), diluted five-fold and incubated in the presence of 10 μM LatA to depolymerize the F-actin. Final concentrations of Tmods and TMs were as indicated, with 20 nM CapZ-capped actin filaments (2 μM actin). The mixtures were ultracentrifuged, fractionated into supernatants and pellets, and separated using SDS-PAGE. A, actin; TM; tropomyosin. (B) Densitometric quantification was performed to calculate the percentage of depolymerized actin in the supernatants. Data shown are mean ± S.D. of three independent experiments. n.s., not significant (Student's t-test).
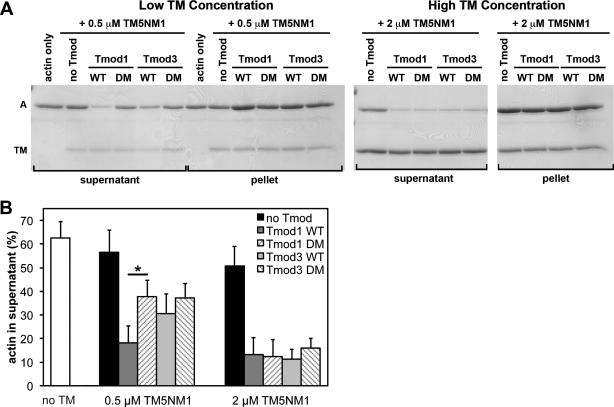
Figure 6. Effects of wild-type and TM-binding-disabled mutant Tmod proteins on LatA-induced depolymerization from the pointed ends of CapZ-capped actin filaments coated with TM5NM1.
(A) CapZ-capped actin filaments (CapZ:actin ratio of 1:100) were incubated with TM5NM1 then mixed with wild-type Tmod1, wild-type Tmod3, Tmod1-L27G/I131D (DM), or Tmod3-L29G/L134D (DM), diluted five-fold and incubated in the presence of 10 μM LatA to depolymerize the F-actin. Final concentrations of Tmods and TMs were as indicated, with 20 nM CapZ-capped actin filaments (2 μM actin). The mixtures were ultracentrifuged, fractionated into supernatants and pellets, and separated using SDS-PAGE. A, actin; TM; tropomyosin. (B) Densitometric quantification was performed to calculate the percentage of depolymerized actin in the supernatants. Data shown are mean ± S.D. of three independent experiments. *, p<0.05 (Student's t-test).
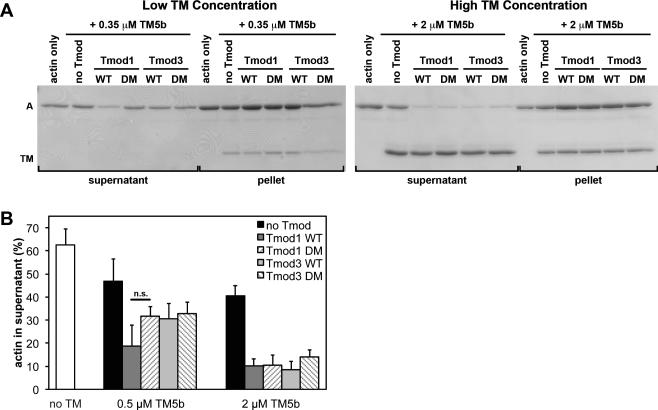
Figure 5. Effects of wild-type and TM-binding-disabled mutant Tmod proteins on LatA-induced depolymerization from the pointed ends of CapZ-capped actin filaments coated with TM5b.
(A) CapZ-capped actin filaments (CapZ:actin ratio of 1:100) were incubated with TM5b then mixed with wild-type Tmod1, wild-type Tmod3, Tmod1-L27G/I131D (DM), or Tmod3-L29G/L134D (DM), diluted five-fold and incubated in the presence of 10 μM LatA to depolymerize the F-actin. Final concentrations of Tmods and TMs were as indicated, with 20 nM CapZ-capped actin filaments (2 μM actin). The mixtures were ultracentrifuged, fractionated into supernatants and pellets, and separated using SDS-PAGE. A, actin; TM; tropomyosin. (B) Densitometric quantification was performed to calculate the percentage of depolymerized actin in the supernatants. Data shown are mean ± S.D. of three independent experiments. n.s., not significant (Student's t-test).
Tmod1 and Tmod3 were equally strong inhibitors of F-actin depolymerization in the presence of either 0.5 μM or 2 μM concentrations of TMs, with only ~10-30% actin in the supernatant, regardless of whether α/βTM (Fig. 4), TM5b (Fig. 5), or TM5NM1 (Fig. 6) was used. Surprisingly, the Tmod3 double mutant with negligible TM-binding activity (Tmod3-L29G/L134D; Fig. 3) was also an equally strong inhibitor of F-actin depolymerization when α/βTM (Fig. 4) or TM5b (Fig. 5), or TM5NM1 (Fig. 6) was used. By contrast, the Tmod1 double mutant with negligible TM-binding activity (Tmod1-L27G/I131D; Fig. 2) inhibited F-actin depolymerization less effectively than wild-type Tmod1 in the presence of 0.5 μM TM5NM1 (Fig. 6). Similar trends were observed for Tmod1-L27G/I131D in the presence of either 0.5 μM α/βTM (Fig. 4) or 0.5 μM TM5b (Fig. 5), but these trends did not achieve statistical significance.
To summarize, a decrease in Tmod-TM binding ability only appears to significantly diminish Tmod1's ability to stabilize TM-bound F-actin when the concentration of TM5NM1 (but not α/βTM or TM5b) is limiting. The most likely explanation for this is that limiting concentrations of TM5NM1 reduce the probability of TM5NM1 molecules binding near the pointed end (Wegner 1979), concomitantly reducing the number of pointed ends with high-affinity Tmod-binding sites at any given time. This would amplify any observed reduction in the Tmod-TM binding affinity and increase the extent of F-actin depolymerization. However, even at these subsaturating TM concentrations, LatA-induced F-actin depolymerization does not appear to be sensitive to the large (~100-fold) reductions in TM binding displayed by Tmod1-L27G/I131D and Tmod3-L29G/L134D, as measured in the binary TM-Tmod binding assays (Figs. 2,3). The fact that LatA-induced depolymerization shows an overall lack of sensitivity to Tmod-TM binding strength may indicate that pointed-end stability relies more on TM-actin interactions than on Tmod-TM interactions. Consistent with this, Tmod1 inhibits depolymerization of TM5a- and αTM-coated actin filaments to similar extents (Kostyukova and Hitchcock-DeGregori 2004), despite Tmod1 binding more strongly to TM5a than to αTM (Greenfield and Fowler 2002). Our findings also agree with the observation that effects of disabling Tmod-TM binding are only apparent with the weakest F-actin-binding TM isoform, TM5NM1 (Yamashiro et al. 2014). Another possibility is that the strength of Tmod binding to pointed ends may depend on TM isoform-specific control of the conformation of TM-actin pointed ends.
TM-binding-disabled Tmod mutants retain ability to inhibit actin pointed-end elongation for some but not all TM isoforms
Previous work has shown that Tmod1 and Tmod3 only show differential effects on the kinetics TM-actin filament pointed-end elongation in the presence of a very weak Tmod-binding TM isoform (recombinant unacetylated αTM2) (Yamashiro et al. 2014). To further investigate the role of Tmod-TM binding strength in Tmods’ ability to inhibit the kinetics of actin filament pointed-end elongation, we tested the ability of Tmods and their TM-binding-disabled mutants to reduce initial polymerization rates from actin filament pointed ends in the presence of various TMs (Fig. 7A,B). When analyzing the resulting data, we compared the initial rate/control rates for pyrene-actin polymerization kinetics in the presence of wild-type and mutant Tmod1 and Tmod3, measuring pyrene-labeled actin pointed-end elongation rates from CapZ-capped, TM-coated actin filament seeds. Rate/control rate ratios were expressed as the initial elongation rate in the presence of Tmod, with respect to a control elongation rate in the absence of Tmod, as described previously (Fowler et al. 2003; Weber et al. 1994; Weber et al. 1999). In these experiments, we varied the Tmod concentration from 2.5–10 nM (at 0.5 μM TMs and 1.8 μM actin) to better reveal potential differences in capping activity.
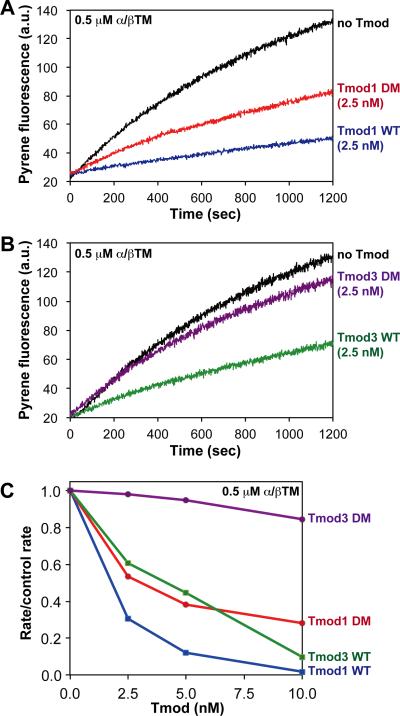
Figure 7. Effects of wild-type and TM-binding-disabled mutant Tmod proteins on pointed-end F-actin elongation in the presence of skeletal muscle α/βTM.
(A) Pyrene-actin polymerization kinetics from the pointed ends of CapZ-capped actin filaments in the presence of 0.5 μM skeletal muscle α/βTM and either no Tmod, 2.5 nM wild-type Tmod1, or 2.5 nM Tmod1-L27G/I131D (DM). (B) Pyrene-actin polymerization kinetics from the pointed ends of CapZ-capped actin filaments in the presence of 0.5 μM skeletal muscle α/βTM and either no Tmod, 2.5 nM wild-type Tmod3, or 2.5 nM Tmod3-L29G/L134D (DM). (C) Comparison of differences in pointed-end capping activity in the presence of 0.5 μM skeletal muscle α/βTM for Tmod1, Tmod3, Tmod1-L27G/I131D (DM), and Tmod3-L29G/L134D (DM). The extent of pointed-end capping is plotted as the initial rate of elongation for each Tmod or mutant, divided by the initial rate of elongation for the “no Tmod” TM-actin control (rate/control rate), versus increasing concentrations of each Tmod or mutant (nM). When rate/control rate = 1.0, no pointed ends are capped; when rate/control rate = 0, all pointed ends are capped. Concentrations of proteins were: 1.8 μM G-actin (8% pyrene), 1.6 nM CapZ-capped actin filaments (1:100; 1.6 nM CapZ, 160 nM actin), and Tmods and TMs as indicated.
In the presence of skeletal muscle α/βTM, both wild-type Tmod1 and Tmod3 strongly capped TM-actin filament pointed-ends, reducing initial elongation rates by 90-95% at 10 nM Tmods (Fig. 7C). However, contrary to our expectations, Tmod1-L27G/I131D was only ~25-30% less effective than wild-type Tmod1 at inhibiting actin polymerization from pointed ends in the presence of skeletal muscle α/βTM (Fig. 7C), despite a ~40-fold decrease in Tmod1-L27G/I131D's TM-binding activity as compared to wild-type Tmod1 (Fig. 2). On the other hand, but in agreement with our expectations, Tmod3-L29G/L134D was almost completely ineffective at blocking elongation of actin polymerization from pointed ends in the presence of skeletal muscle α/βTM (Fig. 7C), demonstrating that disabling both of Tmod3's TM-binding sites, leading to a ~10-fold reduction in Tmod3-α/βTM binding (Fig. 3), almost completely eliminates Tmod3's ability to cap pointed ends when filaments were coated with skeletal muscle α/βTM.
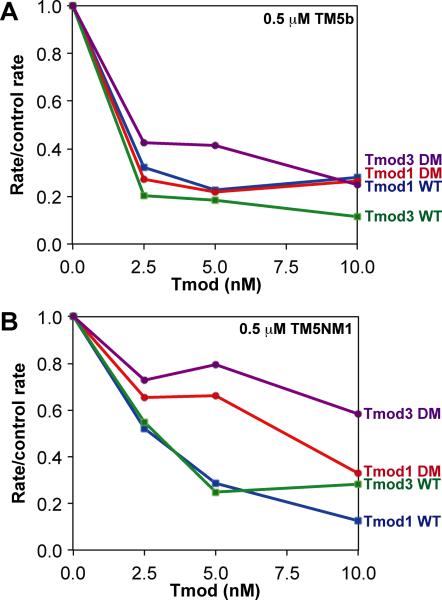
In the presence of TM5b, both Tmod1 and Tmod3 were similarly strong actin filament pointed-end caps, inhibiting initial elongation rates 70-80% at 2.5 nM Tmods and 80-90% at 10 nM Tmods (Fig. 8A). Both Tmod1 and Tmod3 were somewhat better at blocking pointed-end elongation with TM5b than with skeletal muscle α/βTM, as revealed by comparison of the rate/control rate ratios at submaximal Tmod concentrations (Fig. 7C). Moreover, both Tmod1-L27G/I131D and Tmod3-L29G/L134D exhibited unexpectedly strong pointed-end capping activities in the presence of TM5b, similar to wild-type Tmod1 and Tmod3 (Fig. 8A). Thus, with TM5b, a large reduction in Tmod-TM binding for the mutants (~50-fold for Tmod1, and ~10-fold for Tmod3; see Fig. 2,3), does not lead to a significant reduction in either Tmod's TM-actin capping activity. Note that both α/βTM and TM5b bind to F-actin with high affinity (Yamashiro et al. 2014); therefore, the strength of TM-actin binding cannot account for the differential effects of disabling Tmod-TM binding in the presence of these two TMs.
Figure 8. Effects of wild-type and TM-binding-disabled mutant Tmod proteins on pointed-end F-actin elongation in the presence of TM5b or TM5NM1.
The abilities of wild-type Tmod1, wild-type Tmod3, Tmod1-L27G/I131D (DM), and Tmod3-L29G/L134D (DM) to inhibit the initial rates of F-actin elongation from the pointed ends of CapZ-capped actin filaments were evaluated in the presence of either (A) 0.5 μM TM5b or (B) 0.5 μM TM5NM1. The extent of pointed-end capping is plotted as the initial rate of elongation for each Tmod or mutant, divided by the initial rate of elongation for the “no Tmod” TM-actin control (rate/control rate), versus increasing concentrations of each Tmod or mutant (nM). When rate/control rate = 1.0, no pointed ends are capped; when rate/control rate = 0, all pointed ends are capped. Concentrations of proteins were: 1.8 μM G-actin (8% pyrene), 1.6 nM CapZ-capped actin filaments (1:100; 1.6 nM CapZ, 160 nM actin), and Tmods and TMs as indicated.
With TM5NM1, Tmod1 and Tmod3 similarly reduced actin filament pointed-end elongation rates, with ~50% of F-actin elongation inhibited at 2.5 nM Tmods and ~75-85% of elongation inhibited at 10 nM Tmods (Fig. 8B). Such pointed-end capping activities were similar to Tmod1's capping activity in the presence of skeletal muscle α/βTM (Fig. 7C), but not quite as robust as either Tmod1's or Tmod3's pointed-end capping activity in the presence of TM5b (Fig. 8A). This was most evident at the lower concentrations of Tmods, where both Tmod1-L27G/I131D and Tmod3-L29G/L134D showed reduced pointed-end capping activities (~20-30% at 5 nM) compared to wild-type Tmods (~70% at 5 nM) (Fig. 8B). At a higher concentration of Tmods (10 nM), Tmod1-L27G/I131D recovered its normal pointed-end capping activity, approaching that of wild-type Tmod1, while the pointed-end capping activity of Tmod3-L29G/L134D remained only ~50% of that of wild-type Tmod3 (Fig. 8B). Thus, disabling Tmods’ TM-binding sites leads to reduced pointed-end capping activity in the presence of TM5NM1, similar to α/βTM (Fig. 7C), with a stronger effect observed for Tmod3 than Tmod1.
To summarize, in actin pointed-end elongation assays, differences in Tmod1 and Tmod3 binding to TMs correlate with their actin filament pointed-end capping activities with skeletal muscle α/βTM, to a lesser extent with TM5NM1, and not at all with TM5b. Thus, Tmod1 and Tmod3 appear as the strongest caps for F-actin with TM5b (saturating at 2.5 nM Tmod), and disabling the Tmods’ TM-binding sites has little effect. By contrast, wild-type Tmod1 and Tmod3 are somewhat weaker caps for TM5NM1-coated actin filaments (saturating at ~5 nM Tmod), and their actin filament pointed-end capping activities are partially reduced by disabling their TM-binding sites. Finally, for skeletal muscle α/βTM, Tmod1 capping saturates at ~5 nM, and Tmod1 capping is partially reduced for the Tmod1-L27G/I131D mutant; by comparison, capping by wild-type Tmod3 is close to maximal at 10 nM, and its pointed-end capping activity is dramatically reduced for the Tmod3-L29G/L134D mutant. We conclude that differences in Tmod-TM binding do affect high-affinity Tmod isoform capping of the pointed ends of TM-coated actin filaments, but these differences are only detectable when Tmod concentrations are limiting, and only for certain TMs. Furthermore, the unexpectedly small decreases in capping activities due to mutations in the two known TM-binding sites on Tmods suggest that additional regions of Tmods may contribute to TM binding at the pointed end (Moroz et al. 2013; Tsukada et al. 2011). In addition, mechanisms other than direct Tmod-TM binding may enhance Tmods’ ability to inhibit actin elongation at TM-actin pointed ends, including potential effects on actin pointed-end conformation induced by TM binding to F-actin, as proposed above to explain our observations that disabling TM binding has little effect on Tmods’ ability to inhibit LatA-induced depolymerization of TM-actin filaments (Fig. 4-6).
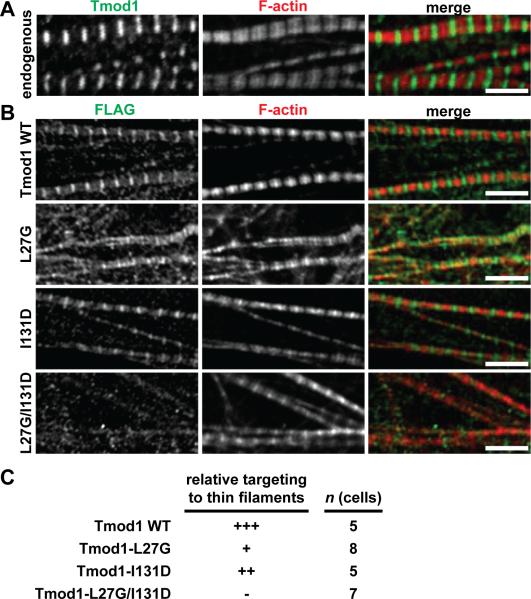
TM-binding-disabled mutant Tmod1 fails to target to the pointed ends of the thin filaments in cardiac myocytes
Our battery of in vitro assays revealed two divergent facets of Tmod1's pointed-end capping activity for α/βTM-actin filaments: (i) TM-binding-independent α/βTM-actin capping activity during depolymerization, as shown by our LatA-induced depolymerization assays (Fig. 4); (ii) TM-binding-dependent α/βTM-actin capping activity during elongation, as shown by our pyrene-actin elongation assays (Fig. 7). However, it remains unclear which of these facets of Tmod1's pointed-end capping activity is more germane to Tmod1's ability to localize to and cap the pointed ends of α/βTM-actin filaments in vivo. Hence, we investigated whether disabling the TM-binding sites on Tmods might affect the targeting of Tmod1 to the pointed ends of α/βTM-containing thin filaments in striated muscle cells. A hallmark feature of striated muscle cells is that the pointed ends of the thin filaments are aligned into periodic stripes along contractile myofibrils, which can be visualized with conventional fluorescence microscopy. Thus, striated muscle cells provide a tractable and useful living “test tube” to test hypotheses about Tmod targeting to pointed ends.
We investigated the targeting of Tmod1 and its single and double mutants to thin filament pointed ends by transfecting cultured embryonic chick cardiac myocytes with FLAG-tagged wild-type and mutant Tmod1 proteins. We observed that wild-type FLAG-Tmod1 localized robustly to the thin filament pointed ends in sarcomeres, while FLAG-Tmod1-L27G/I131D failed to do so (Fig. 9), consistent with findings in a previous study (Tsukada et al. 2011). Moreover, disabling the second TM-binding site in Tmod1 (FLAG-Tmod1-I131D) only slightly reduced targeting to the thin filament pointed ends, while disabling the first TM-binding site (FLAG-Tmod1-L27G) led to a much more dramatic reduction in targeting (Fig. 9). This targeting result is consistent with the results of our binary TM-Tmod binding assays, which showed that Tmod1-L27G binds to α/βTM less strongly than Tmod1-I131D (Fig. 2). However, this result is somewhat different from the targeting results reported previously, which showed that Tmod1-L27E localizes to thin filament pointed ends to a greater extent than does Tmod1-I131D (Tsukada et al. 2011). These differential functional effects of TM-binding site-disabling mutations may depend on the substituted amino acids, since both the L27E and L27G mutations disable the α-helix within the TM-binding site, but only the L27E mutation introduces a negative charge. Alternatively, the targeting of Tmod1-L27E but not Tmod1-L27G may be due to the fact that the retention of α-helical structure in Tmod1-L27E, but not Tmod1-L27G, could permit residual binding to α/βTM and actin at thin filament pointed ends by adjacent regions in the Tmod1 sequence. An additional caveat of our experiments is that the transfected FLAG-tagged Tmod1 mutants may be outcompeted by endogenous wild-type Tmod1, exaggerating the impairment of targeting of the FLAG-tagged Tmod1 mutants; RNAi knockdown of endogenous Tmod1 prior to transfection with the FLAG-tagged Tmod1 mutants would be required to eliminate this possibility. Nevertheless, our results do indicate that Tmod1 localization at the pointed ends of the thin filaments depends on Tmod1 binding to skeletal muscle α/βTM, suggesting that the α/βTM-dependent capping activity of Tmod1 measured in pointed-end elongation assays (Fig. 7) is operative for Tmod1 targeting to thin filament pointed ends in cardiac myocytes.
Figure 9. Localization of FLAG-tagged wild-type and mutant Tmod1 proteins in embryonic chick cardiac myocytes.
(A) Cardiac myocytes were immunostained for endogenous Tmod1 using an anti-Tmod1 antibody and phalloidin-stained for F-actin. Note the localization of Tmod1 at the pointed ends of the phalloidin-stained thin filaments. Bar, 5 μm. (B) Cardiac myocytes expressing FLAG-tagged WT Tmod1, Tmod1-L27G, Tmod1-I131D, or Tmod1-L27G/I131D were immunostained using an anti-FLAG antibody to localize exogenous Tmod1 protein and phalloidin-stained for F-actin. Bars, 5 μm. (C) Semiquantitation of WT Tmod1, Tmod1-L27G, Tmod1-I131D, and Tmod1-L27G/I131D targeting to thin filament pointed ends. Tmod1-L27G shows dramatically impaired localization at the pointed ends of the thin filaments, Tmod1-I131D shows nearly normal localization, and Tmod1-L27G/I131D shows completely abolished localization. In all experiments, cardiac myocytes were also immunostained for α-actinin to identify Z-lines and confirm normal myofibril organization.
Concluding remarks
This study is the first to introduce mutations in the TM-binding sites of full-length Tmod1 and Tmod3 and examine the effects of disrupting Tmod-TM binding on Tmods’ ability to cap and stabilize the pointed ends of TM-coated actin filaments in vitro as well as target to the pointed ends of sarcomeric thin filaments in vivo (summarized in Table 1). Intriguingly, disabling the TM-binding activities of Tmods had different functional effects on actin depolymerization vs. elongation at pointed filament ends. These differences were most stark with our TM-binding-disabled Tmod3 mutant (Tmod3-L29G/L134D), which was indistinguishable from wild-type Tmod3 when stabilizing α/βTM-coated actin filaments in LatA-induced depolymerization assays (Fig. 4), but showed dramatically impaired ability to block α/βTM-actin pointed-end elongation as compared to wild-type Tmod3 in pyrene-actin polymerization assays (Fig. 7). We propose two possible explanations for Tmods’ differential functions in pointed-end depolymerization vs. elongation. First, the rate of pointed-end depolymerization may depend primarily on the strength of the TM-actin interaction, thereby reducing actin subunit off-rates in a manner independent of Tmods (Broschat 1990; Broschat et al. 1989; Weber et al. 1994), while the rate of pointed-end elongation may depend more on the actin subunit association rate at the pointed end, which is inhibited by Tmods binding to pointed ends (Weber et al. 1994; Weber et al. 1999). Second, Tmods’ TM-actin pointed-end capping activities may be sensitive to the nucleotide content of the terminal actin subunit (i.e., ATP or ADP·Pi during elongation vs. ADP during depolymerization (Weber et al. 1999)), and, in turn, the different rate constants of actin subunit association vs. dissociation (Carlier 1991; Pollard 1986).
Table 1.
Summary of the functional activities of wild-type and mutant Tmod1 and Tmod3 proteins with various TM isoforms, as determined in this study.
| TM isoforms | ||||||
|---|---|---|---|---|---|---|
| Tmod functions | Tmod proteins | α/βTM | αslowTM | TM5a | TM5b | TM5NM1 |
| TM binding | Tmod1 WT | - | Yes | Yes | - | Yes |
| Tmod1-L27G | - | Poor | Moderate | - | Yes | |
| Tmod1-I131D | - | Moderate | Yes | - | Poor | |
| Tmod1-L27G/I131D | - | No | No | - | No | |
| Tmod3 WT | Yes | - | - | Yes | Yes | |
| Tmod3-L29G | Poor | - | - | Poor | Moderate | |
| Tmod3-L134D | Moderate | - | - | Yes | Moderate | |
| Tmod3-L29G/L134D | No | - | - | No | Very poor | |
| Inhibition of LatA-induced pointed-end depolymerization | Tmod1 WT | Moderate | - | - | Moderate | Strong |
| Tmod1-L27G/I131D | Moderate | - | - | Moderate | Moderate | |
| Tmod3 WT | Moderate | - | - | Moderate | Moderate | |
| Tmod3-L29G/L134D | Moderate | - | - | Moderate | Moderate | |
| Inhibition of pointed-end elongation | Tmod1 WT | Very strong | - | - | Strong | Strong |
| Tmod1-L27G/I131D | Moderate | - | - | Strong | Moderate | |
| Tmod3 WT | Strong | - | - | Strong | Moderate | |
| Tmod3-L29G/L134D | Very poor | - | - | Strong | Poor | |
| Targeting to thin filament pointed ends | Tmod1 WT | Yes | - | - | - | - |
| Tmod1-L27G | Poor | - | - | - | - | |
| Tmod1-I131D | Moderate | - | - | - | - | |
| Tmod1-L27G/I131D | No | - | - | - | - | |
An unexpected finding of this study was that our TM-binding-disabled Tmod1 mutant (Tmod1-L27G/I131D) could partially inhibit elongation from the pointed ends of α/βTM-coated actin filaments and protect α/βTM-coated actin filaments from LatA-induced pointed-end depolymerization in vitro (Fig. 4 and Fig. 7), while still failing to target to the pointed ends of the thin filaments in cardiac myocytes (Fig. 9). Lack of Tmod1-L27G/I131D targeting to thin filament pointed ends agrees with antibody inhibition experiments, in which a monoclonal antibody inhibiting Tmod1-α/βTM binding (mAb17) causes disappearance of Tmod1 from thin filament pointed ends when microinjected into cultured cardiac myocytes or when incubated with isolated skeletal muscle myofibrils (Mudry et al. 2003; Ochala et al. 2014). However, microinjection of mAb17 also causes thin filament depolymerization and disassembly in cultured cardiac myocytes, as evidenced by loss of striated F-actin staining (Mudry et al. 2003). The differences between these studies suggest that factors in the intracellular environment and/or differences in the actin polymerization mechanism in vitro vs. thin filament assembly in vivo can modulate the functional interplay between TM binding and TM-actin capping by Tmods. Future studies will seek to identify the intracellular factor(s) involved in this phenomenon.
Differences in TM binding and TM-actin filament capping by Tmod1 and Tmod3 can provide insights into the biochemical mechanisms that target these Tmods to different intracellular compartments in diverse cell types. For example, in skeletal muscle fibers, Tmod1 caps the pointed ends of the α/βTM-actin thin filaments, while Tmod3 caps the pointed ends of SR-associated TM5NM1-actin filaments (Gokhin and Fowler 2011a; Gokhin et al. 2010). However, in Tmod1−/− skeletal muscle, Tmod3 translocates from the SR to the thin filament pointed ends (Gokhin and Fowler 2011a; Gokhin et al. 2010). Now, it is also known that Tmod1 binds ~5-fold more strongly than Tmod3 to skeletal muscle α/βTM in binary blot overlay assays (Gokhin et al. 2010; Yamashiro et al. 2014), but Tmod1 and Tmod3 show more modest differences in terms of their ability to cap α/βTM-actin filaments in pyrene-actin polymerization assays (Fig. 7C and (Yamashiro et al. 2014)). Thus, translocation of Tmod3 in Tmod1−/− skeletal muscle is likely driven by the elimination of Tmod1 outcompeting Tmod3 for α/βTM binding and not elimination of Tmod1 outcompeting Tmod3 for capping α/βTM-actin pointed ends.
The differential TM-binding and TM-actin filament capping activities of Tmod1 and Tmod3 also illuminate mechanisms of actin capping in the spectrin-actin membrane skeleton of red blood cells. The short actin filaments of the red blood cell membrane skeleton are normally capped by Tmod1 and associated with a combination of TM5b and TM5NM1 (Fowler 1987; Fowler 1990; Fowler and Bennett 1984; Ursitti and Fowler 1994). In Tmod1−/− red blood cells, Tmod3 (which is normally not present in adult red blood cells) appears at 1/5th of wild-type Tmod1 levels, leading to a compensated spheroelliptocytic anemia with osmotic fragility and reduced deformability due to a disrupted membrane skeleton with abnormally variable actin filament lengths (Moyer et al. 2010). Our data show that Tmod1 and Tmod3 have similar abilities to cap the pointed ends of both TM5b-actin and TM5NM1-actin filaments (Fig. 8 and (Yamashiro et al. 2014)). This suggests that the increased variability of actin filament lengths and alterations in membrane skeleton architecture in Tmod1−/− red blood cells are likely due to the substoichiometric amounts of Tmod3 rather than any intrinsic insufficiency of the TM-actin capping activity of Tmod3. Additional studies are required to elucidate functional differences between Tmod1 vs. Tmod3 and mechanisms of Tmod isoform compensation in cellular contexts.
MATERIALS AND METHODS
Proteins
Actin
Rabbit skeletal muscle actin was prepared from acetone powder as described previously (Pardee and Spudich 1982). Pyrene-labeled rabbit skeletal muscle actin was prepared as described previously (Kouyama and Mihashi 1981).
CapZ
CapZ (capping protein of chicken skeletal muscle) was a gift from Dorothy A. Schafer (University of Virginia, Charlottesville, VA).
TMs
Rabbit skeletal muscle α/βTM was prepared from rabbit skeletal muscle acetone powder as described previously (Smillie 1982). Recombinant skeletal muscle human αslowTM, expressed in baculovirus and purified as described previously (Akkari et al. 2002), was a gift from Sandra T. Cooper (University of Sydney, Sydney, New South Wales, Australia) and Nigel G. Laing (University of Western Australia, Perth, Western Australia). Recombinant rat TM5a was a gift from Sarah E. Hitchcock-DeGregori (Rutgers University, New Brunswick, NJ). Recombinant rat TM5b (TM5b cDNA in the pJC20 vector), expressed in BL21 E. coli and purified as described previously (Maytum et al. 2004), was a gift from Michael A. Geeves (University of Kent, Canterbury, UK). A clone of human TM5NM1 was a gift from Andrea Bacconi (The Scripps Research Institute, La Jolla, CA) and amplified from a pEGFP-TM5NM1 expression vector using PCR primers designed to introduce NcoI and XhoI restriction sites for cloning into the bacterial pET-14b expression vector (Novagen). Recombinant human TM5NM1 was expressed in BL21 E. coli and purified in the same manner as for TM5b. Protein concentrations were determined using a BCA Protein Assay Kit (Pierce). Note that the bacterially expressed short TM isoforms used in this study (TM5a, TM5b, and TM5NM1) all lacked N-terminal acetylation as well as the N-terminal Ala-Ser dipeptide to mimic acetylation (Hitchcock-DeGregori and Heald 1987; Monteiro et al. 1994; Urbancikova and Hitchcock-DeGregori 1994). However, lack of N-terminal acetylation was not expected to adversely impact the functions of TM5a, TM5b, and TM5NM1, as unacetylated recombinant short TM isoforms do not demonstrate deficiencies in their binding avidities for F-actin or Tmods (Greenfield and Fowler 2002; Pittenger and Helfman 1992; Yamashiro et al. 2014).
Tmods
Human Tmod1, mouse Tmod3, and their TM-binding mutants (Tmod1-L27G, -I131D, -L27G/I131D and Tmod3-L29G, -L134D, -L29G/L134D) were expressed as glutathione-S-transferase fusion proteins in BL21 E. coli and purified by thrombin cleavage followed by affinity chromatography using glutathione-sepharose and ion-exchange chromatography, as described previously (Yamashiro et al. 2010). Protein concentrations of purified Tmods and mutants were determined spectroscopically as described previously (Yamashiro et al. 2010). In some experiments, FLAG-tagged human Tmod1 was created using site-directed PCR mutagenesis to insert a FLAG epitope between residues L140 and M141 of human Tmod1, downstream of the second TM-binding site. Insertion of the FLAG tag was verified by sequencing and a slight increase in apparent molecular weight on western blots. FLAG-Tmod1-L27G, -I131D, and -L27G/I131D mutations were introduced into FLAG-tagged Tmod1 using site-directed PCR mutagenesis and verified by sequencing. FLAG-tagged Tmod1 and its mutants were expressed in HEK293FT cells using Lipofectamine 2000, and cell lysates were prepared using CelLytic M buffer (Sigma) supplemented with protease inhibitor cocktail (1:1000, Sigma), according to the manufacturer's instructions. FLAG-tagged Tmod1 proteins and mutants were affinity-purified using an anti-FLAG M2 column (Sigma). Concentrations of Tmod proteins and their mutants were determined spectroscopically, as described previously (Yamashiro et al. 2010).
TM-Tmod binding assays
We determined the relative strengths of TMs binding to Tmods using binary blot overlay assays, as described previously (Gokhin et al. 2010; Ilkovski et al. 2008; Yamashiro et al. 2014). Briefly, increasing amounts of purified wild-type or mutant Tmods, or increasing amounts of FLAG-tagged wild-type or mutant Tmods from HEK293FT cell lysates, were electrophoretically separated by SDS-PAGE on 12% Laemmli mini-gels (Laemmli 1970), and transferred to nitrocellulose in 20% methanol. Lysates from mock-transfected HEK293FT cells were also included as negative controls. Parallel gels were stained with Coomassie blue to control for total protein loading. Blots were incubated with 5 μg/ml of purified TMs. TMs bound to immobilized Tmods were detected using anti-TM antibodies as follows: TM311 (1:1000, Abcam) for skeletal muscle α/βTM, AB5441 (1:1000, Millipore) for TM5b, and AB5447 (1:1000, Millipore) for TM5NM1. After washing, blot overlays were incubated with HRP-conjugated secondary antibodies (1:10,000, Invitrogen). Bands were detected using an Enhanced Chemiluminescence Kit (Pierce), according to the manufacturer's instructions, and exposed to film. Band intensities on gels and blots were densitometrically quantified using ImageJ (http://rsbweb.nih.gov/ij/, National Institutes of Health).
Sedimentation assay for actin filament depolymerization from pointed ends
The abilities of TMs and Tmods to inhibit LatA-induced depolymerization of F-actin was performed as described previously with slight modifications (Morton et al. 2000; Yamashiro et al. 2008). CapZ-capped actin filaments (CapZ:actin = 1:100) were prepared by mixing 15 μM G-actin and 150 nM CapZ in G-buffer, followed by addition of 1/10th volume 10× polymerization buffer and incubation overnight on ice. 100 nM CapZ-capped filaments (100 nM CapZ, 10 μM actin) were incubated with or without 100 nM Tmods and 2.5 μM or 10 μM α/βTM, TM5b, or TM5NM1 for 30 min at 25°C. The mixtures were diluted five-fold by adding four volumes of F-buffer (20 mM Hepes, pH 7.5, 2 mM MgCl2, 0.1 M KCl, 1 mM DTT) in the presence of 10 μM LatA (Enzo Life Sciences) for 90 min at 25°C, and then ultracentrifuged in a Beckman TLA100 rotor at 300,000g for 20 min. Final concentrations of proteins after dilution were: 20 nM Tmods, 0.5 or 2 μM TMs as indicated, and 20 nM CapZ-capped filaments (20 nM CapZ, 2 μM actin). Supernatants and pellets were solubilized in SDS gel sample buffer, adjusted to the same volume, and analyzed by SDS-PAGE on 12% gels. Note that the amount of TMs required to saturate the F-actin was determined in separate experiments for each TM by ultracentrifugation followed by SDS-PAGE and Coomassie blue staining of supernatants and pellets (1 μM for α/βTM and TM5b; 2 μM for TM5NM1) (Yamashiro et al. 2014). Protein bands were densitometrically quantified using ImageJ.
Pyrene-actin polymerization kinetics
F-actin elongation rates at pointed ends were measured as described previously with slight modifications (Fowler et al. 2003; Weber et al. 1994). Briefly, 1.8 μM G-actin (8% pyrene-labeled) was converted to Mg2+-actin as described previously (Young et al. 1990). CapZ-capped actin filament nuclei (CapZ:actin = 1:100) were prepared by mixing 10 μM G-actin and 100 nM CapZ in G-buffer, followed by addition of 1/10th volume 10× polymerization buffer and incubation overnight on ice. Seeds (final concentration = 1.6 nM CapZ:160 nM actin) were incubated with indicated concentrations of Tmod proteins and TMs for 5 min at 25°C, added to the indicated concentrations of G-actin and polymerization was initiated immediately by addition of 1/10th volume of 10× polymerization buffer (200 mM Hepes, pH 7.5, 1 M KCl, 20 mM MgCl2, 2 mM DTT, 5 mM ATP, 10 mM EGTA). Fluorescence measurements (excitation = 366.5 nm, emission = 407 nm) were performed using a Fluoromax 3 fluorimeter (HORIBA Jobin Yvon, Edison, NJ). Pointed-end capping activities for Tmod1, Tmod3, and their TM-binding-disabled mutants were obtained from initial elongation rates, measured directly from the slopes of the polymerization traces over the first 1 min. Initial polymerization rates in the presence of increasing concentrations of Tmods or TMs were divided by the initial polymerization rate for actin in the absence of Tmod, yielding a series of rate/control rate ratios.
Cardiac myocyte cultures, transfection, and confocal imaging
Embryonic chick cardiac myocytes were isolated from E6 embryos as described previously (Gregorio et al. 1995; Littlefield et al. 2001). Cardiac myocytes were plated on glass coverslips at 1×105 cells/ml and transfected using Lipofectamine 2000 or Effectene 24 h after plating, following the manufacturer's instructions, using expression plasmids for FLAG-tagged wild-type Tmod1, Tmod1-L27G, Tmod1-I131D, or Tmod1-L27G/I131D (see above). In some experiments, cardiac myocytes were not transfected in order to visualize endogenous Tmod1. After 3-5 days post-transfection, cells were processed for immunostaining by washing twice in relaxing buffer (150 mM KCl, 5 mM MgCl2, 10 mM MOPS, 1 mM EGTA, 4 mM ATP), fixing for 15 min in 4% paraformaldehyde in relaxing buffer, permeabilizing for 15 min with 0.3% Triton X-100 in PBS, quenching for 15 min with 100 nM NaBH4 in PBS, and blocking for 1 h with 3% BSA and 1% goat serum in PBS.
Endogenous Tmod1 and exogenous FLAG-tagged Tmod1 proteins were localized using indirect immunofluorescence. Cells were first incubated with primary antibodies diluted in blocking buffer for 1 h at 37°C. Primary antibodies were as follows: mouse monoclonal anti-α-actinin (EA53, 1:100, Sigma), mouse monoclonal anti-FLAG (1:100, Sigma), rabbit polyclonal anti-α-actinin (1:250, a gift from Elisabeth Ehler, Kings College London), and rabbit polyclonal anti-Tmod1 (R1749bl3c, 1:75). After washing, cells were incubated with secondary antibodies for 1 h at 37°C. Secondary antibodies were as follows: Alexa 488-conjugated goat anti-mouse IgG (1:500, Invitrogen), Alexa 546-conjugated goat anti-mouse IgG (1:500, Invitrogen), Alexa 546-conjugated goat anti-rabbit IgG (1:500, Invitrogen). Secondary antibodies were supplemented with Alexa 647-conjugated phalloidin (1:250, Invitrogen). In some experiments, a Mouse on Mouse (M.O.M.™) Immunodetection Kit (Vector Laboratories, Burlingame, CA) was used for simultaneous immunolocalization of α-actinin and FLAG, according to the manufacturer's instructions. After washing, cells were mounted onto glass slides with Gel/Mount aqueous mounting medium (Sigma). Images of single optical sections were collected on a Zeiss LSM710 laser scanning confocal microscope mounted on a Zeiss Axio Observer Z1 microscope using a 63×/1.4-n.a. oil-objective lens (zoom 1.0 or 1.5). ZEN software was used for image collection.
Statistics
Data are presented as mean ± S.D. Differences between two groups were detected using Student's t-tests. Differences among more than two groups were detected using one-way ANOVA with post hoc Fisher's PLSD tests. Statistical significance was defined as p<0.05. Statistical analysis was performed in Microsoft Excel.
Supplementary Material
Supplementary Figure 1. Effects of wild-type and TM-binding-incompetent mutant Tmod proteins on LatA-induced depolymerization from the pointed ends of CapZ-capped actin filaments in the absence of TMs. (A) CapZ-capped actin filaments (CapZ:actin ratio of 1:100) were mixed with wild-type Tmod1, wild-type Tmod3, Tmod1-L27G/I131D (DM), or Tmod3-L29G/L134D (DM), diluted five-fold and incubated in the presence of 10 μM LatA to depolymerize the F-actin. Final concentrations of Tmods were as indicated, with 20 nM CapZ:actin seeds (2 μM actin). The mixtures were ultracentrifuged, fractionated into supernatants and pellets, and separated using SDS-PAGE. A, actin. (B) Densitometric quantification was performed to calculate the percentage of depolymerized actin in the supernatants. Data shown are mean ± S.D. of three independent experiments.
ACKNOWLEDGEMENTS
This research was supported by NIH grant R01-HL083464 (to V.M.F.), NIH Ruth L. Kirschstein National Research Service Award postdoctoral fellowship F32-AR055870 (to R.A.L.), and a development grant from the Muscular Dystrophy Association (to D.S.G.).
Footnotes
The authors have no conflicts of interest to disclose.
REFERENCES
- Akkari PA, Song Y, Hitchcock-DeGregori S, Blechynden L, Laing N. Expression and biological activity of Baculovirus generated wild-type human slow alpha tropomyosin and the Met9Arg mutant responsible for a dominant form of nemaline myopathy. Biochem Biophys Res Commun. 2002;296(2):300–4. doi: 10.1016/s0006-291x(02)00852-5. [DOI] [PubMed] [Google Scholar]
- Almenar-Queralt A, Lee A, Conley CA, Ribas de Pouplana L, Fowler VM. Identification of a novel tropomodulin isoform, skeletal tropomodulin, that caps actin filament pointed ends in fast skeletal muscle. J Biol Chem. 1999;274(40):28466–75. doi: 10.1074/jbc.274.40.28466. [DOI] [PubMed] [Google Scholar]
- Babcock GG, Fowler VM. Isoform-specific interaction of tropomodulin with skeletal muscle and erythrocyte tropomyosins. J Biol Chem. 1994;269(44):27510–8. [PubMed] [Google Scholar]
- Broschat KO. Tropomyosin prevents depolymerization of actin filaments from the pointed end. J Biol Chem. 1990;265(34):21323–9. [PubMed] [Google Scholar]
- Broschat KO, Weber A, Burgess DR. Tropomyosin stabilizes the pointed end of actin filaments by slowing depolymerization. Biochemistry. 1989;28(21):8501–6. doi: 10.1021/bi00447a035. [DOI] [PubMed] [Google Scholar]
- Carlier MF. Actin: protein structure and filament dynamics. J Biol Chem. 1991;266(1):1–4. [PubMed] [Google Scholar]
- Coue M, Brenner SL, Spector I, Korn ED. Inhibition of actin polymerization by latrunculin A. FEBS Lett. 1987;213(2):316–8. doi: 10.1016/0014-5793(87)81513-2. [DOI] [PubMed] [Google Scholar]
- Fischer RS, Fritz-Six KL, Fowler VM. Pointed-end capping by tropomodulin3 negatively regulates endothelial cell motility. J Cell Biol. 2003;161(2):371–80. doi: 10.1083/jcb.200209057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler VM. Identification and purification of a novel Mr 43,000 tropomyosin-binding protein from human erythrocyte membranes. J Biol Chem. 1987;262(26):12792–800. [PubMed] [Google Scholar]
- Fowler VM. Tropomodulin: a cytoskeletal protein that binds to the end of erythrocyte tropomyosin and inhibits tropomyosin binding to actin. J Cell Biol. 1990;111(2):471–81. doi: 10.1083/jcb.111.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler VM, Bennett V. Erythrocyte membrane tropomyosin. Purification and properties. The Journal of biological chemistry. 1984;259(9):5978–89. [PubMed] [Google Scholar]
- Fowler VM, Greenfield NJ, Moyer J. Tropomodulin contains two actin filament pointed end-capping domains. J Biol Chem. 2003;278(41):40000–9. doi: 10.1074/jbc.M306895200. [DOI] [PubMed] [Google Scholar]
- Fritz-Six KL, Cox PR, Fischer RS, Xu B, Gregorio CC, Zoghbi HY, Fowler VM. Aberrant myofibril assembly in tropomodulin1 null mice leads to aborted heart development and embryonic lethality. J Cell Biol. 2003;163(5):1033–44. doi: 10.1083/jcb.200308164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhin DS, Fowler VM. Cytoplasmic gamma-actin and tropomodulin isoforms link to the sarcoplasmic reticulum in skeletal muscle fibers. J Cell Biol. 2011a;194(1):105–20. doi: 10.1083/jcb.201011128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhin DS, Fowler VM. Tropomodulin capping of actin filaments in striated muscle development and physiology. J Biomed Biotechnol. 2011b;2011:103069. doi: 10.1155/2011/103069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhin DS, Lewis RA, McKeown CR, Nowak RB, Kim NE, Littlefield RS, Lieber RL, Fowler VM. Tropomodulin isoforms regulate thin filament pointed-end capping and skeletal muscle physiology. J Cell Biol. 2010;189(1):95–109. doi: 10.1083/jcb.201001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield NJ, Fowler VM. Tropomyosin requires an intact N-terminal coiled coil to interact with tropomodulin. Biophys J. 2002;82(5):2580–91. doi: 10.1016/S0006-3495(02)75600-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield NJ, Kostyukova AS, Hitchcock-DeGregori SE. Structure and tropomyosin binding properties of the N-terminal capping domain of tropomodulin 1. Biophys J. 2005;88(1):372–83. doi: 10.1529/biophysj.104.051128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorio CC, Weber A, Bondad M, Pennise CR, Fowler VM. Requirement of pointed-end capping by tropomodulin to maintain actin filament length in embryonic chick cardiac myocytes. Nature. 1995;377(6544):83–6. doi: 10.1038/377083a0. [DOI] [PubMed] [Google Scholar]
- Gunning P, O'Neill G, Hardeman E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol Rev. 2008;88(1):1–35. doi: 10.1152/physrev.00001.2007. [DOI] [PubMed] [Google Scholar]
- Gunning PW, Schevzov G, Kee AJ, Hardeman EC. Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol. 2005;15(6):333–41. doi: 10.1016/j.tcb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Hitchcock-DeGregori SE. Tropomyosin: function follows structure. Adv Exp Med Biol. 2008;644:60–72. doi: 10.1007/978-0-387-85766-4_5. [DOI] [PubMed] [Google Scholar]
- Hitchcock-DeGregori SE, Heald RW. Altered actin and troponin binding of amino-terminal variants of chicken striated muscle alpha-tropomyosin expressed in Escherichia coli. J Biol Chem. 1987;262(20):9730–5. [PubMed] [Google Scholar]
- Ilkovski B, Mokbel N, Lewis RA, Walker K, Nowak KJ, Domazetovska A, Laing NG, Fowler VM, North KN, Cooper ST. Disease severity and thin filament regulation in M9R TPM3 nemaline myopathy. J Neuropathol Exp Neurol. 2008;67(9):867–77. doi: 10.1097/NEN.0b013e318183a44f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyukova AS. Tropomodulin/tropomyosin interactions regulate actin pointed end dynamics. Adv Exp Med Biol. 2008a;644:283–92. doi: 10.1007/978-0-387-85766-4_21. [DOI] [PubMed] [Google Scholar]
- Kostyukova AS. Tropomodulins and tropomodulin/tropomyosin interactions. Cell Mol Life Sci. 2008b;65(4):563–9. doi: 10.1007/s00018-007-7347-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyukova AS, Choy A, Rapp BA. Tropomodulin binds two tropomyosins: a novel model for actin filament capping. Biochemistry. 2006;45(39):12068–75. doi: 10.1021/bi060899i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyukova AS, Hitchcock-DeGregori SE. Effect of the structure of the N terminus of tropomyosin on tropomodulin function. J Biol Chem. 2004;279(7):5066–71. doi: 10.1074/jbc.M311186200. [DOI] [PubMed] [Google Scholar]
- Kostyukova AS, Hitchcock-Degregori SE, Greenfield NJ. Molecular basis of tropomyosin binding to tropomodulin, an actin-capping protein. J Mol Biol. 2007;372(3):608–18. doi: 10.1016/j.jmb.2007.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyukova AS, Rapp BA, Choy A, Greenfield NJ, Hitchcock-DeGregori SE. Structural requirements of tropomodulin for tropomyosin binding and actin filament capping. Biochemistry. 2005;44(12):4905–10. doi: 10.1021/bi047468p. [DOI] [PubMed] [Google Scholar]
- Kouyama T, Mihashi K. Fluorimetry study of N-(1-pyrenyl)iodoacetamide-labelled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur J Biochem. 1981;114(1):33–8. [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Littlefield R, Almenar-Queralt A, Fowler VM. Actin dynamics at pointed ends regulates thin filament length in striated muscle. Nat Cell Biol. 2001;3(6):544–51. doi: 10.1038/35078517. [DOI] [PubMed] [Google Scholar]
- Maytum R, Bathe F, Konrad M, Geeves MA. Tropomyosin exon 6b is troponin-specific and required for correct acto-myosin regulation. J Biol Chem. 2004;279(18):18203–9. doi: 10.1074/jbc.M311636200. [DOI] [PubMed] [Google Scholar]
- McKeown CR, Nowak RB, Gokhin DS, Fowler VM. Tropomyosin is required for cardiac morphogenesis, myofibril assembly, and formation of adherens junctions in the developing mouse embryo. Dev Dyn. 2014;243(6):800–17. doi: 10.1002/dvdy.24115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown CR, Nowak RB, Moyer J, Sussman MA, Fowler VM. Tropomodulin1 is required in the heart but not the yolk sac for mouse embryonic development. Circ Res. 2008;103(11):1241–8. doi: 10.1161/CIRCRESAHA.108.178749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro PB, Lataro RC, Ferro JA, Reinach Fde C. Functional alpha-tropomyosin produced in Escherichia coli. A dipeptide extension can substitute the amino-terminal acetyl group. J Biol Chem. 1994;269(14):10461–6. [PubMed] [Google Scholar]
- Moroz NA, Novak SM, Azevedo R, Colpan M, Uversky VN, Gregorio CC, Kostyukova AS. Alteration of tropomyosin-binding properties of tropomodulin-1 affects its capping ability and localization in skeletal myocytes. J Biol Chem. 2013;288(7):4899–907. doi: 10.1074/jbc.M112.434522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton WM, Ayscough KR, McLaughlin PJ. Latrunculin alters the actin-monomer subunit interface to prevent polymerization. Nat Cell Biol. 2000;2(6):376–8. doi: 10.1038/35014075. [DOI] [PubMed] [Google Scholar]
- Moyer JD, Nowak RB, Kim NE, Larkin SK, Peters LL, Hartwig J, Kuypers FA, Fowler VM. Tropomodulin 1-null mice have a mild spherocytic elliptocytosis with appearance of tropomodulin 3 in red blood cells and disruption of the membrane skeleton. Blood. 2010;116(14):2590–9. doi: 10.1182/blood-2010-02-268458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudry RE, Perry CN, Richards M, Fowler VM, Gregorio CC. The interaction of tropomodulin with tropomyosin stabilizes thin filaments in cardiac myocytes. J Cell Biol. 2003;162(6):1057–68. doi: 10.1083/jcb.200305031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochala J, Gokhin DS, Iwamoto H, Fowler VM. Pointed-end capping by tropomodulin modulates actomyosin crossbridge formation in skeletal muscle fibers. FASEB J. 2014;28(1):408–15. doi: 10.1096/fj.13-239640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee JD, Spudich JA. Purification of muscle actin. Methods Cell Biol. 1982;24:271–89. doi: 10.1016/s0091-679x(08)60661-5. [DOI] [PubMed] [Google Scholar]
- Pittenger MF, Helfman DM. In vitro and in vivo characterization of four fibroblast tropomyosins produced in bacteria: TM-2, TM-3, TM-5a, and TM-5b are co-localized in interphase fibroblasts. J Cell Biol. 1992;118(4):841–58. doi: 10.1083/jcb.118.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J Cell Biol. 1986;103(6 Pt 2):2747–54. doi: 10.1083/jcb.103.6.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie LB. Preparation and identification of alpha- and beta-tropomyosins. Methods Enzymol. 1982;85(Pt B):234–41. doi: 10.1016/0076-6879(82)85023-4. [DOI] [PubMed] [Google Scholar]
- Spector I, Shochet NR, Blasberger D, Kashman Y. Latrunculins--novel marine macrolides that disrupt microfilament organization and affect cell growth: I. Comparison with cytochalasin D. Cell Motil Cytoskeleton. 1989;13(3):127–44. doi: 10.1002/cm.970130302. [DOI] [PubMed] [Google Scholar]
- Sui Z, Nowak RB, Bacconi A, Kim NE, Liu H, Li J, Wickrema A, An XL, Fowler VM. Tropomodulin3-null mice are embryonic lethal with anemia due to impaired erythroid terminal differentiation in the fetal liver. Blood. 2014;123(5):758–67. doi: 10.1182/blood-2013-03-492710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman MA, Baque S, Uhm CS, Daniels MP, Price RL, Simpson D, Terracio L, Kedes L. Altered expression of tropomodulin in cardiomyocytes disrupts the sarcomeric structure of myofibrils. Circ Res. 1998;82(1):94–105. doi: 10.1161/01.res.82.1.94. [DOI] [PubMed] [Google Scholar]
- Sussman MA, Fowler VM. Tropomodulin binding to tropomyosins. Isoform-specific differences in affinity and stoichiometry. Eur J Biochem. 1992;205(1):355–62. doi: 10.1111/j.1432-1033.1992.tb16787.x. [DOI] [PubMed] [Google Scholar]
- Temm-Grove CJ, Jockusch BM, Weinberger RP, Schevzov G, Helfman DM. Distinct localizations of tropomyosin isoforms in LLC-PK1 epithelial cells suggests specialized function at cell-cell adhesions. Cell Motil Cytoskeleton. 1998;40(4):393–407. doi: 10.1002/(SICI)1097-0169(1998)40:4<393::AID-CM7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Tobacman LS. Cooperative binding of tropomyosin to actin. Adv Exp Med Biol. 2008;644:85–94. doi: 10.1007/978-0-387-85766-4_7. [DOI] [PubMed] [Google Scholar]
- Tsukada T, Kotlyanskaya L, Huynh R, Desai B, Novak SM, Kajava AV, Gregorio CC, Kostyukova AS. Identification of residues within tropomodulin-1 responsible for its localization at the pointed ends of the actin filaments in cardiac myocytes. J Biol Chem. 2011;286(3):2194–204. doi: 10.1074/jbc.M110.186924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbancikova M, Hitchcock-DeGregori SE. Requirement of amino-terminal modification for striated muscle alpha-tropomyosin function. J Biol Chem. 1994;269(39):24310–5. [PubMed] [Google Scholar]
- Ursitti JA, Fowler VM. Immunolocalization of tropomodulin, tropomyosin and actin in spread human erythrocyte skeletons. J Cell Sci. 1994;107(Pt 6):1633–9. doi: 10.1242/jcs.107.6.1633. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Shah SP, Gritsyna Y, Hitchcock-DeGregori SE, Kostyukova AS. Systematic analysis of tropomodulin/tropomyosin interactions uncovers fine-tuned binding specificity of intrinsically disordered proteins. J Mol Recognit. 2011;24(4):647–55. doi: 10.1002/jmr.1093. [DOI] [PubMed] [Google Scholar]
- Vera C, Sood A, Gao KM, Yee LJ, Lin JJ, Sung LA. Tropomodulin-binding site mapped to residues 7-14 at the N-terminal heptad repeats of tropomyosin isoform 5. Arch Biochem Biophys. 2000;378(1):16–24. doi: 10.1006/abbi.2000.1802. [DOI] [PubMed] [Google Scholar]
- Vlahovich N, Kee AJ, Van der Poel C, Kettle E, Hernandez-Deviez D, Lucas C, Lynch GS, Parton RG, Gunning PW, Hardeman EC. Cytoskeletal tropomyosin Tm5NM1 is required for normal excitation-contraction coupling in skeletal muscle. Mol Biol Cell. 2009;20(1):400–9. doi: 10.1091/mbc.E08-06-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlahovich N, Schevzov G, Nair-Shaliker V, Ilkovski B, Artap ST, Joya JE, Kee AJ, North KN, Gunning PW, Hardeman EC. Tropomyosin 4 defines novel filaments in skeletal muscle associated with muscle remodelling/regeneration in normal and diseased muscle. Cell Motil Cytoskeleton. 2008;65(1):73–85. doi: 10.1002/cm.20245. [DOI] [PubMed] [Google Scholar]
- Weber A, Pennise CR, Babcock GG, Fowler VM. Tropomodulin caps the pointed ends of actin filaments. J Cell Biol. 1994;127(6 Pt 1):1627–35. doi: 10.1083/jcb.127.6.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Pennise CR, Fowler VM. Tropomodulin increases the critical concentration of barbed end-capped actin filaments by converting ADP.P(i)-actin to ADP-actin at all pointed filament ends. J Biol Chem. 1999;274(49):34637–45. doi: 10.1074/jbc.274.49.34637. [DOI] [PubMed] [Google Scholar]
- Weber KL, Fischer RS, Fowler VM. Tmod3 regulates polarized epithelial cell morphology. J Cell Sci. 2007;120(Pt 20):3625–32. doi: 10.1242/jcs.011445. [DOI] [PubMed] [Google Scholar]
- Wegner A. Equilibrium of the actin-tropomyosin interaction. J Mol Biol. 1979;131(4):839–53. doi: 10.1016/0022-2836(79)90204-3. [DOI] [PubMed] [Google Scholar]
- Yamashiro S, Cox EA, Baillie DL, Hardin JD, Ono S. Sarcomeric actin organization is synergistically promoted by tropomodulin, ADF/cofilin, AIP1 and profilin in C. elegans. J Cell Sci. 2008;121(Pt 23):3867–77. doi: 10.1242/jcs.040477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro S, Gokhin DS, Kimura S, Nowak RB, Fowler VM. Tropomodulins: pointed- end capping proteins that regulate actin filament architecture in diverse cell types. Cytoskeleton. 2012;69(6):337–70. doi: 10.1002/cm.21031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro S, Gokhin DS, Sui Z, Bergeron SE, Rubenstein PA, Fowler VM. Differential actin-regulatory activities of tropomodulin1 and tropomodulin3 with diverse tropomyosin and actin isoforms. J Biol Chem. 2014;289(17):11616–29. doi: 10.1074/jbc.M114.555128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro S, Speicher KD, Speicher DW, Fowler VM. Mammalian tropomodulins nucleate actin polymerization via their actin monomer binding and filament pointed end-capping activities. J Biol Chem. 2010;285(43):33265–80. doi: 10.1074/jbc.M110.144873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young CL, Southwick FS, Weber A. Kinetics of the interaction of a 41-kilodalton macrophage capping protein with actin: promotion of nucleation during prolongation of the lag period. Biochemistry. 1990;29(9):2232–40. doi: 10.1021/bi00461a005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Effects of wild-type and TM-binding-incompetent mutant Tmod proteins on LatA-induced depolymerization from the pointed ends of CapZ-capped actin filaments in the absence of TMs. (A) CapZ-capped actin filaments (CapZ:actin ratio of 1:100) were mixed with wild-type Tmod1, wild-type Tmod3, Tmod1-L27G/I131D (DM), or Tmod3-L29G/L134D (DM), diluted five-fold and incubated in the presence of 10 μM LatA to depolymerize the F-actin. Final concentrations of Tmods were as indicated, with 20 nM CapZ:actin seeds (2 μM actin). The mixtures were ultracentrifuged, fractionated into supernatants and pellets, and separated using SDS-PAGE. A, actin. (B) Densitometric quantification was performed to calculate the percentage of depolymerized actin in the supernatants. Data shown are mean ± S.D. of three independent experiments.