Abstract
Lungfish (Dipnoi) are the closest living relatives to tetrapods, and they represent the transition from water to land during vertebrate evolution. Lungfish are armed with immunoglobulins (Igs), one of the hallmarks of the adaptive immune system of jawed vertebrates, but only three Ig forms have been characterized in Dipnoi to date. We report here a new diversity of Ig molecules in two African lungfish species (Protopterus dolloi and Protopterus annectens). The African lungfish Igs consist of three IgMs, two IgWs, three IgNs, and an IgQ, where both IgN and IgQ originated evidently from the IgW lineage. Our data also suggest that the IgH genes in the lungfish are organized in a transiting form from clusters (IgH loci in cartilaginous fish) to a translocon configuration (IgH locus in tetrapods). We propose that the intraclass diversification of the two primordial gnathostome Ig classes (IgM and IgW) as well as acquisition of new isotypes (IgN and IgQ) has allowed lungfish to acquire a complex and functionally diverse Ig repertoire to fight a variety of microorganisms. Furthermore, our results support the idea that “tetrapod-specific” Ig classes did not evolve until the vertebrate adaptation to land was completed ∼360 million years ago.
Keywords: Lungfish, Immunoglobulin, Heavy chain, Antibody diversity, Evolution
Introduction
As one of the most important arms of adaptive immunity, immunoglobulin (Ig) genes first appeared about 500 million years ago (MYA) when jawed vertebrates emerged. Ig genes are present from cartilaginous fish to mammals (Flajnik 2002). A hallmark in the evolution of the gnathostome Igs is the diversification of heavy chain isotypes, which has been primarily associated with evolution of novel Ig functions. Tetrapods evolved from sarcopterygian fish in the Devonian and were the first vertebrates to colonize land. Lungfish (Dipnoi) are the sister group to living tetrapods (Amemiya et al. 2013; Brinkmann et al. 2004; Liang et al. 2013), the divergence time between lungfish and tetrapods has been estimated to be 382–388 MYA (Hallström and Janke 2009). Therefore, the study of Ig genes in Dipnoi is expected to provide unique insights into the evolution of these immune molecules and allows for the reconstruction of their evolutionary history.
Lungfish have extraordinarily large genome sizes (Gregory 2009), which have been explained by duplication of genome segments or the whole genome (polyploidy) as well as the activity of transposable elements (Gregory 2005). Average rates of molecular evolution decline with increasing genome size in vertebrates (Sclavi and Herrick 2013). Amongst the four species of African lungfish, important differences are found at the genome level. Protopterus dolloi is the only species with a genome size twice as large as the other Protopterus species (Vervoort 1980). Importantly, P. dolloi is a tetraploid organism whereas Protopterus annectens and Protopterus aethiopicus are diploid. The evolutionary distance between P. dolloi and the rest of the African lungfish species is unknown, but it has been suggested that P. dolloi (the only species to aestivate in surface mucus cocoons) emerged ca. 20 million years earlier than the mud burrower species (P. aethiopicus and P. annectens).
IgM and IgD are considered the most primordial Ig isotypes perpetuated in all vertebrates (with the exception of avians and some groups of mammals, which appear to have lost IgD) (Ohta and Flajnik 2006). A previous Ig genetic survey, using the African lungfish (P. aethiopicus) liver RNA as the source for cDNA library construction, found three different Ig heavy-chain isotypes including an IgM-type heavy chain as well as short and long forms of IgW (Ota et al. 2003). Lungfish IgW heavy-chain TM exons/cDNAs have not been identified thus far (Ohta and Flajnik 2006). Additionally, at the protein level, three Ig isotypes designated as high molecular weight (IgM), intermediate molecular weight, and low molecular weight (IgN) have been identified in two species of lungfish (Chartrand et al. 1971; Marchalonis 1969).
With the advent of high throughput sequencing technologies, it is now possible to deep sequence the transcriptome of non-model species such as the lungfish. In the present study, we revisit the investigation of the Ig heavy chain diversity in two different species of African lungfish, P. dolloi and P. annectens. The two species were chosen due to their different genome size and polyploidy, as well as aestivation strategies. By means of 454 and Illumina platforms, we report herein a greater diversity of Igs in lungfish than that previously unraveled by other genetic surveys. We present molecular, genomic, and functional studies that provide a novel and more complete picture of the Ig heavy-chain evolutionary history in vertebrates. Our results show that diversification of IgH genes resulting in new Ig subclasses such as IgY, IgX, or IgA did not take place prior to the emergence of tetrapods since they are absent in Dipnoi.
Materials and methods
Animals
Juvenile Nigerian spotted lungfish (P. dolloi) (22–30 cm in size) were obtained from Segrest farms (Florida, USA) and maintained in 10-gallon aquarium tanks at a constant temperature of 27 °C. Fish were fed with frozen earthworms three times a week. Feeding was terminated 48 h before sacrifice. Fish were acclimated to the laboratory conditions for at least 2 weeks before being used in experiments. West African lungfish (P. annectens) (30–40 cm in size) were obtained from a local market (Beijing, China). All animal studies were reviewed and approved by the Office of Animal Care Compliance at the University of New Mexico and the Animal Care and Use Committee of the China Agricultural University.
High-throughput sequencing of African lungfish transcriptomes
Initial identification of Ig molecules in P. dolloi was done by 454 pyrosequencing of the transcriptome of the pre-pyloric spleen of a P. dolloi individual. Total RNA was isolated from pre-pyloric spleen tissue, and mRNA was isolated from total RNA with the MPG® mRNA Purification Kit (PureBiotech, USA). cDNA was synthesized using the cDNA Synthesis System Kit with random primers (Roche, USA). A cDNA Rapid library was constructed with the GS FLX Titanium Rapid Library Preparation Kit. Emulsion-based polymerase chain reaction (PCR) amplification of the DNA library was carried out with the GS FLX Titanium LVemPCR Lib-L Kit. Pyrosequencing was conducted using a GS FLX Titanium Sequencing Kit XL+ in a GS FLX+ System. All reagents and protocols used were from Roche 454 Life Sciences (Roche, USA).
To sequence the transcriptomes of P. annectens, total RNA was isolated using a TRIzol reagent (CWBio, Beijing, China) following the manufacturer's instructions and treated with RNase-free DNase I (Qiagen, Beijing, China). The total RNA samples isolated from the gut, kidney, liver, and spermary were pooled together. Beads with Oligo(dT) were used to isolate poly(A) mRNA after total RNA was collected, before fragmentation buffer was added for chopping mRNA to short fragments. Using these short fragments as templates, random hexamer-primers were used to synthesize the first-strand cDNA. The second-strand cDNA was subsequently synthesized. Short fragments were purified with QiaQuick PCR extraction kit (Qiagen, Beijing, China). The short fragments were then linked with sequencing adapters, and the suitable fragments were selected for the PCR amplification as templates. At last, the library was sequenced using Illumina HiSeq™ 2000 (BGI, Shenzhen, China).
Analysis of IgH genes
To conduct a thorough analysis of the putative IgH sequences in the transcriptomes, we first used all obtained Unigenes to do BLAST searches against protein databases (Nr SwissProt KEGG COG) to search for putative IgH genes. Then, the previously identified lungfish P. aethiopicus IgM (accession number: AF437735.1), IgW short (accession number: AAO52810), and IgW long (accession number: AAO52811) were also used to BLAST search against the transcriptomes for putative IgH sequences. The putative IgH sequences were further used to perform rapid-amplification of cDNA to their 5′ and 3′ ends (5′ and 3′ RACE) to obtain full cDNA sequences of all putative IgH classes (for primers, see Supplemental Table 1). All the Protopterus IgH genes sequences were deposited in GenBank (http://www.ncbi.nlm.nih.gov).
Experimental infection and tissue samples
The experimental infection was carried out using the Gram negative enterobacterium Edwardsiella ictaluri strain J100 (Santander et al. 2010) as explained elsewhere (Tacchi et al. 2013). Briefly, E. ictaluri was grown in Bacto-Brain Heart Infusion broth at 27 °C for 48 h. An OD590 of 1 corresponded to 2 × 107 cfu/ml as assessed by plate counts. Bacterial cultures were washed three times in PBS, and 100 μl of a 2 × 107 cfu/ml suspension was delivered into the olfactory canal of P. dolloi. Twenty-one days after the first immunization, fish received a secondary immunization following the same protocol. Ten days following the secondary immunization, fish were sacrificed. Control and infected specimens were euthanized by a lethal dose of MS-222 diluted in water. After bleeding from the caudal vein, pre-pyloric spleen, postpyloric spleen, kidney, lung, gut, and skin tissues were collected and placed in RNAlater (Invitrogen, NY, US) for total RNA extraction.
Quantitative real-time PCR (qPCR)
Power SYBR Green PCR master mix (Invitrogen, NY, US) was used in quantitative real-time PCR (qPCR) to determine the abundance of Igs transcripts in different tissues of P. dolloi. In addition, in order to detect the presence of transmembrane (TM) and secreted Ig forms, a universal TM primer (Varriale et al. 2010) and an oligo dT primer were used in separate reactions to generate cDNA from total RNA. The qPCRs were performed in triplicate, and elongation factor EF-1α, was used as control gene for normalization. All primers used in the experiments are shown in Supplemental Table 1.
Southern blotting
Approximately 80 μg of lungfish (P. annectens) muscle genomic DNA was used in each enzymatic digestion reaction. After separated on a 0.8 % agarose gel, the digested DNA was transferred to a nylon membrane for hybridization. The lungfish IgH cDNA containing plasmids were used as templates for PCR labeling of probes with a PCR DIG Probe Synthesis kit (Roche, Beijing, China). Hybridization and detection were performed using the DIGHigh Prime DNA Labeling and Detection Starter Kit II by following the manufacturer's instructions (Roche, Beijing, China). Two different hybridization protocols (low and high stringency) were also used for IgM1 in order to establish the translocon or cluster organization of the lungfish IgH locus. In addition to the IgM1 heavy-chain probe directed against the fourth heavy chain constant region (CH4) domain, a heavy-chain variable region (VH) probe (derived from VH family II) was also designed and used under both low and high stringency conditions.
Sequence analysis
The open reading frame was predicted using the programs BLAST (Altschul et al. 1990) and the ExPASy proteomics server (ca.expasy.org/). Multiple sequence alignments were generated using CLUSTAL W (http://align.genome.jp/) (Chenna et al. 2003). The phylogenetic trees were constructed using MrBayes3.1.2 and were viewed by FigTree or Tree View. To obtain the identity of the sequences, the software MatGat 2.02 (Campanella et al. 2003) was used.
Construction of the all-IgH-isotype and VH phylogenetic tree
Phylogenetic analyses of the P. dolloi and P. annectens IgH genes identified as well as available IgH sequences for most vertebrate groups was conducted in Phylip 3.695 and MrBayes 3.1.2. Three different approaches were used: maximum likelihood (ML), neighbor joining (NJ), and CAT-model Bayesian inference in order to confirm the topology of the phylogenetic trees. Bootstrap support was evaluated with 1,000 replicates. Unless otherwise indicated, all CH domains of Ig classes other than IgD were used. For those species (not including teleost fish) that express IgD/W longer than four CH domains, only the first four domains were used. For sturgeon and teleost fish IgD, domains 2–5 were used. Nurse shark IgM was used as outgroup. The accession numbers of the sequences used, in addition to the here reported lungfish sequences, are listed below: μ, nurse shark, ABW84249.1; skate, S12838; spotted ratfish, AF003844-7; P. aethiopicus, AAO52808.1; zebrafish, AF281480; rainbow trout, AAB27359.2; channel catfish, CAB38072.1; Atlantic cod, CAA41680.1; sturgeon, KC734556; Xenopus laevis, CAA33212; lizard, ABV66128; gecko, ABY74509; axolotl, A46532; Chinese alligator IgM1, AFZ39166.1; chicken, X01613; platypus, AY168639; mouse, CAA24199.1; human, P01871. δ, channel catfish, T18537; trout, AAW66977; mandarin fish, ACO88906.1; orange-spotted grouper, AFI33218.1; fugu, BAD34541.1; Atlantic cod, AAF72565.1; Japanese flounder, BAB41204.1; sturgeon, KC734555; Xenopus tropicalis, DQ350886; Chinese alligator, JQ417418.1; gecko, ABY67439; lizard, ABV66130; platypus, ACD31540; mouse, AAB59654.1; human, BC021276. ζ/τ/π, zebrafish, AY643752; trout, AY872256; and Iberian ribbed newt, CAL25718; ω, sandbar shark, U40560; P. aethiopicus, AF437727; and nurse shark, U51450; coelacanth W1, JX848736.1; coelacanth W2, JX840472.1. γ, human, J00228; mouse, J00453; and platypus, AY055781;ε, human, J00222; mouse, X01857; and platypus, AY055780; υ: chicken, X07175; Xenopus laevis, X15114; axolotl, CAA49247; lizard, ABV66132; gecko ACF60236;Xenopus tropicalis, BC089679; Chinese alligator, AFZ39169.1. α or χ, axolotl, CAO82107; chicken, S40610; human, J00220; mouse, J00475; platypus, AY055778; Xenopus laevis, BC072981; Xenopus tropicalis, AAI57651; Chinese alligator, AFZ39164.1. The accession numbers of the sequences used for VH tree are listed below: human: V1, L22582; V2, X62111; V3, X92206; V4, Z12367; V5, M99686; V6, X92224; V7, AB019437; mouse: V1, AF304557, V2, J00502; V3, K01569; V4, V00774; V5, AJ851868; V6, AJ972404; V7, X03253; V8, AC073939; V9, L14368; V10, AF064445; V11, AJ851868; V12, AC073590; V13, X55935; V14, X55934; V15, U39293; V16, AC073563; chicken: AB233003.1; X. laevis: V1, Y00380; V2, M30025; V3, M24675; V5, M24677; V6, M24678; V7, M24679; V8, M24680; V9, M24681; V10, M27254; V11, M27255; zebrafish: V1-V14, BX649502; snake, V1-V6 (our own data); Chinese alligator, V1-V13 (our own data); nurse shark: V1, M92851; V2, L38965.1.
Statistical analysis
Data are presented as mean± standard error. In qPCR experiments, the relative expression level of the genes was determined using the Pfaffl method (Pfaffl 2001); measurements were analyzed by t-test by comparing values with either the EF-1α control or the non-infected control. One-way ANOVA analysis followed by Tukey's post-hoc test was used to identify expression differences among tissues. P values <0.05 were considered significant.
Results
Overview of the analysis of IgH genes in the transcriptomes of two lungfish species by high throughput sequencing
A total of one million reads were obtained from the 454-sequencing of P. dolloi pre-pyloric spleen transcriptome. The mean read length obtained was of 425 bp. The assembly resulted in 14,072 isotigs and 136,857 contigs plus singletons. Sequence similarity searches were performed against SwissProt and Uniprot databases with 38,753 and 48,199 contigs and singletons annotated, respectively.
The transcriptomes of the gut, kidney, liver, and spermary of a P. annectens were obtained using Illumina HiSeq™ 2000. A total of 110,439,106 raw reads were obtained accounting for approximately 105,000,000 clean reads, which were assembled into 859,969 contigs after removal of short reads and trimming of low-quality sequence regions. These contigs were taken into further process of sequence splicing and redundancy removing with sequence clustering software to acquire non-redundant Unigenes. In this manner, a total of 122,123 Unigenes with 575 bp of mean length were obtained. All-Unigene sequences were used in BLAST against protein databases for gene annotation.
Using these transcriptome databases, we first searched for putative IgH sequences based on the gene annotation results. Meanwhile, the previously identified lungfish P. aethiopicus IgM and IgW were also used as query sequences in BLAST search against the transcriptomes to identify putative IgH sequences. These putative IgH transcripts were further used to design primers for 5′ and 3′ RACE or additional reverse transcription-polymerase chain reactions to obtain their full cDNA sequences. Three P. dolloi Igs: IgM, IgW short, and IgW long were first identified and reported in a previous study, although no sequence analyses were performed at the time (Tacchi et al. 2013). The efforts undertaken in the present study finally led to the discovery of two IgM, three IgW(D), and one IgN genes in P. dolloi, whilst three IgM and two IgW(D), three IgN, and one IgQ molecules were identified in P. annectens (Fig. 1). It is worth pointing out that the IgN nomenclature was chosen according to the IMGT Ig nomenclature, and thus, IgN in the current manuscript does not correspond to the IgN molecule identified by biochemical approaches by (Chartrand et al. 1971; Marchalonis 1969). Since the full sequence for IgQ could not be obtained, its structure is not represented in Fig. 1.
Fig. 1.
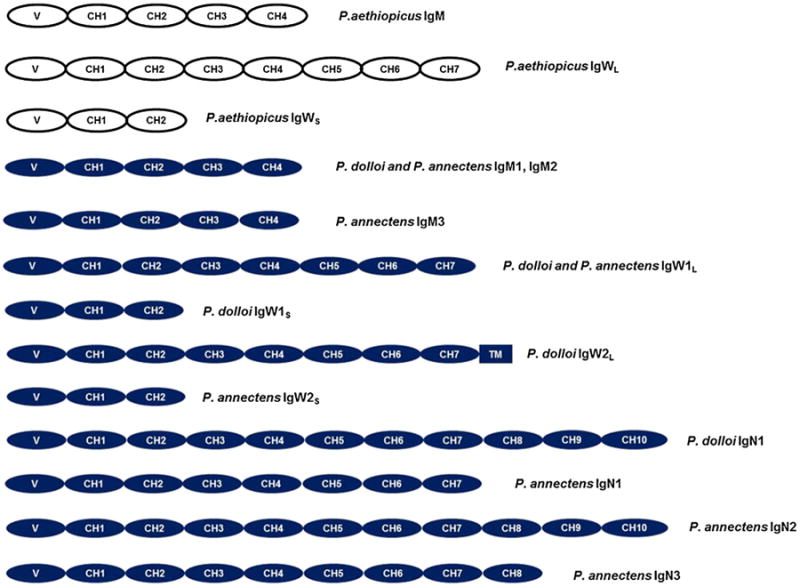
Structural cartoon of IgH-chains identified in P. aethiopicus, P. annectens, and P. dolloi. Both VH and CH domains are shown as ovals. A hinge-like region sequence in P. dolloi IgW2 L is drawn as a thick bar. Cartoons for P. annectens and P. dolloi derived from 454 and Illumina sequencing followed by RACE experiments
African lungfish express up to three different IgM heavy chains
IgM1
Both P. dolloi (accession number: KC812389) and P. annectens (accession number: KC849709) express IgM1, which had already been identified in P. aethiopicus (with protein sequence identities of 99.6 % and 96.2 %, respectively, Supplemental Table 2). Three (Bayesian, ML, and NJ) methods used for phylogenetic analysis converged to roughly the same tree topology: lungfish IgM1 clustered with other tetrapod IgMs as previously observed (Ota et al. 2003). Figure 2 shows the phylogenetic analysis obtained with the Bayesian method. A detailed sequence comparison revealed several interesting observations with regard to the distribution of important amino acid residues and potential N-linked glycosylation sites (Fig. 3). For example, IgM1 lacks a cysteine that is supposed to form an intra-domain disulfide bond in their CH1 domains, whereas in the very N terminus of CH4, P. aethiopicus and P. dolloi IgM1 lack a cysteine that is usually seen in the IgM molecule of nearly all jawed vertebrates. This cysteine is thought to be involved in the formation of IgM polymers (Paul 2012).
Fig. 2.
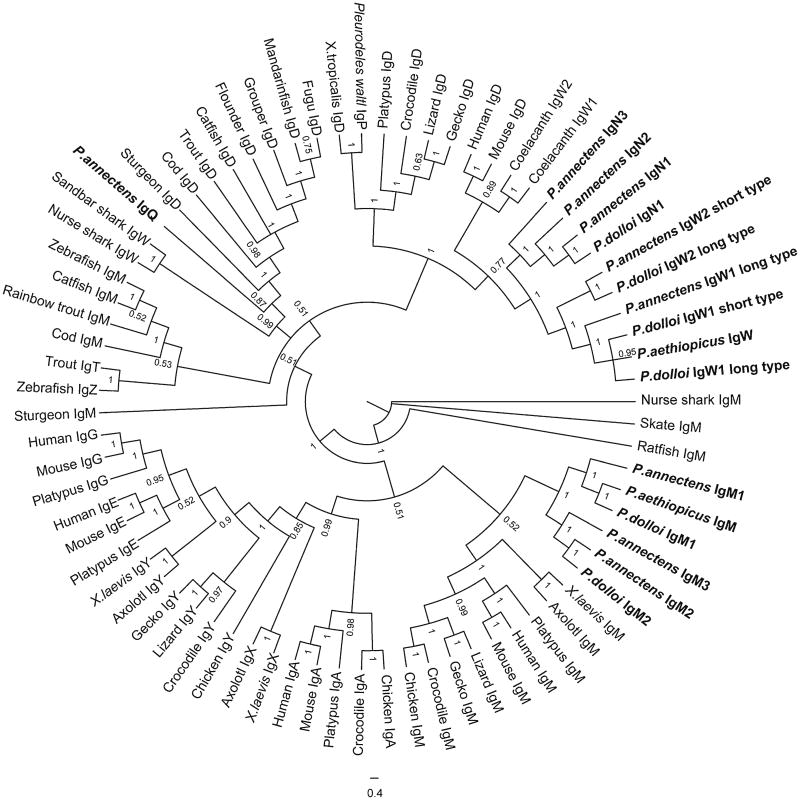
Phylogenetic analysis of the lungfish IgH constant region genes using the Bayesian method. The trees were constructed using MrBayes. The scale bars show the genetic distance, and the credibility value of each node is shown. See “Materials and methods” for accession numbers of the sequences used in the tree construction
Fig. 3.
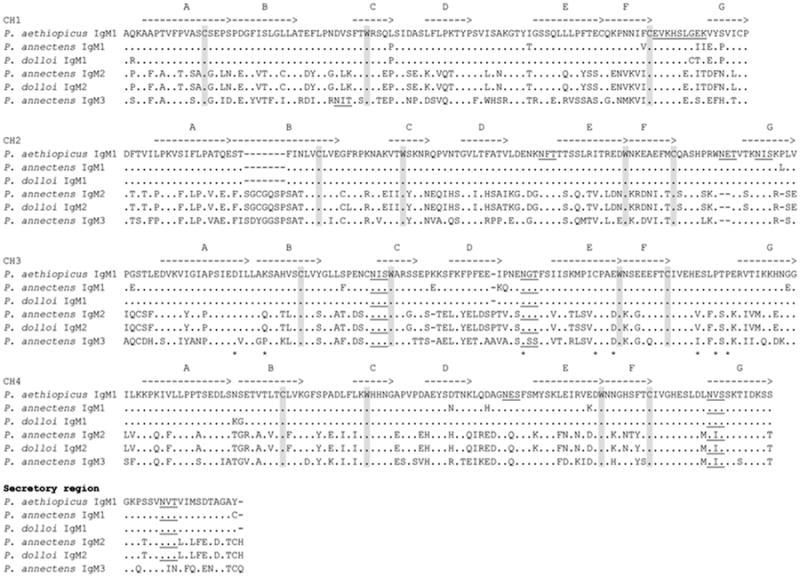
Amino acid sequence alignment of lungfish IgM. Dots denote identical amino acids, whereas dashes indicate gaps. Canonical cysteines and tryptophans are shaded, and the conserved N-linked glycosylation sites are underlined. Potential IgSF strands are indicated as A–G with lines over the sequences
IgM2
Surprisingly, an IgM2 (accession numbers: P. dolloi JX999963 and P. annectens KC849710) was also found in both lungfish species with a 99.1 % protein sequence identity between them (Supplemental Table 2) Regardless of the species, the newly identified IgM2 shows approximately 57 % protein sequence identity with IgM1 and clustered with the other Proptopterus IgM and tetrapod IgM sequences in our phylogenetic analysis.
IgM3
IgM3 was found only in P. annectens, (accession number: KC849711) even if multiple primers sets were tested in PCR reactions using P. dolloi cDNAs from different tissues. IgM3 shows 53 % identity with IgM1 and an approximately 67 % sequence identity with IgM2 (Supplemental Table 2). Three (Bayesian, ML, and NJ) methods used for phylogenetic analysis converged to roughly the same tree topology: lungfish IgMs clustered with other tetrapod IgMs (Fig. 2). A detailed sequence comparison revealed that, similar to IgM1, IgM3 lacks the cysteine that is supposed to form an intra-domain disulfide bond in their CH1 domains. Additionally, a highly conserved potential N-linked glycosylation site among vertebrates was missing in the C terminus of IgM3, whereas most of the residues possibly involved in complement activation are highly conserved in IgM3 as well IgM1 and IgM2 molecules (Fig. 3).
To examine the presence of these different IgM encoding genes in the genome, we conducted Southern blots on digested P. annectens genomic DNA using single CH-exoned probes, which showed that they were highly likely present in the genome as single-copy genes (Fig. 4).
Fig. 4.
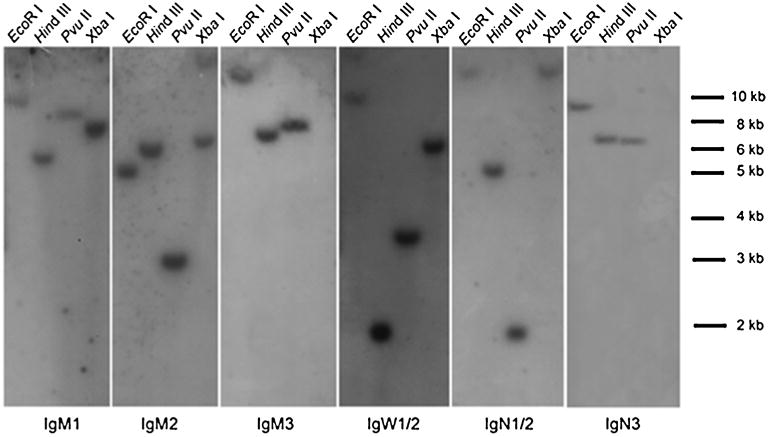
Southern blotting detection of IgH constant region encoding genes in P. annectens. The restriction enzymes used in the experiment are indicated on the top, while the DNA size is shown on the right. Probes used in the detection are as follows: CH4 of all IgM isotypes; CH2 of IgW2 for IgW1/2 detection; CH1 of IgN2 for IgN1/N2 detection; CH2 of IgN3
Lungfish IgW heavy chains
IgW1
Previous to this study, two IgW1 transcripts, IgW1L (long), and IgW1S (short) (derived from a same gene by alternative RNA splicing) were reported in P. aethiopicus. Both IgW1L and IgW1S orthologous genes were observed in P. dolloi (accession numbers: KC152447 and JX999964), which shared 99.3 % protein sequence identity with the P. aethiopicus IgW (the comparison was conducted using all seven CH domains). Only IgW1L (a 7-CH IgW transcript) was expressed in P. annectens (accession number: KC849716) (Fig. 1), which shared 96.6 % protein sequence identity with P. aethiopicus IgW (Supplemental Table 3). IgW1s always consists of two CH domains whereas IgW1L always consists of seven CH domains, and both have a highly conserved in amino acid sequence. The major difference is that the IgW1L in P. annectens shows a missing valine in the C terminus when compared with those of the other two species (Fig. 5).
Fig. 5.
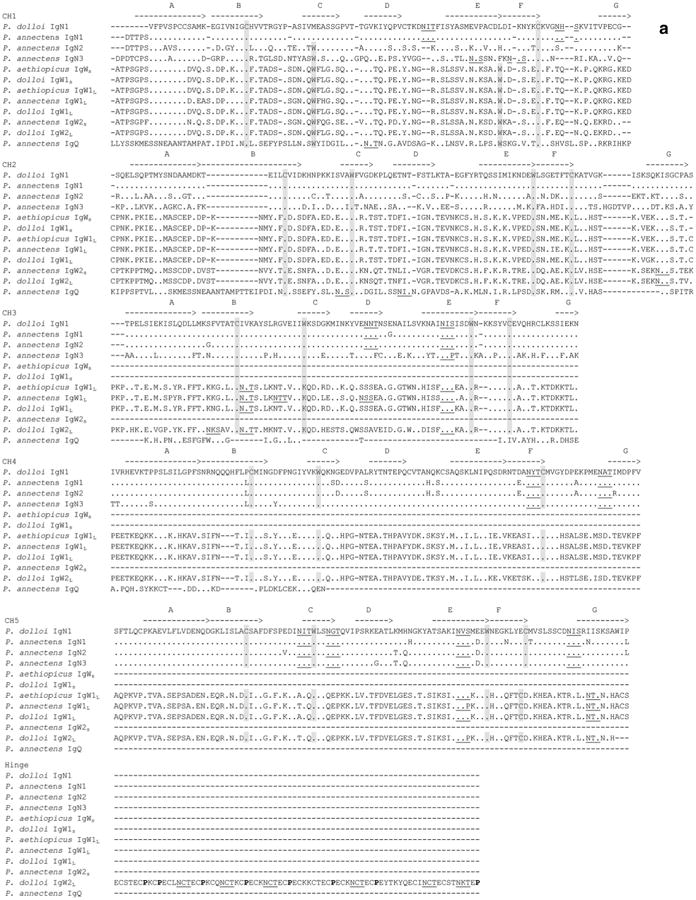
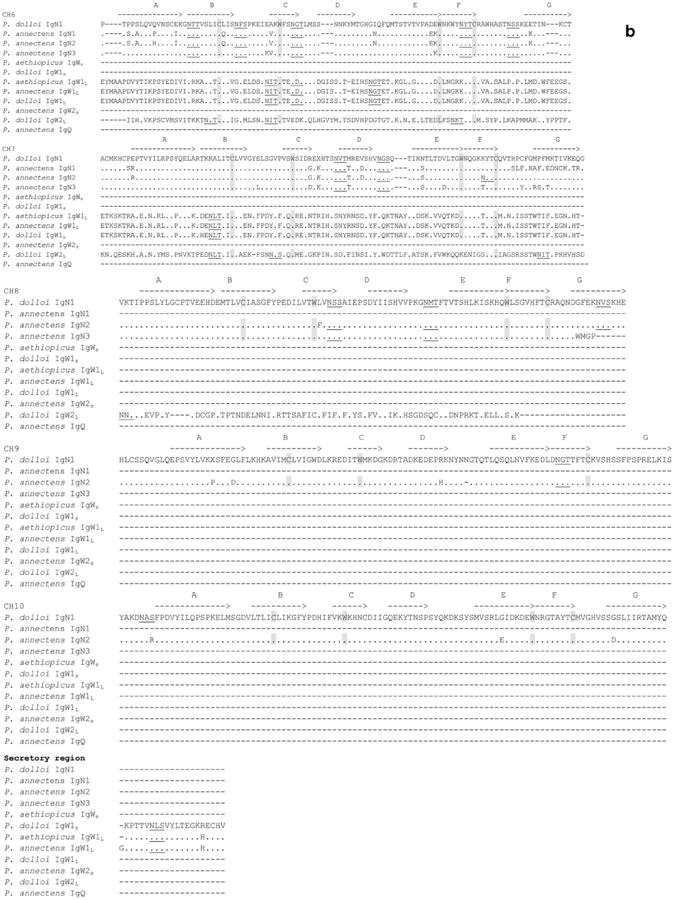
Amino acid sequence alignment of lungfish IgW, IgN, and IgQ. Dots denote identical amino acids, whereas dashes indicate gaps. Canonical cysteines and tryptophans are shaded, and the conserved N-linked glycosylation sites are underlined. Potential IgSF strands are indicated as A–G with lines over the sequences
IgW2
For the second IgW encoding gene, IgW2, we only identified a short secreted form transcript, IgW2S, in P. annectens (accession number: KC849717) but a long membrane-bound form, IgW2L in P. dolloi (accession number: KC812390). Regardless of species, these IgW2 exhibit approximately 66–76 % sequence identity with IgW1 (Supplemental Table 3). Similar to IgW1, the short form of IgW2S in P. annectens consists of two CH domains, while IgW2L in P. dolloi contains seven CH domains. Strikingly, a sequence examination revealed a long, hinge-like, stretch of sequence rich in cysteine and proline between the CH5 and CH6 of P. dolloi IgW2L (Fig. 6a). Apparently, this sequence consists of four repeats of a core amino acid stretch (15–18 amino acids long), and each repeat contains one to two prolines and three or six cysteines, accounting for totally 21 cysteines and 6 prolines in the entire region. These features are reminiscent of the hinge region of human IgG3, which is also abundant in cysteine and proline, and is encoded by four exons with similar sequences (Fig. 6b). Interestingly, this region also contains six potential N-linked glycosylation sites (five NCT, one NKT). Additionally, the cytoplasmic tail in this molecule is long (25 amino acids) and lacks any tyrosine residues.
Fig. 6.

Hinge region-like sequence found in P. dolloi IgW2L. a Hinge region like sequence in the P. dolloi IgW2L constant region. Potential N-glycosylation sites and the hinge-like sequence are underlined and in bold. TM: transmembrane region; CYT: cytoplasmic tail. b Comparison of the P. dolloi IgW2L hinge-like sequence with that of human IgG3. Potential N-glycosylation sites are underlined and in bold. IgW2HL: IgW2L hinge-like sequence; HIgG3H: human IgG3 hinge region
It is worth noting that, as previously observed, all the lungfish IgW transcripts had the conserved double-cysteine feature in the N-terminal C1 domain (Fig. 5), one of which is predicted to associate with light (L) chains, as it is the case in Xenopus IgD and IgY and catfish IgD (Ohta and Flajnik 2006).
Like IgM, IgW Southern blots showed that they were also highly likely present in the genome as single-copy genes (Fig. 4).
A novel Ig isotype exists in African lungfish: IgN
IgN1
IgN1 transcripts are composed of ten CH domains in P. dolloi (accession number: KC174714) and seven CH domains in P. annectens (accession number: KC849711), and both appear to encode secreted antibody forms (Fig. 1). Although IgNs are most closely related to the lungfish IgWs in the phylogenetic tree, amino acid sequence analyses performed using full constant region or corresponding individual CH domains support the new isotype terminology. IgN1 shares only approximately 30 % sequence identity with IgW1 and 27–30 % with IgW2 (Supplemental Table 3).
IgN2 and IgN3
IgN2 and IgN3 transcripts were only observed in P. annectens (accession numbers: KC849712 and KC849714, respectively), and they encode secreted antibody forms. IgN2 is composed of ten CH domains, while IgN3 contains eight CH domains (Fig. 1). IgN2 and IgN3 share 74 % sequence identity. They are quite similar to IgN1 (87.6 % and 70.2 % sequence identity, respectively) but considerably diverged from IgW1 and IgW2 (24–31 % sequence identity) (Supplemental Table 3). Phylogenetic analyses showed that IgN1, IgN2, and IgN3 formed a distinct clade (Fig. 2).
IgN Southern blots support that IgNs are also highly likely present in the genome as single-copy genes (Fig. 4).
P. annectens also expresses IgQ
In addition to IgN1–IgN3, we also found an incomplete Ig sequence (KC849715). We named this Ig IgQ due to its low identity with any of the other IgH previously found. IgQ contains two duplicated CH exons (Fig. 5). We failed to obtain the complete sequence of IgQ by RACE, most likely due to its low expression level and repetitive sequence nature. For this reason, we did not include it in Fig. 1. Phylogenetic analysis revealed that IgQ is more closely related to the teleost IgD clade (Fig. 2). Because teleost IgD is a chimeric Ig consisting of μCH1 exon, we performed a series of PCR reactions using primers derived from CH1 exons of all three IgMs and IgQ-specific primers. These PCRs failed to generate products indicating that the lungfish IgQ is most likely not a chimeric Ig.
Analysis of variable gene segments expressed in the African lungfish
To analyze the VH repertoire expressed in the two lungfish species studied, we conducted 5′ RACE to obtain the variable sequences associated with the identified IgH isotypes. Together with the VH sequences identified in the transcriptome data, we found 17 VH families in P. annectens and 7 in P. dolloi based on a criterion that all family members within a single family should share at least 75 % sequence identity at the nucleotide level (Fig. 7). According to the same criterion, further sequence comparisons revealed that all seven VH families in P. dolloi and eight VH families previously identified in P. aethiopicus could be attributed to the 17 VH families found in P. annectens. For consistency, the VH families found in P. annectens and in P. dolloi that corresponded to the eight VH families in P. aethiopicus were still termed VH family I–VIII, where the remaining were sequentially named VH family IX–XVII.
Fig. 7.

Amino acid sequence alignment of representative VH family members in lungfish. Dashes are used for sequence adjustment. CDRs and FRs are indicated on the top
To analyze the VH gene expression more thoroughly, we designed primers for all 17 VH families, which were used in nested PCR reactions together with constant region-specific primers for all Ig forms in P. annectens. These PCR reactions were performed to confirm the association of VH genes with specific Ig isotypes at the transcriptional level. Together the transcriptome and 5′ RACE data revealed that most VH families (I–XIII) were used by IgM1 (Supplemental Table 4), whereas three families (XIV–XVI) were used by IgM2. IgM3 in P. annectens exhibited a very limited VH diversity with only a single VH family (XVII) identified. Among the 17 VH families, only four (V–IX) were shared by IgM1 and IgW, two (VII, IX) shared by IgM1 and IgN isotypes.
Phylogenetic analysis of these lungfish VH families together with VH genes derived from other vertebrates revealed that 11 lungfish VH families (I-II, V, VI, VIII-X, XII, XIII, XVI, XVII) belonged to Clan II, where the remaining were clustered into Clan V (Fig. 8). Apparently, Clan II consists of VH genes derived from lungfish and all classes of tetrapods, indicating the VH genes in this clan have been conserved from lungfish to mammals. However, Clan V only contains VH genes from cartilaginous fish, lungfish, amphibians, and reptiles but not mammals. The phylogenetic analysis strongly suggests that the VH repertoire in each class of vertebrates has been selectively shaped during the evolutionary process, perhaps by specific environmental factors that animals are confronted with.
Fig. 8.
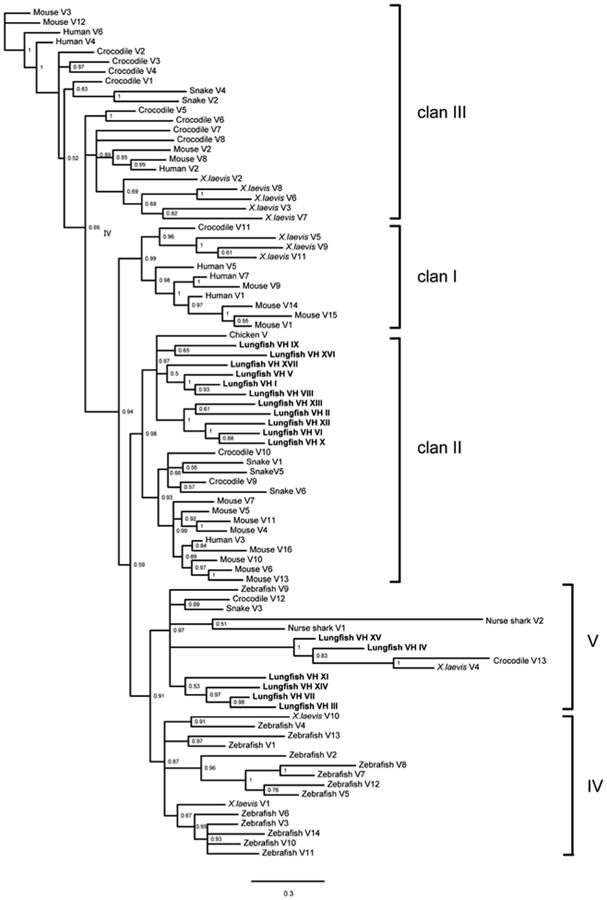
Phylogenetic analysis of the lungfish IgH variable region genes. The trees were constructed using MrBayes. The scale bars show the genetic distance, and the credibility value of each node is shown. See “Materials and methods” for accession numbers of the sequences used in the tree construction
Genomic configuration of the African lungfish IgH loci
Although it had been suggested that lungfish IgH loci are arranged in the translocon configuration as in most vertebrates, conclusive evidence to support this hypothesis was lacking to date. Using IgM1 as an example, we conducted Southern hybridizations under both low and high stringency, using a VH and a CH4 probe, respectively. The VH probe revealed many bands under both low and high stringency conditions. However, the CH4 probe detected multiple bands under low stringency and only a few under high stringency, a result consistent with a translocon but not cluster-like IgH locus configuration (Fig. 9).
Fig. 9.
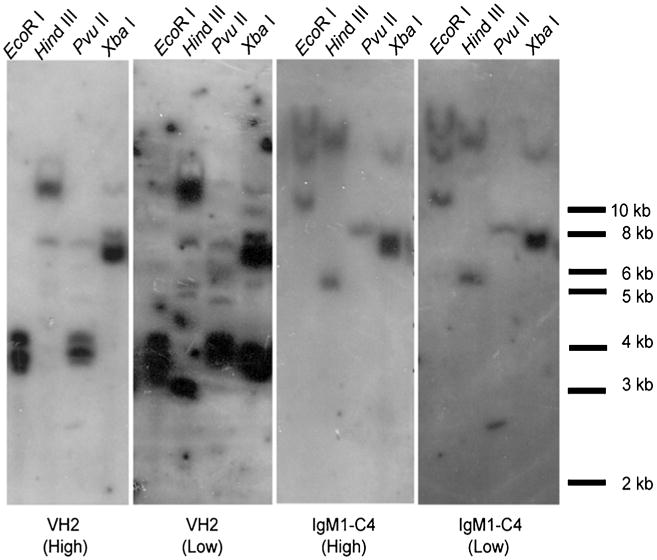
The lungfish IgH has a translocon configuration. Southern blot analysis of VH2 and IgM1-CH4 genes under low and high stringency conditions using four different digestion enzymes. Probe VH2 was derived from a member of VH family II
The results obtained by Southern blots combined with the VH usage results here found indicate that IgM1 encoding gene has a translocon configuration and that all other Ig isotype encoding genes are either organized together with the IgM1 but most likely interrupted by V, D, and J gene segments, or located in different chromosomal positions.
P. dolloi IgH gene expression in tissues and in response to enterobacterial infection
In order to understand the functional role of the newly identified Ig genes in response to a pathogen, we used the recently developed infection model of P. dolloi and E. ictaluri (Tacchi et al. 2013). First, we evaluated the constitutive expression of IgH genes in different tissues by qPCR using the EF-1α as an internal control (Fig. 10a). The primers used in this experiment do not distinguish between TM and secretory forms, so the results reported here refer to both forms. Additionally, due to the similarity between IgW1S and IgW1L, the set of primers designed to amplify IgW1s also detects the first two CH domains of IgW1L (expressed in the graph as IgW1S+L). A second set of primers was therefore designed to amplify the unique domains of IgW1L not present in the short form. The highest expression levels of all IgH transcripts were found to be in the post-pyloric spleen, followed by the pre-pyloric spleen (IgM1, IgM2, IgW1S+L, and IgW1L) and kidney. The expression levels in the post-pyloric spleen were between approximately five- to tenfold compared with the other tissues, indicating that this tissue is a main organ for Ig production expression in lungfish. Interestingly, in the skin, IgN1 was the most expressed of all Igs.
Fig. 10.
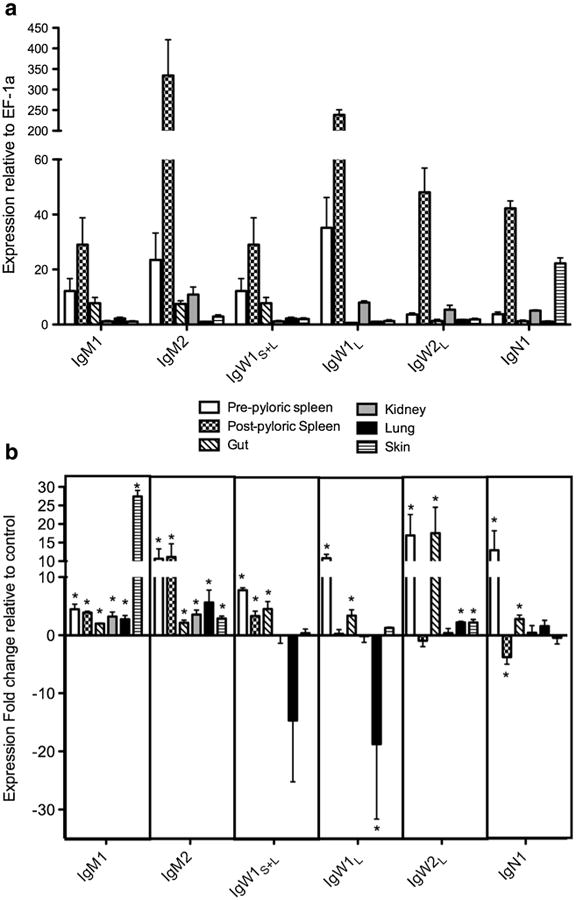
qPCR detection of IgH gene expression in tissues of P. dolloi. a Expression of the P. dolloi IgH genes in different tissues (n=5). b P. dolloi IgH gene expression in response to enterobacterial infection (n=4). The qPCRs were performed in triplicate and elongation factor EF-1α was used as control gene for normalization
The expression of IgH TM forms was confirmed for all isotypes except for IgM2, suggesting that this form is expressed at very low levels in P. dolloi. The same results were confirmed both in pre-pyloric and post-pyloric spleen tissue samples. In order to rule out unspecific amplification during the generation of the TM-cDNA, we examined the expression of EF-1α, a gene that does not contain a TM domain. We could not amplify EF-1α from TM-cDNA supporting the specificity of our assay. Overall, the expression of all TM forms was considerably lower (higher Ct values, not shown) than that of the secreted forms, and even if these experiments are not fully quantitative, we estimate that the expression of TM forms is approximately few hundred-fold lower than that of secreted forms.
The expression of Ig transcripts in response to E. ictaluri oral infection in different tissues is shown in Fig. 10b. Ten days after secondary immunization, statistical analysis revealed that the expression of IgM1 was significantly (p value <0.05) increased in all tissues sampled. Surprisingly, the greatest upregulation of IgM1 occurred in the skin, with a ∼25-fold increase in expression. IgM2 expression was also significantly upregulated in all tissues, but the highest levels of expression were found in the pre and post-pyloric spleen. The expression levels of IgW1L suffered a significant decrease in the lung tissue. However, IgW1L expression was significantly upregulated in the pre-pyloric spleen and gut. The expression levels of IgW1S+L significantly increased upon infection in the pre-pyloric spleen, post-pyloric spleen, and the gut. IgW2L expression showed a ∼17-fold increase in both the pre-pyloric spleen and the gut and a twofold increase in the lung and skin. Finally, IgN1 was upregulated ∼13-fold and ∼2-fold in the pre-pyloric spleen and gut, respectively. In the post-pyloric spleen, in turn, a significant decrease in the expression of IgN1 (∼3-fold) was observed.
Discussion
Discovery of new IgH diversity in Dipnoi
To date, lungfish have been thought to possess only three IgH isotypes, a typical IgM antibody with four CH domains and two secreted IgW forms. However, the large genome sizes in Dipnoi may have obscured the discovery of all IgH isotypes using traditional cDNA libraries. Our studies using high-throughput transcriptome sequencing, indeed, reveal that lungfish express an unprecedented multiplicity of IgH isotypes.
A highly complex Ig repertoire is present in Dipnoi with no further diversification due to polyploidy
We sought to investigate the diversity of IgH genes in two species with different genome sizes and number of chromosomes. Genome-doubling events are common in fish, yet their importance in the evolution of fish remains unclear (Leggatt and Iwama 2003). Whole genome duplications leading to polyploidy have been interpreted as a strategy to diversify antibody diversity in salmonids (Hordvik 1998). We show here that, in both lungfish species studied, a complex repertoire of IgH genes is present. Thus, it appears that tetraploidy (acquired only by P. dolloi) has not led to new diversity of Igs as evidenced by the comparison with the diploid species P. annectens.
Reconstructing the evolutionary history of Igs in vertebrates
With the advent of genome sequencing and deep sequencing of transcriptomes, our view of the evolutionary history of Igs is almost complete. The diversification of Igs into new isotypes during vertebrate evolution has been associated with acquisition of novel antibody functions. Currently, we do not know what these are for IgN and IgQ, but they appear to be present in Dipnoi only. The current dogma regarding the evolution of IgY, IgA(X) postulates that IgA(X) is an IgY/IgM chimera, and thus IgY may have emerged earlier than IgA(X) (Magadán-Mompó et al. 2013; Mashoof et al. 2013).
However, as no IgY-like molecule was found in the present survey, our results confirm the notion that IgY and IgA(X) are tetrapod-restricted Igs (Zhao et al. 2006).
We report here a total of three different IgM transcripts, two IgW transcripts, three IgN transcripts, and one IgQ transcript in P. annectens, and two IgM, three IgW, and one IgN transcript in P. dolloi. Thus, an unprecedented multiplicity of IgM and IgW forms has evolved in lungfish as well as the acquisition of lungfish-specific Ig isotypes. Until now, only one IgM had been identified in lungfish (Ota et al. 2003). The investigation of two new Protopterus species and the use of high-throughput sequencing revealed the presence of at least two more IgM genes. Interestingly, sequence differences indicate that these different IgMs may encode for distinct multimeric and monomeric forms. Multiplicity of IgM is common in cartilaginous and bony fish and points to the capacity of duplicated Ig genes to diverge and evolve new roles (Lee et al. 2008). This may also somehow compensate for the limited number of IgH isotypes in fish. Albeit rare, the duplication of IgM genes has also been found in tetrapods such as crocodiles, strongly suggesting a functional significance for IgM multiplicity (Cheng et al. 2013).
Lungfish IgW TM exons had not been found to date (Ohta and Flajnik 2006). Although the majority of the IgW here described are sec forms, we also identified a transmembrane (TM) IgW using deep sequencing. The presence of sec and TM IgW forms in lungfish therefore resembles the diversity found in cartilaginous fish (Rumfelt et al. 2004). However, Chondrichthyes and the coelacanth appear to have a less diverse repertoire of IgW genes than Dipnoi. Our results support the current dogma of IgD, the IgW orthologue (Ohta and Flajnik 2006), being the most evolutionary labile, structurally diverse, and plastic IgH isotype found in vertebrates (Edholm et al. 2011; Ohta and Flajnik 2006). In ectotherms, IgD varies with respect to the number of unique Cδ domains ranging from 6 to 11. IgW, in turn, can range from 2 (Ota et al. 2003) to 19 (Amemiya et al. 2013). It has been suggested that the lack of a hinge in the IgD in these species is compensated by the length of the IgD H chain that may allow for some flexibility (Edholm et al. 2011). In the case of lungfish IgW, we identified a long IgW form (P. dolloi IgW2L) containing a TM region, a hinge-like region and seven CH domains. Moreover, VH analysis showed that IgW2L uses specific V families not shared with the other Ig molecules at least in P. dolloi. Altogether, the particular characteristics of IgW2L indicate a specialized function for this molecule in the immune response of P. dolloi.
Reconstructing the evolution of IgM and IgW
IgM and IgD/IgW are thought to be primordial Ig classes (Mashoof et al. 2013; Ohta and Flajnik 2006). IgM is the only Ig isotype present in all vertebrates, except for the coelacanth (Amemiya et al. 2013). IgD was present in the ancestor of all jawed vertebrates and arose together with IgM at the time of the emergence of the adaptive immune system, approximately 500 MYA (Chen and Cerutti 2010). This ancient Ig class was, however, lost in avians and some groups of mammals. When lungfish IgW was first discovered in 2003, it was recognized as an orthologue of cartilaginous fish IgW (Ota et al. 2003). Recently, two IgW loci have been identified in the coelacanth (Amemiya et al. 2013). Teleost fish IgM and IgD are co-produced through alternative splicing of a long pre-mRNA containing the VDJ region, the Cμ exons, and the Cδ exons, as in mammals (Edholm et al. 2011). The way fish IgHδ mature transcripts are produced results in chimeric Cμ1/Cδ molecules. We show here that neither lungfish IgWs nor lungfish IgN/IgQ share this feature with teleosts, since no chimeric Cμ1/Cδ molecules were found. The unprecedented diversity of IgW forms within Dipnoi indicates a great expansion of this isotype in a group of vertebrates with very large genome sizes. Thus, it seems that genome size did not impose slow evolutionary speeds on Dipnoi IgH genes, probably due to the strong selective pressures on immune molecules such as antibodies.
Bayesian, ML, and NJ approaches all converged to the same topology. In phylogenetic analysis, repeated tree runnings demonstrated that lungfish IgWs and IgNs were always clustered together with the tetrapod IgD, while IgQ formed a clade with the teleost IgD and cartilaginous IgW (Fig. 2). IgN formed a cluster by itself, and this cluster was most closely related to the lungfish IgWs, suggesting that IgNs may have evolved from a lungfish IgW ancestor. Lungfish IgMs were clustered with the shark and tetrapod IgMs but not with those of teleost. Although this is consistent with a previous analysis (Ota et al. 2003), the bootstrapping value supporting this branch was rather low. To clarify this issue, we made additional phylogenetic trees using full length, CH1 domain, and CH4 domain of IgMs, respectively (data not shown). While both CH1 and C4 trees conform to the previous results, the tree constructed using all IgM CH domains showed that the lungfish IgMs were more close to teleost IgM. These inconsistent results reflect that different IgM CH domains may have been subject to different selection pressures due to functional restrictions (i.e., association with light chain in the case of CH1).
Lungfish IgH genes are organized in a transiting form from clusters (IgH loci in cartilaginous fish) to a translocon configuration (IgH locus in tetrapods)
IgH genes can be found in two different arrangements: the gene cluster arrangement found in cartilaginous fish and the translocon organization found in teleost fish and tetrapods. While all other vertebrates employ the so-called translocontype organization (Vn-Dn-Jn-C), cartilaginous fish Ig genes are encoded in clusters (V-D-J-C)n. Thus, in cartilaginous fish, the IgM and IgW heavy-chain gene loci are encoded by up to 100 separate, paralogous loci (Lee et al. 2008; Cheng et al. 2013; Edholm et al. 2011). From the Southern blot analyses and VH usage data here presented, we show that IgM1 encoding gene in lungfish has a translocon configuration, and all the remaining Ig isotype encoding genes are either in organized together with the IgM1 but are most likely interrupted by V, D, and J gene segments, or located in different chromosomal positions. Apparently, the genomic IgH loci in lungfish show characteristics of cartilaginous fish and tetrapods.
Lungfish are the closest living relative to tetrapods and are the link to the “water to land” transition of vertebrates. We have here revisited the diversity of IgH genes in Dipnoi. This study reveals an untapped, unprecedented, and highly diverse repertoire of IgH genes in two African lungfish species. Importantly, lungfish appear to have diversified their Ig repertoire by expanding the two primordial Ig classes, IgM and IgD/W as well as by acquiring two new Ig isotypes, IgN and IgQ. Our results reconstruct the evolutionary history of IgH molecules in the vertebrate lineage and highlight that the origins of “tetrapod-characteristic Igs” such as IgY, IgA(X), IgG, and IgE lay within the tetrapod lineage and not in their sister group, the lungfish.
Supplementary Material
Acknowledgments
This study was supported by NIH COBRE grant P20GM103452 (I.S), the National Basic Research Program of China (973 Program-2010CB945300), and the China National Natural Science Foundation (31100886).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00251-014-0769-2) contains supplementary material, which is available to authorized users.
Contributor Information
Tianyi Zhang, State Key Laboratory of Agrobiotechnology, College of Biological Sciences, National Engineering Laboratory for Animal Breeding, China Agricultural University, Beijing 100193, People's Republic of China.
Luca Tacchi, Center for Evolutionary and Theoretical Immunology, University of New Mexico, Albuquerque, NM 87131-0001, USA.
Zhiguo Wei, College of Animal Science and Technology, Henan University of Science and Technology, Henan 471003, People's Republic of China.
Yaofeng Zhao, Email: yaofengzhao@cau.edu.cn, State Key Laboratory of Agrobiotechnology, College of Biological Sciences, National Engineering Laboratory for Animal Breeding, China Agricultural University, Beijing 100193, People's Republic of China.
Irene Salinas, Email: isalinas@unm.edu, Center for Evolutionary and Theoretical Immunology, University of New Mexico, Albuquerque, NM 87131-0001, USA.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amemiya CT, Alföldi J, Lee AP, Fan S, Philippe H, MacCallum I, Braasch I, Manousaki T, Schneider I, Rohner N. The African coelacanth genome provides insights into tetrapod evolution. Nature. 2013;496:311–316. doi: 10.1038/nature12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann H, Venkatesh B, Brenner S, Meyer A. Nuclear protein-coding genes support lungfish and not the coelacanth as the closest living relatives of land vertebrates. Proc Natl Acad Sci. 2004;101:4900–4905. doi: 10.1073/pnas.0400609101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanella JJ, Bitincka L, Smalley J. MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinforma. 2003;4:29. doi: 10.1186/1471-2105-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrand S, Litman G, Lapointe N, Good R, Frommel D. The evolution of the immune response XII. The immunoglobulins of the turtle. Molecular requirements for biologic activity of 5.7 S immunoglobulin. J Immunol. 1971;107:1–11. [PubMed] [Google Scholar]
- Chen K, Cerutti A. Vaccination strategies to promote mucosal antibody responses. Immunity. 2010;33:479–491. doi: 10.1016/j.immuni.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G, Gao Y, Wang T, Sun Y, Wei Z, Li L, Ren L, Guo Y, Hu X, Lu Y. Extensive diversification of IgH subclass-encoding genes and IgM subclass switching in crocodilians. Nat Commun. 2013;4:1337. doi: 10.1038/ncomms2317. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edholm ES, Bengten E, Wilson M. Insights into the function of IgD. Dev Comp Immunol. 2011;35:1309–1316. doi: 10.1016/j.dci.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Flajnik MF. Comparative analyses of immunoglobulin genes: surprises and portents. Nat Rev Immunol. 2002;2:688–698. doi: 10.1038/nri889. [DOI] [PubMed] [Google Scholar]
- Gregory TR. Genome size evolution in animals. The evolution of the genome. 2005;1:4–87. [Google Scholar]
- Gregory TR. Macroevolution, hierarchy theory, and the C-value enigma 2009 [Google Scholar]
- Hallström BM, Janke A. Gnathostome phylogenomics utilizing lungfish EST sequences. Mol Biol Evol. 2009;26:463–471. doi: 10.1093/molbev/msn271. [DOI] [PubMed] [Google Scholar]
- Hordvik I. The impact of ancestral tetraploidy on antibody heterogeneity in salmonid fishes. Immunol Rev. 1998;166:153–157. doi: 10.1111/j.1600-065x.1998.tb01260.x. [DOI] [PubMed] [Google Scholar]
- Lee V, Huang JL, Lui MF, Malecek K, Ohta Y, Mooers A, Hsu E. The evolution of multiple isotypic IgM heavy chain genes in the shark. J Immunol. 2008;180:7461–7470. doi: 10.4049/jimmunol.180.11.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggatt RA, Iwama GK. Occurrence of polyploidy in the fishes. Rev Fish Biol Fish. 2003;13:237–246. [Google Scholar]
- Liang D, Shen XX, Zhang P. One thousand two hundred ninety nuclear genes from a genome-wide survey support lungfishes as the sister group of tetrapods. Mol Biol Evol. 2013;30:1803–1807. doi: 10.1093/molbev/mst072. [DOI] [PubMed] [Google Scholar]
- Magadán-Mompó S, Sánchez-Espinel C, Gambón-Deza F. IgH loci of American alligator and saltwater crocodile shed light on IgA evolution. Immunogenetics. 2013:1–11. doi: 10.1007/s00251-013-0692-y. [DOI] [PubMed] [Google Scholar]
- Marchalonis J. Isolation and characterization of immunoglobulin-like proteins of the Australian lungfish (Neoceratodus forsteri) Aust J Exp Biol Med Sci. 1969;47:405–419. doi: 10.1038/icb.1969.46. [DOI] [PubMed] [Google Scholar]
- Mashoof S, Goodroe A, Du C, Eubanks J, Jacobs N, Steiner J, Tizard I, Suchodolski J, Criscitiello M. Ancient T-independence of mucosal IgX/A: gut microbiota unaffected by larval thymectomy in Xenopus laevis. Mucosal Immunol. 2013;6:358–368. doi: 10.1038/mi.2012.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Y, Flajnik M. IgD, like IgM, is a primordial immunoglobulin class perpetuated in most jawed vertebrates. Proc Natl Acad Sci U S A. 2006;103:10723–10728. doi: 10.1073/pnas.0601407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T, Rast JP, Litman GW, Amemiya CT. Lineage-restricted retention of a primitive immunoglobulin heavy chain isotype within the Dipnoi reveals an evolutionary paradox. Proc Natl Acad Sci U S A. 2003;100:2501–2506. doi: 10.1073/pnas.0538029100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul WE. Fundamental Immunology. (seventh) 2012 [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29 doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumfelt LL, Diaz M, Lohr RL, Mochon E, Flajnik MF. Unprecedented multiplicity of Ig transmembrane and secretory mRNA forms in the cartilaginous fish. J Immunol. 2004;173:1129–1139. doi: 10.4049/jimmunol.173.2.1129. [DOI] [PubMed] [Google Scholar]
- Santander J, Xin W, Yang Z, Curtiss R. The aspartate-semialdehyde dehydrogenase of Edwardsiella ictaluri and its use as balanced-lethal system in fish vaccinology. PLoS One. 2010;5:e15944. doi: 10.1371/journal.pone.0015944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclavi B, Herrick J. Slow Evolution of rag1 and pomc genes in vertebrates with large genomes. 2013 arXiv preprint arXiv:1302.2182. [Google Scholar]
- Tacchi L, Larragoite E, Salinas I. Discovery of J chain in African lungfish (Protopterus dolloi, Sarcopterygii) using high throughput transcriptome sequencing: implications in mucosal immunity. PLoS One. 2013;8:e70650. doi: 10.1371/journal.pone.0070650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varriale S, Merlino A, Coscia MR, Mazzarella L, Oreste U. An evolutionary conserved motif is responsible for Immunoglobulin heavy chain packing in the B cell membrane. Mol Phylogenet Evol. 2010;57:1238–1244. doi: 10.1016/j.ympev.2010.09.022. [DOI] [PubMed] [Google Scholar]
- Vervoort A. Tetraploidy in Protopterus (Dipnoi) Experientia. 1980;36:294–296. [Google Scholar]
- Zhao Y, Pan-Hammarström Q, Yu S, Wertz N, Zhang X, Li N, Butler JE, Hammarström L. Identification of IgF, a hinge-region-containing Ig class, and IgD in Xenopus tropicalis. Proc Natl Acad Sci U S A. 2006;103:12087–12092. doi: 10.1073/pnas.0600291103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.