Abstract
When the amount of labeled data are limited, semi-supervised learning can improve the learner's performance by also using the often easily available unlabeled data. In particular, a popular approach requires the learned function to be smooth on the underlying data manifold. By approximating this manifold as a weighted graph, such graph-based techniques can often achieve state-of-the-art performance. However, their high time and space complexities make them less attractive on large data sets. In this paper, we propose to scale up graph-based semisupervised learning using a set of sparse prototypes derived from the data. These prototypes serve as a small set of data representatives, which can be used to approximate the graph-based regularizer and to control model complexity. Consequently, both training and testing become much more efficient. Moreover, when the Gaussian kernel is used to define the graph affinity, a simple and principled method to select the prototypes can be obtained. Experiments on a number of real-world data sets demonstrate encouraging performance and scaling properties of the proposed approach. It also compares favorably with models learned via ℓ1-regularization at the same level of model sparsity. These results demonstrate the efficacy of the proposed approach in producing highly parsimonious and accurate models for semisupervised learning.
Index Terms: Graph-based methods, large data sets, low-rank approximation, manifold regularization, semisupervised learning
I. Introduction
In Many data analysis and mining applications, the amount of unlabeled data available can be huge. However, the amount of labeled data remains scarce, due to the expensive and tedious human labeling process involved. Semisupervised learning [10], which can use unlabeled data together with the labeled data during training, is thus an effective approach to improve the learner's generalization performance.
In semisupervised learning, the label dependencies among the samples are captured by exploiting the intrinsic geometric structure of the data. This can be implemented using the cluster assumption, which encourages the separating hypersurface to pass through low-density regions [11], [19]. Another popular smoothness assumption is the manifold assumption, which assumes that the underlying function is smooth on a low-dimensional manifold formed by the data. Often, this manifold is approximated by a weighted graph defined on all the labeled and unlabeled data, and with the graph's affinity matrix encoding the similarities between the samples. This leads to the development of various graph-based semisupervised learning algorithms [4], [22], [26], [33], [45], [46], [48]. Besides these, techniques based on generative models [2], [27], self-training [39], co-training [7], and conditional random fields [20], [21] have also been used for semisupervised learning. An excellent survey can be found in [47].
In this paper, we will focus on the graph-based approach [4], [6], [48], which has a clear mathematical framework, good empirical performance and has also been widely applied in a variety of real-world problems. In particular, we will consider the manifold regularization framework [4]. It incorporates an additional regularizer to ensure that the learned function is smooth on the manifold; while at the same time the predictions on the labeled data are required to be consistent with the known class labels. In contrast to some other graph-based learning methods, this regularization framework is semisupervised (not transductive) and allows generalization to out-of-sample (unseen) patterns. Besides semisupervised learning, the use of manifolds has also been successfully applied in other learning problems such as manifold learning, dimension reduction, and clustering [3], [25], [29], [31], [34].
However, as graph-based semisupervised learning needs to manipulate the affinity (kernel) matrix of the graph (which is defined on all the available data), its space complexity has to grow (at least) quadratically with the data set size n [4], [45], [48]. On the other hand, its time complexity typically scales as n3. These pose a big challenge in large-scale applications.
In recent years, many efforts have been devoted to scaling up graph-based semisupervised learning. For example, Zhu and Lafferty [49] proposed an elegant framework that combines the generative mixture model with graph-based regularization. Recently, this is further extended for semisupervised feature selection [38]. However, when the data are high-dimensional, the curse of dimensionality sets in and could prevent the mixture model from fitting the data well [49]. In [13], a nonparametric function induction framework is proposed, which performs prediction based on a subset of preselected samples. However, it cannot enforce smoothness of the target function over those unlabeled data not in the preselected sample subset. In [8], the Nyström method [37] is used to numerically approximate the large matrix inverse associated with the graph-based semisupervised learning method of [45]. This leads to an efficient, transductive algorithm. The Nyström low-rank approximation is also used to scale up support vector training recently in [43], and transfer learning in the Hilbert space [44]. Other very recent attempts, such as the primal Laplacian supported vector machine (LapSVM) [24] and the anchor graph [23] will be presented in Section II-B.
Our key observation is that the computational intensiveness of graph-based semisupervised learning arises from the regularization term. On the one hand, this requires manipulation (multiplication, or inverse) of the n × n kernel matrix, which is impractical for large problems. On the other hand, the representer theorem [4] shows that the learned model spans over both labeled and unlabeled samples, making it inefficient in both training and testing.
The main contribution of this paper is in improving the scalability of semisupervised learning algorithms with the use of prototypes. These refer to a small set of points in the input space that can be used as a replacement of the original data in obtaining an efficient yet accurate predictive solution. Specifically, there are two types of prototypes used in the paper. First, low-rank approximation prototypes, which are used to obtain a low-rank approximation of the kernel matrix. This in turn allows graph-based regularization to be performed efficiently without having to manipulate the n × n kernel matrix. Second, label-reconstruction prototypes, which form a set of basis functions to span the predictive model. Since the number of prototypes is much smaller than the sample size, a highly compact model can be obtained, leading to fast training and testing. Moreover, without this functional representation, the learned model can only be transductive, and is unable to handle out-of-sample patterns. It will also be shown that when the graph affinity is defined by the Gaussian kernel, both kinds of prototypes can be selected in a principled and simple manner. Experiments on a number of real-world data sets demonstrate encouraging performance and scaling properties of the proposed approach.
The rest of this paper is organized as follows. In Section II, we first give a brief review on graph-based semisupervised learning algorithms and low-rank approximation algorithms. In Section III, we discuss the two important roles of prototypes in scaling up graph-based semisupervised learning, namely approximating the graph-based regularizer and simplifying the model parametrization. We also show that when the Gaussian kernel is used to define the graph affinity, these two criteria lead to a simple and unified prototype selection scheme via k-means clustering. In Section IV, we demonstrate how to use the prototypes to reduce the size of the optimization problem associated with semisupervised learning.
In Section V, we perform experiments on a number of real-world data sets to demonstrate the performance of the proposed method. Finally, Section VI gives some concluding remarks. A preliminary version of this paper is published in [42].
In the sequel, the transpose of vector/matrix is denoted by the superscript⊤, the identity matrix by I, and the n-dimensional vector of all ones by 1n. Moreover, for matrix A = [aij], we use for its Frobenius norm; and ‖A‖2 for its spectral norm (which is equal to the maximum singular value of A). Besides, diag(a) converts a n-dimensional vector a to a n × n diagonal matrix.
II. Related Work
Section II-A first reviews some standard graph-based semi-supervised learning algorithms. Section II-B then introduces a number of representative recent developments that can handle larger data sets. Finally, the approach of low-rank approximation, which will be a core component of the proposed method, is introduced in Section II-C.
A. Graph-Based Semisupervised Learning
In semisupervised learning, we are given a set of l labeled samples and u unlabeled samples , where xi ∈ ℝd, and yi ∈ {−1, 1} is the class label. For simplicity of exposition, we only consider binary classification problems here. The yi's from all the labeled samples can be concatenated to form the vector yl ∈ {−1, 1}l. For a given learning algorithm, let f be the learned function f, f ∈ ℝn be the vector of f's evaluations on the n = l + u samples. This f can be divided into two sub vectors: fl ∈ ℝl for the labeled samples, and fu ∈ ℝu for the unlabeled ones.
A number of graph-based semisupervised learning algorithms can be formulated as the following optimization problem:
| (1) |
where C1 and C2 are the regularization parameters. The first term in (1) enforces smoothness of the predicted labels with respect to the data's manifold structure, which is captured via the matrix S ∈ ℝn×n. Usually, S is chosen as the graph Laplacian matrix
| (2) |
where K ∈ ℝn×n is the kernel (adjacency) matrix of the graph [with associated kernel function K(·, ·)], and D = diag(K1n) is the corresponding degree matrix. Alternatively, the normalized graph Laplacian matrix
| (3) |
can also be used. The second term in (1) requires the predictions on the labeled samples be consistent with the known class labels, and the discrepancy is penalized by the empirical loss function L(·, ·). Finally, the third term enforces regularization on the learned function f. Popular choices for Ω(f) include the norm of f in some reproducing kernel Hilbert space (RKHS) [4]. Alternatively, Zhou et al. [45] simply uses Ω(f) = ‖fu‖, which regularizes predictions on the unlabeled samples only.
Well-known examples that follow the formulation in (1) include:
the method of learning with local and global consistency (LGC) [45], where S is the normalized graph Laplacian, and the two regularization parameters C1 and C2 are set to be equal;
learning using Gaussian fields and harmonic functions [48], where S is the unnormalized graph Laplacian, C1 approaches infinity, and C2 = 0;
the Laplacian regularized least-squares (LapRLS) and the LapSVM algorithms [4], which will be described in more detail in the following section.
The standard representer theorem in kernel learning can be extended to graph-based semisupervised learning. Specifically, Belkin et al. [4] showed that the learned function in (1) admits a representation as an expansion over all the labeled and unlabeled samples
| (4) |
where αi ∈ ℝ.
1) Laplacian Regularized Least-Squares
In this paper, we will focus on two loss functions commonly used in (1), the square and the hinge loss. The square loss function leads to the LapRLS algorithm [4], which solves the optimization problem
| (5) |
where ‖f‖k is the norm of f in the RKHS induced by the kernel function K, and γA, γI are regularization parameters related to the C1, C2 in (1) as C1 = (l + u)2/γIl and C2 = γA(l + u)2/γI. By plugging in the form of f in (4), we obtain
where α = [α1, …, αl+u]⊤ ∈ ℝl+u, y = [y1, …, yl, 0, …, 0]⊤ ∈ ℝl+u, and . It can be shown that the solution of α is given by
| (6) |
2) Laplacian SVM
Instead of using the square loss in (5), the LapSVM [4] uses the hinge loss, which then leads to
where (1 − yf(x))+ = max(0, 1 − yf(x)). Again, by plugging in (4), we obtain the primal problem
where ξ = [ξ1, …, ξl]⊤ ∈ ℝl. This leads to the solution
| (7) |
where J = [Il0] ∈ ℝl×(l+u) with Il as the l × l identity matrix, Y = diag([y1, …, yl]⊤), and β* ∈ ℝl is obtained by solving the following optimization problem:
and
As can be observed from (6) and (7), both LapRLS and LapSVM require the storing of a number of (l + u) × (l + u) matrices (such as K, J and S), and expensive matrix inversion of an (l + u) × (l + u) matrix. Consequently, this costs O(n3) time and O(n2) space (recall that n = l + u). Besides, testing is also expensive as the representation of f in (4) can involve n terms. In many real-world applications, u can be huge as there are often an abundance of unlabeled samples. Hence, both the algorithms can become impractical on large data sets.
B. Scaling Up Graph-based Semisupervised Learning
In this section, we briefly review some of the recent advances in scaling up graph-based semisupervised learning algorithms.
1) Nyström Method for Learning With LGC
A popular method for graph-based semisupervised learning is based on LGC [45]. Its predictions on the set of n (labeled and unlabeled) samples are given by
| (8) |
where y = [y1, …, yl, 0, …, 0]⊤ ∈ ℝn, Q ∈ ℝn×n is the normalized similarity matrix on the samples, and η is a regularization parameter. As (8) involves the inverse of the n × n matrix I − ηQ, it is expensive when n is large. To alleviate this problem, Camps-Valls et al. [8] used the Nyström method (which will be reviewed in Section II-C) to speed up the computation of (I − ηQ)−1. Specifically, they use the Nyström method to efficiently obtain an approximate eigenvalue decomposition VΛV⊤ [where V ∈ ℝn×m and Λ ∈ ℝm×m] of Q in O(m2n) time. Using the Woodbury formula, (8) can then be written as y − V(ΛV⊤V − η−1I)−1ΛV⊤y, which can be computed in O(m2n) time. By choosing m ≪ n, this approach is thus very suitable for large-scale applications. However, as will be demonstrated in Section V-B, empirically this approximation can hamper the prediction accuracy.
2) Primal LapSVM
As discussed in Section II-A1, traditionally the LapSVM is solved in the dual formulation. Very recently, new strategies are proposed to solve the LapSVM in the primal [24], which can be efficiently performed with preconditioned conjugate gradient [30]. Convergence can be further speed up using an early stopping criterion based on the predictions on the unlabeled data or validation examples. The resultant time complexity is reduced to O(cn2), where c is an empirical constant much smaller than n. Though better than the original O(n3) complexity, this quadratic complexity is still more expensive than other competitive methods.
3) Fast Nonparametric Function Induction
Delalleau et al. [13] proposed a nonparametric approach for estimating the continuous labels of unlabeled examples. For an out-of-sample example x, its prediction
| (9) |
has the same form as Parzen window regressors. Here, f(xj) is the estimated label for training sample xj (which may be unlabeled) obtained in the training phase. A straightforward implementation, however, involves O(n3) time and O(n2) space on training. To reduce these complexities, Delalleau et al. [13] force the summations in (9) to only involve a preselected sample subset S, |S| ≪ n, and do not enforce smoothness among the large number of unlabeled samples not in S. As will be empirically demonstrated in Section V-B, these restrictions can hamper the prediction accuracy.
4) Anchor Graph
Liu et al. [23] borrowed the idea of [13] and [42] and proposed to use the so-called anchor points (which are the same as prototypes in [42]) to build sparse graphs for semisupervised learning. The adjacency matrix is computed by a k-nearest-neighbor graph, or by solving a quadratic program (QP) similar to that in locally linear embedding [29] with additional sparsity constraints. In both cases, the computational cost is very expensive for large-scale applications.
C. Low-Rank Approximation
Given a symmetric, positive semidefinite matrix (such as the kernel matrix) K ∈ ℝn×n, a low-rank approximation algorithm [14], [17] produces an approximation of the form GG⊤, where G ∈ ℝn×m and m ≪ n. If the columns of G are linearly independent, GG⊤ is of rank m. It is well known that the optimal rank-m approximation K(m) of K (with respect to the spectral or the Frobenius norm) can be easily constructed from its k leading eigenvectors and eigenvalues. Specifically, let the eigenvalue decomposition of K be UΣU⊤, where the columns of U contain its eigenvectors, and Σ = diag[(σ1, σ2, …, σn)] contains the corresponding eigenvalues in descending order. Then, K(m) = U(m)Σ(m)U(m)⊤, where U(m) contains the m leading eigenvectors and U(m) contains the m leading eigenvalues. The corresponding low-rank approximation errors with respect to the spectral norm and Frobenius norm are ‖K − K(m)‖2 = σm+1 and , respectively. However, while K(m) can be obtained easily from the leading eigenvectors and eigenvalues of K, directly finding this eigenvalue decomposition takes O(n3) time. For large K, this can be expensive and hence more efficient alternatives have to be sought.
In this paper, we will use an efficient low-rank approximation approach called the Nyström method [15], [16], [37]. It first chooses a subset of m rows/columns from K. Let Knm ∈ ℝn×m be the submatrix of K containing these selected columns, and Kmm ∈ ℝm×m the submatrix containing the intersection of the selected columns and rows. Using the eigenvalue decomposition , the matrix containing the m leading eigenvectors of K is approximated by the Nyström extension as [37]. A useful consequence is that the whole matrix can then be approximated as
| (10) |
Note that, Knm can be computed in O(mn) time, and in O(m3) time. Thus, using (10), K can be written as GG⊤, where , in O(m2n + m3) time, which is linear in the sample size n.
Traditionally, the m columns are sampled randomly from K [37] or by a probabilistic sampling scheme [15]. More generally, they can be chosen as landmark points obtained from summarizing the data. The approximation in (10) is then applied as
| (11) |
where W ∈ ℝm×m [with Wij = K(ui, uj)] is the kernel matrix evaluated on the landmark points, and
| (12) |
is the cross-similarity matrix between the whole data X = [x1, …, xn] and the landmark points. For a number of commonly used kernels (such as the Gaussian, linear, and polynomial kernels), it has been shown that the approximation error ‖K−K̂‖F is bounded by the encoding errors of these landmark points [40], [41]. Based on this error analysis, we can choose the k-means cluster centers of the data as the landmark points. In the implementation, we further fix the number of k-means iteration to a small number (such as five). The complexity of k-means is then linear in the sample size and dimension (assuming that each kernel evaluation takes time linear in the dimension), and is thus suitable for large-scale problems.
III. Approximations via Prototypes
This section discusses the basic idea of using two types of prototypes to enable the optimization of graph-based semisupervised learning more efficient. By prototypes, we mean a small set of points in the input space that can be used as a replacement of the original data in obtaining an efficient yet accurate predictive solution. In this paper, there are two types of prototypes serving different purposes: 1) low-rank approximation prototypes that are used for low-rank approximation of the graph Laplacian matrix (Section III-A) and 2) label-reconstruction prototypes that are used in label-reconstruction for accurate model representation (Section III-B).
A. Low-Rank Approximation Prototypes
Recall that the graph Laplacian matrix S in (2) or (3) is constructed from the kernel matrix K of the graph. In practice, K usually has a rank much smaller than its size [37]. This offers the opportunity of reducing the computational burden of graph-based regularization by performing low-rank approximation (Section II-C).
Using the low-rank approximation EW−1E⊤ of K in (11), the unnormalized graph Laplacian S in (2) can be approximated as
| (13) |
where D̃ = diag(EW−1E⊤ln). The corresponding (approximated) graph-based regularizer is then f⊤Sf ≃ f⊤(D̃ − EW−1E⊤)f. Similarly, when S is the normalized graph Laplacian, it is approximated as
| (14) |
and the corresponding regularizer becomes f⊤Sf ≃ f⊤(I − ED̃−1/2W−1E⊤D̃−1/2)f.
B. Label-Reconstruction Prototypes
1) Motivating Example
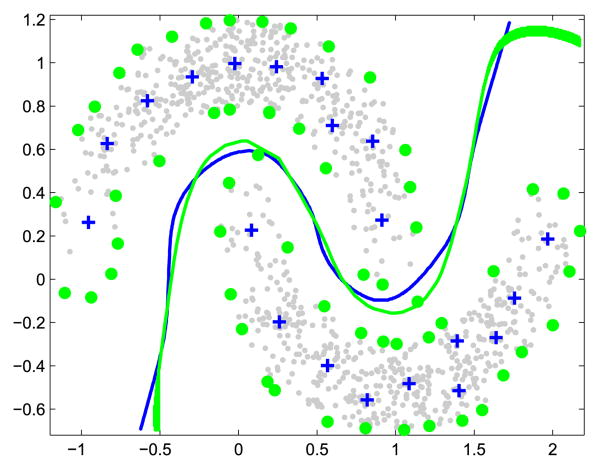
As discussed in Section II-A, the inclusion of a graph-based regularizer results in the model (4) that expands over all the labeled and unlabeled samples. This leads to slow training and testing when n is large. In practice, a classifier seldom needs all the samples as basis functions. Fig. 1 shows a motivating example on a binary classification problem. We first train a standard SVM using the whole data set. The obtained decision boundary (in green) and support vectors (circles) are shown. To construct an alternative classifier, we choose a set of label-reconstruction prototypes (crosses) from each class. To predict the label of a test sample x, we compute its similarities with all the label-reconstruction prototypes, and then linearly combine their labels using the similarities as weights. The resultant decision boundary is shown in blue. As can be observed, the two decision boundaries are very similar. This illustrates that instead of using the whole data set, a small set of properly chosen label-reconstruction prototypes can successfully reconstruct the decision boundary.
Fig. 1.
Decision boundaries of the SVM (green) and a prototype-based model (blue) on a toy data set. Circles are the support vectors and crosses are the prototypes.
2) Formulation
The above idea can be formalized as follows. We assume that the label of any sample x can be reconstructed as a weighted combination of the labels of r label-reconstruction prototypes
| (15) |
where fvi is the label of prototype vi. This resembles the nearest-neighbor classifier in that the predicted label of x is mostly dependent on prototypes vj having large similarities (i.e., K(x, vj) values) with x. Intuitively, as long as there are enough label-reconstruction prototypes filling the input space, the label of any sample can be reconstructed reliably from these nearby prototypes.
To make the model more flexible, we allow each fvi in (15) to be real-valued even for classification problems. Note that, (15) can be written more compactly as
| (16) |
where fυ = [fv1, fv2, …, fvr]⊤ ∈ ℝr and
| (17) |
is the kernel submatrix defined between the whole data set X and prototypes vi's. On large data sets, one can additionally enforce sparsity on H by only computing the similarities between the label-reconstruction prototypes and their k-nearest samples. This has been found useful in the construction of large graphs in semisupervised learning [23].
3) Choosing the Label-Reconstruction Prototypes
We now consider how to choose the label-reconstruction prototypes. Note that, (15) can be deemed as an approximation of the complete model in (4). Hence, we want to minimize
| (18) |
where D(·, ·) measures the difference between the complete model in (4) and the approximate model in (15).
In practice, as αi's, fvi's, and vi's are unknown during training, it is infeasible to directly minimize (18) with respect to all these variables. Recall that forms a basis for the complete model and forms a basis for the approximate model. In order for these two models to have similar representation power, we expect that the two basis can be reliably coded by each other. More specifically, we choose the label-reconstruction prototypes vi's so as to minimize the error of coding each component in with the best element in
| (19) |
Obviously, the choice of vi's depends on the specific kernel K(·, ·) and discrepancy measure D(·, ·). In the following, we consider the use of the Gaussian kernel K(x, y) = exp (−‖x − y‖2/2h2). Since it has the same form as the normal distribution, we use the cross entropy for probability distribution functions (pdfs) as D(·, ·) in (19). Recall that the cross entropy between the two pdfs p and q is
| (20) |
where H(p) = − ∫ p(x)log(p(x))dx is the entropy of p, and DKL(p‖q) = ∫ p(x)log(p(x)/q(x))dx is the Kullback–Leibler (KL) divergence between p and q [18]. Since {x1, …, xl+u} is fixed, minimizing D(K(·, xi), K(·, vj)) = H(K(·, xi), K(·, vj)) in (19) is the same as minimizing their KL-divergence, which can be written as [18]1
| (21) |
The objective in (19) then simplifies as
where Sj is the subset of kernels in whose closest label-reconstruction prototype is K(·, vj). Interestingly, this is exactly the objective of the k-means clustering (apart from the scaling factor 1/4h2). As in Section III-A, the prototypes vi's in (15) can again be chosen as the k-means cluster centers. In other words, the low-rank approximation prototypes in Section III-A and the label-reconstruction prototypes introduced here are indeed the same. Consequently, E in (12) is also the same to H in (17).
For other types of kernels, the cross entropy in (20) may no longer be suitable and other distance measures will be investigated in the future.
IV. Prototype Vector Machine
In Section III, we introduced two types of prototypes: 1) low-rank-approximation prototypes , which avoid the computation of an n × n graph Laplacian matrix and 2) label-reconstruction prototypes , which avoid the use of all the labels in the whole data set. In this section, using the associated simplified equations (13) [or (14)] and (16), we will consider how to realize the proposed prototype vector machine (PVM) with the square and hinge loss functions. As will be seen, since m, r ≪ n (the sample size), the optimization of problem (1) can be made much more efficient.
|
| |
| Algorithm 1 PVM Using the Square Loss | |
|
| |
| 1: | Perform k-means clustering on the given labeled and unlabeled samples, and use the cluster centers as prototypes. |
| 2: | Use (17) to compute H (and thus obtain its submatrices Hl and Hu). |
| 3: | Substitute the approximate graph Laplacian [(13) or (14)] into (23) and obtain . |
| 4: | Prediction |
| 1) on the given unlabeled samples: . | |
| 2) on an unseen . | |
|
| |
A. Training
1) Square Loss
Using the square loss, the objective in (1) can be written as
| (22) |
where Hl ∈ ℝl×r and Hu ∈ ℝu×r are submatrices of H containing the rows corresponding to the labeled and unlabeled samples, respectively, and S is as defined in (13). Replacing S with the normalized graph Laplacian (14) is straightforward. Moreover, as in [45], we have used ‖Hufv‖2 to regularize f. Alternatively, one can use the RKHS norm of f (as in [4]). On using (15), this leads to a slightly different regularizer , where Kυ is the kernel matrix defined on the r label-reconstruction prototypes, and the results in the sequel can be easily extended.
By setting the derivative of the objective in (22) with respect to fυ to zero, we obtain the optimal solution of fυ as
| (23) |
The complete algorithm is shown in Algorithm 1. Note that, if k-means clustering is used as the selection scheme for both the low-rank-approximation prototypes and label-reconstruction prototypes, then we simply have El = Hl in (23).
On using k-means as the unified prototype selection scheme, we have m = r. Step 1 of Algorithm 1 then takes O(ndm) time and O(nm) space. Step 2 takes O(ndr) = O(ndm) time (assuming that each kernel evaluation takes O(d) time) and O(nr) = O(nm) space. In step 3, with the use of the m low-rank approximation prototypes, we have H⊤SH = H⊤DH − (H⊤E)W−1(E⊤H) from (13). Since D̃ is diagonal, H ∈ ℝn×r and E ∈ ℝn×m, H⊤SH can be computed in O(r2n + r2m + m2r + mnr) = O(nr(m + r)) = O(nm2) time and O(n max(r, m)) = O(nm) space. Moreover, the inverse operation in (23) involves a r × r matrix, and takes O(r3) time. Thus, the total time for computing is O(nr(m +r) + r3) = O(nr(m + r)) = O(nm2). Since m ≪ n, the total space and time complexities are both linear in the sample size, which is much faster than the O(n3) time for LapRLS (Section II-A).
2) Hinge Loss
For classification problems (with labels in {±1}), we can use the hinge loss as popularized by the SVM. The other two terms in (22) remain the same, and can be combined together as , where2
| (24) |
Let ek be the transpose of the kth row in Hl
| (25) |
The optimization problem in (1) can then be written as
where the first term performs regularization, and the second term is the hinge loss incurred by approximating the labels of Xl with the reconstructed ones (Hlfυ). The Lagrangian can be written as
By setting its derivative with respect to fυ and ξi's to zero, we obtain C1 = βi + γi, and
| (26) |
on using (25). Here, β = [β1, …, βl]⊤, and ⊙ denotes the element wise product. Plugging these back into the Lagrangian, we obtain the dual problem
| (27) |
where
| (28) |
After solving this QP, labels of the prototypes vi's can be recovered using the Karush–Kuhn–Tucker condition in (26), as
| (29) |
The complete algorithm is shown in Algorithm 2.
As in Section IV-A1, H⊤SH can be computed in O(nm2) time. Hence, matrix A in step 3 can also be computed in O(nr(m + r) + r2u) = O(nr(m + r)) = O(nm2) time, and its inverse in O(r3) time. Moreover, note that the QP in (27) is of the same form as a standard SVM, and so can be solved readily with state-of-the-art SVM solvers. There are only l optimization variables in this QP, and modern SVM implementations typically have a time complexity that scales linearly with l [36]. So, the total time complexity is O(nr(m+r)+r3) = O(nr(m+r)) = O(nm2). Since m ≪ n, this is again much more efficient than the O(n3) time required of the standard LapSVM (Section II-A) and the O(n2) time required of the primal LapSVM (Section II-B2). Moreover, it can be easily seen that its space complexity is O(mn), the same as that when the square loss is used. As a short summary, Table I compares the time and space complexities of a number of semisupervised learning algorithms that will be further studied empirically in Section V-B.
Table I.
Time and Space Complexities for the Training Process of Various Semisupervised Learning Algorithms (Descriptions of the Algorithms Can Be Found in Section V-B). Here, n Is the Total Number of Labeled and Unlabeled Samples, and m Is the Number of Prototypes in the PVM (Which Is Equal to the Sample Subset Size Used in the Other Algorithms)
| LGC [45] | LapRLS [4] | LapSVMp [24] | NYS-LGC [8] | NF1 [13] | Anchor [23] | PVM (sqr) | PVM (hinge) | |
|---|---|---|---|---|---|---|---|---|
| time | O(n3) | O(n3) | O(m2n) | O(n2) | O(m2n) | O(m2n) | O(m2n) | O(n2) |
| space | O(n2) | O(n2) | O(n2)† | O(mn) | O(mn) | O(n2)† | O(mn) | O(mn) |
In the implementation of LapSVMp and Anchor method, the kernel matrix is partitioned into sub-blocks that are loaded into memory sequentially, which can further reduce the O(n2) memory consumption to O(np), where p is a user defined number.
|
| |
| Algorithm 2 PVM Using the Hinge Loss | |
|
| |
| 1: | Perform k-means clustering on the given labeled and unlabeled samples, and use the cluster centers as prototypes. |
| 2: | Use (17) to compute H (and thus obtain its submatrices Hl and Hu). |
| 3: | Use the approximate graph Laplacian [(13) or (14)] to compute A in (24), and subsequently Q in (28). |
| 4: | Solve the QP in (27) to obtain β. |
| 5: | Obtain from (29). |
| 6: | Prediction |
| 1) on the given unlabeled samples: | |
| 2) on an unseen . | |
|
| |
B. Prediction
After obtaining , the predicted labels on the given unlabeled samples can be computed as , and that on an unseen test sample x as . Since it only depends on the r label-reconstruction prototypes, testing takes O(r) time. This is much faster than LapRLS and LapSVM, which take O(n) time.
C. Number of Prototypes
The number of prototypes is an important factor that affects the performance of the proposed method. More prototypes lead to more accurate approximation on both the kernel matrix as well as the class labels. Therefore, this tends to give a better performance. However, the time consumption also grows. Hence, this involves a tradeoff between the accuracy and the efficiency. Empirically, we choose m as 5% of the sample size (or fewer), which gives satisfactory results. In our empirical evaluations (Section V-A.3), we also observe that when m is beyond a certain threshold the performance would reach a plateau.
We are also interested in performance of PVM with respect to choice of the number of prototype vectors as well as the number of labeled and unlabeled samples. First, the larger the number of prototypes, the more complicated concept (e.g., classification boundary) can be potentially delineated. Therefore, more prototypes will lead to a better performance, as has been shown in our empirical evaluations.
In considering the number of labeled samples, note that given a fixed level of intrinsic difficulty of the learning problem (in terms of the complexity of the classification boundaries), more labeled samples will lead to a better prediction performance. In this case, together with more prototypes, we will also expect a better performance.
If the unlabeled data can improve the performance of learning, more prototypes will be able to summarize the distribution of unlabeled data better, therefore the performance will improve with more prototypes and unlabeled data. However, sometimes, the unlabeled data may not help in semisupervised learning. For example, if the low-density separation assumption is violated [11], [19], [28] then it would be hard to predict whether more prototypes will improve performance, since more prototypes will make the effect of unlabeled samples more significant.
In practice, the decision boundary can be determined in a complicated manner by labeled and unlabeled samples together in semisupervised learning. For example, in [32], it is stated that if the complexity of the distribution under consideration is too high to be learned using labeled data points, but is small enough to be learned using unlabeled data points, then semisupervised learning can improve the performance of the supervised learning task. In this case, we may expect a better performance with more prototypes as well as labeled and unlabeled samples. However, in some cases, the needed relation between labeled and unlabeled samples might not be satisfied. For example, Ben-David et al. [5] concluded that using unlabeled data cannot provide sample size guarantees that are better than those using labeled data only, unless one assumes a very strong relationship between the labeled and unlabeled data distribution. Therefore, in case the desired condition is not satisfied, it would be hard to guarantee that more prototypes will lead to improved performance with more labeled and unlabeled samples.
V. Experiments
In Section V-A, we investigate how the performance of the proposed PVM varies with sample size, the number of labeled samples and prototypes. In Section V-B, the PVM is compared with other state-of-the-art semisupervised learning algorithms on a number of real-world data sets. Experiments will be performed on both binary and multiclass classification problems. Note that for PVM using the square loss, the formulation in (22) can be extended for multiclass classification in a straightforward manner. For PVM using the hinge loss, we first decompose a C-class classification problem into C one-versus-rest binary classification problems. Prediction is then made by assigning the sample to the class with the highest decision function value.
PVM is a sparse model, as only a small number of prototypes are involved in (15). Another popular approach for the construction of sparse models is using the ℓ1-regularizer, which encourages model coefficients to approach zero. Recently, this ℓ1 -penalized approach has drawn considerable interest in learning sparse predictive models. For example, lasso [35], the most well-known ℓ1-penalized sparse model, adds the ℓ1-regularizer to the square loss function. Hence, a natural question is how the parsimonious model produced by the PVM compares with that obtained by ℓ1-regularization. This will be addressed in Section V-D.
A. Performance of the PVM
In this section, we study the performance of the PVM by performing experiments on a subset of the MNIST digits data set.3 We use the five digits (represented as 784-D feature vectors) 3, 5, 6, 8, and 9, leading to a total of 29 270 samples. The Gaussian kernel
| (30) |
is used. As discussed in Section III, this leads to a unified prototype selection scheme, namely that the k-means cluster centers can be used as both the low-rank approximation prototypes and the label-reconstruction prototypes.
1) Variation With Sample Size
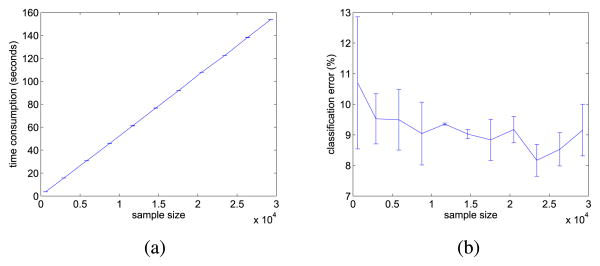
In this experiment, we first study how the performance of PVM (using the square loss) varies when the total number (n) of labeled and unlabeled samples is gradually increased from 1000 to 29270. For each class, 50 samples are labeled. The number of prototypes m is fixed at 200. To reduce statistical variability, results are based on averages over 30 random repetitions.
Fig. 2(a) shows the variation of training time (which includes the time for k-means clustering in step 1 of Algorithms 1 and 2) with n. As can be seen, the time scales linearly with n, which agrees with our analysis in Section IV-A. Fig. 2(b) shows the classification error on the unlabeled samples. Note that although the number of prototypes is fixed, the accuracy remains fairly stable with increasing sample size. This agrees with our intuition that the number of prototypes required for building an accurate model depends largely on the data distribution and class boundary, but not directly on the sample size.
Fig. 2.
Performance of the PVM with different sample sizes. (a) Training time versus sample size (note that the error bars here are very small). (b) Error versus sample size.
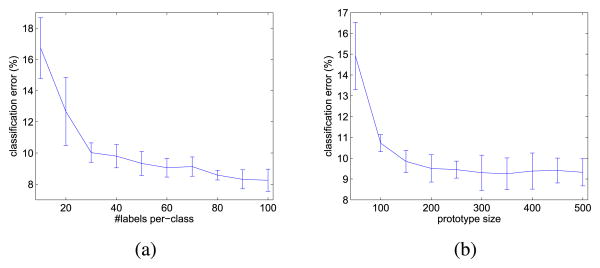
2) Variation With the Number of Labeled Samples
Next, we vary the number of labeled samples used for training, as l = {100, 200, 300, …, 1000}. The number of unlabeled samples is then u = n − l with n = 29270. Fig. 3(a) shows the resultant variation of the classification error (on the unlabeled samples). As can be seen, similar to the other semisupervised learning models, the accuracy increases when more labeled information is available. The improvement in accuracy is particularly prominent when the number of labeled samples is small. This agrees with the observation in [9] that the labeled samples reduce the error exponentially fast.
Fig. 3.
Performance of the PVM at different settings. (a) Different numbers of labeled samples per class. (b) Different numbers of prototypes.
3) Variation With the Number of Prototypes
Finally, Fig. 3(b) examines the variation of the classification error with the number of prototypes m, while fixing the number of labeled samples per class at 50, and the sample size at n = 29720. As can be seen, using more prototypes leads to better performance, though the improvement diminishes when m is beyond a certain value. In this experiment, using m = 200, which is less than 1% of the whole data set size, already suffices to give a good performance.
B. Comparison With the State-of-the-Art
In this section, we compare both versions of PVM: PVM(sqr), using the square loss (Section IV-A1), and PVM(hinge), using the hinge loss (Section IV-A2), with the supervised learner SVM (using only the labeled samples) and a number of state-of-the-art semisupervised learning algorithms, including:
LGC: learning with LGC [45];
LapRLS: Laplacian regularized least squares [4] (Section II-A1);
NYS-LGC: acceleration of LGC using Nyström low-rank approximation [8] (Section II-B1);
LapSVMp: primal LapSVM 4 trained with preconditioned conjugate gradient [24] (Section II-B2);
Nonparametric function induction (NFI): fast nonparametric function induction [13] (Section II-B3);
Anchor5: the method based on anchor graph [23] (Section II-B4).
All codes are in MATLAB and run on an Intel T2400 1.83-GHz laptop with 1-GB memory.
1) Setup
We use a variety of binary and multiclass benchmark data sets from [10]6 and the libsvm data archive.7 The number of labeled samples per class is 50 (except for the two-moon data, which has only one labeled sample per class). A summary of the data sets is shown in Table II.
Table II.
Data Sets Used in the Experiment. l Is the Number of Labeled Samples, u Is the Number of Unlabeled Samples, and m Is the Number of Prototypes
| DATA | DIM | #CLASSES | l | u | m |
|---|---|---|---|---|---|
| G241C | 241 | 2 | 100 | 1,400 | 150 |
| G241D | 241 | 2 | 100 | 1,400 | 150 |
| DIGIT1 | 241 | 2 | 100 | 1,400 | 150 |
| USPS | 241 | 2 | 100 | 1,400 | 150 |
| COIL2 | 241 | 2 | 100 | 1,400 | 150 |
| COIL | 241 | 6 | 300 | 1,200 | 150 |
| BCI | 117 | 2 | 100 | 300 | 40 |
| TEXT | 11,960 | 2 | 100 | 1,400 | 150 |
| SPLICE | 60 | 2 | 100 | 900 | 100 |
| SEGMENT | 29 | 7 | 350 | 1,960 | 231 |
| DNA | 180 | 3 | 150 | 3,036 | 200 |
| SVMGD1A | 4 | 2 | 100 | 6,989 | 200 |
| USPS-FULL | 256 | 10 | 500 | 6,791 | 200 |
| SATIMAGE | 36 | 6 | 300 | 6,135 | 200 |
For PVM, the number of prototypes is chosen as m = 0.1n when n ≤ 3000; and m = 200 otherwise. The same number of samples is used in forming the subset for NFI and NYS-LGC. Other parameters of the various algorithms are chosen by fivefold cross-validation.8 For example, the β parameter of the Gaussian kernel is chosen from {2−5β0, 2−4β0, …, 24β0, 25β0}, where β0 is the reciprocal of average distance between the samples. The regularization parameters in LapRLS are chosen from {10−6, 10−5, …, 1, 10}; while those for the other algorithms are from {10−3, 10−2, …, 104, 105}. For PVM, we simply set the regularization parameter C2 = 0 and only tune C1. To reduce statistical variability, results are based on averages over 30 random selections of the labeled samples.
2) Results
Table III shows the classification errors on the unlabeled samples, and Table IV shows the total training and testing time. The following observations can be made.
Table III.
Classification Errors (%) on the Unlabeled Data Obtained by the Different Algorithms. The Best Results and Those That Are Not Statistically Worse (According to the Two-Sample One-Tailed t-Test, With a p-Value of 0.05) Are in Bold. Note That, LGC and LapRLS Cannot Be Run on the Large Data Sets of Svmgd1a, Usps-Full, and Satimage
| (SUPERVISED) | (SEMI-SUPERVISED) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| DATA | SVM | LGC | LapRLS | LapSVMp | NYS-LGC | NFI | ANCHOR | PVM(sqr) | PVM(hinge) |
| G241C | 22.34±1.41 | 21.92±1.90 | 22.02±1.73 | 25.82±1.75 | 24.19±3.06 | 28.07±2.71 | 35.15±1.45 | 24.50±2.49 | 23.21±1.95 |
| Q241D | 25.14±2.03 | 28.10±3.27 | 22.36±1.78 | 25.89±1.12 | 30.98±4.22 | 30.82±5.36 | 36.61±2.07 | 25.15±2.58 | 24.85±2.70 |
| DIGIT1 | 4.70±1.09 | 5.74±1.09 | 5.74±1.50 | 1.84±0.37 | 6.68±1.63 | 9.83±1.43 | 3.56±0.45 | 4.18±1.17 | 3.72±1.07 |
| USPS | 9.30±2.81 | 4.57±1.27 | 6.11±0.63 | 5.23±2.34 | 9.72±1.82 | 5.49±0.78 | 15.90±3.41 | 5.29±0.73 | 6.35±1.33 |
| COIL2 | 11.17±0.91 | 14.37±3.63 | 10.83±1.94 | 12.74±2.16 | 16.90±3.00 | 13.98±2.48 | 8.11±1.64 | 11.69±2.47 | 14.85±2.36 |
| COIL | 12.21±1.81 | 12.38±1.99 | 21.17±1.71 | 7.79±0.74 | 18.75±1.48 | 30.93±6.22 | 12.41±0.67 | 13.41±1.29 | 12.26±1.01 |
| BCI | 35.17±3.29 | 44.43±2.28 | 29.16±3.56 | 46.23±3.16 | 45.45±2.62 | 45.67±2.61 | 48.37±2.61 | 33.59±3.01 | 31.65±2.86 |
| TEXT | 24.35±2.14 | 23.09±1.43 | 23.99±1.58 | 22.75±1.30 | 34.40±3.63 | 32.54±2.66 | 27.28±1.01 | 30.4±4.46 | 26.29±2.58 |
| SPLICE | 24.33±1.13 | 22.85±1.47 | 19.78±2.41 | 33.76±2.53 | 30.56±3.16 | 34.56±1.97 | 29.11±0.97 | 23.47±1.59 | 25.32±2.48 |
| SEGMENT | 9.91±1.06 | 8.97±1.20 | 9.58±1.73 | 12.15±2.13 | 13.58±1.95 | 15.71±2.15 | 13.75±1.58 | 10.15±1.21 | 9.06±1.15 |
| DNA | 15.88±1.17 | 27.31±2.58 | 17.72±1.29 | 15.27±0.41 | 29.53±2.12 | 43.38±3.94 | 28.60±1.81 | 15.87±1.44 | 14.19±1.28 |
| SVMGD1A | 5.85±1.65 | – | – | 4.23±0.40 | 6.32±1.88 | 14.21±2.92 | 4.S4±0.62 | 5.24±1.09 | 6.08±1.55 |
| USPS-FULL | 6.39±0.50 | – | – | 4.16±0.13 | 17.68±1.57 | 14.43±0.89 | 7.43±0.29 | 7.35±0.62 | 5.88±0.91 |
| SATIMAGE | 14.59±0.71 | – | – | 13.81±0.22 | 16.36±0.64 | 19.27±3.74 | 14.80±0.56 | 14.64±0.50 | 13.27±0.53 |
Table IV. Total Training and Testing Time (s) of the Different Algorithms.
| DATA | SVM | LGC | LapRLS | LapSVMp | NYS-LGC | NFI | ANCHOR | PVM(SQR) | PVM(HINGE) |
|---|---|---|---|---|---|---|---|---|---|
| G241C | 0.54 | 140.84 | 129.86 | 33.47 | 0.86 | 0.48 | 11.75 | 3.30 | 3.19 |
| G241D | 0.55 | 129.78 | 142.65 | 11.95 | 0.84 | 0.49 | 12.33 | 3.31 | 3.16 |
| DIGIT1 | 0.57 | 140.51 | 131.08 | 7.72 | 0.84 | 0.48 | 12.79 | 3.31 | 3.15 |
| USPS | 0.48 | 139.23 | 131.59 | 21.35 | 0.74 | 0.47 | 12.89 | 3.28 | 3.14 |
| COIL2 | 0.57 | 151.36 | 120.48 | 14.57 | 0.87 | 0.48 | 11.51 | 3.26 | 3.47 |
| COIL | 1.58 | 146.92 | 115.22 | 31.54 | 0.79 | 0.49 | 13.01 | 3.35 | 3.51 |
| BCI | 0.08 | 3.08 | 1.94 | 1.12 | 0.53 | 0.22 | 5.89 | 0.71 | 1.09 |
| TEXT | 25.6 | 139.67 | 216.37 | 59.27 | 9.14 | 13.26 | 47.54 | 30.24 | 34.24 |
| SPLICE | 1.69 | 1622.51 | 1439.51 | 33.12 | 2.49 | 0.83 | 23.71 | 4.87 | 4.24 |
| SEGMENT | 4.51 | 1319.67 | 1389.80 | 69.71 | 1.96 | 31.67 | 7.15 | 6.13 | 8.81 |
| DNA | 2.85 | 1566.91 | 1463.75 | 368.91 | 3.07 | 1.22 | 52.91 | 8.92 | 7.57 |
| SVMGD1A | 2.71 | – | – | 1129.44 | 3.22 | 1.66 | 69.21 | 8.06 | 5.38 |
| USPS-FULL | 19.46 | – | – | 8920.13 | 3.96 | 2.87 | 125.70 | 22.48 | 28.21 |
| SATIMAGE | 8.90 | – | – | 2671.56 | 3.34 | 2.57 | 189.56 | 11.56 | 14.32 |
In general, the semisupervised learning algorithms are more accurate than the baseline SVM. In particular, based on the paired student-t test, the proposed PVM(hinge) outperforms SVM on the DIGIT1, USPS, BCI, SEGMENT, DNA, USPS-FULL, and SATIMAGE data sets at the 95% confidence level. It has a comparable performance as the SVM on the G241C, G241D, SPLICE, SVMGD1A data sets, and is only outperformed by the SVM on the two data sets of COIL2 and TEXT.
LGC, LapRLS, and LapSVMp do not employ any approximation to scale up training. Hence, they are usually the most accurate, but also computationally most expensive. Indeed, recall that LGC and LapRLS require O(n2) space. Hence, they cannot be run on the three largest data sets (svmgd1a, usps-full, and satimage). Moreover, though LapSVMp is faster than LapRLS and LGC, it is still computationally expensive on the large data sets as its time complexity is quadratic in n.
LGC-NYS and NFI are computationally very efficient but not as accurate, especially on the more difficult data sets of TEXT and DNA. As discussed in Section II-B.3, we speculate that it is because NFI cannot enforce smoothness of the function on the unlabeled samples not belonging to the basis set [13].
The anchor method is more efficient than LGC, LapRLS, and LapSVMp. However, since it needs to compute the combination coefficients for each sample using QP, its complexity can be huge for large data sets. As can be observed from Table VI, its time consumption is at least several times larger than that of PVM. The larger the data set, the larger the difference.
The accuracies of both PVMs are comparable with LGC, LapRLS, and LapSVMp; while their speeds can be several hundred times faster. On larger data sets, this speedup ratio is expected to be more significant. Overall, PVM achieves a good balance between speed and accuracy.
Table VI. Total Training and Testing Time (s) of CB-LapRLS, CB-LapSVMp, and PVM.
| DATA | CB-LapRLS | CB-LapSVMp | PVM (sqr) | PVM (hinge) |
|---|---|---|---|---|
| G241C | 10.15 | 0.47 | 3.30 | 3.19 |
| G241D | 12.11 | 0.65 | 3.31 | 3.16 |
| DIGIT1 | 12.54 | 0.72 | 3.31 | 3.15 |
| USPS | 13.76 | 0.35 | 3.28 | 3.14 |
| COIL2 | 11.48 | 0.57 | 3.26 | 3.47 |
| COIL | 115.22 | 0.54 | 3.35 | 3.51 |
| BCI | 1.15 | 0.22 | 0.71 | 1.09 |
| TEXT | 62.15 | 28.35 | 30.24 | 34.24 |
| SPLICE | 6.51 | 3.42 | 4.87 | 4.24 |
| SEGMENT | 12.17 | 5.31 | 6.13 | 8.81 |
| DNA | 15.16 | 7.41 | 8.92 | 7.57 |
| SVMGD1A | 23.94 | 5.58 | 8.06 | 5.38 |
| USPS-FULL | 57.14 | 19.85 | 22.48 | 28.21 |
| SATIMAGE | 39.58 | 11.29 | 11.56 | 14.32 |
We are also interested in training the full semisupervised learning algorithms (LapRLS and LapSVMp) using only a small subset of representative points (such as cluster centers) as unlabeled data. Results and discussions are in Table VII. As can be observed, using representative points in such a way can improve the computational efficiency of the algorithm but lead to less accurate results.
Table VII. Comparison of the Total Training and Testing Time (s) Between PVM and L1-Penalized Sparse Model.
| BCI | COIL | COIL2 | DIGIT1 | USPS-2V7 | USPS-5V6 | USPS-3V8 | USPS-FULL | |
|---|---|---|---|---|---|---|---|---|
| L1-LRK | 178.5 | 169.1 | 135.7 | 165.4 | 135.0 | 144.1 | 108.5 | - |
| PVM (SQUARED) | 0.71 | 3.35 | 3.26 | 3.31 | 4.12 | 3.98 | 2.73 | 8.06 |
C. Learning With Only the Cluster Centroids
In the PVM, the prototypes are selected from the centroids of k-means clustering. Recall that both the LapRLS and LapSVM (trained in either the primal or dual) have large computational complexities. Hence, one may consider using a computationally less expensive version that replaces the whole unlabeled set with the set of k-means cluster centroids. On the other hand, all the labeled samples are still used, as they are usually much fewer in number. In this section, we compare the performance of these cluster-based (CB) versions (denoted CB-LapRLS and CB-LapSVMp) with PVM.
Tables V and VI show the comparison in terms of the classification error and time consumption, respectively. For easy reference, results for the PVM are also copied from Tables III and IV. As can be seen, though CB-LapRLS and CB-LapSVMp are much faster than the original LapRLS and LapSVMp, their accuracies are often much inferior because of the loss of unlabeled data information. In contrast, the PVM adopts the representative points (cluster centers) in a systematic way for approximations of the whole kernel matrix and the decision function, thus yielding a much better performance in general.
Table V. Classification Errors (%) on the Unlabeled Data Obtained by CB-LapRLS, CB-LapSVMp, and PVM.
| DATA | CB-LapRLS | CB-LapSVMp | PVM (sqr) | PVM (hinge) |
|---|---|---|---|---|
| G241C | 27.14±2.15 | 28.32±2.91 | 24.50±2.49 | 23.21±1.95 |
| G241D | 28.11±1.78 | 27.57±1.43 | 25.15±2.58 | 24.85±2.70 |
| DIGIT1 | 6.78±1.91 | 6.84±1.72 | 4.18±1.17 | 3.72±1.07 |
| USPS | 19.67±3.46 | 19.45±2.25 | 5.29±0.73 | 6.35±1.33 |
| COIL2 | 14.98±2.15 | 17.16±2.81 | 11.69±2.47 | 14.85±2.36 |
| COIL | 21.17±1.71 | 12.39±1.45 | 13.41±1.29 | 12.26±1.01 |
| BCI | 29.16±3.56 | 46.23±3.16 | 33.59±3.01 | 31.65±2.86 |
| TEXT | 27.18±2.77 | 25.31±1.91 | 30.4±4.46 | 26.29±2.58 |
| SPLICE | 22.35±3.48 | 28.82±3.97 | 23.47±1.59 | 25.32±2.48 |
| SEGMENT | 9.19±1.56 | 11.21±1.81 | 10.15±1.21 | 9.06±1.15 |
| DNA | 16.57±1.41 | 15.87±0.44 | 15.87±1.44 | 14.19±1.28 |
| SVMGD1A | 7.71±0.56 | 6.92±0.55 | 5.24±1.09 | 6.08±1.55 |
| USPS-FULL | 14.15±3.41 | 11.15±2.17 | 7.35±0.62 | 5.88±0.91 |
| SATIMAGE | 17.85±2.15 | 15.34±1.66 | 14.64±0.50 | 13.27±0.53 |
D. Comparison With ℓ1-Penalized Sparse Models
In this section, we compare the sparse models produced by PVM and ℓ1-regularization. For ℓ1-regularization, we use the kernel regressor, which involves the optimization problem
| (31) |
where Kα (for some α ∈ ℝn) is the vector of predictions on all n labeled and unlabeled samples, and Kl ∈ ℝl×n is the kernel submatrix. The (Kα)⊤S(Kα) regularization term encourages smoothness over the data manifold, and the ℓ1-regularizer ‖α‖1 encourages sparseness of the coefficient vector α.
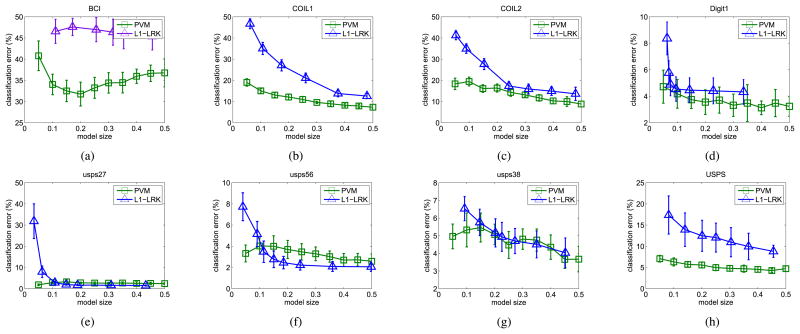
In the following, we compare the performance of PVM and the ℓ1-penalized formulation in (31) at different sparsity levels. Note that how the sparsity is achieved by the PVM is quite different from that of (31). The former is obtained by extracting a highly compact and representative set of prototypes, from which the structure of the kernel matrix can be faithfully preserved. Hence, for the PVM, sparsity can be explicitly controlled by specifying the number of prototypes. On the other hand, (31) enforces sparsity by penalizing via the ℓ1-norm. There is no known algorithm that can produce a model with exactly the desired number of nonzero coefficients. Instead, we can only try different regularization parameter settings to obtain a model with approximately the desired size.9 This parallel the comparison between VQ-based code-book selection and sparse-coding-based codebook selection in dictionary learning [12]. Moreover, to allow (31) to be used on large data sets, low-rank approximation is first applied on the kernel matrix K. In the sequel, the method will be denoted L1-LRK.
The classification errors on the unlabeled data versus sparsity for the two algorithms are reported in Fig. 4. As can be seen, at the same sparsity level, PVM often has better accuracy than L1-LRK. This demonstrates the efficacy of PVM in building highly efficient, parsimonious, and accurate models in the semisupervised learning setting. In particular, PVM tends to perform better than L1-LRK when the data are high dimensional or have a lot of classes.
Fig. 4.
Performance of PVM and the ℓ1-penalized formulation at different levels of model sparsity. The x-axis is the ratio of nonzero coefficients in the predictive model to the sample size. (a) BCI. (b) COIL. (c) COIL2. (d) Digit1. (e) USPS-2v7. (f) USPS-5v6. (g) USPS-3v8. and (h) USPS-FULL.
Table VII compares the time consumption of PVM with L1-LRK. Here, we have chosen the regularization parameters such that the number of nonzero coefficients in the obtained model is around 10% of the sample size. As can be seen, PVM is much more efficient. This is because for the L1-LRK, to transform the problem (31) to a standard LASSO-type problem, O(n2) space and O(n3) time are needed.
VI. Conclusion
In this paper, we proposed the PVM to scale up graph-based semisupervised learning. Using the prototypes to approximate the graph-based regularizer as well as the predictive model, we can drastically reduce the problem size while at the same time guarantee smoothness of the prediction over the data manifold. Experiments on a number of real-world data sets demonstrate that the PVM has appealing scaling behavior (linear in sample size) and competitive performance.
In the future, we will study various extensions of the PVM. For example, we will consider other kernels (besides the Gaussian kernel) and other label reconstruction schemes that can take the local geometrical structure of the samples into account. Moreover, the current prototype selection scheme (k-means clustering) treats the labeled and unlabeled data as equally important. One possibility is to incorporate the label information, and extend the selection scheme to a weighted version so that different importance-based weightings are assigned based on prior knowledge. A similar idea has been successfully used in the context of low-rank approximation [1]. Moreover, we will further study the relationships between the labeled samples, unlabeled samples, and the prototypes.
Acknowledgments
This work was supported in part by the National Institute of Health under Grant R01-CA140663 and in part by the Lawrence Berkeley National Laboratory under Contract DE-AC02-05CH11231. The work of J. T. Kwok was supported by the Hong Kong Competitive Earmarked Research Grant under Project 614012.
Biography

Kai Zhang received the Ph.D. degree in computer science from the Hong Kong University of Science and Technology, Hong Kong, in 2008.
He joined the Life Science Division, Lawrence Berkeley National Laboratory, Berkeley, CA, USA. He is currently a Research Staff Member with NEC Laboratories America, Inc., Princeton, NJ, USA. His current research interests include large scale machine learning, matrix approximation, and applications in bioinformatics, time series, and complex networks.

Liang Lan received the B.S. degree in bioinformatics from the Huazhong University of Science and Technology, Wuhan, China, and the Ph.D. degree in computer and information sciences from Temple University, Philadelphia, PA, USA, in 2007 and 2012, respectively.
He is currently a Researcher with the Huawei Noah's Ark Laboratory, Hong Kong. His current research interests include data mining and machine learning.

James T. Kwok received the Ph.D. degree in computer science from the Hong Kong University of Science and Technology, Hong Kong, in 1996.
He is a Professor with the Department of Computer Science and Engineering, Hong Kong University of Science and Technology. His current research interests include kernel methods, machine learning, and pattern recognition.
Prof. Kwok has been a Program Co-Chair for a number of international conferences, and served as an Associate Editor of the IEEE Transactions on Neural Networks and Learning Systems from 2006 to 2012. He is currently an Associate Editor of the Neurocomputing journal. He was a recipient of the 2004 IEEE Outstanding Paper Award, and the Second Class Award in Natural Sciences from the Ministry of Education, China, in 2008.

Slobodan Vucetic received the B.S. and M.S. degrees in electrical engineering from the University of Novi Sad, Novi Sad, Serbia, and the Ph.D. degree in electrical engineering from Washington State University, Pullman, WA, USA, in 1994, 1997, and 2001, respectively.
He is currently an Associate Professor with the Department of Computer and Information Sciences, Temple University, Philadelphia, PA, USA. His current research interests include data mining and machine learning.

Bahram Parvin is a Principal Scientist with the Lawrence Berkeley National Laboratory, Berkeley, CA, USA, and has an adjunct appointment with the Electrical Engineering Department, University of California, Berkeley. His laboratory focuses on the development of machine learning techniques for realization of pathway pathology, and elucidating molecular signatures of aberrant morphogenesis in normal and engineered matrices.
Prof. Parvin was the General Chair of the IEEE International Symposium on Biomedical Imaging: From Nano to Macro, in 2013. He is also a Steering Committee Member for the IEEE Bioimaging and Signal Processing and the IEEE Bioengineering and Health Care.
Footnotes
This is a special case of the problem considered in [18], where the simplified model has varying bandwidths and the components in the original model are weighted.
If the RKHS norm of f is used as the regularizer instead of ‖Hufυ‖2 as discussed in Section IV-A1, then A becomes H⊤SH + C2Kυ, and the results in Section IV-A2 can also be easily extended.
The MATLAB code is from http://sourceforge.net/projects/lapsvmp/
The MATLAB code is from http://www.ee.columbia.edu/ln/dvmm/downloads/WeiGraphConstructCode/dlform.htm
In each of the 5 repetitions, the semisupervised model is trained using 4/5 of the labeled data and the unlabeled data, and is validated on the remaining 1/5 of the labeled data. The parameter that yields the lowest average validation error is selected. Using this parameter, the model is retrained with all the labeled and unlabeled data.
In the experiments, λ1 and λ2 are selected from {10−4, 10−3, …, 101, 102}.
Contributor Information
Kai Zhang, Email: kzhang@nec-labs.com, NEC Laboratories America, Inc., Princeton, NJ 08540 USA.
Liang Lan, Email: lanliang@temple.edu, Huawei Noah's Ark Laboratory, Hong Kong.
James T. Kwok, Email: jamesk@cs.ust.hk, Department of Computer Science and Engineering, Hong Kong University of Science and Technology, Hong Kong
Slobodan Vucetic, Email: vucetic@temple.edu, Department of Computer and Information Sciences, Temple University, Philadelphia, PA 19122 USA.
Bahram Parvin, Email: b_parvin@lbl.gov, Life Sciences Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720 USA.
References
- 1.Bach F, Jordan M. Predictive low-rank decomposition for kernel methods. Proc. 22nd Int. Conf. Mach. Learn.; Bonn, Germany. Aug. 2005; pp. 33–40. [Google Scholar]
- 2.Baluja S. Probabilistic modeling for face orientation discrimination: Learning from labeled and unlabeled data. Proc Adv NIPS. 1999;11:854–860. [Google Scholar]
- 3.Belkin M, Niyogi P. Laplacian eigenmaps and spectral techniques for embedding and clustering. Proc Adv NIPS. 2002;14:585–591. [Google Scholar]
- 4.Belkin M, Niyogi P, Sindhwani V. Manifold regularization: A geometric framework for learning from labeled and unlabeled examples. J Mach Learn Res. 2006;7(no. 11):2399–2434. [Google Scholar]
- 5.Ben-David S, Lu T, Pal D. Does unlabeled data provably help? Worst-case analysis of the sample complexity of semi-supervised learning. Proc. 21st Annu. Conf. Learn. Theory; 2008. pp. 33–44. [Google Scholar]
- 6.Blum A, Chawla S. Learning from labeled and unlabeled data using graph mincuts. Proc. 18th Int. Conf. Mach. Learn.; San Francisco, CA, USA. 2001. pp. 19–26. [Google Scholar]
- 7.Blum A, Mitchell T. Combining labeled and unlabeled data with co-training. Proc. 11th Annu. Conf. Comput. Learn. Theory; New York, NY, USA. Jul. 1998; pp. 209–214. [Google Scholar]
- 8.Camps-Valls G, Marsheva TVB, Zhou D. Semi-supervised graph-based hyperspectral image classification. IEEE Trans Geosci Remote Sens E. 2007 Oct;45(no. 10):3044–3054. [Google Scholar]
- 9.Castelli V, Cover T. On the exponential value of labeled samples. Pattern Recognit Lett. 1995;16(no. 1):105–111. [Google Scholar]
- 10.Chapelle O, Schölkopf B, Zien A. Semi-Supervised Learning. Cambridge, MA, USA: MIT Press; 2006. [Google Scholar]
- 11.Chapelle O, Zien A. Semi-supervised classification by low density separation. Proc 10th Int Workshop Artif Intell Statist. 2005 Jan; [Google Scholar]
- 12.Coates A, Ng AY. The importance of encoding versus training with sparse coding and vector quantization. Proc. 28th Int. Conf. Mach. Learn.; Bellevue, WA, USA. Jun. 2011; pp. 921–928. [Google Scholar]
- 13.Delalleau O, Bengio Y, Roux NL. Efficient non-parametric function induction in semi-supervised learning. Proc. 10th Int. Workshop Artif. Intell. Statist.; 2005; pp. 96–103. [Google Scholar]
- 14.Drineas P, Kannan R, Mahoney M. Fast Monte Carlo algorithms for matrices II: Computing a low rank approximation to a matrix. SIAM J Comput. 2006;36(no. 1):158–183. [Google Scholar]
- 15.Drineas P, Mahoney MW. On the Nyström method for approximating a Gram matrix for improved kernel-based learning. J Mach Learn Res. 2005 Dec;6:2153–2175. [Google Scholar]
- 16.Fowlkes C, Belongie S, Chung F, Malik J. Spectral grouping using the Nyström method. IEEE Trans Pattern Anal Mach Intell. 2004 Feb;26(no. 2):214–225. doi: 10.1109/TPAMI.2004.1262185. [DOI] [PubMed] [Google Scholar]
- 17.Frieze A, Kannan R, Vempala S. Fast Monte-Carlo algorithms for finding low-rank approximations. Proc. 39th Annu. Symp. Found. Comput. Sci.; Palo Alto, CA, USA. 1998. pp. 370–378. [Google Scholar]
- 18.Goldberger J, Roweis S. Hierarchical clustering of a mixture model. Proc Adv NIPS. 2005;17:505–512. [Google Scholar]
- 19.Joachims T. Transductive inference for text classification using support vector machines. Proc. 16th Int. Conf. Mach. Learn.; San Francisco, CA, USA. 1999. pp. 200–209. [Google Scholar]
- 20.Lafferty J, Zhu X, Liu Y. Kernel conditional random fields: Representation and clique selection. Proc. 21st Int. Conf. Mach. Learn.; Banff, AB, Canada. Jul. 2004; p. 64. [Google Scholar]
- 21.Lee CH, Wang S, Jiao F, Schuurmans D, Greiner D. Proc Adv NIPS. Vol. 19. Cambridge, MA, USA: 2007. Learning to model spatial dependency: Semi-supervised discriminative random fields. [Google Scholar]
- 22.Chen L, Tsang IW, Xu D. Laplacian embedded regression for scalable manifold regularization. IEEE Trans Neural Netw Learn Syst. 2012 Jun;23(no. 6):902–915. doi: 10.1109/TNNLS.2012.2190420. [DOI] [PubMed] [Google Scholar]
- 23.Liu W, He J, Chang SF. Large graph construction for scalable semi-supervised learning. Proc. 27th Int. Conf. Mach. Learn.; Haifa, Israel. Jun. 2010; pp. 679–686. [Google Scholar]
- 24.Melacci S, Belkin M. Laplacian support vector machines trained in the primal. J Mach Learn Res. 2011;12(no. 3):1149–1184. [Google Scholar]
- 25.Ng AY, Jordan MI, Weiss Y. On spectral clustering: Analysis and an algorithm. Proc Adv NIPS. 2001;14:849–856. [Google Scholar]
- 26.Huang Y, Xu D, Nie F. Semi-supervised dimension reduction using trace ratio criterion. IEEE Trans Neural Netw Learn Syst. 2012 Mar;23(no. 3):519–526. doi: 10.1109/TNNLS.2011.2178037. [DOI] [PubMed] [Google Scholar]
- 27.Nigam K, McCallum A, Thrun S, Mitchell T. Text classification from labeled and unlabeled documents using EM. Mach Learn. 2000;39(nos. 2–3):103–134. [Google Scholar]
- 28.Rigollet P. Generalization error bounds in semi-supervised classification under the cluster assumption. J Mach Learn Res. 2007 Dec;8:1369–1392. [Google Scholar]
- 29.Roweis ST, Saul LK. Nonlinear dimensionality reduction by locally linear embedding. Science. 2000;290(no. 5500):2323–2326. doi: 10.1126/science.290.5500.2323. [DOI] [PubMed] [Google Scholar]
- 30.Shewchuk JR. School Comput Sci. Carnegie Mellon Univ; Pittsburgh, PA, USA: 1994. An introduction to the conjugate gradient method without the agonizing pain. Tech. Rep. [Google Scholar]
- 31.Shi J, Malik J. Normalized cuts and image segmentation. IEEE Trans Pattern Anal Mach Intell. 2000 Aug;22(no. 8):888–905. [Google Scholar]
- 32.Singh A, Nowak RD, Zhu X. Unlabeled data: Now it helps, now it doesn't. Proc Adv NIPS. 2008:1513–1520. [Google Scholar]
- 33.Soares RGF, Chen H, Yao X. Semisupervised classification with cluster regularization. IEEE Trans Neural Netw Learn Syst. 2012 Nov;23(no. 11):1779–1792. doi: 10.1109/TNNLS.2012.2214488. [DOI] [PubMed] [Google Scholar]
- 34.Tenenbaum JB, de Silva V, Langford JC. A global geometric framework for nonlinear dimensionality reduction. Science. 2000;290(no. 5500):2319–2323. doi: 10.1126/science.290.5500.2319. [DOI] [PubMed] [Google Scholar]
- 35.Tibshirani R. Regression shrinkage and selection via the lasso. J Roy Statist, Soc B. 1996;58(no. 1):267–288. [Google Scholar]
- 36.Tsang IW, Kwok JT, Cheung PM. Core vector machines: Fast SVM training on very large data sets. J Mach Learn Res. 2005 Dec;6:363–392. [Google Scholar]
- 37.Williams C, Seeger M. Using the Nyström method to speed up kernel machines. Proc Adv NIPS. 2001;13:682–688. [Google Scholar]
- 38.Xu Z, King I, Lyu MRT, Jin R. Discriminative semi-supervised feature selection via manifold regularization. IEEE Trans Neural Netw. 2010 Jul;21(no. 7):1033–1047. doi: 10.1109/TNN.2010.2047114. [DOI] [PubMed] [Google Scholar]
- 39.Yarowsky D. Unsupervised word-sense disambiguation rivaling supervised methods. Proc. 33rd Annu. Meet. Assoc. Comput. Linguistics; Stroudsburg, PA, USA. 1995. pp. 189–196. [Google Scholar]
- 40.Zhang K, Kwok JT. Improved Nyström low rank approximation and error analysis. Proc. 25th Int. Conf. Mach. Learn.; Helsinki, Finland. Jun. 2008; pp. 1232–1239. [Google Scholar]
- 41.Zhang K, Kwok JT. Clustered Nyström method for large scale manifold learning and dimension reduction. IEEE Trans Neural Netw. 2010 Oct;21(no. 10):1576–1587. doi: 10.1109/TNN.2010.2064786. [DOI] [PubMed] [Google Scholar]
- 42.Zhang K, Kwok JT, Parvin B. Prototype vector machine for large scale semi-supervised learning. Proc. 26th Int. Conf. Mach. Learn.; Montreal, QC, Canada. Jun. 2009; pp. 1233–1240. [Google Scholar]
- 43.Zhang K, Lan L, Wang Z, Moerchen F. Scaling up kernel SVM on limited resources: A low-rank linearization approach. Proc Int Conf Artif Intell Statist. 2012:1425–1434. [Google Scholar]
- 44.Zhang K, Zheng V, Wang Q, Kwok J, Yang Q, Marsic I. Covariate shift in Hilbert space: A solution via surrogate kernels. Proc. 30th Int. Conf. Mach. Learn.; Atlanta, GA, USA. Jun. 2013; pp. 388–395. [Google Scholar]
- 45.Zhou D, Bousquet O, Lal TN, Weston J, Schölkopf B. Learning with local and global consistency. Proc Adv NIPS. 2003;16:321–328. [Google Scholar]
- 46.Wang Y, Chen S, Zhou ZH. New semi-supervised classification method based on modified cluster assumption. IEEE Trans Neural Netw Learn Syst. 2012 May;23(no. 5):689–702. doi: 10.1109/TNNLS.2012.2186825. [DOI] [PubMed] [Google Scholar]
- 47.Zhu X. Dept Comput Sci. Vol. 1530. Univ Wisconsin-Madison; Madison, WI, USA: 2008. Semi-supervised learning literature survey. Tech. Rep. [Google Scholar]
- 48.Zhu X, Ghahramani Z, Lafferty J. Semi-supervised learning using Gaussian fields and harmonic functions. Proc. 20th Int. Conf. Mach. Learn.; Washington, DC, USA. Aug. 2003; pp. 912–919. [Google Scholar]
- 49.Zhu X, Lafferty J. Harmonic mixtures: Combining mixture models and graph-based methods for inductive and scalable semi-supervised learning. Proc. 22nd Int. Conf. Mach. Learn.; Bonn, Germany. Aug. 2005; pp. 1052–1059. [Google Scholar]