Abstract
Traditionally, biological probes and drugs have targeted the activities of proteins (such as enzymes and receptors) that can be easily controlled by small molecules. The remaining majority of the proteome has been deemed “undruggable”. By using small molecule modulators of the ubiquitin proteasome, protein levels, rather than protein activities can be targeted instead, increasing the number of druggable targets. While targeting the proteasome itself can lead to a global increase in protein levels, targeting other components of the UPS (e.g., the hundreds of E3 ubiquitin ligases) can lead to an increase in protein levels in a more targeted fashion. Alternatively, multiple strategies for inducing protein degradation with small molecule probes are emerging. With the ability to induce and inhibit the degradation of targeted proteins, small molecule modulators of the UPS have the potential to significantly expand the druggable portion of the proteome beyond traditional targets such as enzymes and receptors.
Keywords: proteasomeubiquitindrug design protein, degradationinhibitors
1. Introduction
Current techniques in molecular biology and genetics allow for virtually any gene to be knocked down or overexpressed in cell culture and in animal models. However, pharmacological control of genes is desirable both for the development of therapeutics and for biological studies due to the improved temporal control and reversibility.[1] Unfortunately, currently available small molecule probes and drugs are much more limited in their possible targets than genetic methods.[2] Traditionally, pharmacological probes have been limited to targeting enzymes and receptors that contain well-defined pockets that tightly bind small molecules. This has left many proteins, including those that act through protein-protein interactions as scaffolds, to be deemed “undruggable”.[3] Finally, while small molecules that act to activate, rather than inhibit, protein activity are known (such as receptor agonists), they act on an even smaller subset of the proteome than inhibitors.
An alternative to small molecule inhibition and activation of protein activity that would more closely mimic the results of molecular biological techniques would be to control protein levels, which can be accomplished at the post-translational level through the modulation of the ubiquitin proteasome system (UPS). Small molecule control of protein levels at the level of transcription is also an exciting area of research with its own set of challenges and is therefore beyond the scope of this review. Small molecule inhibitors of the ubiquitination cascade or of the proteasome offer the possibility of stabilizing proteins targeted by the UPS with varying levels of specificity depending on the target. Inhibitors of deubiquitinases and chaperones that rescue proteins from proteasomal degradation offer the ability to decrease the levels of specific proteins. Additionally, multiple strategies are being developed that seek to highjack the UPS to induce degradation of proteins not already targeted by the UPS.
1.1 Overview of the Ubiquitin Proteasome System
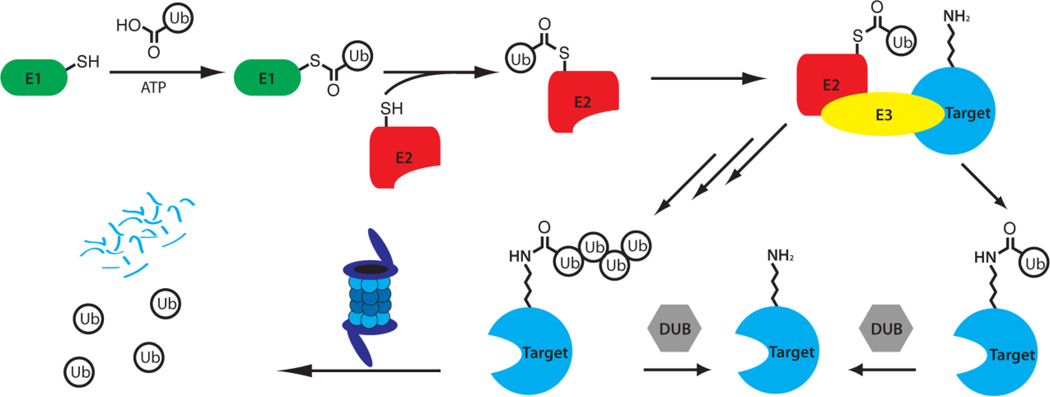
The ubiquitin proteasome system is a complex, coordinated process involving the covalent coupling of the 76 amino acid protein ubiquitin to targeted proteins, leading to their subsequent recognition and degradation by the 26S proteasome.[4] The process begins with the E1 ubiquitin-activating enzyme (UAE or Uba1), which first adenylates the C-terminus of ubiquitin (consuming ATP in the process), forming a reactive thioester bond on a surface cysteine. The E1 then transfers the activated ubiquitin to an E2 ubiquitin-conjugating enzyme, forming a new thioester. One of the over 600 E3 ligases then acts as an adaptor (except in the case of the HECT family of E3 ligases, which involve the formation of a third thioester intermediate), bringing the E2-ubiquitin complex into proximity with the target protein, facilitating the transfer of ubiquitin to a surface lysine to form an isopeptide bond (Figure 1). The addition of ubiquitin is also reversible through the action of various deubiquitinating enzymes (DUBs).[4a, 6]
Figure 1.
Summary of the Ubiquitin Proteasome Pathway (UPS). Ubiquitin is activated by the E1 ubiquitin-activating enzyme and transferred to an E2 ubiquitinating conjugating enzyme. The E2 then transfers the ubiquitin to a target protein with the assistance of an E3 ubiquitin ligase that recognizes the target protein. The process may then be repeated to form polyubiquitin chains, which bind to the regulatory particle of the 26S proteasome, leading to the degradation of the target protein and the recycling of the ubiquitin units.[4a, 5]
The process can then be repeated, by transferring additional ubiquitin units to the N-terminus or one of the seven lysine residues on ubiquitin. The formation of a chain consisting of at least four ubiquitin moieties linked through Lys48 is recognized as being sufficient to target the target protein for degradation by the 26S proteasome.[5] This process involves the removal of the ubiquitin units (which are recycled) and then the processive degradation of the target protein into short peptide fragments (Figure 1).[4a] However, alternate linkages of ubiquitin chains are also possible, leading to a variety of biological consequences in addition to proteasome-mediated degradation. These include formation of Lys63 chains, which have been shown to be involved in the regulation of endocytosis. Mono-ubiquitination of proteins can have diverse biologicaleffects, sometimes through the induction of a conformational change in the target. Additionally, numerous ubiquitin-like proteins (UBLs) have been identified, such as SUMO and NEDD8, which are ligated to proteins through an analogous system but involving distinct enzymes. These UBLs also can have numerous effects, often through the induction of conformational changes in the target proteins.[6]
2. Small Molecule-mediated Protein Stabilization: Inhibitors of Protein Degradation
Due to the multiple effects of ubiquitination on diverse biological processes (including cell cycle progression, apoptosis, oncogenesis, protein quality control and angiogenesis),[7] inhibition of proteasome dependent degradation presents numerous attractive targets for the development of therapeutic treatment for many diseases.Each class of enzymes, ranging from the E1, the roughly 40 E2 enzymes, the over 600 E3 ligases, and the multiple subunits of both the constitutive and immunoproteasomes, has both advantages and disadvantages as drug targets based upon their biological role, specificity and druggability.[6] While other processes (e.g., autophagy) can also control protein degradation, the small molecule control of these systems is beyond the scope of this work but has been recently reviewed elsewhere[8].
2.1. Proteasome Inhibitors
Currently, there are only two FDA approved drugs targeting the UPS, bortezomib/Velcade™ and carfilzomib/Kyprolis™, which both directly inhibit the proteasome.[6b, 9] Numerous classes of proteasome inhibitors have been described[7, 9–10] including peptide aldehydes, peptidyl boronates, epoxyketones, vinyl sulfones[11], β-lactones[12], hydroxyureas,[13] α-keto-aldehydes[14] and others. Many include an electrophile that targets the key nucleophilic Thr1 residues in the catalytic β1, β3 and β5 subunits of the proteasome.[15] These electrophiles are often attached to a linear or cyclic peptidic chain mimicking the substrate protein. Additionally, noncovalent inhibitors have been reported such as TMC-95A[16] and the non-competitive imidazoline class.[17]
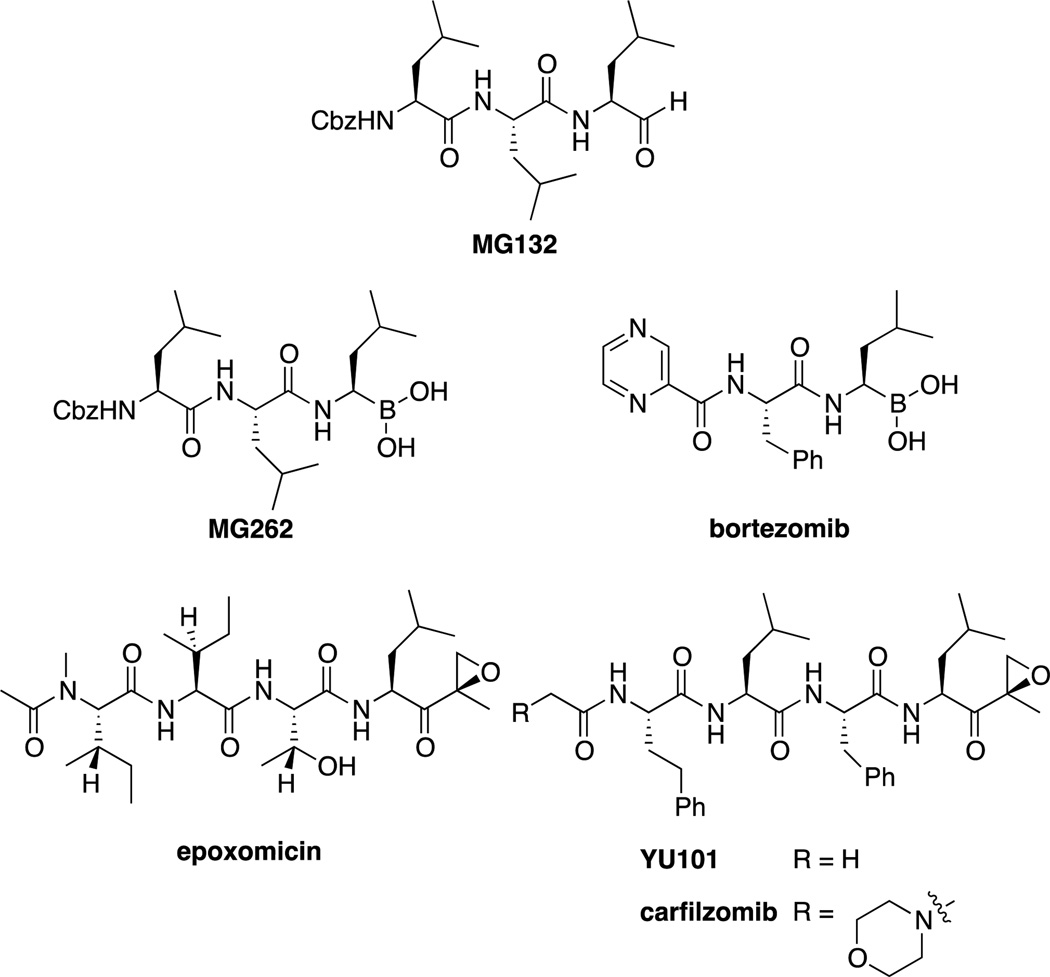
The first class of proteasome inhibitors identified was the peptide aldehydes,[18] of which MG132 (or Z-LLL), developed by Myogenics, is the most studied (Figure 2).[19] These compounds are potent (Ki = 4 nM),[19] covalent inhibitors of the chymotryptic-like activity of the β5 subunit. However, issues related to lack of selectivity towards other proteases (such as calpains and cathepsins), rapid oxidation and the rapid reversibility of the hemiketal formed with Thr1 prevented these compounds from being useful therapeutically. Regardless, the combination of potency and wide availability (at low relatively low cost) has led to MG132 being one of the most used proteasome inhibitors used in biological studies.[7] In addition to synthetic peptide aldehydes, natural products such as fellutamide B have also been shown to inhibit the proteasome.[20]
Figure 2.
Summary of proteasome inhibitors, including the widely used probe compound MG132, as well as bortezomib/Velcade™ and carfilzomib/Kyprolis™, FDA approved drugs for the treatment of multiple myeloma.
The work on peptide aldehydes led directly to discovery of the peptidyl boronates; it was discovered that conversion of the aldehyde of MG132 to a boronic acid (MG262 or Z-LLL-boronate) led to a vast improvement in potency (Ki = 18 pM).[7] This increased potency allowed for the development of smaller, dipeptidyl boronic acids, such as bortezomib, developed by Myogenics/ProScript. This class also had the added benefit of increased specificity compared to peptide aldehydes, as the boronic acid moiety is far less capable of reacting with cysteine proteases such as cathepsins and calpains.[21] Bortezomib reversibly forms a tetrahedral borate with Thr1, which was demonstrated via x-ray crystallography.[22] After being purchased by Millennium Pharmaceuticals (now owned by Takeda), bortezomib entered clinical trials in 1997 and was approved by the FDA in 2003 for use in multiple myeloma. Marketed as Velcade™, it is administered intravenously or subcutaneously and is currently under investigation for treatment of other cancers such as non-Hodgkin’s lymphoma.[6b, 21, 23]
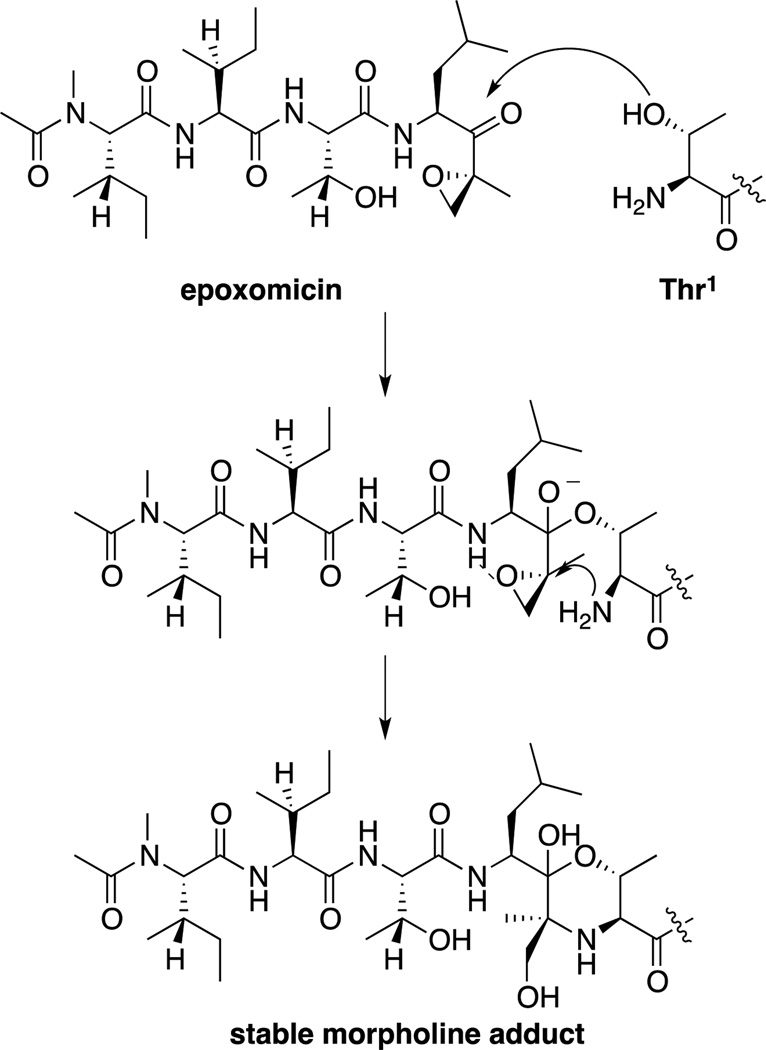
Another class of proteasome inhibitors is the epoxyketones. The first epoxyketones described were eponemycin[24] and epoxomicin(Figure 2),[25] natural products isolated by BMS in the early 1990s based on their activity against melanoma cell lines. However, the mechanism of action of these compounds remained unknown until our lab completed the total syntheses of eponemycin,[26] epoxomicin,[27] and their respective biotinylated affinity reagents, which were used to show that both were potent inhibitors of the proteasome.[28] Our lab then demonstrated that the epoxyketones reacted with Thr1 of the proteasome to first form a hemiketal, followed by attack of the terminal amine on the epoxide to form a stable morpholine ring.[29] This two-step nucleophilic attack on the epoxyketone not only leads to irreversible (as opposed to the slowly reversible peptidyl boronates such as bortezomib) inhibitors of the proteasome, but also leads to specificity, due to the uniqueness of the N-terminal threonine catalytic residue amongst proteases (Scheme 1).
Scheme 1.
Proposed mechanism for the inactivation of the proteasome by epoxomicin. Initial hemiketal formation is followed by epoxide opening by the terminal amine, to form a stable morpholine adduct, which has been observed through x-ray crystallography.[29]
After identifying the proteasome as the target of epoxomicin, our lab sought to optimize the selectivity of epoxyketones for the chymotrypsin-like activity of the proteasome over the other two primary activities, the trypsin-like activity and the caspase-like activity. An increasingly selective inhibitor was desirable as an improved chemical probe, helping to tease out the effects of inhibition of each of the catalytic subunits. Additionally, most proteasome inhibitors (including bortezomib and epoxomicin) already had significant selectivity for the chymotrypsin-like activity of the β5 subunit, suggesting that the therapeutic effects of these inhibitors were primarily due to the inhibition of this activity. The result of this optimization was YU101,[30] which had increased selectivity for the chymotrypsin-like activity over the trypsin-like activity and nearly 8000 fold over the caspase-like activity (Figure 2). We then were able to correlate the Kobs/[I] for the chymotrypsin-like activity with inhibition of cellular proliferation.[31]
These results were then licensed to Proteolix, Inc., which modified the compound by adding a morpholine moiety to increase solubility of the molecule. The resulting compound, carfilzomib[32] then entered clinical trials for multiple myeloma in patients resistant to bortezomib and either thalidomide or lenalidomide. In patients with refractory multiple myeloma, carfilzomib (administered intravenously) had an overall response rate of 23.7%, with a median duration of response of 7.8 months.[33] Based upon this Phase II study, as well as other data showing generally comparable safety profile (compared to bortezomib), as well as significantly improved rates of peripheral neuropathy, carfilzomib was approved by the FDA in 2012, and is currently being marketed by Onyx Pharmaceuticals (which purchased Proteolix in 2009) as Kyprolis™.[34]
In addition to the two FDA approved proteasome inhibitors, there remains a strong interest in the development of improved proteasome inhibitors for clinical use. One avenue of active research is to improve the bioavailability of proteasome inhibitors; currently there are multiple orally bioavailable proteasome inhibitors in clinical trials, such as ixazomib citrate (MLN9708) from Millenium and oprozomib (ONX 912) from Onyx.[35] Another active area of research is the development of inhibitors that are selective for the immunoproteasome, which is expressed in lymphocytes. Inhibition of the immunoproteasome has been suggested to be a possible strategy for the treatment of autoimmune and neurodegenerative disorders. Inhibitors such as ONX 914 have selectivity for the immunoproteasome over the constitutive proteasome.Crystallographic studies have suggested that selectivity can be achieved due to the larger P1 pocket of the immunoproteasome.[36]
One concern about the use of proteasome inhibitors is that the inhibition of protein degradation leads to the formation of protein aggregates. While protein aggregation is linked with neurodegenerative disorders such as Alzheimer’s disease, bortezomib and carfilzomib do not readily cross the blood-brain barrier, their effect on the central nervous system should be limited,[32b, 37] although, as noted above, peripheral neuropathy is a serious side-effect of bortezomib treatment. The toxicity of the polyubiquitylated protein aggregates in non-cancerous cells is undesirable; however, inhibition of other targets of the UPS will avoid this issue, highlighting one of the benefits of more targeted therapies.
2.2. Inhibitors of the E1 Ubiquitin-Activating Enzyme and Related E1s
The E1 ubiquitin-activating enzyme (UAE) carries out its function in two discrete steps. First, it adenylates the C-terminus of ubiquitin,then formsa covalent thioester bond with the active site cysteine. As inhibition at the E1 level would prevent ubiquitination, this would globally disrupt the UPS, and may have similar effects to proteasome inhibition (as opposed to targeting E2, E3 or DUB enzymes, which may have more specific effects). There has been significant interest in developing E1 inhibitors, and the topic has recently been reviewed.[38]
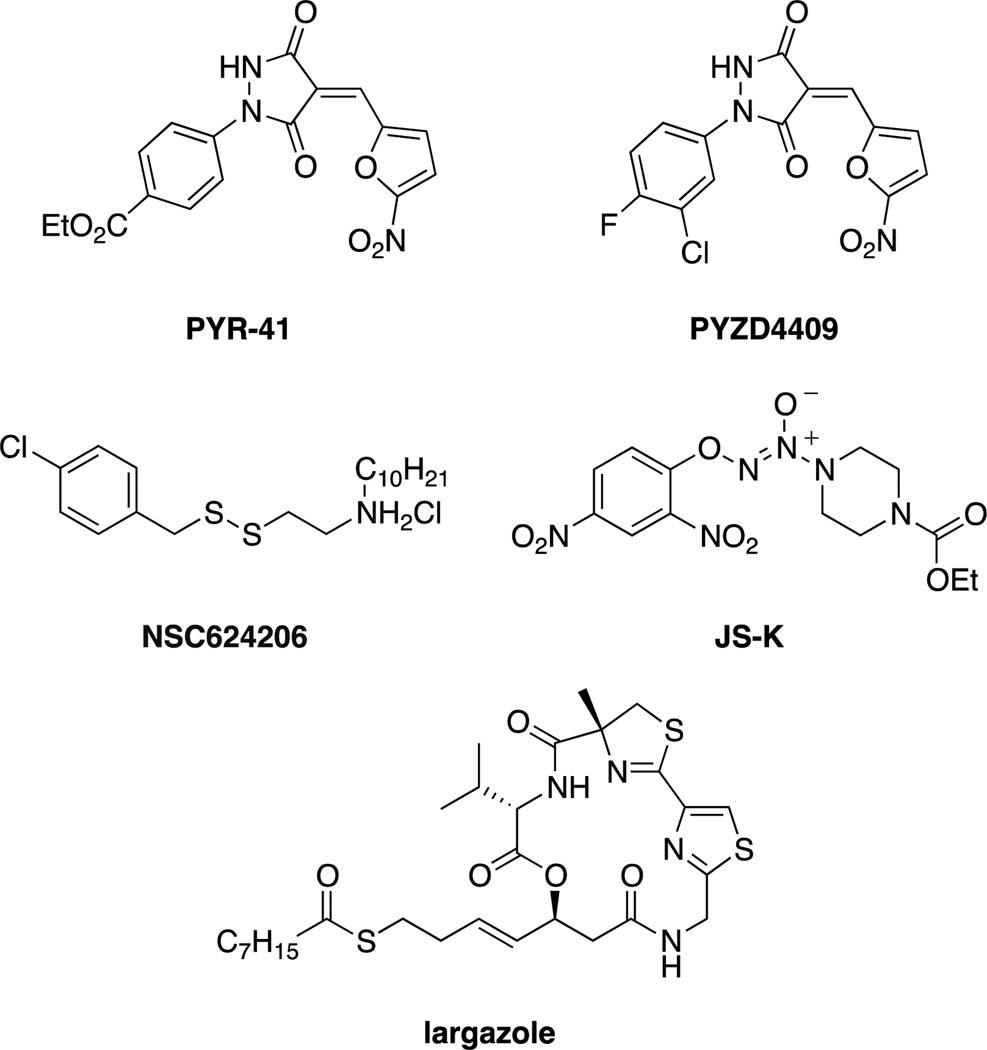
The first cell permeable inhibitor was the pyrazone, PYR41 (Figure 3), that was found to covalently inhibit UAE in cells, preventing ubiquitination and degradation by the proteasome of target proteins such as p53.[39] A similar inhibitor, PYZD4409, was also shown to induce cell death in malignant cells and have activity in a mouse model of leukemia.[40] In addition to the pyrazone UAE inhibitors, other inhibitors reported include the nitric oxide prodrug JS-K[41] and the disulfide NSC624206,[42] both of which act by modification of the catalytic cysteine. The natural product largazole, in addition to its better known affects on histone deacetylases, has also been shown to inhibit UAE by blocking ubiquitin adenylation (but not thioester formation).[43] While numerous UAE inhibitors have been developed, it is likely that new classes of inhibitors with increased specificity and drug-like properties will need to be developed before they are of use therapeutically.[38]
Figure 3.
Small molecule inhibitors of E1 ubiquitin-activating enzyme.
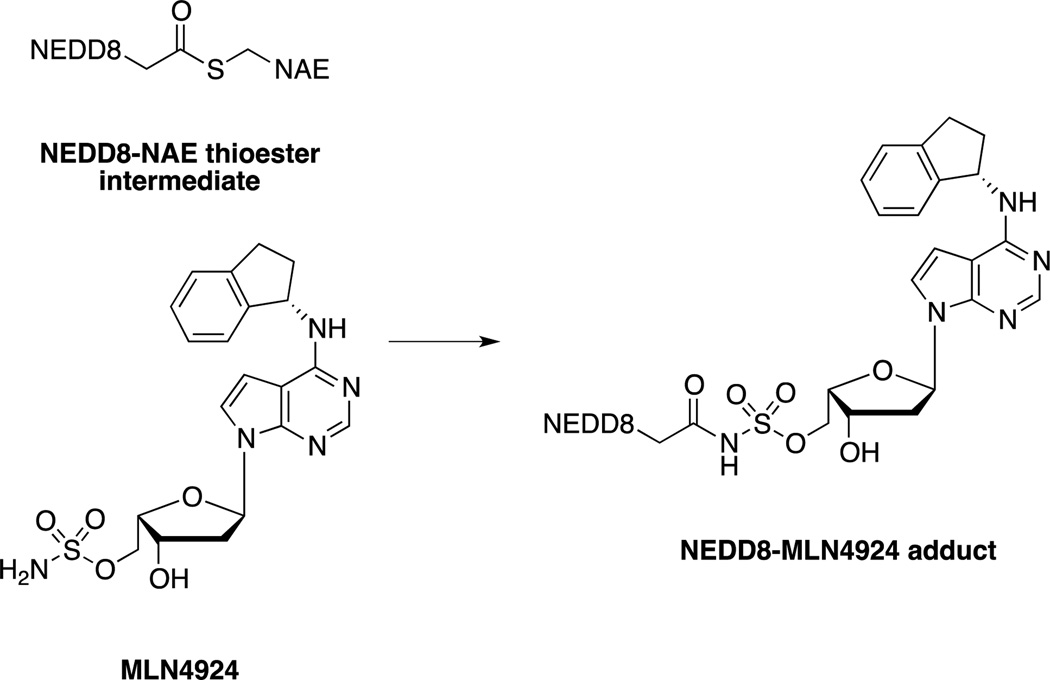
In addition to the ubiquitin-activating enzyme (UAE), there are 7 other E1s that activate other UBLs. Millennium Pharmaceuticals has developed a small molecule inhibitor of the NEDD8 activating enzyme (NAE), MLN4924 that acts through an interesting mechanism termed substrate-assisted inhibition. MLN4924 is an adenine mimic which, in the presence of NAE, forms a covalent bond with NEDD8 through nucleophilic attack by the sulfamate group on the intermediary thioester (Scheme 2). The NEDD8-MLN4924 adduct is then able to act as a potent inhibitor of NAE, as a non-hydrolysable mimic of the adenylated-NEDD8 intermediate.[44] As NEDDylation of cullin is important to the activity of cullin-RING ligases (CRLs), a family of E3 ligases, inhibition of NAE has therapeutic potential. [6a, 44b] With this in mind, MLN4924 has entered into Phase I and I/II trials as a potential treatment for both hematologic and nonhematologic malignancies.[45]
Scheme 2.
MLN4924 is able to react covalently with the activated NEDD8-NAE intermediate to form a non-hydrolysable covalent bond with NEDD8. The MLN4924-NEDD8 adduct then acts as a mimic of the adenylated-NEDD8 substrate, competitively inhibiting NAE.[44]
2.3. Inhibition of E2 Ubiquitin-Conjugating Enzymes
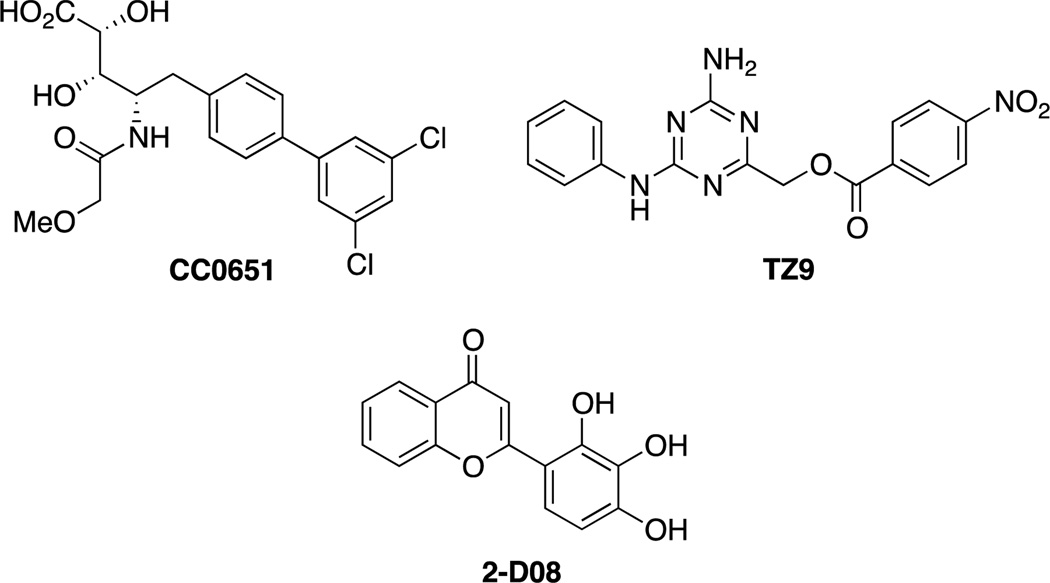
The approximately 40 E2s are responsible for accepting ubiquitin or UBL units from the E1 and transferring them to the target protein in conjunction with the E3 ligase (or for the transfer of ubiquitin to HECT E3 ligases). Despite the existence of a catalytic cysteine residue, development of E2 inhibitors has lagged behind E1 inhibitors, and only two E2 inhibitors have been reported. Ceccarelli et al. described the first inhibitor of an E2 enzyme, which was discovered in a high throughput screen of inhibitors of p27Kip1 ubiquitination, which is important to cell cycle progression and a possible target in cancer. The cell-free assay reconstituted all the components of the ubiquitinylation machinery: biotinylated ubiquitin, the E1 enzyme Uba1, the E2 hCdc34, the E3 complex SCFSkp2 (as well as Cks1) and phosphorylated substrate, p27Kip1. CC0651 (Figure 4) was found to inhibit p27Kip1 ubiquitination. The activity of CC0651 was found to be due to allosteric inhibition of hCdc34, which was confirmed by X-ray crystallography. CC0651 was shown to inhibit proliferation of cells and cause accumulation of p27Kip1.[46]
Figure 4.
Small molecule inhibitors of E2 ligases. CC0651 inhibits hCdc34;[46a] TZ9 inhibits Rad6;[47] 2-D08 inhibits the SUMO E2, UBc-9.[48]
The inhibition of a distinct E2, Rad6 (essential for postreplication DNA repair), was recently reported. TZ9 was developed using a virtual screen and designed to bind to the catalytic site of Rad6. TZ9 was successful at inhibiting histone ubiquitination in vitro and inhibited cell proliferation. Unlike CC0651, which acts through an allosteric mechanism, TZ9 is predicted to block thioester formation, making it the first competitive E2 ligase inhibitor.[47] The SUMO E2, Ubc-9 has also been targeted for inhibition. Schneekloth and co-workers recently reported the identification of the flavonoid 2-D08, which inhibits the transfer of SUMO from Ubc-9 to a model substrate and inhibits SUMOylation of topoisomerase-1 in a cellular assay.[48]
2.4. Small Molecule Inhibitors of E3 Ligases
There are over 600 E3 ligases[6b] (divided into 4 families, HECT domains E3s, U-box E3s, monomeric RING E3s and multisubunit RING E3s)[6a] that catalyze the addition of ubiquitin or UBLs to their target proteins. The majority of substrate specificity of the UPS derives from the selectivity of the E3 ligases for their targets, making them attractive targets for the development of therapeutics. Unfortunately, most E3s lack any enzymatic activity, acting instead by bringing ubiquitin-loaded E2s into proximity with target proteins (the exception being HECT E3s, which form a thioester bond with ubiquitin before transferring it to their substrates). Therefore, inhibition of E3 ligases has generally required the targeting of protein-protein interactions, which are notoriously difficult to modulate using small molecule agents.[3]
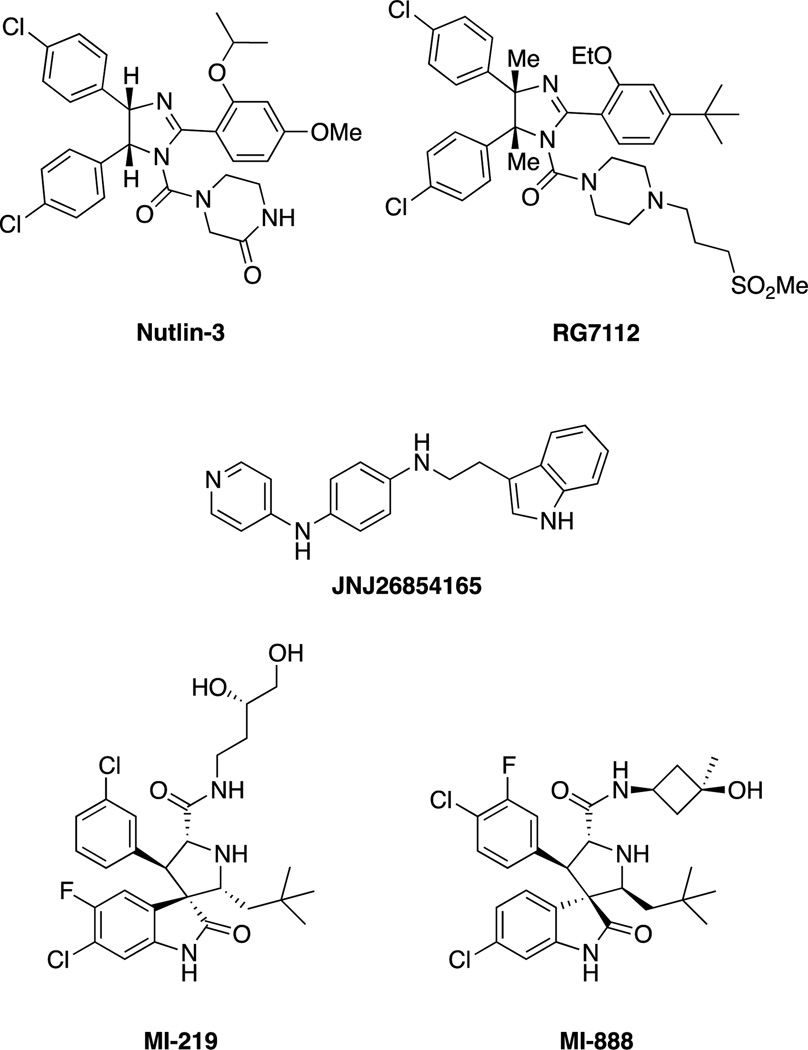
The first E3 ligase successfully targeted was MDM2, which ubiquitinates the tumor suppressor p53. Roche reported the discovery of Nutlins, cis-imidazoline inhibitors of the MDM2-p53 interaction, which stabilize p53 in cells and inhibit growth of tumor xenographs in nude mice.[49] Since then, an orally administered Nutlin derivative, RG7112 (Figure 5),[50] has advanced to Phase I clinical trials for the treatment of solid and hematological tumors.[51] An additional MDM2 inhibitor, the tryptamine JNJ-26854165, is also orally administered and in Phase I clinical trials but appears to act instead by blocking the interaction between the p53-MDM2 complex with the proteasome.[51–52] Numerous other classes of MDM2 inhibitors have been developed,[53] including the spiro-oxindoles, which were discovered through structure-based design[54] and include MI-219[55] and MI-888.[56] However, while p53 is an important tumor suppressor, attempts to stabilize it will have little effect on the large percentage of cancers with mutated p53.[57]
Figure 5.
Nutlin, the first MDM2 inhibitor and other selected MDM2 inhibitors.
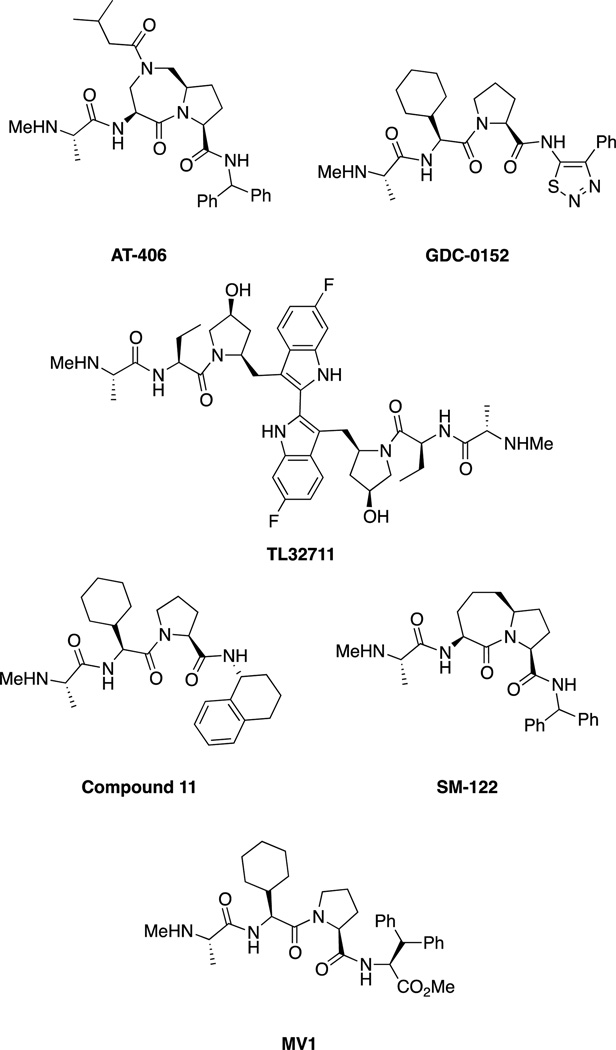
In addition to MDM2 inhibitors, the Inhibitor of Apoptosis Protein (IAP) family of E3 ligases has been extensively targeted, usually with largely peptidic or peptidomimetic inhibitors, inspired by the natural protein inhibitor of the IAPs, SMAC/DIABLO. The IAPs regulate apoptosis through numerous pathways, including caspase 3, 7 and 9 regulation. Inhibitors of IAPs have recently been reviewed extensively[58] and include numerous compounds in clinical trials (Figure 6).[6b] SMAC mimics bind to the BIR3 domains of IAPs, which is distinct from the RING domain. The pioneering work by Abbott led to the optimized peptidic ligand compound 11 (Figure 6).[59] The group of Shaomeng Wang at the University of Michigan optimized this core structure by replacing the cyclohexylglycine-proline motif with a bicyclic structure, such as the one present in SM-122.[60] An additional peptidic IAP antagonist is MV1, developed by Genentech.[61]
Figure 6.
Summary of IAP inhibitors including AT-406 (developed by Ascenta Therapeutics and the University of Michigan),[62] which is administered orally in Phase 1 trials for solid tumors and lymphoma, Genentech/Roche’s GDC-0152 which is administered intravenously and is in Phase I trials for metastatic malignancies,[6b, 63] and the bivalent TL32711 (administered intravenously) developed by Tetralogics Pharma.[6b, 58] LCL161 (Novartis), AEG35156 and AEG40826 (Aegera), and YM155 (Astellas Pharma) are also in clinical trials but are not shown.[6b, 58] Selected SMAC mimics such as SM-122 and MV1 are also shown but are not in clinical trials.
The SCF (SKP1, CUL1, F-box protein) E3 ligases are a subfamily of the multi-subunit cullin-RING ligases (CRLs). They contain various F-box proteins which confer substrate specificity, and often recognize post-translational modifications on their targets, such as phosphorylation (Figure 7).[64] Numerous SCF members have been targeted with small molecule inhibitors. SCFSkp2 ubiquitinates numerous proteins involved in cell cycle control, such as p27Kip1. SCFSkp2 inhibitors were developed through virtual screening to target SCFSkp2-p27 interface. Both the rhodanine C1 and the pyrrolinone C2 inhibit the ubiquitination of p27 in vitro, leading to p27 accumulation in cells and inducing G1/S arrest in cells.[65] Recently, a class of chromones was also reported to inhibit SCFSkp2 by preventing the binding of Skp2 to remainder of the SCF complex. Their lead, compound 25, was found to have antitumor activity in animal models.[66] SMER3 was discovered through the use of a chemical genetics screen of enhancers of rapamycin and found to inhibit Met4 ubiquitination by SCFMet30 through blockage of the interaction between Met30 and the core of the SCF complex.[67] Racemic SCFI2 was reported to inhibit the interaction between SCFCdc4 and its target, phosphorylated Sic1. However, crystallographic studies show that SCFI2 binds the WD40 propeller domain 25 Å away from the substrate-binding site and inhibits SCFCdc4 allosterically. [67b, 68]GS143(Figure 7) was reported as a putative SCFβTrCP1 inhibitor. GS143 stabilizes IκBα in cells. The authors hypothesized that it inhibits the SCFβTrCP1-IκBα interaction after excluding other mechanisms for the stabilization; however, no evidence of direct binding of GS143 to either SCFβTrCP1 or IκBα was reported.[69]
Figure 7.
A) Depiction of the SCF E3 ligase complex. The complex contains a cullin (which is NEDDylated when active) which binds a RING domain as well as the adaptor Skp1. Skp1 also binds various F box proteins (such asβTrCP, Cdc4 and Skp2) which function as the substrate interaction motif, binding target proteins which are ubiquitinated. B) Inhibitors of SCF E3 ligases. The specific E3 inhibited is shown in parentheses.
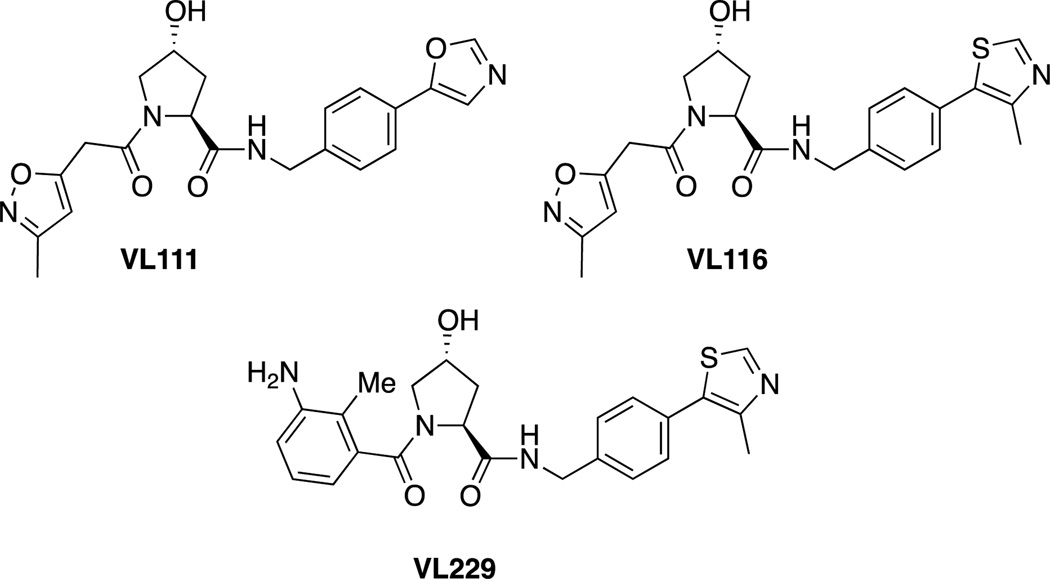
Recently, we reported the design of small molecules that can competitively bind to the primary HIF binding site on VHL, the substrate recognition subunit of a CRL. Drawing inspiration from the key hydroxyproline residue of HIF, we developed VL111. [70] Guided by the x-ray crystal structures of VL111 and other early derivatives bound to VHL,[71] we optimized the VHL ligand, leading to VL116 and VL229 (Figure 8).[72] However, while these compounds were able to inhibit the interaction between VHL and fluorescent peptides derived from HIF in vitro, we have been unable to demonstrate ligand-induced HIF stabilization activity in cell based assays.
Figure 8.
Structures of hydroxyproline-based molecules capable of inhibiting the interaction between VHL and a peptide derived from HIF in vitro.
Thalidomide was originally developed for its sedative properties before it was infamously discovered to be a potent teratogen causing serious birth defects such as phocomelia (limb defects) and amelia (the lack of one or more limbs). While it was discontinued as a sedative, it is still in use today for treatment of serious disorders such as leprosy and multiple myeloma, despite its serious side effects. Recently, a study found that thalidomide bound to and inactivated the E3 ligase cereblon (CRBN), a component of a CRL important to limb development. This strongly indicates a mechanism for the side effects of thalidomide, possibly allowing for the development of thalidomide derivatives that do not target CRBN.[6b, 73]
2.5. Direct Small Molecule Stabilization of Destabilized Proteins and Shield
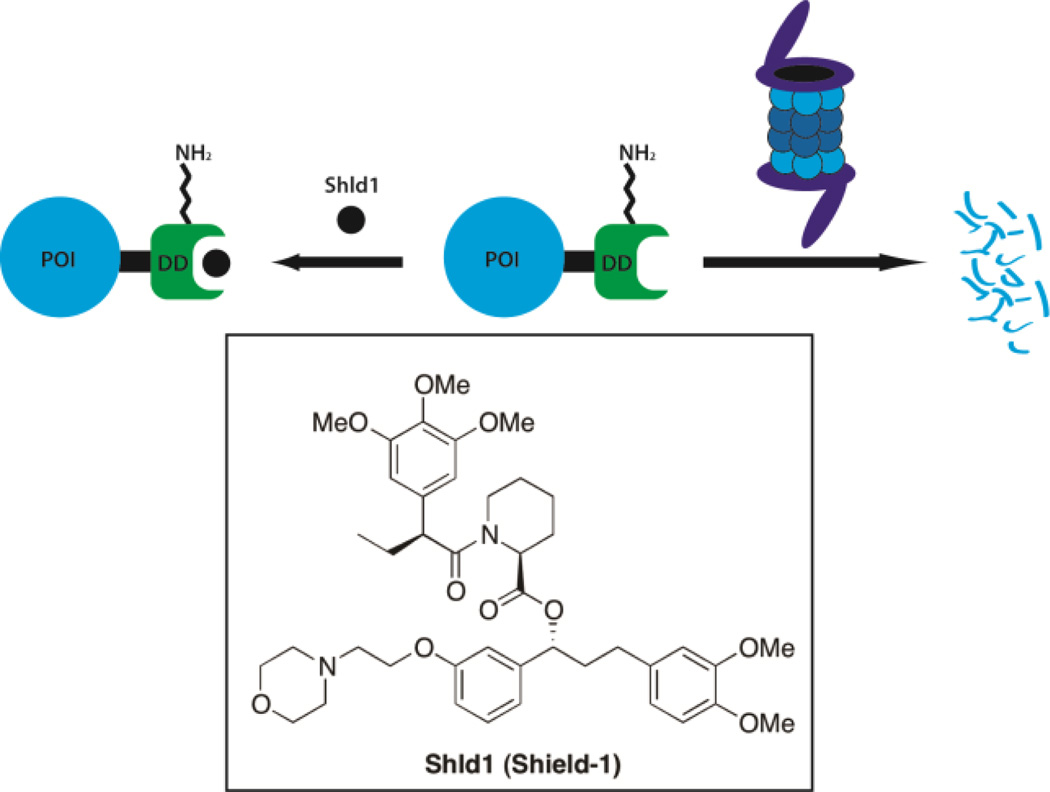
Direct ligand binding can also increase the stability of a protein, through a number of mechanisms. Ligand binding commonly increases the thermal stability of a protein, allowing for the use of differential scanning fluorimetry in high-throughput screening.[75] It can also reduce the protease susceptibility of the target protein, the basis of DARTS, a technique used to identify the cellular targets of small molecules.[76] However, the most striking stabilization in cells results from the use of ligand-responsive degrons.[77]
Building off of earlier work using rapamycin derivatives to stabilize mutated Frb*,[78] Wandless and coworkers mutated anFKBP12-YFP fusion protein using error prone PCR, then analyzed fluorescence in the presence then absence of FKBP12 ligand to screen for destabilized mutants of FKBP12 that are rescued upon addition of ligand. The L106P mutant was then analyzed further using a related ligand Shld1 and was found to rescue the degradation of numerous fusion proteins in cell culture[74] and in mice.[79] This technology was then modified to allow for the cleavage of the degron to yield the native protein in the presence of the stabilizing ligand.[80] An alternative use of Shld1 has the opposite effect and stabilizes proteins fused with mutated FKBP and a cryptic degron. This involves the tethering of FKBP to a degron sequence as well as a proline rich sequence designed to bind to the active site of FKBP. In the absence of Shld1, the degron is masked but upon its addition the degron becomes exposed leading to the degradation of the fusion protein.[81] While these systems require genetic manipulation, they have been widely used in studies allowing for time sensitive control of dendritic proteins[82] and transcription factors.[83]
3. Inducers of Protein Degradation
Inducing protein degradation is also a possible avenue for the development of therapeutics. Unlike enzyme inhibitors and antagonists, which are limited to specific subsets of proteins (i.e. enzymes and receptors), theoretically, any protein can be targeted for degradation. Proteins such as transcription factors or scaffolds that act through protein-protein interactions (PPIs) or protein-nucleic acid interactions have long been considered “undruggable”.[3] Methods such as PROTACs that induce their degradation require only a ligand capable of binding to these targets,not inhibiting interactions that occur across large surface areas. Additionally, while inhibitors and antagonists control only a specific activity of their targets, degradation of the protein would lead to a loss of function of other activities (such as scaffolding functions).[2] Outside of the field of therapeutics, the use of small molecule probes that induce degradation is in many cases preferable to genetic knockdown with techniques such as RNAi due to the better temporal control and reversibility.[1b]
Induced degradation can also be useful in cases where resistance to therapeutics emerges without altering binding to the target. Mechanisms of resistance such as protein overexpression could theoretically be overcome by inducing protein degradation. In the case of the androgen receptor, resistance can emerge to antagonists such as flutamide not by preventing the binding of the antagonist, but by mutations that convert the antagonists into agonists.[84] Small molecule degraders avoid this issue, as the target protein would be degraded, preventing mutations that increase activity from generating resistance.
3.1. PROTACs: Heterobifunctional Molecules that Recruit Specific E3 Ligases to Targeted Proteins
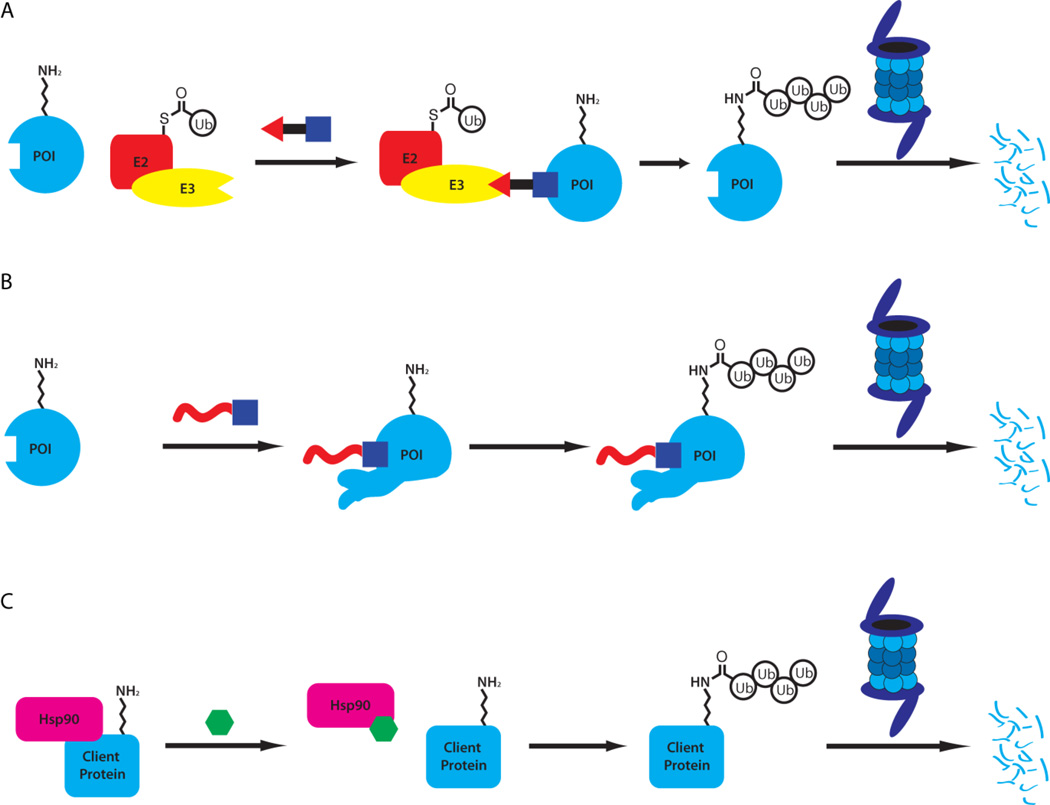
PROteolysis TArgeting Chimeras, or PROTACs, are heterobifunctional molecules[85] that contain a ligand for an E3 ligase, a linker and a ligand for a protein that is to be targeted for degradation. The molecule can then bind to both the E3 ligase and the target, inducing the formation of a ternary complex. This hijacking of the E3 ligase can then lead to the polyubiquitination of the target protein, followed by its degradation by the proteasome (Figure 10).[1]
Figure 10.
Strategies for induced protein degradation include direct recruitment of an E3 ligase with A) PROTACs, B) induced protein misfolding (or mimicking misfolding) with hydrophobic tags (or ligand-mediated degradation) and C) the inhibition of chaperones such as Hsp90.
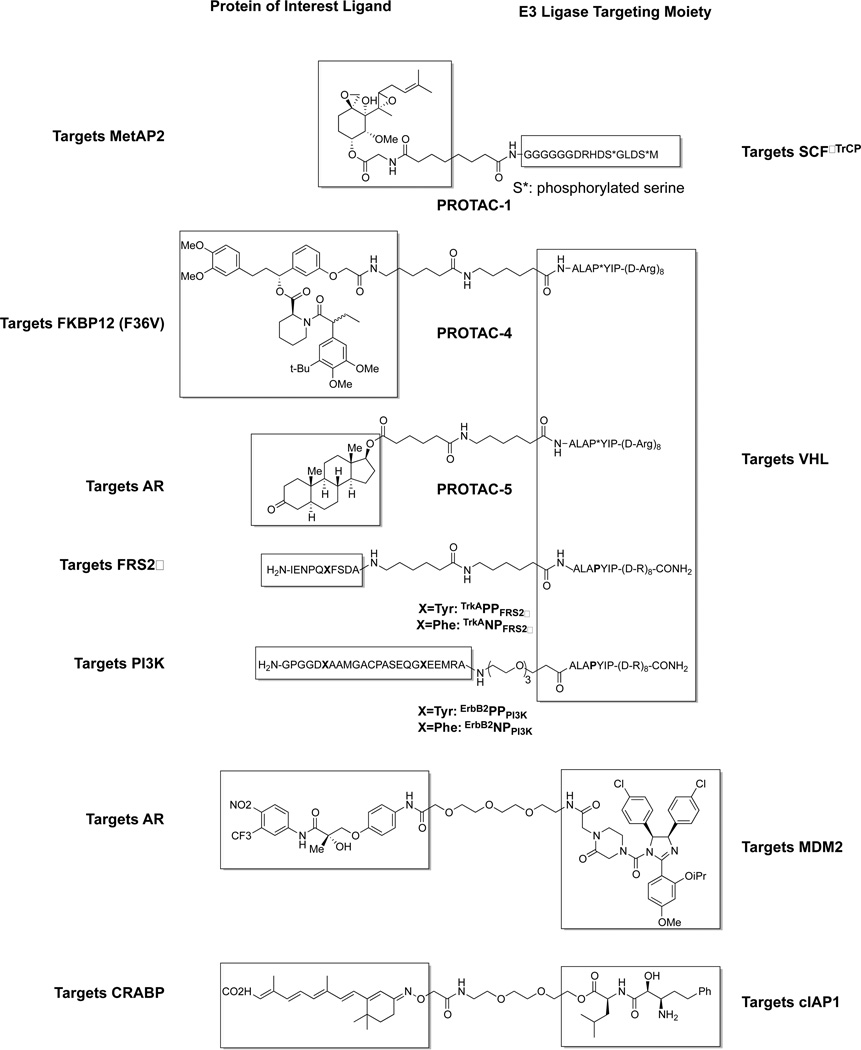
The first PROTAC (PROTAC-1) described contained a phosphopeptide derived from IκBα, which binds to the E3 ligase SCFβTrCP, as well as a moiety derived from ovalicin, which covalently binds MetAP-2 (Figure 11). PROTAC-1 was able to specifically and covalently bind to MetAP-2, recruiting it to SCFβTrCP and leading to its ubiquitination in vitro but lacked cell permeability.[86] Similar PROTACs were synthesized using the same IκBα phosphopeptide targeting both the AR and ER, but also lacked cell permeability.[87]
Figure 11.
PROTACs are heterobifunctional molecules that combine an E3 ligase ligand (shown on the right) with ligands for various proteins of interest (shown on the left). This recruits the E3 ligase to the protein of interest, leading to ubiquitination and degradation. Peptidic ligands have been used to target E3 ligases SCFβTrCP and VHL; small molecule ligands have been used to target MDM2 and cIAP1.
The first cell permeable PROTACs (PROTAC-4 and PROTAC-5) were developed by the incorporation of a peptide derived from HIF (ALAPYIP) that binds to VHL (after hydroxylation by PHD enzymes in situ), the substrate recognition portion of a CRL E3 ligase. A poly-D-arginine chain was also added to aid cell permeability (Figure 11). This was linked to a ligand for FKBP12 (F36V)[88] to give PROTAC-4, which was able to degrade GFP-FKBP12 (F36V) in cells efficiently (at 25 μM). PROTAC-5contained a DHT moiety, and was used to degrade an androgen receptor/GFP fusion protein (at 25 μM) in cells.[89] The same HIF peptide, or related sequences (either as the proline residue which is hydroxylated in situ, or as the hydroxyproline, and of different lengths), has been used repeatedly in numerous PROTACs to target the estrogen receptor[90], the aryl hydrocarbon receptor[91], often achieving degradation in cells without the need of the poly-D-arginine chain.
Recently we reported two peptidic PROTACs with activity dependent upon the cellular response to external stimuli. In response to stimulus by growth factors receptor tyrosine kinases dimerize and undergo transautophosphorylation on specific Tyr residues. This phosphorylation event leads to recruitment of effector/substrates containing PTP and SH2 domains. We designed TrkAPPFRS2α by combining the peptidic phosphorylation sequence from the NGF receptor, TrkA; the VHL binding fragment from previous PROTACs; and the octa-D-Arg sequence to allow for cell permeability. Treatment of NGF stimulated PC12 cells with 60 μM TrkAPPFRS2α led to roughly 90% knockdown of the effector/substrate FRS2α. No degradation was observed in cells that were not treated with NGF, or cells that were treated with the control TrkANPFRS2α.[92]
Based upon these results, a second phosphoPROTAC, ErbB2PPPI3K, was designed based off of a peptidic sequence ErbB3, which is phosphorylated by ErbB2 in response to neuregulin, leading to binding of PI3K. ErbB2PPPI3K causes neuregulin dependent degradation of PI3K and decreased activation of its downstream effector Akt. This leads to dose dependent toxicity of ErbB2PPPI3K in MCF-7 cells, whereas the control, ErbB2NPPI3K, had negligible toxicity. In a mouse xenograft model, daily treatment (10 mg/kg, i.p.) of ErbB2PPPI3K led to a 40% reduction in tumor size compared to the control ErbB2NPPI3K, the first demonstration of PROTAC activity in a mouse model.[92]
While peptidic PROTACs have shown efficacy in mice, there is a strong desire to design non-peptidic PROTACs using small molecule ligands for E3 ligases such as MDM2[93] and cIAP1 (Figure 11).[94] Such PROTACs would theoretically be more drug-like and would lead to more stable probe compounds or possibly be used as future therapeutics. We have previously found that using a Nutlin-based moiety to hijack the E3 ligase MDM2 can lead to the degradation of androgen receptor (AR),[93] albeit much less effectively than our previous work with peptidic VHL ligands.[89] Additionally, Hashimoto and coworkers have exploited bestatin esters, which recruit cellular inhibitor of apoptosis protein 1 (cIAP1) to lead to the degradation of cellular retinoic acid-binding proteins (CRABPs)[94] as well as nuclear receptors such as AR, the estrogen receptor and the retinoic acid receptor.[95] This subclass of PROTACs, termed SNIPERs (Specific and Nongenetic IAPs-dependent Protein ERasers) have been fairly successful but have numerous off-target effects due to the lack of specificity of bestatin (which had been developed as an aminopeptidase inhibitor before its binding to cIAP1 was discovered[96]). These compounds, like bestatin itself (as well as other IAP inhibitors[62]) also lead to the destabilization and degradation of cIAP1. Additionally, the reliance on hydrolytically unstable ester and oxime[97] linkages raises issues with regard to stability. Furthermore, both PROTACs (peptidic and small molecule) and SNIPERs suffer from low potency, often requiring concentrations up to 25 μM to achieve sufficient degradation. These issues highlight the need for improved E3 ligase targeting moieties to allow for more potent and drug-like PROTACs to be developed.
3.2 Hydrophobic Tagging and Ligand Mediated Degradation
In addition to PROTACs, which lead to the degradation of a target protein through the use of a direct E3 ligase ligand, there are a number of examples of ligand-mediated degradation using molecules lacking the ability to directly bind E3 ligases. One such example was discovered in our lab after the observation that the use of hexylchloride tagged ligands for HaloTag2[98] fusion proteins led to their degradation. We then optimized the chloroalkane ligands for their ability to cause degradation of the HaloTag2 fusion proteins, finding that the addition of more hydrophobic led to more potent degradation, leading us to term it hydrophobic tagging. We found the adamantaneacetamide, HyT13to be most successful ligand at inducing this degradation as itled to roughly 75% degradation of numerous proteins (GFP, luciferase, Hras1G12V, Ror2, etc.) fused with HaloTag2 (Figure 12).[99] Furthermore, HyT13 was shown to be effective in degrading HaloTag2 fusion proteins in vivo achieving knockdown of HaloTag–Smad5 zebrafish and of HaloTag-Hras1G12V in mice, leading to reduction of tumor size in a xenograft model.[99] During the course of a small molecule screen, a compound, HALTS, was discovered that stabilized HaloTag2 fusion proteins (in the absence of HyT13) through direct binding to the active site (as determined by crystallography). This stabilization, reminiscent of the Shield system described above, allows for small molecule induced degradation and stabilization of the same system at once.[100]
Figure 12.
Structures of HyT13 and HyT36 and their ability to degrade HaloTag-GFP fusion proteins at 10 μM.[101]
Due in large part to stability issues of HaloTag2, Promega has continued to optimize the HaloTag system to increase stability and decrease the propensity of aggregation of the fusion proteins. Their result was the HaloTag7 protein,[102] which contains 22 point mutations from HaloTag2. We found that HyT13was much less efficacious in inducing degradation of HaloTag7 fusion proteins, resulting in less than 20% degradation of HaloTag7-GFP. After much optimization, we were able to find that related HyT36 (Figure 12) was able to degrade more than half of HaloTag7-GFP.[101]
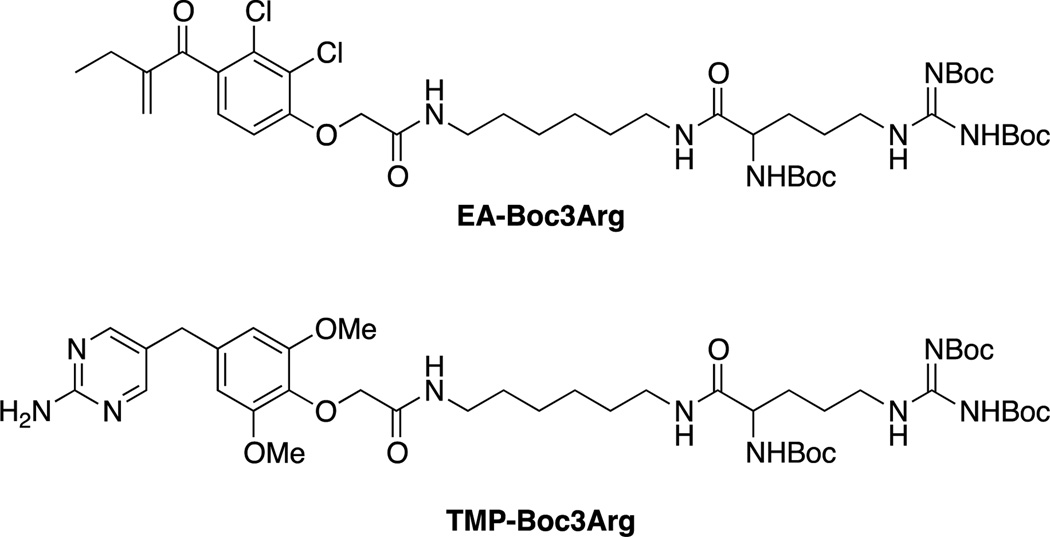
A similar system was recently reported by Hedstrom and coworkers involving the attachment of a Boc3Arg group covalent inhibitors of glutathione-S-transferase and a non-covalent inhibitor of eDHFR. Treatment with EA-Boc3Arg led to the efficient degradation of roughly 80% of GST in lysates and whole cells. The noncovalent TMP-Boc3Arg was less effective, leading to 60% degradation of eDHFR in lysates but only 30% degradation in whole cells.[103]
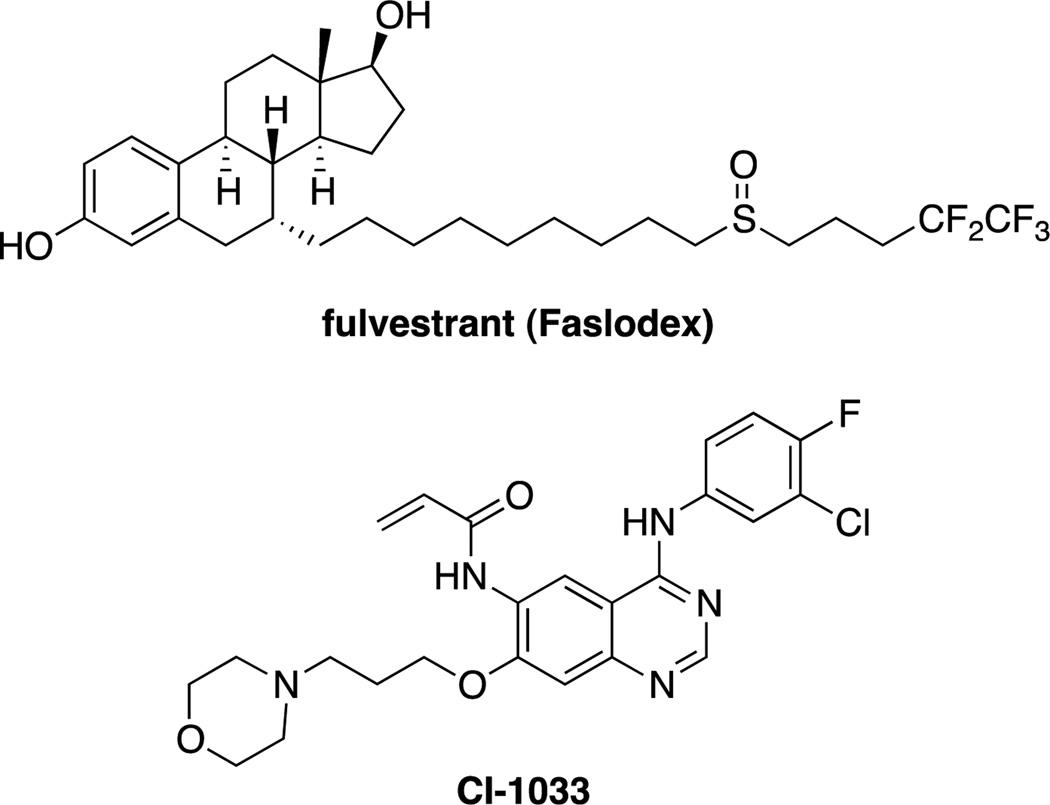
In addition to these methods that have been designed to degrade carefully constructed systems and fusion proteins, similar ligand mediated degradation has been observed serendipitously in the course of traditional medicinal chemistry programs. The most notable is fulvestrant (Figure 14), an FDA approved estrogen receptor (ER) antagonist which works by inhibiting ER dimerization and nuclear localization.[104] However, binding of fulvestrant leads to a conformational change in the ER, forming a less stable complex, leading to its down regulation.[105] Another example is CI-1033, a covalent inhibitor of ErbB2 that induces its degradation by the proteasome.[106] The authors proposed that the covalent modification of the ATP-binding pocket alters the site, and leads to its ubiquitination (and subsequent degradation) by chaperone-mediated destructive system.[106]
Figure 14.
Despite being designed as traditional antagonists or inhibitors, fulvestrant and CI-1033 were discovered to induce the degradation of the ER and ErbB2 respectively.
While these examples of ligand mediated degradation lack the clear degradation signals (i.e. ligands to directly recruit E3 ligases) of PROTACs, they share similar features and may operate by related, albeit distinct mechanisms. A common motif found in many of these degraders is a hydrophobic patch attached via a linker to the ligand, which is often covalently bound to the target. These ligands then can induce a conformational change or disrupt the binding to other members in a multi-protein complex. These non-native states of the target protein may then be recognized by the cell’s machinery to detect misfolded proteins, leading to their degradation and ubiquitination. However, the nature of this proposed mechanism, which is heavily reliant upon the targets ability to adopt a non-native state, likely may prevent rational application of this approach to novel targets.
3.3. Related Systems Requiring Genetic Manipulation
While the above systems have often targeted genetically modified targets, they all have been shown to operate on endogenous proteins as well. A variety of related methods have proven successful at leading to the targeted degradation of proteins, but require genetic manipulation to operate. A notable example involved the fusion of the proteasomal subunit Rpn10 with Fpr1 in yeast.[107] The addition of rapamycin led to chemical induced dimerization with Tor1 fusion proteins, leading to their degradation. The success of this work demonstrated that direct localization to the proteasome was sufficient to degrade some proteins in the absence of polyubiquitination. Another related methodology that lacks small molecule control, but is highly analogous to the concept of PROTACs involves the development of chimeric E3 ligases.[108] Zhou et al. were able to generate a chimeric F-box proteins based upon Cdc4p and βTrCP that contained the N-terminal domain of a viral protein, E7 that binds to the retinoblastoma protein (pRB). Expression of these complexes in yeast and mammalian cells respectively led to the knockdown of pRB.[109] This system was also used to generate a βTrCP-E-cadherin chimera that binds to and degrades mutant β-catenin that had developed resistance to its normal degradation pathway involving the APC.[110]
3.4. Geldanamycin Derivatives and Other Hsp90 Inhibitors
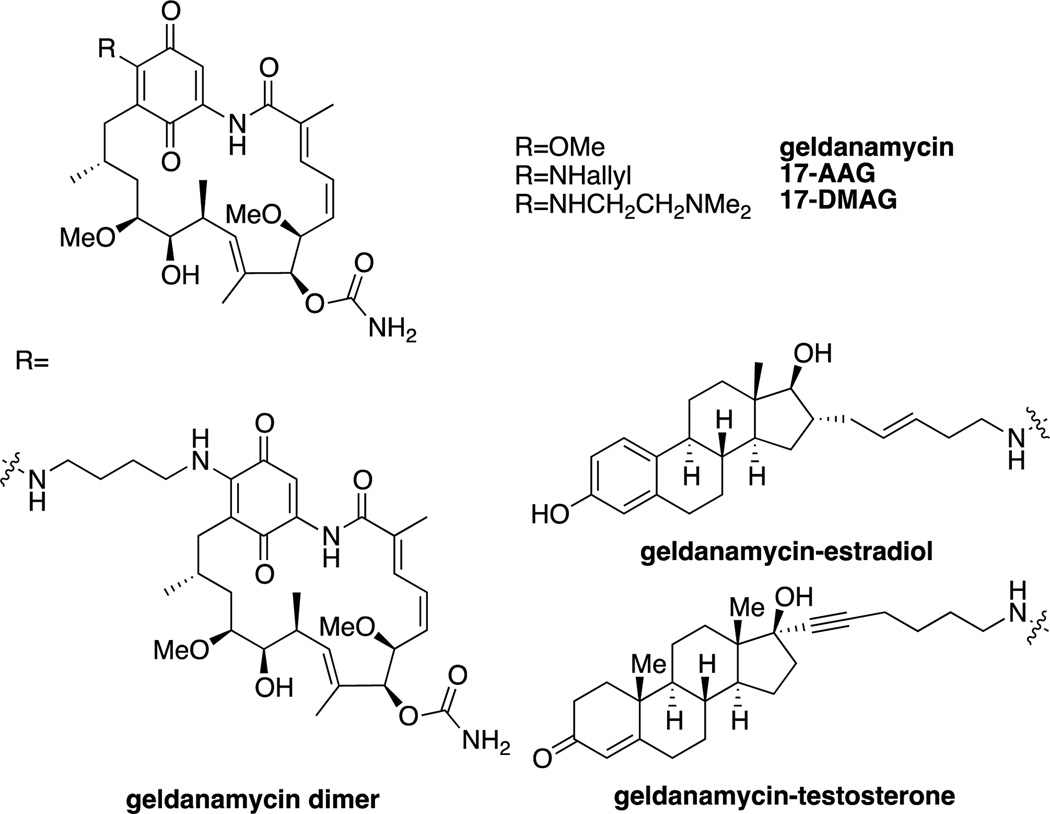
Geldanamycin, an ansamycin antibiotic, is a natural product identified based on its potent growth inhibitory effects on cells transformed with the tyrosine kinase v-src. Although it was originally believed to directly inhibit src, its effects were found in 1994 to be due to its inhibition of Hsp90, a molecular chaperone that assists in the refolding of damaged proteins,[111] leading to the degradation of src.[112] Further investigation showed that geldanamycin treatment led to the degradation of numerous other Hsp90 client proteins, such as the androgen receptor (AR), the estrogen receptor (ER), and kinases such as Raf[113] and HER. As many of these proteins are oncogenic, this has led to the study of Hsp90 inhibitors as chemotherapeutics. The development of these inhibitors has been extensively reviewed,[114] and therefore will only be briefly summarized here.
While geldanamycin itself was too toxic for clinical study, a number of derivatives involving variable substitution on the quinone, such as 17-AAG and 17-DMAG have been studied in prostate and breast cancer models due to degradation of the AR and HER2. 17-AAG progressed through phase I and phase II trials, before being discontinued.[114e] As the quinone moiety was largely responsible for the toxicity and metabolic instability of geldanamycin and its derivatives, purely synthetic Hsp90 inhibitors were developed. An early example was the purine PU3[115], which led to the development of the clinical candidate PU-H71 (administered intravenously).[116] Other examples of synthetic Hsp90 inhibitors in clinical trials include the isoxazole NVP-AUY922[117] and the prodrug SNX-5422 (PF-04929113).[118]
As geldanamycin and other Hsp90 inhibitors lack the ability to selectively degrade specific client proteins, an effort was made to develop geldanamycin hybrids that increased the specificity of the protein knockdown. The creation of a geldanamycin-estradiol hybrid was found to maintain the level of ER knockdown of geldanamycin, while reducing the knockdown of Her2, Raf-1 and IGF1R.[119] In a similar manner, in comparison to geldanamycin, a geldanamycin-testosterone hybrid led to sustained inhibition of cell growth in cells dependent upon the AR but significantly decreased growth inhibition of other cell lines that were AR independent.[120] As HER kinases are particularly sensitive to Hsp90 inhibition and require dimerization to become activated, it was hypothesized that a geldanamycin dimer would have increased selectivity for HER2 degradation. In fact, a dimer with a 4-carbon linker was found to increase selectivity for HER2 degradation over Raf and IGFR.[121]
3.5 Small Molecule Inhibitors of Deubiquitinases (DUBs)
Ubiquitin and UBL units are linked to their protein targets (or to other units to form chains) through stable peptide and isopeptide bonds; however, the formation of these linkages is readily reversible through the action of the roughly 80 human deubiquitinases (DUBs). DUBs are subdivided into 5 families, JAMMs, which are zinc metalloproteases, as well as ubiquitin specific proteases (USPs), UCHs, OTUs and Josephins, which are all cysteine proteases. The purposes of these enzymes vary, including maturation of ubiquitin and other UBLs, removal of ubiquitin chains to prevent degradation and reverse non-degradation UBL signals and the recycling of ubiquitin chains during degradation by the proteasome. The specificity of DUBs varies and can involve recognition of specific target proteins or specific types of ubiquitin linkages. Due to their wide reaching effects within the UPS as well as their relatively druggable enzymatic activity, many DUBs may prove to be attractive therapeutic targets.[122]
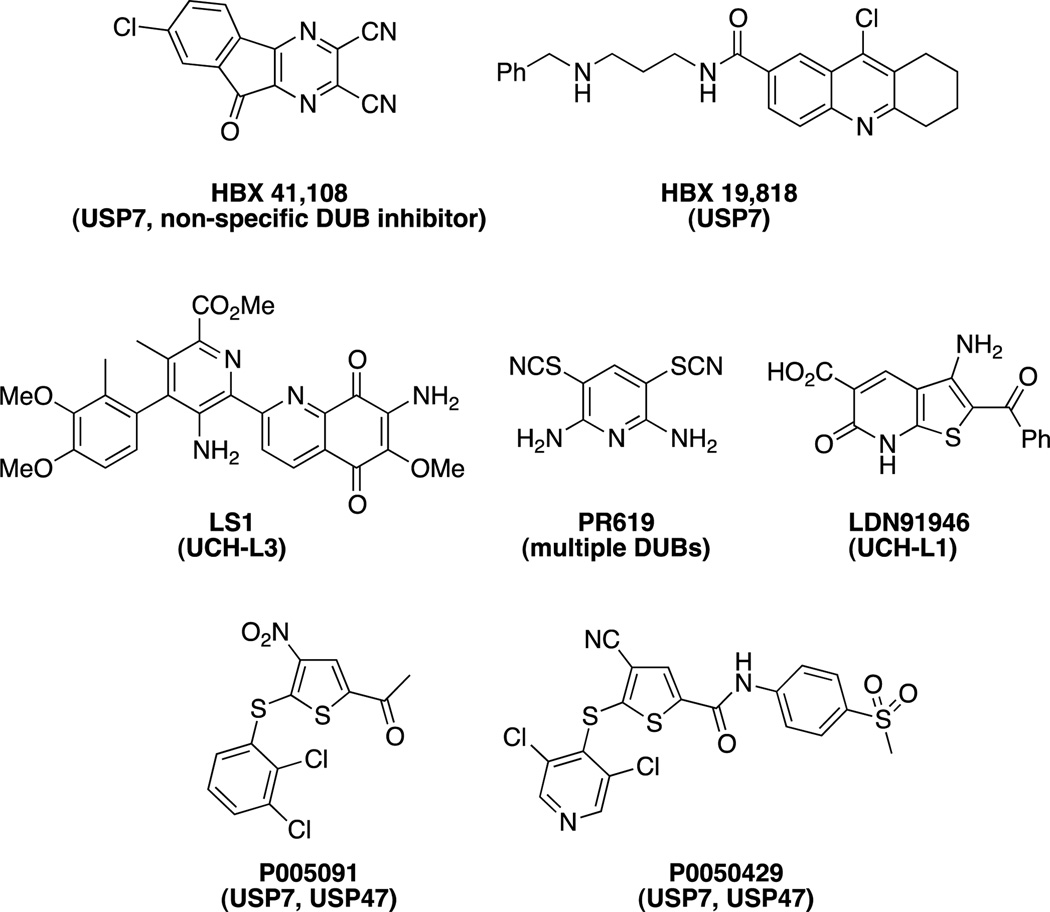
USP7 (Ubiquitin-specific protease 7) is a deubiquitinase that has activity on numerous targets, including the E3 ligase MDM2. As USP7 stabilizes MDM2, downregulating the anti-oncogenic p53, USP7 presents an attractive target for the development of therapeutics for the treatment of cancer. The first small molecule inhibitor of USP7 described was HBX 41,108[123]; however, it was later determined to be a non-specific DUB inhibitor.[124] HBX 19,818 was found to specifically inhibit USP7 (lacking activity against a panel of other USPs and other DUBs) by covalently modifying the active Cys223.[124] Additionally, P005091 and P0050429 were recently reported to have dual activity against USP7 and USP47 in the low micromolar range and leading to inhibition in cells.[125]
Various other DUB inhibitors have been reported. LS1was discovered to inhibit UCH-L3 by screening a 1000 compound library in a FRET assay using fluorescently labeled ubiquitin attached to model substrates with fluorescent quenchers.[126] PR-619 was found to inhibit a broad range of DUBs and had good selectivity over other classes of cysteine proteases.[127] LDN91946 was reported to inhibit UCH-L1, which is potentially involved in Parkinson’s disease.[128]
While inhibitors of DUB enzymes generally lead to increased degradation of their substrates, this is not always the case. The 26S proteasome has deubiquitinase activity, localized to subunits in the 19S regulatory unit. These deubiquitinases remove the polyubiquitin chains of proteins targeted for degradation by the proteasome and their removal of ubiquitin is necessary for the target proteins degradation. Therefore, inhibition of the DUB activity in the 19S regulatory particle should have similar effects to proteasome inhibitors described above. The small molecule b-AP15 was reported to inhibit the 19S associated DUBs UCHL5 (ubiquitin C-terminal hydrolase 5) and USP14, leading to accumulation of polyubiquitin. Using a mouse model, b-AP15 was then shown to inhibit tumor growth in vivo, validating 19S associated DUBs as a target for cancer treatment (Figure 18).[129] However, selective inhibition of USP14 addition with IU1was reported to have the opposite effect, leading to an increase in proteasome activity.[130] These results suggest either opposing effects of USP14 and UCHL5, or a possible redundancy where UCHL5 can compensate for inhibited USP14.
Figure 18.
Inhibitors of 19S regulatory particle associated DUBs. IU1 inhibits USP14 while b-AP15 inhibits USP14 and UCHL5.
4. Summary and Outlook
In the 10 years since the approval of bortezomib/Velcade™, the first marketed drug to target the proteasome, great strides have been made in small molecule inhibitors of the UPS; however, these results have been slow to translate into the clinic. In 2012, the second proteasome inhibitor, carfilzomib/Kyprolis™ was approved for the treatment of relapsed and refractory multiple myeloma.[34] Currently, additional next generation proteasome inhibitors are being developed, including efforts to make an orally bioavailable drug and to selectively target the immunoproteasome.[35b]
The development of inhibitors of targets in the UPS other than the proteasome have also made great strides in the past decade, although none have made it to the market yet. Targeting E3 ligases is likely to allow for more specificity in control of protein stability than proteasome inhibitors but is also more difficult to accomplish due to their lack of defined catalytic residues. In 2004, the first E3 ligase inhibitors were reported targeting MDM2[49] as well as XIAP[59]. Since then, inhibitors of MDM2 and related IAPs have advanced to clinical trials and remain targets for various forms of cancer. The past 5 years have also seen reports of inhibitors of numerous other E3 ligases including various SCF complexes. Despite the current lack of clinically approved drugs targeting E3 ligases, the successes over the past decade should put to rest the once popular idea[57] that E3 ligases could not be targeted by small molecules.
Another approach that is more specific than direct inhibition of the proteasome is the inhibition of the E1 of the Ubl, NEDD8 (NAE). As NEDDylation of cullins is important for the activity of the CRL class of E3 ligases, NAE inhibitors such as MLN4924 (which is currently in phase I/II trials) can regulate a class of E3 ligases at once.[44b, 44c]
Inducers of protein degradation remain an attractive possibility for the development of therapeutics, due to their ability to target all of the functions of a protein (such as scaffolding), as opposed to inhibitors, which only target enzymatic activity.[2] Hsp90 inhibitors are capable of inducing the degradation of a large swath of client proteins relevant to cancer. Due to the large numbers of Hsp90 inhibitors advanced to clinical trials, they remain a promising class of possible therapeutics.[114d]
More research is required to develop general methods to induce targeted degradation. While fulvestrant is capable of inducing degradation of the estrogen receptor, it remains unclear how much of its efficacy is due to its activity as an estrogen receptor antagonist or its activity as an inducer of degradation.[104a] While there are other reports of ligand-mediated degradation,[99] it is unclear which proteins make good targets for such methods and what functionality is required to induce the process. Systems such as PROTACs help solve this problem through the use of bifunctional molecules that can recruit specific E3 ligases to a target protein but they suffer from a lack of drug-likeness and still require a potent ligand for the target protein.[86, 89, 94]
Figure 9.
Shld1 is used to stabilize mutant FKBP proteins (as well as FKBP fusion proteins).[74]
Figure 13.
Structures of Boc3Arg containing degraders of GST and eDHFR.
Figure 15.
Geldanamycin and analogs.
Figure 16.
Selected synthetic Hsp90 inhibitors: PU3,[115] PU-H71[116], NVP-AUY922[117] and SNX-5422[118].
Figure 17.
Inhibitors of DUBs. The specific DUB inhibited is shown in parentheses.
Acknowledgments
We thank Dr. John Hines for helpful input on portions of the manuscript and Eric Buckley for the design of the frontispiece. The frontispiece depicts the crystal structure of the proteasome in complex with fellutamide B (PDB 3D29).[20a]
Abbreviations
- AR
Androgen receptor
- Boc3Arg
an arginine residue containing three Boc protecting groups
- CRABP
Cellular retinoic acid-binding protein
- CRL
Cullin-RING ligases, a multi subunit class of E3 ligases
- DARTS
Drug affinity responsive target stability
- DHT
Dihydrotestosterone
- DUB
Deubiquitinase
- eDHFR
E. coli dihydrofolate reductase
- ER
Estrogen receptor
- FKBP
FK506 binding protein
- Frb
FKBP12-rapamycin binding
- GFP
Green fluorescent protein
- HECT
Homologous to the E6-AP Carboxyl Terminus, a class of E3 ligases
- HIF
Hypoxia-inducible factor, a transcription factor that is ubiquitinated by VHL
- IAP
Inhibitor of apoptosis protein, a family of E3 ligases
- MDM2
Mouse double minute 2 homolog, an E3 ligase
- NAE
NEDD8 activating enzyme
- NEDD8
Neural Precursor Cell Expressed, Developmentally Down-Regulated 8, a UBL
- PPI
Protein-protein interaction
- PROTAC
Proteolysis targeting chimera, heterobifunctional molecules that hijack E3 ligases to degrade proteins
- RING
Really interesting new gene, a domain found in manyE3 ligases
- SCF
Skp, cullin, F-box protein, a subfamily of the CRL E3 ligases that use multiple F-box proteins as adaptors for their substrates
- SMAC
Second mitochondria-derived activator of caspases, a protein that inhibits IAP E3 ligase activity
- SNIPER
Specific and Nongenetic IAPs-dependent Protein
- ERaser
a subclass of PROTACs that hijack an IAP E3 ligase
- SUMO
Small ubiquitin like modifier,a UBL
- TMP
Trimethoprim, an antibiotic that binds DHFR
- UAE
Ubiquitin-activating enzyme
- Ub
Ubiquitin
- UBL
Ubiquitin-like proteins
- UPS
Ubiquitin proteasome system
- VHL
Von Hippel-Lindau tumor suppressor, the substrate recognition subunit of an E3 ligase
- YFP
Yellow fluorescent protein
Biographies

Dennis Buckley completed his undergraduate studies at the State University of New York at Geneseo in 2008. He then joined the lab of Craig Crews at Yale University, where he recently obtained his PhD in Chemistry, focusing on the development of ligands for the E3 ligase, VHL and their use as biological probes.

Craig Crews studied chemistry at the U. Virginia and received his doctorate from Harvard under the supervision of R.L. Erikson. Following a postdoctoral fellowship with S.L.Schreiber, he joined the Yale faculty in 1995.His lab’s research focus on small molecule control of intracellular protein levels culminated in the recent FDA approval of carfilzomib/Kyprolis™, the next generation proteasome inhibitor based on the lead compound YU101 from his lab.
References
- 1.a) Raina K, Crews CM. J. Biol. Chem. 2010;285:11057–11060. doi: 10.1074/jbc.R109.078105. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Schneekloth JS, Jr, Crews CM. ChemBioChem. 2005;6:40–46. doi: 10.1002/cbic.200400274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crews CM. Chem. Biol. 2010;17:551–555. doi: 10.1016/j.chembiol.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Arkin MR, Wells JA. Nat. Rev. Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]; b) Wells JA, Mcclendon CL. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 4.a) Hershko A. Angew. Chem. Int. Ed. 2005;44:5932–5943. doi: 10.1002/anie.200501724. [DOI] [PubMed] [Google Scholar]; b) Hershko A, Ciechanover A. Annu. Rev. Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- 5.Kravtsova-Ivantsiv Y, Sommer T, Ciechanover A. Angew. Chem. Int. Ed. 2013;52:192–198. doi: 10.1002/anie.201205656. [DOI] [PubMed] [Google Scholar]
- 6.a) Bedford L, Lowe J, Dick LR, Mayer RJ, Brownell JE. Nat. Rev. Drug Discov. 2011;10:29–46. doi: 10.1038/nrd3321. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cohen P, Tcherpakov M. Cell. 2010;143:686–693. doi: 10.1016/j.cell.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 7.Kisselev AF, Goldberg AL. Chem. Biol. 2001;8:739–758. doi: 10.1016/s1074-5521(01)00056-4. [DOI] [PubMed] [Google Scholar]
- 8.Zhou M, Wang R. ChemMedChem. 2013;8:694–707. doi: 10.1002/cmdc.201200560. [DOI] [PubMed] [Google Scholar]
- 9.Kim K-B, Crews CM. J. Med. chem. 2008;51:2600–2605. doi: 10.1021/jm070421s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rentsch A, Landsberg D, Brodmann T, Bülow L, Girbig A-K, Kalesse M. Angew. Chem. Int. Ed. 2013;52:5450–5488. doi: 10.1002/anie.201207900. [DOI] [PubMed] [Google Scholar]
- 11.a) Bogyo M, McMaster JS, Gaczynska M, Tortorella D, Goldberg AL, Ploegh HL. Proc. Natl. Acad. Sci. USA. 1997;94:6629–6634. doi: 10.1073/pnas.94.13.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bogyo M, Shin S, McMaster JS, Ploegh HL. Chem. Biol. 1998;5:307–320. doi: 10.1016/s1074-5521(98)90169-7. [DOI] [PubMed] [Google Scholar]; c) Geurink PP, van der Linden WA, Mirabella AC, Gallastegui N, de Bruin G, Blom AEM, Voges MJ, Mock ED, Florea BI, van der Marel GA, Driessen C, van der Stelt M, Groll M, Overkleeft HS, Kisselev AF. J. Med. chem. 2013;56:1262–1275. doi: 10.1021/jm3016987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Corey EJ, Li WD. Chem. Pharm. Bull. 1999;47:1–10. doi: 10.1248/cpb.47.1. [DOI] [PubMed] [Google Scholar]; b) Dick LR, Cruikshank AA, Grenier L, Melandri FD, Nunes SL, Stein RL. J. Biol. chem. 1996;271:7273–7276. doi: 10.1074/jbc.271.13.7273. [DOI] [PubMed] [Google Scholar]; c) Feling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR, Fenical W. Angew. Chem. Int. Ed. 2003;42:355–357. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]; d) Fenteany G, Standaert RF, Lane WS, Choi S, Corey EJ, Schreiber SL. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]; e) Gulder TAM, Moore BS. Angew. Chem. Int. Ed. 2010;49:9346–9367. doi: 10.1002/anie.201000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallastegui N, Beck P, Arciniega M, Huber R, Hillebrand S, Groll M. Angew. Chem. Int. Ed. 2012;51:247–249. doi: 10.1002/anie.201106010. [DOI] [PubMed] [Google Scholar]
- 14.Gräwert MA, Gallastegui N, Stein M, Schmidt B, Kloetzel P-M, Huber R, Groll M. Angew. Chem. Int. Ed. 2011;50:542–544. doi: 10.1002/anie.201005488. [DOI] [PubMed] [Google Scholar]
- 15.a) Löwe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]; b) Marques AJ, Palanimurugan R, Matias AC, Ramos PC, Dohmen RJ. Chem. Rev. 2009;109:1509–1536. doi: 10.1021/cr8004857. [DOI] [PubMed] [Google Scholar]
- 16.Groll M, Koguchi Y, Huber R, Kohno J. J. Mol. Biol. 2001;311:543–548. doi: 10.1006/jmbi.2001.4869. [DOI] [PubMed] [Google Scholar]
- 17.Lansdell TA, Hurchla MA, Xiang J, Hovde S, Weilbaecher KN, Henry RW, Tepe JJ. ACS Chem. Biol. 2013;8:578–587. doi: 10.1021/cb300568r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vinitsky A, Michaud C, Powers JC, Orlowski M. Biochemistry. 1992;31:9421–9428. doi: 10.1021/bi00154a014. [DOI] [PubMed] [Google Scholar]
- 19.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 20.a) Hines J, Groll M, Fahnestock M, Crews CM. Chem. Biol. 2008;15:501–512. doi: 10.1016/j.chembiol.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Schneekloth JS, Jr, Sanders JL, Hines J, Crews CM. Bioorg. Med. Chem. Lett. 2006;16:3855–3858. doi: 10.1016/j.bmcl.2006.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.a) Adams J. Oncologist. 2002;7:9–16. doi: 10.1634/theoncologist.7-1-9. [DOI] [PubMed] [Google Scholar]; b) Adams J, Behnke M, Chen S, Cruickshank AA, Dick LR, Grenier L, Klunder JM, Ma Y-T, Plamondon L, Stein RL. Bioorg. Med. Chem. Lett. 1998;8:333. doi: 10.1016/s0960-894x(98)00029-8. [DOI] [PubMed] [Google Scholar]
- 22.Groll M, Berkers CR, Ploegh HL, Ovaa H. Structure. 2006;14:451–456. doi: 10.1016/j.str.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 23.Adams J. Curr. Opin. Chem. Biol. 2002;6:493–500. doi: 10.1016/s1367-5931(02)00343-5. [DOI] [PubMed] [Google Scholar]
- 24.Sugawara K, Hatori M, Nishiyama Y, Tomita K, Kamei H, Konishi M, Oki T. J. Antibiot. 1990;43:8–18. doi: 10.7164/antibiotics.43.8. [DOI] [PubMed] [Google Scholar]
- 25.Hanada M, Sugawara K, Kaneta K, Toda S, Nishiyama Y, Tomita K, Yamamoto H, Konishi M, Oki T. J. Antibiot. 1992;45:1746–1752. doi: 10.7164/antibiotics.45.1746. [DOI] [PubMed] [Google Scholar]
- 26.Sin N, Meng L, Auth H, Crews CM. Bioorg. Med. Chem. 1998;6:1209–1217. doi: 10.1016/s0968-0896(98)00089-3. [DOI] [PubMed] [Google Scholar]
- 27.Sin N, Kim K-B, Elofsson M, Meng L, Auth H, Kwok BH, Crews CM. Bioorg. Med. Chem. Lett. 1999;9:2283–2288. doi: 10.1016/s0960-894x(99)00376-5. [DOI] [PubMed] [Google Scholar]
- 28.a) Kim K-B, Myung J, Sin N, Crews CM. Bioorg. Med. Chem. Lett. 1999;9:3335–3340. doi: 10.1016/s0960-894x(99)00612-5. [DOI] [PubMed] [Google Scholar]; b) Meng L, Kwok BH, Sin N, Crews CM. Cancer Res. 1999;59:2798–2801. [PubMed] [Google Scholar]; c) Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, Crews CM. Proc. Natl. Acad. Sci. USA. 1999;96:10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Groll M, Kim K-B, Kairies N, Huber R, Crews CM. J. Am. Chem. Soc. 2000;122:1237–1238. [Google Scholar]
- 30.Kim KB, Crews CM. Nat. Prod. Rep. 2013;30:600. doi: 10.1039/c3np20126k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elofsson M, Splittgerber U, Myung J, Mohan R, Crews CM. Chem. Biol. 1999;6:811–822. doi: 10.1016/s1074-5521(99)80128-8. [DOI] [PubMed] [Google Scholar]
- 32.a) Zhou H-J, Aujay MA, Bennett MK, Dajee M, Demo SD, Fang Y, Ho MN, Jiang J, Kirk CJ, Laidig GJ, Lewis ER, Lu Y, Muchamuel T, Parlati F, Ring ER, Shenk KD, Shields J, Shwonek PJ, Stanton T, Sun CM, Sylvain C, Woo TM, Yang J. J. Med. chem. 2009;52:3028–3038. doi: 10.1021/jm801329v. [DOI] [PubMed] [Google Scholar]; b) Demo SD, Kirk CJ, Aujay MA, Buchholz TJ, Dajee M, Ho MN, Jiang J, Laidig GJ, Lewis ER, Parlati F, Shenk KD, Smyth MS, Sun CM, Vallone MK, Woo TM, Molineaux CJ, Bennett MK. Cancer Res. 2007;67:6383–6391. doi: 10.1158/0008-5472.CAN-06-4086. [DOI] [PubMed] [Google Scholar]; c) Kuhn D, Chen Q, Voorhees P, Strader J, Shenk KD, Sun CM, Demo SD, Bennett MK, van Leeuwen FWB, Chanan-Khan A. Blood. 2007;110:3281. doi: 10.1182/blood-2007-01-065888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siegel DS, Martin T, Wang M, Vij R, Jakubowiak AJ, Lonial S, Trudel S, Kukreti V, Bahlis N, Alsina M, Chanan-Khan A, Buadi F, Reu FJ, Somlo G, Zonder J, Song K, Stewart AK, Stadtmauer E, Kunkel L, Wear S, Wong AF, Orlowski RZ, Jagannath S. Blood. 2012;120:2817–2825. doi: 10.1182/blood-2012-05-425934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson JL. Ann. Pharmacother. 2013;47:56–62. doi: 10.1345/aph.1R561. [DOI] [PubMed] [Google Scholar]
- 35.a) http://www.clinicaltrials.gov/; b) Huber EM, Groll M. Angew. Chem. Int. Ed. 2012;51:8708–8720. doi: 10.1002/anie.201201616. [DOI] [PubMed] [Google Scholar]
- 36.Huber Eva M, Basler M, Schwab R, Heinemeyer W, Kirk Christopher J, Groettrup M, Groll M. Cell. 2012;148:727–738. doi: 10.1016/j.cell.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 37.Gilardini A, Marmiroli P, Cavaletti G. Curr. Med. Chem. 2008;15:3025–3035. doi: 10.2174/092986708786848622. [DOI] [PubMed] [Google Scholar]
- 38.da Silva SR, Paiva S-L, Lukkarila JL, Gunning PT. J. Med. chem. 2013;56:2165–2177. doi: 10.1021/jm301420b. [DOI] [PubMed] [Google Scholar]
- 39.Yang Y, Kitagaki J, Dai R-M, Tsai YC, Lorick KL, Ludwig RL, Pierre SA, Jensen JP, Davydov IV, Oberoi P, Li C-CH, Kenten JH, Beutler JA, Vousden KH, Weissman AM. Cancer Res. 2007;67:9472–9481. doi: 10.1158/0008-5472.CAN-07-0568. [DOI] [PubMed] [Google Scholar]
- 40.Xu GW, Ali M, Wood TE, Wong D, Maclean N, Wang X, Gronda M, Skrtic M, Li X, Hurren R, Mao X, Venkatesan M, Zavareh RB, Ketela T, Reed JC, Rose D, Moffat J, Batey RA, Dhe-Paganon S, Schimmer AD. Blood. 2010;115:2251–2259. doi: 10.1182/blood-2009-07-231191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kitagaki J, Yang Y, Saavedra JE, Colburn NH, Keefer LK, Perantoni AO. Oncogene. 2009;28:619–624. doi: 10.1038/onc.2008.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ungermannova D, Parker SJ, Nasveschuk CG, Chapnick DA, Phillips AJ, Kuchta RD, Liu X. J. Biomol. Screen. 2012;17:421–434. doi: 10.1177/1087057111433843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ungermannova D, Parker SJ, Nasveschuk CG, Wang W, Quade B, Zhang G, Kuchta RD, Phillips AJ, Liu X. PLoS ONE. 2012;7:e29208. doi: 10.1371/journal.pone.0029208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.a) Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL, Ma J, Loke H-K, Lingaraj T, Wu D, Hamman KB, Spelman JJ, Cullis CA, Langston SP, Vyskocil S, Sells TB, Mallender WD, Visiers I, Li P, Claiborne CF, Rolfe M, Bolen JB, Dick LR. Mol. Cell. 2010;37:102–111. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]; b) Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, Cullis CA, Doucette A, Garnsey JJ, Gaulin JL, Gershman RE, Lublinsky AR, Mcdonald A, Mizutani H, Narayanan U, Olhava EJ, Peluso S, Rezaei M, Sintchak MD, Talreja T, Thomas MP, Traore T, Vyskocil S, Weatherhead GS, Yu J, Zhang J, Dick LR, Claiborne CF, Rolfe M, Bolen JB, Langston SP. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]; c) Soucy TA, Smith PG, Rolfe M. Clin. Cancer Res. 2009;15:3912–3916. doi: 10.1158/1078-0432.CCR-09-0343. [DOI] [PubMed] [Google Scholar]
- 45.Toth Julia I, Yang L, Dahl R, Petroski Matthew D. Cell Reports. 2012;1:309–316. doi: 10.1016/j.celrep.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.a) Ceccarelli DF, Tang X, Pelletier B, Orlicky S, Xie W, Plantevin V, Neculai D, Chou Y-C, Ogunjimi A, Al-Hakim A, Varelas X, Koszela J, Wasney GA, Vedadi M, Dhe-Paganon S, Cox S, Xu S, Lopez-Girona A, Mercurio F, Wrana J, Durocher D, Meloche S, Webb DR, Tyers M, Sicheri F. Cell. 2011;145:1075–1087. doi: 10.1016/j.cell.2011.05.039. [DOI] [PubMed] [Google Scholar]; b) Harper JW, King RW. Cell. 2011;145:1007–1009. doi: 10.1016/j.cell.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 47.Sanders MA, Brahemi G, Nangia-Makker P, Balan V, Morelli M, Kothayer H, Westwell AD, Shekhar MP. Mol Cancer Ther. 2013 doi: 10.1158/1535-7163.MCT-12-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim Yeong S, Nagy K, Keyser S, Schneekloth John S. Chem. Biol. 2013;20:604–613. doi: 10.1016/j.chembiol.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 50.Gonzalez-Lopez de Turiso F, Sun D, Rew Y, Bartberger MD, Beck HP, Canon J, Chen A, Chow D, Correll TL, Huang X, Julian LD, Kayser F, Lo M-C, Long AM, McMinn D, Oliner JD, Osgood T, Powers JP, Saiki AY, Schneider S, Shaffer P, Xiao S-H, Yakowec P, Yan X, Ye Q, Yu D, Zhao X, Zhou J, Medina JC, Olson SH. J. Med. chem. 2013;56:4053–4070. doi: 10.1021/jm400293z. [DOI] [PubMed] [Google Scholar]
- 51.Yuan Y, Liao Y-M, Hsueh C-T, Mirshahidi HR. J. Hematol. Oncol. 2011;4:16. doi: 10.1186/1756-8722-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kojima K, Burks JK, Arts J, Andreeff M. Mol Cancer Ther. 2010;9:2545–2557. doi: 10.1158/1535-7163.MCT-10-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.a) Vassilev LT. Trends Mol Med. 2007;13:23–31. doi: 10.1016/j.molmed.2006.11.002. [DOI] [PubMed] [Google Scholar]; b) Yang Y, Kitagaki J, Wang H, Hou D-X, Perantoni AO. Cancer Science. 2009;100:24–28. doi: 10.1111/j.1349-7006.2008.01013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding K, Lu Y, Nikolovska-Coleska Z, Qiu S, Ding Y, Gao W, Stuckey J, Krajewski K, Roller PP, Tomita Y, Parrish DA, Deschamps JR, Wang S. J. Am. Chem. Soc. 2005;127:10130–10131. doi: 10.1021/ja051147z. [DOI] [PubMed] [Google Scholar]
- 55.Azmi AS, Philip PA, Beck FWJ, Wang Z, Banerjee S, Wang S, Yang D, Sarkar FH, Mohammad RM. Oncogene. 2011;30:117–126. doi: 10.1038/onc.2010.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao Y, Liu L, Sun W, Lu J, McEachern D, Li X, Yu S, Bernard D, Ochsenbein P, Ferey V, Carry J-C, Deschamps JR, Sun D, Wang S. J. Am. Chem. Soc. 2013;135:7223–7234. doi: 10.1021/ja3125417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garber K. J. Natl. Cancer Inst. 2005;97:166–167. doi: 10.1093/jnci/97.3.166. [DOI] [PubMed] [Google Scholar]
- 58.Fulda S, Vucic D. Nat. Rev. Drug Discov. 2012;11:109–124. doi: 10.1038/nrd3627. [DOI] [PubMed] [Google Scholar]
- 59.Oost TK, Sun C, Armstrong RC, Al-Assaad A-S, Betz SF, Deckwerth TL, Ding H, Elmore SW, Meadows RP, Olejniczak ET, Oleksijew A, Oltersdorf T, Rosenberg SH, Shoemaker AR, Tomaselli KJ, Zou H, Fesik SW. J. Med. chem. 2004;47:4417–4426. doi: 10.1021/jm040037k. [DOI] [PubMed] [Google Scholar]
- 60.a) Lu J, Bai L, Sun H, Nikolovska-Coleska Z, Mceachern D, Qiu S, Miller RS, Yi H, Shangary S, Sun Y, Meagher JL, Stuckey JA, Wang S. Cancer Res. 2008;68:9384–9393. doi: 10.1158/0008-5472.CAN-08-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sun H, Nikolovska-Coleska Z, Lu J, Meagher JL, Yang C-Y, Qiu S, Tomita Y, Ueda Y, Jiang S, Krajewski K, Roller PP, Stuckey JA, Wang S. J. Am. Chem. Soc. 2007;129:15279–15294. doi: 10.1021/ja074725f. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Sun H, Nikolovska-Coleska Z, Yang C-Y, Qian D, Lu J, Qiu S, Bai L, Peng Y, Cai Q, Wang S. Acc. Chem. Res. 2008;41:1264–1277. doi: 10.1021/ar8000553. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Sun H, Nikolovska-Coleska Z, Yang C-Y, Xu L, Liu M, Tomita Y, Pan H, Yoshioka Y, Krajewski K, Roller PP, Wang S. J. Am. Chem. Soc. 2004;126:16686–16687. doi: 10.1021/ja047438+. [DOI] [PubMed] [Google Scholar]
- 61.Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P, Zobel K, Dynek JN, Elliott LO, Wallweber HJA, Flygare JA, Fairbrother WJ, Deshayes K, Dixit VM, Vucic D. Cell. 2007;131:669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 62.Cai Q, Sun H, Peng Y, Lu J, Nikolovska-Coleska Z, McEachern D, Liu L, Qiu S, Yang C-Y, Miller R, Yi H, Zhang T, Sun D, Kang S, Guo M, Leopold L, Yang D, Wang S. J. Med. chem. 2011;54:2714–2726. doi: 10.1021/jm101505d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fulda S. J. Med. chem. 2012;55:4099–4100. doi: 10.1021/jm300475b. [DOI] [PubMed] [Google Scholar]
- 64.a) Petroski MD, Deshaies RJ. Nat. Rev. Mol. Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]; b) Cardozo T, Pagano M. BMC Biochem. 2007;8:S9. doi: 10.1186/1471-2091-8-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Cardozo Timothy J, Pagano M. Nat. Rev. Mol. Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 65.a) Rico-Bautista E, Wolf Dieter A. Chem. Biol. 2012;19:1497–1498. doi: 10.1016/j.chembiol.2012.12.001. [DOI] [PubMed] [Google Scholar]; b) Wu L, Grigoryan Arsen V, Li Y, Hao B, Pagano M, Cardozo Timothy J. Chem. Biol. 2012;19:1515–1524. doi: 10.1016/j.chembiol.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chan C-H, Morrow John K, Li C-F, Gao Y, Jin G, Moten A, Stagg Loren J, Ladbury John E, Cai Z, Xu D, Logothetis Christopher J, Hung M-C, Zhang S, Lin H-K. Cell. 2013;154:556–568. doi: 10.1016/j.cell.2013.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.a) Aghajan M, Jonai N, Flick K, Fu F, Luo M, Cai X, Ouni I, Pierce N, Tang X, Lomenick B, Damoiseaux R, Hao R, Del Moral PM, Verma R, Li Y, Li C, Houk KN, Jung ME, Zheng N, Huang L, Deshaies RJ, Kaiser P, Huang J. Nat. Biotechnol. 2010;28:738–742. doi: 10.1038/nbt.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lydeard JR, Harper JW. Nat. Biotechnol. 2010;28:682–684. doi: 10.1038/nbt0710-682. [DOI] [PubMed] [Google Scholar]
- 68.Orlicky S, Tang X, Neduva V, Elowe N, Brown ED, Sicheri F, Tyers M. Nat. Biotechnol. 2010;28:733–737. doi: 10.1038/nbt.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nakajima H, Fujiwara H, Furuichi Y, Tanaka K, Shimbara N. Biochem. Biophys. Res. Commun. 2008;368:1007–1013. doi: 10.1016/j.bbrc.2008.01.166. [DOI] [PubMed] [Google Scholar]
- 70.Buckley DL, Van Molle I, Gareiss PC, Tae HS, Michel J, Noblin DJ, Jorgensen WL, Ciulli A, Crews CM. J. Am. Chem. Soc. 2012;134:4465–4468. doi: 10.1021/ja209924v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van Molle I, Thomann A, Buckley DL, So Ernest C, Lang S, Crews CM, Ciulli A. Chem. Biol. 2012;19:1300–1312. doi: 10.1016/j.chembiol.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.a) Buckley DL, Gustafson JL, Van Molle I, Roth AG, Tae HS, Gareiss PC, Jorgensen WL, Ciulli A, Crews CM. Angew. Chem. Int. Ed. 2012;51:11463–11467. doi: 10.1002/anie.201206231. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Buckley DL, Gustafson JL, Van Molle I, Roth AG, Tae HS, Gareiss PC, Jorgensen WL, Ciulli A, Crews CM. Angew. Chem. 2012;124:11630–11634. doi: 10.1002/anie.201206231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ito T, Ando H, Suzuki T, Ogura T, Hotta K, Imamura Y, Yamaguchi Y, Handa H. Science. 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- 74.Banaszynski LA, Chen L-C, Maynard-Smith LA, Ooi AGL, Wandless TJ. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Niesen FH, Berglund H, Vedadi M. Nat. Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 76.Lomenick B, Hao R, Jonai N, Chin RM, Aghajan M, Warburton S, Wang J, Wu RP, Gomez F, Loo JA, Wohlschlegel JA, Vondriska TM, Pelletier J, Herschman HR, Clardy J, Clarke CF, Huang J. Proc. Natl. Acad. Sci. USA. 2009;106:21984–21989. doi: 10.1073/pnas.0910040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Banaszynski LA, Wandless TJ. Chem. Biol. 2006;13:11–21. doi: 10.1016/j.chembiol.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 78.a) Bayle JH, Grimley JS, Stankunas K, Gestwicki JE, Wandless TJ, Crabtree GR. Chem. Biol. 2006;13:99–107. doi: 10.1016/j.chembiol.2005.10.017. [DOI] [PubMed] [Google Scholar]; b) Stankunas K, Bayle JH, Gestwicki JE, Lin Y-M, Wandless TJ, Crabtree GR. Mol. Cell. 2003;12:1615–1624. doi: 10.1016/s1097-2765(03)00491-x. [DOI] [PubMed] [Google Scholar]
- 79.Banaszynski LA, Sellmyer MA, Contag CH, Wandless TJ, Thorne SH. Nature Med. 2008;14:1123–1127. doi: 10.1038/nm.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.a) Lau HD, Yaegashi J, Zaro BW, Pratt MR. Angew. Chem. Int. Ed. 2010;49:8458–8461. doi: 10.1002/anie.201003073. [DOI] [PubMed] [Google Scholar]; b) Pratt MR, Schwartz EC, Muir TW. Proc. Natl. Acad. Sci. USA. 2007;104:11209–11214. doi: 10.1073/pnas.0700816104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bonger KM, Chen L-C, Liu CW, Wandless TJ. Nat. Chem. Biol. 2011;7:531–537. doi: 10.1038/nchembio.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Al-Bassam S, Xu M, Wandless Thomas J, Arnold Don B. Cell Reports. 2012;2:89–100. doi: 10.1016/j.celrep.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shoulders MD, Ryno LM, Cooley CB, Kelly JW, Wiseman RL. J. Am. Chem. Soc. 2013 doi: 10.1021/ja402756p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Feldman BJ, Feldman D. Nat. Rev. Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 85.Corson TW, Aberle NS, Crews CM. ACS Chem. Biol. 2008;3:677–692. doi: 10.1021/cb8001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sakamoto K, Kim K-B, Kumagai A, Mercurio F, Crews CM, Deshaies RJ. Proc. Natl. Acad. Sci. USA. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sakamoto K, Kim K-B, Verma R, Ransick A, Stein B, Crews CM, Deshaies RJ. Molecular & Cellular Proteomics. 2003;2:1350. doi: 10.1074/mcp.T300009-MCP200. [DOI] [PubMed] [Google Scholar]
- 88.Yang W, Rozamus LW, Narula S, Rollins CT, Yuan R, Andrade LJ, Ram MK, Phillips TB, van Schravendijk MR, Dalgarno D, Clackson T, Holt DA. J. Med. chem. 2000;43:1135–1142. doi: 10.1021/jm9904396. [DOI] [PubMed] [Google Scholar]
- 89.Schneekloth JS, Jr, Fonseca FN, Koldobskiy M, Mandal AK, Deshaies RJ, Sakamoto K, Crews CM. J. Am. Chem. Soc. 2004;126:3748–3754. doi: 10.1021/ja039025z. [DOI] [PubMed] [Google Scholar]
- 90.Bargagna-Mohan P, Baek S-H, Lee H, Kim K-B, Mohan R. Bioorg. Med. Chem. Lett. 2005;15:2724–2727. doi: 10.1016/j.bmcl.2005.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.a) Lee H, Puppala D, Choi E-Y, Swanson H, Kim K-B. ChemBioChem. 2007;8:2058–2062. doi: 10.1002/cbic.200700438. [DOI] [PubMed] [Google Scholar]; b) Puppala D, Lee H, Kim KB, Swanson HI. Mol. Pharmacol. 2008;73:1064–1071. doi: 10.1124/mol.107.040840. [DOI] [PubMed] [Google Scholar]
- 92.Hines J, Gough JD, Corson TW, Crews CM. Proc. Natl. Acad. Sci. USA. 2013;110:8942–8947. doi: 10.1073/pnas.1217206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schneekloth AR, Pucheault M, Tae HS, Crews CM. Bioorg. Med. Chem. Lett. 2008;18:5904–5908. doi: 10.1016/j.bmcl.2008.07.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Itoh Y, Ishikawa M, Naito M, Hashimoto Y. J. Am. Chem. Soc. 2010;132:5820–5826. doi: 10.1021/ja100691p. [DOI] [PubMed] [Google Scholar]
- 95.Itoh Y, Kitaguchi R, Ishikawa M, Naito M, Hashimoto Y. Bioorg. Med. Chem. 2011;19:6768–6778. doi: 10.1016/j.bmc.2011.09.041. [DOI] [PubMed] [Google Scholar]
- 96.a) Sato S, Tetsuhashi M, Sekine K, Miyachi H, Naito M, Hashimoto Y, Aoyama H. Bioorg. Med. Chem. 2008;16:4685–4698. doi: 10.1016/j.bmc.2008.02.024. [DOI] [PubMed] [Google Scholar]; b) Sekine K, Takubo K, Kikuchi R, Nishimoto M, Kitagawa M, Abe F, Nishikawa K, Tsuruo T, Naito M. J. Biol. chem. 2008;283:8961–8968. doi: 10.1074/jbc.M709525200. [DOI] [PubMed] [Google Scholar]
- 97.Kalia J, Raines RT. Angew. Chem. Int. Ed. 2008;47:7523–7526. doi: 10.1002/anie.200802651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV. ACS Chem. Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 99.Neklesa TK, Tae HS, Schneekloth AR, Stulberg MJ, Corson TW, Sundberg TB, Raina K, Holley SA, Crews CM. Nat. Chem. Biol. 2011;7:538–543. doi: 10.1038/nchembio.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Neklesa TK, et al. ACS Chem. Biol. Article ASAP. 2013 [Google Scholar]
- 101.Tae HS, Sundberg TB, Neklesa TK, Noblin DJ, Gustafson JL, Roth AG, Raina K, Crews CM. ChemBioChem. 2012;13:538–541. doi: 10.1002/cbic.201100793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ohana RF, Encell LP, Zhao K, Simpson D, Slater MR, Urh M, Wood KV. Protein Expression and Purification. 2009;68:110–120. doi: 10.1016/j.pep.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 103.Long MJC, Gollapalli DR, Hedstrom L. Chem. Biol. 2012;19:629–637. doi: 10.1016/j.chembiol.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.a) Howell A. Crit. Rev. Oncol. Hematol. 2006;57:265–273. doi: 10.1016/j.critrevonc.2005.08.001. [DOI] [PubMed] [Google Scholar]; b) Osborne CK, Wakeling A, Nicholson RI. Br. J. Cancer. 2004;90:S2–S6. doi: 10.1038/sj.bjc.6601629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.a) Wijayaratne AL, McDonnell DP. J. Biol. chem. 2001;276:35684–35692. doi: 10.1074/jbc.M101097200. [DOI] [PubMed] [Google Scholar]; b) Wittmann BM, Sherk A, McDonnell DP. Cancer Res. 2007;67:9549–9560. doi: 10.1158/0008-5472.CAN-07-1590. [DOI] [PubMed] [Google Scholar]
- 106.Citri A, Alroy I, Lavi S, Rubin C, Xu W, Grammatikakis N, Patterson C, Neckers L, Fry DW, Yarden Y. EMBO J. 2002;21:2407–2417. doi: 10.1093/emboj/21.10.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Janse DM, Crosas B, Finley D, Church GM. J. Biol. chem. 2004;279:21415–21420. doi: 10.1074/jbc.M402954200. [DOI] [PubMed] [Google Scholar]
- 108.Zhou P. Curr. Opin. Chem. Biol. 2005;9:51–55. doi: 10.1016/j.cbpa.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 109.a) Zhang J, Zheng N, Zhou P. Proc. Natl. Acad. Sci. USA. 2003;100:14127–14132. doi: 10.1073/pnas.2233012100. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhou P, Bogacki R, McReynolds L, Howley PM. Mol. Cell. 2000;6:751–756. doi: 10.1016/s1097-2765(00)00074-5. [DOI] [PubMed] [Google Scholar]
- 110.Cong F, Zhang J, Pao W, Zhou P, Varmus H. BMC Mol. Biol. 2003;4:10. doi: 10.1186/1471-2199-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schneider C, Sepp-Lorenzino L, Nimmesgern E, Ouerfelli O, Danishefsky S, Rosen N, Hartl FU. Proc. Natl. Acad. Sci. USA. 1996;93:14536–14541. doi: 10.1073/pnas.93.25.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Proc. Natl. Acad. Sci. USA. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stancato LF, Silverstein AM, Owens-Grillo JK, Chow Y, Jove R, Pratt WB. J. Biol. chem. 1997;272:4013–4020. doi: 10.1074/jbc.272.7.4013. [DOI] [PubMed] [Google Scholar]
- 114.a) Taldone T, Gozman A, Maharaj R, Chiosis G. Curr. Opin. Pharmacol. 2008;8:370–374. doi: 10.1016/j.coph.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sgobba M, Rastelli G. ChemMedChem. 2009;4:1399–1409. doi: 10.1002/cmdc.200900256. [DOI] [PubMed] [Google Scholar]; c) Taldone T, Sun W, Chiosis G. Bioorg. Med. Chem. 2009;17:2225–2235. doi: 10.1016/j.bmc.2008.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Biamonte MA, Van de Water R, Arndt JW, Scannevin RH, Perret D, Lee W-C. J. Med. chem. 2010;53:3–17. doi: 10.1021/jm9004708. [DOI] [PubMed] [Google Scholar]; e) Neckers L, Workman P. Clin. Cancer Res. 2012;18:64–76. doi: 10.1158/1078-0432.CCR-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chiosis G, Timaul MN, Lucas B, Munster PN, Zheng FF, Sepp-Lorenzino L, Rosen N. Chem. Biol. 2001;8:289–299. doi: 10.1016/s1074-5521(01)00015-1. [DOI] [PubMed] [Google Scholar]
- 116.a) Caldas-Lopes E, Cerchietti L, Ahn JH, Clement CC, Robles AI, Rodina A, Moulick K, Taldone T, Gozman A, Guo Y, Wu N, De Stanchina E, White J, Gross SS, Ma Y, Varticovski L, Melnick A, Chiosis G. Proc. Natl. Acad. Sci. USA. 2009;106:8368–8373. doi: 10.1073/pnas.0903392106. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Immormino RM, Kang Y, Chiosis G, Gewirth DT. J. Med. chem. 2006;49:4953–4960. doi: 10.1021/jm060297x. [DOI] [PubMed] [Google Scholar]
- 117.Eccles SA, Massey A, Raynaud FI, Sharp SY, Box G, Valenti M, Patterson L, de Haven Brandon A, Gowan S, Boxall F, Aherne W, Rowlands M, Hayes A, Martins V, Urban F, Boxall K, Prodromou C, Pearl L, James K, Matthews TP, Cheung K-M, Kalusa A, Jones K, McDonald E, Barril X, Brough PA, Cansfield JE, Dymock B, Drysdale MJ, Finch H, Howes R, Hubbard RE, Surgenor A, Webb P, Wood M, Wright L, Workman P. Cancer Res. 2008;68:2850–2860. doi: 10.1158/0008-5472.CAN-07-5256. [DOI] [PubMed] [Google Scholar]
- 118.Fadden P, Huang KH, Veal JM, Steed PM, Barabasz AF, Foley B, Hu M, Partridge JM, Rice J, Scott A, Dubois LG, Freed TA, Silinski MAR, Barta TE, Hughes PF, Ommen A, Ma W, Smith ED, Spangenberg AW, Eaves J, Hanson GJ, Hinkley L, Jenks M, Lewis M, Otto J, Pronk GJ, Verleysen K, Haystead TA, Hall SE. Chem. Biol. 2010;17:686–694. doi: 10.1016/j.chembiol.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 119.Kuduk SD, Zheng FF, Sepp-Lorenzino L, Rosen N, Danishefsky SJ. Bioorg. Med. Chem. Lett. 1999;9:1233. doi: 10.1016/s0960-894x(99)00185-7. [DOI] [PubMed] [Google Scholar]
- 120.Kuduk SD, Harris CR, Zheng FF, Sepp-Lorenzino L, Ouerfelli Q, Rosen N, Danishefsky SJ. Bioorg. Med. Chem. Lett. 2000;10:1303. doi: 10.1016/s0960-894x(00)00208-0. [DOI] [PubMed] [Google Scholar]
- 121.Zheng FF, Kuduk SD, Chiosis G, Münster PN, Sepp-Lorenzino L, Danishefsky SJ, Rosen N. Cancer Res. 2000;60:2090–2094. [PubMed] [Google Scholar]
- 122.Komander D, Clague MJ, Urbé S. Nat. Rev. Mol. Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 123.Colland F, Formstecher E, Jacq X, Reverdy C, Planquette C, Conrath S, Trouplin V, Bianchi J, Aushev VN, Camonis J, Calabrese A, Borg-Capra C, Sippl W, Collura V, Boissy G, Rain J-C, Guedat P, Delansorne R, Daviet L. Mol Cancer Ther. 2009;8:2286–2295. doi: 10.1158/1535-7163.MCT-09-0097. [DOI] [PubMed] [Google Scholar]
- 124.Reverdy C, Conrath S, Lopez R, Planquette C, Atmanene C, Collura V, Harpon J, Battaglia V, Vivat V, Sippl W, Colland F. Chem. Biol. 2012;19:467–477. doi: 10.1016/j.chembiol.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 125.Weinstock J, Wu J, Cao P, Kingsbury WD, McDermott JL, Kodrasov MP, McKelvey DM, Kumar KGS, Goldenberg SJ, Mattern MR, Nicholson B. ACS Med. Chem. Lett. 2012;3:789. doi: 10.1021/ml200276j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ohayon S, Spasser L, Aharoni A, Brik A. J. Am. Chem. Soc. 2012;134:3281–3289. doi: 10.1021/ja2116712. [DOI] [PubMed] [Google Scholar]
- 127.Altun M, Kramer HB, Willems LI, McDermott JL, Leach CA, Goldenberg SJ, Kumar KGS, Konietzny R, Fischer R, Kogan E, Mackeen MM, Mcgouran J, Khoronenkova SV, Parsons JL, Dianov GL, Nicholson B, Kessler BM. Chem. Biol. 2011;18:1401–1412. doi: 10.1016/j.chembiol.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 128.Mermerian AH, Case A, Stein RL, Cuny GD. Bioorg. Med. Chem. Lett. 2007;17:3729–2732. doi: 10.1016/j.bmcl.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 129.D'arcy P, Brnjic S, Olofsson MH, Fryknäs M, Lindsten K, De Cesare M, Perego P, Sadeghi B, Hassan M, Larsson R, Linder S. Nat. Med. 2011;17:1636–1640. doi: 10.1038/nm.2536. [DOI] [PubMed] [Google Scholar]
- 130.Lee B-H, Lee MJ, Park S, Oh D-C, Elsasser S, Chen P-C, Gartner C, Dimova N, Hanna J, Gygi SP, Wilson SM, King RW, Finley D. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]