Abstract
Toxoplasma gondii is an obligate intracellular parasite of all warm-blooded animals. We previously described a forward genetic screen to identify T. gondii mutants defective in the establishment of a chronic infection. One of the mutants isolated was disrupted in the 3′ untranslated region (3′UTR) of an orthologue of bacterial translation elongation factor G (EFG). The mutant does not have a growth defect in tissue culture. Genetic complementation of this mutant with the genomic locus of TgEFG restores virulence in an acute infection mouse model. Epitope tagged TgEFG localized to the apicoplast, via a non-canonical targeting signal, where it functions as an elongation factor for translation in the apicoplast. Comparisons of TgEFG expression constructs with wild type or mutant 3′UTRs showed that a wild type 3′UTR is necessary for translation of TgEFG. In tissue culture, the TgEFG transcript is equally abundant in wild type and mutant parasites; however, during an animal infection, the TgEFG transcript is increased more than 3-fold in the mutant. These results highlight that in tissue culture, translation in the apicoplast can be diminished, but during an animal infection, translation in the apicoplast must be fully functional.
Keywords: Toxoplasma gondii, apicoplast, translation, virulence
INTRODUCTION
The Apicomplexan parasite Toxoplasma gondii is one of the world’s most successful microbes due to its ability to invade virtually any nucleated cell in all warm-blooded animals, including humans (Dubey, 1994). While infection is normally asymptomatic in healthy adults, T. gondii can cause mental retardation and death in developing fetuses and encephalitis in immunocompromised individuals (Dubey, 1994; Luft et al., 1984). During the acute stage of infection, the parasite divides rapidly and disseminates throughout the host in a form called the tachyzoite. The immune response causes the tachyzoite to convert to a slower growing, encysted stage called the bradyzoite that can persist throughout the life of the host (Dubey, 1998). There are limited therapeutics available against tachyzoites and no therapies that are effective against a chronic bradyzoite infection, making the discovery of new drug targets a critical area of research.
One of the hallmarks of Apicomplexan parasites is the presence of a plastid-like organelle called the apicoplast. The apicoplast has become the focus of many studies to discover putative drug targets (Lizundia et al., 2009; Ralph et al., 2001; Soldati, 1999; Wiesner et al., 2008). Characterization of several essential metabolic pathways that are believed to occur in the apicoplast has also lead to potential novel drug targets (Seeber, 2003). These pathways include isoprenoid biosynthesis (Jomaa et al., 1999; Moreno and Li, 2008; Wiesner and Jomaa, 2007), heme biosynthesis (Sato et al., 2004; Surolia and Padmanaban, 1992), and type II fatty acid biosynthesis (Goodman and McFadden, 2007; Mazumdar et al., 2006). Drugs that are currently known to be effective against T. gondii by blocking apicoplast function result in a phenotype called delayed death (Fichera et al., 1995; Pfefferkorn et al., 1992). Delayed death is a phenomenon in which the parasite is not killed upon addition of the drug. Instead, death of the parasite is delayed until it subsequently invades a new host cell. The exact mechanism of the delayed death phenotype is unknown.
In T. gondii, the apicoplast contains a 35 kilobase genome (plDNA) arranged as a series of linear tandem arrays (Williamson et al., 2001) containing at least 25 copies of plDNA (Matsuzaki et al., 2001). The plDNA encodes a suite of genes involved in the transcription and translation of its genome. These genes include subunits of RNA polymerase, small and large subunit ribosomal RNAs, ribosomal proteins, translation elongation factor Tuf, and a complete set of tRNAs. The only genes encoded in the plDNA not involved in transcription or translation are clpC, a structural chaperone that forms part of the Tic complex (Jackson-Constan et al., 2001) and is thought to function in the import of proteins to the lumen of the apicoplast (Tonkin et al., 2006), and sufB, an orthologue of a protein required for [Fe-S] cluster biosynthesis (Ellis et al., 2001). All of the remaining proteins found in the apicoplast are encoded in the nuclear genome of T. gondii and are imported into the apicoplast following their translation. These proteins have been termed NEAT proteins (Nuclear Encoded, Apicoplast Targeted). NEAT proteins are delivered to the apicoplast due to the presence of a bipartite targeting sequence (Harb et al., 2004). The bipartite targeting sequence begins with a standard signal sequence that targets the protein to the ER. Upon removal of the signal sequence, a transit peptide is exposed at the protein’s N-terminus that directs it to the apicoplast (Waller et al., 2000, DeRocher et al., 2000).
Our previous study used modified signature-tagged mutagenesis to discover genes essential for T. gondii to establish a chronic infection (Frankel et al., 2007). One mutant identified in the screen, called 49E10, has an insertion in the 3′UTR of a putative bacterial translation elongation factor G (EFG). Here we show that TgEFG encodes a functional EFG that is targeted to the apicoplast by a non-canonical targeting sequence. Decrease in TgEFG expression in the 49E10 mutant causes a severe virulence defect. These results suggest that regulation of translation in the apicoplast is essential during an animal infection.
RESULTS
49E10 has reduced lethality during the acute stage of infection
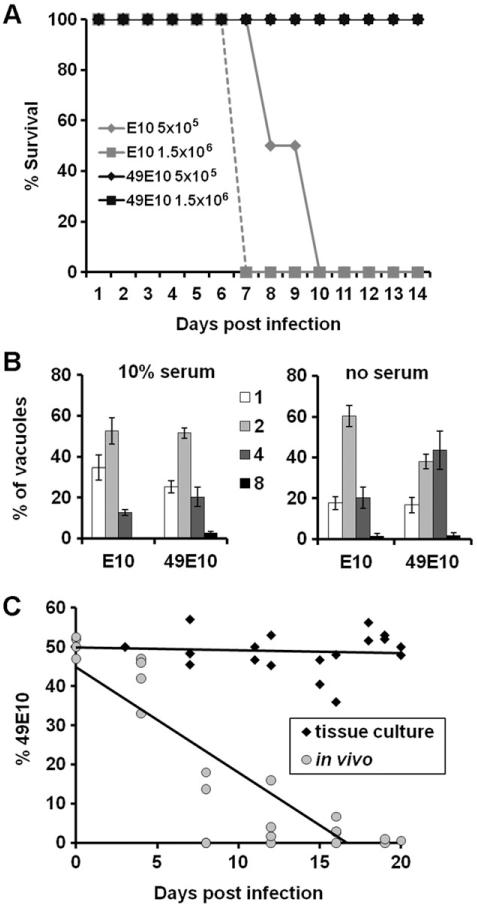
In a previous study, the 49E10 mutant displayed a dramatic decrease in the number of cysts per brain compared to wild-type (WT) parasites (Frankel et al., 2007). This decrease in cyst burden could be due to a defect of 49E10 parasites during acute infection, so we examined the early stages of infection by measuring the lethality of the 49E10 mutant in an acute infection mouse model. Mice were infected with 5×105 or 1.5×106 syringe-lysed tachyzoites of either the 49E10 strain or the E10 parental strain (the signature tag containing strain used to create 49E10 (Frankel et al., 2007)). All of the E10-infected mice succumbed to infection by day ten post-infection while the mice infected with the 49E10 mutant survived to the end of the study (Fig. 1A). These results indicate that the decreased cyst burden seen in mice infected with 49E10 in the initial STM screen is likely due to a defect in 49E10 parasites during acute infection.
Fig. 1.
The 49E10 mutant displays wild type growth in tissue culture but not in vivo.
A. E10 parasites were compared to 49E10 parasites in an acute mouse model by i.p. injection of either 5×105 or 1.5×106 tachyzoites into 4 mice each. Morbidity and mortality were monitored during the course of infection. Shown is a representative experiment of at least three independent experiments at similar doses.
B. Coverslips of confluent HFFs were maintained in 10% serum (left) or changed to no serum media (right) for 16 hours before infection with either E10 or the 49E10 mutant. Parasites were allowed to grow for 24 hours before cells were fixed and stained to visualize parasites. The numbers in the legend indict the number of parasites per vacuole. One hundred vacuoles per coverslip were counted. The experiment was performed in triplicate and the averages are graphed +/− SD.
C. Competitive fitness assay show that the 49E10 mutant grows equally well as the parental E10 strain in standard tissue culture conditions (black diamonds), but not in vivo (gray circles). Each of the shapes represents an independent tissue culture flask or mouse that was serially passaged. R-squared value for the in vivo experiment is 0.7663.
49E10 is not defective for growth or bradyzoite development in fibroblasts
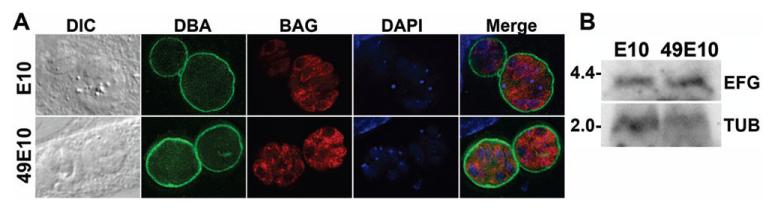
To examine if the reduced lethality of 49E10 parasites during a mouse infection was caused by a defect that could be characterized in tissue culture, we measured the growth and differentiation of 49E10 in fibroblast cells. Replication rates were determined by counting the number of parasites per vacuole 24 hours after infection. No growth defect was seen when we compared the growth of the E10 strain to the 49E10 mutant in standard tissue culture conditions in fibroblasts (Fig. 1B, left panel). Because several essential metabolic pathways occur in the apicoplast, we mimicked the nutrient limited environment of an animal by measuring the replication rate of the 49E10 mutant in serum starved host cells. The 49E10 mutant grew similar to the parental E10 strain in serum starved host cells (Fig. 1B, right panel). We also examined the ability of the 49E10 mutant to differentiate into bradyzoites using alkaline pH on fibroblast cells. Using the bradyzoite-specific markers BAG1 and Dolichos biflorus agglutinin (DBA), we saw that 49E10 developed into bradyzoites similar to the E10 parental strain (Fig. 2A). These findings suggest that the severe defect of the 49E10 mutant in animals cannot be mimicked in tissue culture and is specific to an in vivo environment.
Fig. 2.
49E10 develops into bradyzoites and the disrupted mRNA is not downregulated
A. Immunofluorescent images of E10 and 49E10 parasites grown in bradyzoite inducing conditions for three days. Cells were fixed and stained with DBA to mark the cyst wall (green) and BAG1 (red). All coverslips were mounted with VectaShield mounting medium containing 4′6′-diamidino-2-phenylindole (DAPI, Vector Laboratories) to visualize DNA (blue). Fluorescent images were acquired as described (Mordue et al., 2007).
B. Total RNA from E10 and 49E10 parasites was analyzed by northern blot analysis using a probe upstream of the insertion site of TGME49_023970 in 49E10 mutant (top panel labeled EFG). The blot was striped and reprobed for TUB1 as a loading control (bottom panel labeled TUB). Numbers in the left column are the sizes of the markers in kilobases.
Competition assays between E10 and 49E10 parasites
To directly compare growth differences between parental E10 and 49E10 mutant parasites, we performed a competitive fitness assay (Fohl and Roos, 2003). For these assays, E10 and 49E10 parasites were mixed 1:1, then serially passaged either in tissue culture or mice. At every passage, parasites were removed for plaque assays with or without drug selection. This assay uses the fact that the 49E10 mutant, but no the E10 parental strain, contains the hypoxanthine-xanthine-guanosine phosphoribosyl tranferase (HPT) gene and thus 49E10 is able to survive a mycophenolic acid and xanthine selection in the plaque assay. In tissue culture, the 49E10 was equally competitive with E10 (Fig. 1C, black diamonds). However, in vivo the 49E10 was rapidly out competed by the parental E10 strain regardless if the mice were infected with either 103 (Fig. 1C, gray circles) or 106 (Fig. S1) mixed parasites. These results again show that the 49E10 mutant does not display a defect in tissue culture, but in vivo the mutant does not grow well. These results also show that parental E10 parasites cannot compensate for the 49E10 defect within an animal.
Identification of the gene interrupted in the 49E10 mutant
Within the 49E10 mutant, we mapped the insertion site of the mutagenesis plasmid to be 380 base pairs downstream of the predicted stop codon of the annotated gene TGME49_023970 (www.toxodb.org). The open reading frame (ORF) of the TGME49_023970 gene encodes a 750 amino acid protein. This predicted protein was scanned at the Swiss Institute of Bioinformatics using the BLAST network service (http://www.expasy.org/cgi-bin/blast.pl, (Altschul et al., 1997)) and annotated as translation elongation factor G (EFG) from Escherichia coli (Fig. S2). EFG is a GTPase that is involved in the translocation of the peptidyl-tRNA from the A site of the ribosome to the P site of the ribosome during translation. Two other genes in the nuclear genome of T. gondii are predicted to encode eukaryotic derivatives of EFG. TGME49_005470 is predicted to encode the cytoplasmic form homologous to eukaryotic elongation factor 2 (eEF2) and TGME49_060170 is predicted to encode a mitochondrial form homologous to mitochondrial EFG (mtEFG). Due to its homology to the bacterial form of EFG and the presence of these two other translation elongation factors, we speculated that TGME49_023970 would be located in the apicoplast. Based on its homology to EFG, we designated the TGME49_023970 gene as TgEFG.
The 5′ and 3′ untranslated regions (UTRs) were mapped using rapid amplification of cDNA ends (RACE). The transcription start site of the TgEFG transcript is 1,032 nucleotides upstream of the predicted ORF and the length of the 3′UTR is 933 nucleotides, which places the insertion site of the mutagenesis plasmid within the 3′UTR. The entire transcript was sequenced using cDNA from WT parasites and no introns were detected, making the full length TgEFG transcript 4,217 nucleotides. To examine the TgEFG transcript in the 49E10 mutant, we compared RNA from an equal number of tachyzoites from E10 or 49E10 parasites by Northern hybridization analysis. When this Northern blot was hybridized with a probe from the ORF of TgEFG, i.e. upstream of the insertion site, the TgEFG message was equally abundant in E10 or 49E10 parasites (Fig. 2B). The size of the TgEFG transcript detected in parasites corresponds to the expected size of 4.2 kilobases. To confirm that TgEFG is disrupted in the 49E10 mutant, we compared the mRNA from WT and 49E10 parasites by northern hybridization using a probe to the 3′UTR downstream of the insertion site. Not surprisingly, when hybridized with a probe downstream of the insertion site, the TgEFG transcript was not detectable in the 49E10 mutant (Fig. S3).
TgEFG protein functions as elongation factor G in bacteria
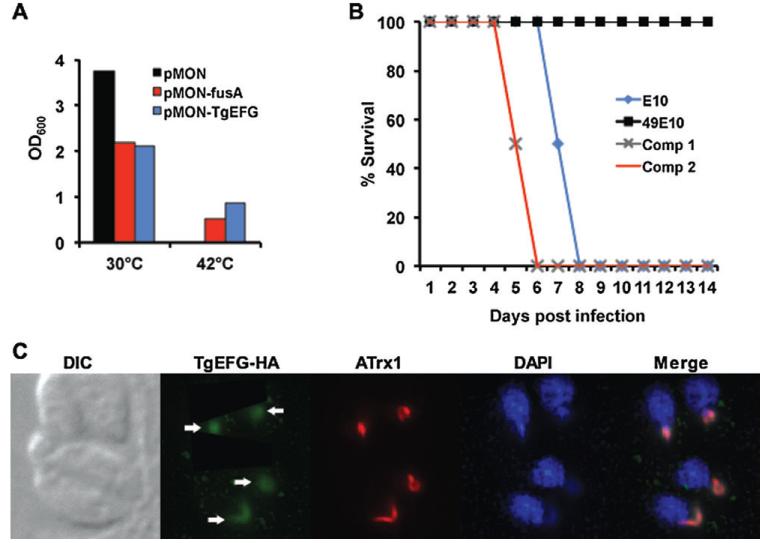
In order to verify that the TgEFG protein functions as EFG, we took advantage of an E. coli strain bearing a temperature-sensitive allele of EFG (PEM100). PEM100 contains a single point mutation in EFG that allows for growth at 30°C but not at 42°C (Hou et al., 1994a). We cloned the predicted TgEFG ORF into an inducible bacterial expression vector (pMON-TgEFG) and transformed it into PEM100 cells. EFG (fusA) from E. coli served as a positive control (pMON-fusA) and transformation of the inducible vector alone served as a negative control (pMON). PEM100 transformed with either pMON, pMON-fusA, or pMON-TgEFG was grown to mid-log phase at the permissive temperature and diluted into fresh broth containing 1mM IPTG to induce expression of fusA or TgEFG. Identically diluted cultures were grown for 16 hours at both the permissive (30°C) and restrictive (42°C) temperatures. At the permissive temperature, PEM100 was able to grow using any of the three vectors, although PEM100 transformed with vector alone grew noticeably better than PEM100 expressing fusA or TgEFG (Fig. 3A). This result was expected because fusA is a highly regulated gene and its overexpression in PEM100 drastically reduces its growth at both the permissive and restrictive temperatures (Hou et al., 1994b). At the restrictive temperature, PEM100 transformed with vector alone was unable to grow; however, the expression of either fusA or TgEFG restored growth of PEM100 at the restrictive temperature. These results confirm that the TgEFG protein can function as an EFG.
Fig. 3.
TgEFG functions as EFG, restores virulence to 49E10 and is localized to the apicoplast
A. A temperature-sensitive EFG mutant strain of E. coli was transfected with either vector alone (pMON, black bar), the vector expressing EFG from E. coli (pMON-fusA, red bar), or the vector expressing TGME49_02370 from T. gondii (pMON-TgEFG, blue bar). All cells were grown at the permissive temperature (30°C) to similar turbidity corresponding to mid-log phase and equivalently diluted into fresh broth containing 1mM IPTG. Identical cultures were grown for 16 hours at both the permissive (30°C) and restrictive (42°C) temperatures. OD600 measurements as an indictor of growth are shown.
B. The virulence of E10 (blue diamond) and 49E10 (black square) parasites was compared to two independent clones of 49E10 complemented with TgEFG expressed from its endogeneous promoter (Comp 1 grey X and Comp 2 red line). For this acute mouse model, 1.5×106 tachyzoites were i.p. injected into four mice per strain. Two independent experiments were combined for a total of eight mice per strain.
C. The cellular localization of TgEFG was examined by IFA using rabbit anti-HA antibody to visualize HA-tagged TgEFG expressed from its endogenous promoter (green), the 11G8 monoclonal to stain ATrx1 (red) and DAPI to visualize nucleic acid (blue). Fluorescent images were acquired as previously described (Mordue et al., 2007). White arrows indicate the TgEFG-HA in the apicoplast.
TgEFG restores acute virulence to the 49E10 mutant
To determine if a reduction in TgEFG expression is responsible for the in vivo defect of 49E10 parasites, we genetically complemented 49E10 parasites with TgEFG. The expression construct included 2 kilobases upstream of the TgEFG transcriptional start site determined by 5′RACE as well as a C-terminal hemagglutinin (HA) epitope tag (TgEFG-HA). The inclusion of an epitope tag allowed us to verify protein expression in complemented strains prior to testing them in the acute mouse model. Mice were infected with 1.5×106 tachyzoites of either E10, 49E10, or two different clones of genetically complemented 49E10 (Comp1 and Comp2). As before, all of the 49E10-infected mice survived throughout the course of infection, while the E10-infected mice succumbed to infection by day 8 post infection (Fig. 3B). Both Comp1 and Comp2 were restored in their lethality during the acute stage of infection with all of the mice succumbing to infection by day 6 post infection (Fig. 3B). Comp1 and Comp2 were consistently more lethal than the E10 parental strain (P<0.001). These data confirm that TgEFG is responsible for the virulence phenotype of 49E10 parasites in the acute infection mouse model.
TgEFG is localized to the apicoplast
We investigated the localization of TgEFG in T. gondii by creating parasites expressing the TGME49_023970 ORF with a C-terminal HA tag expressed with the α-tubulin promoter and 5′UTR. The parasites were examined by immunofluorescence assay (IFA), and the protein distributed throughout the cytoplasm (Fig. S4A). This localization was surprising due to the presence of eEF2 in T. gondii, which should function in translation in the cytoplasm. Although EFG and eEF2 are structurally similar, the two proteins have not been reported to be expressed in the same subcellular location. We then examined the localization using the functional TgEFG-HA construct created for the complementation studies. Once the TgEFG protein was expressed with its endogenous promoter and 5′UTR, it was found exclusively in the apicoplast, co-localizing with ATrx1, a T. gondii thioredoxin family protein that localizes to the apicoplast (DeRocher et al., 2008) and apicoplast DNA (Fig. 3C). These results suggest that either expression of TgEFG from a heterologous promoter causes its mislocalization or that the predicted gene TGME49_023970 initiates translation from an upstream start codon and is missing an apicoplast targeting sequence.
TgEFG has a non-canonical apicoplast targeting sequence
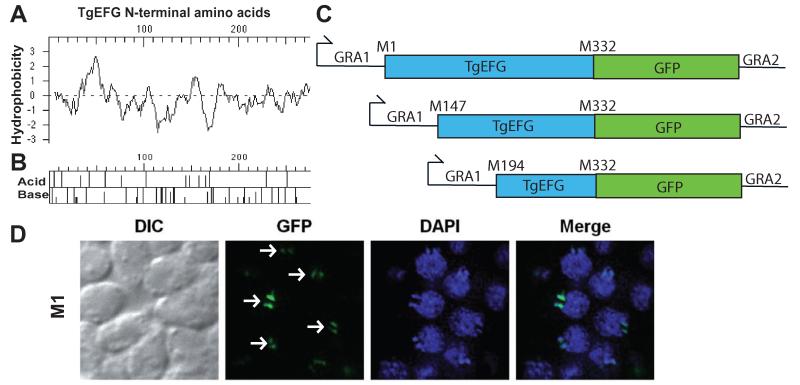
From the predicted initiator methionine of TGME49_023970, a typical apicoplast targeting signal could not be detected. By scanning the 5′UTR, we found three additional in frame ATG codons. The intracellular location of these four potential forms of TgEFG was analyzed using SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) to predict the presence and location of signal peptide cleavage sites (Bendtsen et al., 2004; Nielsen et al., 1997) and MitoProt (http://ihg.gsf.de/ihg/mitoprot.html) to predict targeting sequences (Claros and Vincens, 1996). These programs did not predict any of the four TgEFG forms to traffic to the apicoplast or mitochondrion. SignalP-HMM did predict the protein translated from the first initiator methionine to contain a signal anchor sequence and the protein translated from the fourth initiator methionine to contain a signal peptide (data not shown). Signal anchor peptides are similar to signal peptides but signal anchors result in the peptide remaining embedded in the membrane. We then analyzed from the first methionine to the beginning of the homology with bacterial EFG (i.e., the first 275 amino acids of TgEFG) for hydrophobic and basic amino acids. A Kyte-Doolittle hydropathy plot shows a large hydrophobic batch after the first 40 amino acids, but no other significant hydrophobic areas (Fig. 4A). An acid/base map shows that the N-terminus of TgEFG has an enrichment for basic amino acids (Fig. 4B), which is typical for apicoplast targeting signals.
Fig. 4.
TgEFG localizes to the apicoplast via a non-canonical targeting signal
A. Kyte-Doolittle hydropathy plot of the first 275 amino acids of TgEFG. Positive numbers on the hydrophobicity axis indicate hydrophobic amino acids.
B. Acid/Base graph of the first 275 amino acids of TgEFG.
C. Diagrams of the TgEFG-GFP fusion constructs used to identify the start codon of TgEFG. The fusions started with M1, M147 or M194 of TgEFG, continued until M322 of TgEFG (blue box) and contained the entire GFP coding region (green box). The GRA1 promoter and 5′UTR was used on the 5′end of all constructs while the GRA2 3′UTR was used on the 3′end.
D. Only T. gondii expressing the TgEFG-GFP fusion where TgEFG began at the first in frame methionine showed localization to the apicoplast. The IFA used an anti-GFP antibody to visualize the TgEFG-GFP fusion (green) and DAPI to visualize nucleic acid (blue). White arrows indicate TgEFG-GFP in the apicoplast. Two spots are seen for each apicoplast because the apicoplasts have divided.
To determine which ATG is used for translation initiation, a series of TgEFG-GFP fusions expressed from the dense granule 1 promoter were created and transiently transfected into WT parasites. Each of these constructs initiates translation of TgEFG from a different upstream ATG and continues until M332 of TgEFG, followed by GFP (Fig. 4C). M332 of TgEFG is approximately 50 amino acids into the region of homology with E. coli EFG. The GFP-fusion protein initiating at the first in frame ATG (M1) resulted in targeting of GFP to the apicoplast (Fig. 4D). In contrast, initiation of translation at other potential start codons (M147 and M194) resulted in diffuse staining throughout the parasite (Fig. S4B), similar to the localization seen for the predicted TGME49_023970, which is actually M262 of the mRNA (Fig. S4A). These results indicate that apicoplast targeting of TgEFG can be achieved by expression from a heterologous promoter and that translation of TgEFG starts at the first in-frame ATG codon. Translation initiation at the first in-frame ATG results in a 5′UTR of 248 nucleotides and an ORF of 3,036 nucleotides encoding a protein of 1,011 amino acids. These results also indicate that TgEFG is targeted to the apicoplast without a predicted signal peptide or canonical bipartite leader sequence typically found in apicoplast proteins. This non-canonical apicoplast signal can target reporter proteins such as GFP to the apicoplast.
The 3′UTR of TgEFG affects its expression
Based on our ability to complement the acute virulence defect of 49E10 by endogenous expression of TgEFG, we hypothesized that TgEFG protein expression is decreased in the 49E10 mutant. To test this hypothesis, we used a variety of methods to generate an antibody against TgEFG; however, we were unable to create a specific antibody. As an alternative, we developed a TgEFG-HA expression construct with a “mutant” 3′UTR to examine the effect of the 3′UTR on TgEFG translation. First, we mapped the 3′end of the TgEFG transcript in 49E10 parasites. In 49E10, TgEFG is transcribed into the mutagenesis vector for ~600 nucleotides. This size corresponds to the 553 nucleotides that was removed from the endogenous 3′UTR in the 49E10 mutant and thus the messages of wild type and mutant TgEFG are approximately the same size by northern blot (Fig. 2B). We amplified this “mutant” 3′UTR by PCR and used it to replace the endogenous 3′UTR in the TgEFG-HA expression construct. The TgEFG-mutant 3′UTR construct was stably transfected into 49E10 parasites and Southern analysis was performed to identify unique clones (data not shown).
Parasites expressing the TgEFG-HA construct containing the endogenous TgEFG 3′UTR showed two bands reactive with the anti-HA antibody (Fig. 5A and S5). The dominant band at approximately 90 kilodaltons (kDa) likely corresponds to the post-form of TgEFG found in the apicoplast following removal of the apicoplast targeting signal transit peptide. A higher molecular weight band at approximately 105 kDa likely corresponds to the pro-form of TgEFG found in the ER following removal of the signal peptide but prior to processing of the transit peptide. The pre-pro-form of TgEFG is likely not detectable due to the rapid processing of the signal peptide upon entry to the ER (van Dooren et al., 2002, DeRocher et al., 2005). Even though TgEFG does not contain a canonical apicoplast targeting signal, the presence of two forms of TgEFG shows that it is likely processed similar to other apicoplast proteins.
Fig. 5.
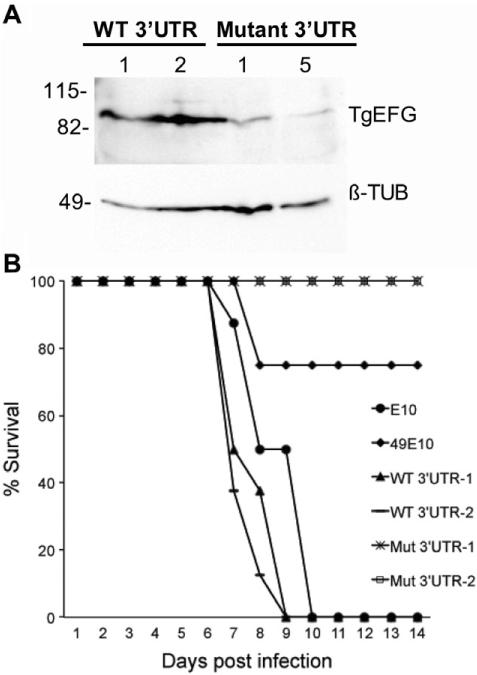
Insertion into the 3′UTR of TgEFG affects protein levels
A. 1×107 tachyzoites of clones expressing TgEFG-HA with either an endogenous 3′UTR (WT 3′UTR) or Mutant 3′UTR were loaded onto a 10% SDS/PAGE gel. Protein was visualized using a mouse anti-HA monoclonal antibody to detect the HA epitope tagged TgEFG (top panel labeled TgEFG). The blot was striped and reprobed for ß-tubulin as a loading control (bottom panel labeled ß-TUB). Size markers are shown to the left in kilodaltons.
B. The virulence of E10 (circle) and 49E10 (diamond) parasites was compared to two independent clones of 49E10 complemented with TgEFG expressed with its endogeneous 3′UTR (WT 3′UTR), or the mutant 3′UTR found in 49E10 (Mut 3′UTR). For this acute mouse model, 1.5×106 tachyzoites were i.p. injected into four mice per strain. Two independent experiments were combined for a total of eight mice per strain.
To analyze the effect of the mutant 3′UTR on TgEFG-HA expression, equal numbers of tachyzoites expressing TgEFG-HA with the mutant 3′UTR were compared to a clone expressing TgEFG-HA with the endogenous 3′UTR. Six unique parasite clones that contained mutant 3′UTR DNA were then analyzed by western immunoblot. TgEFG was only visible in 2 of the 6 clones with the mutant 3′UTR (Fig. S5). The level of TgEFG from the two mutant 3′UTR clones with the highest expression of TgEFG (numbers 1 and 5, Fig. S5) was directly compared with two unique TgEFG endogenous 3′UTR clones. When the blot was reprobed for T. gondii ß-tubulin as a loading control, the level of TgEFG was clearly reduced when the protein was translated from mRNA with a mutant 3′UTR (Fig. 5A). Not surprisingly, this reduced level of TgEFG protein does not complement the virulence defect of the 49E10 mutant (Fig. 5B). To test if restoration of 49E10 virulence was dependent on the level of TgEFG protein or the presence of an intact endogenous 3′UTR, we performed two additional experiments. First, the 3′UTR of GRA2 was exchanged for the endogenous 3′UTR. When expressed with the GRA2 3′UTR, TgEFG complemented 49E10 parasites similar to TgEFG expressed with its endogenous 3′UTR (Fig. S6). Second, the shortened and mistargeted form of TgEFG (M262, Fig. S4A) does not complement 49E10 parasites (Fig S6), even though its mRNA contains the endogenous 3′UTR. Taken together, these results suggest that reduction of TgEFG protein in 49E10 parasites, and not the disruption of the 3′UTR alone, is responsible for the virulence defect of 49E10.
TgEFG mRNA is increased in 49E10 during a mouse infection
Although the TgEFG gene is clearly disrupted in the 49E10 strain (Fig. 1C), further analysis of the TgEFG transcript was necessary as TgEFG mRNA was till detected using a probe upstream of the insertion site (Fig. 2B). Real-time quantitative PCR (qPCR) was performed on cDNA from E10 and 49E10 parasites grown in tissue culture using primers upstream of the insertion site. Calibration curves of serially diluted DNA standards were used to determine the absolute quantity of TgEFG and α-tubulin mRNA molecules in the in vitro cDNA samples. The ratio of TgEFG: α-tubulin was used to compare the steady-state levels of TgEFG mRNA between the two strains. The amount of TgEFG transcript in the 49E10 strain was equivalent to the amount found in E10 (Fig. 6A).
Fig. 6.
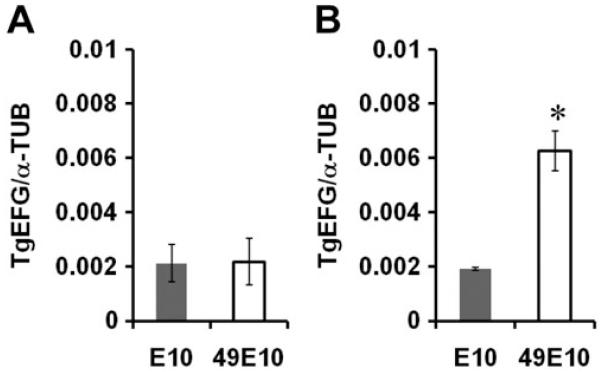
TgEFG mRNA levels in 49E10 are only altered in vivo
A. Quantitative RT-PCR was performed on RNA from in vitro grown E10 and 49E10 parasites using primers against TgEFG and α-tubulin. Standard curves were generated using the same primers to determine the copy number of TgEFG and α-tubulin in each sample and the ratio of the TgEFG copy number to α-tubulin copy number was determined. The average of three independent experiments is shown.
B. Quantitative RT-PCR was performed on RNA from in vivo derived E10 and 49E10 parasites as in A. (* p=0.0042).
Because 49E10 parasites only show a phenotype in vivo, we compared TgEFG transcript levels from E10 and 49E10 parasites harvested from the mouse peritoneum. Mice were infected intraperitoneally with 5×107 tachyzoites, and after six days, parasites were harvested by peritoneal lavage. The cDNA from these in vivo parasites was used in qPCR. Surprisingly, in contrast to the results for the tissue culture cDNA, the TgEFG transcript level was elevated more than 3-fold in the 49E10 mutant compared to E10 parasites (Fig. 6B, p=0.0042). In a mouse, 49E10 parasites are likely compensating for a decrease in TgEFG translation by increasing the expression of TgEFG transcript. We tried to mimic these in vivo results in tissue culture by comparing TgEFG transcript levels from E10 and 49E10 parasites grown in serum starved host cells for three days or macrophages activated with tumor necrosis factor-α (TNF-α) oρ interferon γ (IFN-γ) for two days. Unfortunately, the level of TgEFG transcript was approximately equal in 49E10 and E10 parasites grown in any of these tissue culture conditions (data not shown). Thus the conditions that stimulate the upregulation of TgEFG message in the 49E10 mutant likely cannot be mimicked in tissue culture. These results indicate that the level of TgEFG may not be as critical in tissue culture as it is within an animal.
DISCUSSION
A T. gondii mutant containing an insertion within the 3′UTR of TgEFG showed a dramatic reduction in its lethality in an acute infection mouse model. TgEFG is highly conserved with bacterial EFG and localizes to an endosymbiotic organelle in T. gondii called the apicoplast. The function of TgEFG appears to be conserved based on its ability to functionally complement an EFG deficient bacterial strain. Insertion into the 3′UTR of TgEFG likely reduces the amount of TgEFG in the apicoplast because 1) we were able to restore virulence to the 49E10 mutant upon addition of TgEFG, 2) TgEFG-HA with a mutant 3′UTR showed a dramatic reduction in protein, and 3) the TgEFG transcript is upregulated in the 49E10 mutant only during mouse infection. These results indicate that precise control of translation in the apicoplast is important for T. gondii during an animal infection, but not during tissue culture conditions. To our knowledge, this is the first study that has shown evidence of translational control in a T. gondii 3′UTR and such a dramatic phenotypic difference between tissue culture and animal infection associated with an apicoplast protein.
T. gondii contains three distinct compartments where translation is assumed to occur: the cytoplasm, the apicoplast, and the mitochondrion. Each compartment requires its own set of translational machinery to facilitate protein synthesis. The limited plastid genomes found in the apicoplast and the mitochondrion indicate that the majority of these factors are encoded in the nucleus and subsequently targeted to the proper organelle. While one of these imported proteins appears to be TgEFG, this protein does not contain the traditional bipartite apicoplast targeting signal needed for proper sorting to the apicoplast. TgEFG is the first nuclear-encoded, apicoplast-targeted translation factor to be characterized in T. gondii. The expression and targeting of nuclear-encoded translational machinery to the apicoplast provides further evidence that the T. gondii apicoplast is translationally active.
The apicoplast is essential for proper growth of the parasite. Disruption of the apicoplast leads to a delayed death phenotype that is characterized by the inability of the parasite to invade a new host cell presumably because it is no longer able to synthesize fatty acids sufficiently. It was recently proposed that fatty acid production by the parasite might play a role in reducing the immune response elicited by the parasite by inhibiting TNF-α production by macrophages (Debierre-Grockiego et al., 2007). A lack of immune suppression by the 49E10 mutant caused by a disruption of translation in the apicoplast, and therefore inefficient fatty acid synthesis, would allow for more killing of the parasite by the host and explain its pronounced virulence defect. To test this idea, we measured the cytokines produced by RAW macrophages infected with either E10 or 49E10, then activated three hours later by IFN-γ aλoνε oρ IFN-γ πλνσλlπoπoλψσαχχηαρlδε (∧∏Σ). Thirty-six hours after activation, no differences were seen between the two strains for any of the cytokines tested (TNF-α IFN-γ, interleukin (IL)-12, IL-10, IL-6, monocyte chemoattractant protein, data not shown). The 49E10 mutant was also previously shown to suppress nitric oxide in activated RAW macrophages similar to wild type parasites (Mordue et al., 2007). While 49E10 displays a reduced lethality in a mouse, it does not have a detectable defect in standard or nutrient limiting tissue culture conditions. We hypothesize that translation in the apicoplast may be downregulated without negative consequences in tissue culture, but must be at maximum efficiency during an animal infection. We are currently developing assays to compare apicoplast translation rates in vitro and in vivo.
Regulation of translation is a widespread mechanism for the control of gene expression. In late branching eukaryotes, it is common for translation factors to be translationally regulated themselves (Cully and Downward, 2009). Regulation of translation has previously been shown to occur in T. gondii through the phosphorylation of eukaryotic initiation factor 2 (eIF2) during a stress response (Sullivan et al., 2004). In other eukaryotes, phosphorylation of eIF2 is a key stress response that leads to a decrease in overall protein synthesis. In many organisms, elongation factors play a role in the regulation of translation (Bektas et al., 2005; Ortiz and Kinzy, 2005; Patel et al., 2002). The 3′UTR has also been shown to be involved in the post-transcriptional regulation of protein expression, primarily from the interaction of RNA-binding proteins with elements in the 3′UTR. These mRNA-protein interactions have been shown to both enhance and inhibit translation (Mazumder et al., 2003; Wilkie et al., 2003). It is likely that the latter two-thirds of the TgEFG 3′UTR contain regulatory elements that enhance its translation in vivo. Future studies will identify and define the mechanism of these regulatory elements.
The regulation of translation by elongation factors in chloroplasts has been shown to be essential in the biogenesis of several chloroplast proteins (Berry et al., 1988; Kim and Mullet, 2003; Muhlbauer and Eichacker, 1998; Zhang et al., 2000). In chloroplasts, translation elongation can be stimulated by light and activated via a redox pathway (Trebitsh et al., 2000). Since the apicoplast is derived from an ancient chloroplast, it follows that similar mechanisms of translation regulation may be occurring in the apicoplast. Our results indicate that TgEFG may not be essential in tissue culture, but that TgEFG is absolutely required for survival in an animal infection. We propose that the regulation of translation in the T. gondii apicoplast could occur in a manner similar to the regulation of translation in chloroplasts. A ferredoxin redox system in the apicoplast is presumed to provide electrons that are used in a variety of other metabolic pathways in the apicoplast, such as [Fe-S] cluster biosynthesis, isoprenoid biosynthesis, and lipoic acid biosynthesis (Ralph et al., 2004; Seeber et al., 2005). This redox pathway could also activate translation elongation in the apicoplast in a manner similar to the translation regulation seen in chloroplasts. Furthermore, upon infection in a mouse translation elongation is activated. In 49E10 parasites, the disruption of TgEFG abolishes the ability of these parasites to respond to the stressors of an in vivo environment. These parasites cannot regulate translation in the apicoplast and therefore cannot survive during an animal infection.
MATERIALS AND METHODS
Host cell culture and parasite strains
Strains were maintained as tachyzoites by serial passage on monolayers of human foreskin fibroblasts (HFFs) at 37°C with 5% CO2 in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA), 1% penicillin-streptomycin (Invitrogen, Carlsbad, CA), and 2mM L-glutamine (Invitrogen). For serum starved host cells, the confluent HFFs were placed in the DMEM medium without FBS for 16 hours before infection with T. gondii. RAW macrophages were cultured and activated with either 500 ng/mL TNFα, 100 ng/mL LPS, 100 units/mL IFN-γ, or 100 ng/mL LPS with 100 units/mL IFN-γ (Mordue et al., 2007). Bradyzoite conditions were RPMI1640 supplemented 1% FBS, 1% penicillin-streptomycin, buffered with 50mM HEPES to pH 8 and incubated at 37°C with ambient CO2. All growth curves were performed as described previously (Frankel et al., 2007). The Pru strain of T. gondii with a deletion in the HPT gene (PruΔHPT) was the base strain for creation of the parental and mutant strains (Frankel et al., 2007; Mordue et al., 2007).
Acute mouse model
7-week old female CBA/J mice (National Cancer Institute, Charles River Laboratories, Frederick, MD) were intraperitoneally injected with freshly lysed tachyzoites. Plaque assays were performed immediately following infection to ensure the viability of the tachyzoites used in the infection. Mice were checked a minimum of twice daily to monitor their general condition (including hydration status, activity level, and general physical appearance). Moribund mice (severely hunched, unresponsive to stimulus, not moving) were euthanized and time to death was recorded and statistical significance was analyzed using a student’s t-test.
Competition assays
For tissue culture growth competition, parental E10 and 49E10 mutant parasites were mixed 1:1, then 106 parasites were inoculated into HFF host cells and serially passaged without drugs every 3 days for 20 days. At every passage, parasites were removed for plaque assays with or without selection (50 μg/mL mycophenolic acid and 50 μg/mL xanthine) as the 49E10 mutant contains the HPT gene and is able to survive the selection. For in vivo competition studies, CBA/J female mice (National Cancer Institute, Charles River Laboratories, Frederick, MD) were injected i.p. with either 103 or 106 parasites (1:1 mix of E10 and 49E10). Parasites were harvested by peritoneal lavage every 4 days, for serial passage into another mouse and plaque assays with and without mycophenolic acid/xanthine selection. Relative fitness was assessed by the slope of the line represented by the change over time of the percent of 49E10 in the total population of plagues.
Northern hybridization
Parasite RNA was isolated using ULTRASPEC (Biotecx, Houston, TX) as previously described (Frankel et al., 2007). RNA was run on a 0.8% agarose/6.66% formaldehyde gel and gravity transferred to Zeta-Probe blotting membrane (Bio-Rad, Hercules, CA). The membrane was incubated for four hours in prehybridization solution (50% formamide, 0.12 M Na2HPO4, 0.25 M NaCl, 7% (w/v) SDS, 1mM EDTA) at 42°C. A denatured radiolabeled DNA probe was hybridized overnight at 42°C in prehybridization solution. The blot was washed once with 1× SSC/0.1% SDS followed by two washes with 0.1× SSC/0.1% SDS; all washes were performed for 30 minutes at 42°C.
Immunofluorescence assay
Glass coverslips of confluent monolayers of HFFs were infected for approximately 24 hours with tachyzoites. The monolayer was fixed with 3% formaldehyde, permeabilized and blocked in 3% BSA/0.2% TritonX-100, then stained with anti-HA antibody (1:300, Covance Innovative Antibodies, Princeton, NJ) and Alexafluor 488 secondary antibody (1:1000, Jackson ImmunoResearch, West Grove, PA). The apicoplast was visualized using the 11G8 monoclonal (DeRocher et al., 2008) at a 1:500 dilution and detected with Alexafluor 633 secondary antibody (1:300 dilution, Jackson ImmunoResearch). Apicoplast DNA was stained using 4′6′-diamidino-2-phenylindole (DAPI). Coverslips were mounted onto slides using VectaShield mounting media containing DAPI (Vector Laboratories, Burlingame, CA). Samples were examined using a motorized Zeiss Axioplan 2 equipped with a rear-mounted excitation filter wheel, a triple pass (DAPI/FITC/Texas Red) emission cube, differential interference contrast optics, and a Hamamatsu ORCA-AG CCD camera. Serial image stacks (0.2 micron Z-increment) were collected at 100× (PlanApo oil immersion 1.4 na) and deconvolved and pseudo-colored using OpenLabs 4.0 software (Improvision, Waltham, MA).
Bacterial fusA− strain complementation
The predicted TGME49_023970 ORF was PCR amplified from the second codon of the ORF to an engineered SphI site downstream of the translational stop codon using the primers 5′-GCCTCGCCGTCCGCACTGGGGTCTG-3′ and 5′-TTTCGCGCATGCCCTACCCCTTCA-3′. The fusA gene from E. coli was PCR amplified from the second codon of the ORF to an engineered SphI site downstream of the translational stop codon using the primers 5′-GCTCGTACAACACCCATCGCAC-3′ and 5′-GGAGAGAGCACGGGACTTTGGTAT-3′. These PCR products were digested with SphI and cloned into blunted NcoI/SphI digested pMON5839 (Duronio et al., 1990) creating pTMP62 and pTMP34, respectively. The kanamycin resistance marker of pTMP62, pTMP34 and pMON5839 was replaced with a chloramphenicol resistance marker creating pMON-TgEFG, pMON-fusA and pMON, respectively. pMON, pMON-fusA and pMON-TgEFG were then transformed into PEM100 (Hou et al., 1994a). A single colony of each transformant was grown to mid-log phase in liquid LB-Lennox at 30°C followed by dilution into fresh LB-Lennox containing 1mM IPTG. Identical cultures were grown for 16 hours at both 30°C (permissive temperature) and 42°C (restrictive temperature).
Real-time quantitative PCR
Real-time quantitative PCR (qPCR) was performed using cDNA prepared by reverse transcription of either 1 μg (for in vitro assays) or 5 μg (for in vivo assays) of amplification grade DNase I (Invitrogen) treated RNA using the SuperScript III First-Strand Synthesis System (Invitrogen) primed by random hexamers. For in vitro assays, parasites were grown under standard tachyzoite tissue culture conditions. For in vivo assays, mice were injected intraperitoneally with 5×107 tachyzoites of either the E10 or 49E10 strain. After six days, parasites were harvested by peritoneal lavage with 10 mL of PBS followed by centrifugation. qPCR experiments were performed in triplicate in 96-well plates using the Bio-Rad iCycler IQ v. 3.1.7050 detection system (Bio-Rad) and threshold cycles (Ct) were determined automatically by the software. qPCR reactions contained 1× SYBR Green Supermix (Bio-Rad) and 0.9 μL of each primer (0.3 μM final concentration) as well as either cDNA or DNA template standard. Standard curves were generated for every gene tested using a dilution series of 102-108 copy numbers of DNA amplified using the listed primer sets and plotting the Ct against the log of the copy number. The number of target gene copies can be extrapolated from the linear regression from the standard curve via the following equation:
The coefficient of determination (r2) from the standard curve was used to determine if each assay was optimized and only assays with r2>0.98 were considered valid. Reaction efficiency was determined from the slope of the standard curve using the following equation:
cDNA prepared without addition of Superscript III Reverse Transcriptase were used as negative controls. Each sample (including standard curve samples) was run in triplicate in every experiment and the results of three independent experiments were averaged. The PCR conditions were 1 cycle for 3 min. at 95°C followed by 40 cycles of 30 sec. at 95°C and 30 seconds at 60°C. Following completion of these cycles, a melt curve was performed to confirm primer specificity. Primers were as follows: TgEFG-qRT-F (5′-TCGCGGAAATGTTCGGATAC-3′), TgEFG-qRT-R (5′-TCGGAGTGGGCATGTATTTC-3′), αTUB-qRT-F (5′-GACGACGCCTTCAACACCTTCTTT-3′), and αTUB-qRT-R (5′-AGTTGTTCGCAGCATCCTCTTTCC).
Generation of TgEFG expression constructs and electroporation into T. gondii
To create the TgEFG-HA expression vector, the predicted TGME49_023970 ORF was PCR amplified to engineer an NsiI site at the initiator methionine with an in frame HA tag at the C terminal end followed by an engineered PacI site at the translational stop codon using the primers 5′-ATGCATTCGCCGTCCGCACTGGGGTCT-3′ and 5′-TTAATTAAGGTTGCTTGCAGGTTCCCCCCGT-3′. This PCR product was cloned using pCR2.1-TOPO (Invitrogen) and was subcloned into pT/230-TUB/5CAT (Soldati and Boothroyd, 1995) at the NsiI and PacI sites creating pTMP17. The dihydrofolate reducatase-thymidylate synthase gene from pDHFR-TSc3 (Donald and Roos, 1993) was cloned into pTMP17 by XbaI and ApaI digest creating pTMP18. The promoter and 5′UTR region of the genomic locus construct was created by PCR amplifying genomic DNA using the primers 5′-CAGAAGGGCACCAAGATGATAAAT-3′ and 5′-CGCCGCCGGATCTGCCTCAC-3′. This PCR was product was cloned using pCR2.1-TOPO and subcloned into pTMP18 by NcoI digest creating pTMP19. The 3′UTR region of the genomic construct was created by engineering a PacI site at the translational stop codon and an XbaI site ~470 base pairs downstream of the polyadenylation site using the primers 5′-TTAATTAAAGCTGTACAGCCACCTCAG-3′ and 5′-GGATCTAGAAAAGCACTAAATAATGGG-3′. This product was cloned into pTMP18 by PacI and XbaI digest creating pTMP41. The 3′UTR of GRA2 was substituted for the endogenous 3′UTR on pTMP41 by PacI and XbaI digestion of pGRA1mGFP5 (Kim et al., 2001). The constructs were linearized by restriction digest, ethanol precipitated, and 25 μg of each was electroporated with 1 × 107 T. gondii tachyzoites. Stable transformants were selected as resistant to 1 μM pyrimethamine.
The TgEFG-GFP fusion expression vectors were created by PCR amplifying genomic DNA to engineer NsiI sites at the methionine using the primers 5′-GTCCGATGCATAGGTTAGAAGGC-3′ for M1-EFG-GFP, 5′-GATTCTTCGATGCATTCTTTGTCTGTC-3′ for M147-EFG-GFP, 5′-GGGTGCATGCATGGGTCGTCA-3′ for M194-EFG-GFP, and to engineer an NsiI site in the EFGa ORF using the antisense primer 5′-GCCACAAAATGCATGGTCGCTTCG-3′ for all three constructs. PCR products were cloned using pCR2.1-TOPO and subcloned into NsiI digested pGRA1mGFP5 (Kim et al., 2001). 50 μg of each of the constructs were linearized, ethanol precipitated, and electroporated with 1 × 107 T. gondii tachyzoites. Electroporated parasites were placed directly onto glass coverslips of confluent monolayers of HFFs. Approximately 36 hours post infection, the monolayer was prepared for immunofluorescence assay as described.
Western immunoblotting
Freshly lysed tachyzoites were boiled for 10 minutes in 1× protein loading buffer (40mM Tris pH 6.8, 1% sodium dodecyl sulfate (SDS), 5% glycerol, 0.001% bromophenol blue). Denatured protein samples equivalent to 1×107 tachyzoites were separated in an SDS/PAGE gel and transferred to Immobilon-P membrane (Millipore). All manipulations of the membrane were performed at room temperature with shaking. The membranes were blocked for 1 hour in 1% Tween-20/PBS (PBS-T) supplemented with 5% nonfat dry milk (PBS-T-milk), incubated for 1 hour in PBS-T-milk containing a 1:1000 dilution of a mouse anti-HA monoclonal anitbody (Covance, Princeton, NJ), washed 3× for 5 minutes in PBS-T, incubated for 45 minutes in PBS-T-milk containing a 1:500 dilution of a donkey anti-mouse secondary antibody conjugated to horesradish peroxidase (Jackson ImmunoResearch), and washed 3× for 5 minutes in PBS-T. Protein was visualized by using ECL-Plus Western Blotting Detection Reagents (Amersham Pharmacia Biosciences).
Supplementary Material
ACKNOWLEDGEMENTS
We sincerely thank Rachel Green and Paul March for PEM100 bacteria and advice, Peter Bradley for the 11G8 monoclonal, David Sibley for the ß-tubulin antibody, Jay Bangs for the use of his microscope, Rod Welch for E. coli genomic DNA, and the Knoll lab for editorial assistance. This research was supported by American Heart Association 0840059N (L.J.K.), and NIH National Research Service Award T32 AI007414 (T.M.P.).
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bektas M, Akcakaya H, Aroymak A, Nurten R, Bermek E. Effect of oxidative stress on in vivo ADP-ribosylation of eukaryotic elongation factor 2. Int J Biochem Cell Biol. 2005;37:91–99. doi: 10.1016/j.biocel.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Berry JO, Carr JP, Klessig DF. mRNAs encoding ribulose-1,5-bisphosphate carboxylase remain bound to polysomes but are not translated in amaranth seedlings transferred to darkness. Proc Natl Acad Sci U S A. 1988;85:4190–4194. doi: 10.1073/pnas.85.12.4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Cully M, Downward J. Translational responses to growth factors and stress. Biochem Soc Trans. 2009;37:284–288. doi: 10.1042/BST0370284. [DOI] [PubMed] [Google Scholar]
- Debierre-Grockiego F, Rabi K, Schmidt J, Geyer H, Geyer R, Schwarz RT. Fatty acids isolated from Toxoplasma gondii reduce glycosylphosphatidylinositol-induced tumor necrosis factor alpha production through inhibition of the NF-kappaB signaling pathway. Infect Immun. 2007;75:2886–2893. doi: 10.1128/IAI.01431-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRocher A, Hagen CB, Froehlich JE, Feagin JE, Parsons M. Analysis of targeting sequences demonstrates that trafficking to the Toxoplasma gondii plastid branches off the secretory system. J Cell Sci. 2000;113:3969–77. doi: 10.1242/jcs.113.22.3969. [DOI] [PubMed] [Google Scholar]
- DeRocher A, Gilbert B, Feagin JE, Parsons M. Dissection of brefeldin A-sensitive and -insensitive steps in apicoplast protein targeting. J Cell Sci. 2005;118:565–74. doi: 10.1242/jcs.01627. [DOI] [PubMed] [Google Scholar]
- DeRocher AE, Coppens I, Karnataki A, Gilbert LA, Rome ME, Feagin JE, Bradley PJ, Parsons M. A thioredoxin family protein of the apicoplast periphery identifies abundant candidate transport vesicles in Toxoplasma gondii. Eukaryot. Cell. 2008;7:1518–29. doi: 10.1128/EC.00081-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald RG, Roos DS. Stable molecular transformation of Toxoplasma gondii: a selectable dihydrofolate reductase-thymidylate synthase marker based on drug-resistance mutations in malaria. Proc Natl Acad Sci U S A. 1993;90:11703–11707. doi: 10.1073/pnas.90.24.11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey JP. Toxoplasmosis. J Am Vet Med Assoc. 1994;205:1593–1598. [PubMed] [Google Scholar]
- Dubey JP. Advances in the life cycle of Toxoplasma gondii. Int J Parasitol. 1998;28:1019–1024. doi: 10.1016/s0020-7519(98)00023-x. [DOI] [PubMed] [Google Scholar]
- Duronio RJ, Jackson-Machelski E, Heuckeroth RO, Olins PO, Devine CS, Yonemoto W, Slice LW, Taylor SS, Gordon JI. Protein N-myristoylation in Escherichia coli: reconstitution of a eukaryotic protein modification in bacteria. Proc Natl Acad Sci U S A. 1990;87:1506–1510. doi: 10.1073/pnas.87.4.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis KE, Clough B, Saldanha JW, Wilson RJ. Nifs and Sufs in malaria. Mol Microbiol. 2001;41:973–981. doi: 10.1046/j.1365-2958.2001.02588.x. [DOI] [PubMed] [Google Scholar]
- Fichera ME, Bhopale MK, Roos DS. In vitro assays elucidate peculiar kinetics of clindamycin action against Toxoplasma gondii. Antimicrob Agents Chemother. 1995;39:1530–1537. doi: 10.1128/aac.39.7.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fohl LM, Roos DS. Fitness effects of DHFR-TS mutations associated with pyrimethamine resistance in apicomplexan parasites. Mol Microbiol. 2003;50:1319–27. doi: 10.1046/j.1365-2958.2003.03756.x. [DOI] [PubMed] [Google Scholar]
- Frankel MB, Mordue DG, Knoll LJ. Discovery of parasite virulence genes reveals a unique regulator of chromosome condensation 1 ortholog critical for efficient nuclear trafficking. Proc Natl Acad Sci U S A. 2007;104:10181–10186. doi: 10.1073/pnas.0701893104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CD, McFadden GI. Fatty acid biosynthesis as a drug target in apicomplexan parasites. Curr Drug Targets. 2007;8:15–30. doi: 10.2174/138945007779315579. [DOI] [PubMed] [Google Scholar]
- Harb OS, Chatterjee B, Fraunholz MJ, Crawford MJ, Nishi M, Roos DS. Multiple functionally redundant signals mediate targeting to the apicoplast in the apicomplexan parasite Toxoplasma gondii. Eukaryot Cell. 2004;3:663–674. doi: 10.1128/EC.3.3.663-674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Lin YP, Sharer JD, March PE. In vivo selection of conditional-lethal mutations in the gene encoding elongation factor G of Escherichia coli. J Bacteriol. 1994a;176:123–129. doi: 10.1128/jb.176.1.123-129.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Yaskowiak ES, March PE. Carboxyl-terminal amino acid residues in elongation factor G essential for ribosome association and translocation. J Bacteriol. 1994b;176:7038–7044. doi: 10.1128/jb.176.22.7038-7044.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Constan D, Akita M, Keegstra K. Molecular chaperones involved in chloroplast protein import. Biochim Biophys Acta. 2001;1541:102–113. doi: 10.1016/s0167-4889(01)00148-3. [DOI] [PubMed] [Google Scholar]
- Jomaa H, Wiesner J, Sanderbrand S, Altincicek B, Weidemeyer C, Hintz M, Turbachova I, Eberl M, Zeidler J, Lichtenthaler HK, Soldati D, Beck E. Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science. 1999;285:1573–1576. doi: 10.1126/science.285.5433.1573. [DOI] [PubMed] [Google Scholar]
- Kim J, Mullet JE. A mechanism for light-induced translation of the rbcL mRNA encoding the large subunit of ribulose-1,5-bisphosphate carboxylase in barley chloroplasts. Plant Cell Physiol. 2003;44:491–499. doi: 10.1093/pcp/pcg061. [DOI] [PubMed] [Google Scholar]
- Kim K, Eaton MS, Schubert W, Wu S, Tang J. Optimized expression of green fluorescent protein in Toxoplasma gondii using thermostable green fluorescent protein mutants. Mol Biochem Parasitol. 2001;113:309–313. doi: 10.1016/s0166-6851(01)00212-2. [DOI] [PubMed] [Google Scholar]
- Lizundia R, Werling D, Langsley G, Ralph SA. Theileria apicoplast as a target for chemotherapy. Antimicrob Agents Chemother. 2009;53:1213–1217. doi: 10.1128/AAC.00126-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luft BJ, Brooks RG, Conley FK, McCabe RE, Remington JS. Toxoplasmic encephalitis in patients with acquired immune deficiency syndrome. Jama. 1984;252:913–917. [PubMed] [Google Scholar]
- Matsuzaki M, Kikuchi T, Kita K, Kojima S, Kuroiwa T. Large amounts of apicoplast nucleoid DNA and its segregation in Toxoplasma gondii. Protoplasma. 2001;218:180–191. doi: 10.1007/BF01306607. [DOI] [PubMed] [Google Scholar]
- Mazumdar J, E HW, Masek K, C AH, Striepen B. Apicoplast fatty acid synthesis is essential for organelle biogenesis and parasite survival in Toxoplasma gondii. Proc Natl Acad Sci U S A. 2006;103:13192–13197. doi: 10.1073/pnas.0603391103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder B, Seshadri V, Fox PL. Translational control by the 3′-UTR: the ends specify the means. Trends Biochem Sci. 2003;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- Mordue DG, Scott-Weathers CF, Tobin CM, Knoll LJ. A patatin-like protein protects Toxoplasma gondii from degradation in activated macrophages. Mol Microbiol. 2007;63:482–496. doi: 10.1111/j.1365-2958.2006.05538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno SN, Li ZH. Anti-infectives targeting the isoprenoid pathway of Toxoplasma gondii. Expert Opin Ther Targets. 2008;12:253–263. doi: 10.1517/14728222.12.3.253. [DOI] [PubMed] [Google Scholar]
- Muhlbauer SK, Eichacker LA. Light-dependent formation of the photosynthetic proton gradient regulates translation elongation in chloroplasts. J Biol Chem. 1998;273:20935–20940. doi: 10.1074/jbc.273.33.20935. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Ortiz PA, Kinzy TG. Dominant-negative mutant phenotypes and the regulation of translation elongation factor 2 levels in yeast. Nucleic Acids Res. 2005;33:5740–5748. doi: 10.1093/nar/gki882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J, McLeod LE, Vries RG, Flynn A, Wang X, Proud CG. Cellular stresses profoundly inhibit protein synthesis and modulate the states of phosphorylation of multiple translation factors. Eur J Biochem. 2002;269:3076–3085. doi: 10.1046/j.1432-1033.2002.02992.x. [DOI] [PubMed] [Google Scholar]
- Pfefferkorn ER, Nothnagel RF, Borotz SE. Parasiticidal effect of clindamycin on Toxoplasma gondii grown in cultured cells and selection of a drug-resistant mutant. Antimicrob Agents Chemother. 1992;36:1091–1096. doi: 10.1128/aac.36.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph SA, D’Ombrain MC, McFadden GI. The apicoplast as an antimalarial drug target. Drug Resist Updat. 2001;4:145–151. doi: 10.1054/drup.2001.0205. [DOI] [PubMed] [Google Scholar]
- Ralph SA, van Dooren GG, Waller RF, Crawford MJ, Fraunholz MJ, Foth BJ, Tonkin CJ, Roos DS, McFadden GI. Tropical infectious diseases: metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat Rev Microbiol. 2004;2:203–216. doi: 10.1038/nrmicro843. [DOI] [PubMed] [Google Scholar]
- Sato S, Clough B, Coates L, Wilson RJ. Enzymes for heme biosynthesis are found in both the mitochondrion and plastid of the malaria parasite Plasmodium falciparum. Protist. 2004;155:117–125. doi: 10.1078/1434461000169. [DOI] [PubMed] [Google Scholar]
- Seeber F. Biosynthetic pathways of plastid-derived organelles as potential drug targets against parasitic apicomplexa. Curr Drug Targets Immune Endocr Metabol Disord. 2003;3:99–109. doi: 10.2174/1568008033340261. [DOI] [PubMed] [Google Scholar]
- Seeber F, Aliverti A, Zanetti G. The plant-type ferredoxin-NADP+ reductase/ferredoxin redox system as a possible drug target against apicomplexan human parasites. Curr Pharm Des. 2005;11:3159–3172. doi: 10.2174/1381612054864957. [DOI] [PubMed] [Google Scholar]
- Soldati D, Boothroyd JC. A selector of transcription initiation in the protozoan parasite Toxoplasma gondii. Mol Cell Biol. 1995;15:87–93. doi: 10.1128/mcb.15.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati D. The apicoplast as a potential therapeutic target in and other apicomplexan parasites. Parasitol Today. 1999;15:5–7. doi: 10.1016/s0169-4758(98)01363-5. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan WJ, Jr., Narasimhan J, Bhatti MM, Wek RC. Parasite-specific eIF2 (eukaryotic initiation factor-2) kinase required for stress-induced translation control. Biochem J. 2004;380:523–531. doi: 10.1042/BJ20040262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surolia N, Padmanaban G. de novo biosynthesis of heme offers a new chemotherapeutic target in the human malarial parasite. Biochem Biophys Res Commun. 1992;187:744–750. doi: 10.1016/0006-291x(92)91258-r. [DOI] [PubMed] [Google Scholar]
- Tonkin CJ, Pearce JA, McFadden GI, Cowman AF. Protein targeting to destinations of the secretory pathway in the malaria parasite Plasmodium falciparum. Curr Opin Microbiol. 2006;9:381–387. doi: 10.1016/j.mib.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Trebitsh T, Levitan A, Sofer A, Danon A. Translation of chloroplast psbA mRNA is modulated in the light by counteracting oxidizing and reducing activities. Mol Cell Biol. 2000;20:1116–1123. doi: 10.1128/mcb.20.4.1116-1123.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dooren GG, Su V, D’Ombrain MC, McFadden GI. Processing of an apicoplast leader sequence in Plasmodium falciparum and the identification of a putative leader cleavage enzyme. J Biol Chem. 2002;277:23612–9. doi: 10.1074/jbc.M201748200. [DOI] [PubMed] [Google Scholar]
- Waller RF, Reed MB, Cowman AF, McFadden GI. Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway. Embo J. 2000;19:1794–1802. doi: 10.1093/emboj/19.8.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner J, Jomaa H. Isoprenoid biosynthesis of the apicoplast as drug target. Curr Drug Targets. 2007;8:3–13. doi: 10.2174/138945007779315551. [DOI] [PubMed] [Google Scholar]
- Wiesner J, Reichenberg A, Heinrich S, Schlitzer M, Jomaa H. The plastid-like organelle of apicomplexan parasites as drug target. Curr Pharm Des. 2008;14:855–871. doi: 10.2174/138161208784041105. [DOI] [PubMed] [Google Scholar]
- Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- Williamson DH, Denny PW, Moore PW, Sato S, McCready S, Wilson RJ. The in vivo conformation of the plastid DNA of Toxoplasma gondii: implications for replication. J Mol Biol. 2001;306:159–168. doi: 10.1006/jmbi.2000.4385. [DOI] [PubMed] [Google Scholar]
- Zhang L, Paakkarinen V, van Wijk KJ, Aro EM. Biogenesis of the chloroplast-encoded D1 protein: regulation of translation elongation, insertion, and assembly into photosystem II. Plant Cell. 2000;12:1769–1782. doi: 10.1105/tpc.12.9.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.