Abstract
MicroRNAs (miRNAs) are important regulatory elements for gene expression that are involved in diverse physiological and pathological processes. Canonical miRNA biogenesis consists of a two-step processing, from primary transcripts (pri-miRNAs) to precursor miRNAs (pre-miRNAs) mediated by Drosha in the nucleus and from pre-miRNAs to mature miRNAs mediated by Dicer in the cytoplasm. Various routes of miRNA maturation that are tightly regulated by signaling cascades and specific to an individual or a subclass of miRNAs have been recently identified. Here, we review the current findings in signaling-mediated miRNA processing as well as their potential clinical relevance in cancer.
Introduction
MiRNAs are abundant, short (20–25 nucleotides), single-stranded regulatory RNA molecules that play critical roles in a wide variety of biological processes, including DNA repair and tissue development, and are characterized by the cleavage of their precursor transcripts by one or more RNase III enzymes into mature miRNAs, which are subsequently loaded onto Argonaute proteins to form the RNA-induced silencing complex (RISC), a ribonucleoprotein complex involved in posttranscriptional gene silencing (1, 2). Similar to other classes of Argonaute-bound small RNAs, miRNAs also identify and target messenger RNAs (mRNAs) based on the ~7 nt complementary base-pairing to the “seed sequence” of an miRNA, preferentially nucleotides 2–8 from the 5’ end of a mature miRNA. Consequently, the targeted mRNAs are degraded, destabilized, or translationally suppressed by the Argonaute proteins (1, 3). Computational and experimental studies have provided mounting evidence to support the broad impact of a miRNA on hundreds of mRNA targets, such that a majority of the human transcripts are predicted and proven to carry conserved binding sites for multiple miRNAs (1). Not surprisingly, the deregulation of homeostatic control of miRNA biogenesis is associated with multiple pathological diseases, including cancers (4–6).
A global downregulation of mature miRNA levels as well as upregulation of specific miRNAs that are associated with oncogenic events (oncomiRs) are important features in cancer development and progression (4, 5, 7). The steady level of mature miRNA is determined by the rate of its transcription, biogenesis processing, and turnover (8). Transcriptional regulation, either through activation or silencing, accounts for most of the alterations in miRNA production (9). However, in cancers, a significant portion of mature miRNAs is downregulated in the tumors even though their primary transcripts and/or precursors (pre-miRNAs) are unaltered or even elevated (6, 7,10, 11). Indeed, miRNA maturation is subjected to complex regulations, and defects along this process may significantly contribute to tumorigenesis and cancer progression (12–15). Contrary to the notion that all miRNAs follow a universal linear pathway toward maturation (1, 8), a growing body of evidence indicates that an individual or a cluster of miRNAs can be processed and expressed differentially by miRNA-specific regulatory mechanisms. Such regulations mainly rely on the interplays between miRNA core machineries, RNA-binding proteins (RBPs), and signaling transducers or executors in response to external or internal stimuli, and dynamically shape the extent of miRNA production to maintain robust gene expression under specific physiological and/or pathophysiological conditions. In this review, we discuss the recent progress toward the understanding of the complexity of miRNA processing with specific emphasis on signaling-regulated miRNA maturation and its potential clinical application in cancers.
Classical linear processing of miRNAs
The canonical processing pathway mediated by RNase III enzymes generates the majority of miRNAs in metazoan (1, 16). Biogenesis of miRNA begins with RNA polymerase II-dependent (predominant) or RNA polymerase III-dependent transcription that generates a long primary transcript (pri-miRNA) containing a typical hairpin structure. Like mRNAs, most pri-miRNAs are 5’ 7-methyl-guanosine (m7G) capped and 3’ polyadenylated prior to cleavage by the nuclear microprocessor Drosha/DGCR8 heterodimer (1). DGCR8 (DiGeorge syndrome critical region gene 8; also known as Pasha in invertebrates) functions as a molecular anchor that recognizes pri-miRNA at the stem-single-stranded RNA junction and positions RNase III endonuclease Drosha at the correct catalytic sites to cleave ~11 bp away from the junction, releasing a hairpin-shaped pre-miRNA (1). Alternatively, miRNAs can be generated from short intronic hairpins called mirtrons that are excised by splicing and debranching to mimic a regular pre-miRNA, bypassing the first-step cleavage mediated by Drosha/DGCR8 in the nucleus (2). The secondary double-stranded RNA (dsRNA) stem (>14 bp) along with a short 3’ overhang of the resulting miRNA precursor, ~55–70 nucleotide (nt) in length, is then recognized by exportin-5 (XPO5) in complex with Ran-GTP, enabling its subsequent shuttling to the cytoplasm via GTP hydrolysis (1, 10).
In the cytoplasm, the terminal loop of pre-miRNA is cleaved by another dsRNA-specific RNase III Dicer in cooperation with dsRNA-binding protein TAR RNA-binding protein (TRBP; also known as human immunodeficiency virus transactivation responsive RNA-binding protein 2, TARBP2) or protein activator of PKR (PACT) (1, 8). Both the 5’-phosphorylated end and 3’ overhang of the pre-miRNA are anchored by the PAZ domain of Dicer while the dsRNA stem is placed along the positively charged protein extension to reach the catalytic center of Dicer. This spatial arrangement determines the precise cleavage site in the miRNA precursor at a fixed distance of ~22 nt either from the 3’ (3’ counting model) (1) or 5’ terminus (5’ counting rule) (17). TRBP facilitates Dicer-mediated cleavage through direct binding with the pre-miRNA via its two dsRNA-binding domains (dsRBDs) and concomitantly enhances the stability of Dicer-RNA complex using its third dsRBD (18, 19). Both TRBP and PACT, though not essential for miRNA maturation, participate in the recruitment of Argonaute2 (Ago2) to Dicer, forming the RISC loading complex (RLC) for efficient miRNA processing and subsequent RISC assembly (8). Notably, in vitro reconstitution of RISC loading and activation can be achieved by just Dicer, TRBP, and Ago2 alone (20). Exported miRNA precursors associate with a pre-formed RLC complex independently of ATP hydrolysis (20). After cleavage, miRNA duplex is further unwound based on the rule of asymmetric separation–the strand with less stable base pairs at its 5’ end in dsRNA intermediate is selected and loaded onto RISC (1). Although it remains unclear how the RLC precisely coordinates the sequential events, such as precursor loading, cleavage, duplex unwinding, and subsequent cargo loading onto RISC, evidence suggests that the stable end of the Dicer-cleaved RNA duplex is bound to TRBP in RLC whereas the other end interacts with Ago2 (1, 3), resulting in a functional guide strand (mature miRNA) that is complementary to its mRNA targets and a passenger strand that is quickly degraded. After the dsRNA unwinds, Ago2 (and possibly other Ago proteins) loaded with mature miRNA dissociates from Dicer and TRBP to form the active RISC for target gene silencing (3).
Argonautes are highly specialized binding modules that function as regulators in miRNA maturation and executors in small RNA-mediated gene silencing (8, 21). All Ago proteins are characterized by evolutionarily conserved MID and PAZ domains involved in RNA binding, and an RNase H-like PIWI domain with endonuclease activity (3, 21). In mammals, there are four Ago proteins (Ago1–4) but only Ago2 is catalytically active and functions as an endonuclease (21). Structural studies have indicated that the 3’ end of a mature miRNA is stably bound by the PAZ domain while the 5’ phosphate group of the small RNA is anchored by the MID domain (3). Proper loading and duplex unwinding of the small RNA are further assisted by Argonautes’ N domain (22). In some cases, a processing intermediate is generated by Ago2-dependent cleavage of pre-miRNA to facilitate miRNA maturation and removal of the passenger strand (opposite strand of a mature miRNA). Additionally, Ago proteins are capable of stabilizing mature miRNAs by direct binding and post-transcriptionally regulating miRNA abundance independently of their endonuclease activity (23). Recently, a Dicer-independent miRNA biogenesis pathway utilizing Ago2 slicer catalytic activity for processing, as exemplified by pre-miR-451, was identified (24, 25). The short stem region of pre-miR-451 cannot be recognized by Dicer but is trimmed by Ago2 to generate the corresponding mature miRNA (24). However, the extent to which this Ago-dependent miRNA processing occurs is still unclear and requires further investigation.
What regulates the classical linear miRNA processing?
MiRNAs are often deregulated in cancers (4, 5, 7). In contrast to oncogenes or tumor suppressors, miRNA alteration is rarely caused by gene amplification or disruption (2). Rather, impairment of critical steps in the process of miRNA maturation, either in the nucleus or in the cytoplasm, is likely to be the underlying cause that contributes to the development of malignancies. Notably, recent studies have demonstrated extensive crosstalk between signaling pathways and miRNA processing (Figure 1), suggesting that miRNA biogenesis is under tight signaling control and has been an important part of the large regulatory networks.
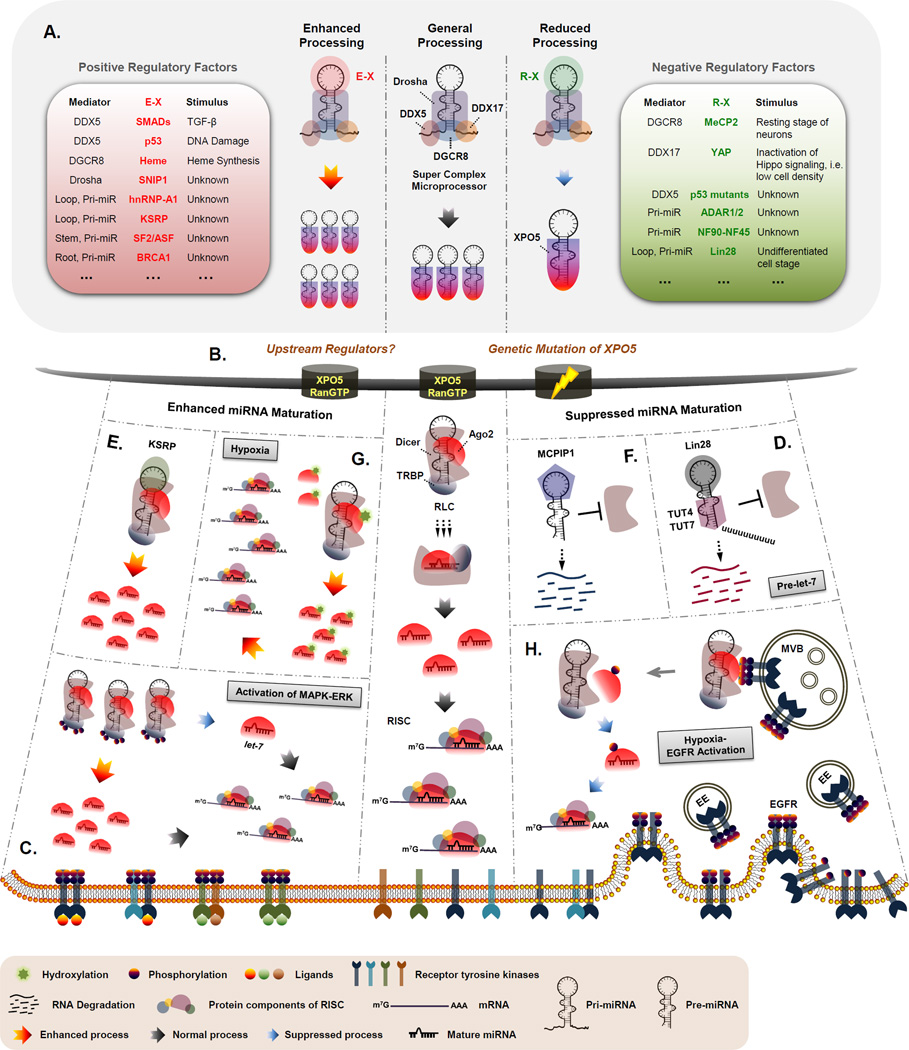
Figure 1. Canonical linear processing and alternative routes of miRNA maturation.
A. Regulatory factors involved in nuclear miRNA processing are summarized as indicated. Mediator, direct interacting partner; E-X, enhancing factor-X; R-X, reducing factor-X. B. The direct upstream regulators for miRNA shuttling have not yet been identified. However, the XPO5 function is compromised in human primary colorectal tumors with microsatellite instability due to frameshift mutations. C. TRBP is phosphorylated and stabilized by activated MAPK-ERK pathway. Phosphorylated TRBP stabilizes Dicer and enhances general miRNA processing with the exception of pre-let-7, which is inhibited by TRBP phosphorylation. D. Lin28 recognizes and binds to the loop of pre-let-7 and subsequently recruits TUT4 and/or TUTase7 to uridylate the miRNA precursor at the 3’ terminus. Polyuridylation inhibits Dicer cleavage and tags the precursor RNA for rapid degradation by nucleases. E. KSRP interacts with Dicer and facilitates miRNA processing by recognizing and anchoring to the conserved region of precursor terminal loop. F. The loop structure of pre-miRNAs is also recognized by MCPIP1, a Dicer antagonist, which binds to and cleaves the loops from precursors for degradation. G. Ago2 is stabilized and functionally potentiated by hydroxylation at P700 mediated by type I collagen hydroxylase whose expression level is elevated under hypoxia. H. In response to hypoxia, Ago2 is phosphorylated by internalized EGFR at Y393 due to enhanced EGFR-Ago2 association in MVBs. Phosphorylation of Ago2 decreases its binding to Dicer, which then suppresses the maturation of a subclass of miRNAs with long-loop structures in their precursors. This in turn reduces the RISC activity due to a decreased loading of mature miRNAs onto RISC under hypoxia. EE, early endosome; MVB, multivesicular body.
Signaling-mediated regulation of miRNA processing in the nucleus
DDX5 and DDX17- hub for signaling transduction
Microprocessor, Drosha-DGCR8 complex, is essential for miRNA processing in the nucleus (1, 16). To maintain homeostatic control of miRNA biogenesis, the stability of this complex is self-regulated such that Drosha-DGCR8 destabilizes DGCR8 mRNA through direct binding and DGCR8 stabilizes Drosha via protein-protein interaction (26). The nuclear processing efficacy of pri-miRNAs is further fine-tuned by other accessory RNA-binding proteins, such as DDX5 (also known as p68) and DDX17 (also known as p72) (1, 10). Both DDX5 and DDX17 interact directly with Drosha/DGCR8 to form a larger nuclear processing complex (also known as super microprocessor) and function as restricted promoting factors that are specifically required for efficient processing of a subset of pri-miRNAs (10, 27, 28). Interestingly, DDX5 and DDX17 also serve as signal mediators that bridge the nuclear microprocessor activity with other signaling pathways under various circumstances. For instance, in DNA damage, tumor suppressor p53 binds with DDX5 to enhance Drosha/DGCR8-mediated processing of a cluster of miRNAs that exert growth suppressive functions. Transcriptionally inactive p53 mutants, frequently identified in human tumors, interfere with the functional assembly between Drosha complex and DDX5, which in turn hinder the nuclear processing of these miRNAs (14). A similar case has been demonstrated for SMAD signal transducers in response to transforming growth factor-β (TGF-β) and bone morphogenetic protein (BMP) growth factor (15) in which the SMADs, in complex with the super microprocessor component DDX5, promote the nuclear processing of miRNA (specifically, miR-21, a well-characterized oncomiR in cancer) via a consensus binding sequence (similar to the known SMAD-response element in gene promoters) in the dsRNA stem of the pri-miRNA. More recently, cell density-dependent control of miRNA biogenesis is linked to the tumor-suppressive Hippo signaling pathway via its downstream target Yes-associated protein 1 (YAP, an oncogene inactivated by Hippo signaling) (13). At low cell density, DDX17 (p72) is bound and sequestered by nuclear YAP and is dissociated from the nuclear microprocessor complex, hence decreasing the processing efficacy of a group of pri-miRNAs in the nucleus. When reaching high cell density, YAP is restricted to the cytoplasm by activated Hippo signaling. In such case, DDX17 associates with Drosha/DGCR8 complex and enhances miRNA processing by binding to a specific sequence motif in the 3’ flanking region of a pri-miRNA.
Notably, DDX5 and DDX17 are also known to function as transcription regulators (27), either as coactivators (predominant) or corepressors. For instance, both DDX5 and DDX17 serve as coactivators of ERα, β-catenin, and MyoD by recruiting other components of transcriptional machinery to the target gene promoters (29–32). More specifically, DDX5 interacts directly with transcription factors p53, AR, RunX2, and NF-kB (p50 subunit) (33–36), and DDX17 upregulates MDM2 transcription in both p53-dependent and -independent manners (27). Although the precise roles of DDX5 and DDX17 in cancer development remain controversial, Ddx5-deficient and Ddx17-deficient mouse embryonic fibroblasts exhibit reduced cell growth and enhanced apoptosis (37). Consistently, silencing both DDX5 and DDX17 inhibits cell proliferation (38) whereas overexpression of DDX5 facilitates cell growth and promotes epithelial-to-mesenchymal (EMT) transition upon growth factor stimulation (32). In human invasive breast carcinomas, overexpression and gene amplification, but rarely mutations, are frequently observed in DDX5 (8.6% amplification vs. 0.3% mutation) (39). Interestingly however, higher expression of DDX17 is associated with increased disease-free and overall survival (40). In terms of prognosis, differential roles of DDX5 and DDX17 have also been reported in other cancer types, including head and neck, prostate, and colorectal cancers (34, 41, 42). It is likely that their functions, as tumor promoters or suppressors, are cell type-dependent or restricted by certain genetic background. Of note, numerous binding partners of DDX5 and DDX17 are involved in both transcriptional regulation and miRNA processing, suggesting that an intimate gene regulatory network exists at transcriptional and posttranscriptional levels. Nonetheless, the detailed functions of DDX5 and DDX17 as components of the Drosha/DGCR8 complex in cancer development remain elusive, necessitating further systematic studies of their specific target miRNAs.
Other regulatory elements of nuclear miRNA processing
Drosha/DGCR8-mediated miRNA processing is also subjected to other regulatory mechanisms, including additional binding partners of microprocessor, posttranscriptional RNA editing, and multiple RNA-binding proteins that recognize and directly interact with a specific subclass of pri-miRNAs (summarized in Figure 1A).
Additional microprocessor partners
DGCR8 is a heme-binding protein, and heme promotes DGCR8 dimerization to trimerize upon pri-miRNA loading which subsequently facilitates nuclear miRNA processing by blocking DGCR8’s auto-inhibition domain (43). In contrast, methyl-CpG binding protein 2 (MeCP2), a transcriptional repressor, which is critical for proper development and function in the central nervous system, interferes with the assembly of Drosha/DGCR8 complex through direct binding to DGCR8 via its C-terminal domain (44). Interestingly, the association between MeCP2 and DGCR8 is tightly regulated by the phosphorylation status of MeCP2 at Ser80, which is continuously phosphorylated in a resting state but rapidly dephosphorylated upon neuronal activation (44). As a protein-binding switch, phosphorylation of MeCP2 at Ser80 maintains it in an open conformation to allow interaction with DGCR8, leading to inhibition of miRNA maturation by dissembling the microprocessor complex (44). Smad nuclear interacting protein 1 (SNIP1), a known inhibitor of the TGF-β and NF-kB signaling pathways, is also involved in the regulation of miRNA biogenesis (45). SNIP1 has direct RNA-binding capability and associates with Drosha to enhance miRNA production. Different from other miRNA regulators, knockdown of SNIP1 downregulates pri- (transcription unaffected), pre- and mature miRNAs, suggesting that SNIP1 regulates the turnover/stability of pri-miRNAs rather than the processing efficacy in the nucleus (45).
Posttranscriptional modification of miRNAs
Intriguingly, the stability and nuclear processing of pri-miRNAs are also regulated by RNA editing. Adenosine deaminases acting on RNA (ADARs), ADAR1 and ADAR2, are involved in the conversion of adenosine to inosine in a subclass of pri-miRNAs. The resulting A to I conversion, suppresses Drosha-mediated cleavage by reducing the overall stability of dsRNA via conformational change. Edited pri-miRNAs are then recognized and degraded by Tudor staphylococcal nuclease, a ribonuclease that specifically targets inosine-containing dsRNAs (46). Of note, the RNA-editing activity of ADAR1 is inhibited by SUMO-1 via SUMOylation at K418 (47), suggesting the presence of an upstream signaling modulator for RNA-editing-regulated miRNA processing.
RNA-binding proteins
Another essential regulatory module in the nuclear miRNA processing belongs to a large family of RNA-binding proteins (RBPs). RBPs, including DDX5 and DDX17, are key players in miRNA biogenesis as they control multi-step processing as well as the localization, degradation and functional activity of miRNAs. Most RBPs recognize specific sequence motifs or secondary structures via their RNA-binding domains. For instance, mRNA splicing factor SRSF1 (also known as SF2/ASF) binds to the stem region of specific pri-miRNAs (48), while heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) and KH-type splicing regulatory protein (KSRP; also known as KHSRP) recognizes the terminal loop of a subclass of pri-miRNAs (49, 50). Nonetheless, they all facilitate cleavage by Drosha/DGCR8 upon pri-miRNA anchoring, with the exception of pri-let-7, which is inhibited by hnRNP A1 (51). Another RBP, Lin28, which is exclusively expressed in the undifferentiated stage, also specifically binds to the terminal loop of pri-let-7, thereby inhibiting its processing in the nucleus (52). Similarly, NF90 and NF45, two known components of the larger Drosha/DGCR8 complex, suppress Drosha-mediated cleavage via competitive loading with certain pri-miRNAs (53). The fact that RBPs share a large number of overlapping RNA targets raises an interesting question of how upstream signals efficiently coordinate different RBP-RNA interactomes, either spatially or temporally, to influence specific miRNA processing in the nucleus.
DNA-binding proteins
Rarely, a DNA-binding protein, as exemplified by the DNA damage repair protein, breast cancer associated gene 1 (BRCA1), promotes the processing of a subclass of miRNAs via direct interaction with their primary transcripts (54). BRCA1 associates with the super microprocessor complex as well as other additional processing modulators, including Smad3, p53, and DHX9. The DNA-binding domain of BRCA1 is required for anchoring the branched structure at the root of a hairpin in pri-miRNAs similar to its DNA-binding preference. Because the regulatory function of BRCA1 in miRNA processing was examined in the absence of DNA damage, it remains unknown whether BRCA1 maintains similar functions in response to DNA damage, and if so, whether the miRNAs regulated by BRCA1 are critical for its functions in DNA repair.
Genetic control of miRNA shuttling
A frameshift mutation was identified in pre-miRNA nuclear export receptor, XPO5, which creates a premature termination codon, results in a truncated protein that lacks a small region in the C-terminus (55). Interestingly, mutant XPO5 fails to form a proper XPO5/Ran-GTP/pre-miRNA ternary complex due to its reduced binding and/or recognition capability with the pre-miRNA cargo. Consequently, the miRNA precursors after the first-step processing are trapped in the nucleus, leading to reduced expression of mature miRNAs (55). Genetic alterations of XPO5 are heterogeneous in human cancers that generally occur at a low frequency. In ovarian serous cystadenocarcinoma and glioblastoma, XPO5 is amplified (with 2.8% and 1.5% incidence, respectively) (56, 57) without mutations, whereas gene amplification and mutations of XPO5 coexist in other cancer types, including breast, prostate and colon cancers (1.4% amplification vs. 0.5% mutation in colorectal carcinomas) (58). In contrast, XPO5 frameshift mutations are present in 22.8% of human primary colorectal tumors with microsatellite instability, a state of genetic hypermutability that results from impaired DNA mismatch repair (55), suggesting that genetic control of miRNA export is critically important in this subpopulation of patients. So far, the upstream signaling regulators for pre-miRNA export, either via XPO5 or Ran-GTP, have not yet been identified (Figure 1B).
Signaling-mediated regulation of miRNA processing in the cytoplasm
Regulation of Dicer
As a haploinsufficient tumor suppressor, mutations and heterozygous deletion of DICER1 are frequently observed in human tumors (59–61). The steady level of Dicer is positively regulated by the Dicer-TRBP complex (18), and the expression of Dicer is further controlled by multiple miRNAs targeting its long 3’ untranslated region (UTR). Both let-7 and miR-103/107 reduce the protein level of Dicer through direct interaction with multiple binding sequences found in the 3’ UTR or the coding region of DICER1 mRNA (62, 63). High expression of miR-103/107 attenuates Dicer-mediated miRNA processing, resulting in downregulation of mature miRNAs. Subsequently, blocked biogenesis, particularly of the miR-200 family, promotes EMT in breast cancer cells (63). More recently, Dicer was shown to be SUMOylated after short exposure of alveolar macrophages to cigarette smoke (64), but the detailed mechanistic nature as well as the potential functional importance of this posttranslational modification in Dicer-mediated miRNA processing remain unclear.
Regulation of TRBP
TRBP, a key component of the RISC-loading complex, is also susceptible to genetic disruptions. Frameshift TRBP mutants with reduced binding affinity with Dicer have been identified in human colorectal tumors with microsatellite instability (65, 66). In general, the mutation rate of TRBP is very low in human cancers, such that less than 1.5% is found in colorectal adenocarcinoma (58). However, TRBP is deleted in 15% of adenoid cystic carcinoma (67), implying a potential pivotal role of TRBP in this specific cancer type. The underlying mechanisms of genetic deletion as well as the corresponding functional consequences of TRBP deletion in these tumors remain to be investigated.
Interestingly, TRBP is rapidly phosphorylated by mitogen-activated protein kinase 1 (Erk) at S142, S152, S283 and S286 in response to growth factor stimulation (68). Phosphorylation of TRBP stabilizes its interaction with Dicer, resulting in enhanced miRNA processing in the cytoplasm. In contrast, expression of the let-7 tumor suppressor family is downregulated in the context of TRBP phosphorylation (Figure 1C), raising the question of how phosphorylated TRBP recognizes and specifically inhibits the maturation of let-7 miRNAs. More recently, additional residues in TRBP at the N-terminal region were reported to be phosphorylated (referred to as “hyperphosphorylation” to distinguish it from the above-mentioned phosphorylation events mediated by Erk) by JNK during cell cycle progression, specifically in M phase (69). Hyperphosphorylated TRBP does not affect its binding affinity for Dicer but enhances its inhibitory activity toward another known binding partner, dsRNA-dependent protein kinase R (PKR), through strengthened direct association between TRBP-PKR during mitosis (69). The mechanistic nature of TRBP hyperphosphorylation, including the specific residues of TRBP phosphorylated by JNK and/or other additional mitosis-specific kinases as well as the detailed biological consequences of this hyperphosphorylation event in cell cycle progression and/or miRNA regulation, is worthy of further investigation.
Specific regulation of the let-7 family
The let-7 miRNAs directly target multiple oncogenes, including RAS (70), myc (71) and HMGA2 (72, 73), which are involved in cell proliferation and invasion. Hence, let-7 family members function as tumor suppressors, and their reduced expression levels in many human cancers are closely associated with cancer progression (74–76). Not surprisingly, it appears that multiple pathways concomitantly control the biogenesis of the let-7 family miRNAs. In the cytoplasm, pre-let-7 bound by Lin28 is frequently uridylated by TUT4 (also known as TUTase4 or ZCCHC11) and/or TUTase7 (also known as ZCCHC6) at the 3’ terminus, which consequently inhibits Dicer cleavage for maturation and tags the precursor RNA for rapid degradation by nucleases (Figure 1D) (77–79). In contrast, KSRP anchors to the conserved region of the let-7 terminal loop and that of the other miRNAs, thereby facilitating their maturation through interaction with Dicer in the cytoplasm (Figure 1E) (50). The loop structure of pre-miRNA is also recognized by the endo-RNase monocyte chemoattractant protein induced protein 1 (MCPIP1; also known as ZC3H12A), a Dicer antagonist, which binds to and cleaves the loop from the target miRNA precursors for degradation (Figure 1F) (80). In particular, both Lin28 and MCPIP1 are upregulated in response to inflammation (80, 81) while KSRP is phosphorylated and functionally inhibited by p38 MAP kinase and the PI3K-AKT pathway (82, 83). All of these suggest that the maturation of the let-7 family is likely under extensive signaling regulation.
Regulation of Argonautes
Another essential regulatory module in miRNA biogenesis is the Argonautes. Particularly, Ago2 is under tight regulation in response to various signals. Ago2 is stabilized and functionally potentiated by type I collagen prolyl-4-hydroxylase-mediated hydroxylation at P700 (Figure 1G) (84, 85). During undifferentiated cell stage, such as in embryonic stem cells and embryocarcinoma cell lines, Ago2 is polyubiquitinated by the E3 ligase mLin-41 (mouse homologue of Lin-41; also known as TRIM71), and subsequently undergoes proteasome-mediated degradation (86). Similar to Lin28, Lin-41 is also targeted and suppressed by let-7 in differentiated cells. To date, the specific E3 ligase responsible for Ago2 ubiquitination in somatic cells has not yet been identified. Nonetheless, studies have indicated that miRNA-loading-free Dicer and Ago2 are marked by polyubiquitination and undergo autophagy-mediated degradation through direct interaction with selective autophagy receptor NDP52 (87). In the context of cellular stress, Ago2 is modified by poly-ADP-ribosylation which consequently relieves miRNA-mediated gene silencing (88). The expression of mature miRNAs under this stress condition was not examined by the authors. Thus, it remains unknown whether the process of miRNA maturation would be similarly affected by this type of posttranslational modification. Besides that, the functionality of Ago2 in miRNA processing is also modulated by its binding partner heat shock protein 90 (Hsp90) which facilitates the loading of RNA duplex from Dicer into Ago2 in an ATP-dependent manner (89–91).
Like most signaling mediators, Ago2 receives signals (e.g., phosphorylation) in response to external or internal stimuli and transmits new signals (functional changes) by orchestrating gene expression programs via miRNA regulation. Human Ago2 is phosphorylated at Ser387 by both p38 MAP kinase and the proto-oncogene Akt3 (92, 93). S387 phosphorylation of Ago2 facilitates its localization to the cytoplasmic processing bodies (P bodies, which are intracellular sites for mRNA turnover and translational repression) and enhances its binding with GW182, a key component of RISC. As a net result, Ago2-mediated translational repression is strengthened while its cleavage activity toward target mRNAs is paradoxically reduced (93). In addition, mass spectrometric analysis identified Ago2-Y529 as a potential phosphorylation site (94). Substitution of Y529 with a negatively charged glutamate reduces the binding ability of Ago2 to small RNAs. Notably, Ago2-Y529 phosphorylation was recently found to be elevated in response to LPS in macrophages, leading to a transient reversal of miRNA-mediated repression that subsequently enhances cytokine expression during the early phase of inflammatory response (95). However, the specific tyrosine kinase that is responsible for this phosphorylation has not yet been identified. More recently, our laboratory reported another phosphorylation of Ago2 at a highly conserved residue Y393, which is located in the linker region between the PAZ and MID domain (12, 94). In response to hypoxia, Ago2-Y393 is phosphorylated by internalized EGFR due to enhanced EGFR-Ago2 association in multivesicular bodies (MVBs) (12). Phosphorylation of Ago2 at Y393 decreases its binding with Dicer, which consequently suppresses the maturation of a subset of miRNAs with long-loop structures in their precursors, leading to a reduced RISC activity under hypoxia (Figure 1H). Interestingly, a majority of the miRNAs regulated by Ago2-Y393 phosphorylation in response to hypoxic stress possesses tumor-suppressive properties, at least in the cell lines examined such as HeLa and MDA-MB-231. Consistently, phospho-Y393-Ago2 enhances cell survival and invasiveness under hypoxia, and is significantly correlated with poorer overall survival in breast cancer patients (12). Moreover, Ago2-Y393 phosphorylation is also regulated by tyrosine phosphatase 1B (PTP1B) and plays a pivotal role in H-RASV12-induced senescence in primary non-transformed cells (96). PTP1B is oxidized and inactivated by oncogene-induced ROS that leads to elevated Ago2-Y393 phosphorylation, which decreases miRNA loading due to its reduced association with Dicer and consequently enhances the expression of p21, resulting in premature senescence (96). It is not clear how one molecular event leads to different biological consequences in cancer and normal cells. A possible explanation is that p53 and RB, two key master regulators in oncogene-induced senescence, are frequently mutated in human breast cancer cells (97, 98), and thus bypassing Ago2 phosphorylation-induced checkpoint in senescence. Alternatively, the role of Ago2-Y393 phosphorylation may be fine-tuned by the expression profile of miRNA precursors and/or target mRNA levels which may vary between different cell types and/or treatments.
From a genetic perspective, Ago2 is very different from Dicer and TRBP. Amplification of AGO2, but not other AGO family members, is frequently observed in human cancers (39, 56, 99), including ovarian serous cystadenocarcinoma (23.4%), breast invasive carcinoma (15.7%), and metastatic prostate adenocarcinoma (14.8%). Intriguingly, amplification and/or upregulation of EGFR and AGO2 as well as TP53 mutation have a strong tendency toward co-occurrence in breast invasive tumors (EGFR and AGO2, P < 0.000001, odds ratio = 11.096; AGO2 and TP53, P < 0.000001, odds ratio = 5.946; EGFR and TP53, P < 0.000001, odds ratio = 19.063; Fisher’s Exact Test, from cBioPortal for Cancer Genomics), suggesting a common regulatory event upstream of EGFR, AGO2, and TP53, or a cross-regulation among them. It remains unclear whether Ago2 itself, via miRNA-dependent and/or -independent functions, is a potential driver in cancer development or whether Ago2 is frequently phosphorylated due to concomitant hyper-activation of kinases (EGFR and possibly other tyrosine kinases), thereby conferring advantages for tumor progression with certain genetic background, such as TP53 mutation. In the latter scenario, complete inhibition of the upstream kinase activities may have potential clinical benefits for breast cancer patients with AGO2 amplification.
Conclusion
Extensive studies in the recent years have demonstrated that miRNA biogenesis is tightly regulated by signaling pathways to achieve robust dynamic control of gene expression. However, the mechanistic nature of signaling-mediated miRNA maturation is only beginning to unfold, particularly how different upstream signals determine the regulation specificity of the general miRNA biogenesis machinery and how important they are as downstream events in response to specific external or internal stimuli. The biological consequences of signaling-mediated regulation of miRNA biogenesis are complicated, likely to be context-dependent, and restricted by specific genetic background and expression profiles of pri- and/or pre-miRNAs. Also, it remains elusive how cancer cells temporally (the sequential order of tumor suppressor silencing, oncogene activation, and dysregulation of miRNA biogenesis) and/or spatially (the potential functional regulation of miRNA biogenesis exerted by signaling-controlled subcellular localization) control the functions of the miRNA biogenesis machinery to shape their miRNA-mediated gene silencing activity toward a more “cancer-prone” background to facilitate tumor development and progression. Systematic investigation and detailed analyses of the crosstalk between miRNA processing and traditional signaling networks under different circumstances, such as certain genetic background and/or specific internal and/or external stimuli, will be a central challenge in miRNA-related research. A better and comprehensive understanding of the regulatory mechanisms of miRNA biogenesis in pathophysiological settings, e.g., in cancers, would be a prerequisite for drug development and clinical applications.
Acknowledgments
We thank Dr. Jennifer L. Hsu for editing the manuscript. We apologize to all scientists whose work could not be cited in this review as a result of space constraints.
Grant Support
This work was funded in part by: National Institutes of Health grants (CA109311, CA099031, and CCSG CA16672); National Breast Cancer Foundation, Inc.; Breast Cancer Research Foundation; Patel Memorial Breast Cancer Endowment Fund; The University of Texas MD Anderson-China Medical University and Hospital Sister Institution Fund; Ministry of Health and Welfare, China Medical University Hospital Cancer Research Center of Excellence (MOHW103-TD-B-111; Taiwan), Program for Stem Cell and Regenerative Medicine Frontier Research (NSC102-2321-B-039; Taiwan), International Research-Intensive Centers of Excellence in Taiwan (NSC103-2911-I-002); and Center for Biological Pathways.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nature reviews Molecular cell biology. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 2.Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Molecular cell. 2011;43:892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meister G. Argonaute proteins: functional insights and emerging roles. Nature reviews Genetics. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 4.Leung AK, Sharp PA. microRNAs: a safeguard against turmoil? Cell. 2007;130:581–585. doi: 10.1016/j.cell.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs--the micro steering wheel of tumour metastases. Nature reviews Cancer. 2009;9:293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 6.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 8.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 9.Choudhry H, Catto JW. Epigenetic regulation of microRNA expression in cancer. Methods Mol Biol. 2011;676:165–184. doi: 10.1007/978-1-60761-863-8_12. [DOI] [PubMed] [Google Scholar]
- 10.van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nature reviews Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 11.Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome biology. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen J, Xia W, Khotskaya YB, Huo L, Nakanishi K, Lim SO, et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497:383–387. doi: 10.1038/nature12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mori M, Triboulet R, Mohseni M, Schlegelmilch K, Shrestha K, Camargo FD, et al. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell. 2014;156:893–906. doi: 10.1016/j.cell.2013.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 15.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nature reviews Molecular cell biology. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 17.Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, et al. Dicer recognizes the 5' end of RNA for efficient and accurate processing. Nature. 2011;475:201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daniels SM, Melendez-Pena CE, Scarborough RJ, Daher A, Christensen HS, El Far M, et al. Characterization of the TRBP domain required for dicer interaction and function in RNA interference. BMC molecular biology. 2009;10:38. doi: 10.1186/1471-2199-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes & development. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nature reviews Molecular cell biology. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 22.Kwak PB, Tomari Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nature structural & molecular biology. 2012;19:145–151. doi: 10.1038/nsmb.2232. [DOI] [PubMed] [Google Scholar]
- 23.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 24.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, et al. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuller-Pace FV, Moore HC. RNA helicases p68 and p72: multifunctional proteins with important implications for cancer development. Future Oncol. 2011;7:239–251. doi: 10.2217/fon.11.1. [DOI] [PubMed] [Google Scholar]
- 28.Slezak-Prochazka I, Durmus S, Kroesen BJ, van den Berg A. MicroRNAs, macrocontrol: regulation of miRNA processing. RNA. 2010;16:1087–1095. doi: 10.1261/rna.1804410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Endoh H, Maruyama K, Masuhiro Y, Kobayashi Y, Goto M, Tai H, et al. Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor alpha. Mol Cell Biol. 1999;19:5363–5372. doi: 10.1128/mcb.19.8.5363. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, et al. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 31.Caretti G, Schiltz RL, Dilworth FJ, Di Padova M, Zhao P, Ogryzko V, et al. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Developmental cell. 2006;11:547–560. doi: 10.1016/j.devcel.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 32.Yang L, Lin C, Liu ZR. P68 RNA helicase mediates PDGF-induced epithelial mesenchymal transition by displacing Axin from beta-catenin. Cell. 2006;127:139–155. doi: 10.1016/j.cell.2006.08.036. [DOI] [PubMed] [Google Scholar]
- 33.Bates GJ, Nicol SM, Wilson BJ, Jacobs AM, Bourdon JC, Wardrop J, et al. The DEAD box protein p68: a novel transcriptional coactivator of the p53 tumour suppressor. The EMBO journal. 2005;24:543–553. doi: 10.1038/sj.emboj.7600550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark EL, Coulson A, Dalgliesh C, Rajan P, Nicol SM, Fleming S, et al. The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Res. 2008;68:7938–7946. doi: 10.1158/0008-5472.CAN-08-0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang R, Jiao Z, Li R, Yue H, Chen L. p68 RNA helicase promotes glioma cell proliferation in vitro and in vivo via direct regulation of NF-kappaB transcription factor p50. Neuro-oncology. 2012;14:1116–1124. doi: 10.1093/neuonc/nos131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jensen ED, Niu L, Caretti G, Nicol SM, Teplyuk N, Stein GS, et al. p68 (Ddx5) interacts with Runx2 and regulates osteoblast differentiation. Journal of cellular biochemistry. 2008;103:1438–1451. doi: 10.1002/jcb.21526. [DOI] [PubMed] [Google Scholar]
- 37.Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 38.Jalal C, Uhlmann-Schiffler H, Stahl H. Redundant role of DEAD box proteins p68 (Ddx5) and p72/p82 (Ddx17) in ribosome biogenesis and cell proliferation. Nucleic Acids Res. 2007;35:3590–3601. doi: 10.1093/nar/gkm058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wortham NC, Ahamed E, Nicol SM, Thomas RS, Periyasamy M, Jiang J, et al. The DEAD-box protein p72 regulates ERalpha-/oestrogen-dependent transcription and cell growth, and is associated with improved survival in ERalpha-positive breast cancer. Oncogene. 2009;28:4053–4064. doi: 10.1038/onc.2009.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Causevic M, Hislop RG, Kernohan NM, Carey FA, Kay RA, Steele RJ, et al. Overexpression and poly-ubiquitylation of the DEAD-box RNA helicase p68 in colorectal tumours. Oncogene. 2001;20:7734–7743. doi: 10.1038/sj.onc.1204976. [DOI] [PubMed] [Google Scholar]
- 42.Haines GK, Ghadge GD, Becker S, Kies M, Pelzer H, Thimmappaya B, et al. Correlation of the expression of double-stranded RNA-dependent protein kinase (p68) with differentiation in head and neck squamous cell carcinoma. Virchows Archiv B, Cell pathology including molecular pathology. 1993;63:289–295. doi: 10.1007/BF02899275. [DOI] [PubMed] [Google Scholar]
- 43.Faller M, Matsunaga M, Yin S, Loo JA, Guo F. Heme is involved in microRNA processing. Nature structural & molecular biology. 2007;14:23–29. doi: 10.1038/nsmb1182. [DOI] [PubMed] [Google Scholar]
- 44.Cheng TL, Wang Z, Liao Q, Zhu Y, Zhou WH, Xu W, et al. MeCP2 Suppresses Nuclear MicroRNA Processing and Dendritic Growth by Regulating the DGCR8/Drosha Complex. Developmental cell. 2014;28:547–560. doi: 10.1016/j.devcel.2014.01.032. [DOI] [PubMed] [Google Scholar]
- 45.Yu B, Bi L, Zheng B, Ji L, Chevalier D, Agarwal M, et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10073–10078. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nature structural & molecular biology. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Desterro JM, Keegan LP, Jaffray E, Hay RT, O'Connell MA, Carmo-Fonseca M. SUMO-1 modification alters ADAR1 editing activity. Mol Biol Cell. 2005;16:5115–5126. doi: 10.1091/mbc.E05-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu H, Sun S, Tu K, Gao Y, Xie B, Krainer AR, et al. A splicing-independent function of SF2/ASF in microRNA processing. Molecular cell. 2010;38:67–77. doi: 10.1016/j.molcel.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nature structural & molecular biology. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 50.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michlewski G, Caceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nature structural & molecular biology. 2010;17:1011–1018. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 53.Sakamoto S, Aoki K, Higuchi T, Todaka H, Morisawa K, Tamaki N, et al. The NF90-NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol Cell Biol. 2009;29:3754–3769. doi: 10.1128/MCB.01836-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kawai S, Amano A. BRCA1 regulates microRNA biogenesis via the DROSHA microprocessor complex. The Journal of cell biology. 2012;197:201–208. doi: 10.1083/jcb.201110008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer cell. 2010;18:303–315. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 56.Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lambertz I, Nittner D, Mestdagh P, Denecker G, Vandesompele J, Dyer MA, et al. Monoallelic but not biallelic loss of Dicer1 promotes tumorigenesis in vivo. Cell death and differentiation. 2010;17:633–641. doi: 10.1038/cdd.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hill DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, Desruisseau D, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science. 2009;325:965. doi: 10.1126/science.1174334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes & development. 2009;23:2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tokumaru S, Suzuki M, Yamada H, Nagino M, Takahashi T. let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis. 2008;29:2073–2077. doi: 10.1093/carcin/bgn187. [DOI] [PubMed] [Google Scholar]
- 63.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 64.Gross TJ, Powers LS, Boudreau RL, Brink B, Reisetter A, Goel K, et al. A microRNA processing defect in smokers' macrophages is linked to SUMOylation of the endonuclease DICER. J Biol Chem. 2014 doi: 10.1074/jbc.M114.565473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nature genetics. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 66.Garre P, Perez-Segura P, Diaz-Rubio E, Caldes T, de la Hoya M. Reassessing the TARBP2 mutation rate in hereditary nonpolyposis colorectal cancer. Nature genetics. 2010;42:817–818. doi: 10.1038/ng1010-817. author reply 8. [DOI] [PubMed] [Google Scholar]
- 67.Ho AS, Kannan K, Roy DM, Morris LG, Ganly I, Katabi N, et al. The mutational landscape of adenoid cystic carcinoma. Nature genetics. 2013;45:791–798. doi: 10.1038/ng.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim Y, Yeo J, Lee JH, Cho J, Seo D, Kim JS, et al. Deletion of Human tarbp2 Reveals Cellular MicroRNA Targets and Cell-Cycle Function of TRBP. Cell reports. 2014;9:1061–1074. doi: 10.1016/j.celrep.2014.09.039. [DOI] [PubMed] [Google Scholar]
- 70.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 71.Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, et al. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 72.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes & development. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 75.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 76.Liu C, Kelnar K, Vlassov AV, Brown D, Wang J, Tang DG. Distinct microRNA expression profiles in prostate cancer stem/progenitor cells and tumor-suppressive functions of let-7. Cancer Res. 2012;72:3393–3404. doi: 10.1158/0008-5472.CAN-11-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 78.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nature structural & molecular biology. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thornton JE, Chang HM, Piskounova E, Gregory RI. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7) RNA. 2012;18:1875–1885. doi: 10.1261/rna.034538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Suzuki HI, Arase M, Matsuyama H, Choi YL, Ueno T, Mano H, et al. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Molecular cell. 2011;44:424–436. doi: 10.1016/j.molcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 81.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Molecular cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Briata P, Forcales SV, Ponassi M, Corte G, Chen CY, Karin M, et al. p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Molecular cell. 2005;20:891–903. doi: 10.1016/j.molcel.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 83.Gherzi R, Trabucchi M, Ponassi M, Ruggiero T, Corte G, Moroni C, et al. The RNA-binding protein KSRP promotes decay of beta-catenin mRNA and is inactivated by PI3K-AKT signaling. PLoS biology. 2006;5:e5. doi: 10.1371/journal.pbio.0050005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 84.Qi HH, Ongusaha PP, Myllyharju J, Cheng D, Pakkanen O, Shi Y, et al. Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature. 2008;455:421–424. doi: 10.1038/nature07186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu C, So J, Davis-Dusenbery BN, Qi HH, Bloch DB, Shi Y, et al. Hypoxia potentiates microRNA-mediated gene silencing through posttranslational modification of Argonaute2. Mol Cell Biol. 2011;31:4760–4774. doi: 10.1128/MCB.05776-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rybak A, Fuchs H, Hadian K, Smirnova L, Wulczyn EA, Michel G, et al. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat Cell Biol. 2009;11:1411–1420. doi: 10.1038/ncb1987. [DOI] [PubMed] [Google Scholar]
- 87.Gibbings D, Mostowy S, Jay F, Schwab Y, Cossart P, Voinnet O. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol. 2012;14:1314–1321. doi: 10.1038/ncb2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Molecular cell. 2011;42:489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Johnston M, Geoffroy MC, Sobala A, Hay R, Hutvagner G. HSP90 protein stabilizes unloaded argonaute complexes and microscopic P-bodies in human cells. Mol Biol Cell. 2010;21:1462–1469. doi: 10.1091/mbc.E09-10-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Iki T, Yoshikawa M, Nishikiori M, Jaudal MC, Matsumoto-Yokoyama E, Mitsuhara I, et al. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Molecular cell. 2010;39:282–291. doi: 10.1016/j.molcel.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 91.Iwasaki S, Kobayashi M, Yoda M, Sakaguchi Y, Katsuma S, Suzuki T, et al. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Molecular cell. 2010;39:292–299. doi: 10.1016/j.molcel.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 92.Zeng Y, Sankala H, Zhang X, Graves PR. Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem J. 2008;413:429–436. doi: 10.1042/BJ20080599. [DOI] [PubMed] [Google Scholar]
- 93.Horman SR, Janas MM, Litterst C, Wang B, MacRae IJ, Sever MJ, et al. Akt-mediated phosphorylation of argonaute 2 downregulates cleavage and upregulates translational repression of MicroRNA targets. Molecular cell. 2013;50:356–367. doi: 10.1016/j.molcel.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rudel S, Wang Y, Lenobel R, Korner R, Hsiao HH, Urlaub H, et al. Phosphorylation of human Argonaute proteins affects small RNA binding. Nucleic Acids Res. 2011;39:2330–2343. doi: 10.1093/nar/gkq1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mazumder A, Bose M, Chakraborty A, Chakrabarti S, Bhattacharyya SN. A transient reversal of miRNA-mediated repression controls macrophage activation. EMBO reports. 2013;14:1008–1016. doi: 10.1038/embor.2013.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang M, Haase AD, Huang FK, Coulis G, Rivera KD, Dickinson BC, et al. Dephosphorylation of tyrosine 393 in argonaute 2 by protein tyrosine phosphatase 1B regulates gene silencing in oncogenic RAS-induced senescence. Molecular cell. 2014;55:782–790. doi: 10.1016/j.molcel.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang NP, To H, Lee WH, Lee EY. Tumor suppressor activity of RB and p53 genes in human breast carcinoma cells. Oncogene. 1993;8:279–288. [PubMed] [Google Scholar]
- 98.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nature medicine. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 99.Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, et al. Multiplatform Analysis of 12 Cancer Types Reveals Molecular Classification within and across Tissues of Origin. Cell. 2014;158:929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]