Abstract
HIV isolates from South Africa are predominantly subtype C. Sporadic isolation of non-C strains has been reported mainly in cosmopolitan cities. HIV isolate j51 was recovered from a rural South African heterosexual female aged 51 years. Near full length amplification of the genome was attempted using PCR with primers targeting overlapping segments of the HIV genome. Analysis of 5593 bp (gag to vpu) at a bootstrap value greater than 70% found that all but the vpu gene was HIV-1 subtype A1. The vpu gene was assigned HIV-1 subtype C. The recombination breaking point was estimated at position 6035+/− 15 bp with reference to the beginning of the HXB2 reference strain. Isolate j51 revealed a unique genome constellation to previously reported recombinant strains with parental A/C backbones from South Africa though a common recombination with subtype C within the vpu gene. Identification of recombinant strains supports continued surveillance of HIV genetic diversity.
Human immunodeficiency virus type 1 (HIV-1) and HIV type 2 (HIV-2) are recognized as the etiology of acquired immunodeficiency syndrome (AIDS). Genetic diversity is the hallmark of HIV infection. HIV-1 group M virus strains are responsible for the global HIV epidemic.1 Globally, different subtypes have previously been shown to dominate certain geographic areas.2 The implications of genetic diversity within HIV strains have been attributed to numerous aspects surrounding the infection including differences in the rate of transmission, pathogenesis, immune response and escape, diagnosis and viral load measurement, response to combined antiretroviral therapy (cART), and drug resistance as well as challenges in vaccine development.3
In Southern Africa, HIV-1 subtype C is the predominant strain in the epidemic,2,4,5 although sporadic identification of non-subtype C strains has been reported.6–8 Although studies have indicated that subtype differences may not have an effect on the response to cART, particularly in HIV-1 group M strains,9,10 recombinant strains may complicate the course of HIV infection.11 Studies have implicated globalization and the movement of people from different geographic areas in the increasing rise in recombinant HIV strains.2,12
In South Africa, HIV-1 subtype C has been reported to be the predominant strain.4,8,13 First reports of non-subtype C strains in South Africa originated from the highly cosmopolitan global tourist city of Cape Town14,15 in men who have sex with men (MSM). Since then, there has been continued identification of non-subtype C strains mainly in studies on HIV drug resistance.8,16–18 Monitoring the genetic diversity of circulating HIV strains has been encouraged to generate information on patient management and even vaccine strategies.3 Recently, Palm et al.11 demonstrated increased risk of AIDS and AIDS-related death in individuals infected with recombinant strain HIV-1 A3/CRF02_AG. The recombinant virus was found to be more virulent causing AIDS and AIDS-related death between 5.0 and 8.0 years from the time of seroconversion.11 This study describes the first unique near full length HIV-1 recombinant strain isolated from a rural South African heterosexual female who was referred to the Dr. George Mukhari Academic Hospital (DGMAH) in Pretoria.
Ethical approval was granted by the University of Limpopo Research and Ethics Committee (MREC/P/37/2010). The study patient was a rural South African heterosexual female [residing in Lephalale (Ellisras), located some 210 km from Polokwane city in Limpopo Province] who was referred to DGMAH in Pretoria. At the time of sample collection, the patient was 51 years of age and had an HIV viral load of log10 6.0 copies/ml. The revelation of a rare subtype in the region prompted near full-length genome analysis of this HIV strain.
Total nucleic acid was extracted from plasma using the high pure nucleic acid extraction kit following the manufacturer's instructions (Roche Diagnostics, Mannheim, Germany).
Near full length HIV genome analysis was conducted in four overlapping fragments spanning the beginning of the gag gene to the end of the vpu gene. The gag, pol [protease (PR) and reverse transcriptase (RT)], and accessory genes (vif, vpr, tat, rev, and vpu) were amplified as single targets as previously described.4 For the integrase gene, primers INFORI (5′-GGA ATC ATT CAA GCA CAA CCA GA-3′) and INREV-I (5′-TCT CCT GTA TGC AGA CCC CAA TAT-3′) for the first round polymerase chain reaction (PCR) and primers V1F (5′-GAA AAG GGG ATC CTC TAT CTG GCA TGG GTA CCA-3′) and V2R (5′-AGC CGT CGA TAG TGG GAT GTG TAC TTC TGA-3′) for the second round PCR were used. The amplification cycles were as described for the amplification of the pol (PR-RT) gene fragment after adjusting the annealing temperature to 54°C for both nested PCR cycles.4
PCR products were sequenced with the ABI 3500XL (Life Technologies, Carlsbad, CA) using gene-specific primers. The HIV pol gene sequences were initially analyzed for genotypic drug resistance-associated mutation and HIV subtype using the online Stanford University HIV drug resistance database interactive tool (http://sierra2.stanford.edu/sierra/servlet/JSierra?action=sequenceInput). The gene positions as assigned by the HIV sequence locator tool (http://hiv.lanl.gov/content/sequence/LOCATE/locate.html) were used to defragment the genome of the suspected recombinant strain into individual HIV genes. The HIV subtypes were further confirmed using jumping profile Hidden Markov Model (jpHMM) analysis (http://jphmm.gobics.de/), the REGA HIV-1 subtyping tool (www.bioafrica.net/subtypetool/html/), and neighbor joining phylogenetic inference in the MEGA 5 software package using reference sequences from GenBank. Previously reported HIV-1 stains with subtypes A and C backbone were downloaded from GenBank and their genome constellations were compared to the isolate in this study. The downloaded sequences were from isolates 98ZADU178,19 TV239,16 and 8519.18
Initial phylogenetic assessment of partial pol (PR-RT) gene sequences isolated from patients at DGMAH indicated the patient sample (isolate j51) as HIV-1 subtype A (Fig. 1a). The sample was assigned 93.6% and 93.5% similarity to HIV-1 subtype A on the PR and RT genes, respectively, by the Stanford HIV drug resistance analysis tool. A phylogenetic tree constructed using PAUP following an HKY85 substitution model with gamma distribution on CRP confirmed the sample as an HIV-1 subtype A1 on the pol gene (data not shown).
FIG. 1.
Phylogenetic analysis of the pol and vpu gene sequences. (A) j51 pol sequence clustered with HIV-1 subtype A reference sequences. (B) j51 vpu clustered with HIV-1 subtype C reference sequences. Color images available online at www.liebertpub.com/aid
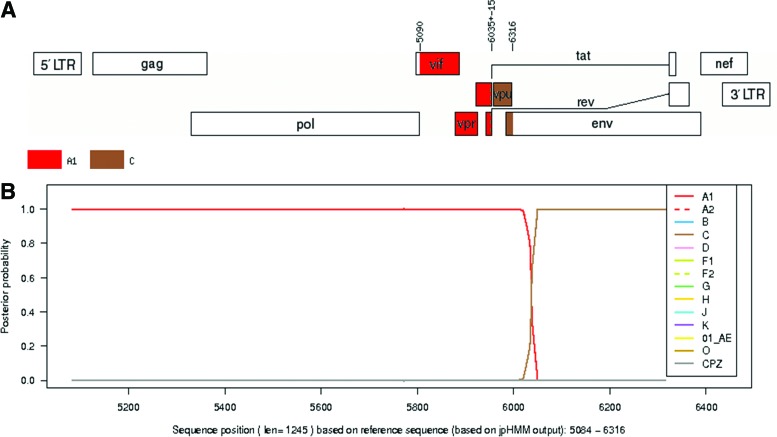
The fragmented overlapping amplification yielded a 5593-bp genomic sequence corresponding to position 723 to 6316 with reference to the start of reference sequence HXB2 (Fig. 2A and B). The genomic sequences were defragmented to individual gene sequences with the aid of the sequence locator tool on the online HIV sequence database (Table 1). Analysis by REGA, jpHMM, and neighbor joining phylogenetic inference revealed that the gag, protease, reverse transcriptase and RNase, integrase, vif, vpr, tat, and rev genes were HIV-1 subtype A1 while the vpu gene was assigned to HIV-1 subtype C (Table 1 and Fig.1b). Approximate recombination breaking point analysis estimated that the recombination activity between the parental A1 strain and C strain occurred around position 6035±15 bp with reference to the start of the HXB2 reference sequence (Fig. 3A). The vpu gene was assigned to HIV-1 subtype C with a posterior probability of 1.0 on jpHMM (Fig. 3B). Bootscan analysis confirmed the recombination between parental HIV-1 subtype A1 and C with bootstrap support greater than 70% (Fig. 4).
FIG. 2.
Sequence analysis of combined gene fragments from the suspected recombinant strain by REGA. (A) Sequence location from the start of HXB2 genome 723→6316 (shown as a red bar in the map between reading frames 1 and 2). (B) The approximate recombination pattern without bootstrap confidence showing an A/C recombination involving the vpu gene. Color images available online at www.liebertpub.com/aid
Table 1.
Summary of Recombinant Sequence HIV Subtype Analysis
| HIV subtyping | ||||
|---|---|---|---|---|
| HIV gene | Positiona | REGA | jpHMM | Phylogenetics |
| Gag | 790–2292 | A1 | A1 | A1 |
| Protease | 2253–2549 | A1 | A1 | A1 |
| Reverse transcriptase | 2550–3869 | A1 | A1 | |
| RNase | 3870–4229 | A1 | A1 | A1 |
| Integrase | 4230–5096 | A1 | A1 | |
| Vif | 5041–5619 | A1 | A1 | A1 |
| Vpr | 5559–5850 | A1 | A1 | |
| Tat | 5831–6045 | A1 | A1 | |
| Rev | 5970–6045 | A1 | A1 | |
| Vpu | 6062–6310 | N/A | C | C |
Nucleotide position relative to HXB2 genome start.
FIG. 3.
Analysis of the accessory genes genome fragment by jpHMM. (A) Fragment positions 5090 to 6035 assigned subtype A1 and positions 6036 to 6316 assigned subtype C. Genome map based on HXB2 numbering showing the estimated recombination breaking point at position 6035±15. (B) Posterior probabilities of the subtypes at each sequence position calculated by jpHMM. Color images available online at www.liebertpub.com/aid
FIG. 4.
Recombination analysis of the amplified 5593-bp genome fragment. Bootscan analysis at greater than 70% shows recombination between HIV subtype A1 and C. Color images available online at www.liebertpub.com/aid
Comparison with previously reported recombinant strains with a parental HIV-1 subtype A and C backbone revealed that the genome constellation of the study isolate j51 was unique. Isolate 98ZADU178 had a subtype C constellation within the gag, pol, vif, and vpr genes with the tat and rev genes revealing a subtype A constellation followed by subtype C in the vpu gene. Isolate TV239 showed the HIV-1 subtype C genome only within the vpu gene in comparison to the same amplified genome segments as isolate j51. Isolate 8519 revealed a recombination pattern consisting of HIV-1 subtype A and C with breaking points within the gag, pol, and vpu genes (Fig. 5).
FIG. 5.
Approximate recombination pattern analysis using the REGA HIV-1 subtyping tool without bootstrap confidence. (i) Genome constellation of study isolate j51 showing recombination between HIV-1 subtype A and C at the vpu gene. (ii) Genome constellation of isolate 8519 showing recombination between HIV subtype A and C within the gag, pol, and vpu genes. Isolate 8519 was recovered from a patient from the same province as the study isolate j51. (iii) Genome constellation of isolate TV 239 showing recombination activity between HIV-1 subtype C and A at the vpu gene. (iv) Genome constellation of isolate 98ZADU178 showing recombination between HIV-1 subtype C and A with reference to the same amplified genome region as study isolate j51. Analysis of the previously reported recombinant strains from parental HIV-1 subtype A and C reveals isolate j51 as a unique virus. Color images available online at www.liebertpub.com/aid
There were no drug resistance-associated mutations on the protease, reverse transcriptase, or integrase genes. The sequences revealed other mutations that do not influence drug susceptibility (data not shown).
This study describes the genetic constellation of a unique HIV-1 recombinant isolate from a heterosexual female from rural South African who was referred to a tertiary hospital. To date, isolation of HIV-1 recombinant and or non-subtype C strains has mainly been reported in cosmopolitan urban cities.8,16 In investigating the importance of HIV-1 subtype differences in drug resistance, Lessells et al.10 concluded that although there is no compelling evidence that the HIV-1 subtype needs to be considered in choosing ART regimens for first-line or second-line therapy, subtype differences may play a role in assessing the response to treatment and surveillance for the transmission of resistance.
In South Africa, HIV subtype reports indicate an almost 100% HIV-1 subtype C dominance.2,4,5,18 However, there have been rare reports of pure HIV-1 non-C and recombinant strains.8,16–18 In drug resistance analysis of the sequences in an HIV genetic diversity study, the isolate from patient j51 was reported to have no drug resistance-associated mutations, but was classified as HIV-1 subtype A1 on both the PR and RT regions. The recombination event was estimated to have occurred around sequence position 6035±15 bp with reference to the beginning of the HIV HBX2 strain (Fig. 3A). The individual genes were then defragmented with the aid of the HIV sequence locator tool on the HIV sequence database. Analysis of the individual gene fragments showed that the vif, vpr, tat, and rev genes were HIV-1 subtype A1 (Table 1). The vpu gene was assigned to HIV-1 subtype C with a posterior probability of 1.0 on jpHMM (Fig. 3B).
Iweriebor et al.18 described an A1/C strain isolated from a patient from Belabela in Limpopo, about 200 km from Lephalale (Ellisras) where the study patient resides. The isolate had six recombination breaking points while the isolate from the patient in this study had 1 recombination breaking point within the amplified genome region (Fig. 5). Comparison with other isolates reported to have an HIV-1 subtype A/C backbone from South Africa16,18,19 revealed that the study isolate was a unique virus. Nonetheless, all the South African A/C isolates revealed a common recombination with HIV-1 subtype C within the vpu gene (Fig. 5). The epidemiological implications of this strain isolation in this region is not known.
Movement of people and interaction with foreign nationals coming from regions dominated by different HIV subtypes have been noted in the emergence of uncommon HIV subtypes and recombinant strains.2,12 The travel history and other possible contributing factors that could expose the patient to non-C strains could not be ascertained from the available patient's records. Furthermore, this study did not establish whether the infection was of a recombinant strain or whether the patient was coinfected with pure HIV subtypes C and A parental strains that recombined in this patient. Nonetheless, since population-based sequencing was used, the recombinant strain appeared to be the dominant isolate.
Although Lessells et al.10 alluded to the fact that there is insufficient evidence to contraindicate the HIV-1 group M subtype difference in the choice of an ART regimen, it is important to note the complications that may arise due to subtype recombination. Palm et al.11 revealed an increased risk of AIDS and AIDS-related deaths in patients infected with an HIV recombinant A3/02 strain rather than their parental strains. This emphasizes the need for continual surveillance of HIV genetic diversity to aid in management, diagnosis, and prevention strategies.
In conclusion, the isolation of a unique recombinant HIV strain in a predominantly HIV-1 subtype C region is indicative of the spread of new HIV strains into rural South Africa. The implications of infection with recombinant HIV strains are poorly understood, emphasizing the need for continued surveillance of circulating strains in endemic regions.
Sequence Data
The genomic sequence of the study isolate has been deposited in GenBank and assigned accession number KM272944.
South Africa sequences downloaded for comparison with the study isolate were retrieved from GenBank using accession numbers AF411965.1, FJ647147.1, and GU201611.1.
Acknowledgments
This project was supported by grants from the Poliomyelitis Research Foundation, National Research Foundation, and National Health Laboratory Service Research Trust (all from South Africa). A.M. Musyoki was supported by bursaries from PRF, NRF, and the University of Limpopo. We thank Ms. Edina Amponsah-Dacosta for proofreading the manuscript.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.www.hiv.lanl.gov accessed June2014
- 2.Hemelaar J, Gouws E, Ghys PD, Osmanov S, and the WHO-UNAIDS Network for HIV Isolation and Characterisation: Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS 2011;25:679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santoro MM. and Perno CF: HIV-1 genetic variability and clinical implications. ISRN Microbiol 2013;481314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Musyoki A, Mothapo K, Rakgole J, et al. : Genetic characterization of HIV before widespread testing of HIV vaccine candidates at a clinical trial site in Pretoria, South Africa. AIDS Res Hum Retroviruses 2012;28:1131–1138 [DOI] [PubMed] [Google Scholar]
- 5.Lihana RW, Ssemwanga D, Abimiku A, and Ndembi N: Update on HIV-1 diversity in Africa: A decade in review. AIDS Rev 2012;14:83–100 [PubMed] [Google Scholar]
- 6.Novitsky VA, Gaolekwe S, McLane MF, et al. : HIV type 1 A/J recombinant with a pronounced pol gene mosaicism. AIDS Res Hum Retroviruses 2000;16:1015–1020 [DOI] [PubMed] [Google Scholar]
- 7.Bredell H, Hunt G, Casteling A, et al. : HIV-1 Subtype A, D, G, AG and unclassified sequences identified in South Africa. AIDS Res Hum Retroviruses 2002;18:681–683 [DOI] [PubMed] [Google Scholar]
- 8.Jacobs GB, Wilkinson E, Isaacs S, et al. : HIV-1 subtypes B and C unique recombinant forms (URFs) and transmitted drug resistance identified in the Western Cape Province, South Africa. PLoS One 2014;9:e90845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geretti AM, Harrison L, Green H, et al. and the UK Collaborative Group on HIV Drug Resistance: Effect of HIV-1 subtype on virologic and immunologic response to starting highly active antiretroviral therapy. Clin Infect Dis 2009;48:1296–1305 [DOI] [PubMed] [Google Scholar]
- 10.Lessells RJ, Katzenstein DK, and de Oliveira T: Are subtype differences important in HIV drug resistance? Curr Opin Virol 2012;2:636–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palm AA, Esbjörnsson J, Månsson F, et al. : Faster progression to AIDS and AIDS-related death among sero incident individuals infected with recombinant HIV-1 A3/CRF02_AG compared with sub-subtype A3. J Infect Dis 2014;209:721–728 [DOI] [PubMed] [Google Scholar]
- 12.Tebit DM. and Arts EJ: Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect Dis 2011;11:45–56 [DOI] [PubMed] [Google Scholar]
- 13.Bessong PO, Mphahlele J, Choge IA, et al. : Resistance mutational analysis of HIV type 1 subtype C among rural South African drug-naive patients prior to large-scale availability of antiretrovirals. AIDS Res Hum Retroviruses 2006;22:1306–1312 [DOI] [PubMed] [Google Scholar]
- 14.Becker ML, De Jager G, and Becker WB: Analysis of partial gag and env gene sequences of HIV type 1 strains from southern Africa. AIDS Res Hum Retroviruses 1995;11:1265–1267 [DOI] [PubMed] [Google Scholar]
- 15.Engelbrecht S, Laten JD, Smith TL, and van Rensburg EJ: Identification of env subtypes in fourteen HIV type 1 isolates from South Africa. AIDS Res Hum Retroviruses 1995;11:1269–1271 [DOI] [PubMed] [Google Scholar]
- 16.Wilkinson E. and Engelbrecht S: Molecular characterization of non-subtype C and recombinant HIV-1 viruses from Cape Town, South Africa. Infect Genet Evol 2009;9:840–846 [DOI] [PubMed] [Google Scholar]
- 17.Papathanasopoulos MA, Vardas E, Wallis C, et al. : Characterization of HIV type 1 genetic diversity among South African participants enrolled in the AIDS Vaccine Integrated Project (AVIP) study. AIDS Res Hum Retroviruses 2010;26:705–709 [DOI] [PubMed] [Google Scholar]
- 18.Iweriebor BC, Bessong PO, Mavhandu LG, et al. : Genetic analysis of the near full-length genome of an HIV type 1 A1/C unique recombinant form from northern South Africa. AIDS Res Hum Retroviruses 2011;27(8):911–915 [DOI] [PubMed] [Google Scholar]
- 19.Papathanasopoulos MA, Cilliers T, Morris L, et al. : Full-length genome analysis of HIV-1 subtype C utilizing CXCR4 and intersubtype recombinants isolated in South Africa. AIDS Res Hum Retroviruses 2002;18:879–886 [DOI] [PubMed] [Google Scholar]